Abstract
Background:
A key requirement for a useful power calculation is that the calculation mimic the data analysis that will be performed on the actual data, once that data is observed. Close approximations may be difficult to achieve using analytic solutions, however, and thus Monte Carlo approaches, including both simulation and bootstrap resampling, are often attractive. One setting in which this is particularly true is cluster-randomized trial designs. However, Monte Carlo approaches are useful in many additional settings as well. Calculating power for cluster-randomized trials using analytic or simulation-based methods is frequently unsatisfactory due to the complexity of the data analysis methods to be employed and to the sparseness of data to inform the choice of important parameters in these methods.
Methods:
We propose that among Monte Carlo methods, bootstrap approaches are most likely to generate data similar to the observed data. In bootstrap approaches, real data are resampled to build complete data sets based on real data that resemble the data for the intended analyses. In contrast, simulation methods would use the real data to estimate parameters for the data, and would then simulate data using these parameters. We describe means of implementing bootstrap power calculation.
Results:
We demonstrate bootstrap power calculation for a cluster-randomized trial with a censored survival outcome and a baseline observation period.
Conclusions:
Bootstrap power calculation is a natural application of resampling methods. It provides a relatively simple solution to power calculation that is likely to be more accurate than analytic solutions or simulation-based calculations, in the sense that the bootstrap approach does not rely on the assumptions inherent in analytic calculations. This method of calculation has several important strengths. Notably, it is simple to achieve great fidelity to the proposed data analysis method and there is no requirement for parameter estimates, or estimates of their variability, from outside settings. So, for example, for cluster-randomized trials, power calculations need not depend on intracluster correlation coefficient estimates from outside studies. In contrast, bootstrap power calculation requires initial data that resemble data that are to be used in the planned study. We are not aware of bootstrap power calculation being previously proposed or explored for cluster-randomized trials, but it can also be applied for other study designs. We show with a simulation study that bootstrap power calculation can replicate analytic power in cases where analytic power can be accurately calculated. We also demonstrate power calculations for correlated censored survival outcomes in a cluster-randomized trial setting, for which we are unaware of an analytic alternative. The method can easily be used when preliminary data are available, as is likely to be the case when research is performed in health delivery systems or other settings where electronic medical records can be obtained.
Keywords: 2014 Group Health Seattle Symposium; Power and sample size; cluster randomized trials, bootstrap; resampling
Introduction
Statistical Power
“Statistical power” is defined as “the probability of rejecting the null hypothesis, given that some particular alternative hypothesis (“the alternative”) is true.” Power is particularly important from the perspectives of ethics and of allocating scarce resources. It is often ethically unjustifiable to randomize more subjects than are required to yield sufficient power, and it is a waste of resources to invest time or money in studies that have little chance of rejecting the null or when power is far greater than necessary.
In many settings, the question of how to calculate power is reasonably well addressed by closed-form equations or easily tractable mathematical methods. For instance, the power for an ordinary least squares regression is described in basic textbooks.1 Power for logistic regression can use iterative techniques or relatively simple formulae.2,3 Major statistical packages such as SAS (SAS Institute, Cary NC) contain routines for power calculation, and both functions and packages for power calculation are available for the free and open-source R environment.4 There are also several stand-alone packages that simplify the calculation of power, for example, PASS (NCSS Inc., Kaysville, Utah).
However, there are many settings in which these simple solutions are unsatisfactory. To see how, it is helpful to understand a primary principle of power calculations: the methods used must conform reasonably well to the planned analysis. If we plan to study a confounded relationship using a linear regression, the power assessment must include the confounder. Assessing power using an unconfounded linear regression calculation will misrepresent the power obtained in the actual analysis. If we know the outcome-predictor relationship is heteroscedastic, we should not use closed-form solutions that depend on homoscedasticity. Again, the power obtained in the actual study will not be well represented in that closed-form solution. If our study design includes a baseline period, we should not use a post-only comparison for estimating the power. These simple solutions must be rejected in favor of methods that acknowledge the real-world data settings in which the analysis is to be performed, not the restrictions of the existing power calculation solutions.
Cluster-Randomized Trials
One setting in which power assessment is not simple is cluster-randomized trials. In this design, a relatively small number of administrative clusters, such as hospitals, classrooms, or physician practices, are recruited. Each cluster may contain a large numbers of individuals upon whom outcomes will be measured. Rather than randomize subjects individually to treatment arms, all of the individuals within a cluster are randomized to the same treatment arm, and in practice we say that the cluster itself is randomized to one treatment arm or another.
This study design often reduces cost considerably, and in many settings it is the best way to get estimates of pragmatic effects—the effects of an intervention in a typical clinical population and in settings like those that nontrial patients are likely to encounter. For example, interventions on doctors to affect prescribing practices could hardly generate generalizable results if we randomize patients. We must randomize doctors, but examine the impact on patients.
Randomization by cluster leads to complications in data analysis that have long been recognized by statisticians.5,6 This is due to the likelihood of patients within a cluster to resemble each other, or, more formally, a lack of independence between subjects. This can be parameterized as the covariance or correlation between subjects within a cluster (the “intracluster correlation coefficient” or ICC) or as the variance of cluster-specific parameters ( ). Valid approaches for estimating and testing treatment effects include calculating summary statistics by cluster in a first step and then comparing cluster summaries by treatment arm in a second step, and mixed effects models that incorporate all individual observations in a single model.5,6
There are several existing analytic approaches to calculating the power for cluster-randomized trials. Many of these rely on the “design effect”, 1+(m−1) ρ, where m is the number of observations per cluster and ρ is the ICC.5–8 The “effective sample size” is calculated by dividing the actual number of subjects by the design effect. Power assessment can then continue using methods for uncorrelated data, based on the effective sample size. While this approach can be similar to the analytic power, we do not recommend using it in practice, because it’s equally simple to find the analytic power with modern power calculation software. We mention the approach here because it helps clarify the importance of the ICC: with as few as 1,000 subjects per cluster, increasing the ICC from 0.001 to 0.002 results in a 33 percent loss of effective sample size. In contrast, the confidence limits for estimated ICC are likely to be much broader than 0.001. Cluster sizes of 1,000 or greater are common in trials involving health delivery systems or communities.9,10
While the effective sample size approach is an approximation, accurate analytical approaches also depend on the design effect, and are similarly dramatically affected by the ICC. However, many approaches based on the design effect require that each cluster has an equal number of subjects, which may well not be the case. Several investigations into the impact of this have been performed, though their results are not general.11–14 Approximate methods of incorporating the impact of variable cluster size have been proposed, however.15–17
These analytic and approximate options for power assessment become difficult or untenable when more complex study designs are used. For example, it is often possible to record a baseline period in which neither the treatment clusters nor the control clusters receive the intervention followed by an intervention period in which only clusters so randomized receive the intervention. This design is much stronger than an “intervention period only” design, since it can account for some pre-existing or baseline differences among the clusters. Power calculation via analytic methods are known for normal-distributed outcomes in this design, (see, e.g., Murray pp. 368–369;6 Teerenstra et al.18). A Stata add-on due to Hemming and Marsh provides approximate power and sample size estimation with variable cluster size and can accommodate a baseline observation period.19 For, for example, dichotomous, count, or survival outcomes, or for more complex designs with normal outcomes, analytic results may be unknown.
Another option useful in any difficult setting and in cluster randomized trials in particular is to use simulation, as follows. First, generate data resembling the data anticipated for the study under the specific alternative hypothesis for which a power estimate is required, then perform the planned test on that data. Repeat this process many times: the proportion of the simulated data sets in which the null hypothesis is rejected is an estimate of the power. The precision of the estimate is controlled by the number of time the process is repeated. This approach is very powerful, and has been implemented for cluster-randomized trials with baseline observation periods in at least one package for R.20,21 The package also accommodates more general crossover trials.
But despite the robustness of simulation-based methods to some design issues, they share one key weakness with the analytic approach: it is often extremely difficult to obtain credible estimates of the ICC or . Assessments of the variability of the ICC or are even harder to find, and small differences in these parameters can lead to large differences in the estimated power, as was demonstrated using the effective sample size approximation. The difficulty of obtaining estimates has led to reliance on rules of thumb and to articles that report ranges of ICCs, to serve as reference.22 While perhaps better than no estimate at all, estimates from unrelated areas may lead to poor estimates of power.
In addition, covariate imbalance between arms is likely when few units are randomized. Though there remains debate among trialists about whether covariate adjustment is ever appropriate, it may be thought desirable in the case of a cluster-randomized trial. If so, the adjustment should also be incorporated into the power assessment. As the planned analysis gets more complex and parameters multiply, we should have less confidence in power estimates that depend on simplifying assumptions such as a lack of covariate effects, or on external ICC estimates.
Goal
In the current article, we propose a means of avoiding these problems and maintaining the greatest possible faithfulness to the planned analysis when performing the power calculation. In the Methods section, we discuss the general approach to power assessment using resampling methods, and outline two distinct settings in which they are likely to be useful. In the Results section, we describe a simulation assessment of a simple setting, as well as an application in which we implemented the method.
Methods
General Idea
We propose using bootstrapped samples to assess statistical power, modifying the samples as necessary to generate the desired alternative. This is a natural approach. Bootstrapping for power calculation has been described previously in a few specific applications, but its generality, flexibility, benefits, and heuristic motivation have not been fully explored, to the best of our knowledge.23–26 Nor has the application or its unique advantages in cluster-randomized trials been described.
Bootstrap resampling is simply sampling from observed data, with replacement. Heuristically, the idea is that the observed data represent the population from which they were drawn, and sampling an observation from among the observed data can thus be substituted for sampling from the population itself.
For estimating the power in a typical medical study, the method requires having relatively detailed data before the power must be calculated. This data should be as similar to the prospective study data as possible; one example would be baseline data that would also then be used in the study itself. Another setting where the method may be possible is in laboratory studies, where new experiments may be quite similar to completed experiments. We describe how the approach might be implemented in each of these cases.
A Simple Example: Laboratory Experiment
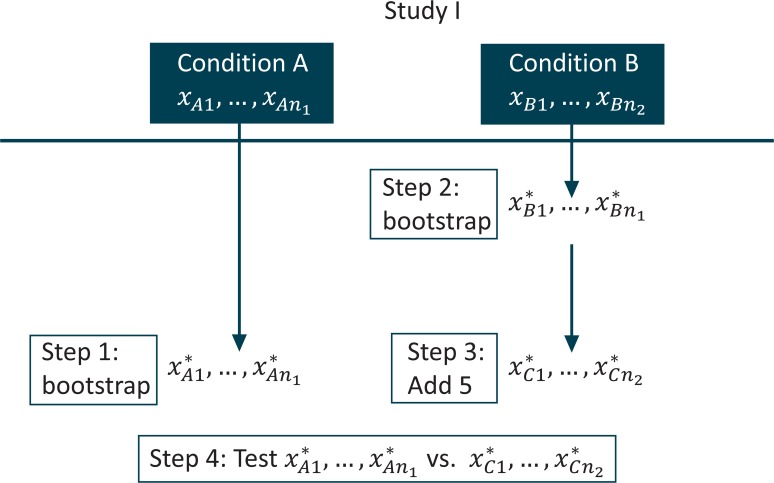
We begin with the laboratory experiment, a simple nonclustered setting, to introduce the idea. Suppose conditions “A” and “B” were compared in “Study I,” which has been completed. Now we wish to assess the power for a new experiment, “Study II,” where we will compare condition A to condition C, a modification of condition B. Let us assess the power under the alternative that the mean of condition C in Study II is 5 units greater than was observed for condition B in study I. We denote the data observed under condition A in Study I as xA1,…,xAn1 and the data observed under condition B in Study I as xB1,…,xBn2. Our bootstrap power calculation for Study II proceeds as follows:
Sample n1values from xA1,…,xAn1 using simple random sampling with replacement; denote these values x*A1,…,x*An1.
Sample n2 values from xB1,…,xBn2 using simple random sampling with replacement; denote these values x*B1,…,x*Bn1.
Add 5 to each of the values from step 2 and denote the values thus modified as x*C1,…,x*Cn2; this is how we include the alternative in the calculation.
Perform the test comparing x*A1,…,x*An1 to x*C1,…,x*Cn2, record whether the null was rejected or not.
Repeat steps 1–4 many times.
The proportion of rejections is the estimated power.
A diagram of steps 1 through 4 is presented in Figure 1.
Figure 1.
Diagram for Bootstrap Power Calculation in Laboratory Experiment
Some advantages to this approach present themselves immediately. Suppose the distribution in the second group is exponential, while that of the first is normal. An analytic approach to the power that accurately incorporates this difference in outcome distribution is not likely to be available. The choice of test with such distributions might be nontrivial, but the above routine will quickly generate the estimated power regardless of the chosen test; the algorithm above does not even specify a test. If we assume the new second condition will change the scale of the outcome, instead of or in addition to the location, we could easily modify step 3 in the above algorithm and still generate the desired result. Note also that in a Monte Carlo power estimation, as in an analytic power calculation, the Type I error plays a role. In a Monte Carlo power estimation, the Type I error level enters through step 4, above. To change the Type I error level for which the power is to be estimated, the α level for rejection can be changed.
A Complex Example: Cluster-Randomized Trial
Next, let us consider a cluster-randomized trial with a baseline observation period. Suppose we have collected baseline data on the presence or absence of an outcome, among individuals at several sites or clusters. We might be able to do this before the study was fully funded by using electronic medical records, for example. Each site may have a different number of subjects. We plan to use the collected data as a baseline against which we will compare data collected on other subjects while an intervention is applied to a random subset of sites. Suppose we need to know how much power we would have, given the intervention increases the odds of an outcome at each site by a factor of 2.
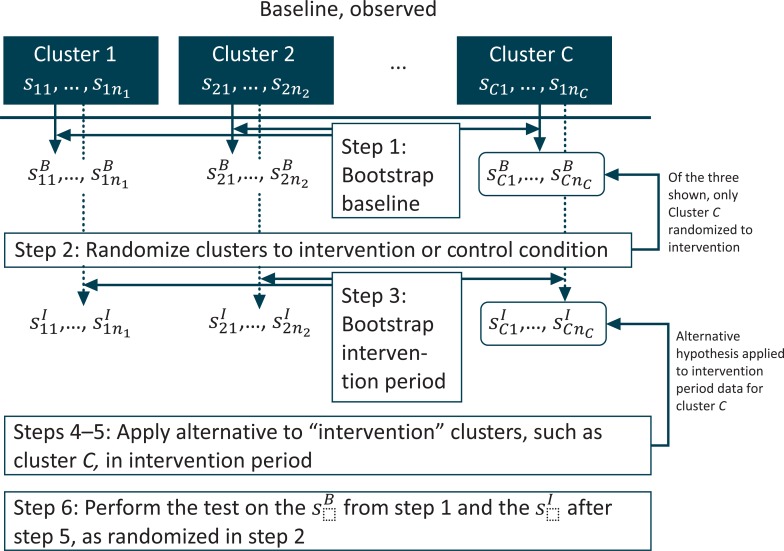
Denote each subject seen at cluster c in the baseline period as scj, = 1,…,nc, and the outcome for subject scj as ycj =1 if the outcome is observed and 0 otherwise. Our bootstrap power calculation would proceed along the following steps:
Within each cluster, resample nc observations, call these bootstrapped subjects . These will serve as the baseline data, thus the “B” in the superscript.
Randomize clusters to the control or intervention condition using whatever randomization strategy is planned for the actual study; this step may involve stratification by features of the “observed” .
Again, within each cluster, resample nc observations, call these subjects . These will serve as the intervention period data, thus the “I” in the superscript. There will be new individuals in the second period; this is why we resample again.
For clusters assigned to the intervention condition, calculate the modified within-cluster probability in the intervention period: Calculate the proportion where , denote this proportion , and from this calculate the odds within the cluster, . Multiply by 2, and solve for the probability implied. Call this , that is, or .
For clusters assigned to the intervention condition, set all , and randomly assign them to have with probability . Optionally, a more sophisticated approach would be to calculate the difference ; this is the additional probability of the outcome in cluster c in the intervention period. Under this approach we would retain the original values of . Then, among subjects with , we would randomly reassign their outcome value so that with probability .
Perform the planned analysis (say, a generalized, linear mixed logistic regression model) on the data set comprising the and the modified from step 5; record whether the null hypothesis was rejected or not.
Repeat steps 1–6 several times.
The proportion of rejections is the power.
A diagram of steps 1 through 6 is presented in Figure 2.
Figure 2.
Diagram for Bootstrap Power Calculation in a Cluster-Randomized Trial with a Baseline Observation Period
Some advantages are clear. There is no concern about how to incorporate the variable cluster size into the design effect and analytic approaches. There is no need to estimate the ICC or , which would be necessary for other approaches. Similarly, we need not estimate the precision of our estimated ICC or . Though it might be possible to estimate the ICC or using the baseline data, and then proceed with a simulation or analytic approach, it might be awkward to incorporate the variability of the estimate.
Note that the resampling is within cluster, so that the correlation within cluster is maintained, and so that independence across the bootstrapped items is also possible. The observed distribution of cluster sizes is also quite naturally maintained. For example, a cluster with 200 observed subjects will have 200 bootstrapped subjects in each iteration, and a cluster with 250 observed subjects will have 250 bootstrapped subjects in each iteration, and so forth, so that the exact observed distribution of cluster sizes is replicated. The correlation within cluster is represented by differential probabilities of the outcome by cluster, and these will be maintained by the bootstrap as well: a cluster with an observed rate of 0.2 will have approximately 0.2 in the bootstrapped sample, while a cluster with an observed rate of 0.4 will have a rate of approximately 0.4 in the bootstrapped sample. Also note that the randomization to study condition is placed within the power assessment process so that it can depend on, e.g., covariate values in the resampled baseline data. The somewhat complex optional formulation of step 5 is intended to retain the actual observations where the outcome is observed, and only add to them. This would possibly facilitate the incorporation of covariates.
Results
Simulation Experiment
We begin with a confirmation that the approach can replicate an analytic power calculation, when one is feasible. Consider a simple laboratory experiment such as that described above, except where the outcome is known to have a normal distribution with mean 0 and standard deviation 1 in the control group and normal distribution with mean 0.12535 and standard deviation 1 in the experimental group. In such a case, the analytic power would be exactly 0.8. To assess whether this power would be estimated correctly by the bootstrapping procedure described above, we simulated 1,000 data sets of 1,000 pseudo-random normal deviates each. For each data set, we followed the above procedure, bootstrapping a sample to serve as the control, another to serve as the experimental group, to which we added 0.12535, and then performing the t-test on these two bootstrapped samples. This bootstrap procedure was repeated 100 times for each of the 1,000 data sets. Since the precision of the estimate depends on the number of bootstrap samples performed, we must also calculate a confidence interval for each estimate; here we calculated exact 95 percent confidence limits. If the bootstrap procedure is correct, we would expect that 95 percent of the 95 percent confidence intervals enclose the analytic power of 0.8. In fact, we found that 95.5 percent of the 1,000 confidence intervals enclosed the value of 0.8, suggesting that the method can reproduce analytic results admirably.
Real Application
Next we present a real application. In 2012, we received a planning grant from the United States National Institutes of Health (NIH)—NIH Health Care Systems Research Collaboratory, Pragmatic Clinical Trials Demonstration Projects—to plan a trial of decolonization to reduce clinical cultures of certain drug-resistant organisms and bloodstream infections in hospitals. The intervention was to involve daily bathing of all patients with an antiseptic soap, plus a nasal antibiotic ointment for patients who harbored antibiotic-resistant bacteria. For cost and practicality reasons, this intervention was to be implemented at the hospital level, rather than at the unit or patient level. Thus, a cluster-randomized trial was planned. Data collection would use the hospitals’ routine electronic records, so a baseline observation period was feasible. During the planning year, we recruited 55 hospitals. We were then invited to pursue funding to actually perform the trial. That trial has since been funded and is ongoing as of this writing. Further details are available from clintrials.gov, identifier NCT02063867.
All data used in the analysis presented below were collected retrospectively from hospital medical and billing records, prior to the funding and performance of the trial. Data were anonymized by removing all information other than an arbitrary hospital and patient identifier, time of infection and type of organism if an infection was recorded, and time of hospital discharge. These data comprise all the information used in the analyses presented below. All data were collected as part of the usual business and medical practice of the hospitals and were retained by them; the hospitals graciously allowed the authors access to the data on their computers for the purposes of the work described. All work presented was approved by the Harvard Pilgrim Health Plan institutional review board (IRB). Informed consent was not obtained, but all data were analyzed anonymously and no experimentation was performed. All work was conducted in accordance with the principles expressed in the Declaration of Helsinki.
Due to variable length-of-stay in the hospital, the planned trial evaluation will treat the outcomes as time-to-event or survival data, censored at hospital discharge if no infection has occurred by that time. We plan to use a proportional hazards model, or Cox model, to assess the effectiveness of the intervention, with shared frailties to account for randomization by cluster.27–29 To assess power, we performed a version of the bootstrap power calculation described above. We know of no analytic approach useful in this setting. The primary outcome is time elapsed until a clinical culture with methicillin-resistant Staphylococcus aureus or vancomycin-resistant Enterococcus is found, that is, a “clinical culture”. These are both important antibiotic-resistant bacteria. Secondary outcomes include time until a clinical culture with a gram negative multi-drug resistant organism and time until bacteremia resulting from any pathogen. Fortunately these organisms and infections are currently rare, and the event rate per 1,000 attributable days for them respectively, is 2.2, 0.6, and 1.1.
The consensus among study planners was that we should assess power assuming there would be 20 percent fewer infections with the intervention. Our bootstrap power routine resembles that described above for a dichotomous outcome. As before, denote each subject seen at cluster c in the baseline as scj, j=1,…,nc. In the baseline period, we observed the time of event for subject scj, which we denote tcj, and the censoring indicator δcj =1 if the event is the outcome of interest and 0 if it is censored, which includes discharge from the hospital with no infection, for example. We also observed the time of discharge for patients who actually were infected. We’ll denote this time as t′cj. Our routine looks something like the following:
Within each cluster, resample nc observations. Call these subjects . These will serve as the baseline data in our assessment.
Randomize clusters to the control or intervention condition using the randomization strategy we plan to employ in the actual study.
Again, within each cluster, resample nc observations, call these subjects . These will serve as the intervention period data in our power assessment. In this design, there are new individuals in the second period; this is why we resample again instead of reusing the same subjects sampled to be the .
For clusters assigned to the intervention condition, randomly reassign 20 percent of the resampled event cases, where δcj =1, to instead be censored and have δcj =0. For these subjects, replace the observed event time tcj with t′cj, the discharge time.
Fit the frailty model to the data set comprising the and the modified , record whether the null hypothesis was rejected or not.
Repeat steps 1–5 several times.
The proportion of rejections is the power.
An example using simulated data in SAS is shown in the Appendix.
We note that the substitution of t′cj for tcj, replacing the event time with the discharge time when an event is removed in step 4, is not perfect. Ideally, we would substitute the date of discharge that would have occurred had there been no event, but of course this is unknowable. The date of discharge after an event might well be later than this unknowable value, since the event itself may delay the time of discharge relative to the unknowable value, for example. It is possible that a better choice would be to leave the time of event unchanged, while changing it to a censoring rather than event time, effectively discharging the patient at the event time, although this would almost certainly censor nonevent time. In our application, however, the event rate is very small, so the difference between these imperfect choices is unlikely to be meaningful.
There was a further complication. The baseline data in hand included 4 months of recruitment, but the planned total baseline accrual period was 12 months. The intervention period was scheduled for 18 months. We resolved this issue by referring back to the heuristic behind the bootstrap: the observed sample represents the original population. Why not bootstrap more than nc samples from among the nc observed subjects in each cluster in the baseline period? In traditional uses of the bootstrap, for example, to obtain confidence limits for statistics that have difficult asymptotic properties, this idea would lead to biased results—narrower confidence limits than appropriate. But in our setting this argument is not relevant. We sampled 3 * nc observations from each cluster for the baseline in step 1 and 4.5 * nc from each cluster in step 3.
Results based on 1,000 bootstrap iterations are shown in Table 1. Power for the primary outcome and two selected secondary outcomes were assessed; we repeated the above process for each of these. We also considered four values of the intervention effect by varying the percent selected to have their events removed in step 4.
Table 1.
Power and Exact 95 Percent CI for Power for Primary and Select Secondary Outcomes*
| INTERVENTION EFFECT | MRSA OR VRE CLINICAL CULTURES | GRAM NEGATIVE MULTI-DRUG RESISTANT CLINICAL CULTURES | ALL-PATHOGEN BACTEREMIA |
|---|---|---|---|
| 0% | 5.6% (4.3–7.2%) | 4.1% (3.0–5.5%) | 5.2% (3.9–6.8%) |
| 10% | 35% (32–38%) | 14% (12–16%) | 22% (19–24%) |
| 20% | 93% (91–95%) | 44% (41–47%) | 67% (63–69%) |
| 30% | 100% (99.6–100%) | 83% (81–86%) | 97% (96–98%) |
Notes: CI = confidence interval; MRSA = methicillin-resistant Staphylococcus aureus; VRE = vancomycin-resistant Enterococcus.
Based on 1,000 bootstrap samples for each effect size and outcome.
The results show that for the primary outcome, there is ample power to detect the anticipated effect of preventing 20 percent of events. There is notably less power for the secondary outcomes. Note that we also present an effect of 0 percent. A 0 percent effect is implemented by not altering the outcomes for any subjects, in step 4. In this case the null hypothesis is true, which may strike some readers as meaning we are assessing something other than power. We include this effect as a face validity check to ensure that the power assessment process and implementation are correct: the probability of rejecting the null hypothesis must be the same as our rejection level—5 percent in this case. The results show that the face validity test was passed.
As is demonstrated by the table, the results of the bootstrap power calculation, as with simulation-based power procedures, are estimates. The result of the calculation is the proportion of rejections, and the precision of the estimate—controlled by the number of bootstrap cycles in step 6—should be reflected by providing confidence limits based on the properties of the binomial distribution. Here we use exact limits.
Discussion
We describe the application of bootstrap resampling to the problem of power calculation. Like the bootstrap itself, the approach is very general. It can be used in laboratory settings and in any application where fairly extensive preliminary data can be obtained before power calculations are necessary. The application of the bootstrap to power calculation has been proposed previously in a handful of specific applications.23–26 Our contribution is to describe its application and to explore the nuances of use and particular advantages in the setting of cluster-randomized trials. In addition, we emphasize that the approach can be used to assess power for any proposed project, an important point apparently missed in previous work. We also demonstrate that the bootstrap approach can replicate analytic power assessments and show a real example of estimating power for a cluster-randomized trial for infection prevention in hospitals. The application to censored survival outcomes allows power estimation for a setting in which analytic results are not available.
The bootstrap power approach offers several advantages beyond its ability to account for arbitrary complexity in the structure of the data and the fact that it does not rely on estimates from the literature. Primary among these is that it can use the precise analysis method contemplated for the planned study, an advantage shared with simulation approaches, but without the requirement of verisimilitude in the simulation. For example, covariates can easily be incorporated without making assumptions about their joint distribution. In place of this requirement, it substitutes the requirement that the preliminary data be sufficiently similar to that of the intended study. The bootstrap power approach also incorporates variability in key parameters without further consideration of the analyst. Another advantage is the ease of implementing the thinking of study planners. For example, in the shared frailty proportional hazards application, the expert consensus was that 20 percent of the infections might be prevented by the intervention. Using the bootstrap power approach, we were able to implement that effect directly, without having to consider implications for model parameters such as the hazard ratio.
Additionally, while we demonstrated simple alternative hypotheses, it would be trivial to implement complex ones. For example, we could change the mean in the laboratory experiment by the same amount by leaving half the experimental subjects unchanged and doubling the effect in the other half. In our real example, we could change the shape of the survival curve by removing events preferentially among early events. Another advantage is that it is entirely generic—any analytic method can be inserted into the data analysis step and the power assessment algorithm will be unchanged. This suggests that bootstrap power calculations could easily be used to compare the power of two competing analyses in a particular data setting.
The bootstrap approach to power calculation is likely to be more accurate than analytic or simulation-based calculations, in the following sense. In order to make analytic calculations or simulation-based power assessment, we typically rely on parametric assumptions about the outcome and covariates, if any. To the extent that these are not true, they will introduce inaccuracies to the power calculations. These will be present, for example, even for approximately normal outcomes for which we calculate power assuming a normal distribution, if the outcome is not in fact normal. In cluster-randomized trial settings, we may have the added assumptions of fixed cluster sizes, or of regularity required to usefully represent the variability of the cluster size through the coefficient of variation, as well as the intracluster coefficient issues outlined above. Each of these introduces at least potentially, and often practically, inaccurate assumptions that are avoided by the Monte Carlo approach, provided that the preliminary data exhibit the features that make simulation or analytic approaches unsatisfactory.
The primary weakness of the approach is the reliance on the availability of detailed data. We provide the example of laboratory experiments as a case where detailed data may well be available, and demonstrate a real example of a large human trial in which it is possible. On the one hand, we believe it will frequently be possible in pragmatic projects using established care delivery systems, where detailed data are often easily available through existing electronic records. On the other hand, in such settings, it may also be possible to estimate key parameters, such as the ICC, from the preliminary data, diminishing the advantage of the bootstrap. We believe the bootstrap power approach is still markedly superior, however, in that it incorporates the variability of these parameters as compared with fixed estimates, or at best, large variability for estimates of second moments.
When data available are not sufficiently similar to the desired power calculation setting to allow the use of the bootstrap, simulation may be the best strategy. This approach is likely to allow incorporation of more idiosyncratic aspects of each particular study. Simulation requires that only modest information be acquired before undertaking the power assessment. In our opinion, purely analytic power calculation is best reserved for simple situations or situations so novel that very little data are available to aid in power assessment.
Note that unlike other power calculation tools, the bootstrap method is particularly tied to the available data. While the approach is very general in that it can be applied whenever sufficient data exists, each solution will be unique to that existing preliminary data and, indeed, to the proposed analysis for that data set. It may be most helpful to retain the bootstrap method in your toolbox for the times when data sufficient to implement it are available and when exploration of that data call the assumptions needed for analytic power calculations into question.
A few other items bear some attention. One is the particular utility of being able to bootstrap more than the observed number of subjects in each cluster. This allowed us to expand the time scale of the available baseline in the cluster-randomized trial application. It also suggests that we can assess the number of subjects needed to achieve a given power, as is often desirable. Another is the unique feature that there is no need to explicitly estimate parameters from the collected data. Thus for the cluster-randomized trial example, we did not calculate the baseline rate or survival curve for each event, or need to know the sample size available at each cluster. These are features that play heavily into analytic and simulation-based power assessments. As mentioned above, it is incumbent upon the user to repeat the bootstrap process many times and to report a confidence interval for the power calculation. The exact number of “many” depends on the application: in some cases, as few as 20 iterations may be sufficient, if the null is rejected in all or in none of them. More typically, 100 or 200 are often sufficient, while for grant applications, we sometimes use 1,000. Finally, as noted above, power estimates from bootstrap and simulation methods are explicitly estimates, and can and should be accompanied by confidence limits. Ironically, power calculations from analytic methods treat their inputs as fixed and offer no formal means of assessing uncertainty. At best, we may vary parameters informally to demonstrate the effects of uncertain inputs to the formulae.
Conclusion
Power calculation by bootstrap is the simple proposal to use resampling techniques to generate data under the alternative hypothesis and to use replication to assess power under that hypothesis. Bootstrap power calculation is a powerful tool that offers unique advantages compared to analytic calculation or simulation. It allows power to be driven by detailed baseline data and avoids weaknesses common to other approaches to power, including the need to assume that literature-based estimates apply to the population under study and the need to find viable estimates of all parameters in the analysis. It should be particularly useful, as demonstrated, in application to cluster-randomized trials.
Acknowledgments
We appreciate the assistance of Taliser Avery in implementing the proposed method as presented in the results. Each author’s effort on the manuscript was supported by NIH grant 1UH2AT007769-01.
Appendix: SAS code
Footnotes
Disciplines
Biostatistics | Statistical Methodology
References
- 1.Neter J, Wasserman W, Kutner M. Applied Linear Regression Models. Chicago: Richard D. Irwin; 1983. [Google Scholar]
- 2.Self SG, Mauritsen RH. Power/sample size calculations for generalized linear models. Biometrics. 1988;44(1):79–86. [Google Scholar]
- 3.Hsieh FY, Block DA, Larsen MD. A Simple Method of Sample Size Calculation for Linear and Logistic Regression. Statistics in Medicine. 1998;17:1623–1634. doi: 10.1002/(sici)1097-0258(19980730)17:14<1623::aid-sim871>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 4.R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna; Austria: 2013. ISBN 3-900051-07-0, URL http://www.R-project.org/. [Google Scholar]
- 5.Donner A, Klar N. Design and analysis of cluster randomization trials in health research. London: Arnold; 2000. [Google Scholar]
- 6.Murray D. Design and Analysis of Group-Randomized Trials. New York: Oxford University Press; 1998. [Google Scholar]
- 7.Donner A, Klar N. Statistical Considerations in the Design and Analysis of Community Intervention Trials. The Journal of Clinical Epidemiology. 1996;49(4):435–439. doi: 10.1016/0895-4356(95)00511-0. [DOI] [PubMed] [Google Scholar]
- 8.Kish L. Survey Sampling. New York: John Wiley & Sons; 1965. [Google Scholar]
- 9.Huang SS, Septimus E, Kleinman K, Moody J, Hickok J, Avery TR, Lankiewicz J, Gombosev A, Terpstra L, Hartford F, Hayden MK, Jernigan JA, Weinstein RA, Fraser VJ, Haffenreffer K, Cui E, Kaganov RE, Lolans K, Perlin JB, Platt R. Targeted versus universal decolonization to prevent ICU infection. New England Journal of Medicine. 2013;368:2255–2265. doi: 10.1056/NEJMoa1207290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finkelstein JA, Huang SS, Kleinman K, Rifas-Shiman SL, Stille CJ, Daniel J, Schiff N, Steingard R, Soumerai SB, Ross-Degnan D, Goldmann D, Platt R. Impact of a 16-community trial to promote judicious antibiotic use in Massachusetts. Pediatrics. 2008;121:e15–e23. doi: 10.1542/peds.2007-0819. [DOI] [PubMed] [Google Scholar]
- 11.van Breukelen GJ, Candel MJ, Berger MP. Relative efficiency of unequal versus equal cluster sizes in cluster randomized and multicentre trials. Statistics in Medicine. 2007;26(13):2589–603. doi: 10.1002/sim.2740. [DOI] [PubMed] [Google Scholar]
- 12.Candel MJ, Van Breukelen GJ. Sample size adjustments for varying cluster sizes in cluster randomized trials with binary outcomes analyzed with second-order PQL mixed logistic regression. Statistics in Medicine. 2010;29(14):1488–501. doi: 10.1002/sim.3857. [DOI] [PubMed] [Google Scholar]
- 13.Hemming K, Girling AJ, Sitch AJ, Marsh J, Lilford RJ. Sample size calculations for cluster randomised controlled trials with a fixed number of clusters. BMC Med Res Methodol. 2011;11:102. doi: 10.1186/1471-2288-11-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.You Z, Williams OD, Aban I, Kabagambe EK, Tiwari HK, Cutter G. Relative efficiency and sample size for cluster randomized trials with variable cluster sizes. Clin Trials. 2011;8(1):27–36. doi: 10.1177/1740774510391492. Epub 2010 Dec 16. [DOI] [PubMed] [Google Scholar]
- 15.Manatunga AK, Hudgens MG, Chen SD. Sample size estimation in cluster randomized studies with varying cluster size. Biometrical Journal. 2001;43(1):75–86. [Google Scholar]
- 16.Kong S-H, Ahn CW, Jung S-H. Sample Size Calculation for Dichotomous Outcomes in Cluster Randomization Trials with Varying Cluster Size. Therapeutic Innovation & Regulatory Science. 2003;37(1):109–114. [Google Scholar]
- 17.Eldridge SM, Ashby D, Kerry S. Sample size for cluster randomized trials: effect of coefficient of variation of cluster size and analysis method. Int. J. Epidemiol. 2006;35(5):1292–1300. doi: 10.1093/ije/dyl129. [DOI] [PubMed] [Google Scholar]
- 18.Teerenstra S, Eldridge S, Graff M, de Hoop E, Borm GF. A simple sample size formula for analysis of covariance in cluster randomized trials. Statistics in Medicine. 2012;31(20):2169–78. doi: 10.1002/sim.5352. [DOI] [PubMed] [Google Scholar]
- 19.Hemming K, Marsh J. A menu-driven facility for samplesize calculations in cluster randomized controlled trials. The Stata Journal. 2013;13(1):114–135. [Google Scholar]
- 20.Reich NG, Myers JA, Obeng D, Milstone AM, Perl TM. Empirical power and sample size calculations for cluster-randomized and cluster-randomized crossover studies. PLoS ONE. 2012;7(4):e35564. doi: 10.1371/journal.pone.0035564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reich NG, Obeng D. clusterPower: Power calculations for cluster-randomized and cluster-randomized crossover trials. R package version 0.4-2 [Internet] 2013 [updated 2015]. Available from : http://CRAN.R-project.org/package=clusterPower.
- 22.Gulliford MC, Ukoumunne OC, Chinn S. Components of variance and intraclass correlations for the design of community-based surveys and intervention studies. American Journal of Epidemiology. 1999;149:867–83. doi: 10.1093/oxfordjournals.aje.a009904. [DOI] [PubMed] [Google Scholar]
- 23.Tsodikov A, Hasenclever D, Loeffler M. Regression with bounded outcome score: evaluation of power by bootstrap and simulation in a chronic myelogenous leukaemia trial. Statistics in Medicine. 1998;17:1909–1922. doi: 10.1002/(sici)1097-0258(19980915)17:17<1909::aid-sim890>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 24.Troendle JF. Approximating the power of Wilcoxon’s rank-sum test against shift alternatives. Statistics in Medicine. 1999;18:2673–2773. doi: 10.1002/(sici)1097-0258(19991030)18:20<2763::aid-sim197>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 25.Walters SJ. Sample size and power estimation for studies with health related quality of life outcomes: a comparison of four methods using the SF-36. Health and Quality of Life Outcomes. 2004;2:26. doi: 10.1186/1477-7525-2-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walters SJ, Campbell MJ. The use of bootstrap methods for estimating sample size and analyzing health-related quality of life outcomes. Statistics in Medicine. 2005;24:1075–1102. doi: 10.1002/sim.1984. [DOI] [PubMed] [Google Scholar]
- 27.Cox DR. Regression Models and Life Tables. Journal of the Royal Statistical Society, Series B. 1972;20:187–220. [Google Scholar]
- 28.Ripatti S, Palmgren J. Estimation of Multivariate Frailty Models Using Penalized Partial Likelihood. Biometrics. 2000;56:1016–1022. doi: 10.1111/j.0006-341x.2000.01016.x. [DOI] [PubMed] [Google Scholar]
- 29.Hayes RJ, Moulton LH. Cluster Randomized Trials. Boca Raton: Chapman&Hall/CRC; 2009. [Google Scholar]