Abstract
Clear cell Renal Cell Carcinoma (ccRCC) is characterized by VHL inactivation1,2. Because no other gene is mutated as frequently, and VHL mutations are truncal3, VHL inactivation is regarded as the governing event4. VHL loss activates HIF-2, and constitutive HIF-2 restores tumorigenesis in VHL-reconstituted ccRCC cells5. HIF-2 is implicated in angiogenesis and multiple other processes6–9, but angiogenesis is the main target of drugs like sunitinib10. HIF-2, a transcription factor, has been regarded as undruggable11. A structure-based design approach identified a selective HIF-2 antagonist (PT2399) that we evaluate using a tumorgraft (TG)/PDX platform12,13. PT2399 dissociated HIF-2 (an obligatory heterodimer [HIF-2α/HIF-1β])14 in human ccRCC suppressing tumorigenesis in 56% (10/18) lines. PT2399 had greater activity than sunitinib, was active in sunitinib-progressing tumors, and was better tolerated. Unexpectedly, some VHL-mutant ccRCCs were resistant. Resistance occurred despite HIF-2 dissociation in tumors and evidence of Hif-2 inhibition in the mouse as determined by suppression of circulating erythropoietin, a HIF-2 target15 and possible pharmacodynamic marker. We identified a HIF-2-dependent gene signature in sensitive tumors. Illustrating drug specificity, gene expression was largely unaffected by PT2399 in resistant tumors. Sensitive tumors exhibited a distinguishing gene expression signature, and generally higher HIF-2α levels. Prolonged PT2399 treatment led to resistance. We identified a binding site and second site suppressor mutation in HIF-2α and HIF-1β respectively. Both mutations preserved HIF-2 dimers despite treatment with PT2399. Finally, an extensively pretreated patient with a sensitive TG had disease control for >11 months with the close analogue PT2385. We validate HIF-2 as a target in ccRCC, show that some ccRCC are, unexpectedly, HIF-2 independent, and set the stage for biomarker-driven clinical trials.
Keywords: RCC, renal cancer, kidney cancer, patient-derived xenograft, HIF-2α (G323E), HIF-1β (F446L), EPAS1, ARNT
The discovery of a 280Å3 cavity within the PAS-B domain of HIF-2α16,17 and subsequent identification by UT Southwestern investigators of compounds that bound this cavity and dissociated HIF-2α from HIF-1β18 led to the founding of Peloton Therapeutics, who through an iterative structure-based program identified selective potent HIF-2α antagonists such as PT2399 (described in Cho et al.)19 and PT238520.
To evaluate PT2399 in renal cancer, we tested a panel of 22 independently-generated TGs12,13 (Extended Data Table 1). To assess tolerability, we evaluated its effects on mouse weight and blood counts in mice bearing those tumorgrafts. PT2399 did not induce weight loss, whereas sunitinib, at doses matching human exposures13, did (Fig. 1a). However, PT2399 caused modest anemia and leukopenia (Fig. 1b and Extended Data Fig. 1a).
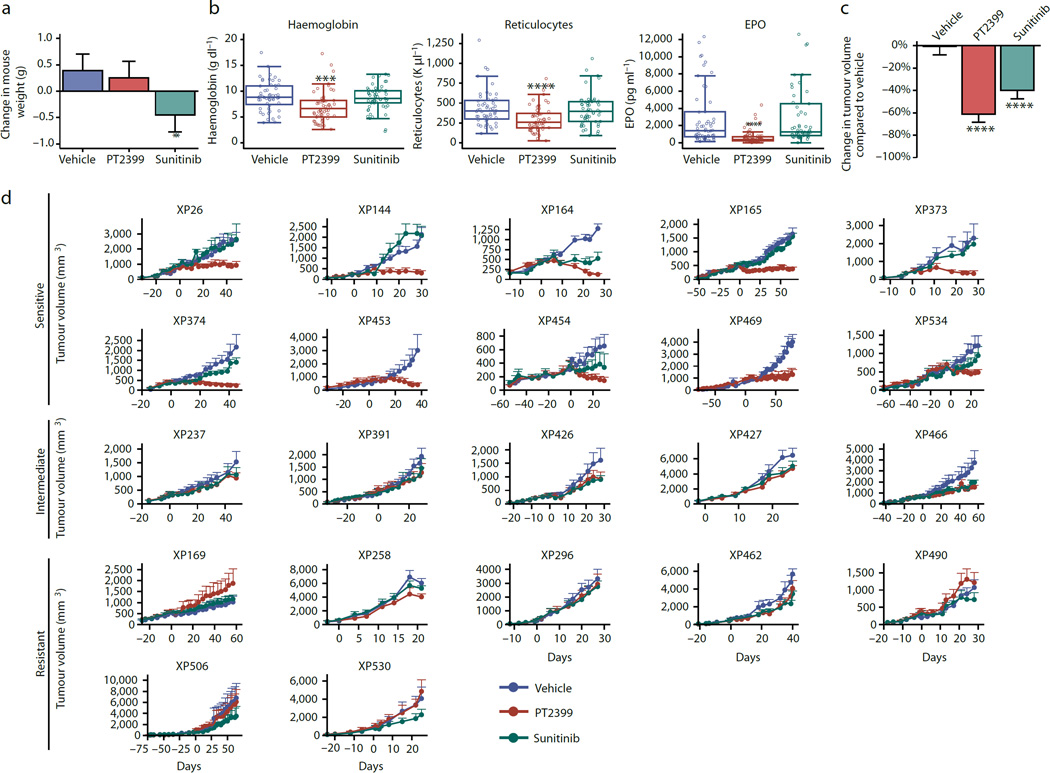
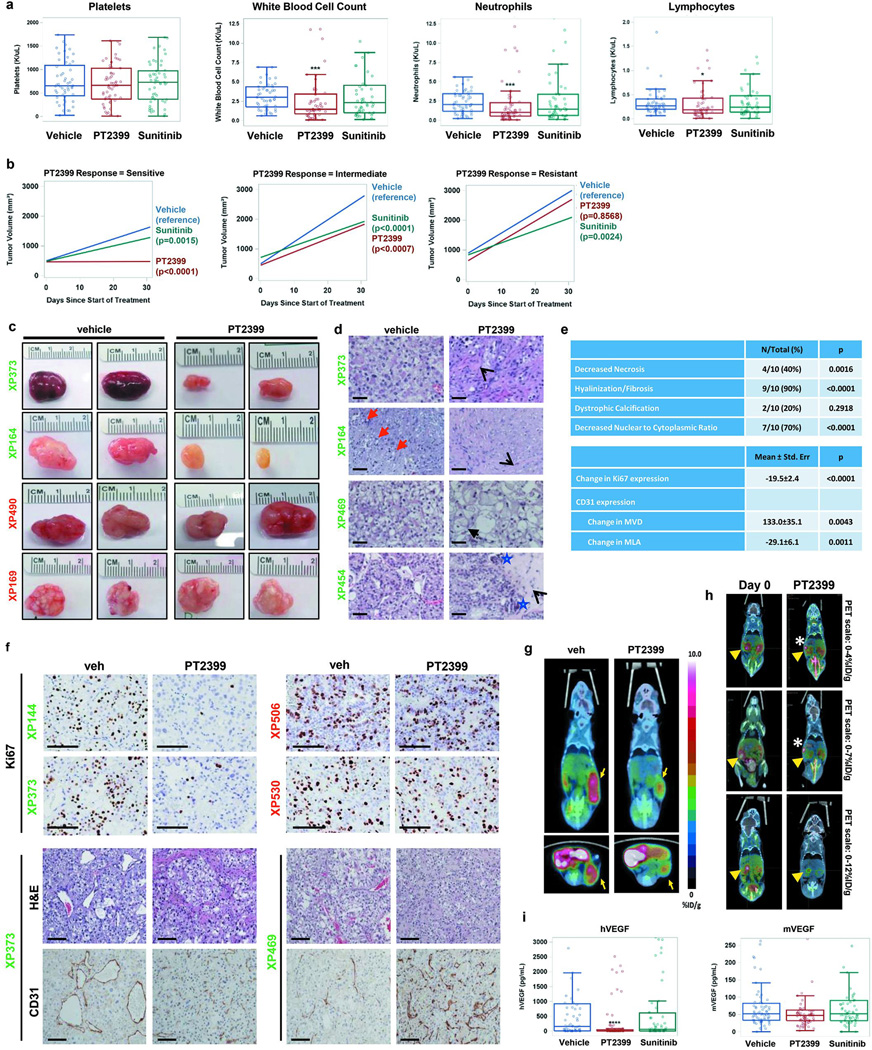
Fig. 1. Evaluation of PT2399 in RCC tumorgraft-bearing mice.
a, Mean change in mouse body weights after treatment with vehicle (veh; n = 89), PT2399 (100 mg/kg) by oral gavage every 12 hours (PT; n = 96), or sunitinib (10 mg/kg) by oral gavage every 12 hours (Sun; n = 82). b, Hemoglobin levels, reticulocyte counts, and erythropoietin (EPO) levels in mice treated as indicated. (Hemoglobin and reticulocytes vehicle n = 52, PT2399 n = 58, sunitinib n = 53; EPO vehicle n = 63, PT2399 n = 74, sunitinib n = 61). c, Mean percent change in tumor volume in mice treated with vehicle (n = 89), PT2399 (n = 96), or sunitinib (n = 82). d, Growth curves of each tumorgraft line grouped according to PT2399 responsiveness into sensitive (GI [growth inhibition] at end of trial >80%), intermediate (GI=40%-80%), or resistant (GI<40%). Treatment starts on day 0 and values represent mean tumor volume +/− s.e.m. To minimize bias (despite overestimation) volumes calculated as length×width×height. Each XP had n ~ 3–5 tumors per treatment group (vehicle n = 89, PT2399 n = 96, sunitinib n = 82). a–c, Tests completed using a mixed model with compound symmetrical covariance structure for mice in the same tumourgraft line using vehicle as the reference group. **, p < 0.01; ***, p < 0.001; and ****, p < 0.0001.
We hypothesized that the reduction in hemoglobin (2.0 g/dL; p=0.0001) was due to decreased erythropoietin (EPO), which is regulated by HIF-215. Consistent with this notion, red blood cell precursors were decreased by 35% (p<0.0001; Fig. 1b) and EPO, which may serve as a pharmacodynamic marker, was suppressed by 75% (p<0.0001; Fig. 1b).
PT2399 decreased tumor growth by 60% across TGs (p<0.0001) (Fig. 1c). According to their responsiveness, TGs were classified into sensitive (tumor growth inhibition at last measurement >80%), intermediate (40–80%), and resistant (<40%) (Extended Data Table 1). Forty-five percent of TGs were sensitive (10/22), 23% intermediate, and 32% resistant (Fig. 1d; Extended Data Fig. 1b and c). Sensitive tumors included tumors with aggressive sarcomatoid and rhabdoid features (Extended Data Table 1). Among ccRCCs, 56% (10/18) were sensitive. Unexpectedly, 4 ccRCCs were resistant, including 3 with VHL mutations (Extended Data Table 1).
PT2399 was more active than sunitinib (p=0.0126) (Fig. 1c and Extended Data Fig. 1b) and inhibited tumor growth in several sunitinib-resistant tumors (Fig. 1d). There was no bias in treatment allocation, as treatment groups were balanced (pre-trial: tumor size, p=0.11; tumor growth rate, p=0.22; and mouse weight, p=0.34). PT2399 reduced tumor cell density and increased fibrosis (Extended Data Fig. 1d and 1e). By Ki67 immunohistochemistry (IHC), PT2399 inhibited tumor cell proliferation 3.5 fold (mean value change of −19.5 ±2.4; p<0.0001; Extended Data Fig. 1e and 1f). Inhibition of cell proliferation was also observed in live mice using 3'-[18F]fluoro-3'-deoxythymidine PET/CT (Extended Data Fig. 1g and 1h). In addition, PT2399 collapsed the tumor vasculature, decreasing vascular area 3-fold (p=0.0011) (Extended Data Fig. 1e and 1f). To determine whether changes in vascular area were due to inhibition of tumor VEGF, we exploited the species difference between graft (human) and host (mouse). PT2399 suppressed circulating human VEGF by 93%, but mouse VEGF was unaffected (Extended Data Fig. 1i). Thus, tumor VEGF production, but not extratumoral VEGF, is HIF-2-dependent and inhibited by PT2399. This tumor selectivity represents a marked improvement over current angiogenesis inhibitors. VEGF production was also inhibited in sensitive tumors progressing on sunitinib in which PT2399 retained activity (Extended Data Fig. 2).
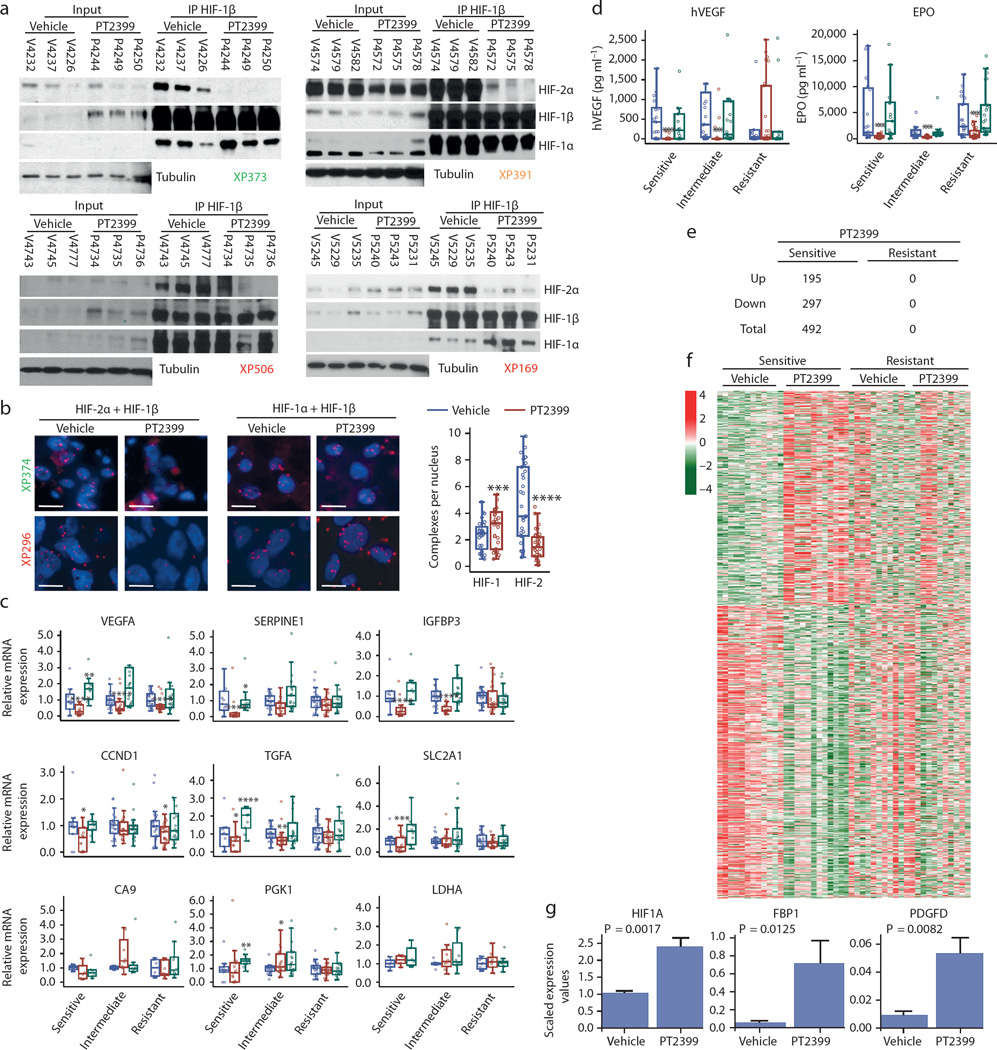
We evaluated the effects of PT2399 on HIF-2 in tumors. We immunoprecipitated (IP) the HIF-1β subunit, shared by both HIF-2α and HIF-1α. PT2399 specifically disassembled HIF-2 but not HIF-1 complexes (Fig. 2a). Similar results were observed using a proximity ligation assay (Fig. 2b). Correspondingly, PT2399 reduced the expression of HIF-2 target genes (VEGF, SERPINE1 [encoding PAI-1], IGFBP3, CCND1 [encoding Cyclin D1], TGFA, and SLC2A1 [encoding GLUT1]) (all comparisons, p<0.05; Fig. 2c), but not HIF-1 targets (CA9, PGK1, and LDHA).
Fig. 2. PT2399 dissociates HIF-2 complexes in both sensitive and resistant RCCs and induces changes in gene expression in sensitive tumors.
a, Immunoprecipitation of HIF-1β from tumor lysates of sensitive (XP373), intermediate (XP391), and resistant (XP506 and XP169) tumors from mice treated with either vehicle (Veh) or PT2399. (Samples are labeled with “V” for vehicle-treated or “P” for PT2399-treated followed by the mouse identifier.) b, Proximity ligation assay detecting either HIF-2α + HIF-1β or HIF-1α + HIF-1β heterodimers from vehicle- or PT2399-treated sensitive (XP374) or resistant (XP296) tumors and summary of results across responsive and resistant tumorgrafts. (Images representative of quantitative data shown in graph.) Summary includes analyses from 11 vehicle-treated tumors and 11 PT2399-treated tumors (3 fields were analyzed for each sample) in 5 sensitive, 3 intermediate, and 3 resistant tumorgraft trials. Scale bars = 20 µM. c, qRT-PCR for the indicated HIF-2 target genes in PT2399 sensitive, intermediate, and resistant tumors treated with vehicle (blue), PT2399 (red), or sunitinib (green). HIF-1 target genes CA9, PGK1, and LDHA included as negative controls. Excepting PGK1 and LDHA, samples were available for n = 58 vehicle-treated tumors (Sensitive: n = 11; Intermediate: n = 21; Resistant: n = 26), n = 62 PT2399-treated tumors (Sensitive: n = 15; Intermediate: n = 21; Resistant: n = 26), and n = 52 sunitinib-treated tumors (Sensitive: n = 10; Intermediate: n = 23; Resistant: n = 19). PGK1 and LDHA were available for 24 tumors for each treatment group (Sensitive: n = 6; Intermediate: n = 8; Resistant: n = 10). d, Circulating tumor-produced hVEGF as well as mouse EPO levels in mice with sensitive, intermediate, and resistant tumors treated with vehicle (blue), PT2399 (red), and sunitinib (green). ELISA data was generated for 63 vehicle-treated tumors (Sensitive: n = 21; Intermediate: n = 19; Resistant: n = 23), 74 PT2399-treated tumors (Sensitive: n = 27; Intermediate: n = 21; Resistant: n = 26), and 61 sunitinib-treated tumors (Sensitive: n = 15; Intermediate: n = 23; Resistant: n = 23). e, Number of RNAs upregulated and downregulated genes by PT2399 in sensitive and resistant tumors. f, Heatmap representation from RNAseq analysis showing differentially-regulated genes by PT2399 in sensitive compared to resistant tumors. Removal of an unclassified tumor (XP169) from the resistant group, did not affect conclusions. g, RNAseq analyses showing increased expression of selected genes by PT2399 in sensitive tumors. b–d, g: Tests completed using a mixed model with compound symmetrical covariance structure for mice in the same tumorgraft line using vehicle as the reference group. qRT-PCR levels were log-transformed for analysis; EPO and hVEGF levels were Box-Cox transformed; RNAseq levels were log2-transformed; Raw values depicted in all graphs. All bar charts depict the mean with the error bar representing s.e.m., while all boxplots have median centre values. *, p < 0.05; **, p < 0.01; ***, p < 0.001; and ****, p < 0.0001. See Supplementary Fig. 1 for gel source data.
Notably, PT2399 did not affect the majority of HIF-2 target genes in resistant tumors (Fig. 2c). A modest decrease in VEGF mRNA did not translate into lower circulating VEGF (Fig. 2d). However, as determined by reduced EPO (p=0.0002; Fig. 2d), Hif-2 was indeed inhibited by PT2399 in mice with resistant tumors. Furthermore, IP experiments showed that Hif-2 complexes were dissociated in resistant tumors (Fig. 2a). Thus, somewhat unexpectedly PT2399 disassembled HIF-2 in resistant tumors, but HIF-2 target genes were largely unaffected.
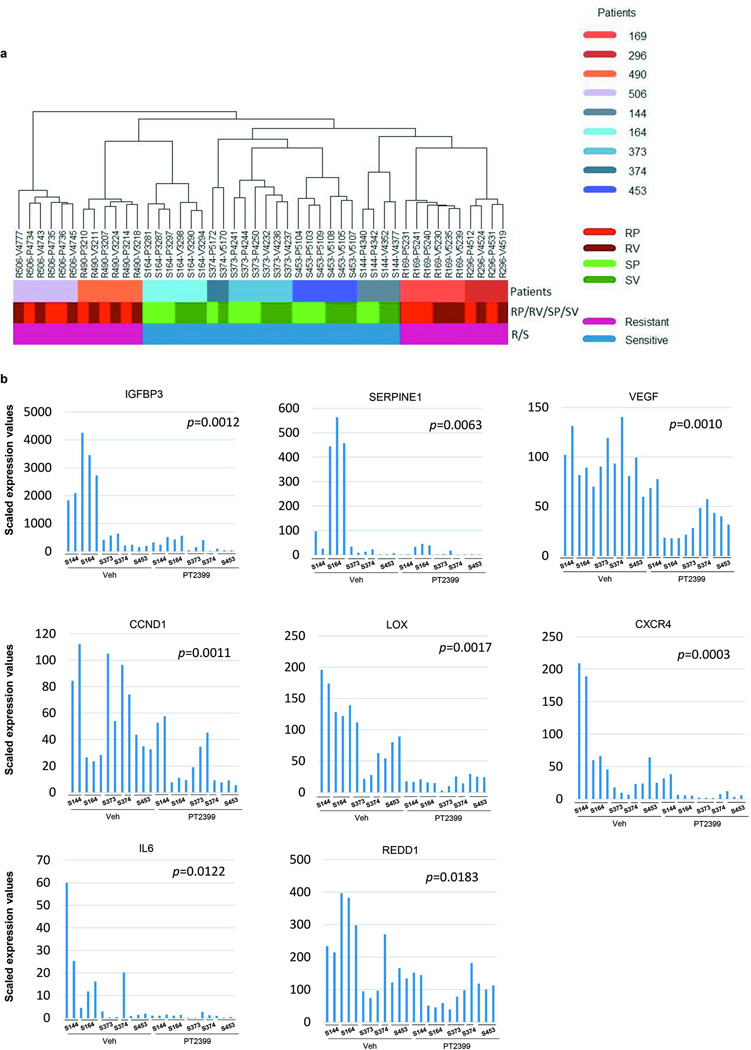
To better characterize the effects of PT2399, we performed RNA sequencing (RNAseq) on 46 tumors (Extended Data Table 1 and 2). In sensitive tumors, we identified 492 RNAs deregulated by PT2399 (FDR≤0.05) (Fig. 2e, Extended Data Table 3). In contrast, the same analysis in resistant tumors found 0 genes deregulated by PT2399 (Fig. 2e and f). Similar results were obtained by an independent, blinded analysis (H.G and C.R.). The selective changes of PT2399 in sensitive tumors suggest that PT2399 sensitivity is linked to its ability to alter gene expression. Furthermore, the lack of gene expression changes in resistant tumours suggests that PT2399 is highly specific. Consistent with this notion, PT2399 had a lesser impact on overall gene expression than subtle differences across patients’ tumors (Extended Data Fig. 3a).
Extensive studies have evaluated HIF-2 target genes in ccRCC21–23. However, by leveraging (i) PT2399 specificity; (ii) RCC tumorgrafts, with minimal human stroma24; and (iii) an RNAseq algorithm excluding contaminating mouse (stromal) transcripts, we were uniquely enabled to define the HIF-2 program. Among the 492 deregulated RNAs in sensitive tumors, 439 were protein coding, and 271 were downregulated including canonical HIF-2 targets previously examined (IGFBP3, SERPINE1, VEGF, CCND1) as well as LOX, CXCR4, IL6 and REDD1 (Extended Data Fig. 3b). Pathway and gene set enrichment analyses showed downregulation of cell cycle, DNA replication, cell cycle checkpoint, and DNA repair processes (Extended Data Table 4). Interestingly, regulation of DNA repair genes by HIF-2, previously observed in cell lines6, may explain ccRCC radioresistance. PT2399 increased the expression of 168 protein-coding genes, including fibrosis-related genes, such as PDGFD, HIF1A (previously shown to be induced by HIF-2α knockdown22), and FBP1, a gluconeogenic gene recently reported to suppress RCC progression25 (Fig. 2g; Extended Data Table 4).
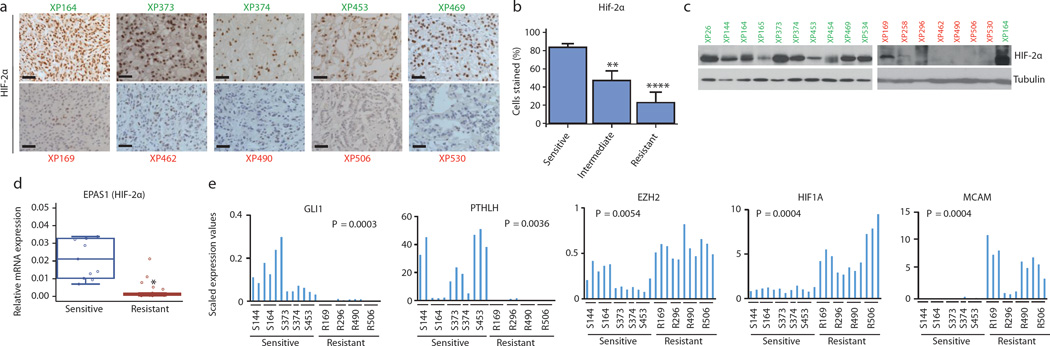
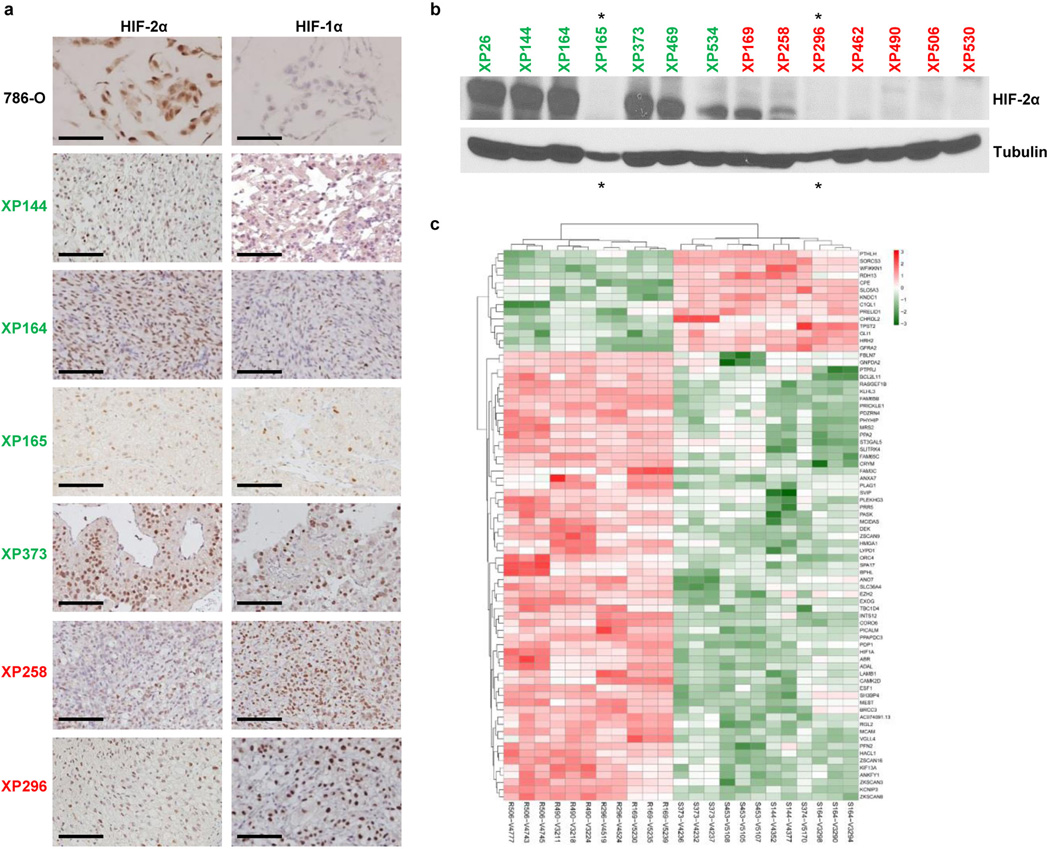
We sought to identify a biomarker distinguishing sensitive from resistant tumors. We found that HIF-2α protein was expressed in 83% of cells in sensitive tumors compared to 23% in resistant (p<0.0001; Fig. 3a and b; Extended Data Fig. 4a). While there were differences even within tumors, higher HIF-2α expression in sensitive tumors was observed by western (Fig. 3c; Extended Data Fig. 4b) and RT-PCR (Fig. 3d). Lower, at times undetectable, HIF-2α levels in resistant tumors may explain the lack of PT2399 effects on gene expression in this group.
Fig. 3. Sensitive and resistant tumors can be distinguished by HIF-2α levels and gene expression signature.
a, HIF-2α expression by immunohistochemistry (IHC) in sensitive (green) and resistant (red) tumors. Scale bars = 50 µM. (Images representative of quantitative data shown in 3b.) b, Quantification of HIF-2α-positive cells as determined by IHC in sensitive, intermediate, and resistant tumors from all 22 tumorgraft lines (Sensitive: n = 10; Intermediate: n = 5; Resistant: n = 7). c, Western blot analysis of sensitive (green) and resistant (red) tumorgraft lines. XP164 lysate loaded twice as a reference for comparison between the two membranes. d, qRT-PCR of EPAS1 (HIF-2α) expression in sensitive (n = 11) versus resistant (n = 26) vehicle-treated tumorgrafts. e, Candidate genes from RNAseq analysis differentially expressed in sensitive and resistant tumors. b: An ANOVA test was used to determine if sensitive tumors were different from intermediate or resistant. Bar chart depicts the mean with the error bar representing s.e.m. d, e: Tests completed using a mixed model analysis with compound symmetrical covariance structure for mice in the same tumorgraft line. RNAseq values were log2-transformed for analysis; Raw values depicted in all graphs. Bar charts depict individual RNA-seq values, while all boxplots have median centre values. **, p < 0.01; and ****, p < 0.0001. See Supplementary Fig. 1 for gel source data.
Next, we compared RNAseq datasets between vehicle-treated sensitive and resistant tumors. Using a rigorous Wilcox test, we identified 1,327 differentially expressed RNAs (Extended Data Table 3) including 94 (76 mRNAs) uniformly over- or under-expressed transcripts across every sensitive vs. resistant tumor sample (Extended Data Figure 2D; Extended Data Table 3). GLI1, a transcription factor of the sonic hedgehog family, and PTHLH (Parathyroid hormone-like hormone), a neuroendocrine peptide implicated in epithelial-mesenchymal interactions and calcium ion transport, were uniformly overexpressed in sensitive tumors (Fig. 3e). Notably, HIF1A was increased in the resistant group (Fig. 3e). Increased expression of HIF-1α protein was also observed by IHC in some, but not all, resistant tumors (Extended Data Fig. 4a). EZH2 and MCAM were also overexpressed in resistant tumors (Fig. 3e).
Overall, our data show that ccRCC can be classified into HIF-2 dependent and independent tumors and that these tumors differ in HIF-2α (and possibly, HIF-1α) levels and in their gene expression. These subtypes did not correlate with BAP1 and PBRM1 status26 in this small series (Extended Data Table 1). Our results point to different mechanisms of tumorigenesis downstream of VHL and may underlie differences in behavior or responsiveness to therapy.
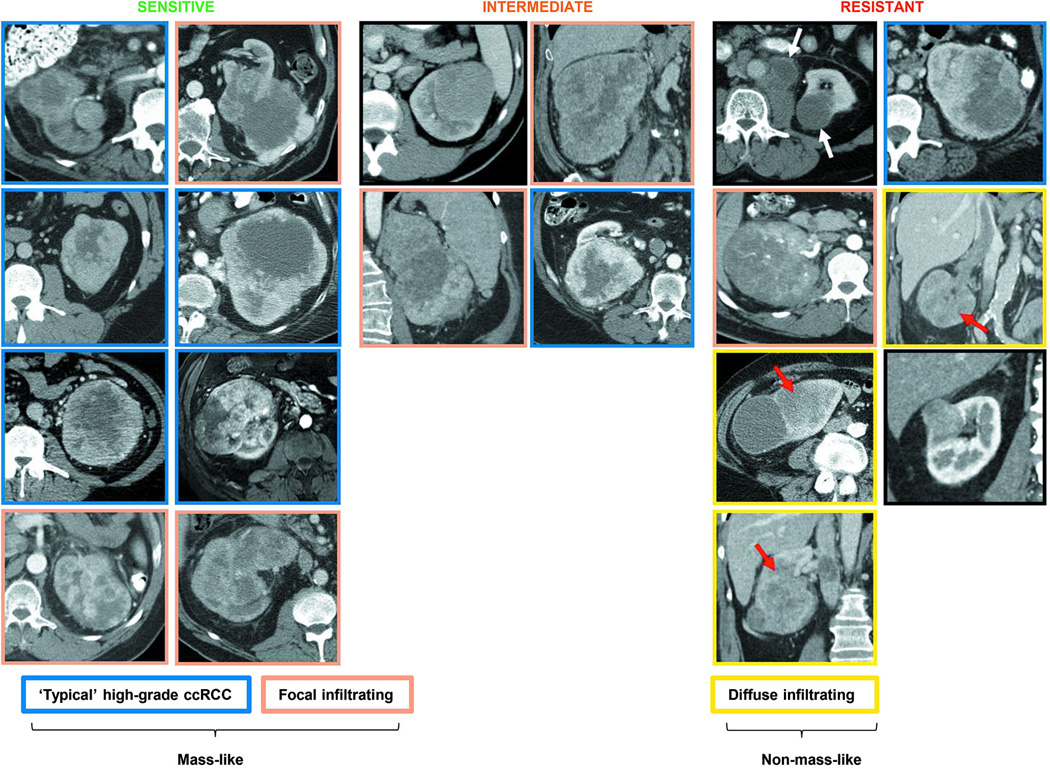
Given differences in gene expression, we asked whether sensitive and resistant tumors diverged in imaging characteristics. We obtained CT scan images from patient tumors giving rise to tumorgrafts prior to surgery. The sensitive group was characterized by tumors with peripheral hypervascularity and a central non-enhancing area (typical of high-grade ccRCC27) and if present, tumor infiltration was focal (Extended Data Fig. 5). The resistant group was more heterogeneous, but several tumors were relatively hypovascular and diffusely infiltrating (Extended Data Fig. 5).
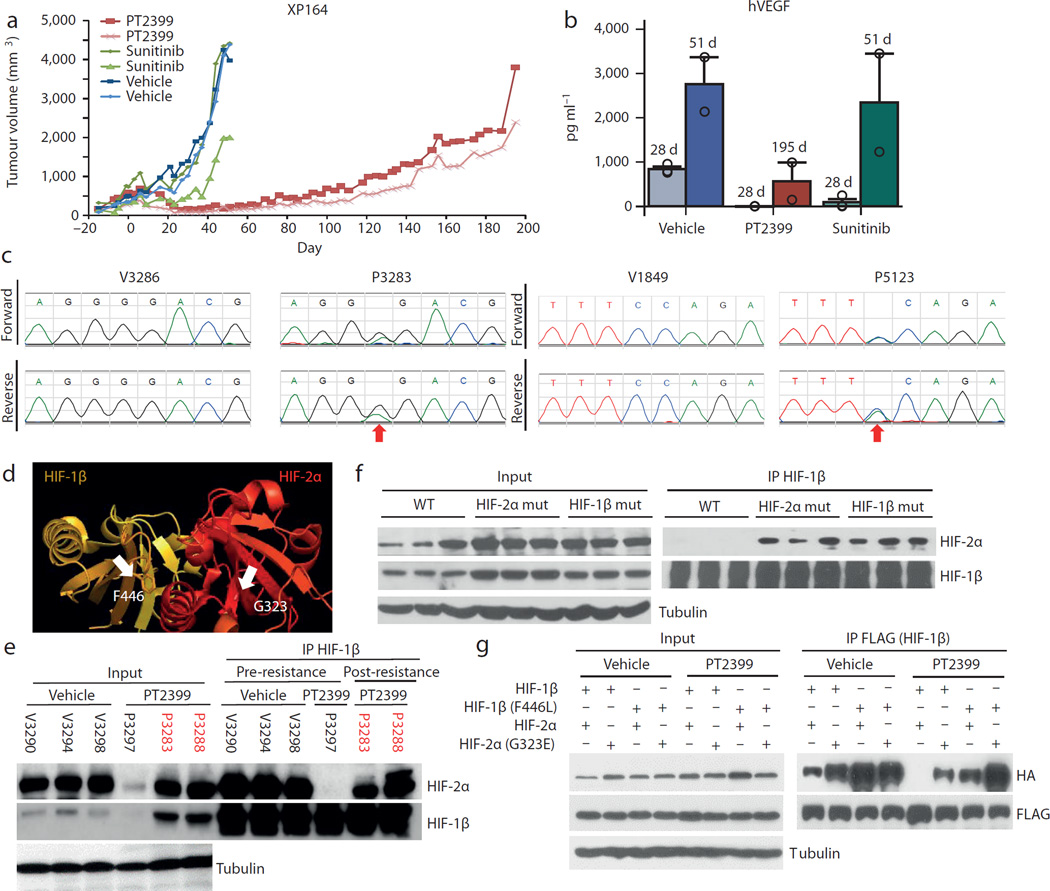
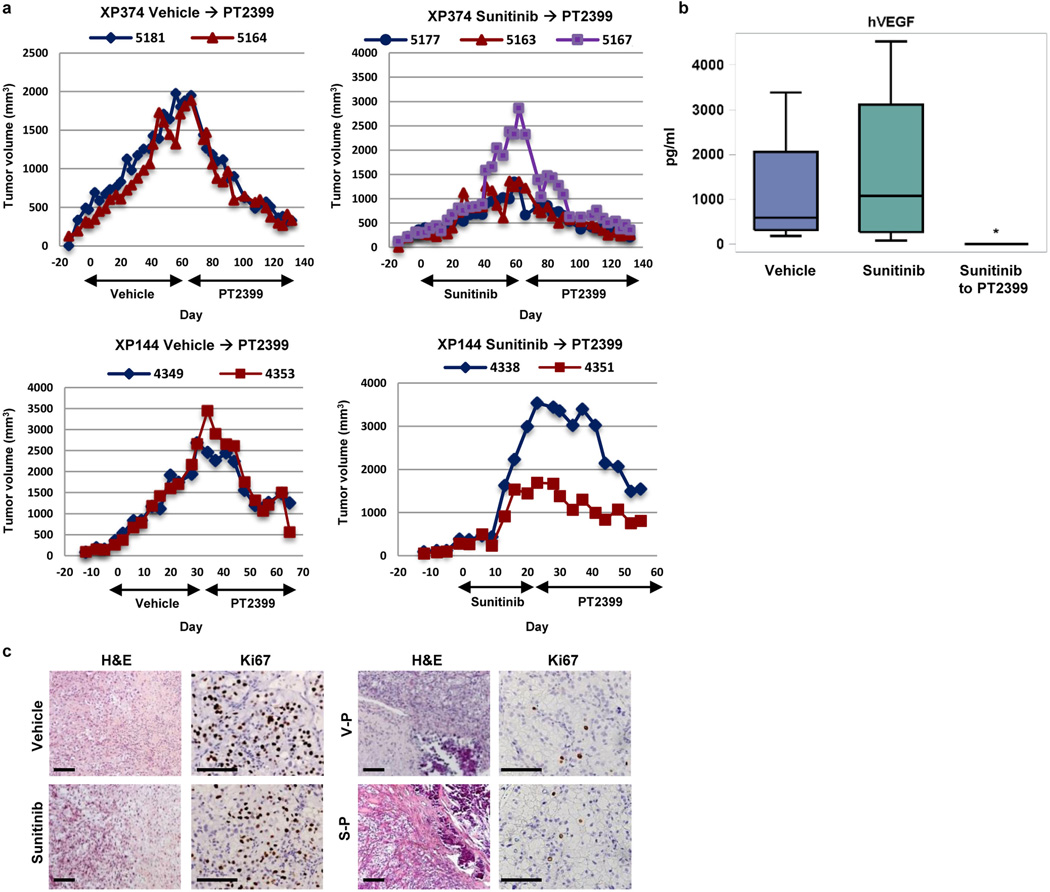
We asked whether sensitive tumors would acquire resistance. We exposed mice bearing tumours formed from a sensitive tumourgraft (XP164) to prolonged treatment with PT2399 or sunitinib. Sunitinib resistance developed within 60 days (Fig. 4a; compare to Fig. 1D), but resistance to PT2399 took >100 days (Fig. 4a). PT2399 resistance was associated with increased tumor vascularity and higher tumor VEGF production (Fig. 4b). We sequenced the HIF-2α gene (EPAS1) and identified a c.968G>A heterozygous mutation resulting in a G323E substitution in one tumor (Fig. 4c). The mutation was absent in a vehicle-treated tumor and in the second resistant tumor (despite originating from the same parental tumor). Structural analyses of HIF-2 quaternary structure28 showed that G323 is at the entrance of the cavity, where PT2399 binds (Fig. 4d). Akin to engineered mutations19,29, a glutamate side chain would prevent PT2399 access. Consistent with this notion, PT2399 failed to dissociate HIF-2 complexes in mutant tumors (Fig. 4e).
Fig. 4. Acquired resistance following prolonged PT2399 exposure.
a, Tumor volumes from a cohort of mice of the XP164 tumorgraft line treated with vehicle (blue lines, n=2; V3286 and V3299); sunitinib until the development of resistance (green lines, n = 2; S3295 and S3296; compare to Fig. 1d); or PT2399 (red lines, n = 2; P3283 and P3288). b, Circulating human VEGF levels in mice treated for the indicated number of days (d) showing increased tumor-produced VEGF with development of resistance (all bars, n = 2). c, Bidirectional chromatograms from tumorgrafts developing resistance compared to controls: P3283 (c.968G>A in EPAS1 [HIF2A] leading to a G323E) and P5123 (derived from P3288; c.1338C>A in ARNT [HIF1B] leading to a F446L). d, Crystal structure of PAS-B domains from HIF-2α bound to HIF-1β (PDB entry 4ZP428) highlighting side chains of G323 (lining opening of PT2399 binding pocket in HIF-2α) and F446 (in HIF-1β at the interface with HIF-2α). In another structure (PDB entry 4GHI18) quaternary arrangement between HIF-2α and HIF-1β PAS-B domains differs, but F446 remains at the interface. e, HIF-1β IP from XP164 tumorgrafts before and after (red) development of resistance showing reformation of HIF-2α/HIF-1β complexes following the acquisition of resistance (V, vehicle; P, PT2399). f, HIF-1β IP from tumors of mice with HIF-2α or HIF-1β mutations (or wild-type controls) treated with PT2399 (n=3 mice per group). g, FLAG IP from HEK293T cells transfected with plasmids encoding FLAG-tagged HIF-1β (FLAG-HIF-1β; FLAG-HIF-1β-F446L) or HA-tagged HIF-2α (HA-HIF-2α; HA-HIF-2α-G323E) and treated with either vehicle or PT2399. See Supplementary Fig. 1 for gel source data.
We then sequenced HIF-1β from the second resistant tumor, and failed to identify a mutation. Nevertheless, HIF-2 complexes had reformed (Fig. 4e). The tumor was passaged in mice, which were maintained on PT2399, and remained resistant. IP experiments again showed dimeric HIF-2 complexes (Fig. 4f). Interestingly, sequencing of passaged tumors revealed a heterozygous c.1338C>A mutation resulting in a F446L substitution in the HIF-1β PAS-B domain (Fig. 4c). F446 is at the interface between HIF-1β and HIF-2α (Fig. 4d). We postulate that F446L functions as a second-site suppressor mutation and that a more flexible side chain at the complex interface accommodates conformational changes induced by PT2399 allowing drug-bound HIF-2α to bind to HIF-1β. Both HIF-1β (F446L) and HIF-2α (G323E), when expressed in cells, were sufficient to preserve HIF-2 dimers despite PT2399 and the effects appeared additive (Fig. 4g). Overall, these results pave the way for second generation inhibitors and/or complementary approaches leveraging other potential drug-binding pockets recently revealed28.
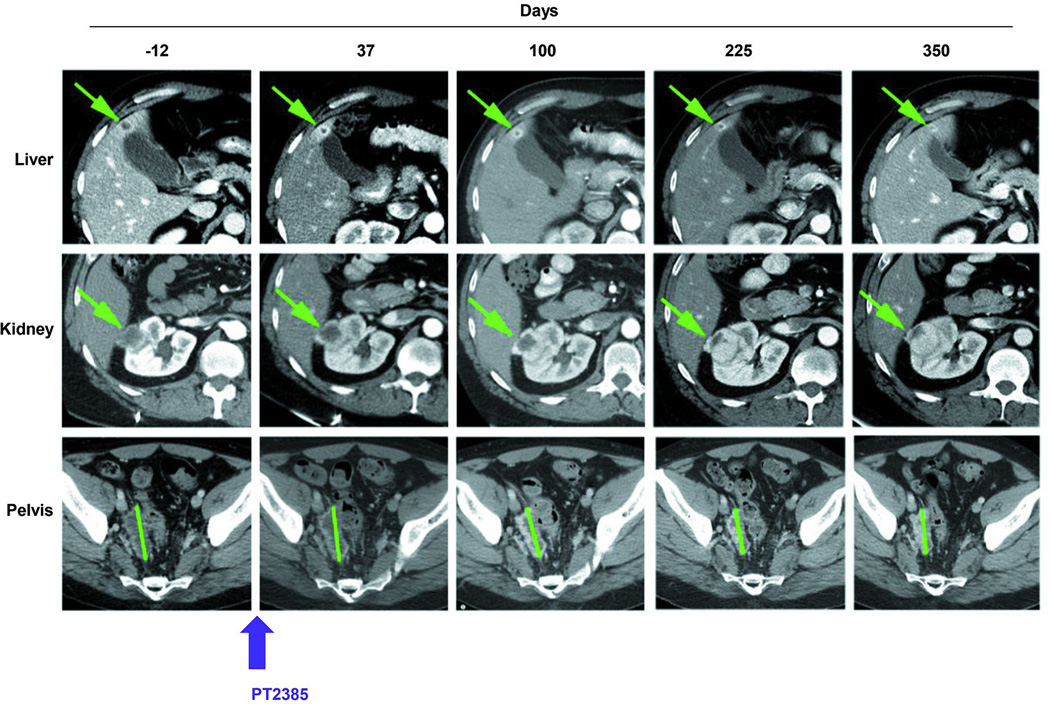
Interestingly, a patient with metastatic ccRCC, whose tumour gave rise to a sensitive tumourgraft (XP165) enrolled in a phase 1 trial with PT238530 (NCT02293980). The patient, a 47 year old male, had originally presented with omental and abdominal wall metastases following a radical nephrectomy of a stage III ccRCC of high grade. After a failed attempt at surgical removal, he had received high-dose IL2, bevacizumab, sorafenib, everolimus, sunitinib, pazopanib, and axitinib. Despite extensive pretreatment, he remained free of progression on PT2384 for more than 11 months (Extended Data Fig. 6). These data validate HIF-2 as a target for ccRCC, provide insight into HIF-2-mediated tumorigenesis, establish variable tumour dependency on HIF-2 identifying different ccRCC subtypes and associated biomarkers that may be incorporated in future clinical trials, showcase the specificity of PT2399, and anticipate mechanisms of resistance.
Materials and Methods
Nomenclature
Throughout the manuscript and figures, “XP” refers to the particular tumorgraft line. “V” refers to vehicle, “S” refers to sunitinib, and “P” refers to PT2399. Numbers following V, S, or P refer to the particular mouse identifier (ear tag) of that sample.
Drug trials
Drug trials in tumorgraft mice were done as previously described12,13. Briefly, ~64 mm3 fragments of tissue from stably growing orthotopic tumorgrafts were implanted subcutaneously in 4–6 week old female and male NOD/SCID mice. When tumor volumes reached ~300–600 mm3, mice were segregated into treatment groups (3–5 mice/group) based on (i) tumor volume, (ii) growth rate, and (iii) mouse weight. A sample size of five mice per treatment arm gave us 80% power to detect a significant tumor volume differential at the 28th day after treatment between the reference arm and a treatment arm using a two-sample t-test, assuming a true 600 mm3 tumor volume difference with a standard deviation of 250 and attrition margin of ~20%. Since the mixed model analysis uses ~8 repeated measures from each mouse, even with a few more covariates included in the model, the power will be similar or even higher. Vehicle (10% EtOH, 30% PEG400, 60% MCT [0.5% methyl cellulose, 0.5% Tween 80 (aq)]) was administered by gavage every 12 h. Sunitinib (LC Laboratories) was administered by oral gavage every 12 h at 10 mg/kg in 0.5% CMC in D5W. PT2399 (Peloton Therapeutics, Inc.) was administered at 100 mg/kg by oral gavage in 10% EtOH, 30% PEG400, 60% MCT. Mouse weights were taken weekly and treatment doses were adjusted weekly. Tumors were generally measured twice a week using a digital caliper. While leading to an overestimation in tumor volumes, to minimize bias12, tumor volume was calculated by the formula: tumor volume = l × w × h, where l is the largest dimension of the tumor, w is the largest diameter perpendicular to l, and h is maximal height of the tumor. Trials typically lasted 4 weeks, but this varied depending upon tumor growth rates. Overall, >14,000 measurements were obtained. Assuming a digital caliper measurement error rate up to 10%, 99.8% of measurements where within protocol limits. Consideration was given to tumor growth rates, curve separation and the foreseeable need for additional mice for repeat experiments. Mice were monitored during treatment and provided appropriate veterinary care. In accordance with UT Southwestern’s Institutional Animal Care and Use Committee (IACUC) policies, animals were euthanized within timeframes specified by the veterinary staff once tumor diameters were greater than 2 cm. Mice were also euthanized if they exhibited signs of adverse clinical health. A total of n = 22 tumorgraft trials were completed with n = 89 vehicle-treated tumors (Sensitive: n = 39; Intermediate: n = 22; Resistant: n = 28), n = 96 PT2399-treated tumors (Sensitive: n = 42; Intermediate: n = 24; Resistant: n = 30), and n = 82 sunitinib-treated tumors (Sensitive: n = 32; Intermediate: n = 22; Resistant: n = 28).
Blood cell counts and hemoglobin measurements
Complete Blood Counts (CBC) (platelets, white blood cells, neutrophils, lymphocytes, and hemoglobin) were measured at the end of ~28 day trials and run on an IDEXX ProCyte Dx analyzer. CBCs were available for 17 tumorgraft trials, with 52 vehicle-treated mice, 58 PT2399-treated mice (Sensitive: n = 13; Intermediate: n = 19; Resistant: n = 26), and 53 sunitinib-treated mice (Sensitive: n = 8; Intermediate: n = 22; Resistant: n = 23).
PET/CT
3'-[18F]fluoro-3'-deoxythymidine ([18]FLT) was synthesized at 160°C for 10 min using a GE FXN module through the nucleophilic substitution reaction between 2,3’-Anhydro-5’-O-benzoyl-2’-deoxythymidine and [18F]KF in DMSO, followed by deprotecting the benzoyl group in 1N NaOH solution. The product was separated and purified by HPLC. The injection dose of [18]FLT was prepared in saline containing 10% ethanol. Small animal positron emission tomography/computerized tomography (PET/CT) imaging studies were performed on a Siemens Inveon PET/CT Multimodality System. PET/CT were conducted on mice with both orthotopic and subcutaneous tumors. Orthotopic tumorgrafts were implanted using 2–3 pieces of 2×2×2 mm tissue underneath the left renal capsule of NOD/SCID mice. Once tumors became palpable, a baseline PET/CT scan was performed, and within 72h, PT2399 treatment was started. PT2399 was continued for 8–10 days, when a second PET/CT was performed to assess tumor response. After injection with 0.12 mCi of [18]FLT via the tail-vein, and a 60 min wait period to allow for the radiotracer’s distribution and uptake, mice were anesthetized using 3% isoflurane, which was decreased to 2% during imaging. CT imaging was acquired at 80 kV and 500 μA with a focal spot of 58 μm. The PET imaging was acquired for 500 s directly following the acquisition of CT data. CT images were reconstructed with Cobra Reconstruction Software, and PET images were reconstructed using the OSEM3D algorithm. Reconstructed CT and PET images were fused and analyzed using the manufacturer’s software. For quantification, regions of interest were drawn aided by CT images and then quantitatively expressed as percent injected dose per gram of tissue (%ID/g).
Immunohistochemistry
Immunohistochemistry (IHC) was performed using Dako Autostainer Link 48. The HIF-1α and HIF-2α immunohistochemical procedures and interpretations were standardized based on expression profiles in well-characterized cell lines (786-O, 786-O empty vector, and 786-O VHL-reconstituted cell lines) and human ccRCC tissue with known expression for these two proteins by western blot. Multiple commercially available antibodies were evaluated and the antibodies with most consistent results were selected for further studies. Briefly, for HIF-1α and HIF-2α staining, after hydration, antigen retrieval was accomplished with EnVision™ FLEX Target Retrieval Solution, Low pH (K800521, Dako) in a Dakocytomation Pascal pressure cooker; Ki67 and CD31 antigen retrieval was done using a Dako PT Link. Slides were incubated in 3% hydrogen peroxide for 10 min. Primary antibodies were added and incubated for 40 min at RT. Primary antibodies: HIF-1α (1:500, NB100-105, Novus), HIF-2α (1:200, sc-46691, Santa Cruz), Ki67 (ready-to-use, IR-626, Dako) and CD31 (1:200, LS-B1932, LifeSpan BioSciences). After rinsing with wash buffer, EnVision FLEX mouse/rabbit linker (K802121/K800921, Dako) was applied to the tissue and incubated for 10 min. Secondary antibody, EnVision™ FLEX/HRP (K800021, Dako), was incubated for 20 min. Sections were then processed using the Envision™ FLEX Substrate Working Solution for 10 min followed by dehydration in a standard ethanol/xylene series, and mounting media (8310-4, Thermo Scientific). IHC of HIF-1α and HIF-2α was performed on pre-treated tumorgraft tissue for n = 22 tumorgraft lines. Appropriate positive and negative controls were used with each run of immunostaining. The percentage of tumor cells in the entire section examined was recorded by a pathologist blinded to the western blot results. Only a 2 or 3+ nuclear positive reaction was considered as positive expression (staining scale: 0 = no staining, 1 = weak, 2 = moderate, 3 = strong).
Proliferation index and microvessel quantitation
To assess tumor proliferation index, we performed immunostaining for Ki67, and to assess tumor microvessels we performed CD31 immunostaining on tumors following treatment with vehicle or PT2399. IHC was completed on n = 10 sensitive tumorgraft lines, with n = 28 vehicle-treated tumors and n = 31 PT2399-treated tumors. Slides were digitally scanned using an Aperio Scanscope AT Turbo and reviewed using the Aperio eSlide Manager (ver. 12.0.0.5040) and Imagescope (ver. 12.1.0.5029) systems (Leica Biosystems, Melbourne, Australia). For Ki67, Aperio Genie (ver. 11.2) pattern recognition software was used to identify and select tumor areas for quantitative analysis with the Aperio Nuclear algorithm (version 11.2), yielding a percentage of tumor nuclei positive for Ki67. In a small subset of tumors where Genie inadequately identified tumor cells, representative tumor regions were manually selected (tumor necrosis areas were avoided) and reanalyzed. Quantitative measurements of microvessels including density, and average lumen area were obtained using the Aperio Microvessel algorithm (version 11.2) from manually-selected representative tumor regions.
Real-time PCR
RT-PCR data was generated for 16 tumorgraft trials, except for CA9 and LDHA, which were evaluated in 12 tumorgraft trials. Three samples were run concurrently for each tumor. Total RNA was isolated as described previously31. cDNA was synthesized using iScript™ Reverse Transcription Supermix for RT-qPCR (170-8841, Bio-Rad). qRT-PCR was performed on a Bio-Rad CFX96 Real-Time PCR system using iTaq Universal SYBR Green SMX (1725124, Bio-Rad). Primers were synthesized by Invitrogen. Primers available upon request.
VHL Methylation
HIF2-I sensitive ccRCC tumorgrafts that had wild-type VHL status (XP164, XP373, XP453, and XP454) were tested for VHL methylation using the Affymetrix Promoter Methylation PCR Kit (MP1100).
Immunoprecipitation and Western Blot
Tumor tissue was lysed in IP buffer containing 25 mM Tris•HCl pH 7.4, 150 mM NaCl, 1% NP-40, 1 mM EDTA, 5% glycerol, 0.5 mM DTT, with 3–4 freeze/thaw cycles. 10–20% of the lysate was saved for input; 40 µg was mixed with 3× loading buffer (10% SDS, 33.3% glycerol, 300 mM DDT, 0.2% Bromophenol Blue) for input. After pre-clearing the lysate with 50 μl of a 1:1 solution of recombinant Protein G-Sepharose 4B (101242, Life Technologies) for 1 hour, 1 mg protein was mixed with 20 μl of ARNT/HIF-1β antibody (sc-55526, Santa Cruz) and rocked overnight at 4°C. 30 μl of Protein G-Sepharose 4B equilibrated with IP buffer were then added, rocked for 1 h at 4° C, and spun at 3,000 rpm for 10 seconds. The supernatant was removed and the beads washed 3 times with IP buffer containing DTT. 20 μl of 1× loading buffer was added to the beads and vortexed gently, then boiled for 5 minutes and spun at max speed for 5 minutes. The entire sample was loaded for western blot analysis. For western blot analysis, both HIF-1α antibody (A300-286A, Bethyl) and HIF-2α antibody (NB100-122, Novus) were diluted at 1:1,000 in 5% BSA and incubated overnight at 4° C. Tubulin antibody (T5168, Sigma) was diluted at 1:5,000. Primary antibodies were detected using horseradish peroxidase-conjugated secondary antibodies (31430, 31460, Pierce) followed by exposure to enhanced chemiluminescence substrate [mixing 1:1 solution 1 (2.5 mM luminol, 0.4 mM pCoumaric acid, 0.1 M Tris-HCl) and solution 2 (0.015% H2O2, 0.1 M Tris-HCl).
Sanger sequencing
Mutations in the PAS-B domain of HIF2A were identified with primers: Forward: GTGGTGCACACCCCTGCCCC; Reverse: CTGGGAAGCTTGGGCACCCCC; and in HIF1B with: Forward: GTCTCTGAAAGGAAGCATGAG; Reverse: CACATAGGGCATCAGAAGTG
Transfections
HEK293T cells (ATCC; no perceived need for authentication or mycoplasma testing) were cotransfected with the indicated expression plasmids using Lipofectamine 2000 (Invitrogen) following manufacturer’s instructions. After 36 h, cells were treated with PT2399 (10 µM) at 37° C for 5 h, harvested for immunoprecipitation with anti-FLAG beads (A2220-1ML, Sigma) and then subjected to western blot analysis. Plasmids laboratory database ID #206 (HIF-1α) and #207 (HIF-2α) were used for in vitro translation. Plasmids #930 (pcDNA3.1 Flag-HIF1β), #931 (pcDNA3.1 Flag-HIF1β [F446L]), #932 (pLVX HA-HIF-2α-IRES-zsGreen), and #933 (pLVX-HA-HIF-2α [G323E]-IRES-zsGreen).
In Silico Structural Analysis
The G323E and F446L mutations were evaluated using PyMOL and Protein Data Bank 4ZP428.
ELISA
Mouse VEGF (MMV00), Human VEGF (DVE00), and Mouse Erythropoietin (MEP00B) ELISA kits were from R&D Systems. Briefly, 50 μl of Assay Diluent was added to each well. 50 μl of either standard, control, or sample was then added to a well. For mEPO and mVEGF ELISA, a 2-fold and 5-fold dilution, respectively, of the serum was made with Calibrator Diluent. For hVEGF, no dilution was performed. Plates were incubated for 2 h at RT on a horizontal orbital microplate shaker, then aspirated and washed for a total of five washes. After the last wash, 100 μl of mEPO/mVEGF/hVEGF conjugate was added to each well and incubated for 2 h at RT on a shaker. Plates were washed five times with wash buffer and 100 μl of Substrate Solution was added to each well and incubated for 30 min at RT during which time the plates were covered to protect from the light. Stop Solution was then added to each well, with gentle tapping to ensure thorough mixing. The optical density of each well was determined using a microplate reader set to 450 nm. Wavelength correction was set to 540 nm. The final OD value was obtained by subtracting readings at 540 nm from the readings at 450 nm. ELISA data was generated for a total of 20 tumorgraft trials.
Proximity Ligation Assay
Mouse anti-HIF-1α (NB100-105, Novus), mouse anti-HIF-2α (sc-46691X, Santa Cruz) and Rabbit anti-ARNT/HIF-1β (A302-765A, Bethyl) were used. Primary antibodies were concentrated and buffers were exchanged using a Vivaspin 500 Centrifugal Concentrator (VS0131, Fisher Scientific). Antibodies were diluted to 1 mg/mL in PBS. Primary antibody conjugation was done with a Duolink® In Situ Probemaker MINUS/PLUS kit (DUO92010 & DUO92009, Sigma-Aldrich). Briefly, 2 µl of Conjugation Buffer was added to 20 µl of the antibody (1 mg/ml), mixed gently, transferred to one vial of lyophilized oligonucleotide (PLUS or MINUS), and incubated at RT overnight. 2 µl of Stop Reagent was then added to the reaction and incubated at RT for 30 min. 24 µl of Storage Solution was added and the conjugation stored at 4°C. Tumor tissue was blocked with PBS-T (0.1% Triton X-100) + 1% BSA for 30 min after antigen retrieval. Conjugated HIF1-α-MINUS, HIF2-α-MINUS and ARNT-PLUS were diluted in blocking buffer containing 1× Assay Reagent (20×) at a dilution of 1:50, 1:50, and 1:200, respectively. The mixture was allowed to sit for 20 min at RT before diluted primary antibody was added to each sample. Slides were incubated in a humidity chamber overnight 4°C. Duolink® In Situ Detection Reagents Red (DUO92008, Sigma-Aldrich) were used for signal detection. Briefly, slides were washed with Wash Buffer A, ligation solution containing ligase at a 1:40 dilution was added, and slides were incubated in a pre-heated humidity chamber for 30 min at 37°C. After washing in 1× Wash Buffer A with gentle agitation, amplification solution containing polymerase was added at a 1:80 dilution, and slides were then incubated in a pre-heated humidity chamber for 100 min at 37°C. After washing slides in 1× Wash Buffer B and then 0.01× Wash Buffer B, slides were dried at RT in the dark, and mounted with a cover slip using a minimal volume of Duolink In Situ Mounting Medium with DAPI (DUO82040, Sigma-Aldrich). After approximately 15 min, slides were analyzed by fluorescence microscopy (Olympus) using a 40× objective. Image analysis was done with the Image J 1.48V program. Pictures of 3 fields for each sample were used. At least 100 cells of each sample were counted.
RNA sequencing
23 vehicle- and 23 PT2399-treated tumor RNA samples, including 5 sensitive XPs (XP144, XP164, XP373, XP374, and XP453) and 4 resistant XPs (XP169, XP296, XP490, and XP506) underwent RNA sequencing at the New York Genome Center. RNA sequencing libraries were prepared using the Illumina TruSeq Stranded mRNA Sample Preparation Kit. Briefly, 500 ng of total RNA was purified by oligo-dT beads selecting for polyadenylated RNA species and fragmented by divalent cations under elevated temperature. The fragmented RNA underwent first strand synthesis using reverse transcriptase and random primers. Second strand synthesis created the cDNA fragments using DNA polymerase I. Following RNaseH treatment, the cDNA fragments went through end repair, adenylation of the 3’ ends, and ligation of adapters. The cDNA library was enriched using 8 cycles of PCR and purified. Quality control consisted of assaying the final library size using the Agilent Bioanalyzer and quantifying the final library by RT- PCR and PicoGreen (fluorescence) methods. A single peak between 250–350 bp indicated a properly constructed and amplified library ready for sequencing. Sequencing was performed on a HiSeq 2500 using v4 SBS chemistry according to the Illumina protocol, as described32. Sequencing libraries were loaded onto the HiSeq 2500 flowcell for clustering on the cBot using the instrument-specific clustering protocol. Given HiSeq 2500 capabilities (200–250M passed filter 2×50bp sequencing reads per flow cell lane), we sequenced 5 samples per lane in order to obtain a minimum of 50M PF reads per sample. With one exception, >100 million reads were obtained per sample (median 146,644,355; 95% distribution-free CI: 142,380,928 – 151,324,826; Extended Data Table 1). Any gene with more than 50 reads in any sample was kept; only genes that had low reads in all of the samples were removed. This left 20,667 genes after removal of pseudogenes. cDNA sequences were aligned to a combined index of mouse and human reference sequences with STAR v 2.4.0c. Mouse reads were filtered out and the remaining reads were re-mapped to the NCBI hg37 using STAR aligner (v2.3.1z)33. Quantification of genes annotated in Gencode v19 was performed using HTSeq34. Picard and RSeQC35 were used to collect QC metrics (http://broadinstitute.github.io/picard/). Differential gene expression analysis was measured using edgeR36. A false discovery rate (FDR) cutoff of 0.05 was applied to identify the statistically significant genes between comparison groups. FDR was calculated using Benjamini and Hochberg method37 for adjusting for multiple hypothesis testing.
RNA Seq data was deposited into the Sequence Read Archive (SRP073253). For RNAseq data, the TG number is preceded by an “S” for sensitive or “R” for resistant followed by treatment and ear tag. Sample1 (S144-P4340), Sample2 (S144-P4342), Sample3 (S164-P3281), Sample4 (S164-P3287), Sample5 ( S164-P3297), Sample6 (S373-P4241), Sample7 (S373-P4244), Sample8 (S373-P4250), Sample9 (S374-P5172), Sample10 (S453-P5103), Sample11 (S453-P5104), Sample12 (S453-P5109), Sample13 (S144-V4352), Sample14 (S144-V4377), Sample15 (S164-V3290), Sample16 (S164-V3294), Sample17 (S164-V3298), Sample18 (S373-V4232), Sample19 (S373-V4236), Sample20 (S373-V4237), Sample21 (S374-V5170), Sample22 (S453-V5105), Sample23 (S453-V5107), Sample24 (S453-V5108), Sample25 (R169-P5231), Sample26 (R169-P5240), Sample27 (R169-P5241), Sample28 (R296-P4512), Sample29 (R296-P4531), Sample30 (R490-P3207), Sample31 (R490-P3210), Sample32 (R490-P3214), Sample33 (R506-P4734), Sample34 (R506-P4735), Sample35 (R506-P4736), Sample36 (R169-V5230), Sample37 (R169-V5235), Sample38 (R169-V5239), Sample39 (R296-V4519), Sample40 (R296-V4524), Sample41 (R490-V3211), Sample42 (R490-V3218), Sample43 (R490-V3224), Sample44 (R506-V4743), Sample45 (R506-V4745), Sample46 (R506-V4777).
Statistical Analyses
Apart from the RNAseq analysis, all reported p-values were obtained from two-tailed tests at the 0.05 significance level. All bar charts depict the mean with the error bar representing s.e.m., while all boxplots have median center values with fences extending to the greatest value inside the upper and lower fences (1.5[IQR] away from the 75th and 25th percentiles, respectively). Transformations were used where indicated to meet normality assumptions for analysis. These tests were completed using SAS 9.4 (SAS Institute Inc., Cary, NC). Except where indicated, the experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Regulatory
Written informed consent was obtained from the patient participating in the phase 1 clinical trial “A Phase 1, Dose-Escalation Trial of PT2385 Tablets In Patients With Advanced Clear Cell Renal Cell Carcinoma” (NCT02293980). UT Southwestern IACUC-approved animal protocol, APN 2015-100932, includes all live vertebrate experimental procedures.
Extended Data
Extended Data Fig. 1. Effects of PT2399 on human RCC-bearing mice.
a, Platelet, white blood cell, neutrophil, and lymphocyte counts from tumorgraft-bearing mice treated with vehicle (n = 52), PT2399 (n = 58), or sunitinib (n = 53) at the end of drug trial period (~28 days). (Low lymphocyte levels throughout consistent with expected levels in age and sex matched NOD/SCID mice.) b, Tumor growth trend lines for sensitive, intermediate, and resistant groups after controlling for baseline tumor volume (refer to Fig. 1d for individual curves). c, Representative gross images of tumors from sensitive (XP373 and XP164; green) and resistant (XP490 and XP169; red) lines at the end of drug trial. d, Representative H&E images illustrating different effects of PT2399 on sensitive tumors including patchy intercellular fibrosis and hyalinization (open arrow heads), reduced tumor necrosis (red arrows), decreased tumor cell density (XP164 and XP469), reduced nuclear to cytoplasmic ratio (XP469), cell ballooning (filled arrow), and dystrophic calcification (blue stars). Scale bars = 50 µM. e, Summary of histopathological changes induced by PT2399 in 10 sensitive tumorgrafts represented as number of tumors (N) compared to the total or as mean ± s.e. in 28 vehicle-treated tumors compared to 31 PT2399-treated tumors. MVD, microvessel density per mm2; MLA, mean lumen area (μm2). PT2399 collapsed tumor vasculature without decreasing number of CD31-expressing endothelial cells. f, (Upper panel) Immunohistochemistry (IHC) for Ki67 in tumors harvested from sensitive (XP144 and XP373) or resistant (XP530 and XP506) tumors following treatment with vehicle or PT2399. (Lower panel) H&E staining and IHC for CD31 in sensitive tumors (XP373 and XP469) treated with vehicle or PT2399. Scale bars = 100 µM. g, Representative [18F]FLT-PET/CT images of mice with subcutaneous tumorgrafts treated with either vehicle or PT2399. Yellow arrows point to tumors where there is uptake of [18F]fluoro-3'-deoxythymidine. h, Representative [18F]FLT-PET/CT images of XP144 mice with orthotopic tumors before and after treatment with PT2399 for 10 days. Yellow arrowheads, kidney tumors. White asterisks, intestine. FLT uptake in tumor compared to normal kidney reduced by 19% after 10-day treatment (n = 3; paired t-test p=0.0010). i, Human and mouse VEGF levels in plasma as determined by ELISA in different treatment groups (Vehicle: n = 63; PT2399: n = 74; Sunitinib: n = 61). a, i: Tests completed using a mixed model analysis with compound symmetrical covariance structure for mice in the same tumorgraft line using vehicle as the reference group. b: Trend lines were obtained from a mixed model analysis for each response group using an autoregressive (1) covariance structure for the longitudinal measurements on each mouse, compound symmetry for mice within the same tumorgraft line, and controlled for baseline volume. e: Continuous measures were analyzed using a mixed model with compound symmetrical covariance structure for mice in the same tumorgraft line and using vehicle treatment as the reference group. Specifically for categorical variables, a binomial test was used to test if the proportion of tumors affected by PT2399 compared to vehicle was different than 10%. hVEGF, and mVEGF levels were Box-Cox transformed; Raw values depicted in all graphs. All boxplots have median centre values. *, p < 0.05; ***, p < 0.001; and ****, p < 0.0001.
Extended Data Fig. 2. Evaluation of the effects of PT2399 on tumors progressing on sunitinib.
a, Tumor volumes in mice from sensitive lines (XP374 or XP144) switched from vehicle or sunitinib to PT2399 as indicated (bottom black arrows). b, Circulating tumor-produced hVEGF levels in mice treated with vehicle, sunitinib, or sunitinib followed by PT2399. The Wilcoxon rank-sum test was used to determine if sunitinib (n = 4) or sunitinib followed by PT2399 (n = 6) were different than vehicle (n = 4). *, p<0.05. Boxplots have median centre values. c, Representative images of H&E and Ki67 staining of tumors from mice (XP144) treated with vehicle or sunitinib (left panel) and from tumors following a switch to PT2399 (right panel). Scale bars = 100 µM.
Extended Data Fig. 3. RNAseq analyses of vehicle and PT2399-treated tumorgrafts.
a, Unsupervised clustering analyses of all tumorgraft samples (sensitive and resistant, both vehicle- and PT2399-treated) showing clustering by tumorgraft line. b, RNA sequencing in sensitive tumorgrafts evaluating the effects of PT2399 on selected HIF-2 target genes. All tests completed using a mixed model analysis with compound symmetrical covariance structure for mice in the same TG line. Values were log2-transformed for analysis; Raw values depicted in all graphs as individual bars.
Extended Data Fig. 4. HIF-2α and HIF-1α levels in sensitive and resistant tumorgrafts.
a, HIF-2α and HIF-1α immunohistochemistry. 786-O cells, which express high levels of HIF-2α, shown as controls. Scale bars, 100 µm. b, Western blot analyses showing heterogeneity within tumors but with overall similar results (compare to Fig. 3c). Green, sensitive; Red, resistant. Asterisks, underloaded samples. c, Heatmap from RNA-seq analysis showing differentially expressed genes in sensitive (S) versus resistant (R) tumourgrafts based on uniform cutoff (see Extended Data Table 3). See Supplementary Fig. 1 for gel source data.
Extended Data Fig. 5. Evaluation of imaging characteristics of tumors in patients corresponding to sensitive, intermediate, and resistant tumorgrafts.
CT scan images from patient tumors that gave rise to tumorgrafts according to TG sensitivity to PT2399. Tumors were classified into masses with peripheral hypervascularity and a central non-enhancing area (blue outline), focally infiltrating (brown outline) and diffuse infiltrating (yellow outline). Three of the seven resistant tumors presented as non-mass-like, infiltrative neoplasms (red arrows) whereas another tumor presented with both a largely necrotic renal mass and retroperitoneal lymph nodes (black outline; white arrows).
Extended Data Fig. 6. Extended disease control in heavily pretreated patient with metastatic ccRCC with sensitive (XP165) tumorgraft.
CT images of selected lesions in patient treated with highly related HIF-2 inhibitor (PT2385) in phase 1 clinical trial showing overall stability in the size of lesions over time. Start of treatment, day 0.
Extended Data Table 1. Tumorgraft features.
Fuhrman grade of primary tumor and stage at presentation (metachronous metastasis may have developed); Tissue, engrafted tissue; IHC, immunohistochemistry from tumorgrafts; GI, growth inhibition; ccRCC, clear-cell renal cell carcinoma; tRCC, translocation renal cell carcinoma; Abd, abdominal; Tu Thr, tumor thrombus; LN, lymph node; mut, mutant; wt, wild-type.
| Response | XP NO. | Histology | Fuhrman Grade |
Tissue | Stage at presentation |
VHL status |
BAP1 (IHC) |
PBRM1 (IHC) |
Relative GI% (p value) |
RNA seq |
|---|---|---|---|---|---|---|---|---|---|---|
| Sensitive | XP26 | ccRCC | 2 | Adrenal | pT1aNxMx | mut | mut | wt | 87 (0.0003) | |
| XP144 | ccRCC | 4 | Kidney | pT2aN1Mx | mut | wt | wt | 98 (<0.0001) | Y | |
| XP164 | ccRCC* | 4 | Kidney | pT4NxMx | wt‡ | wt | wt | 134 (<0.0001) | Y | |
| XP165 | ccRCC | 3 | Abd wall | pT3bNxMx | mut | wt | mut | 112 (<0.0001) | ||
| XP373 | ccRCC | 4 | Tu Thr | pT3aN1M1 | wt† | mut | wt | 103 (<0.0001) | Y | |
| XP374 | ccRCC*# | 4 | Kidney | pT4NxMx | mut | wt | wt | 109 (<0.0001) | Y | |
| XP453 | ccRCC | 3 | Tu Thr | pT3bN0Mx | wt† | wt | wt | 110 (<0.0001) | Y | |
| XP454 | ccRCC | 3 | Kidney | pT3aN0Mx | wt‡ | mut | wt | 156 (0.0032) | ||
| XP469 | ccRCC | 3 | Kidney | pT3bN0Mx | mut | mut | mut | 91 (<0.0001) | ||
| XP534 | ccRCC | 4 | Kidney | pT3aN0Mx | mut | mut | n/a | 129 (0.0007) | ||
| Intermediate | XP237 | tRCC | n/a | LN | pT3aN1M1 | n/a | wt | wt | 43 (0.0144) | |
| XP391 | ccRCC | 4 | Tu Thr | pT3bN0Mx | mut | wt | mut | 45 (0.0018) | ||
| XP426@ | ccRCC# | 4 | Kidney | pT4N1Mx | mut | wt | wt | 44 (0.0273) | ||
| XP427@ | ccRCC# | 4 | LN | pT4N1MX | mut | wt | wt | 54 (0.0206) | ||
| XP466 | ccRCC | 3 | Kidney | pT3aN0M1 | wt | mut | n/a | 67 (0.0030) | ||
| Resistant | XP169 | Unclassified | n/a | Kidney | pT4N1M1 | wt | wt | wt | 0 (0.0119$) | Y |
| XP258 | ccRCC* | 4 | Kidney | pT3aN0M1 | mut | mut | wt | 39 (0.11) | ||
| XP296 | ccRCC* | 4 | Kidney | pT3aNxM1 | mut | wt | wt | 29 (0.30) | Y | |
| XP462 | Unclassified | n/a | Kidney | pT3aN0M1 | wt | wt | mut | 29 (0.11) | ||
| XP490 | ccRCC*# | 4 | Kidney | pT3aN1M1 | mut | wt | wt | 39 (0.89) | Y | |
| XP506 | ccRCC | 3 | Ascites | pT3aN1Mx | wt‡ | wt | wt | 20 (0.76) | Y | |
| XP530 | Unclassified | n/a | Kidney | pT3bN0Mx | wt | n/a | n/a | 2 (0.68) |
Independent tumors from same patient,
Sarcomatoid differentiation,
Rhabdoid features,
PT2399-treated mice had greater relative growth than vehicle-treated mice,
promoter methylation,
promoter not methylated
Extended Data Table 2. RNA sequencing read data.
Samples are labeled as S (sensitive) or R (resistant) followed by the tumorgraft (XP) line, a “–“ linking to treatment type (P, PT2399 or V, vehicle) and mouse identifier (ear tag).
| Samples | Read Count | Samples | Read Count |
|---|---|---|---|
| S144-P4340 | 131,078,351 | R169-P5231 | 150,980,881 |
| S144-P4342 | 127,945,953 | R169-P5240 | 146,751,739 |
| S164-P3281 | 121,045,606 | R169-P5241 | 144,959,159 |
| S164-P3287 | 128,070,443 | R296-P4512 | 151,324,826 |
| S164-P3297 | 138,586,535 | R296-P4531 | 144,982,512 |
| S373-P4241 | 162,092,320 | R490-P3207 | 142,380,928 |
| S373-P4244 | 146,116,441 | R490-P3210 | 164,412,241 |
| S373-P4250 | 140,629,410 | R490-P3214 | 169,970,555 |
| S374-P5172 | 88,374,928 | R506-P4734 | 165,472,466 |
| S453-P5103 | 120,921,569 | R506-P4735 | 154,474,148 |
| S453-P5104 | 108,148,316 | R506-P4736 | 173,988,590 |
| S453-P5109 | 117,009,388 | R169-V5230 | 159,863,685 |
| S144-V4352 | 128,119,810 | R169-V5235 | 146,783,488 |
| S144-V4377 | 148,456,002 | R169-V5239 | 144,377,378 |
| S164-V3290 | 144,464,174 | R296-V4519 | 146,536,970 |
| S164-V3294 | 161,750,684 | R296-V4524 | 148,798,769 |
| S164-V3298 | 152,823,172 | R490-V3211 | 162,273,604 |
| S373-V4232 | 156,310,574 | R490-V3218 | 123,559,977 |
| S373-V4236 | 150,155,973 | R490-V3224 | 151,672,989 |
| S373-V4237 | 148,496,505 | R506-V4743 | 181,173,536 |
| S374-V5170 | 130,903,402 | R506-V4745 | 156,598,756 |
| S453-V5105 | 123,966,544 | R506-V4777 | 164,427,358 |
| S453-V5107 | 123,347,998 | ||
| S453-V5108 | 112,341,672 |
Extended Data Table 3. Number of differentially-regulated genes across tumorgraft groups by RNAseq analysis.
| SP vs. SV | RV vs. SV | RP vs. SP | ||||
|---|---|---|---|---|---|---|
| (Cutoff) | Up | Down | Up | Down | Up | Down |
|
Original (FDR < 0.05) |
195 | 297 | 1776 | 1766 | 1640 | 1815 |
|
T-Test (P-value < 0.01) |
99 | 213 | 852 | 584 | 798 | 695 |
|
Wilcox (P-value < 0.01) |
90 | 207 | 829 | 498 | 760 | 621 |
|
Uniform (all Δ>0 or Δ<0) |
2 | 5 | 78 | 16 | 61 | 15 |
S = sensitive; R = resistant; V = vehicle; P = PT2399; FDR = False Discovery Rate
Extended Data Table 4. Top 15 downregulated and top 15 upregulated pathways in sensitive tumors treated with PT2399.
| Ingenuity Canonical Pathways | Molecules | P-Value |
|---|---|---|
| Top 15 Downregulated pathways | ||
| Cell Cycle Control of
Chromosomal Replication |
CDC7,ORC1,MCM7,CDC45,MCM2,CDK2,ORC6,MCM3,MCM6,MCM4,MCM5,CDC6 | 7.94328E-16 |
| Mitotic Roles of Polo-Like Kinase | KIF23,ESPL1,PRC1,CDC20,CCNB2,PPP2R2A,CDC7,PLK4,PTTG1,PLK1,CDK1,CCNB1,FBXO5 | 3.98107E-12 |
| Cell Cycle: G2/M DNA Damage Checkpoint Regulation |
TOP2A,BRCA1,AURKA,CKS1B,CKS2,CCNB2,PLK1,CDK1,CCNB1 | 2.51189E-08 |
| Cyclins and Cell Cycle Regulation | CCNE2,SUV39H1,CCND1,CDK2,E2F1,CCNA2,CCNB2,PPP2R2A,CDK1,CCNB1 | 1.31826E-07 |
| ATM Signaling | JUN,BLM,BRCA1,FANCD2,CDK2,RAD51,CCNB2,CDK1,CCNB1 | 1.38038E-07 |
| GADD45 Signaling | CCNE2,BRCA1,CCND1,CDK2,CDK1,CCNB1 | 1.77828E-07 |
| DNA damage-induced 14-3-3σ Signaling | CCNE2,BRCA1,CDK2,CCNB2,CDK1,CCNB1 | 1.77828E-07 |
| Estrogen-mediated S-phase Entry | CCNE2,CCND1,CDK2,E2F1,CCNA2,CDK1 | 8.31764E-07 |
| Role of BRCA1 in DNA Damage Response |
BLM,BRCA1,FANCB,FANCD2,E2F1,FANCA,BRCA2,RAD51,PLK1 | 1.58489E-06 |
| Hereditary Breast Cancer Signaling | BLM,BRCA1,FANCB,FANCD2,CCND1,E2F1,FANCA,BRCA2,RAD51,CDK1,CCNB1 | 1.8197E-06 |
| Role of CHK Proteins in Cell Cycle Checkpoint Control |
BRCA1,CDK2,E2F1,CLSPN,PPP2R2A,PLK1,CDK1 | 1.20226E-05 |
| Ovarian Cancer Signaling | BRCA1,VEGFA,SUV39H1,LEF1,PTGS2,CCND1,E2F1,PRKAG2,BRCA2,RAD51 | 1.65959E-05 |
| Pancreatic Adenocarcinoma Signaling | VEGFA,SUV39H1,PTGS2,CCND1,BIRC5,CDK2,E2F1,BRCA2,RAD51 | 1.99526E-05 |
| Aryl Hydrocarbon Receptor Signaling | JUN,CCNE2,MCM7,CCND1,IL1A,CDK2,E2F1,CCNA2,IL6,ALDH1A3 | 2.29087E-05 |
| Small Cell Lung Cancer Signaling | CCNE2,SUV39H1,PTGS2,CCND1,CDK2,E2F1,CKS1B | 6.45654E-05 |
| Top 15 Upregulated pathways | ||
| Hepatic Fibrosis / Hepatic Stellate
Cell Activation |
COL19A1,COL4A6,FGFR2,IGFBP4,TGFB3,MYL4,PDGFD,COL21A1,MYL3,IGF1 | 8.13E-06 |
| Axonal Guidance Signaling | GLIS2,CXCL12,BMP5,MYL4,PDGFD,EPHB3,SRGAP3,SMO,SEMA3F,IGF1,ADAMTS1,SEMA3G,BMP4, MYL3 |
2.09E-05 |
| Human Embryonic Stem Cell Pluripotency |
FGFR2,TGFB3,BMP5,PDGFD,BMP4,LEFTY2,SMO | 0.000145 |
| PAK Signaling | MYL4,PDGFD,EPHB3,MYLK,MYL3 | 0.001 |
| Basal Cell Carcinoma Signaling | GLIS2,BMP5,BMP4,SMO | 0.00302 |
| RhoA Signaling | MYL4,MYLK,SEMA3F,MYL3,IGF1 | 0.00389 |
| Agranulocyte Adhesion and Diapedesis | CXCL12,MYL4,MYL3,CCL11,CXCL14,MMP24 | 0.004266 |
| Cellular Effects of Sildenafil (Viagra) | MYL4,ADCY5,MYLK,MYL3,PDE5A | 0.004467 |
| TR/RXR Activation | THRSP,HIF1A,G6PC,AKR1C1/AKR1C2 | 0.00631 |
| Regulation of Actin-based Motility by Rho | DIRAS3,MYL4,MYLK,MYL3 | 0.006918 |
| Factors Promoting Cardiogenesis
in Vertebrates |
TGFB3,BMP5,BMP4,SMO | 0.007413 |
| CXCR4 Signaling | DIRAS3,CXCL12,MYL4,ADCY5,MYL3 | 0.010233 |
| Cardiomyocyte Differentiation via
BMP Receptors |
BMP5,BMP4 | 0.011482 |
| Cardiac Hypertrophy Signaling | DIRAS3,TGFB3,MYL4,ADCY5,MYL3,IGF1 | 0.012303 |
| Molecular Mechanisms of Cancer | DIRAS3,TGFB3,BMP5,BCL2L11,BMP4,ADCY5,HIF1A,SMO | 0.013183 |
Acknowledgments
We thank the patients who generously provided tissues for our studies. PT2399 was kindly provided by Peloton Therapeutics, Inc. Funding was provided by Peloton Therapeutics, Inc. (OTD-105466), CPRIT (RP160440) and philanthropy, including Tom Green memorial. W.C. is supported by grants from the National Natural Science Foundation of China (No. 811011934) and the Science and Technology Program of Guangzhou, China (No. 2012J5100031). M.S.K. and H.Z. are supported by a grant from CPRIT (RP150596). I.P. is supported by grants from NIH (RO1CA154475, P50CA196516). X.S. is supported by a grant from CPRIT (RP110771). J.B. is a Virginia Murchison Linthicum endowed scholar and is supported by grants from NIH (RO1CA175754, P50CA196516, P30CA142543) and CPRIT (RP130603). R.K.B. is the Michael L. Rosenberg Scholar in Medical Research and was supported by CPRIT (RP130513). Histology equipment was purchased with funding from the National Center for Advancing Translational Sciences (Center for Translational Medicine UL1TR001105).
Footnotes
Author contributions: W.C. designed and performed biochemical experiments; H.H. performed tumorgraft experiments; A.C. performed extensive statistical analyses with the supervision of X.-J. X.; M.S.K. performed RNAseq analyses under the supervision of T.H.H. and Y.X., who also supervises H.Z. on sequencing analysis; E.H., A.P.J., Y.M., N.P., Q.Y. and A.J. performed experiments; F.H. and P.Y. performed histological analyses under the supervision of P.K.; G.H. performed PET studies under the supervision of X.S.; I.P. performed patient imaging analyses; H.G. and C.R. performed RNAseq validation studies at the New York Genome Center; J.C. is the clinical research coordinator of the PT2385 phase 1 trial overseen by K.C. and N.Z.; K.H.G., R.K.B., and E.F. participated in discussions; T.W., J.P.R., E.M.W., and J.A.J. oversaw the development and characterization of PT2399 and provided the drug; R.M.M. assisted with manuscript preparation, writing and submission; and J.B. conceived and supervised the project, and wrote the manuscript with input from R.M.M. and the other authors.
Competing interests: T.W, J.P.R., E.M.W., N.Z. and J.A.J. are employees and own equity in Peloton Therapeutics, Inc. and K.H.G. and R.K.B. have licensed IP, consult for and own equity. M.S.K., T.H., Y.X. and J.B. are authors on a filed patent pertaining to a biomarker of PT2399. J.B. is a member of the advisory board for Bethyl Laboratories.
Data and materials availability: RNAseq was released to NCBI Sequence Read Archive (SRA). ID: SRP073253
References
- 1.Gnarra JR, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nature genetics. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 2.Herman JG, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:9700–9704. doi: 10.1073/pnas.91.21.9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG., Jr Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer cell. 2002;1:237–246. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 6.Bertout JA, et al. HIF2alpha inhibition promotes p53 pathway activity, tumor cell death, and radiation responses. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14391–14396. doi: 10.1073/pnas.0907357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Covello KL, et al. HIF-2alpha regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes & development. 2006;20:557–570. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ema M, et al. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1alpha regulates the VEGF expression and is potentially involved in lung and vascular development. Proceedings of the National Academy of Sciences of the United States of America. 1997;94:4273–4278. doi: 10.1073/pnas.94.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer cell. 2007;11:335–347. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mendel DB, et al. In vivo antitumor activity of SU11248, a novel tyrosine kinase inhibitor targeting vascular endothelial growth factor and platelet-derived growth factor receptors: determination of a pharmacokinetic/pharmacodynamic relationship. Clinical cancer research. 2003;9:327–337. [PubMed] [Google Scholar]
- 11.Koehler AN. A complex task? Direct modulation of transcription factors with small molecules. Current opinion in chemical biology. 2010;14:331–340. doi: 10.1016/j.cbpa.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pavia-Jimenez A, Tcheuyap VT, Brugarolas J. Establishing a human renal cell carcinoma tumorgraft platform for preclinical drug testing. Nature protocols. 2014;9:1848–1859. doi: 10.1038/nprot.2014.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sivanand S, et al. A validated tumorgraft model reveals activity of dovitinib against renal cell carcinoma. Science translational medicine. 2012;4:137ra175. doi: 10.1126/scitranslmed.3003643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes & development. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 15.Rankin EB, et al. Hypoxia-inducible factor-2 (HIF-2) regulates hepatic erythropoietin in vivo. J Clin Invest. 2007;117:1068–1077. doi: 10.1172/JCI30117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erbel PJ, Card PB, Karakuzu O, Bruick RK, Gardner KH. Structural basis for PAS domain heterodimerization in the basic helix--loop--helix-PAS transcription factor hypoxia-inducible factor. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:15504–15509. doi: 10.1073/pnas.2533374100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheuermann TH, et al. Artificial ligand binding within the HIF2alpha PAS-B domain of the HIF2 transcription factor. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:450–455. doi: 10.1073/pnas.0808092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scheuermann TH, et al. Allosteric inhibition of hypoxia inducible factor-2 with small molecules. Nature chemical biology. 2013;9:271–276. doi: 10.1038/nchembio.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cho H, et al. On-target efficacy of a HIF2α antagonist in preclinical kidney cancer models. Nature. 2016 doi: 10.1038/nature19795. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wallace EM, et al. A small molecule antagonist of HIF-2a is efficacious in preclinical models of renal cell carcinoma. Cancer research. 2016 doi: 10.1158/0008-5472.CAN-16-0473. In press. [DOI] [PubMed] [Google Scholar]
- 21.Hu CJ, et al. Differential regulation of the transcriptional activities of hypoxia-inducible factor 1 alpha (HIF-1alpha) and HIF-2alpha in stem cells. Mol Cell Biol. 2006;26:3514–3526. doi: 10.1128/MCB.26.9.3514-3526.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raval RR, et al. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Biol. 2005;25:5675–5686. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keith B, Johnson RS, Simon MC. HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nature reviews. Cancer. 2012;12:9–22. doi: 10.1038/nrc3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pena-Llopis S, et al. BAP1 loss defines a new class of renal cell carcinoma. Nature genetics. 2012;44:751–759. doi: 10.1038/ng.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li B, et al. Fructose-1,6-bisphosphatase opposes renal carcinoma progression. Nature. 2014;513:251–255. doi: 10.1038/nature13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brugarolas J. Molecular genetics of clear-cell renal cell carcinoma. J Clin Oncol. 2014;32:1968–1976. doi: 10.1200/JCO.2012.45.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pedrosa I, et al. MR classification of renal masses with pathologic correlation. European radiology. 2008;18:365–375. doi: 10.1007/s00330-007-0757-0. [DOI] [PubMed] [Google Scholar]
- 28.Wu D, Potluri N, Lu J, Kim Y, Rastinejad F. Structural integration in hypoxia-inducible factors. Nature. 2015;524:303–308. doi: 10.1038/nature14883. [DOI] [PubMed] [Google Scholar]
- 29.Rogers JL, et al. Development of inhibitors of the PAS-B domain of the HIF-2alpha transcription factor. Journal of medicinal chemistry. 2013;56:1739–1747. doi: 10.1021/jm301847z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Courtney KD, et al. A phase I dose escalation trial of PT2385, a first-in-class oral HIF-2a inhibitor, in patients with advanced clear cell renal cell carcinoma. J Clin Oncol. 2016;34(Suppl) doi: 10.1200/JCO.2017.74.2627. Abstract 2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pena-Llopis S, Brugarolas J. Simultaneous isolation of high-quality DNA, RNA, miRNA and proteins from tissues for genomic applications. Nature protocols. 2013;8:2240–2255. doi: 10.1038/nprot.2013.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- 36.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society. 1995;Series B 57:289–300. [Google Scholar]