Abstract
In fungi, heterotrimeric guanine-nucleotide binding proteins (G-proteins) are key elements of signal transduction pathways, which control growth, asexual and sexual development, as well as virulence. In this study, we have identified two genes encoding heterotrimeric G protein alpha subunits, named Gvm2 and Gvm3, from Valsa mali, the causal agent of apple Valsa canker. Characterization of Gvm2 and Gvm3 mutants indicates that Gvm3 may be a crucial regulator of vegetative growth. Deletion of the corresponding gene results in a 20% reduction in growth rate. Besides, Gvm2 and Gvm3 seem to be involved in asexual reproduction, and mutants are hypersensitive to oxidative and cell membrane stresses. Interestingly, both G protein alpha subunits were most probably involved in V. mali virulence. In infection assays using Malus domestica cv. ‘Fuji’ leaves and twigs, the size of lesions caused by deletion mutants △Gvm2, or △Gvm3 are significantly reduced. Furthermore, many genes encoding hydrolytic enzymes—important virulence factors in V. mali—are expressed at a lower level in these deletion mutants. Our results suggest that Gvm2 and Gvm3 play an important role in virulence probably by regulation of expression of cell wall degrading enzymes. △Gvm2, and △Gvm3 mutants were further analyzed with respect to their impact on the transcript levels of genes in the cAMP/PKA pathway. The expression of the genes encoding adenylate cyclase VmAC, protein kinase A (PKA) regulatory subunit VmPKR, and PKA catalytic subunit VmPKA1 are down-regulated in both mutants. Further analyses indicated that intracellular cAMP level and PKA activity are down-regulated in the △Gvm3 mutant, but are basically unchanged in the △Gvm2 mutant. Overall, our findings indicate that both Gvm2 and Gvm3 play diverse roles in the modulation of vegetative growth, asexual development, and virulence in V. mali.
Introduction
Signal transduction cascades are the primary means by which external stimuli are communicated to the nuclei of eukaryotic organisms. In fungi, heterotrimeric guanine-nucleotide binding proteins (G-proteins) are key elements of signal transduction pathways, which control growth, asexual and sexual development, and virulence [1]. G-proteins are composed of three subunits: α, β and γ, which remain inactive in the heterotrimeric state with GDP bound to the Gα subunit [2]. Heterotrimeric G-proteins are activated by members of the seven-transmembrane-spanning family of receptors [3]. Binding of signal ligands to such receptors promotes an exchange of GDP to GTP on the Gα subunit, which then triggers a conformational change and dissociation from the Gβγ heterodimer [4]. Either Gα, or Gβγ, or both, are then free to activate downstream targets such as phospholipases, protein kinases, adenylate cyclases, or ion channels [5–8]. Activated G-proteins are later desensitized by the intrinsic GTPase activity of the Gα subunit, followed by re-association with the Gβγ complex. Therefore, the guanine nucleotide state of the Gα subunit seems to play a crucial role in controlling G-protein signaling [4].
Multiple alignments of fungal Gα genes revealed three major groups (I-III), based on amino acid sequence identity and functional similarities [9]. Magnaporthe oryzae for example possesses three Gα subunits (MagA, MagB, and MagC) with sequence similarity to mammalian Gαs, Gαi superfamily proteins, and the fungal-specific GαII subfamily, respectively [10]. Deletion of magA has no effect on vegetative growth, conidiation, or appressorium formation. Deletion of magC reduces conidiation, but does not affect vegetative growth, or appressorium formation. However, disruption of magB significantly reduces vegetative growth, conidiation, and appressorium formation [10]. In Saccharomyces cerevisiae Gpa2 regulates growth and pseudohyphal development via a cAMP-dependent mechanism [11,12]. Previous research showed that GanB plays a role during asexual conidiation and germination through regulating the cAMP/PKA pathway in response to glucose in Aspergillus nidulans [13,14]. Further fungal Gα subunit homologs that are well characterized include CPG-2 of Cryphonectria parasitica [15], and GNA-2 of Neurospora crassa [16], as well as FfG2 and FfG3 of Fusarium fujikuroi [17].
Valsa canker caused by the Ascomycete Valsa mali, is one of the most destructive diseases on apple in Eastern Asia [18], especially in China [19,20], Japan [21] and Korea [22], and leads to heavy damage to apple production [23,24]. Since the pathogen penetrates extensively into the host phloem and xylem [25], chemical treatment cannot effectively cure or control Valsa canker [23]. Once inside the host tissue the fungus can induce tissue maceration and cell death. Cell wall degrading enzymes have been shown to play an important role in the infection process of V. mali [25–28]. Meanwhile, it has been reported that G-protein signaling is implicated in the regulation of cellulase genes in C. parasitica and Trichoderma reesei [29–31]. Other studies have shown that Gα subunit homologs are important for regulating asexual development and virulence in Botrytis cinerea [32]. Therefore, a better understanding of the role of Gα subunits in pathogenesis and their regulatory pathways in V. mali seems crucial for developing more effective disease management strategies.
The genome sequence of V. mali opened new opportunities and perspectives for basic research to study its mechanism of plant infection [27]. To explore the roles of Gα subunits in V. mali, three Gα genes were cloned from the fungus, two of which were functionally characterized in the present study. We used Gα subunit mutants to elucidate their role in controlling development and virulence. We found that Gvm2 has a role in regulating conidiation and virulence. Gvm3 on the other hand seems to be involved in the regulation of vegetative growth, asexual development, and virulence. In addition, Gvm2 and Gvm3 also play a positive role in regulating cell wall degrading enzymes.
Materials and methods
Fungal strains, culture conditions
The wild type V. mali strain 03–8 [27] was obtained from the Laboratory of Integrated Management of Plant Diseases in College of Plant Protection, Northwest A&F University, PRC. All strains were routinely preserved in 20% glycerol at -80°C. Potato Dextrose Broth (PDB) was used to grow mycelium for DNA and RNA extraction. Fungal genomic DNA was extracted using the CTAB method [33]. The binary vector pBIG2RHPH2-GFP-GUS [34] was provided by Dr. Fengming Song at Zhejiang University, PRC. This vector carries the hygromycin B resistance cassette.
Gene deletion and complementation analysis
To examine the biological function(s) of Gα proteins, we generated deletion mutants. To construct gene knockout cassettes the double joint PCR approach was used [35]. The hygromycin-phosphotransferase (hph) cassette, was amplified from PBIG2RHPH2-GFP-GUS with primers HYG/F and HYG/R (S1 Table). Upstream and downstream gene flanking sequences were amplified with primer pairs Gvm1-1F/Gvm1-2R, Gvm1-3F/Gvm1-4R, Gvm2-1F/Gvm2-2R, Gvm2-3F/Gvm2-4R, and Gvm3-1F/Gvm3-2R, Gvm3-3F/Gvm3-4R (S1 Table), respectively. After ligation with the hph cassette, the ligation product was transformed into V. mali strain 03–8 [36]. Hygromycin B (Roche, Mannheim, Germany) was added to a final concentration of 100 μg/ml for selection. Putative knockout mutants were identified by screening with primers GvmX-5F/GvmX-6R and H850/H852 (S1 Table), and further analyzed by PCR with primers GvmX-7F and H855R, and primers H856F and GvmX-8R (S1 Table). Southern Blot analyses were used to confirm gene replacement events (S2 Fig). Genomic DNA was labeled with digoxigenin (DIG)-dUTP using the DIG DNA Labeling and Detection Kit II (Roche). Hybridization and detection were carried out according to manufacturer’s instructions. All mutants generated in this study were preserved in 20% glycerol at -80°C.
For generation of complemented strains, fragments containing the entire Gα genes and their native promoter regions were amplified by PCR using primers Gvm2-GFP-CF/CR and Gvm3-GFP-CF/CR, respectively (S1 Table). The resulting PCR products were co-transformed into S. cerevisiae strain XK1-25 together with XhoI-digested pFL2 vector [37,38]. PGvm2-GFP and PGvm3-GFP fusion constructs were identified by PCR with primers Gvm2-GFP-CF/CR and Gvm3-GFP-CF/CR (S1 Table) and confirmed by sequencing. Fusion constructs were transformed into protoplasts of V. mali ΔGvm2 and ΔGvm3 mutants, respectively.
Vegetative growth and pycnidia production
Small agar blocks were cut from the edge of 3-day-old cultures and placed onto Potato Dextrose Agar (PDA) in the dark at 25°C. Size and morphology of colonies were examined after 24 h, 48 h, and photographed after 48 h. For pycnidia production assays, cultures were grown in the dark for seven days, then transferred to an incubator with 12 h illumination per day at 25°C, and examined and photographed after 40 days. All treatments were performed with at least eight replicates, and all experiments were repeated three times. Data were analyzed by Student’s t-test using the SAS software package (SAS Institute, Cary, USA), p<0.05.
Measuring stress responses
Fungal strains from glycerol stocks were inoculated onto PDA and placed in the dark for 3 d at 25°C. In order to test for sensitivity to osmotic stress, free radical stress, and cell membrane integrity, culture blocks of wild-type and the two mutants were inoculated onto PDA containing 0.1 M NaCl, 0.03% H2O2, or 0.01% SDS, respectively. Size and morphology of colonies were examined each day for three consecutive days. All treatments were performed with at least eight replicates, and the experiment was repeated three times. Data were analyzed by Student’s t-test using the SAS software package (SAS Institute), p<0.05.
Quantitative RT-PCR
Infected twigs (0.2 g) were collected 6 h, 12 h, 18 h, 24 h, 36 h, and 48 h post inoculation (hpi), frozen in liquid nitrogen, and stored at -80°C. Mycelium was collected after 4 days incubation in PDB. Total RNA was extracted using the RNeasy Micro kit (Qiagen, Shenzhen, PRC). cDNA synthesis was performed using the StrataScript qPCR cDNA synthesis kit (Stratagene, La Jolla, U.S.A.) following the manufacturer’s instructions. Primers G6PDHF and G6PDHR [39] were used to amplify the 6-phosphogluconate dehydrogenase, decarboxylating (G6PDH) gene of V. mali. Relative changes in transcript level of target genes were calculated by the 2–ΔΔCt method [40] with G6PDH as endogenous reference. Data from three biological replicates were used to calculate the mean and standard deviation. Primers used for qRT-PCR were listed in S2 Table.
Pathogenicity tests
For leaf pathogenicity assays, leaves of Malus domestica Borkh. cv. ‘Fuji’ from the green house were collected and inoculated according to Wei et al. [41]. Leaves were washed with tap water, immersed in 0.6% sodium hypochlorite for 3 min and rinsed with sterile water three times. The basal parts of petioles were wrapped with moistened cotton. Four little wounds were made on a leaf using a sterile needle. A 5-mm PDA culture block was placed upside down onto each wound. Leaves were placed in a plastic box and the box was immediately covered with a vinyl film to retain humidity and placed in the dark at 25°C. Examination and photo documentation took place after 72 h. Assays were repeated eight times with three biological replicates each.
For twig pathogenicity assays, biennial intact apple twigs of Malus domestica Borkh. cv. ‘Fuji’ from the green house were collected and inoculated according to Wei et al. [41]. Twigs were cut into 40 cm long segments and washed with tap water, immersed in 1% sodium hypochlorite for 10 min and rinsed with sterile water three times. The top of the twigs was sealed with wax. Four wounds were made on each segment using a flat iron (5 mm diameter), wounds were 8 cm apart. Twigs were inserted into sand in a plastic basin. A 5-mm PDA culture block was used to inoculate each wound. Then the basin was immediately covered with a vinyl film to retain humidity and placed in the dark at 25°C. Examination and photo documentation took place after eleven days. All treatments were performed with at least eight replicates, and all experiments were repeated three times. Data were analyzed by Student’s t-test using the SAS software package (SAS Institute), p<0.05.
cAMP and PKA activity assays
Three-day-old YEPD liquid mycelial cultures were harvested and frozen in liquid nitrogen. cAMP levels of the samples were measured using an HPLC [42,43]. Data were analyzed by Student’s t-test using the SAS software package (SAS Institute), p<0.05.
PKA activity was measured from 3-day-old YEPD liquid cultures. Sample (0.3g) were ground in liquid nitrogen. PKA activity was detected using PepTag® Non-Radioactive Protein Kinase Assays kit (Promega, Madison, USA). Samples were separated on a 1.2% agarose gel at 160 V for 15 minutes.
Results
Identification of Gα subunit genes in V. mali
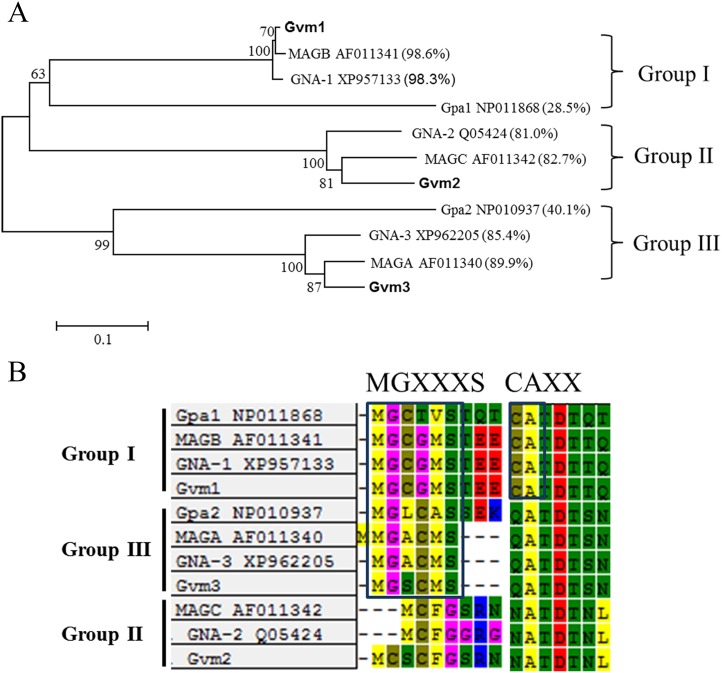
Based on sequence information from the V. mali genome, we cloned three Gα subunit genes termed Gvm1 (VM1G_00876), Gvm2 (VM1G_09956), and Gvm3 (VM1G_04248) [27]. The Gvm1 gene contains three introns and encodes a polypeptide of 353 amino acids, which shows 98.6% identity to M. oryzae MAGB (AF011341), and 98.3% to N. crassa GNA-1 (XP957133). Gvm2 contains four introns and encodes a polypeptide of 357 amino acids, which shows 82.7% identity to M. oryzae MAGC (AF011342), and 81.0% to N. crassa GNA-2 (Q05424). Gvm3 contains five introns and encodes a polypeptide of 355 amino acids, which shows 89.9% identity to M. oryzae MAGA (AF011340), 85.4% identity to N. crassa GNA-3 (XP962205), and 40.1% identity to S. cerevisiae Gpa2 (NP010937) (Fig 1A).
Fig 1. Three Gα subunits in Valsa mali.
A: Phylogenetic analysis with G protein sequences from V. mali, N. crassa, S. cerevisiae, and M. oryzae. Protein sequences were aligned, and the Neighbor-Joining phylogenic tree was drawn using MEGA 5.0. B: Sequence alignment of the predicted active sites of the Gα subunits from V. mali, N. crassa, S. cerevisiae, and M. oryzae. MGXXXS: myristoylation site; CXXX: pertussis toxin-labeling site.
A phylogenetic tree of different Gα subunits shows three distinct groups: Group I, Group II, and Group III (Fig 1A), which correspond to the three groups proposed by Bӧlker [9]. Gvm1 contains a consensus myristoylation site (MGXXXS) [44], and pertussis toxin-labeling site (CXXX) [45] at its N and C termini, respectively. Gvm3 groups together with homologs in Group III. Gvm3 shows high similarity to mammalian Gαs and contains a potential myristoylation site at its N terminus, but does not have a pertussis toxin-labeling site at its C terminus (Fig 1B). Gvm2 belongs to the fungal Gα subunit Group II and does not contain either site (Fig 1B).
Deletion of Gvm2 and Gvm3
Our results show that we successfully obtained knockout mutants for Gvm2 and Gvm3 (S1 Fig). All putative knockout mutants were also verified by Southern blot (S2 Fig). We obtained at least two deletion mutants for each gene with similar phenotypes, as described later in Table 1. For Gvm1, we failed to identify true knockout mutants after screening over one thousand transformants from at least four independent transformation experiments, indicating that deletion of this gene may be lethal.
Table 1. Wild type and mutant strains of V. mali used in this study.
| Strains | Brief description | Reference |
|---|---|---|
| 03–8 | Wild-type | [27] |
| G2M-1 | gvm2 deletion mutant of 03–8 | This study |
| G2M-2 | gvm2 deletion mutant of 03–8 | This study |
| G2C-1 | gvm2/Gvm2 complemented transformant | This study |
| G2C-2 | gvm2/Gvm2 complemented transformant | This study |
| G3M-1 | gvm3 deletion mutant of 03–8 | This study |
| G3M-2 | gvm3 deletion mutant of 03–8 | This study |
| G3C-1 | gvm3/Gvm3 complemented transformant | This study |
| G3C-2 | gvm3/Gvm3 complemented transformant | This study |
Gvm3 is involved in vegetative growth and asexual reproduction, whereas Gvm2 only plays a role in asexual reproduction
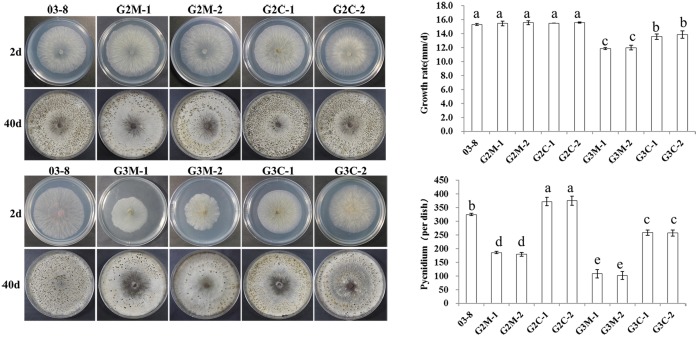
Gvm3 was found to play an important role in vegetative growth. Deletion of this gene results in an over 20% reduction in growth rate (Fig 2). Compared with the wild type strain (15.3 mm/day), growth rates of △Gvm3 mutants G3M-1 (11.9 mm/day) and G3M-2 (12.0 mm/day) are significantly reduced (p = 0.05). Both complemented strains (G3C-1 and G3C-2) exhibit at least partially restored growth rates. However, both △Gvm2 mutants (G2M-1 and G2M-2) show unaltered growth (Fig 2). In addition, Gvm2 and Gvm3 seem to be involved in asexual reproduction. Compared with the wild type strain, the amount of conidiation of △Gvm2 and △Gvm3 mutants is significantly decreased. Complementation (G2C-1, G2C-2 and G3C-1, G3C-2) could at least partially restore normal conidiation.
Fig 2. Colony morphology and conidiation of △Gvm2 and △Gvm3 deletion mutants.
Colony morphology was assessed by incubating cultures in the dark for 48 h at 25°C, followed by measuring colony diameters. For the pycnidia production assays, cultures were placed in the dark at 25°C for 7 d, then transferred to the light, examined and photographed at 40 days. Bars indicate standard deviation of the mean of eight individual plates. All experiments were performed in triplicate. Different letters indicate statistical significance.
Susceptibility of △Gvm2 and △Gvm3 mutants to abiotic stresses
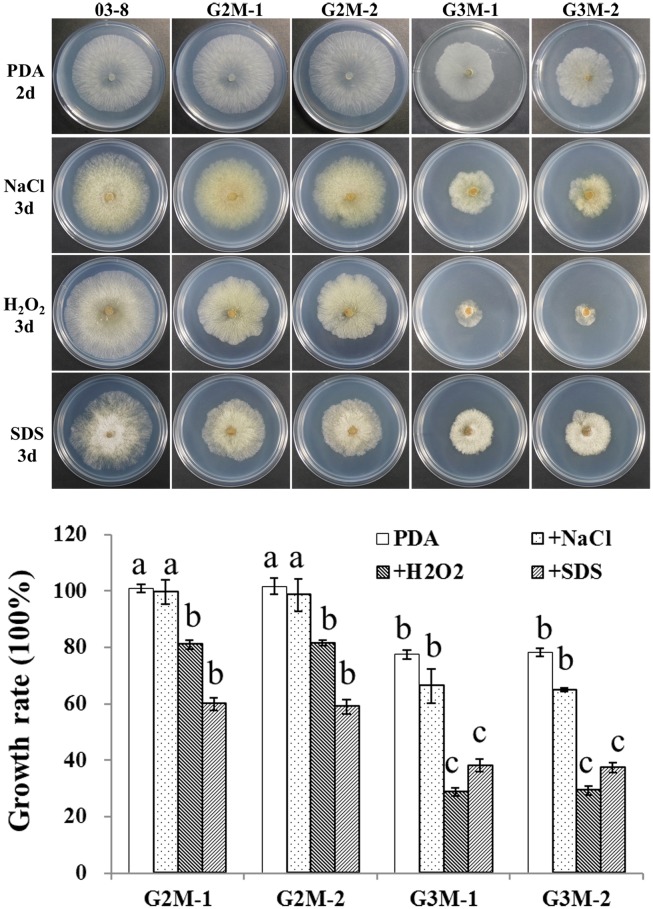
As results from growth on PDA supplemented with 0.03% H2O2 suggest, △Gvm2 and △Gvm3 mutants seem to be more sensitive to Reactive Oxygen Species (ROS) (Fig 3). When assayed for growth on PDA plates supplemented with 0.01% SDS, to simulate membrane stress, both mutants, especially △Gvm3 mutants, showed drastically reduced growth. By contrast, △Gvm2 and △Gvm3 mutants do not seem to be affected by osmotic stress, simulated by inclusion of 0.1 M NaCl to PDA (Fig 3). These results indicate that Gvm2 and Gvm3 may play the same role with respect to tolerance to abiotic stresses in V. mali.
Fig 3. Responses of △Gvm2 and △Gvm3 mutants to hyperosmotic, oxidative, and membrane stresses.
Colony diameters of wild-type strain 03–8, △Gvm2 mutants G2M-1, G2M-2 and △Gvm3 mutants G3M-1, G3M-2 on PDA with 0.1 M NaCl, 0.03% H2O2, or 0.01% SDS were measured after incubation in the dark for 3 d at 25°C. The percentage of the growth rate of the G2M-1, G2M-2, G3M-1, and G3M-2 mutants compared to that of the wild-type (set at 100%) on PDA cultures with or without different stresses. Different letters indicate statistically significant differences (P < 0.05, T-test). Bars indicate standard deviation of the mean of eight individual plates. All experiments were performed in triplicate.
Deletion of Gvm2, or Gvm3 leads to reduced virulence
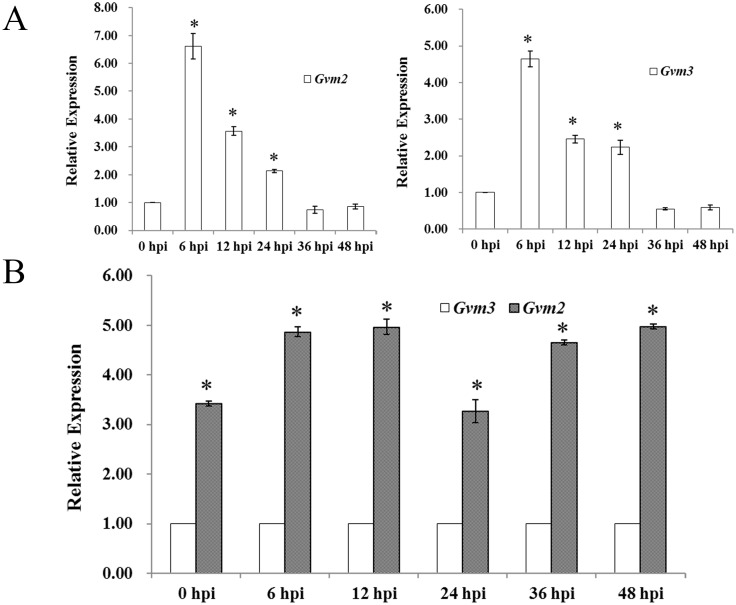
To gain insight into a possible function of Gvm2 and Gvm3 in pathogenicity, we examined their transcription profiles during infection using quantitative real-time PCR (qRT-PCR). Compared to axenically grown mycelium, transcript levels of Gvm2 are 6.6-fold, 3.6-fold, and 2.1-fold higher at 6 hpi, 12 hpi, and 24 hpi, respectively. However, expression of Gvm2 is not significantly changed at 36 hpi and 48 hpi (Fig 4A). Similarly, transcript levels of Gvm3 are 4.7-fold, 2.5-fold, and 2.2-fold increased at 6 hpi, 12 hpi, 24 hpi, respectively (Fig 4A). These results confirm that transcripts of Gvm2 and Gvm3 are up-regulated during early stages of infection. Gvm3 has a transcript profile similar to Gvm2. However, its transcript levels in general are lower than those of Gvm2 (Fig 4B).
Fig 4. Transcript levels of Gvm2 and Gvm3 assayed by qRT-PCR.
A: RNA samples were isolated from mycelium of strain 03–8 cultured in PDB medium at 25°C for 48 h. 0 hpi: axenic culture. Infected twigs were collected 6, 12, 24, 36 and 48 hpi. Relative transcript levels of Gvm2 and Gvm3 were calculated with G6PDH as internal control using the 2–ΔΔCT method. Transcript levels of Gvm2 or Gvm3 at the mycelium stage were set to 1. B: Relative transcript levels of Gvm2 in comparison with Gvm3. The transcript level of Gvm3 was set to 1 for all samples. Data from three biological replicates were used to calculate the mean and standard deviation.
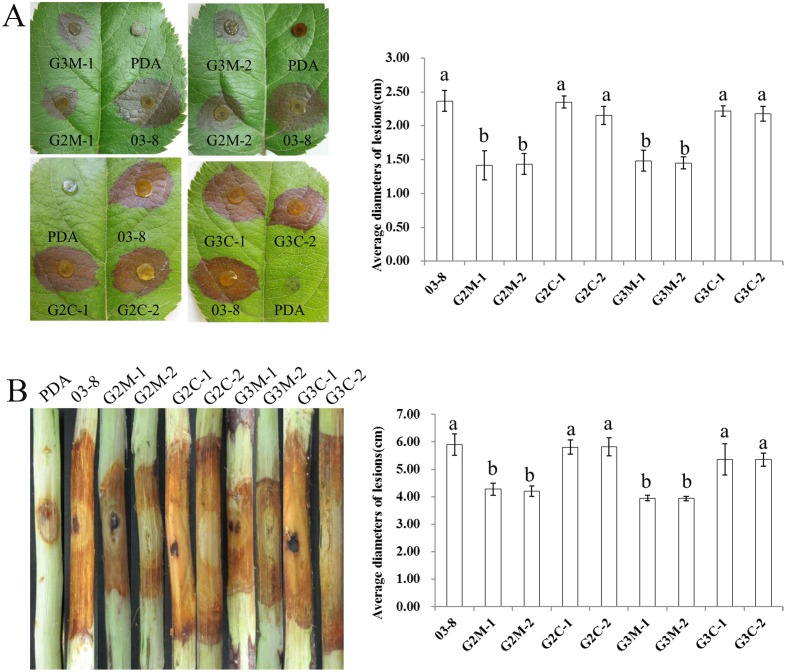
To further characterize the function of Gvm2 and Gvm3 in pathogenesis, △Gvm2 and △Gvm3 mutants were inoculated onto leaves and twigs. In infection assays with apple leaves, virulence of both deletion mutants is significantly reduced. Compared to the wild type strain 03–8, the average diameter of lesions caused by △Gvm2, or △Gvm3 mutants are significantly decreased. Compared to the wild type (23.7 mm), △Gvm2 mutants show a reduction by 40.1%, and △Gvm3 mutants exhibit a reduction of 35.0% (Fig 5A). Similarly, lesion lengths caused by △Gvm2 and △Gvm3 mutants are also smaller on twigs. The lesion length of wild type 03–8 strain was 59.0 mm. Compared to the wild type, both of △Gvm2 mutants G2C-1 and G2C-2 show a reduction of 27.5% and 28.8%, and △Gvm3 mutants G3C-1 and G3C-2 exhibit a reduction of 33.1% and 33.2% (Fig 5B).
Fig 5. Assay for plant infection of the △Gvm2 and △Gvm3 mutants.
5 mm circular agar plugs were inoculated onto (A) leaves or (B) twigs, examined and photographed 3, and 11 dpi, respectively. Three biological replicates and eight technical replicates were performed. Bars indicate standard deviation of the mean of eight individual plants.
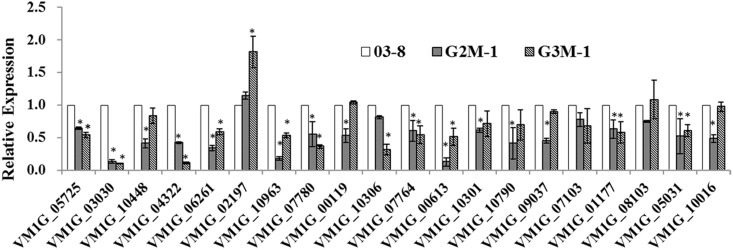
Cell wall degrading enzymes have been shown to constitute important virulence factors in V. mali [25–28]. A role for G protein signaling in cellulase gene expression was described in C. parasitica and T. reesei [29–31]. To further analyze the influence of Gvm2, or Gvm3 on cell wall degrading enzymes, transcript levels of different genes, including nine pectinase, six cellulase and five hemicellulase genes (S3 Table) encoded in the V. mali genome were checked in △Gvm2 and △Gvm3 mutants. RNA samples were isolated from vegetative mycelium of wild type strain 03–8, △Gvm2, and △Gvm3 mutants inoculated on twigs for 3 days at 25°C. Our results show that many hydrolytic enzyme encoding genes including pectinase, cellulase and hemicellulase genes are expressed at much lower levels in △Gvm2 and △Gvm3 mutants (Fig 6). These results clearly indicate that Gvm2 and Gvm3 are involved in regulating cell wall degrading enzyme genes.
Fig 6. Transcript levels of Cell Wall Degrading Enzyme (CWDE) genes in the △Gvm2 and △Gvm3 mutants of V. mali.
RNA samples were isolated from infected twigs of WT, G2M-1, and G3M-1 three days after inoculated at 25°C. Relative transcript levels were calculated with G6PDH as internal control using the 2–ΔΔCT method. Transcript levels of WT were set to 1 for all samples.
Gvm2 and Gvm3 mutants’ influence on interrelated downstream genes
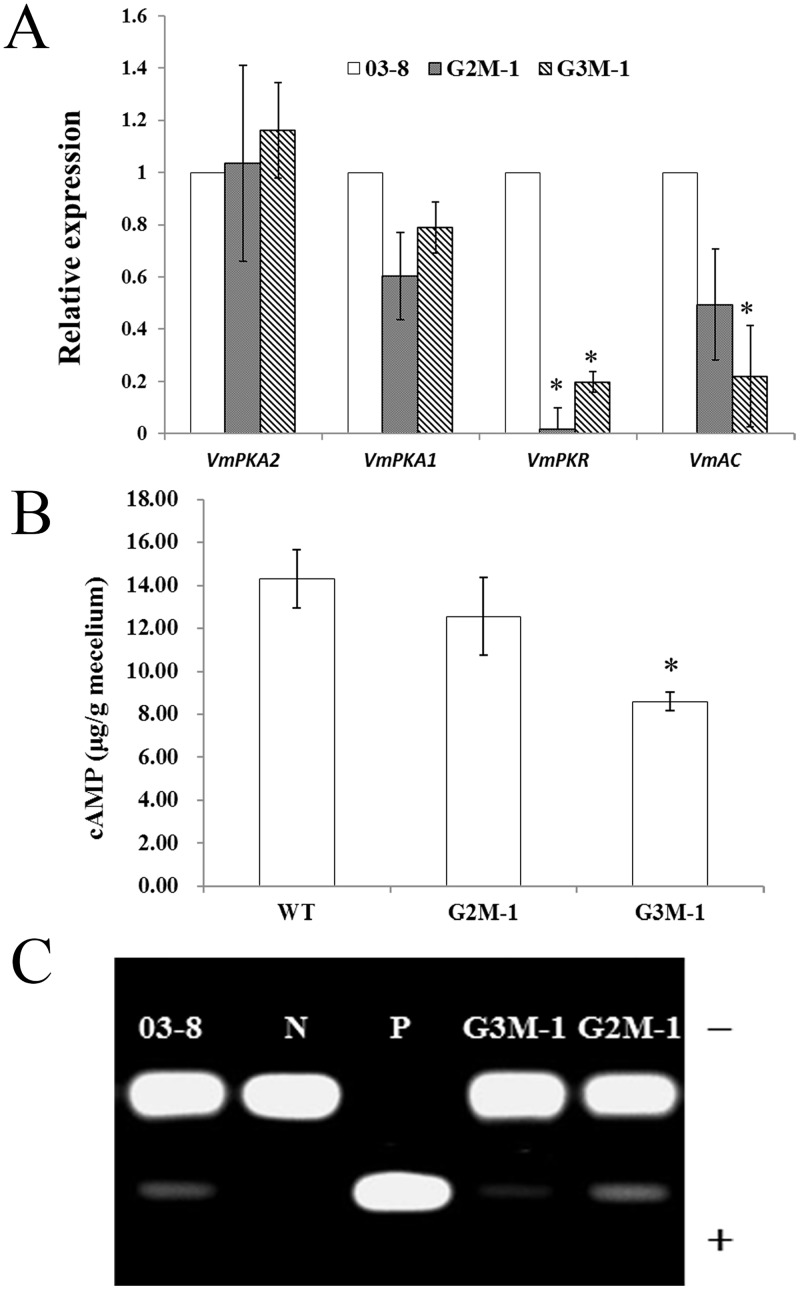
To further analyze the effects of a deletion of Gvm2 or Gvm3 on the expression of genes of the cAMP/PKA pathway, we checked the transcript levels of different genes, including one adenylate cyclase gene (VmAC, VM1G_01407), two PKA catalytic subunits genes (VmPKA1, VM1G_00266; and VmPKA2, VM1G_08687), and one PKA regulatory subunit gene (VmPKR, VM1G_08329) encoded in the V. mali genome. RNA samples were isolated from vegetative mycelium of wild type strain 03–8, △Gvm2 and △Gvm3 mutants cultured in PDB medium for 4 d at 25°C. Our results show that VmAC, and VmPKR are down-regulated in both mutants (Fig 7A). The expression of VmPKA1 is also down-regulated, however, VmPKA2 is not significantly affected in the two mutants (Fig 7A).
Fig 7. Assay for cAMP/PKA signaling pathway of △Gvm2 and △Gvm3 mutants.
A: Transcript levels of genes related to the cAMP/PKA signaling pathway. Relative transcript levels of VmPKA1, VmPKA2, VmPKR and VmAC genes in the wild type strain were set to 1. B: Intracellular cAMP levels in the wild-type and mutants. Bar chart showing quantification of intracellular cAMP in the mycelia of the indicated strains following three days of culturing in Yeast Extract Peptone Dextrose Medium (YEPD). The standard deviation represent SD of three replicates. C: PKA activity in the wild-type and mutants. Phosphorylated (+), nonphosphorylated (-), cAMP-Dependent Protein Kinase, Catalytic Subunit was the PKA positive control (P); water insteaded of PKA added as the negative control (N).
Effects of V. mali Gvm2 and Gvm3 on cAMP level and PKA activity
It was reported that Group III Gα proteins could influence cAMP levels through the regulation of adenylate cyclase [9]. To determine whether Gvm2 and Gvm3 are also involved in this process, we measured intracellular cAMP levels in the mutants G2M-1 and G3M-1. Our results indicate that the △Gvm3 mutant strain G3M-1 accumulates somewhat lower levels of cAMP than the wild-type strain. Compared with the wild-type, G3M-1 shows a 1.7-fold lower intracellular cAMP level, while the intracellular cAMP level in G2M-1 did not obviously decrease (Fig 7B). These results suggest that Gvm3 protein plays a role in regulating the intracellular cAMP level.
Similarly, PKA activity is reduced in the △Gvm3 mutant strain G3M-1 (Fig 7C). However, PKA activity in the △Gvm2 mutant strain G2M-1 shows no obvious change (Fig 7C). These results suggest that Gvm3 is also involved in regulating PKA activity.
Discussion
The importance of G protein signaling in regulating diverse biological processes in fungi has already been demonstrated [46]. In this study, we have identified genes encoding two heterotrimeric Gα subunits, Gvm2 and Gvm3, from V. mali. We found that Gvm2 and Gvm3 play various roles in the modulation of vegetative growth, asexual development, and virulence possibly via the cAMP/PKA pathway in this pathogenic fungus. Reduced virulence of △Gvm2 and △Gvm3 mutants may be due to a lower expression level of cell wall degrading enzymes.
Except for yeasts, which contain two Gα proteins, most characterized filamentous fungi possess three Gα proteins belonging to distinct groups [9,47]. Group I Gα proteins are highly conserved in most filamentous fungi, containing a consensus sequence for myristoylation (MGXXXS) at the amino terminus [44,48] and a site for ADP-ribosylation by pertussis toxin (CAAX) at the carboxy terminus [45]. Gvm1 seems to be a Group I Gα subunit, but unfortunately, we were not able to obtain a deletion mutant. Gvm2 is a Group II Gα subunit, and Gvm3 is a Group III Gα subunit similar to M. oryzae MAGC, and MAGA, respectively. Group II Gα subunits are not as well conserved as Group I, or III Gα subunits. Group III Gα subunits are highly conserved and most possess a myristoylation sequence [49]. We found that Gvm3 contains a potential consensus myristoylation site (MMGXXXS) at its N terminus, while Gvm2 does not contain such a site.
Gvm3 was found to play an important role in the regulation of vegetative growth. Similarly, deletion of ffg3 in F. fujikuroi caused reduces growth rates on minimal as well as complete medium [17]. Defects in growth rate upon deletion of Gα subunit genes has also been shown for A. nidulans, and B. cinerea [13,14,32]. However, deletion of magA in M. oryzae has no effect on vegetative growth [10]. It remains to be clarified if there is a species-specific pattern in the influence of Group III Gα subunits on the growth of different fungi. Our △Gvm2 mutants on the other hand exhibit no significant effect on fungal growth. It is reported that the role of Group II Gα subunits in vegetative growth is less explicit. For example, deletion of magC does not affect vegetative growth in M. oryzae [10].
Gα subunits have also been reported to be involved in asexual reproduction. Deletion of magA has no effect on conidiation, but a mutation in magC considerably reduces conidiation in M. oryzae [10]. In this study, we found that conidiation of V. mali was negatively influenced by mutations in both Gα subunit genes, Gvm2 and Gvm3. Despite the different growth rate phenotypes exhibited by these mutants, both Gα subunits seem to contribute to the regulation of asexual reproduction in V. mali.
We also found that mutants defective in Gvm2, or Gvm3 are more sensitive to free radicals. Some reports have indicated that G protein-coupled signaling components are involved in H2O2-induced responses [50]. Besides, it has been reported that Gαi and Gαo are critical components of oxidative stress responses, e.g. for activation of extracellular signal-related kinase (ERK) [50]. Based on our findings, we suggest that Gvm2 as well as Gvm3 play important roles in oxidative stress responses.
It has also been shown that G proteins may play a significant role in pathogenesis [49]. The results from our qRT-PCR analyses show that transcript levels of Gvm2 and Gvm3 are up-regulated during early stages of infection. Both proteins may therefore play an important role in the early stages of infection in V. mali. Many reports indicate that Group III Gα subunits may be involved in pathogenesis. In Ustilago maydis, Gpa3 seems to be involved in the invasion of corn [51]. Group III Gα subunits in B. cinerea (Bcg3), Cryptococcus neoformans (Gpa1), F. fujikuroi (Ffg3) and Fusarium oxysporum (Fga2) also seem to be required for full virulence [17,32,52–55]. However, deletion of magA in M. grisea does not appear to affect the ability to infect and spread within host tissue [10]. In infection assays with apple leaves and twigs, virulence of Gvm3 deletion mutants is significantly reduced (Fig 5). We cannot totally exclude that the growth defect of △Gvm3 mutants may contribute to the reduced virulence, at least partially. However, △Gvm3 mutants show a 30% to 35% reduction in lesion length in leaves or twigs (Fig 5)—this is not proportional to the 20% reduction in growth rate (Fig 2). In addition, cell wall degrading enzymes have been described as important virulence factors in V. mali [25–28]. It is likely that the defect in virulence of △Gvm3 mutants may be related to its reduced expression of cell wall degrading enzyme genes (Fig 6). G protein signaling in cellulase gene expression has also been described for C. parasitica and T. reesei [29–31]. Interestingly, △Gvm2 mutants, which do not show a growth defect, also show a significant reduction in virulence. Reduced virulence of △Gvm2 mutants is consistent with reduced expression of cell wall degrading enzymes, indicating that Gvm2 also plays an important role in virulence. Transcript levels of genes encoding cell wall degrading enzymes in △Gvm2 mutants are lower compared to △Gvm3 mutants. This may be the reason why △Gvm2 mutants show the same reduction in virulence as △Gvm3 mutants, though △Gvm2 mutants do not exhibit a growth defect. Similarly, it has been found that Gpa3 from C. neoformans is also involved in pathogenesis [54]. However, in most organisms, the function of Group II Gα proteins is less significant than that of Group III Gα proteins. For example, deletion of magC in M. oryzae, ffg3 in F. fujikuroi, and gpa2 in U. maydis has no effect on virulence [10,17,51] and B. cinerea bcg-2 mutants only show slightly reduced virulence [32].
Both Gβγ and Gα subunits are able to trigger downstream signaling pathways by interacting with various targets such as phosphodiesterases, protein kinases, and adenylate cyclases [6,56,57]. In this study, we analyzed transcript levels of VmAC, VmPKR, VmPKA1, and VmPKA2 in △Gvm3 and △Gvm2 mutants. Transcript levels of VmAC, VmPKR, and VmPKA1 are down-regulated in both mutants. Gvm2 and Gvm3 may therefore be involved in the cAMP/PKA pathway. To further determine whether Gvm2 and Gvm3 proteins are involved in this process, we measured intracellular cAMP levels. Our results indicate that △Gvm3 mutants accumulate somewhat lower levels of cAMP, while the intracellular cAMP level in △Gvm2 mutants seems unchanged (Fig 7B). We also measured PKA activity in △Gvm2 and △Gvm3 mutants. Results show that PKA activity is reduced in △Gvm3 mutants (Fig 7C). However, PKA activity in △Gvm2 mutants show no change (Fig 7C). These results suggest that Gvm3 plays a more important role in regulating the cAMP-PKA signaling pathway. It has been reported that Group III Gα proteins could influence cAMP levels through the regulation of adenylate cyclase [9]. Gα proteins, including A. nidulans GanB, U. maydis Gpa3, F. fujikuroi Ffg3 and C. neoformans Gpa1, hae been implicated in the regulation of cAMP signaling [14,17,52,53,58,59].
In conclusion, two heterotrimeric Gα subunits, Gvm2 and Gvm3, were functionally characterized in V. mali. Both seem to be important for virulence. Gvm3 also seems to be involved in regulating vegetative growth. Both, Gvm2 and Gvm3, seem to be involved in the response to different abiotic stresses in V. mali.
Supporting information
Verification of mutants by PCR was done using four pairs of primers. Primers GvmX-5F and GvmX-6R (1) are located within the ORFs for negative screening, primers H852 and H850 (2) are located within the hygromycin-resistant gene, primers GvmX-7F and GvmX-8R are located beyond the gene flanking sequences, primers H856F and H855R are located within the hygromycin-resistance conferring gene, primers GvmX-7F/H855R (3) for positive screen (upstream), and primers H856F/ GvmX-8R (4) for positive screen (downstream). M: 2,000 bp marker.
(TIF)
Arrows indicate orientations of the Gα and hygromycin phosphotransferase (hph) genes. Thin lines below the arrows indicate the probe sequence for each gene (Probe 1), or the hph gene (Probe 2). A: Southern blot analyses of Gvm2 knockout mutants. Genomic DNA was digested with restriction enzymes HindIII (H), EcoRI (EI), or XbaI (X). When hybridized with a Gvm2 fragment amplified with primers Gvm2-5F/Gvm2-6R (Probe 1), the wild-type strain 03–8 shows the expected 4.4 kb band. gvm2 mutants show no corresponding hybridization signal. When hybridized with an hph probe (Probe 2) amplified with primers H850/H852, the wild-type strain shows no hybridization signal. gvm2 mutants on the other hand exhibit the expected 4.6 kb, or 9.0 kb bands in the respective XbaI, or EcoRI digests. B: Southern blot analyses of Gvm3 knockout mutants. Genomic DNA was digested with EcoRV (EV), or BamHI (B). When hybridized with a Gvm3 fragment amplified with primers Gvm3-5F/Gvm3-6R (Probe 1), the wild-type strain 03–8 shows the expected 4.1 kb band. gvm3 mutants show no hybridization signal. When hybridized with an hph probe (Probe 2) amplified with primers H850/H852, the wild-type strain shows no hybridization signal, whereas the gvm3 mutants show the expected 4.0 kb band.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We thank Fengming Song (Zhejiang University, PRC) for providing the pBIG2RHPH2-GFP-GUS plasmid and Dr. Hao Feng and Liangsheng Xu at Northwest A&F University for proofreading this manuscript. This work was financially supported by the National Natural Science Foundation of China (No. 31471732; 31671982).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was financially supported by the National Natural Science Foundation of China (No. 31471732; 31671982).
References
- 1.Jacqueline AS, Campbell AJ, Borkovich KA (2012) G protein signaling components in filamentous fungal genomes Biocommunication of Fungi: Springer; pp. 21–38. [Google Scholar]
- 2.Neves SR, Ram PT, Iyengar R (2002) G protein pathways. Science 296: 1636–1639. 10.1126/science.1071550 [DOI] [PubMed] [Google Scholar]
- 3.Malbon CC (2005) G proteins in development. Nat Rev Mol Cell Biol 6: 689–701. 10.1038/nrm1716 [DOI] [PubMed] [Google Scholar]
- 4.Dohlman HG, Thorner J (2001) Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu Rev Biochem 70: 703–754. 10.1146/annurev.biochem.70.1.703 [DOI] [PubMed] [Google Scholar]
- 5.Gilman AG (1987) G proteins: transducers of receptor-generated signals. Annual Review of Biochemistry Annu Rev Biochem 56: 615–649. 10.1146/annurev.bi.56.070187.003151 [DOI] [PubMed] [Google Scholar]
- 6.Simon MI, Strathmann MP, Gautam N (1991) Diversity of G proteins in signal transduction. Science 252: 802–808. [DOI] [PubMed] [Google Scholar]
- 7.Clapham DE, Neer EJ (1997) G protein βγ subunits. Annu Rev Pharmacol Toxicol 37: 167–203. 10.1146/annurev.pharmtox.37.1.167 [DOI] [PubMed] [Google Scholar]
- 8.Hamm HE (1998) The many faces of G protein signaling. J Biol Chem 273: 669–672. [DOI] [PubMed] [Google Scholar]
- 9.Bölker M (1998) Sex and crime: heterotrimeric G proteins in fungal mating and pathogenesis. Fungal Genet Biol 25: 143–156. 10.1006/fgbi.1998.1102 [DOI] [PubMed] [Google Scholar]
- 10.Liu S, Dean RA (1997) G protein α subunit genes control growth, development, and pathogenicity of Magnaporthe grisea. Mol Plant Microbe Interact 10: 1075–1086. 10.1094/MPMI.1997.10.9.1075 [DOI] [PubMed] [Google Scholar]
- 11.Kübler E, Mösch HU, Rupp S, Lisanti MP (1997) Gpa2p, a G-protein α-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J Biol Chem 272: 20321–20323. [DOI] [PubMed] [Google Scholar]
- 12.Lorenz MC, Heitman J (1997) Yeast pseudohyphal growth is regulated by GPA2, a G protein α homolog. EMBO J 16: 7008–7018. 10.1093/emboj/16.23.7008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang M, Chae K, Han D, Jahng K (2004) The GanB Galpha-protein negatively regulates asexual sporulation and plays a positive role in conidial germination in Aspergillus nidulans. Genetics 167: 1305 10.1534/genetics.103.025379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lafon A, Seo J-A, Han K-H, Yu J-H, d'Enfert C (2005) The heterotrimeric G-protein GanB(α)-SfaD(β)-GpgA(γ) is a carbon source sensor involved in early cAMP-dependent germination in Aspergillus nidulans. Genetics 171: 71–80. 10.1534/genetics.105.040584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao S, Nuss DL (1996) Distinct roles for two G protein α subunits in fungal virulence, morphology, and reproduction revealed by targeted gene disruption. Proc Natl Acad Sci USA 93: 14122–14127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krystofova S, Borkovich KA (2005) The heterotrimeric G-protein subunits GNG-1 and GNB-1 form a Gβγ dimer required for normal female fertility, asexual development, and Gα protein levels in Neurospora crassa. Eukaryot Cell 4: 365–378. 10.1128/EC.4.2.365-378.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Studt L, Humpf H-U, Tudzynski B (2013) Signaling governed by G proteins and cAMP is crucial for growth, secondary metabolism and sexual development in Fusarium fujikuroi. PloS one 8: e58185 10.1371/journal.pone.0058185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones AL, Aldwinckle H (1990) Compendium of apple and pear diseases and Pests: APS Press, St. Paul. [Google Scholar]
- 19.Cao K, Guo L, Li B, Sun G, Chen H (2009) Investigations on the occurrence and control of apple canker in China. Plant Protect 35: 114–116. [Google Scholar]
- 20.Wang L, Zang R, Huang LL, Xie FQ, Gao XN (2005) The investigation of apple tree Valsa canker in Guanzhong region of Shaanxi province. J Northwest Sci-Tech Univer Agricult and Forest 33: 98–100. [Google Scholar]
- 21.Abe K, Kotoda N, Kato H, Soejima J (2011) Genetic studies on resistance to Valsa canker in apple: genetic variance and breeding values estimated from intra-and inter-specific hybrid progeny populations. Tree Genet Genom 7: 363–372. [Google Scholar]
- 22.Lee DH, Lee SW, Choi KH, Kim DA, Uhm JY (2006) Survey on the occurrence of apple diseases in Korea from 1992 to 2000. Plant Pathol J 22: 375–380. [Google Scholar]
- 23.Abe K, Kotoda N, Kato H, Soejima J (2007) Resistance sources to Valsa canker (Valsa ceratosperma) in a germplasm collection of diverse Malus species. Plant Breed 126: 449–453. [Google Scholar]
- 24.Chen C, Li M, Shi X, Wang J (1987) Studies on the infection period of Valsa mali Miyabe et Yamada, the causal agent of apple tree canker. Act Phytopathol Sin 17: 3–6. [Google Scholar]
- 25.Ke XW, Huang LL, Han QM, Gao XN, Kang ZS (2013) Histological and cytological investigations of the infection and colonization of apple bark by Valsa mali var. mali. Australas Plant Path 42: 85–93. [Google Scholar]
- 26.Ke XW, Yin ZY, Song N, Dai QQ, Voegele RT, Liu YY, et al. (2014) Transcriptome profiling to identify genes involved in pathogenicity of Valsa mali on apple tree. Fungal Genet Biol 68: 31–38. 10.1016/j.fgb.2014.04.004 [DOI] [PubMed] [Google Scholar]
- 27.Yin ZY, Liu HQ, Li ZP, Ke XW, Dou DL, Gao XN, et al. (2015) Genome sequence of Valsa canker pathogens uncovers a potential adaptation of colonization of woody bark. New Phytol 208: 1202–1216. 10.1111/nph.13544 [DOI] [PubMed] [Google Scholar]
- 28.Xu CJ, Dai QQ, Li ZP, Gao XN, Hang LL (2016) Function of Polygalacturonase Genes Vmpg7 and Vmpg8 of Valsa mali. Scientia Agricultura Sinica 49: 1489–1498. [Google Scholar]
- 29.Schmoll M, Schuster A, do Nascimento Silva R, Kubicek CP (2009) The G-alpha protein GNA3 of Hypocrea jecorina (anamorph Trichoderma reesei) regulates cellulase gene expression in the presence of light. Eukaryot cell 8: 410–420. 10.1128/EC.00256-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seibel C, Gremel G, do Nascimento Silva R, Schuster A, Kubicek CP, Schmoll M. (2009) Light-dependent roles of the G-protein α subunit GNA1 of Hypocrea jecorina (anamorph Trichoderma reesei). BMC Biol 7: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang P, Nuss DL (1995) Induction of a Cryphonectria parasitica cellobiohydrolase I gene is suppressed by hypovirus infection and regulated by a GTP-binding-protein-linked signaling pathway involved in fungal pathogenesis. Proc Natl Acad Sci USA 92: 11529–11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doehlemann G, Berndt P, Hahn M (2006) Different signalling pathways involving a Gα protein, cAMP and a MAP kinase control germination of Botrytis cinerea conidia. Mol Microbiol 59: 821–835. 10.1111/j.1365-2958.2005.04991.x [DOI] [PubMed] [Google Scholar]
- 33.Hallen HE, Watling R, Adams GC (2003) Taxonomy and toxicity of Conocybe lactea and related species. Mycol Res 107: 969–979. [DOI] [PubMed] [Google Scholar]
- 34.Fang L (2005) Agrobacterium tumefaciens-mediated transformation of Fusarium graminearum and Colletotrichum lagenarium and preliminary screening for pathogenicity mutants. Master’s Thesis, Zhejiang University, Zhejiang.
- 35.Yu JH, Hamari Z, Han KH, Seo JA, Reyes-Domínguez Y, Scazzocchio C. (2004) Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet Biol 41: 973–981. 10.1016/j.fgb.2004.08.001 [DOI] [PubMed] [Google Scholar]
- 36.Gao J, Li Y, Ke X, Kang Z, Huang L (2011) Development of genetic transformation system of Valsa mali of apple mediated by PEG. Acta microbiologica Sinica 51: 1194–1199. [PubMed] [Google Scholar]
- 37.Bruno KS, Tenjo F, Li L, Hamer JE, Xu JR (2004) Cellular localization and role of kinase activity of PMK1 in Magnaporthe grisea. Eukaryot cell 3: 1525–1532. 10.1128/EC.3.6.1525-1532.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou X, Li G, Xu JR (2011) Efficient approaches for generating GFP fusion and epitope-tagging constructs in filamentous fungi. Methods Mol Biol 722, 199–212. 10.1007/978-1-61779-040-9_15 [DOI] [PubMed] [Google Scholar]
- 39.Yin ZY, Ke XW, Huang DX, Gao XN, Voegele RT, Kang ZS, et al. (2013) Validation of reference genes for gene expression analysis in Valsa mali var. mali using real-time quantitative PCR. World J Microbiol Biotechnol 29: 1563–1571. 10.1007/s11274-013-1320-6 [DOI] [PubMed] [Google Scholar]
- 40.Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 41.Wei JL, Hang LL, Gao XN, Ke XW, Kang ZS (2010) Laboratory evaluation methods of apple Valsa canker disease caused by Valsa ceratosperma sensu Kobayashi. Act Phytopathol Sin 40: 14–20. [Google Scholar]
- 42.Duan YX (2013) Separation and purification of cAMP from red jujube and research of its oral liquid. Master’s Thesis, Tianjin University of Commerce, Tianjin.
- 43.You FH (2007) Studies on absorption and separation for cAMP from ziziphus jujube. Master’s Thesis, Hefei University of Technology, Hefei.
- 44.Buss JE, Mumby SM, Casey PJ, Gilman AG, Sefton BM (1987) Myristoylated alpha subunits of guanine nucleotide-binding regulatory proteins. Proceedings of the National Academy of Sciences 84: 7493–7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.West R, Moss J, Vaughan M, Liu T, Liu TY (1985) Pertussis toxin-catalyzed ADP-ribosylation of transducin. Cysteine 347 is the ADP-ribose acceptor site. Journal of Biological Chemistry 260: 14428–14430. [PubMed] [Google Scholar]
- 46.D'Souza CA, Heitman J (2001) Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol Rev 25: 349–364. [DOI] [PubMed] [Google Scholar]
- 47.Kays A, Borkovich K (2004) Signal transduction pathways mediated by heterotrimeric G proteins Biochemistry and Molecular Biology: Springer; pp. 175–207. [Google Scholar]
- 48.Turner G, Borkovich K (1993) Identification of a G protein alpha subunit from Neurospora crassa that is a member of the Gi family. Journal of Biological Chemistry 268: 14805–14811. [PubMed] [Google Scholar]
- 49.Li L, Wright SJ, Krystofova S, Park G, Borkovich KA (2007) Heterotrimeric G protein signaling in filamentous fungi. Annu Rev Microbiol 61: 423–452. 10.1146/annurev.micro.61.080706.093432 [DOI] [PubMed] [Google Scholar]
- 50.Nishida M, Maruyama Y, Tanaka R, Kontani K, Nagao T, Kurose H. (2000) Gαi and Gαo are target proteins of reactive oxygen species. Nature 408: 492–495. 10.1038/35044120 [DOI] [PubMed] [Google Scholar]
- 51.Regenfelder E, Spellig T, Hartmann A, Lauenstein S, Bölker M, Kahmann R. (1997) G proteins in Ustilago maydis: transmission of multiple signals? EMBO J 16: 1934–1942. 10.1093/emboj/16.8.1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alspaugh JA, Pukkila-Worley R, Harashima T, Cavallo LM, Funnell D, Cox GM, et al. (2002) Adenylyl cyclase functions downstream of the Gα protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot Cell 1: 75–84. 10.1128/EC.1.1.75-84.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alspaugh JA, Perfect JR, Heitman J (1997) Cryptococcus neoformans mating and virulence are regulated by the G-protein α subunit GPA1 and cAMP. Genes Dev 11: 3206–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li L, Shen G, Zhang Z-G, Wang Y-L, Thompson JK, Wang P. (2007) Canonical heterotrimeric G proteins regulating mating and virulence of Cryptococcus neoformans. Mol Biol Cell 18: 4201–4209. 10.1091/mbc.E07-02-0136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jain S, Akiyama K, Takata R, Ohguchi T (2005) Signaling via the G protein α subunit FGA2 is necessary for pathogenesis in Fusarium oxysporum. FEMS Microbiol Lett 243: 165–172. 10.1016/j.femsle.2004.12.009 [DOI] [PubMed] [Google Scholar]
- 56.Kaziro Y, Itoh H, Kozasa T, Nakafuku M, Satoh T (1991) Structure and function of signal-transducing GTP-binding proteins. Ann Rev Biochem 60: 349–400. 10.1146/annurev.bi.60.070191.002025 [DOI] [PubMed] [Google Scholar]
- 57.Neer EJ (1995) Heterotrimeric C proteins: Organizers of transmembrane signals. Cell 80: 249–257. [DOI] [PubMed] [Google Scholar]
- 58.Krüger J, Loubradou G, Regenfelder E, Hartmann A, Kahmann R (1998) Crosstalk between cAMP and pheromone signalling pathways in Ustilago maydis. Mol Gen Genet 260: 193–198. [DOI] [PubMed] [Google Scholar]
- 59.D'Souza CA, Alspaugh JA, Yue C, Harashima T, Cox GM, Heitman J. (2001) Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol Cell Biol 21: 3179–3191. 10.1128/MCB.21.9.3179-3191.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Verification of mutants by PCR was done using four pairs of primers. Primers GvmX-5F and GvmX-6R (1) are located within the ORFs for negative screening, primers H852 and H850 (2) are located within the hygromycin-resistant gene, primers GvmX-7F and GvmX-8R are located beyond the gene flanking sequences, primers H856F and H855R are located within the hygromycin-resistance conferring gene, primers GvmX-7F/H855R (3) for positive screen (upstream), and primers H856F/ GvmX-8R (4) for positive screen (downstream). M: 2,000 bp marker.
(TIF)
Arrows indicate orientations of the Gα and hygromycin phosphotransferase (hph) genes. Thin lines below the arrows indicate the probe sequence for each gene (Probe 1), or the hph gene (Probe 2). A: Southern blot analyses of Gvm2 knockout mutants. Genomic DNA was digested with restriction enzymes HindIII (H), EcoRI (EI), or XbaI (X). When hybridized with a Gvm2 fragment amplified with primers Gvm2-5F/Gvm2-6R (Probe 1), the wild-type strain 03–8 shows the expected 4.4 kb band. gvm2 mutants show no corresponding hybridization signal. When hybridized with an hph probe (Probe 2) amplified with primers H850/H852, the wild-type strain shows no hybridization signal. gvm2 mutants on the other hand exhibit the expected 4.6 kb, or 9.0 kb bands in the respective XbaI, or EcoRI digests. B: Southern blot analyses of Gvm3 knockout mutants. Genomic DNA was digested with EcoRV (EV), or BamHI (B). When hybridized with a Gvm3 fragment amplified with primers Gvm3-5F/Gvm3-6R (Probe 1), the wild-type strain 03–8 shows the expected 4.1 kb band. gvm3 mutants show no hybridization signal. When hybridized with an hph probe (Probe 2) amplified with primers H850/H852, the wild-type strain shows no hybridization signal, whereas the gvm3 mutants show the expected 4.0 kb band.
(TIF)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.