Abstract
Non-syndromic cleft lip with or without cleft palate (NSCL/P) is the most common craniofacial birth defect in humans, affecting 1 in 700 live births. This malformation has a complex etiology where multiple genes and several environmental factors influence risk. At least a dozen different genes have been confirmed to be associated with risk of NSCL/P in previous studies. However, all the known genetic risk factors cannot fully explain the observed heritability of NSCL/P, and several authors have suggested gene-gene (G×G) interaction may be important in the etiology of this complex and heterogeneous malformation. We tested for G×G interactions using common SNPs derived from targeted sequencing in 13 regions identified by previous studies spanning 6.3 MB of the genome in a study of 1,498 NSCL/P case-parent trios. We used the R-package trio to assess interactions between polymorphic markers in different genes, using a 1 degree of freedom (1df) test for screening, and a 4 degree of freedom (4df) test to assess statistical significance of epistatic interactions. To adjust for multiple comparisons, we performed permutation tests. The most significant interaction was observed between rs6029315 in MAFB and rs6681355 in IRF6 (4df p=3.8×10−8) in case-parent trios of European ancestry, which remained significant after correcting for multiple comparisons. However, no significant interaction was detected in trios of Asian ancestry.
Keywords: Oral clefts, gene-gene interaction, case-parent trios
Introduction
Oral clefts include three distinct anatomical malformations: cleft lip (CL), cleft palate (CP) and cleft lip and palate (CLP), and combined these represent the most common craniofacial birth defects in humans, affecting 1.7 per 1000 live births [Rahimov et al. 2012]. Since CL and CLP share similar epidemiologic distributions and develop during the same embryologic periods, CL and CLP are often grouped together as cleft lip with or without cleft palate (CL/P). The majority of oral cleft cases are “non-syndromic” because they occur as an isolated anomaly with no other structural abnormality or developmental disability in the child. Among all cases, approximately 70% of CL/P cases and 50% of CP cases are non-syndromic [Jugessur et al. 2009], while the remaining cases have another congenital anomaly or developmental delay representing some malformation syndrome. While the overall prevalence of oral clefts is high, this varies by type of cleft: the prevalence of CL/P ranges between 3.4 and 22.9 per 10,000 live births, while the prevalence of CP is 1.3–25.3 per 10,000 live births [Mossey and Castilla, 2003]. There are substantial differences in prevalence of CL/P across racial groups and populations: Asians and Native Americans have the highest rate of 2 per 1,000 live births, Caucasians have a prevalence around 1 per 1,000 and African populations have the lowest prevalence rate of 1 per 2,500 live births [Dixon et al. 2011; Mossey et al. 2009]. Gender is also related to risk to oral clefts, CL/P is more common in males with a 2:1 ratio of males: females, while CP is more frequent in females [Mossey et al. 2009; Matthews et al. 2015].
Oral clefts represent a complex and heterogeneous group of malformations where both genetic and environmental risk factors control risk [Leslie and Marazita, 2013]. Since 2009, genome-wide association studies (GWAS) using both case-control and case-parent trio designs have identified more than a dozen genes achieving genome-wide significance as influencing risk to oral clefts, and other possible genomic regions have been identified through genome-wide linkage studies using multiplex cleft families (i.e. those with more than one affected individual). As the list of putative causal genes expands, it is logical to ask if some of these genes may interact with one another in biological pathways that can be identified through tests for statistical interaction. Detecting such gene-gene (G×G) interactions on a large scale is a daunting challenge since the number of potential hypotheses and the incurred multiple comparisons burden limit the power to detect interaction even in studies with large sample sizes [Cordell, 2002]. Li et al. [2015] tested for G×G interaction involving the WNT signaling pathway using CL/P case-parent trios from the GWAS by Beaty et al. [2010] and identified an interaction between markers in WNT5B and MAFB among both Asian and European case-parent trios, as well as interactions between markers in WNT5A and IRF6 in Asian trios, and markers in the 8q24 region and WNT5B in European trios.
Here we present a step-wise strategy for testing for G×G interaction in 1,409 case-parent trios of European or Asian descent, using common variants identified from targeted sequencing of 13 recognized candidate genes/regions (8q24, ARHGAP29, BMP4, FGFR2, FOXE1, IRF6, MAFB, MSX1, NOG, NTN1, PAX7, PTCH1, VAX1, see Table I). We focused on common tagging SNPs available through the study described by Leslie et al. [2015]. Each of these 13 regions was previously shown to be associated with NSCL/P in either previous GWAS or genome-wide linkage studies. We limited our tests for interaction to common variants since low-frequency and rare variants do not provide sufficient power to detect epistasis with the number of trios available. To assure scalability and accuracy, we selected tagging SNPs, and carried out a two-step procedure to detect SNP-SNP interactions. We first used a very fast 1 degree of freedom (1df) Wald test in a simplified statistical model to generate a list of candidate SNP pairs. Since departures from the null in these 1df tests can also indicate violations of model specifications, we analyzed the candidate pairs using a 4df test based on a more general statistical model proposed by Cordell [2002] to comprehensibly assess epistatic SNP-SNP interactions. The overall significance in the context of multiple comparisons was then assessed using a permutation test.
Table I.
Candidate genes or regions sequenced in this study.
| Gene | Targeted Region (GRCh37) | Total (kbp) | Number of common SNPs |
|---|---|---|---|
| IRF6 | chr1:209837199-210468406 | 631.2 | 50 |
| MAFB | chr20:38902646-39614513 | 711.9 | 111 |
| ARHGAP29 | chr1:94324660-95013109 | 688.4 | 102 |
| 8q24 | chr8:129295896-130354946 | 1059.1 | 97 |
| PAX7 | chr1:18772300-19208054 | 435.8 | 172 |
| VAX1 | chr10:118421625-119167424 | 745.8 | 82 |
| NTN1 | chr17:8755114-9266060 | 510.9 | 175 |
| NOG | chr17:54402837-54957390 | 554.6 | 73 |
| FOXE1 | chr9:100357692-100876841 | 519.1 | 52 |
| MSX1 | chr4:4825126-4901385 | 76.3 | 57 |
| BMP4 | ch14:54382690-54445053 | 62.4 | 12 |
| FGFR2 | chr10:123096374-123498771 | 402.4 | 42 |
| PTCH1 | chr9:98133647-98413162 | 279.5 | 48 |
Methods
Study Population
A total of 1,498 cleft case-parent trios were recruited from different sites in China, the Philippines, the United States and Europe (see Leslie et al. 2015 for a full description of the sample). These were used for targeted sequencing of 13 genes and regions considered to be prime candidates for containing genes or regulatory elements important in controlling risk to oral clefts (Table I). After quality control (QC), 1,409 case-parent trios remained available for analyses. We analyzed the case-parent trio data in two separate groups (shown in Table II): an Asian group which contained the Filipino and Chinese families (1,034 case-parent trios), and a smaller European group composed of European and European-American families (375 trios).
Table II.
Number of case-parent trios, by population, available for analysis after quality control.
| Population | Country | Total Trios |
|---|---|---|
| Asian | China | 401 |
| Philippines | 633 | |
| Asian TOTAL | 1,034 | |
| European | USA | 266 |
| Denmark | 9 | |
| Hungary | 65 | |
| Spain | 26 | |
| Turkey | 9 | |
| European TOTAL | 375 | |
| TOTAL | 1,409 | |
Sequencing
Details of the sequencing protocol are available in Leslie et al. [2015]. In brief, 1µg of native genomic DNA was used to construct Illumina multiplexed libraries. Reads were mapped to the GRCh37-lite reference sequence using BWA [Li and Durbin, 2010]. Picard was used to merge alignments and mark duplicates, and Polymutt was used to perform germline and de novo variant calling. We used bam-readcount to identify and flag potential artifact variants [Leslie et al. 2015]. The single nucleotide variant calls were combined into a variant call file (VCF) file. All variants with a depth (DP) less than 7 or genotype quality (GQ) less than 20 were removed. Variants located within 75bp of indels or dinucleotide polymorphisms occurring in more than 5% of samples were included in analyses, but were flagged as potential artifacts.
Data processing
To evaluate the family relationship between members of the case-parent trios, we used BEAGLE’s fast-IBD to calculate identity by descent (IBD) between parents and their offspring. If a parent-child pair shared less than 40% of the targeted region, the trio was dropped from all analysis. To increase the power to detect G×G interaction, we only selected highly polymorphic variants with a minor allele frequency (MAF) larger than 20%. We also excluded all SNPs with missing genotype rate larger than 1%. We tested for Hardy-Weinberg equilibrium (HWE) in parents separately within Asian and European groups, and excluded SNPs that showed substantial departure from HWE (p<1×10−5). We used Haploview [Barrett et al., 2005] to choose tagging SNPs (defined as r2 >0.8) within the Asian and European groups. Haplotype phasing required for the permutation tests was carried out using BEAGLE [Browning and Yu, 2009].
Screening step
To assure scalability, we implemented a screening step to generate candidate interactions among all pairwise tagging SNP combinations in the 13 genomic regions of interest (excluding pairs of SNPs from the same region), using a 1df Wald test for G×G interaction based on a conditional logistic regression model implemented in the open source Bioconductor package trio [Schwender et al. 2014]. As in the genotypic TDT [Schaid 1999], each trio is represented as the four possible Mendelian offspring given the parental genotypes, i.e. where the affected proband is the case and the other three possible genotypes serve as “pseudo-controls”. The conditional model (with grouping by family) assumes an additive mode of inheritance for both bi-allelic SNPs, and contains one parameter for interaction between the two SNPs. This model is considerably simpler than the more general modeling approach proposed by Cordell [2002], and allows for the rapid assessment of all pairwise interactions. The Cordell 4df LRT (see next section) requires numerical estimation of four and eight parameters respectively in conditional logistic regression models using an iterative procedure.
General model to assess interaction
Significant results based on the 1df Wald test can indicate epistasis, but also simply a violation of model specifications, in particular the interaction term itself. To avoid such biases, we selected the 500 most significant marker pairs from each of the pairwise combinations and comprehensibly assessed the potential SNP-SNP interactions using the model proposed by Cordell [2002]. Again representing each trio as the four possible offspring based on the parental genotypes (one affected “case” and three “pseudo-controls”), this model can for the i-th trio be written as
where XAa and XAA are orthogonal representations of the genotype at locus A, and XBb and XBB are orthogonal representations of the genotype at locus B (Cordell [2002]). The four parameters (i11, i12, i21, i22) encode the possible departure from additive genotype effects. To assess SNP-SNP interaction, we use a 4df LRT to compare the likelihood of the full model above to the likelihood of the reduced model without these epistatic parameters:
Permutation tests
We carried out permutation test to assess statistical significance in the light of multiple hypothesis testing. Specifically, we aimed to answer the following questions: (1) whether any region pair(s) showed evidence for interaction, i.e. is there any signal in the data? And (2) if so, which region/gene pairs and which SNPs within such a region pair interact. As our permutation data set, we created 1000 “shuffled” data sets using the phased parental haplotypes, to maintain the correlation structure between genotypes within a region. To create simulated genotypes for the children, we randomly chose one haplotype from each of the parents as the transmitted haplotype, yielding a simulated case and three simulated pseudo-controls for each trio.
For each of the 1,000 permutation data sets, we repeated our analytic procedure separately for each of the pairs of regions, for each region pair selecting the 500 most significant SNP pairs in the screening 1df Wald test, and evaluating these 500 candidates using the 4df Cordell LRT. For each permutation data set, we recorded the maximum test statistic from the LRT across all SNP-SNP interactions in the respective region pair. This yielded a set of 78×1,000 (maximum) test statistics representing results under the null for each of the 78 possible region pairs, across the 1000 permutations.
To assess whether any region pair(s) showed evidence for interaction, i.e. whether any signal is present in the data, we compared the maximum 4df test statistic observed for any SNP pair across all region pairs to the 1,000 corresponding values obtained in the same fashion from the permutation data. The p-value for this test was estimated by the fraction of permuted test statistics exceeding the observed. To assess which of the 78 region pairs (and which SNP pairs) show deviation from randomness, we compared for each region pair the maximum 4df test statistic observed for any SNP pair within the respective region pairs to the 1,000 corresponding values obtained in the same fashion from the permutation data. The p-value for each region pair was estimated by the fraction of permuted test statistics exceeding the observed. Significance under family-wise error rate protection for the 78 region pairs was assessed using a Bonferroni correction.
Due to the computational expense of our procedure, we considered the region-pair selection as a hypothesis-generating step, with the assessment of a particular SNP-SNP combination as the step requiring strict multiple-testing correction to control the family-wise error rate. Hence any SNP-SNP combination should have a permutation adjusted p-value less than 0.05/78=0.00064. Since 1000 permutations is not adequate to reach this level of significance, for any region pair that passed the first selection process, we generated an additional 1000 permuted data sets to further assess significance.
Results
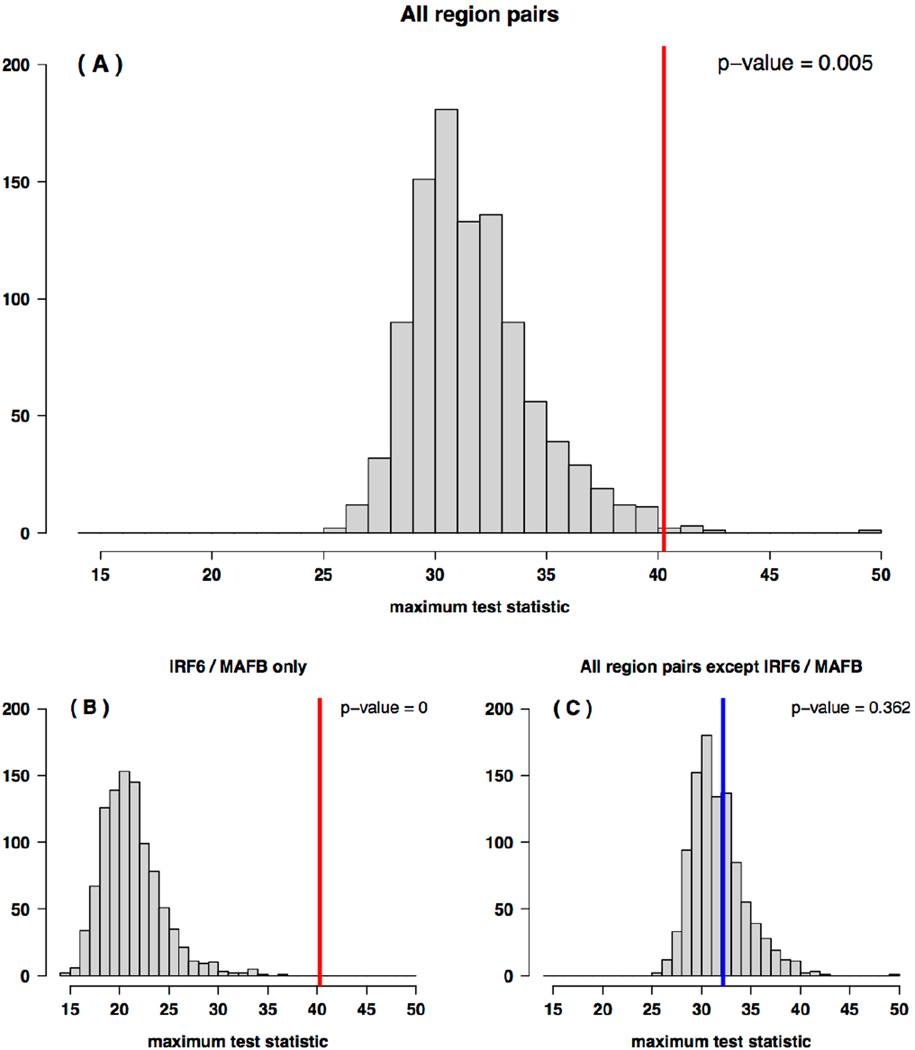
We obtained 1,075 and 1,016 tag SNPs in European and Asian groups, respectively, after applying quality control filters to the common SNPs from the targeted sequencing data and selecting tagging markers. We only investigated interactions between different genes/regions, resulting in 78 different gene/region combinations for a total of 519,086 and 468,037 hypothesis tests among European and Asian trios, respectively. The most significant interaction based on the 4df LRT among Europeans was between rs6681355 in IRF6 and rs6029315 in MAFB (LRT=40.25, p=3.8×10−08, Table III). Among the 1,000 permutations, the maximum test statistic across all SNP-SNP interactions in all 78 region pairs for Europeans exceeded this test statistic only 5 times (Figure 1A), yielding an empirical p-value of 0.005 with an upper bound for the 95% exact Binomial confidence interval of 0.0116 for this first hypothesis-generating stage. The maximum test statistic across all SNP-SNP interactions in the IRF6 / MAFB pair never exceeded the LRT of 40.25 (Figure 1B), yielding an empirical p-value < 0.001. The same was true in an additional independent 1,000 permutations, yielding a permutation p-value < 1/2,000 = 0.0005, and thus resulting in a p-value below the Bonferroni threshold of 0.05/78 = 0.00064.
Table III.
Top 10 most significant results from the 4 degree of freedom Likelihood Ratio Test for SNP-SNP interactions in 375 European case-parent trios (observed test statistic, nominal p-value, and nominal permutation p-value).
| First Gene | Second Gene |
Marker 1 | Marker 2 | Test Statistic |
p-value | Permutation p-value |
|---|---|---|---|---|---|---|
| IRF6 | MAFB | rs6681355 | rs6029315 | 40.25 | 3.83E-08 | <0.0005* |
| MAFB | NTN1 | rs6029421 | rs8081873 | 32.17 | 1.76E-06 | 0.02 |
| MAFB | PAX7 | rs6029182 | rs11584404 | 31.25 | 2.72E-06 | 0.02 |
| NOG | NTN1 | rs8074637 | rs2315286 | 29.62 | 5.84E-06 | 0.04 |
| ARHGAP29 | MSX1 | rs4147848 | rs730575 | 29.01 | 7.80E-06 | 0.03 |
| 8q24 | MAFB | rs6470670 | rs3092775 | 28.05 | 1.22E-05 | 0.05 |
| MSX1 | NTN1 | rs2968669 | rs9892906 | 27.97 | 1.27E-05 | 0.05 |
| MAFB | MSX1 | rs6029145 | rs6851263 | 26.75 | 2.23E-05 | 0.06 |
| IRF6 | PAX7 | rs2484030 | rs10907314 | 25.72 | 3.60E-05 | 0.10 |
| 8q24 | FOXE1 | rs72730212 | rs16923269 | 25.35 | 4.27E-05 | 0.10 |
The permutation p-value for the IRF6/MAFB pair was based on 2,000 permutations to assess whether its value is below the Bonferroni threshold of 0.05/78 = 0.00064.
Figure 1.
Distributions of the maximum 4 degree-of-freedom likelihood ratio test statistics, across all pairwise SNP-SNP interactions, for 1,000 permutation case-parent trio data sets of European ancestry. (A) The maximum test statistic across all SNP-SNP interactions in all 78 region pairs. The largest observed test statistic (LRT=40.25) is indicated by the red vertical line. (B) The maximum test statistic across all SNP-SNP interactions in the IRF6 / MAFB pair. (C) The maximum test statistic across all SNP-SNP interactions in all region pairs except the IRF6 / MAFB pair. The largest observed test statistic (LRT=32.17) is indicated by the blue vertical line.
No other region pair however showed significant interaction after multiple comparisons correction in Europeans. Removing the IRF6 / MAFB pair from consideration in the permutation test yielded a p-value of 0.362 for the maximum LRT statistic across all SNP-SNP interactions in the remaining 77 region pairs for the Europeans (Figure 1C). With the exception of this IRF6 / MAFB pair, no permutation p-value was observed below 0.05/78 = 0.00064.
Although there were more case-parent trios of Asian than European descent, we found no indication of SNP-SNP interactions among the Asians. The most significant interaction based on the 4df LRT among Asians was between rs3761910 in ARHGAP29 and rs2149722 in PTCH1 (nominal p=7.7×10−06, Table IV), which did not retain significance after correcting for multiple comparisons.
Table IV.
Top 10 most significant results from the 4 degree of freedom Likelihood Ratio Test for SNP-SNP interactions in 1,034 Asian case-parent trios (observed test statistic, nominal p-value, and nominal permutation p-value).
| First Gene | Second Gene | Marker 1 | Marker 2 | Test Statistic |
p-value | Permutation p-value |
|---|---|---|---|---|---|---|
| ARHGAP29 | PTCH1 | rs3761910 | rs2149722 | 29.04 | 7.67E-06 | 0.03 |
| NOG | NTN1 | rs17821518 | rs12452003 | 28.79 | 8.63E-06 | 0.04 |
| MAFB | NTN1 | rs13041631 | rs72809908 | 28.16 | 1.16E-05 | 0.05 |
| BMP4 | FGFR2 | rs2738265 | rs2936861 | 27.41 | 1.64E-05 | 0.03 |
| FGFR2 | PAX7 | rs12763463 | rs11488726 | 26.23 | 2.84E-05 | 0.18 |
| 8q24 | ARHGAP29 | rs873232 | rs3789398 | 25.82 | 3.44E-05 | 0.10 |
| 8q24 | IRF6 | rs10111530 | rs6540559 | 24.94 | 5.16E-05 | 0.14 |
| NOG | PAX7 | rs227723 | rs2236832 | 24.74 | 5.67E-05 | 0.22 |
| 8q24 | NTN1 | rs10956419 | rs2429370 | 24.39 | 6.66E-05 | 0.26 |
| MSX1 | NOG | rs9291153 | rs7222986 | 24.34 | 6.81E-05 | 0.07 |
Discussion
Compared to other large scale studies searching for evidence of gene-gene (G×G) interactions, our study implemented an efficient screening strategy to screen pairwise combinations of all highly polymorphic SNVs and focused more specific tests on the most promising pairs of markers. The 4df interaction model proposed by Cordell [2002] can detect a variety of epistatic interactions even if the individual markers (or the genes they tag) do not display detectable marginal gene effects. Moreover, to account for correlations between markers within a region due to LD between SNPs, we performed permutation testing which can control for multiple comparisons more effectively than a Bonferroni correction.
We detected evidence of a possible G×G interaction between markers in and around IRF6 and MAFB (rs6681355:rs6029315; empirical p<0.0005) in the 375 trios in the European group. This evidence of statistical interaction between SNPs in IRF6 and MAFB is especially interesting, because IRF6 is one of a few recognized NSCL/P loci showing consistent evidence of association with risk to NSCL/P across different populations, and some of its functional activities have been identified [Leslie and Marazita, 2013]. For instance, in humans mutations in IRF6 cause Van der Woude syndrome, the most common Mendelian syndrome involving oral clefts [Kondo et al. 2002], while common variants in regulatory elements confer risk of NSCL/P [Rahimov et al. 2008]. Studies in animal models have characterized IRF6 expression patterns (Knight et al. 2006) and elucidated the identities of several members of the regulatory network for IRF6 [Kousa and Schutte, 2015]. Less is known about MAFB, which resides on 20q12 and was first identified in a GWAS study [Beaty et al. 2010]. Expression studies in the mouse support some role for MAFB in palatal development [Beaty et al. 2010]. Because little else is known about the role of MAFB in craniofacial development, identifying a statistical interaction between MAFB and IRF6 is an important step. Interestingly, interactions between WNT5B and MAFB and between WNT5A and IRF6 have also been identified [Li et al. 2015], which could represent the beginnings of a new interaction network for palatal development.
We failed to detect significant G×G interaction in the Asian group of case-parent trios, despite its larger sample size. Many factors could limit our ability to detect G×G interaction between these same SNPs in this larger Asian group. Although we have a large sample of 1,034 Asian case-parent trios, markers can have different MAF across ancestral groups and some key genotypes might be under-represented in Asian populations, making it hard to fit the full interaction model for G×G interaction. In our study, we used tagging SNPs to reduce the number of multiple comparisons; however, relying on the most highly polymorphic tagging SNPs could also make it impossible to identify critical G×G interaction effects.
One of the limitations of our study was its modest sample size and low power to detect G×G interaction. Compared to detecting a marginal effect for any single SNP, detecting pairwise or 2-way G×G interaction requires much larger sample sizes, and it becomes hard to fit the 4 df G×G interaction model with its total of 8 parameters representing individual gene effects and their interactions. Even in large data sets with 2000–3000 individuals, it is difficult to detect epistasis for low frequency markers (i.e. those with MAF<0.1) [Emily et al., 2009]. Therefore our approach will only be powerful in detecting G×G interaction between highly polymorphic SNPs. Although we relied on an efficient two-stage screening strategy, the number of tests was still large. Another limitation of our study is that we only used parametric logistic regression models to test for G×G interaction. A major challenge of using traditional regression models to detect interaction is correctly specifying both the full and reduced models. Additionally, analyzing high-dimensional data, which often contains many potential interacting predictor variables, can lead to very sparse contingency tables with empty cells. Machine-learning or data-mining methods represent an alternative approach that do not rely solely a pre-specified parametric model.
Finally, the scope of our analysis was limited to targeted sequencing data on these 13 regions (see Table I) which were all previously shown by other studies to be associated with NSCL/P. Variants in other regions (i.e. those without significant marginal effects) could also be important in G×G interaction, but our study would have missed these completely. Interaction between SNPs in these regions and elements elsewhere in the genome will require more comprehensive genotyping or sequencing studies. Nonetheless, our evidence of significant G×G interaction between polymorphic markers in the IRF6 and MAFB genes in a group of case-parent trios of European ancestry is especially intriguing and should be explored more thoroughly.
Supplementary Material
Acknowledgments
We wish to acknowledge support the contributions of the CleftSeq Consortium (HG005925 (J.C.M., M.L.M.)) in generating data for this study. Parts of this work were supported by grants from the NIH (DE008559 (J.C.M., M.L.M.), DE009886 (M.L.M.), DE016930 (M.L.M.), DE016148 (M.L.M.), DE014581 (T.H.B.), DE018993 (T.H.B.), DE025060 (E.J.L.). We thank the families who participated in this study and the staff at each recruitment site around the world.
References
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, Liang KY, Wu T, Murray T, Fallin MD, et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet. 2010;42(6):525–529. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Yu Z. Simultaneous genotype calling and haplotype phasing improves genotype accuracy and reduces false-positive associations for genome-wide association studies. Am J Hum Genet. 2009;85(6):847–861. doi: 10.1016/j.ajhg.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell HJ. Epistasis: what it means, what it doesn't mean, and statistical methods to detect it in humans. Hum Mol Genet. 2002;11(20):2463–2468. doi: 10.1093/hmg/11.20.2463. [DOI] [PubMed] [Google Scholar]
- Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nat Rev Genet. 2011;12(3):167–178. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emily M, Mailund T, Hein J, Schauser L, Schierup MH. Using biological networks to search for interacting loci in genome-wide association studies. Eur J Hum Genet. 2009;17(10):1231–1240. doi: 10.1038/ejhg.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jugessur A, Farlie PG, Kilpatrick N. The genetics of isolated orofacial clefts: from genotypes to subphenotypes. Oral Dis. 2009;15(7):437–453. doi: 10.1111/j.1601-0825.2009.01577.x. [DOI] [PubMed] [Google Scholar]
- Knight AS, Schutte BC, Jiang R, Dixon MJ. Developmental expression analysis of the mouse and chick orthologues of IRF6: the gene mutated in Van der Woude syndrome. Dev Dyn. 2006;235(5):1441–1447. doi: 10.1002/dvdy.20598. [DOI] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, Howard E, de Lima RL, Daack-Hirsch S, Sander A, et al. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet. 2002;32(2):285–289. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kousa YA, Schutte BC. Toward an orofacial gene regulatory network. Developmental dynamics. 2015 doi: 10.1002/dvdy.24341. (epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Marazita ML. Genetics of cleft lip and cleft palate. Am J Med Genet C Semin Med Genet. 2013;163C(4):246–258. doi: 10.1002/ajmg.c.31381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie EJ, Taub MA, Liu H, Steinberg KM, Koboldt DC, Zhang Q, Carlson JC, Hetmanski JB, Wang H, Larson DE, et al. Identification of functional variants for cleft lip with or without cleft palate in or near PAX7, FGFR2, and NOG by targeted sequencing of GWAS loci. Am J Hum Genet. 2015;96(3):397–411. doi: 10.1016/j.ajhg.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Kim Y, Suktitipat B, Hetmanski JB, Marazita ML, Duggal P, Beaty TH, Bailey-Wilson JE. Gene-Gene Interaction Among WNT Genes for Oral Cleft in Trios. Genet Epidemiol. 2015;39(5):385–394. doi: 10.1002/gepi.21888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26(5):589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig KU, Mangold E, Herms S, Nowak S, Reutter H, Paul A, Becker J, Herberz R, AlChawa T, Nasser E, et al. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nat Genet. 2012;44(9):968–971. doi: 10.1038/ng.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita ML, Murray JC, Lidral AC, Arcos-Burgos M, Cooper ME, Goldstein T, Maher BS, Daack-Hirsch S, Schultz R, Mansilla MA, et al. Meta-analysis of 13 genome scans reveals multiple cleft lip/palate genes with novel loci on 9q21 and 2q32–35. Am J Hum Genet. 2004;75(2):161–173. doi: 10.1086/422475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita ML, Lidral AC, Murray JC, Field LL, Maher BS, Goldstein McHenry T, Cooper ME, Govil M, Daack-Hirsch S, Riley B, et al. Genome scan, fine-mapping, and candidate gene analysis of non-syndromic cleft lip with or without cleft palate reveals phenotype-specific differences in linkage and association results. Hum Hered. 2009;68(3):151–170. doi: 10.1159/000224636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita ML. The evolution of human genetic studies of cleft lip and cleft palate. Annu Rev Genomics Hum Genet. 2012;13:263–283. doi: 10.1146/annurev-genom-090711-163729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews JL, Oddone-Paolucci E, Harrop RA. The Epidemiology of Cleft Lip and Palate in Canada, 1998 to 2007. Cleft Palate Craniofac J. 2015;52(4):417–424. doi: 10.1597/14-047. [DOI] [PubMed] [Google Scholar]
- Mossey PA, Castilla EE. Global registry and database on craniofacial anomalies: Report of a WHO Registry Meeting on Craniofacial Anomalies. 2003 [Google Scholar]
- Mossey PA, Little J, Munger RG, Dixon MJ, Shaw WC. Cleft lip and palate. Lancet. 2009;374(9703):1773–1785. doi: 10.1016/S0140-6736(09)60695-4. [DOI] [PubMed] [Google Scholar]
- Rahimov F, Jugessur A, Murray JC. Genetics of nonsyndromic orofacial clefts. Cleft Palate Craniofac J. 2012;49(1):73–91. doi: 10.1597/10-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaid DJ. Likelihoods and TDT for the case-parents design. Genet Epidemiol. 1999;16(3):250–260. doi: 10.1002/(SICI)1098-2272(1999)16:3<250::AID-GEPI2>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Schwender H, Taub MA, Beaty TH, Marazita ML, Ruczinski I. Rapid testing of SNPs and gene-environment interactions in case-parent trio data based on exact analytic parameter estimation. Biometrics. 2012;68(3):766–773. doi: 10.1111/j.1541-0420.2011.01713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchero TM, Cooper ME, Maher BS, Daack-Hirsch S, Nepomuceno B, Ribeiro L, Caprau D, Christensen K, Suzuki Y, Machida J, et al. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med. 2004;351(8):769–780. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.