Abstract
Background/Objectives
The combination of energy dense diets and reduced energy expenditure in modern society has escalated the prevalence of obesity and obesity-related comorbidities. Among these disease-states, type-2 diabetics (T2D) are disproportionately associated with obesity, suggesting a shared etiology. In conjunction with defects in hormonal and inflammatory states, obesity and T2D are also characterized by dysbiosis.
Methods
We have recently described the beneficial effects of duodenal nutrient exclusion, as induced by the duodenal endoluminal sleeve (DES); including body weight loss, prevented fat mass accumulation, and improved glucose tolerance in the ZDF rat, a rodent model of obesity and type-2 diabetes (T2D). To assess the relative role of DES on hindgut microbiota in the context of these metabolic changes, we analyzed cecal samples from rats implanted with a duodenal endoluminal sleeve (DES), or a sham control of this procedure. A group of pair-fed (pf) sham controls was also included to account for changes induced by reduced body weight and food intake.
Results
Analysis of hind gut microbiota following DES in the ZDF rat elucidated discrete changes in several microbial populations including a reduction in Paraprevotella family members of the Clostridiales order along with an increase in Akkermansia muciniphila and species of the Allobaculum and Bifidobacterium genera.
Conclusions
Together these observations suggest that like Roux-en Y gastric bypass (RYGB) and Metformin, regulation of gut microbiota may be a contributing factor to the therapeutic effects of DES.
Keywords: Bariatric Surgery, Duodenal Nutrient Exclusion, Microbiota, Glucose tolerance
Introduction
The combination of energy dense diets and reduced energy expenditure in modern society has produced a dramatic shift in the number of obese and overweight individuals worldwide(1). Far from a benign accumulation of excess adipose, the escalating prevalence of obesity and obesity-related comorbidities has rapidly become a worldwide epidemic(1). Paramount among these disease-states is the disproportionate population of type-2 diabetics (T2D)(1). Thus intensive investigation over the last decade has focused on the underlying mechanisms for these sister syndromes.
Emerging evidence suggests that in conjunction with the long-known defects in hormonal and inflammatory states, obesity and T2D are also characterized by dysbiosis(2, 3). Once thought to be simple parasitic passengers, we are now aware of the complex regulatory role that gut microbiota play in our digestion, immune response, metabolism, and even mental status(4, 5). Moreover, therapeutics such as Roux-en Y gastric bypass (RYGB) and Metformin, may induce beneficial shifts in a patient’s gut microbiota(6, 7).
We have recently described the beneficial effects of duodenal nutrient exclusion, as induced by the duodenal endoluminal sleeve (DES)(8). This simple barrier device prevents contact of chyme with the duodenum, allowing for transit of food from the stomach to the jejunum. This temporary separation of chyme and digestive enzymes, mimics a portion of the RYGB procedure, and presumably invokes a dramatic change in the environmental niche of duodenal microbiota. We found that this simple device stimulated body weight loss and prevented fat mass accumulation in the ZDF rat, a rodent model of obesity and glucose dysregulation(8). This device also dramatically improved glucose tolerance, even when compared to sham-operated control rats pair-fed to match for caloric intake, body, and fat mass(8). Since this procedure mimics various aspects of RYGB, and RYGB is known to rectify components of dysbiosis(6, 9, 10), we set out to investigate the role of duodenal nutrient exclusion on gut microbiota in our DES rat model.
Materials and Methods
Animals, DES surgery, and Phenotypic analysis
DES or sham surgeries were conducted in age (10 w/old), weight, and fat mass matched ZDF rats as previously described(8). All rats received liquid diet for 120 h postoperatively. Chow diet (Teklad LM-485, 5.6% fat) was reintroduced during the last 24 h of liquid diet feeding and was then provided ad libitum to sham and DES groups. Sham-pf rats were provided chow diet in daily feedings (1 h prior to dark-phase), isocaloric to the average intake in DES rats. Subcutaneous injections of Metacam (0.25 mg/100 g BW once daily for 4 days), gentamicin (0.8 mg/100 g BW on the day of surgery), Buprenex (0.3 mL 2× per day for 5 days), and warm saline (10 and 5 mL 2× per day for days 0–3 and 4–5, respectively) were administered to all surgical groups. All procedures were approved by the University of Alabama at Birmingham Institutional Animal Care and Use Committee.
Isolation of microbial DNA and creation of 16S V4 amplicon library
In 4–8 rats per experimental group (n = 7 sham, 8 pf-sham, and 4 DES), microbial genomic DNA was isolated as previously described(11) via Fecal DNA isolation kit (Zymo Research). An amplicon library from each sample was created from the variable region 4 (V4) region of the 16S rDNA gene via PCR(12, 13).
MiSeq DNA sequencing, bioinformatics, and statistical analysis
Sequencing and bioinformatics were conducted as we have previously described(11). Briefly, we sequenced 251-base single-end reads using Illumina MiSeq(12, 13) followed by FASTQ conversion of the raw data files. We assessed FASTQ file quality via FASTQC(14) and filtered using the FASTX toolkit(15). The last base was trimmed from all reads due to low quality of at the 3’ end. Reads with Q scores of < 20 or unknown bases (“N”) were discarded. The Quantitative Insight into Microbial Ecology (QIIME) suite was utilized to group sequences into operational taxonomic units (OTUs) using the clustering program UCLUST at a similarity threshold of 97%(16). Taxonomic assignments for all OTUs at confidence threshold of 80% (0.8)(17) were made via the Ribosomal Database Program (RDP) classifier trained using the Greengenes (v13.8) 16S rRNA database(18) and OTUs whose average abundance was less than 0.005% were removed. OTUs were then grouped together to summarize taxon abundance and multiple sequence alignments of OTUs were performed with PyNAST(19). Alpha (within sample, calculated using Shannon and Chao1 metrics) and beta (between sample, using Bray-Curtis and weighted and unweighted UniFrac metrics) diversity were assessed via QIIME (20).
All data are represented as mean and standard error of the mean (SEM) from biological replicates. Where identified by ROUT (Q=10%), statistical outliers were removed. QIIME was used to conduct principal coordinates analysis (PCoA) between all samples. One-way ANOVA with Tukey’s multiple comparison post-test and ROUT performed using GraphPad Prism 6 software (San Diego, CA) In all cases, statistical significance was assumed when p < 0.05.
Results
Metabolic improvement following DES in ZDF rats
ZDF rats were matched for age, body and fat mass and then administered either the DES or a sham procedure as previously described(8). An additional group of matched, sham-operated rats was pair-fed to the DES cohort. DES reduced food intake, body-weight, and fat mass over a 28 d period as previously described(8) and re-drawn in Supplementary Table 1. The reduction in body weight, fat mass, and fasting insulin after DES was matched by the pair-fed sham controls suggesting that caloric intake was responsible for these improvements. However, intraperitoneal and oral glucose challenge revealed an enhanced glucose tolerance compared to either sham or pf-sham controls (redrawn in Supplementary Table 1) and therefore independent of the changes in body mass and composition(8). Analysis of circulating factors revealed elevated fasting glucagon, circulating bile acids, and a trend for hypersecretion of glucagon-like peptide 1 (GLP1) in response to a mixed meal challenge (redrawn in Supplementary Table 1 and (8)). Altogether these data suggest that duodenal nutrient exclusion recues multiple components of the metabolic syndrome in the ZDF rat model.
DES alters taxonomic distribution of distal gut microbiota
Following the 28 d study rats were euthanized and cecal contents collected. Microbial DNA was isolated from the cecal contents and then subjected to 16S V4 region microbiome analysis(13, 21).
We first determined whether the microbiota of the Sham, Sham-pf and DES microbiota clustered into distinct groups. Three different metrics, Bray-Curtis, Weighted and Un-weighted Unifrac revealed no significant differences in clustering of the three different groups (PERMANOVA p > 0.05, data not shown).
We next focused on the most abundant species, and utilized statistical analyses based on general linear models (two –sample test, two factor ANOVA, and multivariable regression). Evaluation of 564 independent sequences identified altered phylogenic distribution of distal gut microbiota as defined by operational taxonomic units (OTUs) following duodenal nutrient exclusion. The Firmicutes (60–64%) and Bacteroidetes (27–35%) phyla dominated gut microbiota in all rats, with smaller contributions (1–2%) from the Proteobacteria, Cyanobacteria, and Verrucomicrobia phyla (Figures 1 and 2). DES in rats was associated with a suppression of the Bacteroidetes as compared to either ad libitum or pair-fed sham groups. Conversely, we observed a relative increase in the Verrucomicrobia phyla after duodenal nutrient exclusion as compared to either sham control, with a similar trend observed in Tenericutes (Figure 1). Previous reports suggest that improved metabolic states are associated with larger Bacteroidetes to Firmicutes ratios(22, 23). In our ZDF rats this ratio was unchanged in the reduced obesity stimulated by caloric restriction (Figure 3j; ad libitum vs. pf-shams). Moreover, duodenal nutrient exclusion was associated with reduced Bacteroidetes (Figure 3b) rather than Firmicutes abundance (Figure 3a), resulting in a trend for a decrease in this ratio (Figure 3j).
Figure 1.
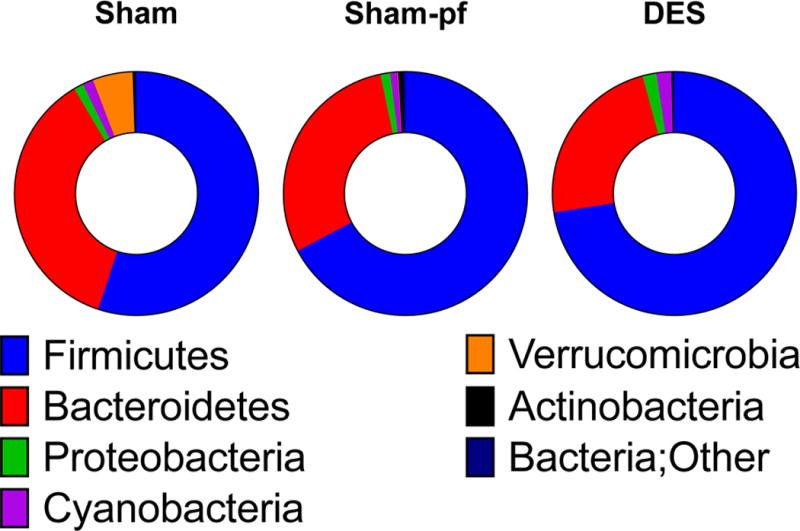
Phylum distribution in hindgut microbiota of Sham, Sham-pf, and DES rats. All data are represented as mean of 4–8 rats/group.
Figure 2.
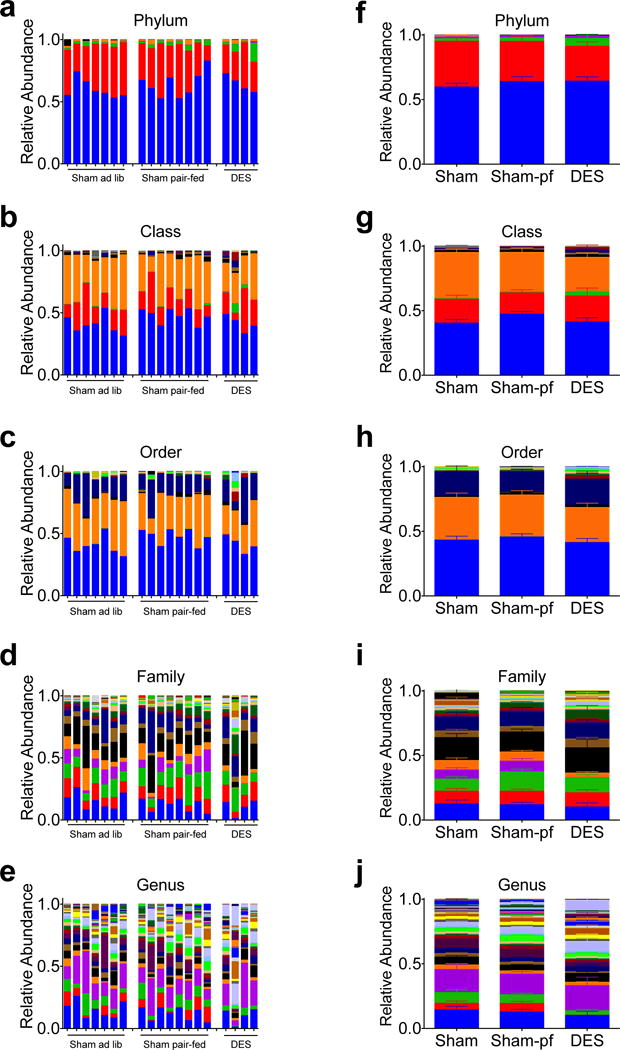
Taxonomic distribution of hindgut microbiota in individual rats (a–e) and by surgical group (f–j) following 28 d study. Data in (f–j) represented as mean +/− SEM (n=4–8 rats/group).
Figure 3.
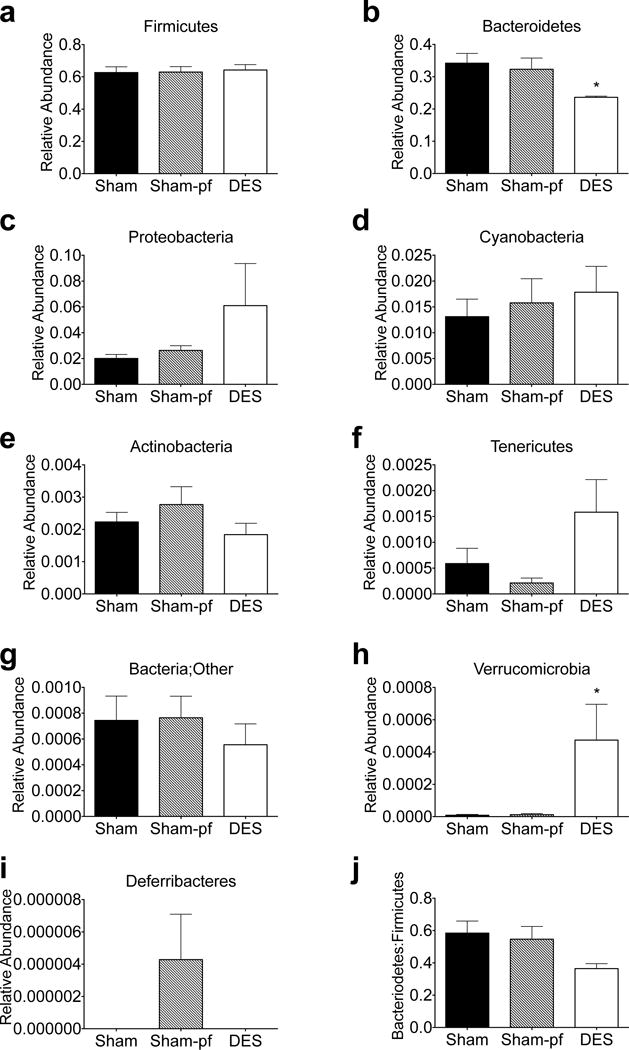
Relative regulation of hindgut microbiota by phyla in DES rats (a–i) and Bacteroidetes to Firmicutes ratio (j) following 28 d study. All data represented as mean +/− SEM (n=4–8 rats/group). *p< 0.05 vs. Sham control.
DES preferentially favors discrete microbiota of the distal gut
The relative abundance of many species, across multiple phyla, are modulated by DES; thus, we first focused on those species that comprised the largest proportion of the microbial population. Within the top 25 most abundant OTU, DES suppressed species of the Lactobacillus, Streptococcus, and the Prevotella genera, with a similar trend in a species of the Paraprevotella genus as compared to their weight- and fat mass- matched Sham-pf controls (Figure 4a–d). Likewise, we observed that DES induced a suppression of species from the Candidatus Arthromitus and Rothia genera as compared to ad libitum sham controls (Figure 4e–f). Conversely, DES stimulated a species-specific increase in the abundance of Clostridium (C.) perfringens and species of the Enterococcus genus (Figure 4g–h).
Figure 4.
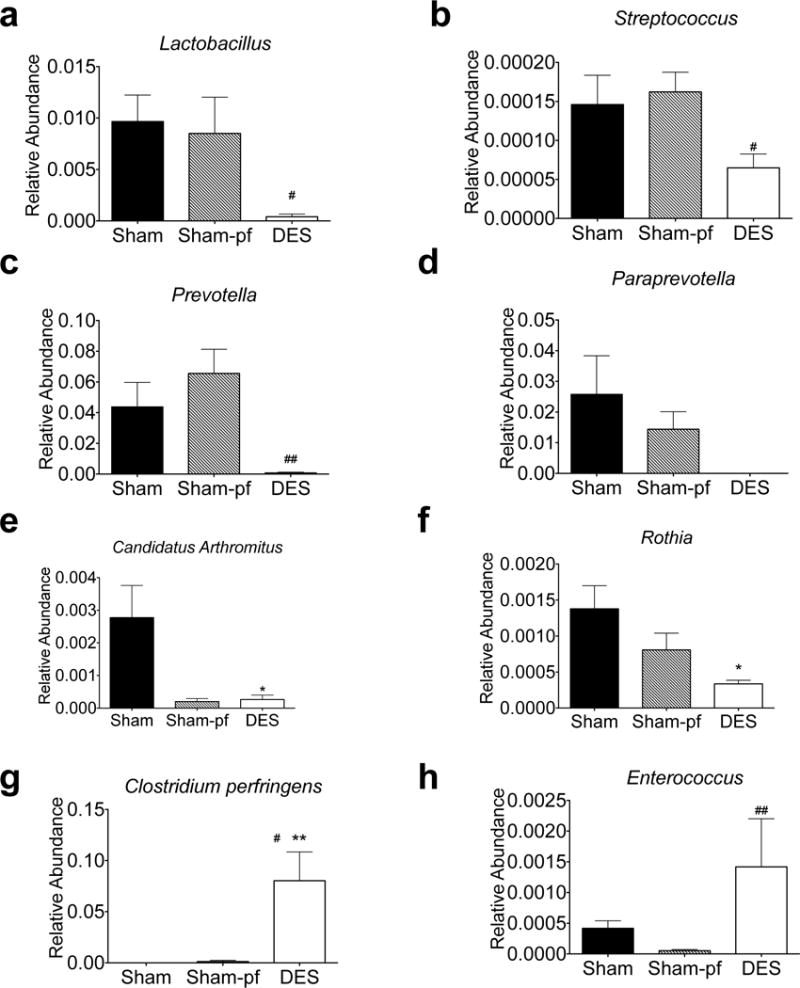
Relative regulation of the most abundant hindgut microbiota by genera in DES rats (a–h) following 28 d study. All data represented as mean +/− SEM (n=4–8 rats/group). *p< 0.05, **p< 0.01, vs. Sham control, #p< 0.05, ##p< 0.01 vs. Sham-pf control.
Beyond these most abundant OTUs, DES rats were characterized by a 30% reduction in the relative abundance of the Bacteroidetes phyla and a 27% reduction in the Bacteroidia class (p<0.05) as compared to weight and fat mass matched sham-pf rats (Figure 2). This suppression was also observed at the level of order (bacteroidales; Figure 5a) and was characterized by an almost complete abrogation of the Paraprevotellaceae family (Figure 5b), which normally comprises 7–8% of the total gut microbiota abundance (Figure 2g–h; Sham). Within this family are two genera, Prevotella and Paraprevotella, where we observed a universal suppression of 5 independent species. However, not all Bacteroidales families were suppressed. We observed a 70% increase in the genus Parabacteroides of the Porphyromonadaceae family in the DES rats as compared to sham controls (Figure 5i). Moreover, we observed elevated levels of several species within the S24-7 family of Bacteriodales in DES animals (Figure 5j).
Figure 5.
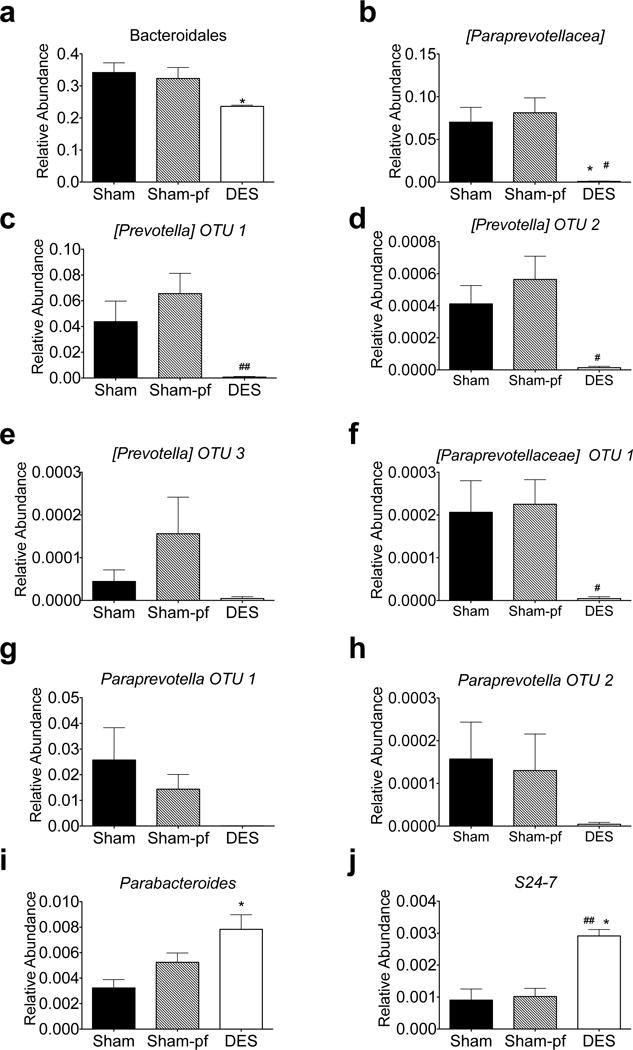
Relative suppression of the Paraprevotellaceae family of Bacteroidia (a–h) in hindgut microbiota of DES rats following 28 d study. Relative increase in the Porphyromonadaceae and S24-7 families (i–j) induced by DES following 28 d study. All data represented as mean +/− SEM (n=4–8 rats/group). *p< 0.05 vs. Sham control, #p< 0.05, ##p< 0.01 vs. Sham-pf control.
Firmicutes comprise the vast majority of gut microbiota in DES and control rats, yet duodenal nutrient exclusion had no effect on relative Firmicutes abundance. However, several families of this phylum were significantly altered by the DES procedure. As mentioned above, DES stimulated a considerable elevation in species of the Enterococcus genus (Figure 4h), while stimulating inverse effects on the Candidatus Arthromitus and C. perfringens species (Figure 4e and g). We also observed an increase in the Erysipelotrichi class of Firmicutes (Figure 6a) as compared to either ad libitum or pf sham controls and this increase in abundance was driven by the Allobaculum genus of the Erysipelotrichaceae family (Figure 6b).
Figure 6.
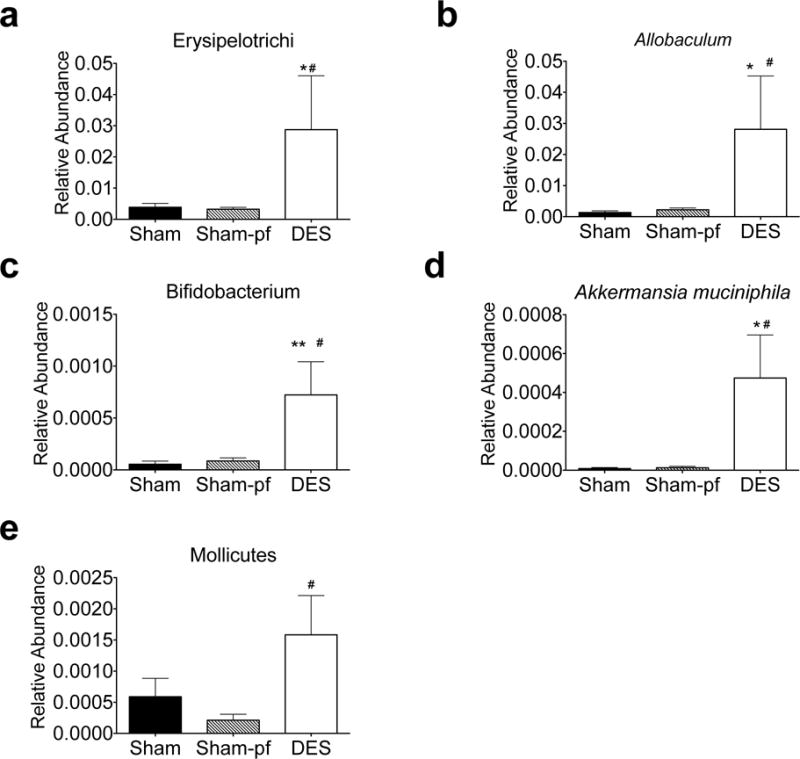
Relative abundance of hindgut microbiota in the Firmicutes phyla of DES rats following 28 d study. All data represented as mean +/− SEM (n=4–8 rats/group). *p< 0.05, **p< 0.01 vs. Sham control, #p< 0.05 vs. Sham-pf control.
Aside from these prominent changes in the Bacteriodetes and Firmicutes phyla, we also observed changes in members of several smaller phyla. Specifically, we uncovered a considerable increase in the Bifidobacterium genus of the Actinobacteria phylum (Figure 6c) and Akkermansia muciniphila of the Verrucomicrobia phylum (Figure 6d), as well as the Mollicutes class in the Tenericutes phylum (Figure 6e). Taken together, these data suggest that like RYGB(6, 9, 10) and Verticle Sleeve Gastrectomy (VSG) (24, 25), DES alters the distal gut microbiota.
Discussion
Gut microbiota have emerged as a potential regulator of energy balance, glucose and lipid metabolism(26). Furthermore, bariatric interventions such as RYGB, VSG, and ileal transposition are all known to rescue components of the metabolic syndrome and are associated with pronounced modifications of gut microbiota(6, 24, 27). Thus, we set out to test the hypothesis that duodenal nutrient exclusion via duodenal endoluminal sleeve would likewise modulate gut microbiota.
Our studies were conducted in the ZDF rat model of obesity and T2D. These rats are characterized by hyperphagia, obesity, dyslipidemia, and β cell failure leading to eventual hyperglycemia. While diet and obesity are both known to influence the gut microbiota(26), these metabolic defects develop in ZDF rats maintained on standard chow, excluding high-fat feeding from the experimental variables. Moreover, our experimental design included a pair-fed (pf) sham group which was body weight and fat mass matched(8) to the DES cohort. In our prior studies, we observed that the weight loss effects of DES were matched via isocaloric pair-feeding of sham rats(8). However, DES stimulated enhanced glucose tolerance as compared to these otherwise matched rats(8). Thus we are able to identify the interactions between gut microbiota and obesity (sham vs. sham-pf), and the enhanced glucose handling induced by DES (sham-pf vs. DES). It must be noted that this model of T2D and obesity is based on defects in leptin action. Thus this model may be characterized by changes in gut microbiota related to defects in leptin signaling(28), obesity, hyperglycemia, or any combination of these variables. Moreover, the extensive inbreeding necessary to create this strain likely influences the relative homogeneity of the microbiota in these rats, and by extension our studies. Furthermore, this background may contribute to differential effects as compared to human studies as well as studies in mice and other rat models.
When comparing our DES intervention to the ad libitium sham, or more appropriately, the body weight and fat mass matched pf-shams, we were surprised to find an increased Firmicutes:Bacteriodetes ratio. A greater ratio of these two largest microbiota phyla has been previously implicated as a marker of obesity and T2D. Moreover, interventions known to improve components of the metabolic syndrome (e.g. RYGB, VSG, IT, BD, metformin) reduce this ratio. However, the causality of this shift in phyla as component of the organism’s health, or even as a useful biomarker, has recently come into question(29). A recent review suggests that is rather the targeted manipulation of specific microbiota (i.e. Bifidobacteria, Bacillus, and Lactobacillus as well as the ratio of Bacteriodes-Prevotella to C. coccoides-E. rectale) which results in divergent metabolic outcomes(29). Thus, it is important to evaluate these interventions and their induced changes in gut microbiota beyond this simple ratio. It should also be noted that both the small number of samples (4) and the genetic background of the model may contribute to this surprising finding.
Prior evidence for microbiota associated with the beneficial effects of bariatric surgeries has identified changes in several key genera and even specific species. Specifically, investigation of RYGB patients has uncovered elevated abundance of species in the Gammaproteobacteria class (e.g. E. coli) and reduced abundance of Clostridium members of the Firmicutes phyla(25). Studies in high-fat fed mouse models of RYGB confirmed conserved elevations of Gammaproteobacteria (Escherichia) and identified similar effects in the Verrucomicrobia (Akkermansia) class(6). With regard to our studies, we found that like RYGB, DES stimulates an increase in the abundance of these classes. Among those microbes previously identified in RYGB, we observed a dramatic reduction in multiple members of the Prevotellaceae family. These gram-negative anaerobes are enriched in obesity, impair IL-18 production, disrupt mucosal barrier function(30), and suppressed following RYGB(22). Conversely, we found the increase in Akkermansia muciniphila of particular interest. This mucin-degrading bacterium is has been proposed to regulate gut health, energy balance, and glucose homoeostasis(31, 32). Its abundance is elevated by RYGB(6) and Metformin(7) therapies alike, yet it has also been linked to exacerbated gut inflammation (33). However, VSG, which stimulates RYGB-like changes in glucose metabolism an energy balance, is associated with suppression of this genus(24) in high-fat fed mice. The comparison of these divergent bariatric interventions suggests that duodenal nutrient exclusion may be selectively beneficial for Akkermansia muciniphila, but it also suggests that this microbe may not be solely responsible for the benefits of these procedures. A similar comparison can be drawn with respect to the Bifidobacterium genus. This gram-positive anaerobe is depleted in T1D(34) and conversely, prebiotic-stimulated increases in this genus reduce metabolic deficits associated with high-fat-diet-induced diabetes in mice(35). Consistent with these beneficial actions, we observed a considerable increase in the abundance of the Bifidobacterium genus following DES. This microbe is suppressed following RYGB(9, 10) and remains unchanged after VSG(25), suggesting a key divergence between these procedures. The Erysipelotrichaceae family of Firmicutes is similarly elevated in obesity, associated with inflammatory bowel disease (30), and suppressed to control levels following RYGB in man(22). However, we observed a clear increase in Erysipelotrichaceae abundance that was primarily associated with an increase in the Allobaculum genus. This genus of the Firmicutes is specifically enriched in low- vs. high-fat fed mice, in spite of the relative increase in all other Firmicutes observed in high-fat feeding(28).
While DES shares a greater anatomical and microbiota similarity with RYGB, we were able to identify similar regulation of DES and VSG with regard to the Porphyromonadaceae family. These gram-negative anaerobes are negatively correlated with T1D(36) and display suppressed abundance in high-fat fed mice(37). Furthermore, a genus in the Porphyromonadaceae family was increased in high-fat fed, VSG mice in an FXR-dependent manner(24). Likewise, we observed an increase in the Parabacteroides genus of Porphyromonadaceae following DES. Coupled with the shared modulation of bile acids observed in these procedures, these data suggest that Parabacteroides abundance in DES may be similarly regulated via FXR activation.
Beyond those previously associated with bariatric interventions, we also identified several microbes associated with obesity, diabetes, or both disease states. One of the most abundant examples is the relative increase in C. perfringens species observed after DES. Reduced C. perfringens has previously been associated with obesity in man(38), yet our data suggest that this microbe is dramatically increased as compared to either ad libitum or pair-fed sham groups, implying that C. perfringens may contribute to the beneficial effects of DES on glucose metabolism. An increase in Clostridia has also been observed in low-fat fed, ZDF rats that have undergone RYGB (39). C. perfringens is readily found in the intestines of animals and humans but it is not clear whether this increase is due to favorable growth conditions after surgery. Although the potential exists for pathogenesis as a result of the increased C. perfringins (40), we did not observe any illness in the animals. Conversely, DES implant in ZDF rats stimulated a suppression of Candidatus Arthromitus abundance. This finding stands in contrast to prior reports suggesting that early-life microbiota disruption stimulated defects in body composition and glucose tolerance and was associated with reduced Candidatus Arthromitus abundance(41).
Bariatric interventions such as RYGB and VSG are currently the most effective therapies for obesity and T2D. However, the molecular and systemic mechanisms responsible for the beneficial physiology observed after these surgeries have yet to be fully elucidated. The DES procedure mimics RYGB in the context of duodenal nutrient exclusion, yet preserves overall gut anatomy and is fully reversible. Our data suggest that a similar regulation of gut microbiota is associated with this overlapping physiology. During the preparation of this manuscript novel associations between small intestine bacterial overgrowth and enhanced weight loss following RYGB were described (42). Of note, these effects were not observed in patients who received adjustable gastric banding, suggesting that gut perturbation is an essential component of this effect. While not tested in our model, this observation adds to the growing list of potential microbiota;gut interactions that must be addressed. Moreover, although we have identified the regulation of several intriguing microbes, these observations warrant further critical investigation before causality or implication as potential therapeutic targets can be made.
Supplementary Material
Acknowledgments
Peter Eipers for sample processing and 16S analysis.
Support:
The project described was supported by Award Number 5K01DK098319 (KMH) from the National Institute Of Diabetes And Digestive And Kidney Diseases.
The following are acknowledged for their support of the Microbiome Resource at the University of Alabama at Birmingham: School of Medicine, Comprehensive Cancer Center (P30AR050948), Center for AIDS Research (5P30AI027767), Center for Clinical Translational Science (UL1TR000165) and Heflin Center.
Footnotes
Conflict of interest:
The authors declare no conflict of interest.
Author contributions
TK and KMH were responsible for study conception and design, data analyses and interpretation, and drafting the article; TK and CLH generated experimental data; TP and CDM advised study concept, data analyses, and critical revision of the article. KMH is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Supplementary information is available at IJO’s website
References
- 1.Smith KB, Smith MS. Obesity Statistics. Prim Care. 2016;43(1):121–35. doi: 10.1016/j.pop.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Anhe FF, Varin TV, Le Barz M, Desjardins Y, Levy E, Roy D, et al. Gut Microbiota Dysbiosis in Obesity-Linked Metabolic Diseases and Prebiotic Potential of Polyphenol-Rich Extracts. Curr Obes Rep. 2015;4(4):389–400. doi: 10.1007/s13679-015-0172-9. [DOI] [PubMed] [Google Scholar]
- 3.Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 4.Dore J, Simren M, Buttle L, Guarner F. Hot topics in gut microbiota. United European Gastroenterol J. 2013;1(5):311–8. doi: 10.1177/2050640613502477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rios-Covian D, Ruas-Madiedo P, Margolles A, Gueimonde M, de Los Reyes-Gavilan CG, Salazar N. Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Front Microbiol. 2016;7:185. doi: 10.3389/fmicb.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liou AP, Paziuk M, Luevano JM, Jr, Machineni S, Turnbaugh PJ, Kaplan LM. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med. 2013;5(178):178ra41. doi: 10.1126/scitranslmed.3005687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee H, Ko G. Effect of metformin on metabolic improvement and gut microbiota. Appl Environ Microbiol. 2014;80(19):5935–43. doi: 10.1128/AEM.01357-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Habegger KM, Al-Massadi O, Heppner KM, Myronovych A, Holland J, Berger J, et al. Duodenal nutrient exclusion improves metabolic syndrome and stimulates villus hyperplasia. Gut. 2014;63(8):1238–46. doi: 10.1136/gutjnl-2013-304583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furet JP, Kong LC, Tap J, Poitou C, Basdevant A, Bouillot JL, et al. Differential adaptation of human gut microbiota to bariatric surgery-induced weight loss: links with metabolic and low-grade inflammation markers. Diabetes. 2010;59(12):3049–57. doi: 10.2337/db10-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong LC, Tap J, Aron-Wisnewsky J, Pelloux V, Basdevant A, Bouillot JL, et al. Gut microbiota after gastric bypass in human obesity: increased richness and associations of bacterial genera with adipose tissue genes. Am J Clin Nutr. 2013;98(1):16–24. doi: 10.3945/ajcn.113.058743. [DOI] [PubMed] [Google Scholar]
- 11.Kumar R, Maynard CL, Eipers P, Goldsmith KT, Ptacek T, Grubbs JA, et al. Colonization potential to reconstitute a microbe community in patients detected early after fecal microbe transplant for recurrent C. difficile. BMC Microbiol. 2016;16:5. doi: 10.1186/s12866-015-0622-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Applied and environmental microbiology. 2013;79(17):5112–20. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar R, Eipers P, Little RB, Crowley M, Crossman DK, Lefkowitz EJ, et al. Getting started with microbiome analysis: sample acquisition to bioinformatics. Curr Protoc Hum Genet. 2014;82:18 8 1–29. doi: 10.1002/0471142905.hg1808s82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.FASTQC. [Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 15.FASTX-Toolkit. [Available from: http://hannonlab.cshl.edu/fastx_toolkit/
- 16.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics (Oxford, England) 2010;26(19):2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and environmental microbiology. 2007;73(16):5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6(3):610–8. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics (Oxford, England) 2010;26(2):266–7. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lozupone C, Hamady M, Knight R. UniFrac–an online tool for comparing microbial community diversity in a phylogenetic context. BMC bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brawner KM, Morrow CD, Smith PD. Gastric microbiome and gastric cancer. Cancer J. 2014;20(3):211–6. doi: 10.1097/PPO.0000000000000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci U S A. 2009;106(7):2365–70. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryan KK, Tremaroli V, Clemmensen C, Kovatcheva-Datchary P, Myronovych A, Karns R, et al. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature. 2014;509(7499):183–8. doi: 10.1038/nature13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tremaroli V, Karlsson F, Werling M, Stahlman M, Kovatcheva-Datchary P, Olbers T, et al. Roux-en-Y Gastric Bypass and Vertical Banded Gastroplasty Induce Long-Term Changes on the Human Gut Microbiome Contributing to Fat Mass Regulation. Cell Metab. 2015;22(2):228–38. doi: 10.1016/j.cmet.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hartstra AV, Bouter KE, Backhed F, Nieuwdorp M. Insights into the role of the microbiome in obesity and type 2 diabetes. Diabetes Care. 2015;38(1):159–65. doi: 10.2337/dc14-0769. [DOI] [PubMed] [Google Scholar]
- 27.Cummings BP, Bettaieb A, Graham JL, Kim J, Ma F, Shibata N, et al. Bile-acid-mediated decrease in endoplasmic reticulum stress: a potential contributor to the metabolic benefits of ileal interposition surgery in UCD-T2DM rats. Dis Model Mech. 2013;6(2):443–56. doi: 10.1242/dmm.010421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ravussin Y, Koren O, Spor A, LeDuc C, Gutman R, Stombaugh J, et al. Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity (Silver Spring) 2012;20(4):738–47. doi: 10.1038/oby.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown K, DeCoffe D, Molcan E, Gibson DL. Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients. 2012;4(8):1095–119. doi: 10.3390/nu4081095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagao-Kitamoto H, Kitamoto S, Kuffa P, Kamada N. Pathogenic role of the gut microbiota in gastrointestinal diseases. Intest Res. 2016;14(2):127–38. doi: 10.5217/ir.2016.14.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013;110(22):9066–71. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dao MC, Everard A, Aron-Wisnewsky J, Sokolovska N, Prifti E, Verger EO, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut. 2016;65(3):426–36. doi: 10.1136/gutjnl-2014-308778. [DOI] [PubMed] [Google Scholar]
- 33.Ganesh BP, Klopfleisch R, Loh G, Blaut M. Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PLoS One. 2013;8(9):e74963. doi: 10.1371/journal.pone.0074963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murri M, Leiva I, Gomez-Zumaquero JM, Tinahones FJ, Cardona F, Soriguer F, et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. 2013;11:46. doi: 10.1186/1741-7015-11-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cani PD, Neyrinck AM, Fava F, Knauf C, Burcelin RG, Tuohy KM, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50(11):2374–83. doi: 10.1007/s00125-007-0791-0. [DOI] [PubMed] [Google Scholar]
- 36.Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455(7216):1109–13. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parks BW, Nam E, Org E, Kostem E, Norheim F, Hui ST, et al. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 2013;17(1):141–52. doi: 10.1016/j.cmet.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuo HJ, Xie ZM, Zhang WW, Li YR, Wang W, Ding XB, et al. Gut bacteria alteration in obese people and its relationship with gene polymorphism. World J Gastroenterol. 2011;17(8):1076–81. doi: 10.3748/wjg.v17.i8.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo Y, Liu CQ, Shan CX, Chen Y, Li HH, Huang ZP, et al. Gut microbiota after Roux-en-Y gastric bypass and sleeve gastrectomy in a diabetic rat model: Increased diversity and associations of discriminant genera with metabolic changes. Diabetes Metab Res Rev. 2016 doi: 10.1002/dmrr.2857. [DOI] [PubMed] [Google Scholar]
- 40.Hatheway CL. Toxigenic clostridia. Clin Microbiol Rev. 1990;3(1):66–98. doi: 10.1128/cmr.3.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cox LM, Blaser MJ. Antibiotics in early life and obesity. Nat Rev Endocrinol. 2015;11(3):182–90. doi: 10.1038/nrendo.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabate JM, Coupaye M, Ledoux S, Castel B, Msika S, Coffin B, et al. Consequences of Small Intestinal Bacterial Overgrowth in Obese Patients Before and After Bariatric Surgery. Obes Surg. 2016 doi: 10.1007/s11695-016-2343-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


