Abstract
Since most analysis software for genome-wide association studies (GWAS) currently exploit only unrelated individuals, there is a need for efficient applications that can handle general pedigree data or mixtures of both population and pedigree data. Even data sets thought to consist of only unrelated individuals may include cryptic relationships that can lead to false positives if not discovered and controlled for. In addition, family designs possess compelling advantages. They are better equipped to detect rare variants, control for population stratification, and facilitate the study of parent-of-origin effects. Pedigrees selected for extreme trait values often segregate a single gene with strong effect. Finally, many pedigrees are available as an important legacy from the era of linkage analysis. Unfortunately, pedigree likelihoods are notoriously hard to compute. In this paper we re-examine the computational bottlenecks and implement ultra-fast pedigree-based GWAS analysis. Kinship coefficients can either be based on explicitly provided pedigrees or automatically estimated from dense markers. Our strategy (a) works for random sample data, pedigree data, or a mix of both; (b) entails no loss of power; (c) allows for any number of covariate adjustments, including correction for population stratification; (d) allows for testing SNPs under additive, dominant, and recessive models; and (e) accommodates both univariate and multivariate quantitative traits. On a typical personal computer (6 CPU cores at 2.67 GHz), analyzing a univariate HDL (high-density lipoprotein) trait from the San Antonio Family Heart Study (935,392 SNPs on 1388 individuals in 124 pedigrees) takes less than 2 minutes and 1.5 GB of memory. Complete multivariate QTL analysis of the three time-points of the longitudinal HDL multivariate trait takes less than 5 minutes and 1.5 GB of memory. The algorithm is implemented as the Ped-GWAS Analysis (Option 29) in the Mendel statistical genetics package, which is freely available for Macintosh, Linux, and Windows platforms from http://genetics.ucla.edu/software/mendel.
Keywords: genome-wide association study, pedigree, kinship, score test, fixed-effects models, multivariate traits
1 Introduction
Genome-wide association studies (GWAS) are now at a crossroads. After the discovery of thousands of genes influencing hundreds of common traits [Hindorff et al. 2009], much of the low-hanging fruit has been plucked [Ku et al. 2010, Visscher et al. 2012]. Because of the enormous sample sizes of current studies, new trait genes are still being uncovered. Unfortunately, most entail small effects. Is it possible that inheritance is predominantly polygenic, and a law of diminishing returns has set in? The push to exploit rare variants is one response to this dilemma. The previous generation of geneticists relied on linkage to map rare variants. Linkage mapping fell from grace because of its poor resolution. Reducing a genome search to a one or two megabase region leaves too large an expanse of DNA to sift. The real gold of linkage mapping may well be its legacy pedigrees [Ott et al. 2011]. Pedigree data is particularly attractive in association studies because it permits control of population substructure and study of parent-of-origin effects. Related affecteds are also more likely to share the same disease predisposing gene than unrelated affecteds. Even in population-based association studies, taking into account estimated identity-by-descent (IBD) information is apt to reduce false positives and increases power. The recent availability of dense marker data from genotyping chips enables quick and accurate estimation of global and even local IBD [Day-Williams et al. 2011].
Geneticists turned to random sample and case-control data because of the relative ease of collecting population data and the computational challenges posed by pedigrees. The tide of computational complexity is now beginning to turn. To handle pedigree data in association testing, statistical geneticists have proposed semiparametric methods such as the generalized linear mixed model (GLMM) [Amin et al. 2007, Aulchenko et al. 2007] and generalized estimating equations (GEE) [Chen and Yang 2010, Chen et al. 2011]. Although such methods work for both quantitative and binary traits, they are compromised by current restrictions that reduce power. The GEE approach requires input of a working correlation structure for each pedigree. The kinship coefficient matrix is a natural candidate. However, current implementations require the same working correlation matrix across all clusters, which implicitly requires all pedigrees to have the same structure [Chen et al. 2011]. This is a dubious and restrictive assumption. In the limited context of case-control studies, recent methods such as MQLS [Thornton and McPeek 2007], ROAD-TRIPS [Thornton and McPeek 2010], and FPCA [Zhu and Xiong 2012] correct for pedigree and ethnically induced correlations by exploiting dense marker data. Other authors attack the same issues more broadly from the GLMM perspective [Kang et al. 2010, Zhang et al. 2010, Lippert et al. 2011]. Korte et al. [2012] generalizes GLMM to multivariate traits. Models based on the transmission-disequilibrium test (TDT) [Spielman and Ewens 1998] and its generalization, the family-based association test (FBAT) [Laird et al. 2000, Lange and Laird 2002, Van Steen and Lange 2005, Won et al. 2009a; b], are promising but ignore covariates and polygenic background. See Van Steen [2011] for a recent overview of FBAT methods for GWAS. We treat all of these extensions in a unified framework consistent with exceptionally fast computing.
The present paper re-examines the computational bottlenecks encountered in association mapping with pedigree data. It turns out that the previous objections to pedigree GWAS can be overcome. Kinship coefficients can be based on explicitly provided pedigree structure or estimated from dense markers when genealogies are missing or dubious. Frequentist hypothesis testing usually operates by comparing maximum likelihoods under the null and alternative hypotheses. Maximization of the alternative likelihood must be conducted for each and every marker. Score tests constitute a more efficient strategy than likelihood ratio tests. This is the point of departure taken by Chen and Abecasis [2007], but they use approximations that we avoid. The glogs program [Stanhope and Abney 2012] makes similar approximations in the case-control setting. Here we consider arbitrary pedigrees and multivariate quantitative traits. Score tests require no additional iteration under the alternative model. All that is needed is evaluation of a quadratic form combining the score vector and the expected information matrix at the maximum likelihood estimates under the null model. Although it takes work to assemble these quantities, a careful analysis of the algorithm shows that fast testing is perfectly feasible.
In our implementation of score testing, the few SNPs with the most significant score-test p-values are automatically re-analyzed by the slightly more powerful, but much slower, likelihood ratio test (LRT). Our fixed effects (mean component) model assumes Gaussian variation of the trait; the two alleles of a SNP shift trait means. There is no confounding of association and linkage. This framework carries with it several advantages. First, it applies to random sample data, pedigree data, or a mix of both. Second, it enables covariate adjustment, including correction for population stratification. Third, it accommodates additive, dominant, and recessive SNP models. Fourth, it also accommodates both univariate and multivariate traits. And fifth, as just mentioned, it fosters both likelihood ratio tests and score tests. The mean component model is now implemented in our software package Mendel for easy use by the genetics community. In addition, Mendel provides a complete suite of tools for pedigree analysis, including GWAS data preparation and manipulation, pedigree genotype simulation (gene dropping), trait simulation, genotype imputation, local and global kinship coefficient estimation, and pedigree-based GWAS (ped-GWAS) [Lange et al. 2005; 2013].
The competing software packages EMMAX [Kang et al. 2008], MMM [Pirinen et al. 2013], FaST-LMM [Lippert et al. 2011, Listgarten et al. 2012], GEMMA [Zhou and Stephens 2012; 2014], and GWAF [Chen and Yang 2010] already implement variance component models for quantitative trait locus (QTL) analysis. Exhaustive comparison of Mendel to each of these programs is beyond the scope of the current paper. We limit our comparisons of Mendel to the state-of-art packages FaST-LMM and GEMMA, arguably the fastest and most sophisticated of the competition. Table 1 summarizes some of the qualitative features of these packages. Our numerical examples also demonstrate an order of magnitude advantage in speed of Mendel over FaST-LMM, GEMMA, and GWAF. This advantage stems from our careful formulation of the score test and our exploitation of the multicore processors resident in almost all personal computers and computational clusters.
Table 1.
Comparison of features in Mendel, FaST-LMM, and GEMMA for GWAS of QTLs.
| Mendel | FaST-LMM | GEMMA | |
|---|---|---|---|
| Multi-threaded operation | Yes | Yes | No |
| Can estimate kinships via SNPs | Yes | Yes | Yes |
| Imports & exports kinship estimates | Yes | Yes | Yes |
| Allows retained co-variates | Yes | Yes | Yes |
| Allows linear constraints on co-variates | Yes | No | No |
| Can use either LRT or score test | Yes | No | Yes* |
| Allows multivariate analysis | Yes | No | Yes |
| Can perform multiple univariate analyses | Yes | No | No |
| Allows > 2 variance components | Yes | No | No |
| Analyzes X-linked loci | Yes | No | No |
| Automatic SNP filtering on MAF | Yes | No | Yes |
| Allows non-additive SNP models | Yes | No | No |
| Detects outlier pedigrees | Yes | No | No |
| Detects outlier individuals | Yes | No | No |
| Can simulate genotype/phenotype data | Yes | No | No |
| Reads in fractional genotype values | No | Yes | Yes |
GEMMA can use the likelihood ratio, score, or Wald test.
2 Methods
2.1 QTL Association Mapping with Pedigrees
QTL association mapping typically invokes the multivariate Gaussian distribution to model the trait values y = (yi) over a pedigree. The observed trait value yi of person i can be either univariate or multivariate. For simplicity we first assume yi is univariate and later indicate the necessary changes for multivariate yi. The standard model [Lange 2002] collects the corresponding trait means into a vector ν and the corresponding covariances into a matrix Ω and represents the loglikelihood of a pedigree as
| (1) |
where det denotes the determinant function and the covariance matrix is typically parametrized as
| (2) |
Here the variance component Φ is the global kinship coefficient matrix capturing additive polygenic effects, and Δ7 is a condensed identity coefficient matrix capturing dominance genetic effects. When pedigree structure is explicitly given, these genetic identity coefficients are easily calculated [Lange 2002]. With unknown or dubious genealogies, the global kinship coefficient can be accurately estimated from dense markers [Day-Williams et al. 2011]. The household effect matrix H has entries hij = 1 if individuals i and j belong to the same household and 0 otherwise. Individual environmental contributions and trait measurement errors are incorporated via the identity matrix I. Mendel‘s implementation of this model can include both the two standard variance classes, additive and environmental, as well as the two extra variances classes, dominance and household. Inclusion of additional variance classes has no significant effect on Mendel‘s speed of computation.
In general, a mixed model for QTL association mapping captures polygenic and other random effects through Ω and captures QTL fixed effects through ν. Let β denote the full vector of regression coefficients parameterizing ν. In a linear model one postulates that ν = Aβ for some predictor matrix A incorporating relevant covariates such as age, gender, and diet. In testing association against a given SNP, A is augmented by an extra column whose entries encode genotypes according to one of the models (additive, dominant, and recessive) shown in see Table 2. To accommodate imprecise imputation in an additive model, these encodings can be made fractional. The corresponding component of β, βSNP, is the SNP effect size. In likelihood ratio association testing one contrasts the null hypothesis βSNP = 0 with the alternative hypothesis βSNP ≠ 0. In testing a univariate trait, the likelihood ratio statistic asymptotically follows a distribution. In testing a multivariate trait with T > 1 components, each row of A must be replicated T times. The likelihood ratio statistic then asymptotically follows a distribution. To implement likelihood ratio testing, iterative maximum likelihood estimation must be undertaken for each and every SNP under the alternative hypothesis. This unfortunate requirement is the major stumbling block retarding pedigree analysis.
Table 2.
Genotype encodings for the major gene models.
| Genotype | Additive | Dominant | Recessive |
|---|---|---|---|
| 1/1 | –1 | –1 | –1 |
| 1/2 | 0 | –1 | +1 |
| 2/2 | +1 | +1 | +1 |
The additive model is the default choice. In the genotype column, “1” and “2” represent the first and second alleles for each SNP. An effect size estimate reflects the change in trait values due to each positive unit change in the encodings. For example, the default additive model estimates the mean trait difference in moving from a 1/2 genotype to a 2/2 genotype.
Score tests serve as convenient substitutes for likelihood ratio tests. The current paper describes how to implement ultra-fast score tests for screening SNPs. Only SNPs with the most significant score test p-values are further subjected to the more accurate likelihood ratio test. An advantage of the likelihood ratio method is that it estimates effect sizes. In contrast, the score test only requires parameter estimates under the null hypothesis and involves no iteration beyond fitting the null model. The score vector is the gradient ∇L(θ) of the loglikelihood L(θ), where the full parameter vector θ includes variance components such as the additive genetic variance in addition to the regression coefficient vector β. The transpose dL(θ) of the score is a row vector called the first differential of L(θ). The expected information J(θ) is the covariance matrix of the score vector. It is well known that the expected value of the observed information matrix (negative second differential) −d2L(θ) coincides with J(θ) [Rao 2009]. The score statistic
is evaluated at the maximum likelihood estimates under the null hypothesis with the parameter βSNP of the alternative hypothesis set to 0.
2.2 Fast Score Test for Individual SNPs
Under the multivariate model, the expected information matrix J(θ) for a single pedigree can be written in the block diagonal form
| (3) |
where σ denotes the vector of variance parameters [Lange 2002]. For independent pedigrees, the log-likelihoods (1) and corresponding score vectors and expected information matrices add. Hence, the block diagonal form of J(θ) is preserved. Because the inverse of a block diagonal matrix is block diagonal, the score statistic splits into a piece contributed by the variance components plus a piece contributed by the mean components. The maximum likelihood estimate θ̂ = (β̂,σ̂) under the null model is a stationary point of the loglikelihood. Thus, the variance components segment ∇σL(θ̂) of the score vector vanishes. We therefore focus on the mean components segment of the score vector.
If the pedigrees are labeled 1,…, n, then the pertinent quantities for implementing the score test are
where ri = yi − Aiβ̂ is the residual for pedigree i and the covariance matrix Ωi for pedigree i is determined by equation (2). See Chapter 8 of Lange [2002] for a detailed derivation of the score and expected information. Since the score statistic is calculated from estimated parameters under the null model, residuals do not change when we expand the null model to the alternative model keeping βSNP = 0. Calculation of the maximum likelihood estimate θ̂ under the null is accomplished by a quasi-Newton algorithm whose initial step reduces to Fisher scoring [Lange et al. 1976, Lange 2002].
For pedigree i under the alternative hypothesis, the design matrix Ai can be written as (ai, Ni), where Ni is the design matrix under the null hypothesis and ai conveys the genotypes at the current SNP. In testing a univariate trait, the entries of ai are taken from Table 2. If allele counts are imputed under the additive model, then the entries of ai may be fractional numbers drawn from the interval [−1,1]. In testing a multivariate trait with T > 1 components, each row of Ai = (ai, Ni) must be replicated T times. The only exceptions to this rule occur for people missing some but not all component traits; otherwise, the covariance matrix Ωi for pedigree i decomposes into a sum of Kronecker products [Lange 2002]. Regardless of whether the trait is univariate or multivariate, one must compute the quantities
At the maximum likelihood estimates under the null model, the partial score vector vanishes. Hence, the score statistic for testing a SNP can be expressed as
where
In the score statistic S, the covariance matrices and residual vectors ri are evaluated at the maximum likelihood estimates under the null model. Large sample theory says that S asymptotically follows a distribution.
These formulas suggest that we precompute and store the quantities for each pedigree i and the overall sum at the maximum likelihood estimates under the null hypothesis. From these parts, the basic elements of the score statistic can be quickly assembled. The most onerous quantity that must be computed on the fly as each new SNP is encountered is . If there are pi people in pedigree i, then computation of the quadratic form requires arithmetic operations. This looks worse than it is in practice since the entries of ai are integers (−1, 0, and 1) in the absence of fractional imputation. This simplification allows one to avoid a fair amount of arithmetic. Assembling the remaining parts of the score statistic requires O(pi) arithmetic operations.
Individuals missing univariate trait values are omitted from analysis. Individuals missing some but not all components of a multivariate trait are retained in analysis. The proper adjustments for missing data are made automatically in the score statistic because sections of Gaussian random vectors are Gaussian.
SNPs with minor allele counts below a user-designated threshold are also omitted from analysis. Note that if the minor allele count across a study is 0, then the given SNP is mono-allelic and worthless in association testing. Mendel‘s default threshold of 3 is motivated by the rule of thumb in contingency table testing that all cells have an expected count of at least 3. For a multivariate trait, a SNP may fall below the threshold for some component traits but not for others. This situation can occur when each trait displays a different pattern of missing data across individuals. Mendel retains such anomalous SNPs only for those component traits with a sufficient number of minor alleles. Again, proper adjustments are made automatically within the score test statistic to account for partial data.
Mendel‘s analysis yields a score test p-value for each SNP. For the user-designated most significant SNPs, Mendel‘s subsequent likelihood ratio test outputs an estimated SNP effect size, a standard error of that estimate, and the fraction of the total variance explained by that SNP. For a multivariate trait, Mendel outputs a SNP effect size and associated standard error for each component trait. In the initial analysis under the null model with no SNPs, Mendel provides estimates with standard errors of all mean and variance components included in the model. Finally, an estimate of heritability with standard error is also provided.
The extension of the score test to the multivariate t-distribution is straightforward [Lange et al. 1989]. Suppose η equals the degrees of freedom of the t-distribution and mi equals the number of observed person-trait combinations for pedigree i. The sections of the score and expected information pertinent to the mean components for the pedigree reduce to
where ri is the residual and is the associated Mahalanobis distance. A sensible choice for η is its estimate under the null model.
2.3 Kinship Estimation From SNPs
Mendel can either calculate the global kinship coefficient matrix Φ from the provided pedigree structures or estimate it from dense genotypes. In global kinship estimation Mendel‘s default uses an evenly spaced 20% of the available SNPs, and only compares pairs of individuals within defined pedigrees. Hence, Φ is block diagonal. Users can trivially elect to exploit a larger fraction of the available SNPs or estimate kinship for all pairs of individuals. Given S selected SNPs, Mendel estimates the global kinship coefficient of individuals i and j based on either the genetic relation matrix (GRM) method
or the method of moments (MoM) [Day-Williams et al. 2011, Lange et al. 2014]
where pk is the minor allele frequency at SNP k, xik is the number of minor alleles in i’s genotype at SNP k, and
is the observed fraction of alleles identical by state (IBS) between i and j. The GRM method is Mendel‘s default. In general, one can think of the GRM method centering and scaling each genotype, while the MoM method uses the raw genotypes and then centers and scales the final result.
2.4 Other Utilities for Handling Pedigree Data
To encourage thorough testing of new statistical methods, such as the current Ped-GWAS score test, we have implemented both genotype and trait simulation in our genetic analysis program Mendel [Lange et al. 2013]. Mendel does genotype simulation (gene dropping) subject to prescribed allele frequencies, a given genetic map, and Hardy-Weinberg and linkage equilibrium. If one fixes founder haplotypes and simulates conditional on these, then the unrealistic assumption of linkage equilibrium can be relaxed. Missing data patterns are respected or imposed by the user. It is also possible to set the rate for randomly deleting data and to simulate genotypes for people of mixed ethnicity by defining different ancestral populations, each with its own allele frequencies. If this feature is invoked, then each pedigree founder should be assigned to a population.
Trait simulation can be layered on top of genotype simulation. Mendel simulates either univariate traits determined by generalized linear models or multivariate Gaussian traits determined by variance component models. The biggest limitations are the restriction to a single major locus and the generalized linear model assumption that trait correlations are driven solely by this locus. Variance component models enable inclusion of environmental effects and more complicated correlations among relatives. In the variance component setting, univariate as well as multivariate Gaussian traits can be simulated. Most variance component models are built on Gaussian distributions, but Mendel allows one to replace these by multivariate t-distributions. Thus, users can investigate robust statistics less prone to distortion by outliers. More theoretical and implementation details appear in the Mendel documentation [Lange et al. 2013].
3 Results
3.1 Simulated Data Examples
We performed a variety of simulations to evaluate the score test’s computational efficiency, type I error, power, and treatment of multivariate traits. Run times in this section were recorded on a standard laptop computer with a 2.6 GHz Intel i7 CPU.
SNP Data Preparation
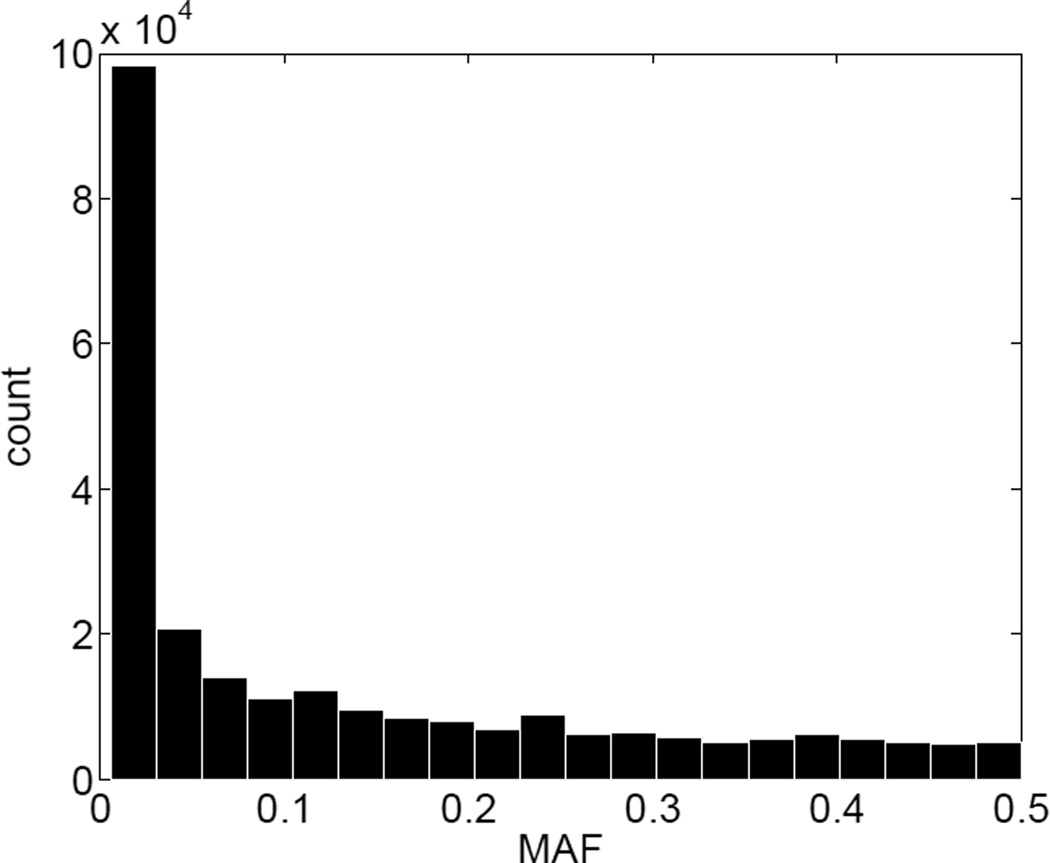
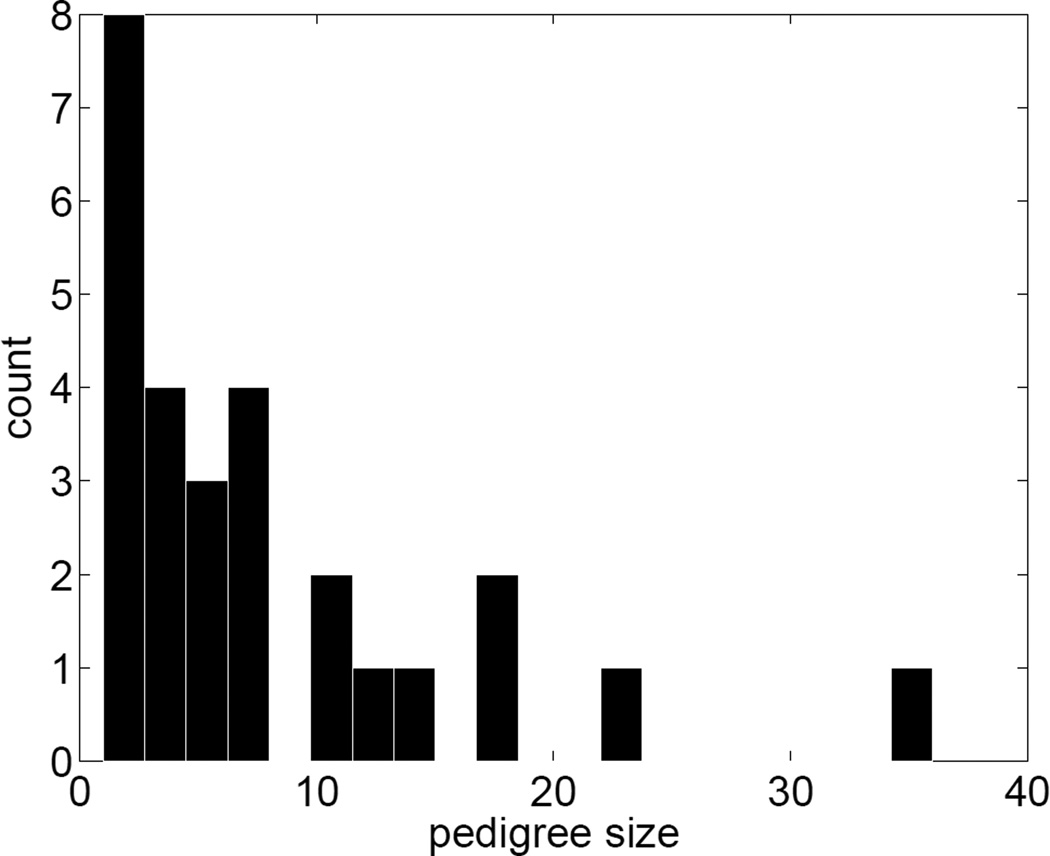
To simulate data with realistic linkage disequilibrium (LD) structure, we took advantage of phased sequence data from chromosome 19 on 85 individuals of northern and western European ancestry (originally from the CEPH sample) made publicly available in the 1000 Genomes Project [The 1000 Genomes Project Consortium 2010]. After we used the VCFtools software [Danecek et al. 2011] to remove markers that were mono-allelic in this set of individuals, 253,141 SNPs remained. Figure 1 displays the histogram of the minor allele frequencies (MAF) in these individuals. Almost half of the SNPs have MAFs below 5%. The haplotype pairs attributed to the 85 CEPH members were reassigned to the 85 founders of 27 pedigree structures selected from the Framingham Heart Study (FHS). The selected Framingham pedigrees were chosen to reflect the kind of pedigrees commonly collected in family-based genetic studies. The 27 pedigrees encompass 212 people, range in size from 1 to 36 people and from 1 to 5 generations, and contain sibships of 1 to 5 children. Figure 2 shows the histogram of the pedigree sizes. The genotypes of non-founders were simulated conditional on the haplotypes imposed on the founders and recorded as unordered for subsequent analysis purposes.
Figure 1.
Histogram of minor allele frequencies (MAF) of 253,141 SNPs on chromosome 19 in 85 individuals.
Figure 2.
Histogram of pedigree sizes.
Univariate Trait QTL Mapping
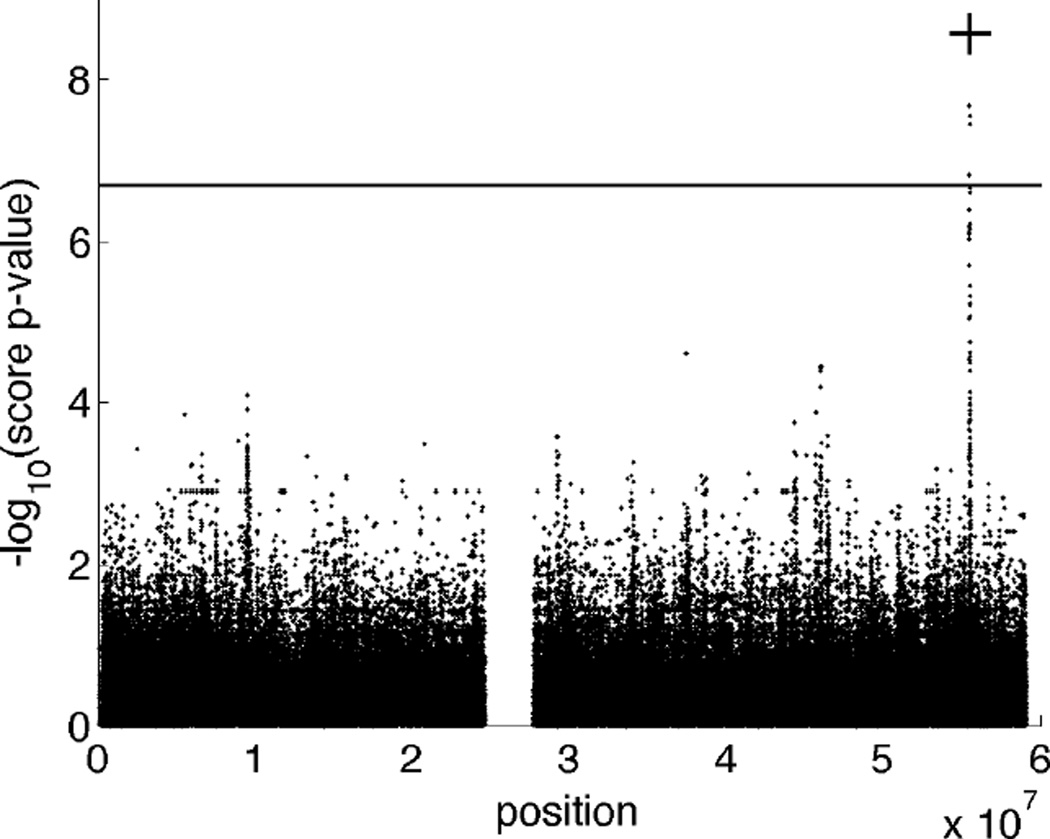
We simulated a univariate quantitative trait with a major locus at SNP rs10412915 (MAF = 0.259; position 55,494,740 on chromosome 19) using the trait simulation option of Mendel. The mean effects included the intercept µ = 40, the regression coefficients βsnp = 2 and βsex = 6, and the variance components . (See equation (2) and the subsequent description of the model for the definition of these parameters.) Power under other effect sizes is explored in a later experiment. Figure 3 displays a Manhattan plot of the p-values generated by the score tests. The signal emanating from the major locus is clearly discernible and is the only significant finding. Mendel took about 6.5 seconds for initialization, which includes reading the data, checking for gross errors, performing standard quality control (QC) procedures such as filtering of SNPs and individuals with low genotyping rates, and computing summary statistics. Using all 27 pedigrees, Mendel then required 5.9 seconds to compute the score test p-values at all 253,141 SNPs. Total run time was less than 13 seconds.
Figure 3.
Manhattan plot of the score test p-values for 253,141 SNPs on chromosome 19. Trait values were simulated based on a major locus at SNP rs10412915 (position 55,494,740) in the NLRP2 gene. The −log10(score p-value) at this SNP is marked with a plus sign. The horizontal line represents the significance threshold for this data set. See the text for the detailed simulation model.
Score test vs LRT
Mendel allows users to specify how many of the most significant score-test SNPs are reanalyzed using a likelihood ratio test (LRT). In the current example we told Mendel to calculate the LRTs on the 50 most significant SNPs flagged by the score test. It took Mendel an additional second to perform these LRTs. This translates into a total run time for data input, QC, and analysis of less than 14 seconds. When we told Mendel to perform LRTs on all SNPs, it took 53 minutes and 37 seconds. The almost 500-fold speedup of the score test over the LRT demonstrates the dramatic gains in computational efficiency possible. In large-scale sequencing studies, we expect an order of magnitude increase in both study individuals and typed SNPs. In later sections we discuss more fully efficiency and power for various models and data sets.
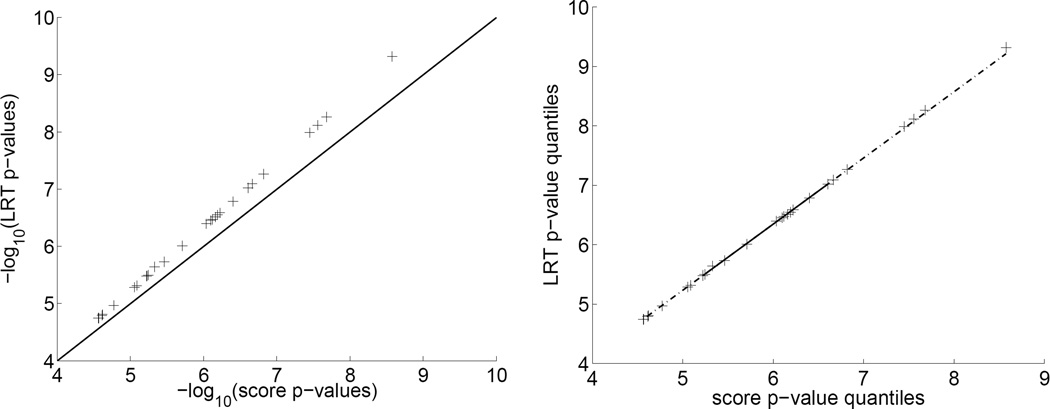
To alleviate concerns about the loss of power in substituting the score test for the LRT, we plot in Figure 4 the top 50 score test and LRT p-values. The two top-50 SNP sets coincide. The scatter plot (left panel) shows extremely high correlation (r = 0.9999). That all points lie above the 45-degree line indicates that the LRT has uniformly more power (smaller p-values) than the score test. The ranking of SNPs is of interest in many pilot studies. The Q-Q plot (right panel) shows that these two tests produce virtually identical rankings. Kendall’s τ correlation is 0.9983, and Spearman’s correlation is 0.9998.
Figure 4.
Comparisons of the score and LRT p-values. Left: A scatter plot of the top 50 score and LRT p-values demonstrates extremely high correlation (r = 0.9999) between the two sets of p-values and a uniformly higher power for the LRT. Right: A Q-Q plot of the top 50 score and LRT p-values shows that the two tests produce virtually identical rankings. The simulation model is the same as in Figure 3.
Discarding Versus Estimating Pedigree Information
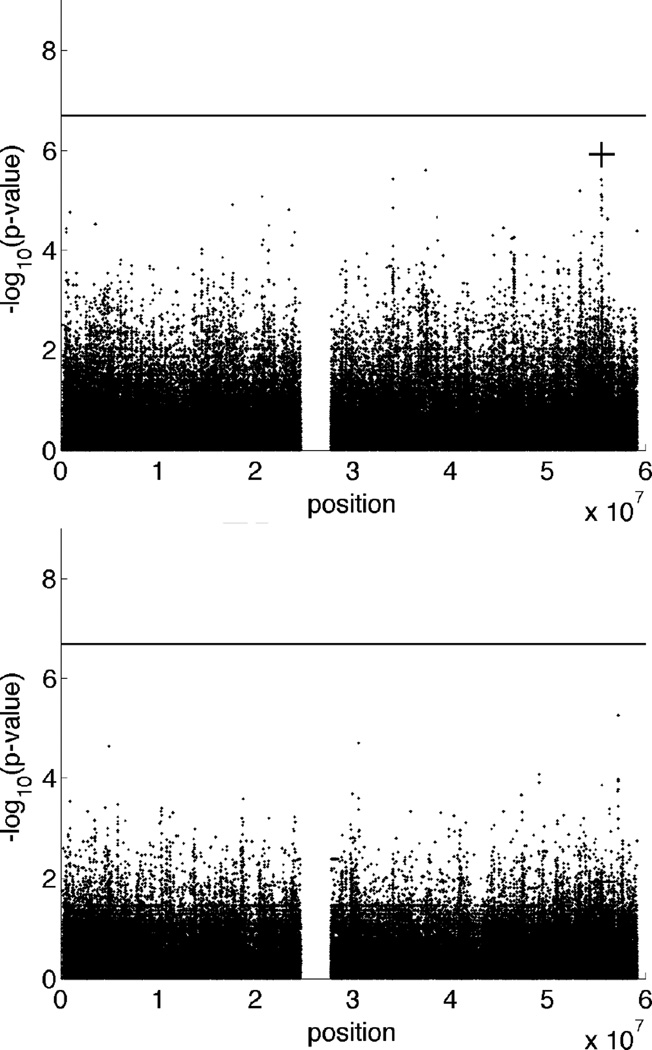
We performed two experiments to evaluate the impact of discarding pedigree information in association testing. In the first, we treated all 212 individuals as unrelated and tested all SNPs by linear regression with sex as a covariate. This is the same mean effects model employed in the previous example. It took Mendel about 6.5 seconds for initialization and 5.3 seconds for analysis. In the second experiment, we discarded the non-founders and carried out the same association testing on just the 85 founders. This took Mendel 4.3 seconds for initialization and 4.5 seconds for analysis. The top two panels of Figure 5 display the Manhattan plots of the two experiments discarding pedigree information. As expected, particularly for the second experiment, ignoring pedigree structure leads to a significant loss of power. Inspection of Figure 5 shows that no SNPs pass the significance threshold in the altered data sets.
Figure 5.
GWAS results suffer when pedigree structure is ignored. Upper: Manhattan plot of GWAS that treats all 212 individuals as unrelated. Lower: Manhattan plot of GWAS that includes only the 85 founders. Both show a loss of power due to discarding pedigree information. A plus sign marks the −log10(score p-value) at the SNP used to simulate the trait. The horizontal line represents the significance threshold for this data set. The simulation model is the same as in Figure 3.
Fortunately, when genealogies are missing or dubious, the method of Day-Williams et al [Day-Williams et al. 2011] implemented in Mendel allows fast and accurate estimation of global kinship coefficients from dense markers. It took Mendel18.8 seconds to estimate the global kinship coefficients from the 253,141 SNPs. The third panel in Figure 5 shows the Manhattan plot of the pedigree GWAS based on the estimated kinship coefficients. There is little difference from the results using exact pedigree structures.
Multivariate Trait QTL Mapping
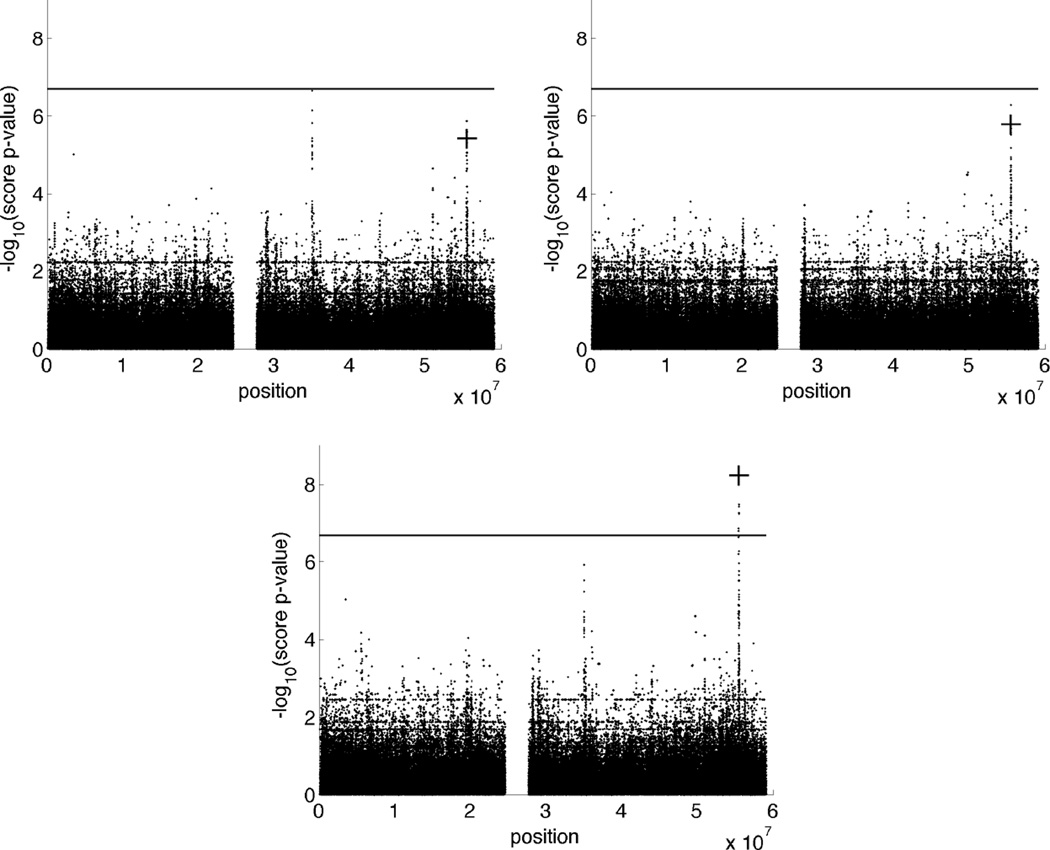
To assess the ability of our ped-GWAS method to detect a pleiotropic effect at the selected major locus rs10412915, we simulated two correlated quantitative traits on the previously constructed pedigrees. Trait 1 has mean effects µ1 = 40, βsex, 1 = 6, βsnp, 1 = 1.5 and variance components . Trait 2 has mean effects µ2 = 20, βsex,2 = 4, βsnp,2 = 1.5 and variance components . The additive and environmental covariances between the two traits are . Compared to our earlier univariate trait simulation, SNP effects are reduced for each trait while variance components are held fixed. Figure 6 displays Manhattan plots for testing trait 1 alone, trait 2 alone, and both traits 1 and 2 together. When both traits are tested simultaneously, it takes Mendel about 6.9 seconds for initialization and 9.9 seconds for analysis. Despite the reduction in SNP effect sizes, testing both traits simultaneously boosts power significantly. The benefits diminish when the traits are more highly correlated, for example by taking . For the sake of brevity, these further results are not graphed.
Figure 6.
Bivariate QTL mapping. Upper left: Manhattan plot for testing trait 1. Upper right: Manhattan plot for testing trait 2. Lower: Manhattan plot for testing traits 1 and 2 together. Bivariate QTL mapping demonstrates better power than testing each univariate trait separately. The −log10(score p-value) at the major locus rs10412915 (position 55,494,740) is marked with a plus sign. The horizontal line represents the significance threshold for this data set. See the text for the simulation model.
Comparison to current methods
In this section we compare the score test to the competing generalized estimating equation (GEE) and variance component model (linear mixed model, LMM) approaches implemented in the R package GWAF [Chen and Yang 2010]. Our comparison criteria include computational efficiency, memory usage, type I error, and power. Table 3 shows run times for testing the first 100, 1000, 10,000, and 100,000 SNPs on chromosome 19. Simulation parameters coincide with those used in Figure 3. Mendel-LRT lists runs in which the 50 most significant SNPs were further subjected to an LRT. The table lists the total wall clock times for the initialization and analysis phases. In testing 100,000 SNPs, Mendel shows a roughly 1000-fold speed-up over the GWAF-GEE and GWAF-LMM approaches. This fact validates our initial premise that the score test would offer large gains in speed. When testing 100,000 SNPs, Mendel never used more than 76 MB of RAM. In contrast, GWAF had a memory footprint larger than 500 MB, a serious concern for testing large-scale GWAS data.
Table 3.
Comparison of total run times (in seconds on a standard laptop computer) with GWAF.
| # SNPs | Mendel-Score | Mendel-LRT | GWAF-GEE | GWAF-LMM |
|---|---|---|---|---|
| 100 | 4.69 | 5.32 | 0.71 | 8.83 |
| 1,000 | 4.75 | 5.48 | 7.71 | 87.06 |
| 10,000 | 5.28 | 6.05 | 207.60 | 894.82 |
| 100,000 | 10.28 | 11.07 | 26,486.92 | 11,703.88 |
Run times are based on testing the first 100, 1000, 10,000 and 100,000 SNPs on chromosome 19. The column labeled Mendel-LRT displays the total run times after adding likelihood ratio tests for the top 50 SNPs identified by the score test. The simulation model is the same as in Figure 3.
Next we compared the type I error and power of the four methods. In the alternative model, we simulated trait values according to the settings pertinent to Figure 3 with the major locus rs10412915 retained but with varying effect sizes βsnp. In the null model, we discarded the major locus effect and kept the other simulation parameters. All results represent averages across 100 replicates per model. Table 4 tallies the empirical type I error (proportion of replicates with p-values less than 0.05 under the null model) and power (proportion of replicates with p-values less than 0.05 under the alternative model), along with their standard errors. We observe inflated type I error and lowest power in the GEE results, especially at medium to large effect sizes. This is possibly due to the imposition in the current implementation of GWAF-GEE of a uniform working correlation structure across all pedigrees. Although standard semi-parametric theory states that main effects can be consistently estimated even under misspecification of the correlation structure, the sample sizes in real genetic studies are rarely sufficient for such asymptotics to hold. Table 4 suggests that Mendel and GWAF-LMM possess similar operating characteristics. Unfortunately, the extremely low computational efficiency of GWAF-LMM makes it an unattractive choice for GWAS. Modern genetic studies such as those in Framingham and San Antonio often involve at least an order of magnitude more people and (imputed) SNPs than we have simulated here.
Table 4.
Empirical power and type I error for various major-locus effect sizes.
| Mendel-Score | Mendel-LRT | GWAF-GEE | GWAF-LMM | |
|---|---|---|---|---|
| (βsnp = 2.0) | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 | 1.00 ± 0.00 |
| (βsnp = 1.5) | 1.00 ± 0.00 | 1.00 ± 0.00 | 0.97 ± 0.02 | 1.00 ± 0.00 |
| (βsnp = 1.2) | 0.98 ± 0.01 | 0.98 ± 0.01 | 0.89 ± 0.03 | 0.98 ± 0.01 |
| Power (βsnp = 1.0) | 0.92 ± 0.03 | 0.92 ± 0.03 | 0.80 ± 0.04 | 0.92 ± 0.03 |
| (βsnp = 0.8) | 0.75 ± 0.04 | 0.75 ± 0.04 | 0.54 ± 0.05 | 0.75 ± 0.04 |
| (βsnp = 0.5) | 0.38 ± 0.05 | 0.39 ± 0.05 | 0.29 ± 0.05 | 0.40 ± 0.05 |
| (βsnp = 0.3) | 0.14 ± 0.03 | 0.15 ± 0.04 | 0.16 ± 0.04 | 0.15 ± 0.04 |
| Type I Error (βsnp = 0.0) | 0.04 ± 0.02 | 0.04 ± 0.02 | 0.09 ± 0.03 | 0.04 ± 0.02 |
The simulation model is the same as in Figure 3. The empirical power is the proportion of replicates with p-values less than 0.05 under the alternative model with the listed major-locus effect size. The empirical type I error is the proportion of replicates with p-values less than 0.05 under the null model with no major locus. All results represent averages across 100 replicates per model; standard errors appear to the right of each average. The column labeled Mendel-LRT displays the results after adding likelihood ratio tests for the top 50 SNPs identified by the score test.
3.2 The San Antonio Family Heart Study
We analyzed a real data set collected by the San Antonio Family Heart Study (SAFHS) [Mitchell et al. 1996]. The data consist of 3637 individuals in 211 Mexican American families. High-density lipoprotein (HDL) levels were measured at up to three time points for each of the 1429 phenotyped individuals. These traits are denoted HDL1, HDL2, and HDL3, measured at corresponding ages AGE1, AGE2, and AGE3. Some of the phenotyped individuals have HDL measurements at only one or two of the time points. Of the 1429 phenotyped individuals, 1413 were genotyped at 944,427 genome-wide SNPs. The genotyping success rate exceeded 98% in 1388 of these individuals over 124 pedigrees. The largest family contains 247 individuals; five others also contain more than 90 individuals. The smallest pedigree was a singleton. Genotyping success rates were above 98% for 935,392 SNPs.
3.3 Comparison with FaST-LMM and GEMMA
For fair comparisons, we directed Mendel to estimate SNP-based global kinship coefficients for all pairs of individuals ignoring the input pedigrees. This is the default in FaST-LMM and GEMMA. In addition, we ran Mendel’s default in which the coefficients are estimated only for pairs of individuals within the same input pedigree. We also slightly adjusted some of the default quality control thresholds so the programs would be analyzing roughly the same set of SNPs and individuals. For example, by default Mendel filters SNPs with fewer than three occurrences of the minor allele in the data; in contrast, FaST-LMM only filters SNPs with zero occurrences of the minor allele, and GEMMA filters SNPs with minor allele frequency (MAF) < 0.01. All other defaults were observed throughout. Users can easily adjust the Mendel analysis parameters via its control file and the FaST-LMM and GEMMA analysis parameters via their command line.
We first carried out three univariate QTL analyses of HDL1, HDL2, and HDL3, using SEX and AGE1, AGE2, or AGE3 as covariates. We then ran a multivariate QTL analysis of HDL1, HDL2, and HDL3 jointly, which we refer to as HDLJoint. For the multivariate analysis, the most appropriate configuration is to constrain the effects of the SEX and AGE covariates to be the same on all three HDL measurements. Such linear constraints are imposed in Mendel via a few simple lines in its control file. FaST-LMM and GEMMA do not allow constraints on covariates. Therefore, we also ran a multivariate analysis with only the SEX covariate and no constraints. With no constraints, SEX will have a slightly different effect on each component phenotype in the multivariate analysis. For example, Mendel’s default run estimated a female effect of 2.5 ± 0.3 on HDL1, 2.1 ± 0.4 on HDL2, and 2.7 ± 0.4 on HDL3. FaST-LMM cannot do multivariate analyses.
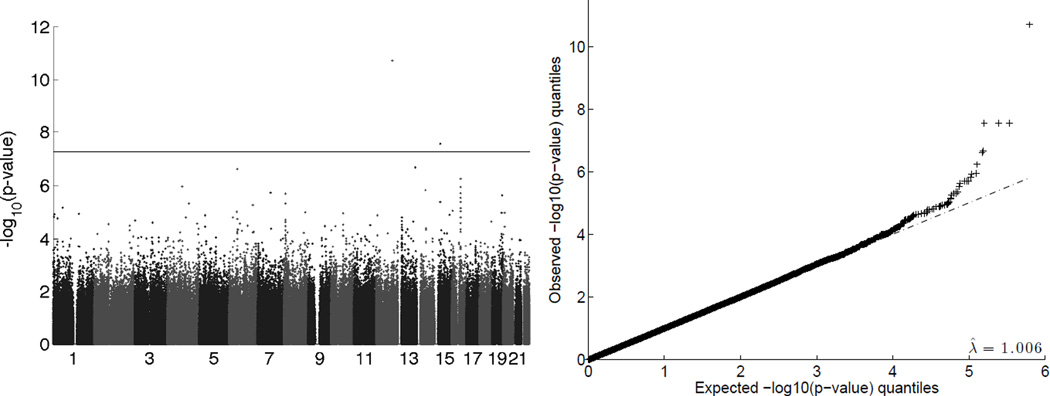
Table 5 reports all SNPs with MAF > 0.01 that achieve genome-wide significance (p-values less than 5 × 10−8) as reported by at least one software package. For the univariate analyses, each software package found the same set of significant SNPs, except that one of GEMMA’s p-values was slightly short of the significance threshold. Figure 7 shows a Manhattan plot and a Q-Q plot from the HDL1 analysis by Mendel given kinship estimates for all pairs of individuals. The results for the other analyses, both univariate and multivariate, were similar. Each Mendel all-pairs univariate analysis had genomic control λ in the range 1.002 to 1.006; in default mode, λ was in the range 0.992 to 1.022. The various Q-Q plots and associated λ values show there is no systematic biases in the data or analysis. In the all-pairs Mendel HDL1 analysis, the grand mean (intercept) was 49.0 ± 0.8. The SEX covariate was significant in all null models. For example, in the all-pairs Mendel HDLJoint analysis with constrained covariates, the SEX effect was 2.4 ± 0.3 for females and, by design, the opposite for males. The AGE covariate was not significant in any run. For example, again in the all-pairs HDLJoint analysis with parameter constraints, the AGE effect was 0.04 ± 0.02. In the null model for the all-pairs Mendel HDL1 analysis, the additive variance was estimated as 78.8 ± 9.9, and the environmental variance was estimated as 78.1 ± 7.2. This gives an overall heritability estimate for HDL1 of 0.50 ± 0.04. Similar variance estimates were seen in other null models.
Table 5.
All SNPs with minor allele frequency (MAF) above 0.001 that reach genome-wide significance in any of the analyses of the HDL traits from the San Antonio Family Heart Study (SAFHS).
| Trait | SNP | Chr. | Base Pair Position |
MAF | − log10(p-val) Mendel default |
− log10(p-val) Mendel all-pairs |
− log10(p-val) FaST-LMM |
− log10(p-val) GEMMA |
|---|---|---|---|---|---|---|---|---|
| HDL1 | rs7303112 | 12 | 97,596,023 | 0.00455 | 10.21 | 10.71 | 7.63 | 7.24 |
| rs8040647 | 15 | 32,304,988 | 0.00147* | 7.44 | 7.56 | 7.35 | 7.45 | |
| rs9972594 | 15 | 32,421,102 | 0.00147* | 7.44 | 7.56 | 7.37 | 7.46 | |
| rs7167103 | 15 | 32,830,477 | 0.00147* | 7.44 | 7.56 | 7.35 | 7.44 | |
| HDL2 | rs7100957 | 10 | 28,207,332 | 0.00183* | 8.84 | 8.95 | 8.88 | 8.82 |
| HDL3 | rs17060933 | 8 | 22,510,029 | 0.00382 | 8.23 | 8.28 | 8.61 | 8.59 |
| HDLJoint with constrained covariates |
rs7303112 | 12 | 97,596,023 | 0.00644 | 9.89 | 9.94 | Not Available |
Not Available |
| rs16925210 | 10 | 25,308,103 | 0.00217 | 8.15 | 8.33 | |||
| rs7091416 | 10 | 25,318,381 | 0.00217 | 8.15 | 8.33 | |||
| rs10075658 | 5 | 148,911,957 | 0.00144* | 8.16 | 8.21 | |||
| rs7733139 | 5 | 145,977,990 | 0.00217 | 7.36 | 7.34 | |||
| rs7100957 | 10 | 28,207,332 | 0.00870 | 7.20 | 7.30 | |||
| HDLJoint without constrained covariates |
rs7303112 | 12 | 97,596,023 | 0.00644 | 9.82 | 9.88 | Not Available |
11.08 |
| rs16925210 | 10 | 25,308,103 | 0.00217 | 8.04 | 8.23 | 3.53 | ||
| rs7091416 | 10 | 25,318,381 | 0.00217 | 8.04 | 8.23 | 3.52 | ||
| rs10075658 | 5 | 148,911,957 | 0.00144* | 8.12 | 8.17 | 3.47 | ||
| rs7733139 | 5 | 145,977,990 | 0.00217 | 7.41 | 7.40 | 3.47 | ||
| rs7100957 | 10 | 28,207,332 | 0.00870 | 7.19 | 7.30 | 4.48 | ||
| rs10083226 | 13 | 104,434,452 | 0.00219 | 7.10 | 7.31 | 2.14 | ||
All default parameters were used except for minor changes to the quality control thresholds (see text). Also, Mendel was run in both default and all-pairs modes. Mendel‘s default mode estimates non-zero global kinship coefficients only for pairs of individuals within the same input pedigree; Mendel in all-pairs mode, FaST-LMM, and GEMMA estimate coefficients for all pairs of individuals. Genome-wide significance was declared for p-values < 5 × 10−8 ⇒ −log10(p-value) > 7.3. The SAFHS has 1413 genotyped and phenotyped individuals in 124 pedigrees. The genotypes include roughly 1 million SNPs. The phenotypes include the subjects’ high-density lipoprotein (HDL) level and age at three time points. The HDLJoint runs are multivariate analyses of HDL1, HDL2, and HDL3 jointly; all other runs are univariate analyses. See the text for a list of the covariates used in each analysis. Note that in the multivariate analysis, Mendel is able to use roughly twice as many individuals as GEMMA (see text and Table 6), which may explain the less significant findings for GEMMA. Each MAF is based on the pedigree founders, except where marked by an asterisk.
In these cases the minor allele did not appear in the genotyped founders, and its frequency was estimated from all genotyped individuals.
Figure 7.
The results of Mendel‘s HDL1 univariate analysis in the SAFHS data set with global kinship coefficients estimated for all pairs of individuals. Upper: The Manhattan plot graphs roughly one million SNPs against their −log10(p-value). The horizontal line is the genome-wide significance threshold, 7.3 = −log10(5 × 10−8). Lower: The Q-Q plot graphs the observed −log10(p-value) quantiles versus their expectations. The genomic control value of λ̂ = 1.006 derived from this comparison suggests no systematic biases in the data or analysis.
For the multivariate analysis without parameter constraints, Mendel is able to include almost twice as many individuals in the analysis as GEMMA (see Table 6). GEMMA only includes individuals phenotyped at all component traits and covariates. This probably explains why Mendel finds several more SNPs with significant p-values than GEMMA.
Table 6.
Comparison of run times and memory (RAM) usage on a typical computer but with adequate RAM to accommodate FaST-LMM (6 CPU cores at 2.67 GHz, with 48 GB total RAM).
| Program | Trait | Analyzed Samples |
Analyzed SNPs |
RunTime (min:sec) |
RAM (GB) |
|---|---|---|---|---|---|
| Mendel default | HDL1 | 1357 | 935,392 | 1:51 | 1.2 |
| Mendel all-pairs | 1357 | 935,392 | 7:49 | 1.2 | |
| FaST-LMM | 1397 | 941,546 | 76:11 | 30.0 | |
| GEMMA | 1397 | 919,050 | 206:54 | 0.4 | |
| Mendel default | HDL2 | 818 | 935,392 | 1:33 | 1.1 |
| Mendel all-pairs | 818 | 935,392 | 3:25 | 1.1 | |
| FaST-LMM | 840 | 934,216 | 49:44 | 18.0 | |
| GEMMA | 840 | 914,051 | 180:21 | 0.3 | |
| Mendel default | HDL3 | 914 | 935,392 | 1:38 | 1.1 |
| Mendel all-pairs | 914 | 935,392 | 3:54 | 1.1 | |
| FaST-LMM | 939 | 937,208 | 54:58 | 20.0 | |
| GEMMA | 939 | 918,626 | 182:26 | 0.3 | |
| Mendel default | HDLJoint with constrained covariates |
1388 | 935,392 | 4:08 | 1.2 |
| Mendel all-pairs | 1388 | 935,392 | 83:24 | 1.2 | |
| FaST-LMM | Not Available | ||||
| GEMMA | Not Available | ||||
| Mendel default | HDLJoint without constrained covariates |
1388 | 935,392 | 3:49 | 1.2 |
| Mendel all-pairs | 1388 | 935,392 | 80:04 | 1.2 | |
| FaST-LMM | Not Available | ||||
| GEMMA | 712 | 912,318 | 630:37 | 0.6 | |
The listed run times include reading the data set, performing quality checks, estimating the kinship coefficients, and calculating the association test p-values. All default parameters were used except for minor changes to the quality control thresholds (see text). Also, Mendel was run in both default and all-pairs modes. Mendel’s default mode estimates non-zero global kinship coefficients only for pairs of individuals within the same input pedigree; Mendel in all-pairs mode, FaST-LMM, and GEMMA estimate coefficients for all pairs of individuals. For the multivariate analysis, Mendel includes roughly twice as many individuals as GEMMA because GEMMA only analyzes individuals phenotyped at all component traits and covariates. Mendel performs score tests for all SNPs and LRTs for the top SNPs; FaST-LMM performs LRTs; and GEMMA by default performs Wald tests, but the user can change this to LRTs or score tests. Using score tests in GEMMA would make it faster (see text).
Table 6 tallies the run times and memory footprints from each analysis on a typical personal computer with adequate RAM to accommodate FaST-LMM (6 CPU cores at 2.67 GHz, with 48 GB total RAM). Even when estimating the global kinship coefficients for all pairs of individuals, each univariate QTL run took Mendel less than 8 minutes to read, quality check, and analyze the data for kinship estimates and association tests, roughly 10% of the time required for FaST-LMM and 5% of the time required by GEMMA. (For GEMMA, the kinship estimation and association tests are run separately. The run times reported here are their total.)
The three programs use different association test strategies: Mendel performs score tests for all SNPs and LRTs for the top SNPs; FaST-LMM performs LRTs; and GEMMA by default performs Wald tests, but the user can change this to LRTs or score tests. For the univariate analyses on a six-core computer, excluding estimation of kinship coefficients, GEMMA’s run times under the Wald test and LRT options were roughly similar to FaST-LMM’s; GEMMA’s run time under the score test option was roughly double Mendel’s in all-pairs mode. This is impressive given GEMMA’s lack of multithreading. It is kinship estimation, which in practice can be done once per data set, that is substantially slower in GEMMA (running roughly 135 minutes) than in FaST-LMM or Mendel (less than 1 minute).
Each trivariate QTL run took Mendel less than 90 minutes. Mendel required roughly one-eighth the time of GEMMA while analyzing almost twice as many individuals. Mendel is also memory efficient. The univariate and multivariate runs each required less than 1.5 GB of memory, which is well below the amount of RAM in a typical computer. FaST-LMM’s memory usage is more than 15 times larger than Mendel’s. GEMMA uses even less memory than Mendel but is considerably slower.
4 Discussion
We have implemented an ultra-fast algorithm for QTL analysis of pedigree data or mix of population and pedigree data. In our opinion Mendel’s comprehensive environment for genetic data analysis is a decided advantage. In addition to its exceptional speed and memory efficiency, Mendel can handle multivariate quantitative traits and detect outlier trait values and pedigrees. Most competing programs ignore multivariate traits and outliers altogether.
A recent review of univariate QTL analysis packages for family data [Eu-ahsunthornwattana et al. 2014] shows that all the explored packages obtain similar results, leaving speed, features, and ease of use as the important factors in choosing between them. Once the current version of Mendel came out, the authors of the review were kind enough to add a comment (www.plosgenetics.org/annotation/listThread.action?root=81847) to their article observing that Mendel was now the fastest and one of the easiest to use packages they reviewed.
In the SAFHS example data set we used with HDL phenotypes, all the significant SNPs we found had MAF < 0.01. Due to these low MAFs, we do not claim these SNPs are strong candidates for further study. However, the key point here is that all four methods found the same SNPs, at least for the univariate analyses. We also note that the p-values are quite similar regardless of whether one uses kinship estimates between all individuals ( Mendel’s all-pairs mode) or only between individuals within the same input pedigrees ( Mendel’s default mode). This suggests that the input pedigree structures for this data set are substantially correct and complete, with few mistaken or hidden relationships. Obviously, this may not be true for other data sets. By supplying good kinship estimates ignoring pedigree structures, the currently reviewed packages make the hard fieldwork of relationship discovery superfluous.
A future version of Mendel will address its failure to read fractional genotype values. This is simply a logistical issue, as all Mendel’s internal genotype computations are already handled as floating point operations. Another imminent feature is a fourth style of kinship coefficient estimation that allows the user to force theoretical kinship coefficients for pairs of individuals within the same pedigree and estimated kinships for all other pairs.
By supplying a comprehensive, fast, and easy to use package for GWAS on quantitative traits in general pedigrees, we hope to encourage exploitation of family-based data sets for gene mapping. A gene mapping study should collect as large a sample as possible consistent with economic constraints and uniform trait phenotyping. If the sample includes pedigrees, all the better. One should not let the choice of statistical test determine the data collected; on the contrary, the data should determine the test. Here we have argued that score tests can efficiently handle unrelated individuals, pedigrees, or a mixture of both. For human studies, where controlled breeding is forbidden, nature has provided pedigrees segregating every genetic trait. Many of these pedigrees are known from earlier linkage era studies and should be treasured as valuable resources.
Let us suggest a few directions for future work. The current method works marker by marker and is ill equipped to perform model selection. Lasso penalized regression is available to handle model selection for case-control and random sample data [Wu and Lange 2008, Wu et al. 2009, Zhou et al. 2010; 2011] and can be generalized to variance component models. Although we have generalized the score test to distributions such as the multivariate t, extending it to discrete traits may be out of reach. For likelihood based methods, there simply are no discrete analogues of the Gaussian distribution that lend themselves to graceful evaluation of pedigree likelihoods. Treating case/control data as a 0/1 quantitative variable is a possibility that has been explored by Pirinen et al. [2013]. The GEE method is another fallback option because it does not depend on precise distributional assumptions.
In rare variant mapping, grouping related SNPs in a variance component may be a good alternative to the mean component models used here. Each variant may be too rare to achieve significance in hypothesis testing. Fortunately, aggregating genotype information within biological units such as genes or pathways offer better power than marginal testing of individual SNPs. See Asimit and Zeggini [2010] for a recent review of aggregation strategies. Kwee et al. [2008] have successfully applied a variance component model for association testing of SNP sets in a sample of unrelated subjects. Rönnegård et al. [2008] consider score tests for random effects models in the context of experimental line crosses. Score tests may well be the key to implementing random effect models in pedigrees. However, the computational demands are apt to be more formidable than those encountered here with fixed effects models. In particular, if tests are based simply on local identity-by-descent (IBD) sharing, then the boundaries between pedigrees disappear, and the entire sample collapses to one large pedigree. The required local kinship coefficients can again be well estimated from dense markers, but this demands more computation than the estimation of global kinship coefficients under the mean components model advocated here [Day-Williams et al. 2011]. Since inversion of a pedigree covariance matrix scales as the cube of the number of individuals in the pedigree, treating the entire sample as a single pedigree will put a practical upper limit on sample size. There are other issues in implementing variance component models such as assigning p-values and dealing with multivariate traits that are best left to a separate paper.
Acknowledgments
The authors gratefully acknowledge the NIH grants GM053275 (EMS and KL), HG006139 (HZ, EMS, and KL), MH059490 (JB, TDD, EMS, and KL), and GM105785 (HZ) and NSF grant DMS1310319 (HZ) supporting this research. KKC also gratefully acknowledges the fellowship support from the Burroughs Wellcome Fund Inter-school Training Program in Metabolic Diseases.
References
- Amin N, van Duijn CM, Aulchenko YS. A genomic background based method for association analysis in related individuals. PLoS ONE. 2007;2(12):e1274. doi: 10.1371/journal.pone.0001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asimit J, Zeggini E. Rare variant association analysis methods for complex traits. Annual Review of Genetics. 2010;44(1):293–308. doi: 10.1146/annurev-genet-102209-163421. [DOI] [PubMed] [Google Scholar]
- Aulchenko YS, de Koning D-J, Haley C. Genomewide rapid association using mixed model and regression: A fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics. 2007;177(1):577–585. doi: 10.1534/genetics.107.075614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M-H, Liu X, Wei F, Larson MG, Fox CS, Vasan RS, Yang Q. A comparison of strategies for analyzing dichotomous outcomes in genome-wide association studies with general pedigrees. Genetic Epidemiology. 2011;35(7):650–657. doi: 10.1002/gepi.20614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M-H, Yang Q. GWAF: an R package for genome-wide association analyses with family data. Bioinformatics. 2010;26(4):580–581. doi: 10.1093/bioinformatics/btp710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W-M, Abecasis GR. Family-based association tests for genome-wide association scans. American Journal of Human Genetics. 2007;81(5):913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R 1000 Genomes Project Analysis Group. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day-Williams AG, Blangero J, Dyer TD, Lange K, Sobel EM. Linkage analysis without defined pedigrees. Genetic Epidemiology. 2011;35(5):360–370. doi: 10.1002/gepi.20584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eu-ahsunthornwattana J, Miller EN, Fakiola M, Jeronimo SMB, Blackwell JM, Cordell HJ Wellcome Trust Case Control Consortium 2. Comparison of methods to account for relatedness in genome-wide association studies with family-based data. PLoS Genetics. 2014;10(7):e1004445. doi: 10.1371/journal.pgen.1004445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong S-Y, Freimer NB, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nature Genetics. 2010;42(4):348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte A, Vilhjalmsson BJ, Segura V, Platt A, Long Q, Nordborg M. A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nature Genetics. 2012;44(9):1066–1071. doi: 10.1038/ng.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku CS, Loy EY, Pawitan Y, Chia KS. The pursuit of genome-wide association studies: where are we now? Journal of Human Genetics. 2010;55(4):195–206. doi: 10.1038/jhg.2010.19. [DOI] [PubMed] [Google Scholar]
- Kwee LC, Liu D, Lin X, Ghosh D, Epstein MP. A powerful and flexible multilocus association test for quantitative traits. American Journal of Human Genetics. 2008;82(2):386–397. doi: 10.1016/j.ajhg.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genetic Epidemiology. 2000;19(Suppl 1):S36–S42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Lange C, Laird NM. On a general class of conditional tests for family-based association studies in genetics: the asymptotic distribution, the conditional power, and optimality considerations. Genetic Epidemiology. 2002;23(2):165–180. doi: 10.1002/gepi.209. [DOI] [PubMed] [Google Scholar]
- Lange K. Statistics for Biology and Health. 2nd. New York: Springer-Verlag; 2002. Mathematical and Statistical Methods for Genetic Analysis. [Google Scholar]
- Lange K, Little RJA, Taylor JMG. Robust statistical modeling using the t distribution. Journal of the American Statistical Association. 1989;84(408):881–896. [Google Scholar]
- Lange K, Papp JC, Sinsheimer JS, Sobel EM. Next-generation statistical genetics: Modeling, penalization, and optimization in high-dimensional data. Annual Review of Statistics and Its Application. 2014;1(1):279–300. doi: 10.1146/annurev-statistics-022513-115638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange K, Papp JC, Sinsheimer JS, Sripracha R, Zhou H, Sobel EM. Mendel: The Swiss army knife of genetic analysis programs. Bioinformatics. 2013;29(12):1568–1570. doi: 10.1093/bioinformatics/btt187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange K, Sinsheimer JS, Sobel E. Association testing with Mendel. Genetic Epidemiology. 2005;29(1):36–50. doi: 10.1002/gepi.20073. [DOI] [PubMed] [Google Scholar]
- Lange K, Westlake J, Spence MA. Extensions to pedigree analysis iii. variance components by the scoring method. Annals of Human Genetics. 1976;39(4):485–491. doi: 10.1111/j.1469-1809.1976.tb00156.x. [DOI] [PubMed] [Google Scholar]
- Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, Heckerman D. FaST linear mixed models for genome-wide association studies. Nature Methods. 2011;8(10):833–835. doi: 10.1038/nmeth.1681. [DOI] [PubMed] [Google Scholar]
- Listgarten J, Lippert C, Kadie CM, Davidson RI, Eskin E, Heckerman D. Improved linear mixed models for genome-wide association studies. Nature Methods. 2012;9(6):525–526. doi: 10.1038/nmeth.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell BD, Kammerer CM, Blangero J, Mahaney MC, Rainwater DL, Dyke B, Hixson JE, Henkel RD, Sharp RM, Comuzzie AG, VandeBerg JL, Stern MP, MacCluer JW. Genetic and environmental contributions to cardiovascular risk factors in Mexican Americans: The San Antonio Family Heart Study. Circulation. 1996;94(9):2159–2170. doi: 10.1161/01.cir.94.9.2159. [DOI] [PubMed] [Google Scholar]
- Ott J, Kamatani Y, Lathrop M. Family-based designs for genome-wide association studies. Nature Reviews Genetics. 2011;12(7):465–474. doi: 10.1038/nrg2989. [DOI] [PubMed] [Google Scholar]
- Pirinen M, Donnelly P, Spencer CCA. Efficient computation with a linear mixed model on large-scale data sets with applications to genetic studies. Annals of Applied Statistics. 2013;7(1):369–390. [Google Scholar]
- Rao C. Linear Statistical Inference And Its Applications. 2nd. Wiley; 2009. [Google Scholar]
- Rönnegård L, Besnier F, Carlborg O. An improved method for quantitative trait loci detection and identification of within-line segregation in f2 intercross designs. Genetics. 2008;178(4):2315–2326. doi: 10.1534/genetics.107.083162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman RS, Ewens WJ. A sibship test for linkage in the presence of association: the sib transmission/disequilibrium test. American Journal of Human Genetics. 1998;62(2):450–458. doi: 10.1086/301714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanhope SA, Abney M. GLOGS: a fast and powerful method for GWAS of binary traits with risk covariates in related populations. Bioinformatics. 2012;28(11):1553–1554. doi: 10.1093/bioinformatics/bts190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton T, McPeek MS. Case-control association testing with related individuals: A more powerful quasi-likelihood score test. American Journal of Human Genetics. 2007;81(2):321–337. doi: 10.1086/519497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton T, McPeek MS. ROADTRIPS: Case-control association testing with partially or completely unknown population and pedigree structure. American Journal of Human Genetics. 2010;86(2):172–184. doi: 10.1016/j.ajhg.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Steen K. Perspectives on genome-wide multi-stage family-based association studies. Statistics in Medicine. 2011;30(18):2201–2221. doi: 10.1002/sim.4259. [DOI] [PubMed] [Google Scholar]
- Van Steen K, Lange C. PBAT: a comprehensive software package for genome-wide association analysis of complex family-based studies. Human Genomics. 2005;2(1):67–69. doi: 10.1186/1479-7364-2-1-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. American Journal of Human Genetics. 2012;90(1):7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won S, Bertram L, Becker D, Tanzi R, Lange C. Maximizing the power of genome-wide association studies: A novel class of powerful family-based association tests. Statistics in Biosciences. 2009a;1(2):125–143. doi: 10.1007/s12561-009-9016-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won S, Wilk JB, Mathias RA, O’Donnell CJ, Silverman EK, Barnes K, O’Connor GT, Weiss ST, Lange C. On the analysis of genome-wide association studies in family-based designs: A universal, robust analysis approach and an application to four genome-wide association studies. PLoS Genetics. 2009b;5(11):e1000741. doi: 10.1371/journal.pgen.1000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TT, Chen Y, Hastie T, Sobel EM, Lange K. Genome-wide association analysis by lasso penalized logistic regression. Bioinformatics. 2009;25(6):714–721. doi: 10.1093/bioinformatics/btp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TT, Lange K. Coordinate descent algorithms for lasso penalized regression. Annals of Applied Statistics. 2008;2(1):224–244. doi: 10.1214/10-AOAS388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ersoz E, Lai C-Q, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, Buckler ES. Mixed linear model approach adapted for genome-wide association studies. Nature Genetics. 2010;42(4):355–360. doi: 10.1038/ng.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Alexander D, Sehl M, Sinsheimer JS, Sobel EM, Lange K. Penalized regression for genome-wide association screening of sequence data. Pacific Symposium on Biocomputing. 2011;2011:106–117. doi: 10.1142/9789814335058_0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Sehl ME, Sinsheimer JS, Lange K. Association screening of common and rare genetic variants by penalized regression. Bioinformatics. 2010;26(19):2375–2382. doi: 10.1093/bioinformatics/btq448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nature Genetics. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Stephens M. Efficient multivariate linear mixed model algorithms for genome-wide association studies. Nature Methods. 2014;11(4):407–409. doi: 10.1038/nmeth.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Xiong M. Family-based association studies for next-generation sequencing. American Journal of Human Genetics. 2012;90(6):1028–1045. doi: 10.1016/j.ajhg.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]