Abstract
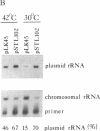
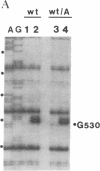
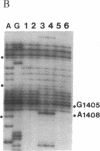
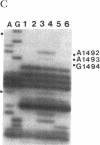
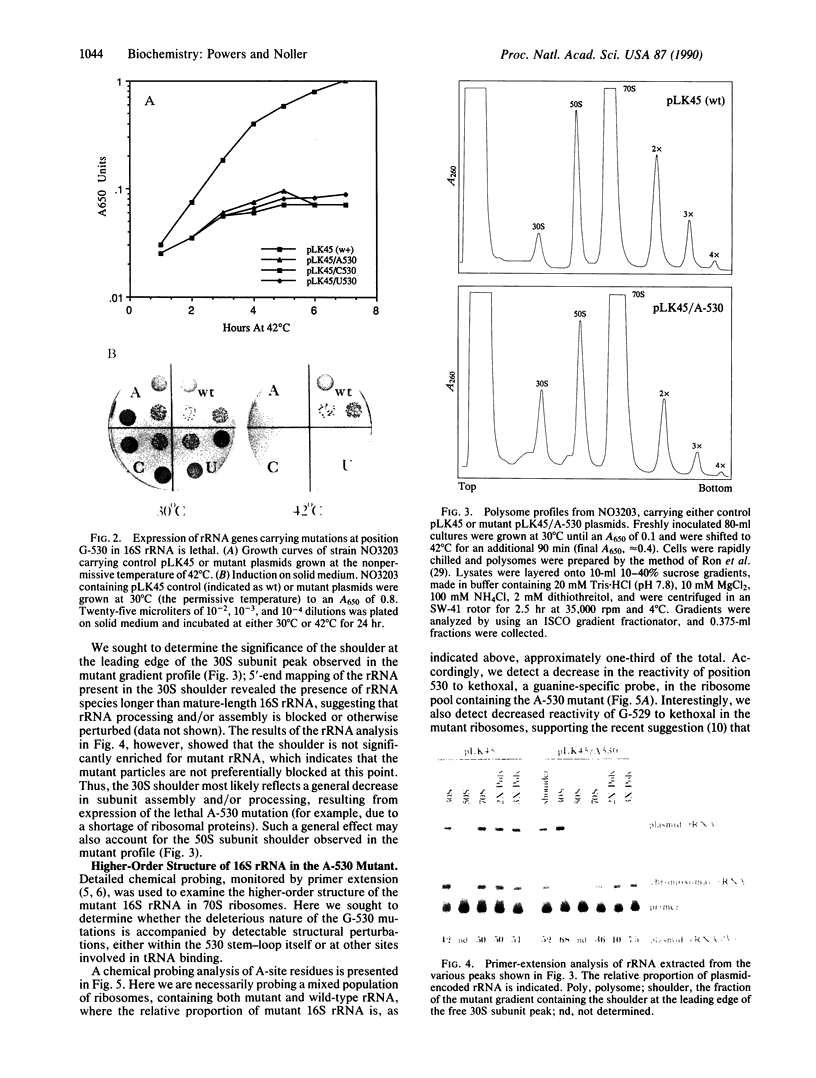
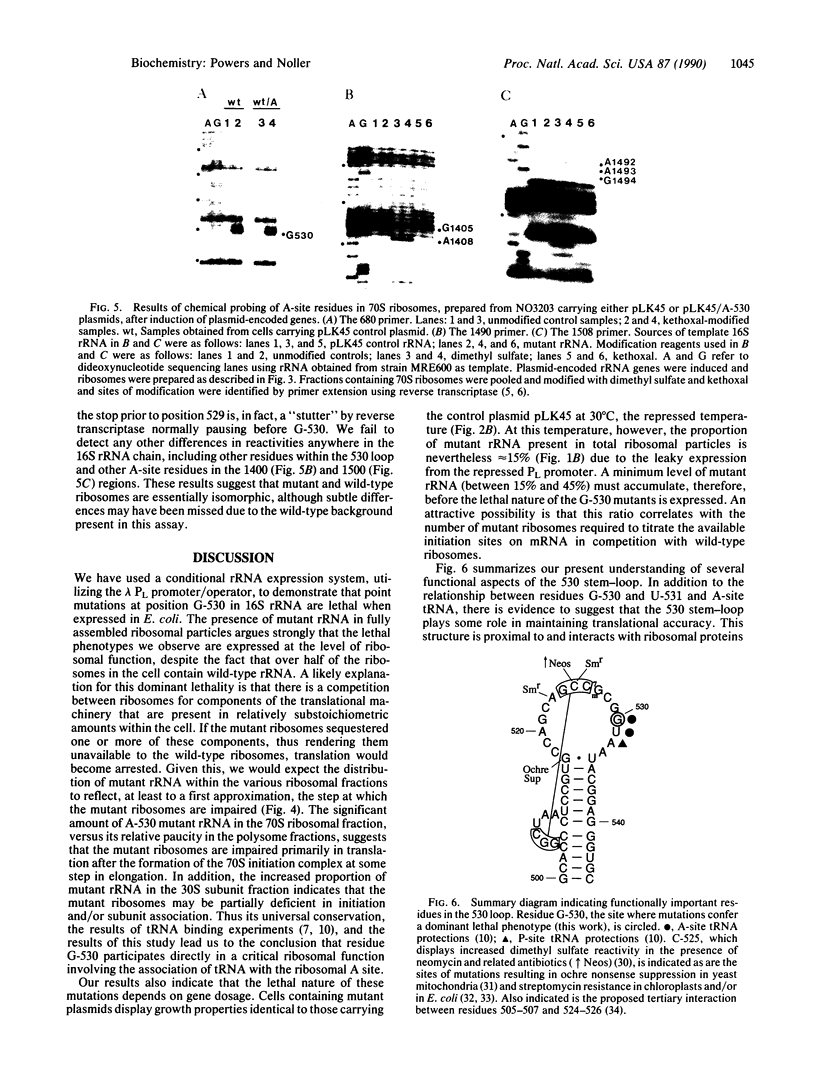
The 530 stem-loop region in 16S rRNA is among the most phylogenetically conserved structural elements in all rRNAs, yet its role in protein synthesis remains mysterious. G-530 is protected from kethoxal attack when tRNA, or its 15-nucleotide anticodon stem-loop fragment, is bound to the ribosomal A site. Based on presently available evidence, however, this region is believed to be too remote from the decoding site for this protection to be the result of direct contact. In this study, we use a conditional rRNA expression system to demonstrate that plasmid-encoded 16S rRNA genes carrying A, C, and T point mutations at position G-530 confer a dominant lethal phenotype when expressed in Escherichia coli. Analysis of the distribution of plasmid-encoded 16S rRNA in ribosomal particles, following induction of the A-530 mutation, shows that mutant rRNA is present both in 30S subunits and in 70S ribosomes. Little mutant rRNA is found in polyribosomes, however, indicating that the mutant ribosomes are severely impaired at the stage of polysome formation and/or stability. Detailed chemical probing of mutant ribosomal particles reveals no evidence of structural perturbation within the 16S rRNA. Taken together, these results argue for the direct participation of G-530 in ribosomal function and, furthermore, suggest that the dominant lethal phenotype caused by these mutations is due primarily to the mutant ribosomes blocking a crucial step in protein synthesis after translational initiation.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Brow D. A., Noller H. F. Protection of ribosomal RNA from kethoxal in polyribosomes. Implication of specific sites in ribosome function. J Mol Biol. 1983 Jan 5;163(1):27–46. doi: 10.1016/0022-2836(83)90028-1. [DOI] [PubMed] [Google Scholar]
- Dahlberg A. E. The functional role of ribosomal RNA in protein synthesis. Cell. 1989 May 19;57(4):525–529. doi: 10.1016/0092-8674(89)90122-0. [DOI] [PubMed] [Google Scholar]
- Douthwaite S., Powers T., Lee J. Y., Noller H. F. Defining the structural requirements for a helix in 23 S ribosomal RNA that confers erythromycin resistance. J Mol Biol. 1989 Oct 20;209(4):655–665. doi: 10.1016/0022-2836(89)93000-3. [DOI] [PubMed] [Google Scholar]
- Evnin L. B., Craik C. S. Development of an efficient method for generating and screening active trypsin and trypsin variants. Ann N Y Acad Sci. 1988;542:61–74. doi: 10.1111/j.1749-6632.1988.tb25808.x. [DOI] [PubMed] [Google Scholar]
- Gornicki P., Nurse K., Hellmann W., Boublik M., Ofengand J. High resolution localization of the tRNA anticodon interaction site on the Escherichia coli 30 S ribosomal subunit. J Biol Chem. 1984 Aug 25;259(16):10493–10498. [PubMed] [Google Scholar]
- Gourse R. L., Takebe Y., Sharrock R. A., Nomura M. Feedback regulation of rRNA and tRNA synthesis and accumulation of free ribosomes after conditional expression of rRNA genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1069–1073. doi: 10.1073/pnas.82.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Weiser B., Woese C. R., Noller H. F. Comparative anatomy of 16-S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- Hui A. S., Eaton D. H., de Boer H. A. Mutagenesis at the mRNA decoding site in the 16S ribosomal RNA using the specialized ribosome system in Escherichia coli. EMBO J. 1988 Dec 20;7(13):4383–4388. doi: 10.1002/j.1460-2075.1988.tb03337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob W. F., Santer M., Dahlberg A. E. A single base change in the Shine-Dalgarno region of 16S rRNA of Escherichia coli affects translation of many proteins. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4757–4761. doi: 10.1073/pnas.84.14.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melançon P., Lemieux C., Brakier-Gingras L. A mutation in the 530 loop of Escherichia coli 16S ribosomal RNA causes resistance to streptomycin. Nucleic Acids Res. 1988 Oct 25;16(20):9631–9639. doi: 10.1093/nar/16.20.9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Interaction of antibiotics with functional sites in 16S ribosomal RNA. Nature. 1987 Jun 4;327(6121):389–394. doi: 10.1038/327389a0. [DOI] [PubMed] [Google Scholar]
- Moazed D., Noller H. F. Transfer RNA shields specific nucleotides in 16S ribosomal RNA from attack by chemical probes. Cell. 1986 Dec 26;47(6):985–994. doi: 10.1016/0092-8674(86)90813-5. [DOI] [PubMed] [Google Scholar]
- Moazed D., Stern S., Noller H. F. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J Mol Biol. 1986 Feb 5;187(3):399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Chaires J. B. Functional modification of 16S ribosomal RNA by kethoxal. Proc Natl Acad Sci U S A. 1972 Nov;69(11):3115–3118. doi: 10.1073/pnas.69.11.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Woese C. R. Secondary structure of 16S ribosomal RNA. Science. 1981 Apr 24;212(4493):403–411. doi: 10.1126/science.6163215. [DOI] [PubMed] [Google Scholar]
- Politz S. M., Glitz D. G. Ribosome structure: localization of N6,N6-dimethyladenosine by electron microscopy of a ribosome-antibody complex. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1468–1472. doi: 10.1073/pnas.74.4.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron E. Z., Kohler R. E., Davis B. D. Polysomes extracted from Escherichia coli by freeze-thaw-lysozyme lysis. Science. 1966 Sep 2;153(3740):1119–1120. doi: 10.1126/science.153.3740.1119. [DOI] [PubMed] [Google Scholar]
- Shen Z. H., Fox T. D. Substitution of an invariant nucleotide at the base of the highly conserved '530-loop' of 15S rRNA causes suppression of yeast mitochondrial ochre mutations. Nucleic Acids Res. 1989 Jun 26;17(12):4535–4539. doi: 10.1093/nar/17.12.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigmund C. D., Ettayebi M., Borden A., Morgan E. A. Antibiotic resistance mutations in ribosomal RNA genes of Escherichia coli. Methods Enzymol. 1988;164:673–690. doi: 10.1016/s0076-6879(88)64077-8. [DOI] [PubMed] [Google Scholar]
- Sigmund C. D., Ettayebi M., Morgan E. A. Antibiotic resistance mutations in 16S and 23S ribosomal RNA genes of Escherichia coli. Nucleic Acids Res. 1984 Jun 11;12(11):4653–4663. doi: 10.1093/nar/12.11.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S., Moazed D., Noller H. F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- Stern S., Powers T., Changchien L. M., Noller H. F. Interaction of ribosomal proteins S5, S6, S11, S12, S18 and S21 with 16 S rRNA. J Mol Biol. 1988 Jun 20;201(4):683–695. doi: 10.1016/0022-2836(88)90467-6. [DOI] [PubMed] [Google Scholar]
- Stern S., Weiser B., Noller H. F. Model for the three-dimensional folding of 16 S ribosomal RNA. J Mol Biol. 1988 Nov 20;204(2):447–481. doi: 10.1016/0022-2836(88)90588-8. [DOI] [PubMed] [Google Scholar]
- Thomas C. L., Gregory R. J., Winslow G., Muto A., Zimmermann R. A. Mutations within the decoding site of Escherichia coli 16S rRNA: growth rate impairment, lethality and intragenic suppression. Nucleic Acids Res. 1988 Aug 25;16(16):8129–8146. doi: 10.1093/nar/16.16.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe M. R., Ohgi K., Glitz D. G. Ribosome structure. Localization of 7-methylguanosine in the small subunits of Escherichia coli and chloroplast ribosomes by immunoelectron microscopy. J Biol Chem. 1982 Aug 25;257(16):9822–9829. [PubMed] [Google Scholar]
- Triman K., Becker E., Dammel C., Katz J., Mori H., Douthwaite S., Yapijakis C., Yoast S., Noller H. F. Isolation of temperature-sensitive mutants of 16 S rRNA in Escherichia coli. J Mol Biol. 1989 Oct 20;209(4):645–653. doi: 10.1016/0022-2836(89)92000-7. [DOI] [PubMed] [Google Scholar]
- Vester B., Garrett R. A. The importance of highly conserved nucleotides in the binding region of chloramphenicol at the peptidyl transfer centre of Escherichia coli 23S ribosomal RNA. EMBO J. 1988 Nov;7(11):3577–3587. doi: 10.1002/j.1460-2075.1988.tb03235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Fox G. E., Zablen L., Uchida T., Bonen L., Pechman K., Lewis B. J., Stahl D. Conservation of primary structure in 16S ribosomal RNA. Nature. 1975 Mar 6;254(5495):83–86. doi: 10.1038/254083a0. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Gutell R. R. Evidence for several higher order structural elements in ribosomal RNA. Proc Natl Acad Sci U S A. 1989 May;86(9):3119–3122. doi: 10.1073/pnas.86.9.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]