Abstract
Using Cell Surface Capture Technology, the cell surface N-glycoproteome of human induced pluripotent stem cell derived hepatic endoderm cells was assessed. Altogether, 395 cell surface N-glycoproteins were identified, represented by 1273 N-glycopeptides. This study identified N-glycoproteins that are not predicted to be localized to the cell surface and provides experimental data that assist in resolving ambiguous or incorrectly annotated transmembrane topology annotations. In a proof-of-concept analysis, combining these data with other cell surface proteome datasets is useful for identifying potentially cell type and lineage restricted markers and drug targets to advance the use of stem cell technologies for mechanistic developmental studies, disease modeling, drug discovery, and regenerative medicine.
Keywords: N-glycoproteins, Cell surface proteins, Plasma membrane, Hepatic endoderm
Human pluripotent stem cells (hPSC), including embryonic (hESC) and induced (hiPSC), are characterized by an ability to continuously self-renew and differentiate into any cell type in the body. Using modern culturing and differentiation protocols, hPSC can be effectively differentiated into cell types of each developmental lineage - endoderm, ectoderm, and mesoderm. For these reasons, hPSC are a valuable and unlimited source of cells for drug discovery, disease modeling, studies of early embryonic development, and regenerative medicine [1]. However, for most hPSC derivatives, there remain challenges to directing differentiation to a homogeneous population of cells with a desired phenotype, driving differentiation to an adult-like phenotype, and determining the appropriate phenotype and maturation stage most apt for various downstream applications [2]. Consequently, new tools to define, select, and study specific cellular phenotypes, especially approaches that enable single cell level analyses, would facilitate use of hPSC derivatives in research and clinical applications.
Cell surface proteins participate in inter- and intracellular communication, cellular structure, adhesion, and are a major gateway for sending and receiving exogenous signals. Therefore, the collection of proteins localized to the cell surface (i.e. surfaceome) is a rich source of accessible targets for developing new tools and strategies to address current challenges outlined above. Defining the proteins localized to the cell surface at a particular developmental stage or phenotype should contribute to a greater understanding of the cellular mechanisms that are involved in lineage and cell fate specification and could be exploited to drive differentiation to a selected endpoint. Furthermore, extracellular epitopes of cell surface proteins provide a means for accessible, non-genetic cell identification and tracking through the use of immunophenotyping. This will enable selection of live cells for clinical applications and also facilitate single cell level analysis of molecular events that regulate critical cell fate decisions during very early development. This type of immunophenotyping has enabled the systematic study of molecular events regulating hematopoietic stem cell lineage specification [3].
In this study, cell surface N-glycoproteins on hiPSC-derived specified hepatic endoderm cells were identified using Cell Surface Capture Technology (CSC-Technology) [4]. CSC-Technology is an antibody-independent strategy that uses affinity enrichment and mass spectrometry to identify cell surface N-glycoproteins while simultaneously determining N-glycosite occupancy and membrane topology. Extracellular oligosaccharides on live cells are biotinylated using membrane-impermeable reagents and biotinylated glycopeptides are captured with immobilized streptavidin. Peptide-N-glycosidase F cleaves the oligosaccharide from the peptide backbone, releasing the now formerly N-glycosylated peptide while simultaneously modifying the mass of the asparagine residue at the site of N-glycosylation (asparagine → aspartic acid). We have previously applied CSC-Technology to mouse (m) ESC, miPSC, mESC-derived cardiomyocytes, mouse C2C12 myoblasts, human fibroblasts, hiPSC, hESC, and primary human hepatocytes [5–9]. Others have applied the strategy to more than 40 human cell types [10–12] and many of these data are part of the Cell Surface Protein Atlas (CSPA [13]), a public resource containing 1492 experimentally determined human cell surface N-glycoproteins that has already proven valuable for identification of new cell type specific makers [5, 6]. Therefore, we expect these data will add to our knowledge of proteins important during endoderm differentiation and hepatic development and will be useful for comparative purposes whereby CSC-Technology data from a wide variety of cell types is used as a first step in identifying potentially cell type and lineage restricted markers [5, 6, 14].
Human iPSC (iPSK3; [15]) that were derived from foreskin fibroblasts were cultured and differentiated towards the hepatic lineage as described [16]. Cells on day 10 of differentiation, corresponding to specified hepatic endoderm, were used for proteomic analyses. Approximately 1×108 cells from independent differentiations (biological replicates; n=3) were taken through the CSC-Technology workflow as reported [4, 7, 8]. Experimental details are provided in the supplemental methods. To be included in the final dataset, proteins were required to be ranked as “high confidence” by at least one of the two search algorithms used and identified by at least one unique peptide sequence containing a deamidated asparagine within the sequence motif for N-glycosylation (NxS/T/C). Altogether, 395 cell surface N-glycoproteins, including 83 cluster of differentiation (CD) antigens, were identified on day 10 hiPSC-derived hepatic endoderm cells (Table S1–S4). Of these, 358 were identified by three or more MS/MS spectra and 37 were identified by two spectra. 286 N-glycoproteins were identified in at least two biological replicates and are thus classified as high confidence identifications while an additional 109 were identified in a single biological replicate and are considered as lower confidence and indicated as such in Tables S1, S2. The 395 N-glycoproteins were identified by 1319 peptides, of which 1273 (96%) contain a deamidated asparagine within the sequence motif NxS/T/C (i.e. formerly N-glycosylated peptide; Table S2). In this approach, the number of peptides identified per protein is dependent upon the number of occupied N-glycosylation sites, and 36 N-glycoproteins identified here contain a single predicted N-glycosylation site in the extracellular domain. Of these, proteins are ranked based on whether they were identified by two or more peptide spectrum matches (Table S1). Further detailed analyses are summarized in Figure 1.
Figure 1.
Detailed analyses of the N-glycopeptides and N-glycoproteins identified in hiPSC-derived hepatic endoderm cells. (A) The number of unique peptide sequences per protein, (B) number of N-glycosites identified per N-glycopeptide, (C) number of times each N-glycosite sequence motif was observed, (D) distribution of transmembrane, GPI-anchored, and secreted/extracellular matrix N-glycoproteins, and (E) distribution of the proteins reported vs. not reported in the bioinformatic surfaceome [17].
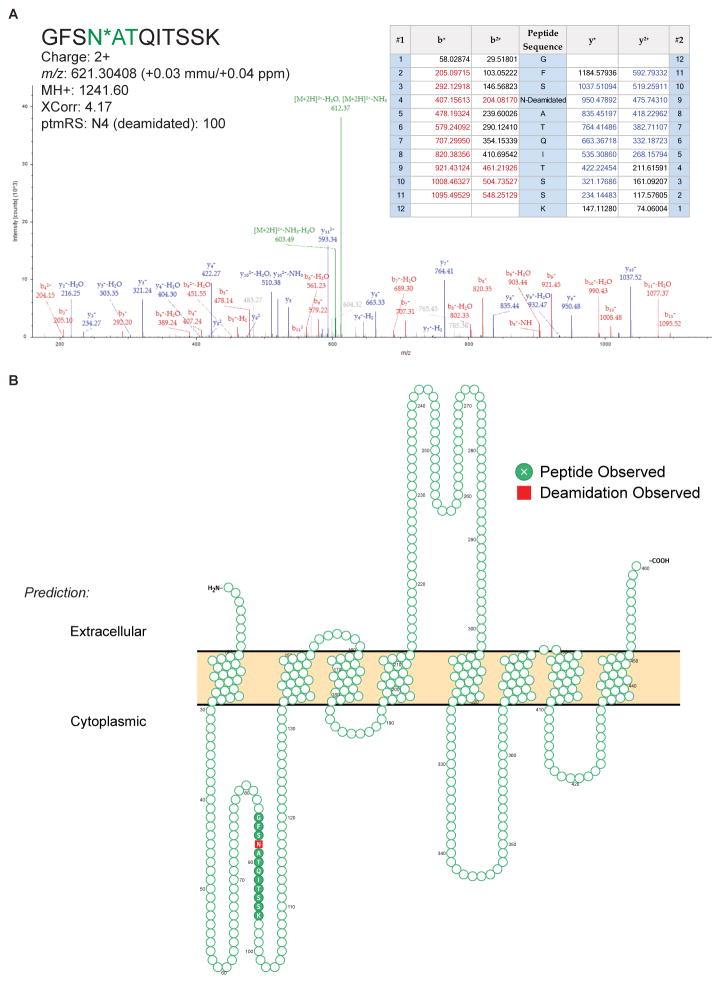
Benefits of CSC-Technology include its high specificity for surface-accessible proteins and its ability to directly verify extracellular domains by identifying sites of N-glycosylation, thereby avoiding reliance on database annotations and/or prediction algorithms to determine protein localization [4, 8]. While the presence of a signal peptide at the protein’s N-terminus can predict its likelihood to translocate to the plasma membrane, alternative pathways do not require a signal peptide and can be more difficult to predict. Overall, 107 N-glycoproteins in this data set do not contain a predicted signal peptide (Supplemental Table S1). Of these, 49 are predicted to be non-canonically translocated, leaving 58 not predicted to be translocated to the cell surface. In an alternative approach, a previous study by da Cunha et al. reported a predicted human cell surfaceome based on transmembrane domain analysis [17]. 95 (25%) of the N-glycoproteins identified here are not among the 3702 genes predicted in this bioinformatic surfaceome (Figure 1E). While intracellular glycoproteins may contaminate the CSC-Technology data if the integrity of the plasma membrane is compromised at the time of labeling, many of the N-glycoproteins identified here, but not otherwise predicted to be in the surfaceome by da Cunha et al., are known cell surface proteins (e.g. CD317, ENPP1, S1PR2). Finally, as CSC-Technology identifies N-glycopeptides from the extracellular domain, these data can be used to support transmembrane protein topology predictions, and in some cases, provide new evidence for proteins whose orientation is ambiguous or incorrectly predicted. Overall, CSC-Technology data are consistent with current UniProt annotations for 361 transmembrane proteins, clarify the orientation for five proteins whose annotations are ambiguous, and specifies extracellular orientation for SLC39A8, which is incorrectly annotated in UniProt (Table S1; Figure 2). It is noted that alternative prediction algorithms do correctly predict the orientation of this protein [18]. However, while advanced bioinformatic tools and gene ontology play critical roles in modern proteomic analyses, there remains value in applying technologies that can provide experimental evidence of a protein’s subcellular localization and transmembrane topology, as not all cell surface proteins fit into predictive models or are well characterized. Moreover, predictions cannot assert which proteins are present on a specific cell type at a specific time/condition, which is further confounded when proteins are sequestered within the cell and cycled to the cell surface in response to selected stimuli.
Figure 2.
(A) Annotated MS/MS spectrum of a formerly N-glycosylated peptide from SLC39A8. (B) Predicted membrane topology (UniProt) of SLC39A8 showing that the experimentally observed N-glycopeptide identified via CSC-Technology spans a predicted intracellular region. Image was generated using Protter [25].
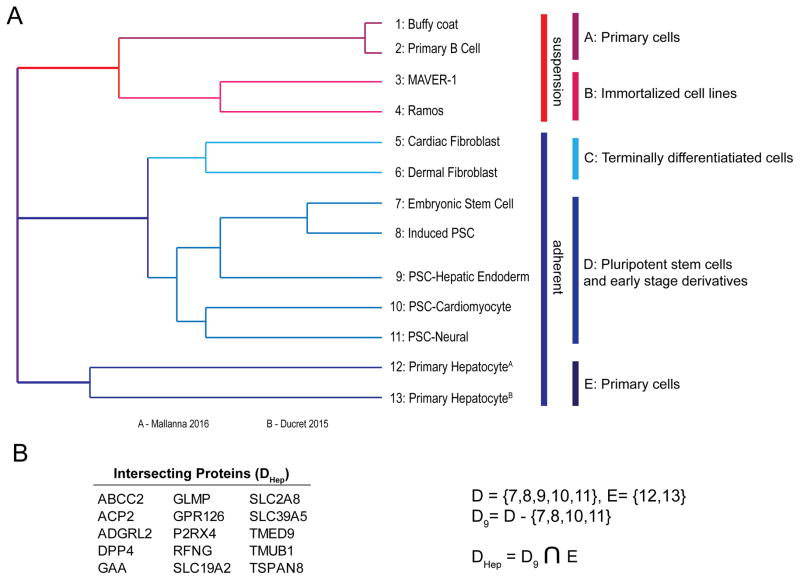
As the CSPA has already proven valuable for the identification of cell type and lineage-restricted proteins, these data were compared to 12 other human cell types in a proof-of-concept study to determine their utility for distinguishing cell types. Important caveats include that these data are qualitative, generated among different laboratories, the resulting datasets vary in size often in relation to the number of replicates and amount of starting material, and a failure to identify a protein in a particular dataset is not confirmation of its absence. Nevertheless, hierarchical clustering based on 1239 N-glycoproteins identified via CSC-Technology successfully clustered related cell types into distinct clusters, where terminally differentiated cell types are separate from pluripotent stem cells and their early derivatives. Moreover, of the 75 proteins that distinguish hPSC-derived hepatic endoderm from other related cell types in cluster D, 15 of these are also identified in primary human hepatocytes (cluster E; Figure 3). Among these, ABCC2, SLC2A8 (GLUT8), and TMUB1 have known roles in hepatocyte biology [19–21], which altogether lends support to the utility of this approach for filtering a large dataset down to a manageable set of candidates for subsequent functional studies.
Figure 3.
(A) Hierarchical clustering of 13 human cell types using 1239 cell surface N-glycoproteins identified via CSC-Technology. (B) The 15 N-glycoproteins that are shared between the hiPSC-hepatic endoderm and Cluster E, but are absent from the other cell types in Cluster D.
In summary, these data provide a snapshot view of the N-glycoprotein surfaceome on day 10 hepatic endoderm. As with any approach that provides an average view of a heterogeneous cell population, further efforts are required to determine the cell type specificity and distribution of these proteins among the cell population. If the goal is to develop cell surface protein markers or “barcodes” capable of identifying specific cell types, multiple approaches are possible. In an iterative strategy we recently described [5, 14], CSC-Technology and CSPA are used to discover and prioritize candidates. In the refinement phase, targeted quantitation by mass spectrometry can rapidly quantify tens to hundreds of targets among a broader range of cellular phenotypes or experimental conditions, including cell types that are in limited supply and therefore precluded from the discovery process. Subsequently, monoclonal antibodies are generated for the highest priority candidates and used for single cell level analyses to determine distribution of the protein(s) among the population. Additional rounds of analysis can be performed until a combination of markers with sufficient specificity and sensitivity is achieved. Although antibody-based screening approaches could be used in place of proteomics for the discovery phase, antibody arrays may only or preferentially target CD antigens. Of the 395 N-glycoproteins identified here, only 83 are human CD antigens. Consequently, future efforts to develop cell surface marker barcodes will benefit from expanding beyond CD molecule profiling. While CSC-Technology is valuable, current limitations include that detection by this approach may be affected by alterations in N-glycosylation status, quantitation is challenging, and the relatively large amount of starting material required may prohibit its application to rare or primary cell types. As approaches in sample handling and MS technology improve, we expect to apply the approach to smaller sample sizes in a quantitative manner (e.g. 10 million cells; Wollscheid, Gundry, personal communication). Finally, as this and other similar approaches [22–24] are applied to more cell types, the growing resource of experimentally identified cell surface proteins will continue to gain statistical power for identifying proteins that are lineage, cell type, or disease state specific.
Supplementary Material
Supplemental Table S1. Annotated list of 395 cell surface N-glycoproteins identified in hiPSC-derived hepatic endoderm. For each protein, information from UniProt (CD antigen, transmembrane orientation, number of possible N-glycosites, number of transmembrane domains, whether it is a predicted or known GPI-anchored protein or secreted/extracellular matrix protein), whether it was predicted in the Surfaceome by de Cunah, et al., PNAS 2009, whether it has predicted signal peptides based on Phobius or Signal P or is secreted based on Secretome P. Also listed are the number of other cell types within the human CSPA it was identified, and whether it was in our previously reported N-glycoprotein surfaceome studies of primary human hepatocytes or undifferentiated hPSC, and the number of biological replicates it was observed in the current study.
Supplemental Table S2. Protein-level MS data for each cell surface N-glycoprotein reported. Listed are the gene symbol, protein description, number of unique peptides, number of total peptides, number of peptide-spectrum-matches (PSM). For each protein, the confidence of its detection among each biological and technical replicate is indicated (based on MS search criteria in Proteome Discoverer 2.1).
Supplemental Table S3. Peptide-level data for each cell surface N-glycoprotein reported. For each protein entry (blue header = protein name followed by gene symbol), peptide-level information from ProteomeDiscoverer is reported, including peptide sequence, modifications, ptmRS site probabilities for post-translational modifidations, # PSMs, # missed cleavages, and scores from Sequest HT and MS Amanda search algorithms and corresponding Percolator q-values.
Supplemental Table S4. MS/MS spectrum level data for each peptide reported. For each protein entry (blue header = protein name foled by gene symbol), peptide-spectrum match (PSM)-level information from ProteomeDiscoverer is reported. For a single PSM, results for each search algorithm are listed on separate rows. It is noted that the “Ions Matched Column” is only populated for MSAmanda searches.
Acknowledgments
This research was supported by gifts from the Marcus Family, the Phoebe R. and John D. Lewis Foundation, the Sophia Wolf Quadracci Memorial Fund, the Advancing a Healthier Wisconsin Fund and by NIH grants DK102716, HG006398, and HD082570 to SAD; NIH grants HL094708, HL126785, and HL134010 and the Paul G. Allen Family Foundation (Grant Award 11715) to RLG. We thank Dr. Kate Noon, Michael Pereckas, and Xioagang Wu at the MCW Mass Spectrometry Facility for assistance with data collection.
Abbreviations
- CSC-Technology
Cell Surface Capture Technology
- hiPSC
Human induced pluripotent stem cells
- hPSC
Human pluripotent stem cells
- hESC
Human embryonic stem cells
- CSPA
Cell surface protein atlas
- CD
Cluster of differentiation
Footnotes
Conflict of Interest:
The authors have declared no conflict of interest.
References
- 1.Grskovic M, Javaherian A, Strulovici B, Daley GQ. Induced pluripotent stem cells--opportunities for disease modelling and drug discovery. Nature reviews Drug discovery. 2011;10:915–929. doi: 10.1038/nrd3577. [DOI] [PubMed] [Google Scholar]
- 2.Zeltner N, Studer L. Pluripotent stem cell-based disease modeling: current hurdles and future promise. Curr Opin Cell Biol. 2015;37:102–110. doi: 10.1016/j.ceb.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Shizuru JA, Negrin RS, Weissman IL. Hematopoietic stem and progenitor cells: clinical and preclinical regeneration of the hematolymphoid system. Annual review of medicine. 2005;56:509–538. doi: 10.1146/annurev.med.54.101601.152334. [DOI] [PubMed] [Google Scholar]
- 4.Wollscheid B, Bausch-Fluck D, Henderson C, O’Brien R, et al. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nature biotechnology. 2009;27:378–386. doi: 10.1038/nbt.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mallanna SK, Cayo MA, Twaroski K, Gundry RL, Duncan SA. Mapping the Cell-Surface N-Glycoproteome of Human Hepatocytes Reveals Markers for Selecting a Homogeneous Population of iPSC-Derived Hepatocytes. Stem cell reports. 2016;7:543–556. doi: 10.1016/j.stemcr.2016.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boheler KR, Bhattacharya S, Kropp EM, Chuppa S, et al. A human pluripotent stem cell surface N-glycoproteome resource reveals markers, extracellular epitopes, and drug targets. Stem cell reports. 2014;3:185–203. doi: 10.1016/j.stemcr.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gundry RL, Raginski K, Tarasova Y, Tchernyshyov I, et al. The mouse C2C12 myoblast cell surface N-linked glycoproteome: identification, glycosite occupancy, and membrane orientation. Molecular & cellular proteomics : MCP. 2009;8:2555–2569. doi: 10.1074/mcp.M900195-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gundry RL, Riordon DR, Tarasova Y, Chuppa S, et al. A cell surfaceome map for immunophenotyping and sorting pluripotent stem cells. Molecular & cellular proteomics : MCP. 2012;11:303–316. doi: 10.1074/mcp.M112.018135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kropp EM, Bhattacharya S, Waas M, Chuppa SL, et al. N-glycoprotein surfaceomes of four developmentally distinct mouse cell types. Proteomics Clinical applications. 2014;8:603–609. doi: 10.1002/prca.201400021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ducret A, Kux van Geijtenbeek S, Roder D, Simon S, et al. Identification of six cell surface proteins for specific liver targeting. Proteomics Clinical applications. 2015;9:651–661. doi: 10.1002/prca.201400194. [DOI] [PubMed] [Google Scholar]
- 11.DeVeale B, Bausch-Fluck D, Seaberg R, Runciman S, et al. Surfaceome profiling reveals regulators of neural stem cell function. Stem Cells. 2014;32:258–268. doi: 10.1002/stem.1550. [DOI] [PubMed] [Google Scholar]
- 12.Danzer C, Eckhardt K, Schmidt A, Fankhauser N, et al. Comprehensive description of the N-glycoproteome of mouse pancreatic beta-cells and human islets. J Proteome Res. 2012;11:1598–1608. doi: 10.1021/pr2007895. [DOI] [PubMed] [Google Scholar]
- 13.Bausch-Fluck D, Hofmann A, Bock T, Frei AP, et al. A mass spectrometric-derived cell surface protein atlas. PloS one. 2015;10:e0121314. doi: 10.1371/journal.pone.0121314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boheler KR, Gundry RL. Concise Review: Cell Surface N-Linked Glycoproteins as Potential Stem Cell Markers and Drug Targets. Stem Cells Transl Med. 2016 doi: 10.5966/sctm.2016-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Si-Tayeb K, Noto FK, Sepac A, Sedlic F, et al. Generation of human induced pluripotent stem cells by simple transient transfection of plasmid DNA encoding reprogramming factors. BMC Dev Biol. 2010;10:81. doi: 10.1186/1471-213X-10-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mallanna SK, Duncan SA. Differentiation of hepatocytes from pluripotent stem cells. Curr Protoc Stem Cell Biol. 2013;26(Unit 1G):4. doi: 10.1002/9780470151808.sc01g04s26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.da Cunha JP, Galante PA, de Souza JE, de Souza RF, et al. Bioinformatics construction of the human cell surfaceome. Proc Natl Acad Sci U S A. 2009;106:16752–16757. doi: 10.1073/pnas.0907939106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dobson L, Remenyi I, Tusnady GE. CCTOP: a Consensus Constrained TOPology prediction web server. Nucleic Acids Res. 2015;43:W408–412. doi: 10.1093/nar/gkv451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konig J, Nies AT, Cui Y, Leier I, Keppler D. Conjugate export pumps of the multidrug resistance protein (MRP) family: localization, substrate specificity, and MRP2-mediated drug resistance. Biochim Biophys Acta. 1999;1461:377–394. doi: 10.1016/s0005-2736(99)00169-8. [DOI] [PubMed] [Google Scholar]
- 20.Gorovits N, Cui L, Busik JV, Ranalletta M, et al. Regulation of hepatic GLUT8 expression in normal and diabetic models. Endocrinology. 2003;144:1703–1711. doi: 10.1210/en.2002-220968. [DOI] [PubMed] [Google Scholar]
- 21.Liu M, Liu H, Wang X, Chen P, Chen H. IL-6 induction of hepatocyte proliferation through the Tmub1-regulated gene pathway. Int J Mol Med. 2012;29:1106–1112. doi: 10.3892/ijmm.2012.939. [DOI] [PubMed] [Google Scholar]
- 22.Rugg-Gunn PJ, Cox BJ, Lanner F, Sharma P, et al. Cell-surface proteomics identifies lineage-specific markers of embryo-derived stem cells. Dev Cell. 2012;22:887–901. doi: 10.1016/j.devcel.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Hoof D, Dormeyer W, Braam SR, Passier R, et al. Identification of cell surface proteins for antibody-based selection of human embryonic stem cell-derived cardiomyocytes. J Proteome Res. 2010;9:1610–1618. doi: 10.1021/pr901138a. [DOI] [PubMed] [Google Scholar]
- 24.Parker BL, Palmisano G, Edwards AV, White MY, et al. Quantitative N-linked glycoproteomics of myocardial ischemia and reperfusion injury reveals early remodeling in the extracellular environment. Molecular & cellular proteomics : MCP. 2011;10:M110 006833. doi: 10.1074/mcp.M110.006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Omasits U, Ahrens CH, Muller S, Wollscheid B. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2013 doi: 10.1093/bioinformatics/btt607. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table S1. Annotated list of 395 cell surface N-glycoproteins identified in hiPSC-derived hepatic endoderm. For each protein, information from UniProt (CD antigen, transmembrane orientation, number of possible N-glycosites, number of transmembrane domains, whether it is a predicted or known GPI-anchored protein or secreted/extracellular matrix protein), whether it was predicted in the Surfaceome by de Cunah, et al., PNAS 2009, whether it has predicted signal peptides based on Phobius or Signal P or is secreted based on Secretome P. Also listed are the number of other cell types within the human CSPA it was identified, and whether it was in our previously reported N-glycoprotein surfaceome studies of primary human hepatocytes or undifferentiated hPSC, and the number of biological replicates it was observed in the current study.
Supplemental Table S2. Protein-level MS data for each cell surface N-glycoprotein reported. Listed are the gene symbol, protein description, number of unique peptides, number of total peptides, number of peptide-spectrum-matches (PSM). For each protein, the confidence of its detection among each biological and technical replicate is indicated (based on MS search criteria in Proteome Discoverer 2.1).
Supplemental Table S3. Peptide-level data for each cell surface N-glycoprotein reported. For each protein entry (blue header = protein name followed by gene symbol), peptide-level information from ProteomeDiscoverer is reported, including peptide sequence, modifications, ptmRS site probabilities for post-translational modifidations, # PSMs, # missed cleavages, and scores from Sequest HT and MS Amanda search algorithms and corresponding Percolator q-values.
Supplemental Table S4. MS/MS spectrum level data for each peptide reported. For each protein entry (blue header = protein name foled by gene symbol), peptide-spectrum match (PSM)-level information from ProteomeDiscoverer is reported. For a single PSM, results for each search algorithm are listed on separate rows. It is noted that the “Ions Matched Column” is only populated for MSAmanda searches.