Abstract
Targeting the dysregulated BRaf-MEK-ERK pathway in cancer has increasingly emerged in clinical trial design. Despite clinical responses in specific cancers using inhibitors targeting BRaf and MEK, resistance develops often involving non-genomic adaptive bypass mechanisms. Inhibition of MEK1/2 by trametinib in triple negative breast cancer (TNBC) patients induced dramatic transcriptional responses, including upregulation of receptor tyrosine kinases (RTKs) comparing tumor samples before and after one week of treatment. In preclinical models MEK inhibition induced genome-wide enhancer formation involving the seeding of BRD4, MED1, H3K27 acetylation and p300 that drives transcriptional adaptation. Inhibition of P-TEFb associated proteins BRD4 and CBP/p300 arrested enhancer seeding and RTK upregulation. BRD4 bromodomain inhibitors overcame trametinib resistance, producing sustained growth inhibition in cells, xenografts and syngeneic mouse TNBC models. Pharmacological targeting of P-TEFb members in conjunction with MEK inhibition by trametinib is an effective strategy to durably inhibit epigenomic remodeling required for adaptive resistance.
Keywords: adaptive transcriptional response, P-TEFb complex, MEK inhibition, epigenomic landscape, enhancer remodeling
INTRODUCTION
Mutations and genomic amplification of components and regulators of the Ras-BRaf-MEK-ERK pathway are common in cancer. Activating mutations in Ras have been found in up to 30% of all tumors that have been sequenced, with the mutant Ras isoform (KRAS, NRAS, HRAS) depending on the specific cancer (1). BRAF is mutated in approximately 50% of metastatic melanomas and 55% of advanced thyroid carcinomas and at a lower frequency in colorectal, ovarian, and lung carcinomas (2–7). Sequencing initiatives including The Cancer Genome Atlas (TCGA) are rapidly expanding the tumor sequencing database with additional tumor types having lower frequencies but still significant numbers of activating BRAF mutations (2). Other cancers such as TNBC are different, with activating mutations in Ras and protein kinases being rare, but instead have gene amplification of BRAF or upstream regulators of the MAPK pathway (2,8). Approximately 80% of basal-like TNBC have genomic amplification of members of the EGFR-KRas-BRaf signaling network correlating with the BRaf-MEK-ERK pathway being activated in basal-like breast cancers (2,8,9).
BRaf inhibitors such as vemurafenib or dabrafenib in combination with the MEK inhibitor trametinib have proven to have a significant beneficial response for BRAF mutant melanoma, including a higher incidence of complete response as well as longer progression-free survival (10,11). MEK inhibition has also shown benefit in mutant NRAS driven melanoma (12). Even though BRaf and MEK inhibitors produce initial clinical responses in melanoma, resistance ultimately occurs due to acquired or selected mutations or upregulation of adaptive bypass pathways (13–15). Non-genomic adaptive bypass mechanisms, for example, involving transcriptional upregulation of receptor tyrosine kinases (RTKs), are not limited to melanoma, but rather are increasingly observed as major mechanisms of clinical resistance in many cancers (16,17). In TNBC, trametinib inhibition of MEK to block the MEK-ERK pathway causes an initial potent growth arrest that is overcome by an adaptive bypass response (9). Inhibition of MEK-ERK elicits upregulation of alternative kinase pathways contributing to escape from growth inhibition. This adaptive kinome remodeling underscores the difficulty of attaining successfully targeted kinase inhibitor treatments and suggests that even combination therapies with multiple kinase inhibitors will have a poor likelihood of success.
Mechanistically, one consequence of MEK inhibition is the loss of ERK-catalyzed Myc Ser62 phosphorylation causing rapid ubiquitination and proteasomal degradation of Myc (18). Myc turnover is required for multiple steps in transcriptional activation including histone acetylation and recruitment of BRD4 and P-TEFb to chromatin (19), and inhibiting Myc degradation (e.g. by proteasome inhibition) blocks transcriptional activation of Myc target genes. Herein, we show that MEK inhibition results in a rapid degradation of Myc and dramatic transcriptomic changes resulting in resistance. Surprisingly, the transcriptomic changes promoting resistance are driven epigenetically with de novo enhancer formation and dramatic genome-wide enhancer and promoter remodeling. Enhancers are distal regulatory elements comprised of Mediator complex members that regulate transcription in cis by recruiting transcriptional activators and by looping to promoter regions (20,21). Quiescent, but poised enhancers are enriched for histone H3 lysine 4 monomethylation (H3K4me1), while co-occupancy of H3K4me1 and H3K27 acetylation is a hallmark of active enhancers (22). The dynamic nature of enhancers has become increasingly apparent whereby diverse stimuli have been shown to induce de novo enhancer formation or to reorganize the existing enhancer landscape (23–28). We demonstrate targeting P-TEFb complex members with small molecule inhibitors or RNAi blocks enhancer remodeling and the MEK inhibitor adaptive transcriptomic response. Combining MEK inhibitor with P-TEFb complex inhibitors sustains MEK inhibition and alleviates adaptive resistance by reversing the upregulation of adaptive response genes including RTKs.
RESULTS
MEK inhibition induces a transcriptomic/kinome response in patient TNBC tumors
To examine the clinically relevant occurrence of extensive kinome reprogramming following therapy with a potent MEK inhibitor, a seven day window-of-opportunity clinical trial was used to assess the adaptive response to trametinib in TNBC patients. Pretreatment needle core biopsies and surgical tumor resections following 7 day trametinib treatment were subjected to RNAseq. Similar to the TCGA, we used RNAseq by Expectation-Maximization (RSEM) (29) to quantify transcript abundances from RNAseq data. PAM50 subtype calls were made from RNAseq profiles for post and pre-treatment tumor samples for each of the six patients in the window trial (30). Five tumors profiled as basal-like in both post- and pre-trametinib treatment (referred to as BL;BL for post/pre subtype) and one as claudin-low (CL;CL). Claudin-low tumors represent less than 10% of TNBC consistent with only one in six tumors profiling as this subtype (8,31,32). Increased transcript ratios of post to pre-treatment RSEM transcript abundance (≥2 fold) ranged from 1.7% (Patient 5, referred to as Pt. 5) to 7.8% (Pt. 2) of total expressed transcripts (Fig. 1A). Decreased transcript ratios of post to pre-treatment RSEM transcript abundance ranged from 0.8% (Pt. 4) to 16.1% (Pt. 3). Examination of the tyrosine kinome showed that trametinib induced up to 26% (23 TKs) of the tyrosine kinome (Pt. 2, Pt. 4) among BL;BL patient tumors and in the sole CL;CL tumor, 16% (14 TKs) was upregulated (Fig. 1B). DESeq2 differential expression analysis (33) was used for detection of differentially expressed genes between post and pre-treatment tumors. Focusing on the kinome, four patient BL;BL pretreatment biopsies matched to post-treatment surgical specimens showed overall concordance of the transcriptional response to trametinib, with FRK exhibiting the highest mean increase and BMX exhibiting the highest mean decrease among patients 1, 2, 3 and 5 (Fig. 1C). Patient 4 clustered separately from the other patients with an enhanced immune kinase upregulation, while Pt. 6 responded differently from the five BL tumors.
Figure 1.
Response to MEK inhibition in TNBC patient tumors from window-of-opportunity clinical trial. (A) Transcriptional response to trametinib in TNBC patient tumors in pre-treatment needle core biopsies (NCBs) and in corresponding surgical resections following 7 day trametinib treatment. Total number of expressed genes are indicated in black; percentage of transcripts induced (red) or suppressed (green) > 2 fold after trametinib treatment are indicated. (B) Tyrosine kinome transcriptional response (> 1.5 fold) to trametinib treatment in BL;BL patient tumors (blue) or CL;CL patient tumor (red). (C) Differential Expression-Seq2 (DESeq2) analysis comparing pre-trametinib and post-trametinib BL;BL tumors. Shown are differentially expressed kinases using 0.05 FDR for significance. Pt. 4 was excluded from the DESeq2 analysis because of high immune kinase expression but presented in the heat map for comparison. CL;CL tumor 6 is presented in the heat map for comparison to the BL;BL tumor response. (D) Adaptive response RTK protein upregulation in BL;BL patient tumors. (E) Scatterplot of RSEM transcript abundance values vs. MIB/MS TK MIB binding as a ratio of trametinib-treated surgical resection:pre-treatment NCB in BL;BL Pt. 5 (blue) and CL;CL Pt. 6 (red). Non-TKs are indicated with black circles. Arrows highlight decreased MEK1/2 MIB binding following trametinib.
On average, patient samples expressed ~2500 more transcripts than BL or CL cell lines (excluding transcripts with <50 RSEM pre- and post-treatment, Supplementary Fig. S1A). Comparing the baseline transcriptomes of two TNBC cell lines (BL, HCC1806 and CL, SUM-159PT) to the pretreatment transcriptomes from BL patient 2 and CL patient 6 respectively revealed tumor transcripts enriched for immune and inflammatory response genes absent in the cell lines (Supplementary Fig. S1B), indicating a stromal contribution in tumors absent in cell lines. The stromal transcripts responded to MEK inhibition with a higher percentage of transcripts suppressed than induced (Supplementary Fig. S1C).
Immunoblot analysis showed upregulation at the protein level in post-treatment tumors of FGFR2, KIT, IGF1R, and DDR1 relative to pretreatment biopsies (Fig. 1D). Multiplexed inhibitor bead (MIB) chromatography was used to capture expressed protein kinases that were assayed using mass spectrometry (MIB/MS) (9,34) as a measure of change in the functional kinome in pre- and post-trametinib treated tumors. In BL;BL Pt.5, 3 of 8 transcriptionally upregulated TKs were concordantly enriched as functional protein kinases binding to MIB affinity columns, while in CL;CL Pt. 6, 7 of 14 induced TKs were found by MIB/MS, demonstrating functional TK expression increases in patient tumors in response to MEK inhibition (Fig. 1E). In addition, MIB/MS patient tumor analysis showed MEK1/2 was inhibited by trametinib, observed by loss of MIB binding (Fig. 1E); immunoblots also showed decreased pERK1/2 levels (Fig. 1D), demonstrating that trametinib inhibited the MEK-ERK pathway in patient tumors. The window trial demonstrated one week of trametinib induced a strong adaptive bypass response with upregulation of several TKs that upon sustained trametinib treatment would contribute to resistance to MEK inhibition.
MEK1/2 inhibition induces differential transcriptomic responses in basal-like and claudin-low TNBC cells
Having established through RNAseq and MIB/MS analysis that MEK inhibitor-induced TK reprogramming occurred in patients, we sought to understand the mechanism. Myc turnover is required for histone acetylation, recruitment of BRD4 to chromatin and transcriptional elongation (19). Inhibition of MEK1 and 2 (referred to as MEK) leads to rapid ERK activity loss and Myc degradation (Fig. 2A) (18). We assessed transcriptional responses to MEK inhibition by RNAseq in biological duplicate using TNBC cell lines representing BL and CL intrinsic molecular subtypes (Fig. 2B). MEK inhibition by trametinib induced large transcriptional responses varying between 2.5 to 28.6% of expressed transcripts. DESeq2 and Pearson correlation analysis of three biological replicates confirmed the transcriptional responses to trametinib in SUM-159PT cells (Supplementary Fig. S2A). Gene Set Enrichment Analysis (GSEA) (35,36) revealed global downregulation of Myc target genes following trametinib consistent with loss of Myc due to MEK-ERK inhibition (Supplementary Fig. S2B). KRas signaling was also inhibited (Supplementary Fig. S2B). Collectively, the magnitude of transcriptional changes across TNBC cell lines was striking with loss of Myc and KRas regulated transcription by diminishing MEK-ERK activity with a highly selective, allosteric MEK inhibitor.
Figure 2.
Trametinib elicits a subtype-specific transcriptional response in TNBC cells. (A) Myc protein loss and RTK upregulation in SUM-159PT CL cells and HCC1806 BL cells after 48 h 10 nM trametinib treatment. (B) RNAseq results (mean RSEM of biological duplicates) in the indicated BL or CL human cell lines following 24 h 500 nM trametinib. (C) DESeq2 differential expression analysis of the kinome response to 24 h 500 nM trametinib comparing basal-like cell lines (HCC1806, SUM-149PT EpCAM+/CD49f+, MDA-MB-468) or claudin-low cell lines (SUM-159PT, Hs 578T, WHIM12). Log2 fold trametinib changes for kinases significant with a threshold of 0.05 FDR (Supplementary Table S1) are displayed in heat map following hierarchical clustering using (1 − Spearman Correlation)/2 as the distance metric and row scaling. (D) BL;BL patient tumor trametinib-upregulated kinases (Fig. 1C) overlapping with basal-like cell line trametinib-upregulated kinases (Supplementary Table S2). Hypergeometric test p value for overlap = 0.007 (patient FDR = 0.05, cell line FDR = 0.01). (E) Mean tyrosine kinome response to 24 h 500 nM trametinib of biological duplicates. TK transcripts upregulated > 1.5 fold are displayed and Log2 magnitude of response plotted for BL (HCC1806, SUM-149PT EpCAM+; blue) and CL (SUM-159PT, Hs 578T, WHIM12; red) cells. TK transcripts upregulated > 1.3 fold are displayed for MDA-MB-468 cells due to low relative transcriptome-wide responsiveness to trametinib (B). (F) Sorting SUM-229PE parental cells into distinct BL EpCAM+/CD49f+ and CL EpCAM−/CD49f− populations using flow cytometry. (G) Top: Commonly (grey) and uniquely (blue, red) induced (> 2 fold) transcripts following 24 h 30 nM trametinib treatment of EpCAM+ or EpCAM− subpopulations. Bottom: mRNAseq in EpCAM+ or EpCAM− subpopulations showing response to 24 h 30 nM trametinib treatment. (H) Kinases induced > 1.5 fold or suppressed > 1.5 fold after 24 h 30 nM trametinib treatment that are either unique to EpCAM+ cells (blue) or EpCAM− cells (red), or both subpopulations (grey). (I) Upregulation of BL adaptive response RTK FGFR2 in EpCAM+ cells and CL adaptive response RTK PDGFRB in EpCAM− cells following 24 h 30 nM trametinib.
Distinct BL and CL kinome signatures induced by MEK inhibition
Having determined the magnitude of response induced by trametinib at the level of the transcriptome we next employed DESeq2 differential expression analysis to assess the response of the kinome. The transcriptional effects on the kinome clearly clustered by subtype by DESeq2 using a FDR of 0.05 for significance, indicating that the basal-like and claudin-low adaptive kinome responses elicited by MEK inhibition are unique. (Fig. 2C, Supplementary Table S1). In addition to this differential expression analysis cross-comparing adaptive responses by subtype, we analyzed the adaptive response to MEK inhibition by DESeq2 with each subtype in isolation (Supplementary Table S2). This allowed trametinib-responsive kinases like FGFR2 and PDGFRB that are expressed exclusively in one subtype to be considered by DESeq2 for significance that otherwise would be excluded in the subtype comparative analysis. There was a strong TK response concordance between the BL;BL patient tumors and BL cell lines. Upregulated kinases common in the BL;BL patients (Fig. 1C) and BL cell line signature (Supplementary Table S2) included FRK, ERBB2, DDR1, CDC42BPG, CDKL5, and CDK19, consistent with an adaptive response in patient tumors similar to that observed with in vitro cell models of TNBC (Fig. 2D). The TNBC adaptive response to MEK inhibition requires upregulation and activation of tyrosine kinases (TKs), bypassing of MEK-ERK inhibition and restoring cell growth (9). Each of the BL and CL human cell lines responded with upregulation of TKs (Fig. 2E). For the cell lines tested SUM-149PT EpCAM+ BL and SUM-159PT CL cells displayed the largest number of induced TKs, 21 and 20, respectively. TKs selectively induced by trametinib treatment in BL cells included FGFR2 and CSF1R, with PDGFRB and ERBB4 being upregulated in the CL cells (Fig. 2E, Supplementary Table S2). ROS1, FRK, DDR1, and ERBB2 were common to both BL and CL subtype signatures. In addition to human TNBC cell lines including the WHIM12 PDX-derived line (37), we assessed trametinib TK transcriptional response in cells derived from CL T11 orthotopic syngeneic transplant (OST) tumors (38) and the mixed BL/CL C3(1)/Tag GEMM (39). In T11 cells, PDGFRB and DDR1 were upregulated after trametinib treatment (Supplementary Fig. S2C), concordant with the CL cell line response (Fig. 2E, Supplementary Table S2). Given the mixed BL/CL nature of the C3(1)/Tag tumor, in addition to upregulation of PDGFRB and DDR1, trametinib induced expression of CSF1R of the BL cell line TK signature (Supplementary Fig. S2C). Subtype-specific adaptive TK transcriptional signatures were confirmed by western blotting (Supplementary Fig. S2D) and proteomically with MIB/MS profiles (Supplementary Fig. S2E), showing that adaptive response TKs are functionally expressed following MEK inhibition.
SUM-229PE subpopulations display distinct adaptive responses to MEK inhibition
SUM-229PE TNBC cells provided a unique system to study the BL vs CL subtypes of TNBC. SUM-229PE cells have two intrinsic subpopulations with differential expression of EpCAM and CD49f (Fig. 2F). The epithelial-like EpCAM+/CD49f+ (229 EpCAM+) subpopulation exhibits a BL gene expression signature while the more mesenchymal EpCAM−/CD49f− (229 EpCAM−) subpopulation profiles as CL and is enriched for epithelial to mesenchymal transition genes (40–42). Whole exome sequencing of the two subpopulations identified 8373 common nucleotide variants with no subpopulation-unique variants (Supplementary Table S3). Additionally, we performed a model-based assessment to probe for copy number alterations (CNAs). No clonal CNAs (>50% of the cells experiencing an event) were detected (Supplementary Fig. S2F). Thus, SUM-229PE cells provide an isogenic system to study the BL and CL subtype specificity of the adaptive response.
RNAseq profiling of the FACS-sorted EpCAM+ or EpCAM- cells following trametinib treatment revealed distinct adaptation profiles. EpCAM+ cells were more responsive to trametinib than EpCAM− cells, with 15% vs. 9% of their transcriptomes modulated (Fig. 2G). In response to MEK inhibition, EpCAM+ cells upregulated 594 unique transcripts and EpCAM− cells upregulated 369 unique transcripts, while 280 transcripts were upregulated in common.
Kinome analysis showed a greater induction (107 kinases) vs. suppression (70 kinases) response to trametinib. 37 kinase transcripts unique to EpCAM+ cells and 28 EpCAM− kinases were specifically upregulated, while 42 kinases were commonly induced (Fig. 2H). MEK inhibition resulted in the downregulation of 29 kinases in common between subpopulations, 29 EpCAM+ specific, and 12 kinases specific to EpCAM− cells (Fig. 2H). TK and TK-like kinase families were enriched in upregulated transcripts while cell cycle and mitotic checkpoint kinases were enriched in the downregulated cohort, consistent with MEK inhibition and growth arrest.
Eleven TKs were in the shared upregulated kinases between EpCAM+ and - cells. Members of BL and CL TK transcriptional signatures (Fig. 2E) were modulated concordantly with EpCAM status in SUM-229PE subpopulations, including BL cell line signature TKs in EpCAM+ cells and CL cell line signature TKs in EpCAM− cells (Supplementary Fig. S2G). Immunoblotting showed the CL signature RTK PDGFRB specifically upregulated in EpCAM− CL subpopulation (Fig. 2I) and FGFR2 increased selectively in the BL subpopulation. EpCAM and vimentin protein expression validated the cells’ epithelial BL vs. mesenchymal CL phenotype (Fig. 2I).
Genome-wide methylation was assessed for the SUM-229PE EpCAM+ and SUM-229PE EpCAM− subpopulations. Methylation fraction (β) at all probes (within 200 bp of TSS) for subpopulation-specific induced TKs (Fig. 2H and Supplementary Fig. S2G) did not significantly differ between EpCAM+ and EpCAM− populations and (Supplementary Fig. S2H and Supplementary Table S4). We tested if trametinib-induced differential cytosine methylation changes contributed to transcriptional adaptation. Principal component analysis (PCA) of SUM-159PT, SUM-149PT EpCAM+/−, or SUM-229PE EpCAM+/− cell populations of genome-wide Illumina 450k methylation data in the presence or absence of trametinib revealed a lack of segregation in the top components due to drug (Supplementary Figure S3A and Supplementary Table S4). The lack of cytosine methylation variation, exonic nucleotide variants and CNAs indicates that the distinct adaptive response signatures between SUM-229PE EpCAM+ and EpCAM− subpopulations are not rooted in genomic differences but rather represent modulation of chromatin beyond that of baseline DNA methylation.
Trametinib induces the dynamic formation of an epigenomic landscape
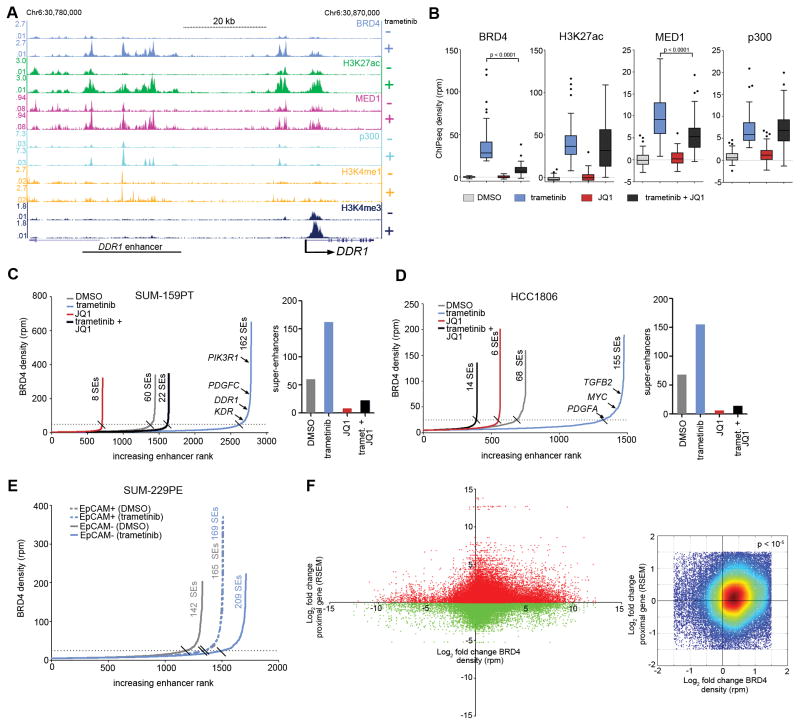
We next specifically assessed the density of a series of enhancer and promoter marks at the DDR1 locus, a prototypical adaptive response kinase using ChIPseq. In the MEK inhibitor-responsive DDR1 locus in SUM-159PT cells at baseline, H3K4me3 occupancy defined a core promoter at the TSS, while at a region 50 kb 5′ there was modest density of BRD4, MED1, p300, H3K4me1 and H3K27ac markers indicative of an enhancer. Trametinib caused a striking increase in occupancy of BRD4 and the other enhancer marks assayed (Fig. 3A). MEK inhibition also resulted in increased BRD4 and H3K27ac density at the DDR1 core promoter while MED1, p300, H3K4me1 marks displayed a relatively minimal degree of promoter change. The induction of de novo BRD4 and associated marker density positively correlated with the 8-fold transcriptional induction of DDR1 (Supplementary Table S5).
Figure 3.
Remodeling of epigenomic landscape induced by MEK inhibition. (A) SUM-159PT ChIPseq density tracks at the DDR1 adaptive response RTK locus in the presence or absence of 24 h 100 nM trametinib. (B) Response of BRD4, H3K27ac, MED1, and p300 ChIPseq density to 24 h 100 nM trametinib alone, or the combination of 300 nM JQ1 at the highest 50 ranking BRD4 peaks defined by trametinib induction magnitude. Quantification of enhancers and super-enhancers by BRD4 density following 24 h 100 nM trametinib, 300 nM JQ1 or the combination in SUM-159PT (C) or HCC1806 (D) cells. (E) Enhancer quantification by BRD4 density following 24 h 30 nM trametinib in SUM-229PE EpCAM+/CD49f+ (dotted lines) or EpCAM−/CD49f− (solid lines) cells. (F) Left: Fold change of genome-wide BRD4 stitched peak ChIPseq density vs. transcriptional fold change of genes whose TSS resides +/− 200 kb from the BRD4 peak density in SUM-159PT cells with 24 h 100 nM trametinib. Right: Zoom of plot on left with warmer colors representing higher density of points showing enrichment in upper right quadrant. Empirical p value (< 10−5) from randomization test indicates that in each of 10,000 cycles of data randomization the number of points under the null hypothesis of no enrichment did not exceed the number of observed counts for this quadrant.
To gain insight into MEK inhibitor-induced regions of BRD4 chromatin occupancy, we determined H3K27ac, p300, and MED1 ChIPseq density at the 50 highest ranking BRD4 ChIPseq peaks induced by trametinib. Concomitant with BRD4 density increase, trametinib increased chromatin occupancy of H3K27ac, p300, and MED1 (Fig. 3B). The small molecule JQ1 binds the acetyl-lysine binding pocket of BET family bromodomains and thus is capable of displacing BRD4 from chromatin by interfering with BRD4 interaction with acetylated histones and acetylated non-histone proteins (43). As BRD4 bound the DDR1 enhancer de novo upon trametinib treatment, we tested the effect of JQ1 on the integrity and composition of this enhancer. BET bromodomain inhibition and trametinib significantly reduced BRD4 and MED1 chromatin occupancy, while H3K27ac and p300 density was not significantly altered, suggesting BRD4 regulates MED1 association with enhancers without affecting p300 association. We hypothesized that analysis of DNA sequence motifs enriched in BRD4-induced regions would suggest other transcriptional regulatory factors involved in the epigenomic remodeling mediated by trametinib. Multiple EM for Motif Elicitation (MEME) (44) analysis of all trametinib-induced BRD4 peaks predicted CEBPB and CEBPD enrichment at these loci (Supplementary Fig. S3B). Consistent with this prediction, ChIPseq studies showed gain of CEBPB density at DDR1 and PIK3R1 enhancers upon trametinib treatment, mirroring BRD4 density dynamics (Supplementary Fig. S3C). Like H3K27ac and p300, trametinib-induced CEBPB density was not altered by JQ1 co-treatment, suggesting CEBPB chromatin occupancy is BET bromodomain independent (Supplementary Fig. S3D).
MEK inhibition induces formation of genome-wide enhancers
The formation of a putative DDR1 enhancer enriched for BRD4 prompted us to quantify the genome-wide extent of enhancer formation mediated by trametinib. BRD4 peaks within 12.5 kb of each other were stitched and designated as putative enhancers for a given gene if they resided either within 200 kb 5′ of the TSS or 200 kb 3′ of the 3′-most exon, but did not reside within promoter territory defined as +/− 5 kb of a TSS. Using these criteria, at baseline (vehicle DMSO-treated), SUM-159PT cells had 1445 BRD4 enriched enhancers (Fig. 3C, Supplementary Table S6). Trametinib robustly remodeled the enhancer landscape, with de novo enhancer induction almost doubling, reaching 2782 defined enhancers. Co-treatment of SUM-159PT cells with trametinib + JQ1 disrupted the trametinib-induced BRD4 enhancer landscape returning it to near-baseline (1632 enhancers).
Large, multi-kilobase expanses of enhancer density termed super-enhancers have been shown to regulate genes important for development and show enhanced sensitivity to BET bromodomain inhibition relative to classical enhancers (25,45,46). Ranking enhancers by BRD4 density allowed us to define super-enhancer formation and dissolution during drug treatments. 162/2782 enhancers were categorized as super-enhancers following trametinib treatment, and JQ1+trametinib co-treatment squelched the number of super-enhancers to 22, even below that of the 60 constitutive super-enhancers found at baseline. As expected, JQ1 treatment alone displaced BRD4 density reducing the 60 baseline super-enhancers to 8. The HCC1806 BL cell line similarly displayed remarkable genome-wide enhancer induction which was muted by JQ1+trametinib co-treatment (Fig. 3D, Supplementary Table S7). SUM-229PE sorted EpCAM+ or EpCAM− subpopulations also displayed remarkably distinct enhancer dynamics. SUM229PE EpCAM− cells responded to trametinib treatment with a near 30% increase in enhancer number (1328 to 1712) while, strikingly, the SUM229PE EpCAM+ enhancer number remained constant (1505 to 1510) (Fig. 3E). The paucity of enhancer remodeling in the EpCAM+ subpopulation suggested that these cells may not mount as an effective adaptive response relative to the negative population, consistent with their enhanced sensitivity to trametinib (Supplementary Fig. S3E).
We determined if de novo BRD4 density positively modulated transcription of genes closest to the density. In SUM-159PT cells, following computational stitching of BRD4 density peaks, trametinib-induced fold change of BRD4 peaks was compared to trametinib-induced fold change of transcription of the genes with TSSs +/− 200 kb from the peaks. There was a bias for association of induced transcripts with induced BRD4 density (Fig. 3F and Supplementary Table S5), suggesting a large fraction of the MEK inhibitor-induced transcriptome is regulated by BRD4 commissioning. To a lesser extent MEK inhibitor-repressed transcripts correlated with gain of BRD4 density. This anti-correlation is consistent with the presence of both positive and negative BRD4 transcriptional regulatory paradigms as well as the possibility that induced BRD4 density is regulating more distal genes in addition to the gene most proximal to the density. In the SUM-229PE model of BL and CL breast cancer differential baseline enhancer density in the EpCAM+ or EpCAM− cells correlated with transcription of subpopulation-specific adaptive response kinase genes (Supplementary Fig. S3F). Prominent BRD4 enhancer density was detected at RIPK4 and LIMK2, EpCAM+ specific trametinib-responsive genes, in the EpCAM+ but not EpCAM− cells. Conversely, EpCAM− specific trametinib-responsive genes MAP2K6 and PDK4 displayed prominent BRD4 enhancer density only in the EpCAM− subpopulation, together suggesting that BRD4 enhancer density contributes to defining subtype specificity of the TNBC adaptive transcriptional response.
Dynamics of kinase enhancer formation and blockade by proteasomal or BET bromodomain inhibition
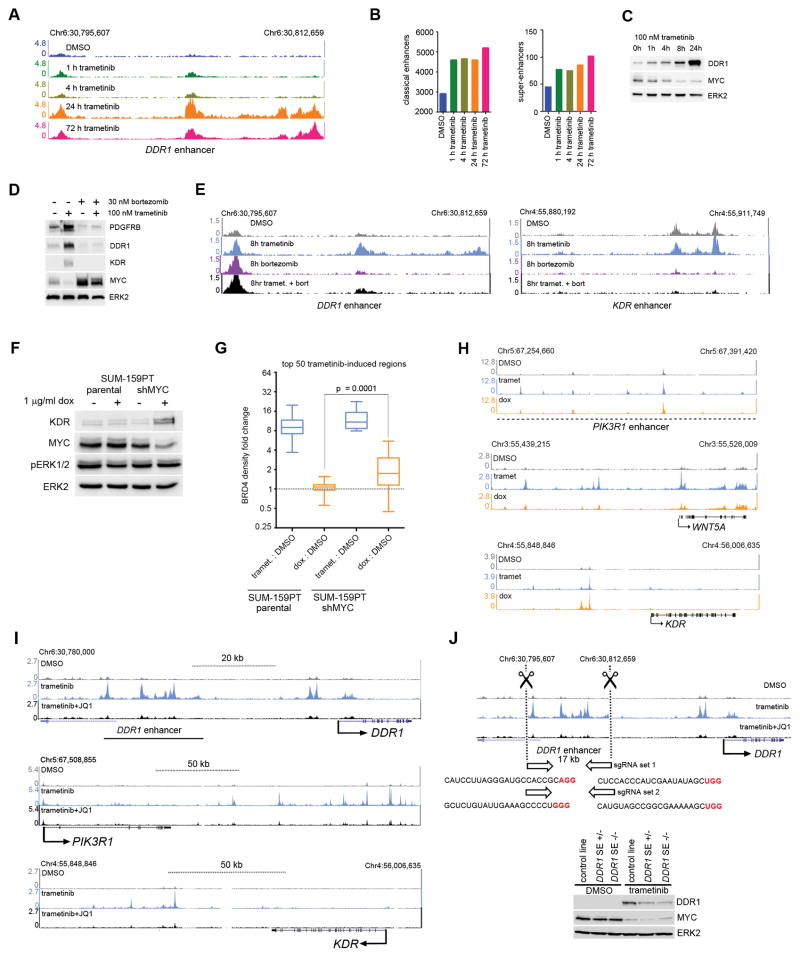
Time course ChIPseq for SUM-159PT cells following trametinib showed rapid BRD4 recruitment within 1–4 h, at which point the majority of de novo enhancers have formed in response to trametinib, and was maximal by 72 h (Figs. 4A and B). Myc protein levels were inversely correlated with the kinetics of DDR1 protein upregulation (Fig. 4C) and BRD4 enhancer density formation upon MEK inhibition (Figs. 4A and B). Proteasome inhibition blocked Myc degradation (19) and expression of adaptive RTKs (Fig. 4D), as well as blocked BRD4 seeding at DDR1 and KDR enhancers (Fig. 4E). More globally, proteasome inhibition significantly attenuated the highest ranking trametinib-induced regions in terms of fold change in BRD4 density (Supplementary Fig. S4A). We sought to corroborate the effects mediated by proteasomal inhibition with Myc loss-of-function studies. An inducible Myc shRNA SUM-159PT cell line displayed upregulation of the adaptive RTK KDR concurrent with Myc protein loss upon doxycycline induction (Fig. 4F). At the 50 highest ranking regions of trametinib-induced BRD4 density, Myc knockdown alone significantly increased BRD4 density following 48 h of doxycycline induction, albeit a partial phenocopy of trametinib in terms of magnitude of BRD4 density change (Fig. 4G). At PIK3R1, WNT5A, KDR adaptive response loci, 48 h of doxycycline induction of Myc shRNA phenocopied the genomic location of BRD4 peak induction by trametinib (Fig. 4H). Thus Myc shRNA clearly alters enhancer regulation that overlaps with MEK inhibition, and combined with proteasomal inhibition effects on Myc stability and RTK adaptive expression, our data are consistent with loss of Myc contributing to enhancer seeding required for the adaptive transcriptome response to MEK inhibition.
Figure 4.
Proteasome or BET bromodomain inhibition attenuates trametinib-induced enhancers at kinase loci. (A) Time course of BRD4 density induced by 100 nM trametinib treatment at the DDR1 enhancer. (B) Classical enhancer (left) or super-enhancer (right) quantification by BRD4 density over 100 nM trametinib time course. (C) Time course of Myc protein levels following trametinib treatment showing anticorrelation of DDR1 protein induction and BRD4 density (A) or enhancer induction (B). (D) Western blot showing Myc stabilization and loss of PDGFRB, DDR1, and KDR upregulation with 8 h co-treatment of 100 nM trametinib and 30 nM bortezomib. (E) Loss of trametinib-induced DDR1 (left) and KDR (right) BRD4 enhancer density upon co-treatment with 30 nM bortezomib. (F) Upregulation of adaptive response RTK KDR upon doxycycline induction of Myc shRNA in stable SUM-159PT cells. (G) BRD4 density change at the highest ranking 50 trametinib-induced regions in response to 48 h 100 nM trametinib or 1 μg/ml doxycycline induction of Myc shRNA. (H) BRD4 density induction following 48 h 100 nM trametinib or 1 μg/ml doxycycline induction of Myc shRNA at PIK3R1, WNT5A or KDR1 adaptive response loci. (I) BRD4 ChIPseq density tracks depicting enhancer formation following 24 h 100 nM trametinib and enhancer blockade following co-treatment with 300 nM JQ1 at the DDR1, PIK3R1, and KDR SUM-159PT adaptive response genes. (J) Top: CRISPR/Cas9 deletion of the SUM-159PT DDR1 trametinib-induced enhancer. Bottom: Attenuation of DDR1 protein induction following 24 h 100 nM trametinib in stable SUM-159PT cell lines either heterozygous or homozygous for the enhancer deletion.
The de novo DDR1 enhancer (Fig. 3A and Fig. 4A, E) was categorized as a super-enhancer by our genome-wide analysis of BRD4 ChIPseq data (Fig. 3C). Genes for the SUM-159PT adaptive response RTK KDR and PI3 kinase regulatory subunit PIK3R1 were also identified as loci harboring MEK inhibitor-induced super-enhancers. BET bromodomain inhibition in each case squelched the induced BRD4 density to near baseline (Fig. 4I). Transcriptional induction by MEK inhibitor and transcriptional suppression by JQ1 correlated with trametinib-induced and JQ1-disrupted BRD4 chromatin occupancy (Supplementary Table S5).
Functional validation of DDR1 induced enhancer density
To assess the function of drug-induced BRD4 enhancer density we deleted the 17 kb putative DDR1 super-enhancer (Fig. 3A and Figs 4A,E,I) by CRISPR-Cas9 (Fig. 4J and Supplementary Fig. S4B) and established clonal SUM-159PT cell lines for the deletion in either a heterozygous or homozygous state. Both the heterozygous and the homozygous deletion strongly attenuated the MEK inhibitor-induced DDR1 upregulation (Fig. 4J), validating the functional relevance of the induced density as an adaptive response enhancer.
BET bromodomain inhibition synergizes with MEK inhibition for growth suppression
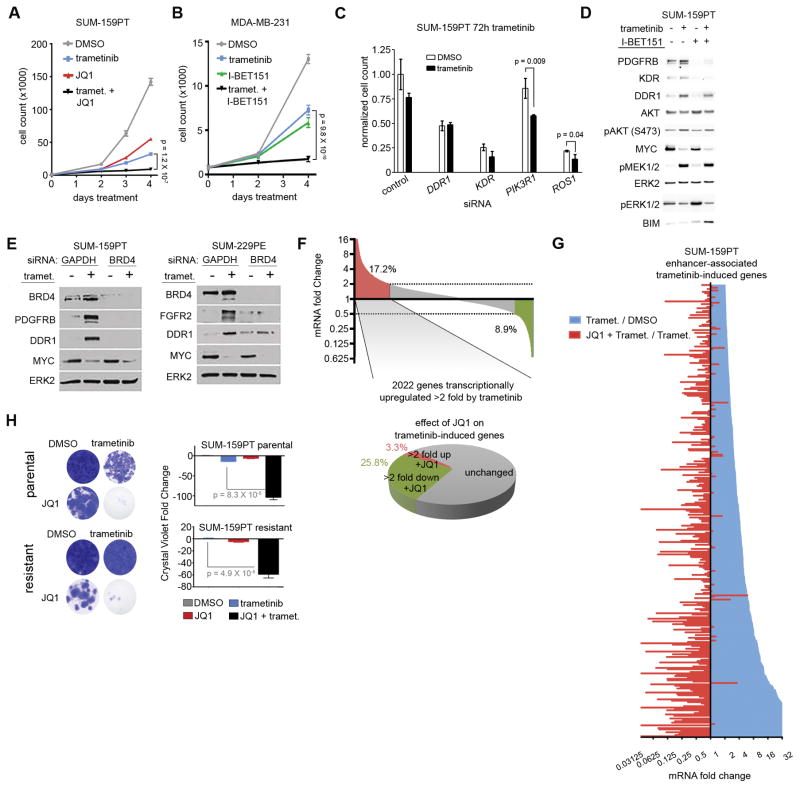
JQ1 and I-BET151, both selective BET bromodomain inhibitors, strongly enhanced trametinib-induced growth inhibition in short-term (4-day) growth assays in claudin-low cell lines SUM-159PT and MDA-MB-231, respectively (Fig. 5A and B). It should be noted growth assays used a lower trametinib dose (30 nM) relative to those employed in our RNAseq studies (100–500 nM) to maximize adaptive responses without eliciting apoptotic signatures resulting from high dose 4 day trametinib treatments. SiRNA knockdown of super-enhancer regulated adaptive response genes DDR1, KDR, PIK3R1, and ROS1 each enhanced growth suppression during 72h trametinib treatment while knockdown of all genes except PIK3R1 yielded significant growth suppression in the absence of drug—establishing a growth promoting role for the proteins whose MEK inhibitor induced super-enhancer is disrupted by JQ1 (Fig. 5C). I-BET151 co-treatment with trametinib blocked the adaptive upregulation of PDGFRB, DDR1, and KDR (Fig. 5D) and the combination increased BIM, indicative of pro-apoptotic priming that does not occur with single agents. SiRNA knockdown of BRD4 phenocopied the BET bromodomain inhibitors, blocking trametinib-induced PDGFRB and DDR1 (SUM-159PT CL cells) and FGFR2 and DDR1 (BL SUM-229PE) (Fig. 5E).
Figure 5.
BET bromodomain inhibition enhances growth suppression elicited by MEK inhibition. (A) SUM-159PT four day growth curve with 30 nM trametinib, 300 nM JQ1, or the combination. (B) MDA-MB-231 (CL) four day growth curve with 30 nM trametinib, 500 nM I-BET151, or the combination. (C) Cell counting assay showing growth suppression in SUM-159PT cells in the presence or absence of 72 h 1 nM trametinib and the indicated siRNAs, normalized to non-targeting control siRNA. In the DMSO condition, all super-enhancer associated siRNAs yielded significantly different (p < 0.05) growth suppression relative to control siRNA except for PIK3R1. P values are indicated for siRNAs that showed significantly different growth suppression between DMSO and trametinib conditions. (D) Westerns with indicated antibodies in SUM-159PT cells treated 24 h with 100 nM trametinib, 500 nM I-BET151, or the combination. (E) Westerns showing loss of adaptive response RTKs in SUM-159PT cells (left) or SUM-229PE parental cells (right) in cells after 48 h 10 nM trametinib and BRD4 siRNA. (F) Top: SUM-159PT RNAseq showing percentage of genes induced (red) or suppressed (green) > 2 fold by 100 nM trametinib. Bottom: Percentage of trametinib-induced genes further induced (red) or suppressed (green) > 2 fold by co-treatment with 300 nM JQ1. (G) Long tail plot of trametinib mRNA induction or JQ1 mRNA suppression (100 nM trametinib : DMSO, or 100 nM trametinib + 300 nM JQ1 : 100 nM trametinib) for enhancer-associated genes with > 2 fold trametinib-induced expression change. (H) Four-week crystal violet assays in SUM-159PT parental cells (top) or SUM-159R (bottom) cells in the presence or absence of 30 nM trametinib or 300 nM JQ1.
Trametinib treatment induced >2000 SUM-159PT transcripts > 2 fold; the JQ1/trametinib combination suppressed 26% of the induced transcripts > 2 fold (Fig. 5F). This fraction of JQ1-suppressed transcripts increased when considering enhancer regulation only, whereby JQ1/trametinib co-treatment nearly exclusively resulted in the suppression of enhancer-associated transcripts induced > 2 fold by trametinib (Fig. 5G). Trametinib + JQ1 synergy was also observed for inhibiting KRas signaling-associated molecules and Myc targets identified as being regulated transcriptome-wide by trametinib (Supplementary Fig. S2B). GSEA indicated enhanced suppression of both KRas signaling and Myc target transcripts upon trametinib + JQ1 treatment relative to trametinib treatment alone (Supplementary Fig. S4C). BET bromodomain and MEK inhibition synergize for growth suppression by attenuating enhancer-regulated adaptive response transcription. In parental cells, co-treatment of trametinib+JQ1 resulted in a durable synergistic growth suppression in long-term (4 week) crystal violet colony formation assays (Fig. 5H). Continual passage of SUM-159PT cells in low-dose trametinib established a trametinib-resistant cell line, SUM-159R, with an IC50 of 18.4 nM relative to the parental IC50 of 1.2 nM (Supplementary Fig. S4D). Addition of JQ1 resulted in the re-sensitization of the resistant cell line to trametinib (Fig. 5H), indicating BRD4 inhibition sensitizes SUM-159R cells to trametinib growth inhibition.
Trametinib and I-BET151 synergize in vivo for tumor growth inhibition
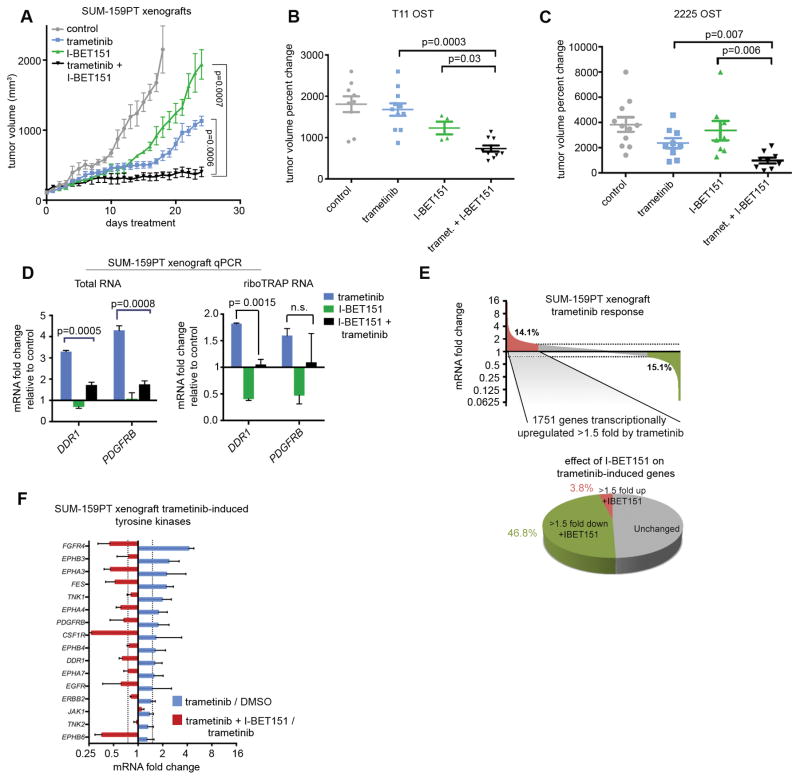
To examine synergistic effects using an in vivo model, we used orthotopic xenografts. Orthotopic SUM-159PT cell xenografts were allowed to grow until tumors reach a volume of 100 mm3 at which point 4 treatment groups were established: vehicle control, trametinib, I-BET151, and trametinib/I-BET151 combination. Mice receiving the combination treatment displayed a pronounced difference in tumor volume relative to single-agents (Fig. 6A). Tumors in the combination treatment group remained static, whereas the single-agent cohorts displayed rapid tumor growth by day 15 of treatment. We also assessed the effects of dual trametinib/I-BET151 treatment in orthotopic syngeneic transplant (OST) murine models of triple-negative breast cancer (38,47). In both T11 (CL) and 2225 (BL) OST tumors the combination treatment significantly inhibited tumor growth relative to single agents (Fig. 6B,C), validating the cell culture growth suppression in vivo in 3 TNBC tumor models.
Figure 6.
MEK inhibition and BET bromodomain inhibition synergy in vivo. (A) Tumor volume in SUM-159PT xenografts: vehicle, 2 mg/kg daily trametinib, 30 mg/kg daily I-BET151, or combination treatment. Percent change in tumor volume from T11 (B) or 2225 (C) orthotopic serial transplant (OST) models following 2 week treatment of 1.0 mg/kg (chow) trametinib, 30 mg/kg (3X weekly, IP) I-BET151, or the combination. Error bars show +/− SEM. (D) Trametinib-induced mRNA upregulation and I-BET151-mediated suppression of DDR1 and PDGFRB as assayed from total RNA (left) or riboTRAP RNA (right) isolated from SUM-159PT xenografts (n=3). Error bars show SD from mean. (E) Top: SUM-159PT xenograft mean (n=3) transcriptome showing percentage of genes induced (red) or suppressed (green) > 1.5 fold by trametinib treatment. Bottom: Percentage of trametinib-induced genes further induced (red) or suppressed (green) > 1.5 fold by co-treatment with 300 nM JQ1. (F) mRNA fold change of SUM-159PT xenograft tyrosine kinases induced > 1.5 fold by trametinib treatment and corresponding JQ1-mediated suppression. Data are mean +/− SD, n=3 tumors.
Suppression of trametinib-induced transcription in vivo by BET bromodomain inhibition
qRT-PCR analysis from xenograft total RNA or from actively translated riboTRAP RNA showed that DDR1 and PDGFRB were induced by trametinib at 48 hours; co-treatment with I-BET151 suppressed induction (Fig. 6D). Loss of expression of G2/M checkpoint genes and MYC targets was observed by GSEA in trametinib-treated xenografts (Supplementary Fig. S4E). Of the 1751 genes transcriptionally upregulated by trametinib in SUM-159PT xenografts, I-BET151 suppressed 47%, with only 4% being upregulated (Fig. 6E). Notably, the trametinib-induced TK family exhibited robust I-BET151-mediated suppression (Fig. 6F).
Depletion of P-TEFb complex associated proteins attenuates the adaptive response to MEK inhibition
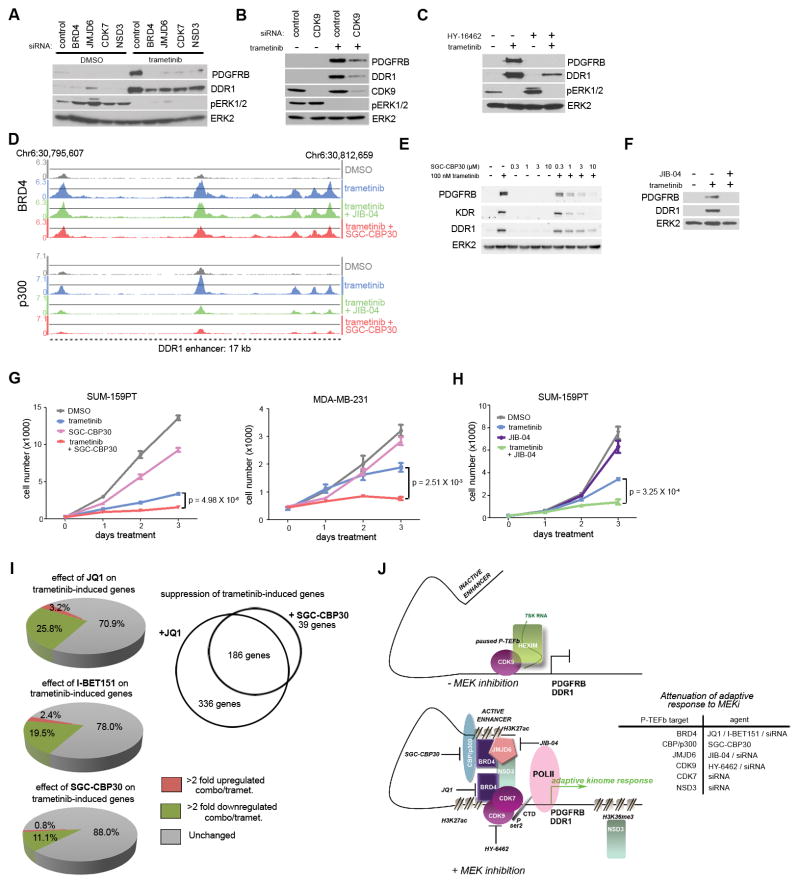
We hypothesized that targeting components of the P-TEFb transcriptional elongation regulatory complex would block the adaptive response. The extra-terminal (ET) domain of BRD4 interacts with NSD3, an H3K36me3 histone methyltransferase, as well as with JMJD6, a Jmj-C family demethylase—both of which are components of P-TEFb-associated transcriptional regulatory complexes (48,49). Knockdown of NSD3 or JMJD6 attenuated the trametinib-mediated upregulation of PDGFRB and DDR1 to the same magnitude as BRD4 siRNA (Fig. 7A). CDK9, comprising the core P-TEFb complex along with cyclin T, and CDK7, are known to phosphorylate BRD4 and the CTD of RNA Pol II for control of transcriptional pause-release (50). CDK7 siRNA (Fig. 7A) and CDK9 siRNA (Fig. 7B) both blocked the induction of PDGFRB and DDR1 by MEK inhibitor. Small molecule inhibitor of CDK9, HY-16462, similarly blocked adaptive PDGFRB and DDR1 upregulation (Fig. 7C). Thus, targeting members of P-TEFb regulatory complexes results in the blockade of adaptation to MEK inhibitor.
Figure 7.
Attenuation of adaptive response to MEK inhibition by P-TEFb complex perturbation. (A) PDGFRB and DDR1 western blot of SUM-159PT cells treated with the indicated P-TEFb complex siRNAs for 48 h followed by 24 h 100 nM trametinib. (B) Western with the indicated antibodies of SUM-159PT cells treated with CDK9 siRNA for 48 h followed by 24 h 100 nM trametinib. (C) Attenuation of 24 h 100 nM trametinib-induced PDGFRB and DDR1 upregulation by co-treatment with 100 nM HY-16462. (D) BRD4 (top) and p300 (bottom) ChIPseq density tracks at the DDR1 enhancer upon 24 h 100 nM trametinib alone, or in combination with either 300 nM JIB-04 or 1 μM SGC-CBP30. (E) Dose-dependent blockade of PDGFRB, KDR, and DDR1 upregulation to 24 h 100 nM trametinib by SGC-CBP30 co-treatment in SUM-159PT cells. (F) Loss of PDGFRB and DDR1 upregulation to 100 nM 24 h trametinib by co-treatment with 300 nM JIB-04 in SUM-159PT cells. (G) Enhancement of SUM-159PT (left) or MDA-MB-231 (right) growth suppression with 10 nM trametinib treatment with 5 μM SGC-CBP30. (H) Enhancement of SUM-159PT growth suppression with 5 nM trametinib and 500 nM JIB-04. (I) Left: Percentage of SUM-159PT transcripts further upregulated > 2 fold (red) or downregulated > 2 fold (green) with either 300 nM JQ1, 500 nM I-BET151, or 3 μM SGC-CBP30 in combination with 24 h 100 nM trametinib. Right: Trametinib-induced genes commonly suppressed by JQ1 and SGC-CBP30. (J) Model of dynamic enhancer formation in the adaptive response to MEK inhibition (left) and targeting strategies for different P-TEFb complex members to attenuate the response (right).
Pharmacological p300 or jumonji-C demethylase inhibition disrupts enhancer composition and increases MEKi-mediated growth suppression
As an established member of P-TEFb regulatory complexes, we predicted CBP/p300 lysine acetyl transferases (KATs) are primary H3K27 KATs of the MEK inhibitor adaptive response to MEK inhibitor. CBP30 is a small molecule inhibitor of the CBP/p300 bromodomains with 40-fold selectivity over the tandem bromodomains of BRD2/3/4 (51). We performed BRD4 and p300 ChIPseq in SUM-159PT cells in the presence or absence of trametinib or CBP30 to assess the consequences of CBP/p300 inhibition on MEK inhibitor adaptation. Co-treatment of CBP30 and trametinib reduced MEK inhibitor-induced density of p300 to near baseline at the CRISPR-Cas9 functionally-validated DDR1 super-enhancer (Fig. 7D). In contrast, MEK inhibitor-induced BRD4 density was only modestly reduced. Assessing p300 chromatin occupancy at the 50 top-ranking trametinib-induced BRD4 peaks (in addition to the DDR1 super-enhancer) revealed similar enhanced CBP30 suppression of induced p300 relative to BRD4 density (Supplementary Fig. S4F). CBP30 co-treatment with trametinib was sufficient to block adaptive RTK upregulation of PDGFRB, KDR, and DDR1 protein levels (Fig. 7E), showing that CBP/p300 inhibition can block the adaptive response by loss of p300 from induced transcriptional regulatory complexes.
JIB-04 is a pan jumonji-C family demethylase inhibitor that prolonged survival in a murine model of breast cancer (52). JIB-04 + trametinib, like CBP30 + trametinib, depleted induced p300 but not induced BRD4 density from the DDR1 super-enhancer and at the highest ranking regions of trametinib-induced BRD4 chromatin occupancy (Fig. 7D and Supplementary Fig. S4F). JIB-04, like other inhibitors of P-TEFb complex members JQ1/I-BET151, HY-16462, and CBP30, blocked trametinib-induced upregulation of PDGFRB and DDR1 (Fig. 7F).
CBP30 co-treatment with trametinib showed strong enhanced growth suppression in SUM-159PT and MDA-MB-231 cells relative to single-agents (Fig. 7G). JIB-04/trametinib co-treatment also enhanced growth suppression in SUM-159PT cells relative to single agents (Fig. 7H). Transcriptomically, CBP30 was capable of attenuating 11% of trametinib-induced SUM-159PT transcripts compared to the ability of JQ1 to suppress 26%. CBP30 suppressed 186 transcripts in common with JQ1 (Fig. 7I), which defines a cohort of critical transcripts regulated by BRD4/p300 whose loss reverses adaptive reprogramming and inhibits cell growth.
DISCUSSION
Our studies demonstrate TNBC patients treated with trametinib for seven days resulted in their tumors having an inhibition of MEK-ERK and a robust transcriptional response that included a significant reprogramming of the tyrosine kinome. Although our patient number is low due to the inherent difficulty in doing such window trials, our study serves as proof-of-concept that the adaptive bypass response occurs in patients. A similar transcriptional response and reprogramming of the tyrosine kinome is seen in TNBC cell lines and mouse xenografts. TNBC has been characterized to have two primary subtypes, basal-like and claudin-low (8,31,41), based on their differing transcriptional profiles in patient tumors that is also seen in cell lines derived from human tumors and mouse models of TNBC. We determined basal-like and claudin-low human TNBC cells and mouse tumor subtypes have different adaptive transcriptional responses to MEK-ERK inhibition. The basal (EpCAM+) and claudin-low (EpCAM−) SUM-229PE subpopulations are isogenically similar and do not have significant differences in DNA methylation, copy number or exonic nucleotide variants, indicating that chromatin regulation independent of baseline DNA methylation is responsible for subtype-specific gene expression signatures. In SUM-229PE subpopulations inhibition of MEK-ERK led to differential subtype-specific genome-wide enhancer formation and enhancer/promoter remodeling driving the basal versus claudin-low adaptive transcriptional response.
We previously showed that MEK inhibition upregulated PDGFRB, KDR and PDGFB in SUM-159PT cells (9). This could be blocked by siRNA to Myc, proteasome inhibitor or mutation of Myc Thr58 to Ala to suppress Myc degradation. Our current study indicates that Myc turnover contributes to the modulation of the adaptive response to MEK inhibition by stimulating dynamic enhancer formation and remodeling. There has been increasing evidence for stimulation-dependent enhancer dynamics. For example, de novo enhancers are formed in macrophages in response to LPS activation of the toll receptor 4 (23,24), and proinflammatory TNFα stimulation drives the formation and reorganization of enhancers rich in NFκB and BRD4 (25). Endocrine responses in estrogen receptor (ER) positive breast cancer are dependent on the transcription factor FOXA1, as it regulates ER/enhancer interactions and transcriptional activity (26). In the context of oncogenic kinase signaling, perturbation of ERK signaling by Sprouty deletion or G12V HRas expression remodel histone modifications at both super-enhancers and classical enhancers by distinct mechanisms (27). Looping of the MET enhancer leading to the binding of the transcriptional start site in a 3C assay was shown to be regulated in response to BRaf inhibition (28). These findings together with our current study demonstrate the integration of signaling networks with the epigenetic control of transcription that identifies potential targets for pharmacological intervention.
P-TEFb is recruited by BRD4 to promoters and associates with Mediator complex members to facilitate chromatin looping of enhancer sequences to promoters for pause-release and transcriptional elongation (53). Myc turnover is required for enrichment of histone acetylation and recruitment of P-TEFb/CDK9 to promoters (19). We reasoned that targeting P-TEFb complex associated proteins would block the transcriptional induction of the adaptive bypass response both by disruption of induced or remodeled enhancer complex architecture, and by preventing promoter recruitment and function of core P-TEFb components. Using RNAi and small molecule inhibitors we found that targeting P-TEFb associated proteins JMJD6, NSD3, p300, CDK7 or CDK9 as well as BRD4 inhibited the transcriptional upregulation of the adaptive bypass response. Importantly, cells that had become resistant to trametinib were made sensitive to the drug by JQ1 inhibition of BRD4, effectively reversing resistance to trametinib by suppressing the adaptive upregulation of RTKs. Combination trametinib plus I-BET151 gave synergistic growth inhibition in vitro and in vivo for different TNBC mouse models. In lapatinib-resistant HER2+ breast cancer cells we observed a similar result where we could reverse resistance to a targeted kinase inhibitor with a BET bromodomain inhibitor (34). Cumulatively, our studies define P-TEFb complex associated proteins as validated targets to block adaptive resistance produced by MEK inhibition.
The importance of blocking adaptive bypass resistance to kinase inhibitors at its epigenetic root has significant clinical implications for making therapeutic responses more durable. The relevance of such an approach is evident in our TNBC studies. If the adaptive response to single kinase inhibitors such as trametinib could be blocked epigenetically, by targeting enhancer formation/remodeling by inhibiting P-TEFb constituents such as BRD4, p300, JMJD6, CDK7 or CDK9 adaptive resistance could be prevented and our results suggest resistance could be possibly reversed. Rather than pursuing kinase inhibitor combinations that will invariably lead to adaptive bypass, combining a kinase inhibitor with an epigenetic inhibitor is a novel approach to arrest adaptive reprogramming. Pharmacological targeting of P-TEFb complex associated proteins, localized at de novo enhancer/promoters seeded by MEK inhibition, is an effective strategy to durably inhibit adaptive resistance to MEK inhibition (Fig. 7J).
METHODS
Window-of-opportunity clinical trial
The window trial “Defining the Triple Negative Breast Cancer Kinome Response to GSK1120212” is registered under the ClinicalTrials.gov Identifier NCT01467310. GlaxoSmithKline and Novartis generously provided trametinib for the window trial. Eligible women included those with stage I–IV newly diagnosed and previously untreated triple negative breast cancer that was accessible for biopsy and surgery; stage I–IIIc subjects could not be candidates for therapeutic neoadjuvant treatment. Triple negative status was based upon the clinical assays and defined by ASCO/CAP criteria including estrogen receptor (ER) and progesterone receptor (PR) <1% staining by immunohistochemistry (IHC) and HER2-negative by IHC or fluorescence in situ hybridization. Study subjects provided written informed consent that included details of the nontherapeutic nature of the trial, and the study was approved by the UNC Office of Human Research Ethics and conducted in accordance with the Declaration of Helsinki. After enrollment, study subjects underwent core biopsy of the breast tumor, and then received trametinib for the 7 consecutive days prior to the scheduled surgery date. The last dose of trametinib was taken ≤ 24 hours before surgery. At surgery a post trametinib tumor specimen was reserved for research. Patients were monitored for toxicity during and up to several weeks after treatment, until any evidence of toxicity had resolved. The dosing schedule of trametinib was determined by the surgery date; delays in standard therapy for trial purposes were not permitted. Given the nontherapeutic nature of the trial, the dose was deliberately set low at 1.5 mg orally daily under fasting conditions; this dose was increased per protocol to 2 mg daily after interim analysis of pharmacodynamic endpoints in the pre- and post-treatment tumor samples from, and toxicity assessment of, the first 3 patients enrolled.
Biopsy and surgical specimens were immediately placed into liquid nitrogen. Both pre- and post- trametinib tumor tissue was analyzed for baseline kinome profile and for the dynamic effects of MEK inhibition on the whole kinome. Frozen tissue intrinsic subtyping was performed by gene expression profiling using Agilent DNA microarrays and the PAM50 algorithm (54); claudin-low subtyping used a centroid-based predictor (41).
Cell culture
SUM-159PT and MDA-MB-231 cells were maintained in DMEM/F12 medium (Gibco, ThermoFisher Scientific) supplemented with 5% FBS, 5 μg/ml insulin, 1 μg/ml hydrocortisone, and antibiotic:antimycotic cocktail (Gemini Bio Products). HCC1806, MDA-MB-468, WHIM12, Hs 578T, T11, and T2 C3(1)/Tag cells were maintained in RPMI 1640 (Gibco, ThermoFisher Scientific) supplemented with 10% FBS and antibiotic:antimycotic cocktail (Gemini Bio Products). SUM-149PT EpCAM+ cells were maintained in HuMEC medium (with defined HuMEC supplements, Gibco, ThermoFisher Scientific) supplemented with 5% FBS and antibiotic:antimycotic cocktail (Gemini Bio Products). SUM-229PE parental cells and FACS subpopulations were maintained in F12 medium (Gibco, ThermoFisher Scientific) supplemented with 5% FBS, 5 μg/ml insulin, 1 μg/ml hydrocortisone, 10 mM HEPES, and penicillin/streptomycin cocktail (Gibco, ThermoFisher Scientific).
Trametinib-resistant SUM-159PT (SUM-159R) cells were established by pooling all resistant subclones following continual passage in 30 nM trametinib for > 1 month. Doxycycline-inducible MYC shRNA SUM-159PT cells were created by pooling all resistant subclones following 2.5 μg/ml puromycin selection for pLKO.1 tet-on driven shRNA TRCN0000327647.
Cell line authentication
Cell lines were obtained from the UNC Lineberger Comprehensive Cancer Center Tissue Culture Facility or collaborating labs within the last five years. All established cell lines used in these studies have been authenticated by the Johns Hopkins Genetics Core Resources Facility using their short tandem repeat (STR) profiling service. New cell lines generated from PDXs are whole exome sequenced and RNA sequenced for reference. Cells are routinely checked for mycoplasma.
Compounds
Trametinib, JQ1, I-BET151, SGC-CBP30, JIB-04, and bortezomib were obtained from Selleck Chemicals. HY-16462 was obtained from MedChem Express.
Antibodies
ChIP
The following ChIP-grade antibodies were used: BRD4 (Bethyl Laboratories A301-985A), CEBPβ (Santa Cruz Biotechnology sc-150 X), histone H3K27ac (Active Motif 39133), histone H3K4me1 (Active Motif 39297), histone H3K4me3 (EMD Millipore 07-473), MED1/CRSP1/TRAP220 (Bethyl Laboratories A300-793A), p300 (Santa Cruz Biotechnology sc-585 X).
Western blotting
Abcam: EpCAM. Bethyl Laboratories: BRD4. Cell Signaling Technology: AKT, BIM, CDK9, DDR1, IGF1R, KDR, KIT, MEK1/2, MYC, pAKT (S473), pAKT (T308), PDGFRB, pERK1/2 (T202,Y204), pSRC (Y416), vimentin. Santa Cruz Biotechnology: ERK2, FGFR2.
RNAseq
Cell lines
For SUM-229PE and T11 OST, C3(1)Tag - T2 datasets
2 μg total RNA isolated using Qiagen RNeasy Plus kit was used for library construction by the UNC Lineberger Comprehensive Cancer Center Genomics Core using Illumina TruSeq RNA Library Prep Kit v2 with 15 cycles of amplification. 1×50 Illumina HiSeq2000 sequencing was performed by the UNC High Throughput Sequencing Facility.
For all other RNAseq datasets
4 μg total RNA isolated using Qiagen RNeasy Plus kit was used for library construction with KAPA Stranded mRNAseq kits and Illumina TruSeq indexed adapters following the manufacturer’s recommended protocol with the following exception: 10 cycles of PCR were used with 0.5 X the recommended template DNA. 12-plex, single-indexed, 1×75 bp Illumina NextSeq500 sequencing was employed for RNAseq libraries to yield an average of 3.5–4.0 × 107 reads per sample.
Patient tumors
Total RNA was isolated with Qiagen RNeasy Mini kit. mRNAseq libraries were made with Illumina TruSeq RNA Sample Prep Kit with 0.5–1 μg of total RNA. Libraries were sequenced using an Illumina HiSeq2000, producing 48×7×48 bp paired-end reads with multiplexing.
Cell line raw and processed RNAseq is deposited in GEO as SuperSeries GSE87424. Patient tumor RNAseq data deposition to dbGaP is in progress.
ChIPseq
Chromatin immunoprecipitation
Formaldehyde fixation and chromatin immunoprecipitations were performed as described for BRD4 (46) with the following modifications:. ~ 1 × 107 cells were used per IP, a Bioruptor Pico (Diagenode) chilled water bath sonicator was used for chromatin shearing (15 cycles: 30 sec pulse, 30 sec cooling), and Qiagen MinElute PCR Purification columns were used for ChIP DNA purification after decrosslinking and RNase/proteinase treatment. Buffers: Lysis buffer 1: 50 mM HEPES pH 7.3, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, and 0.25% Triton X-100; Lysis buffer 2: 10 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM EDTA pH 8.0 and 0.5 mM EGTA pH 8.0; LiCl wash buffer: 20 mM Tris pH 8.0, 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate.
Amount of antibody per IP: 10 μg BRD4, 5 μg H3K27ac, 10 μg CEBPB, 10 μg p300, 10 μg MED1, 10 μl H3K4me1, 4 μg histone H3K4me3 (EMD Millipore 07-473).
Library preparation and sequencing
10–50 ng ChIP DNA was used for library construction using KAPA HyperPrep kit and Illumina TruSeq indexed adapters. Dual size selection was performed after 18 cycles of PCR amplification according to KAPA’s recommended protocol. 12-plex, single-indexed, 1×75 bp Illumina NextSeq500 sequencing was employed for ChIPseq libraries to yield an average of 3.5–4.0 × 107 reads per sample.
Library preparation and sequencing for H3K4me3 ChIPseq was performed by the UNC High Throughput Sequencing Facility. H3K4me1 ChIPseq libraries were constructed using DNA SMART ChIP Seq kit (Clontech) with 10 ng ChIP DNA, 18 cycles of amplification, and double size selection post-PCR following the manufacturer’s suggested protocol. For the DNA SMART ChIPseq libraries, the first three bases of the sequencing read, corresponding to the template switching oligo, were trimmed prior to mapping.
Raw and processed ChIPseq data is deposited in GEO as SuperSeries GSE87424. Python code generated in the laboratory for ChIPseq analysis is available at GitHub (55).
SUM-229PE subpopulation cell sorting
SUM-229PE cells were trypsinized and resuspended in Hank’s Balanced Salt Solution containing 2% fetal bovine serum (HF media). For analytical flow cytometry, cells were fixed with 3% paraformaldehyde. Cells were stained for 30 minutes at 4°C with the fluorescently-labeled primary antibodies EpCAM-FITC (Stem Cell Technologies) and CD49f-PE-Cy5 (BD Biosciences), washed twice with HF media and filtered with a 30 μm filter. Fixed cells were analyzed using a Beckman-Coulter CyAn Cytometer, and live cells were sorted using a Sony iCyt Reflection Cytometer. Sorting was analyzed with FlowJo v7.6.5 software.
SUM-229PE subpopulation whole exome sequencing
Genomic DNA was isolated from EpCAM+/Cd49f+ and EpCAM−/Cd49f− FACS populations using Qiagen DNeasy Blood and Tissue Kit and subsequently treated with RNase A (Sigma). 300 ng genomic DNA was submitted to the UNC High Throughput Sequencing Core Facility for Nextera Rapid Capture exome enrichment (Illumina) followed by 2X100 HighSeq2500 sequencing.
Exome sequencing data is deposited in GEO as SuperSeries GSE87424.
siRNA transfection
siGENOME SMARTpools (GE Dharmacon) were transfected using Dharmafect 1 reagent (GE Dharmacon) at 25 nM final concentration. Unless otherwise noted, siGENOME Non-Targeting siRNA pool #2 was used as a negative control. Cells were incubated 48 h post-siRNA transfection to ensure knockdown prior to the addition of drug, except for the cell counting analysis shown in Fig. 5C whereby drug was added concurrently with transfection
Cell lysis – western blotting
Cells were harvested in RTK array lysis buffer containing 20 mM Tris-HCl (pH 8.0), 1% NP-40, 10% glycerol, 137 mM NaCl, 2 mM EDTA, 1X EDTA-free protease inhibitor cocktail (Roche), and 1% each of phosphatase inhibitor cocktails 1 and 2 (Sigma).
CRISPR/Cas9 enhancer deletion
CRISPR/Cas9 sgRNAs were identified using the MIT CRISPR Design tool. Two sets of sgRNAS where chosen; sgRNA Set 1 was cloned into lentiCRISPRv2 (Addgene #52961) and the sgRNA Set 2 was cloned into pSpCas9(BB)-2A-GFP (Addgene #48138). SUM-159PT cells were first infected with lentiCRISPRv2-DDR1-Set 1. Single cells were sorted into 96-well plates and selected with 2.5 μg/ml puromycin. sgRNA Set 2 was delivered to SUM-159PT-DDR1-SE+/− cells by electroporation using the Neon (ThermoFisher Scientific) electroporation system. Cells expressing GFP were sorted in 96-well plates and tested for biallelic deletion. To detect monoallelic and biallelic deletion of the DDR1 super-enhancer, genomic DNA was extracted using QIAamp DNA extraction kit (Qiagen) and then used as template for PCR with Expand High-Fidelity DNA polymerase (Roche) with the primers: DDR15FV2 (TGAGTCAGAACCCAACAGGC), DDR15RV2 (ATTGCAAAGGAGGCACCACT), DDR13FV2 (GCAAGGAAGACAGCTCACCT) and DDR13RV2 (GGCTCTTAGACTTGGGCCAG). PCR products were gel purified with Qiagen QIAquick PCR purification kit prior to sequencing.
Cell line growth assays
96 h and 8-day growth assays were performed in 96-well plates. Cells were plated one day prior to first treatment. Media containing fresh drug was changed every 24 h unless otherwise noted. Live cells were stained with Hoescht in PBS for 20 min at 37°C and imaged/counted with a Thermo Cellomics ArrayScan VTI at 25 frames per well.
Crystal violet colony formation assays
Crystal violet assays were performed in 6-well plates, with three technical replicates per condition. Drug-containing media was changed every 3 days for 4 weeks. Cells were rinsed with PBS, fixed in methanol (10min, −20°C), and stained with 0.5% crystal violet for 20min. Crystal violet was solubilized with 30% acetic acid and quantified by absorbance at 600nm.
In vivo tumorigenesis experiments
SUM-159PT xenografts
Female NOD/SCID mice (Jackson Labs) were given orthotopic mammary fat pad injections of 2×106 SUM-159PT cells suspended in 50% matrigel. Mice were housed and treated in accordance with protocols approved by the Institutional Care and Use Committee for animal research at the University of North Carolina. Once tumor volume reached approximately 100 mm3 mice were treated daily with 2.0 mg/kg trametinib by oral gavage (vehicle: 0.5% hydroxypropylmethylcellulose, 0.2% tween 80 in diH2O) or 30mg/kg I-BET151 by IP injection (vehicle: 5% tween 80, 5% DMSO in saline) as single agents or in combination. Tumor volume was calculated daily by caliper measurements ((width)2 × length))/2 until tumors reached maximum size of 2,000 mm3 or at the end of treatment. Tumors used for long-term growth study were snap-frozen in liquid nitrogen and stored at −80°C. Tumors used for riboTRAP and RNA sequencing were treated 48 hours and fresh tissue was harvested for downstream analysis. Tumor number for Fig. 6A: vehicle: n=5, trametinib: n=6, I-BET151: n=4, trametinib+I-BET151 combination: n=3.
OST models
BALB/c females (Jackson Labs strain 000651) aged 6–8 weeks old were inoculated in the mammary gland with 5X105 cells of “T11”or “2225”, both p53 null cell lines described previously (38,47). Mice were housed and treated in accordance with protocols approved by the Institutional Care and Use Committee for animal research at the University of North Carolina. Once inoculated, mice were examined for tumors weekly until a palpable mass was found. Treatment began the same day. Tumor size was assessed once weekly by caliper measurements of tumor areas ((width)2 × length))/2 until reaching tumor burden. Percent change of tumor volume was calculated using (Final volume – Initial Volume)/Initial Volume. Trametinib was incorporated into the diet (Research Diets, New Brunswick, NJ) of mice to achieve a daily dose of 1.0 mpk and I-BET151 was delivered IP at 30 mpk thrice weekly. Food was provided ab libitum and the amount of daily food intake was pre-determined using Jackson Labs Phenome Database. Tumors at harvest were cut in half and either snap-frozen in liquid nitrogen and stored at −80°C or placed in neutral buffered 10% formalin solution.
riboTRAP analysis of SUM-159PT xenograft tumors
Translating ribosome affinity purification (riboTRAP) was performed on fresh tissue from SUM-159PT xenograft tumors harvested in ice-cold dissection buffer (1X HBSS, 2.5 mM HEPES-KOH (pH7.3), 35 mM glucose, 4 mM NaHCO3, 100 μg/ml cycloheximide). SUM-159PT cells were infected with lentivirus to express L10a-GFP (construct kindly provided by Marc Caron). Tumors were processed as described (56) with minor modifications. Briefly, fresh tissue was homogenized in 15-second pulses using a tissue-tearor homogenizer at approximately 15,000 rpm on ice in lysis buffer (20mM HEPES-KOH (pH 7.3), 150 mM KCl, 10 mM MgCl2, 1% NP-40, Roche EDTA-free protease inhibitor tablet (1 per 10 ml), 0.5mM DTT, 100 μg/ml cycloheximide). Lysate was centrifuged at 4.7 rpm for 10 minutes at 4°C. Supernatant was treated with 1/9 volume of 300mM DHPC and 10% NP-40 and incubated on ice 5 minutes before centrifugation for 10 minutes at max speed. The resulting supernatant was used for immediate IP overnight at 4°C with GFP antibodies (Memorial Sloan Kettering Monoclonal Antibody Facility; clone names: Htz-GFP-19C8 and Htz-GFP-19F7) bound to 200 μl protein G magnetic beads. A small aliquot of total supernatant was saved at this point for comparison. After IP samples were washed four times with high salt buffer (20mM HEPES-KOH (pH 7.3), 350 mM KCl, 10 mM MgCl2, 1% NP-40, Roche EDTA-free protease inhibitor tablet (1 per 10 ml), 0.5mM DTT, 100 μg/ml cycloheximide). RNA was eluted from the beads in 100 μl Qiagen RNeasy lysis buffer with β-mercaptoethanol added and RNA was purified for downstream analysis according to the manufacturer’s instructions.
SUM-159PT xenograft qPCR
Total RNA was isolated from fresh SUM-159PT xenograft tumors using Qiagen RNeasy Plus kit. First strand cDNA was synthesized from 2.0 μg total RNA or riboTRAP RNA (see riboTRAP analysis of SUM-159PT xenograft tumors) using High-Capacity cDNA Reverse Transcription kit (Applied Biosystems/ThermoFisher Scientific) according to the manufacturer’s recommended protocol. The following TaqMan (Applied Biosystems/ThermoFisher Scientific) primers were used: DDR1: Hs01058430_m1, PDGFRB: Hs01019589_m1, ERK2: Hs_01046830_m1.
MIB/MS
Tumor tissue and cell lines were processed in lysis buffer (50 mM HEPES, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.5% Triton X-100, at pH 7.5 containing inhibitors [10 mM NaF, 2.5 mM NaVO4, Sigma phosphatase inhibitor cocktails 2+3, and Roche protease inhibitor tablets]) and gravity-flowed over MIBs (multiplexed kinase inhibitor beads; Sepharose conjugated to VI-16832, CTx-0294885, PP58, Purvalanol B, UNC8088A, UNC21474). MIBs were sequentially washed with lysis buffer without inhibitors containing 1M NaCl, then 150 mM NaCl+0.1% SDS. Bound kinases were eluted by boiling in 100 mM Tris-HCl, 0.5% SDS, and 1% beta-mercaptoethanol, pH 6.8, then treated with 5mM DTT at 60°C and 15 mM iodoacetamide at RT. Proteins were concentrated on Amicon Ultra-4 (10K cutoff) spin columns, purified by methanol/chloroform extraction, then trypsinized overnight in 50 mM HEPES, pH 8. Triton was removed by extraction with hydrated ethyl acetate and peptides were de-salted by C-18 spin column (Pierce, ThermoFisher Scientific).
For patient biopsies and tumor tissue, 50% of the peptides were loaded onto a Thermo Easy-Spray 75μm × 25cm C-18 column with an Easy nLC-1000. Peptides were separated on a 300-minute (5–40% ACN) gradient as a single fraction and identified by a Thermo Q-Exactive orbitrap mass spectrometer. Parameters are as follows: 3e6 AGC MS1, 80 ms MS1 max inject time, 1e5 AGC MS2, 100 ms MS2 max inject time, 20 loop count, 1.8 m/z isolation window, 45 s dynamic exclusion.
Spectral data was searched against the Uniprot/Swiss-prot protein database using MaxQuant 1.5. Default parameters were used with the following exceptions: a minimum of 2 unique peptides was required for quantitation, razor peptides were excluded, peptide matching between runs was included, and peptides containing phospho-STY, acetylation, oxidation, and deamidation modifications were included. Label-free quantification (LFQ) values for each kinase were used for comparison between pre/post-treatment.
DNA methylation
An Illumina Infinium HumanMethylation450 BeadChip array was employed to assess cytosine methylation at baseline and upon trametinib treatment. Genomic DNA was isolated using Qiagen DNeasy Blood & Tissue kit from SUM-159PT or SUM-149PT EpCAM+/− cells treated with either 24 h 100 nM trametinib or DMSO vehicle control or SUM-229PE EpCAM+/− cells treated with either 24 h 30 nM trametinib or DMSO vehicle. Bisulfite conversion of genomic DNA was performed using EZ DNA Methylation kit (Zymo Research) following the manufacturer’s recommended conditions. Hyper-methylated and unmethylated genomic DNA (Human WGA Methylated & Non-methylated DNA kit, Zymo Research) were included on the array as controls. BeadChip hybridization and imaging was performed by the UNC Mammalian Genotyping Core. See Supplementary Table S4 for normalized (to Illumina internal controls) methylation fraction (β) values output from GenomeStudio (Illumina). Principal component analysis was performed using R version 3.2.3 on the normalized beta values.
DNA methylation data is deposited in GEO as SuperSeries GSE87424.
For analysis of RNAseq, ChIPseq, and exome sequencing datasets and statistical analyses, see Supplementary Methods.
Supplementary Material
Kinases significant (FDR 0.05) by DESeq2 differential expression analysis comparing basal-like cell line (HCC1806, SUM-149PT EpCAM+, MDA-MB-468) and claudin-low cell line (SUM-159PT, Hs 578T, WHIM12) group response to 24h 500 nM trametinib treatment. (Excel spreadsheet) Listed is mean log2 fold expression change of biological duplicates for the indicated basal-like (blue) or claudin-low (red) cell lines. Kinases are ordered by their clustering in the heatmap presented in Fig. 2C.
Upregulated kinases significant (FDR 0.05) by DESeq2 differential expression analysis of basal-like cell line (HCC1806, SUM-149PT EpCAM+, MDA-MB-468) or claudin-low cell line (SUM-159PT, Hs 578T, WHIM12) subtypes in isolation. (Excel spreadsheet) Basal-like unique kinases (blue), claudin-low unique (red) or significant kinases common to both subtypes (grey) are indicated along with adjusted p value. Kinases overlapping with patient tumor upregulated kinases (Fig. 2D) are indicated with an asterisk.
Nucleotide variant analysis comparing whole exome sequencing of SUM-229PE subpopulations. (Excel spreadsheet) The “common” tab of the spreadsheet contains nucleotide variants common between the EpCAM+ and EpCAM− SUM-229PE subpopulations, while the “unique” tab of the spreadsheet contains those variants unique to one subpopulation. Q score, Minor Allele Frequency (MAF) and COSMIC database overlap are indicated for each variant.
Cell line methylation fraction (β) values. (Excel spreadsheet) The spreadsheet shows the normalized average methylation fraction β for each Illumina Infinium 450K BeadChip CpG locus for the indicated cell lines upon 24 h DMSO vehicle or 30 nM (SUM-229PE EpCAM+/−, SUM-149PT EpCAM+/−) or 100 nM (SUM-159PT) trametinib treatment. “Methcontrol” column indicates hyper-methylated and “unmethcontrol” column indicates unmethylated genomic DNA controls (see Supplementary Methods).
SUM-159PT Transcriptional correlation with trametinib-induced BRD4 ChIPseq density. (Excel spreadsheet) The spreadsheet contains the merge of SUM-159PT BRD4 stitched ChIPseq peaks and RNAseq data of peak-proximal genes +/− 24 h 100 nM trametinib (see Supplementary Methods). ChIPseq density units are reads per million mapped reads (rpm) and RNAseq data are normalized RSEM transcript abundance estimates. The spreadsheet contains unfiltered ChIPseq data prior to density (rpm) cutoffs and manipulations performed for Fig. 3F.
SUM-159PT ChIPseq data and genic categorization. (Excel spreadsheet) ChIPseq density values for union peaks in reads per million mapped reads (rpm) for the indicated antibodies +/− 24 h trametinib at the indicated doses or +/− 24 h 300 nM JQ1. The “enhancer” spreadsheet classification indicates a putative 5′ enhancer while “3 prime” indicates a putative 3′ enhancer. See Supplementary Methods for union peak, genic category, and “nearby” gene definitions. Spreadsheet contains unfiltered data prior to manipulations for figure construction as described in Supplementary Methods.
HCC1806 ChIPseq data and genic categorization. (Excel spreadsheet) BRD4 ChIPseq density values for union peaks (see Supplementary Methods) in reads per million mapped reads (rpm). The “enhancer” spreadsheet classification indicates a putative 5′ enhancer while “3 prime” indicates a putative 3′ enhancer. See Supplementary Methods for union peak, genic category, and “nearby” gene definitions. Spreadsheet contains unfiltered data prior to manipulations for figure construction as described in Supplementary Methods.
STATEMENT OF SIGNIFICANCE.
Widespread transcriptional adaptation to pharmacological MEK inhibition was observed in triple negative breast cancer patient tumors. In preclinical models MEK inhibition induces dramatic genome-wide modulation of chromatin, in the form of de novo enhancer formation and enhancer remodeling. Pharmacological targeting of P-TEFb complex members at enhancers is an effective strategy to durably inhibit such adaptation.
Acknowledgments
FINANCIAL SUPPORT: Funded by CA058223 (GLJ, HSE, CMP, LAC), Susan G. Komen Foundation IIR12-225201 (GLJ, LAC, HSE), GM101141 (GLJ), GM116534 (SMB), Komen Foundation PDF15331014 (SHV), UNC Junior Faculty Development Award (JSZ), University Research Council Small Grant Award (JSZ), University Cancer Research Fund (TJS, GLJ).
Footnotes
CONFLICT OF INTEREST STATEMENT: GLJ and LMG are co-founders of KinoDyn. HSE is a co-founder of Meryx. CMP is an equity stock holder and Board of Directors Member of BioClassifier LLC and University Genomics and is an inventor on a patent application on the PAM50 molecular assay. No potential conflicts of interest were disclosed by the other authors.
AUTHORS’ CONTRIBUTIONS
Conceptualization, JSZ, GLJ; Methodology, JSZ, SMB, DRG, GLJ; Software, DS, JSP, NS; Formal Analysis, JSZ, SMB, DS, JSP, NS, NUR; Investigation, JSZ, SMB, DRG, TJS, ASB, JFO-Q, XC, JSD, MCW, SPA, SHV, BTG, XP, CS, DBD; Resources, KG, LAC; Data Curation, JSZ, DS, NS; Writing-Original Draft, JSZ, GLJ; Writing-Review and Editing, JSZ, HSE, SMB, TJS, LAC, LMG, CMP; Visualization, JSZ, SMB, TSJ, GLJ; Supervision, GLJ, CMP, LAC; Project Administration, GLJ, LAC, CMP, HSE; Funding Acquisition, JSZ, GLJ, LAC, CMP, HSE
References
- 1.Zhou B, Der CJ, Cox AD. The role of wild type RAS isoforms in cancer. Semin Cell Dev Biol. 2016;58:60–9. doi: 10.1016/j.semcdb.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Genomic Data Commons Data Portal. Available from: https://gcd-portal.nci.nih.gov.
- 3.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat J-P, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 5.Kimura ET, Nikiforova MN, Zhu Z, Knauf JA, Nikiforov YE, Fagin JA. High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res. 2003;63:1454–7. [PubMed] [Google Scholar]
- 6.Xing M. BRAF mutation in papillary thyroid cancer: pathogenic role, molecular bases, and clinical implications. Endocr Rev. 2007;28:742–62. doi: 10.1210/er.2007-0007. [DOI] [PubMed] [Google Scholar]
- 7.Paik PK, Arcila ME, Fara M, Sima CS, Miller VA, Kris MG, et al. Clinical characteristics of patients with lung adenocarcinomas harboring BRAF mutations. J Clin Oncol Off J Am Soc Clin Oncol. 2011;29:2046–51. doi: 10.1200/JCO.2010.33.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–21. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eroglu Z, Ribas A. Combination therapy with BRAF and MEK inhibitors for melanoma: latest evidence and place in therapy. Ther Adv Med Oncol. 2016;8:48–56. doi: 10.1177/1758834015616934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ascierto PA, Schadendorf D, Berking C, Agarwala SS, van Herpen CM, Queirolo P, et al. MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study. Lancet Oncol. 2013;14:249–56. doi: 10.1016/S1470-2045(13)70024-X. [DOI] [PubMed] [Google Scholar]
- 13.Long GV, Stroyakovskiy D, Gogas H, Levchenko E, de Braud F, Larkin J, et al. Combined BRAF and MEK inhibition versus BRAF inhibition alone in melanoma. N Engl J Med. 2014;371:1877–88. doi: 10.1056/NEJMoa1406037. [DOI] [PubMed] [Google Scholar]
- 14.Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–95. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villanueva J, Infante JR, Krepler C, Reyes-Uribe P, Samanta M, Chen H-Y, et al. Concurrent MEK2 mutation and BRAF amplification confer resistance to BRAF and MEK inhibitors in melanoma. Cell Rep. 2013;4:1090–9. doi: 10.1016/j.celrep.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tricker EM, Xu C, Uddin S, Capelletti M, Ercan D, Ogino A, et al. Combined EGFR/MEK Inhibition Prevents the Emergence of Resistance in EGFR-Mutant Lung Cancer. Cancer Discov. 2015;5:960–71. doi: 10.1158/2159-8290.CD-15-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahadevan D, Cooke L, Riley C, Swart R, Simons B, Della Croce K, et al. A novel tyrosine kinase switch is a mechanism of imatinib resistance in gastrointestinal stromal tumors. Oncogene. 2007;26:3909–19. doi: 10.1038/sj.onc.1210173. [DOI] [PubMed] [Google Scholar]
- 18.Sears R, Leone G, DeGregori J, Nevins JR. Ras enhances Myc protein stability. Mol Cell. 1999;3:169–79. doi: 10.1016/s1097-2765(00)80308-1. [DOI] [PubMed] [Google Scholar]
- 19.Jaenicke LA, von Eyss B, Carstensen A, Wolf E, Xu W, Greifenberg AK, et al. Ubiquitin-Dependent Turnover of MYC Antagonizes MYC/PAF1C Complex Accumulation to Drive Transcriptional Elongation. Mol Cell. 2016;61:54–67. doi: 10.1016/j.molcel.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 20.Kuras L, Borggrefe T, Kornberg RD. Association of the Mediator complex with enhancers of active genes. Proc Natl Acad Sci U S A. 2003;100:13887–91. doi: 10.1073/pnas.2036346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrenko N, Jin Y, Wong KH, Struhl K. Mediator Undergoes a Compositional Change during Transcriptional Activation. Mol Cell. 2016;64:443–54. doi: 10.1016/j.molcel.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–6. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–71. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 24.Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, et al. Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell. 2013;51:310–25. doi: 10.1016/j.molcel.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown JD, Lin CY, Duan Q, Griffin G, Federation AJ, Paranal RM, et al. NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol Cell. 2014;56:219–31. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nabet B, Ó Broin P, Reyes JM, Shieh K, Lin CY, Will CM, et al. Deregulation of the Ras-Erk Signaling Axis Modulates the Enhancer Landscape. Cell Rep. 2015;12:1300–13. doi: 10.1016/j.celrep.2015.06.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Webster DE, Barajas B, Bussat RT, Yan KJ, Neela PH, Flockhart RJ, et al. Enhancer-targeted genome editing selectively blocks innate resistance to oncokinase inhibition. Genome Res. 2014;24:751–60. doi: 10.1101/gr.166231.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell. 2015;163:506–19. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8:R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herschkowitz JI, Zhao W, Zhang M, Usary J, Murrow G, Edwards D, et al. Comparative oncogenomics identifies breast tumors enriched in functional tumor-initiating cells. Proc Natl Acad Sci U S A. 2012;109:2778–83. doi: 10.1073/pnas.1018862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stuhlmiller TJ, Miller SM, Zawistowski JS, Nakamura K, Beltran AS, Duncan JS, et al. Inhibition of Lapatinib-Induced Kinome Reprogramming in ERBB2-Positive Breast Cancer by Targeting BET Family Bromodomains. Cell Rep. 2015;11:390–404. doi: 10.1016/j.celrep.2015.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 37.Li S, Shen D, Shao J, Crowder R, Liu W, Prat A, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013;4:1116–30. doi: 10.1016/j.celrep.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Usary J, Zhao W, Darr D, Roberts PJ, Liu M, Balletta L, et al. Predicting drug responsiveness in human cancers using genetically engineered mice. Clin Cancer Res Off J Am Assoc Cancer Res. 2013;19:4889–99. doi: 10.1158/1078-0432.CCR-13-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Green JE, Shibata MA, Yoshidome K, Liu ML, Jorcyk C, Anver MR, et al. The C3(1)/SV40 T-antigen transgenic mouse model of mammary cancer: ductal epithelial cell targeting with multistage progression to carcinoma. Oncogene. 2000;19:1020–7. doi: 10.1038/sj.onc.1203280. [DOI] [PubMed] [Google Scholar]
- 40.Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, et al. Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146:633–44. doi: 10.1016/j.cell.2011.07.026. [DOI] [PubMed] [Google Scholar]
- 41.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res BCR. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Proia TA, Keller PJ, Gupta PB, Klebba I, Jones AD, Sedic M, et al. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell. 2011;8:149–63. doi: 10.1016/j.stem.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–73. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Machanick P, Bailey TL. MEME-ChIP: motif analysis of large DNA datasets. Bioinforma Oxf Engl. 2011;27:1696–7. doi: 10.1093/bioinformatics/btr189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parker SCJ, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110:17921–6. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–34. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pfefferle AD, Agrawal YN, Koboldt DC, Kanchi KL, Herschkowitz JI, Mardis ER, et al. Genomic profiling of murine mammary tumors identifies potential personalized drug targets for p53-deficient mammary cancers. Dis Model Mech. 2016;9:749–57. doi: 10.1242/dmm.025239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rahman S, Sowa ME, Ottinger M, Smith JA, Shi Y, Harper JW, et al. The Brd4 extraterminal domain confers transcription activation independent of pTEFb by recruiting multiple proteins, including NSD3. Mol Cell Biol. 2011;31:2641–52. doi: 10.1128/MCB.01341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu W, Ma Q, Wong K, Li W, Ohgi K, Zhang J, et al. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell. 2013;155:1581–95. doi: 10.1016/j.cell.2013.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Devaiah BN, Singer DS. Cross-talk among RNA polymerase II kinases modulates C-terminal domain phosphorylation. J Biol Chem. 2012;287:38755–66. doi: 10.1074/jbc.M112.412015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hay DA, Fedorov O, Martin S, Singleton DC, Tallant C, Wells C, et al. Discovery and optimization of small-molecule ligands for the CBP/p300 bromodomains. J Am Chem Soc. 2014;136:9308–19. doi: 10.1021/ja412434f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang L, Chang J, Varghese D, Dellinger M, Kumar S, Best AM, et al. A small molecule modulates Jumonji histone demethylase activity and selectively inhibits cancer growth. Nat Commun. 2013;4:2035. doi: 10.1038/ncomms3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Itzen F, Greifenberg AK, Bösken CA, Geyer M. Brd4 activates P-TEFb for RNA polymerase II CTD phosphorylation. Nucleic Acids Res. 2014;42:7577–90. doi: 10.1093/nar/gku449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parker JS, Mullins M, Cheang MCU, Leung S, Voduc D, Vickery T, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol Off J Am Soc Clin Oncol. 2009;27:1160–7. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.https://github.com/darshansinghunc/chippeakanalysis
- 56.Heiman M, Kulicke R, Fenster RJ, Greengard P, Heintz N. Cell type-specific mRNA purification by translating ribosome affinity purification (TRAP) Nat Protoc. 2014;9:1282–91. doi: 10.1038/nprot.2014.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Kinases significant (FDR 0.05) by DESeq2 differential expression analysis comparing basal-like cell line (HCC1806, SUM-149PT EpCAM+, MDA-MB-468) and claudin-low cell line (SUM-159PT, Hs 578T, WHIM12) group response to 24h 500 nM trametinib treatment. (Excel spreadsheet) Listed is mean log2 fold expression change of biological duplicates for the indicated basal-like (blue) or claudin-low (red) cell lines. Kinases are ordered by their clustering in the heatmap presented in Fig. 2C.
Upregulated kinases significant (FDR 0.05) by DESeq2 differential expression analysis of basal-like cell line (HCC1806, SUM-149PT EpCAM+, MDA-MB-468) or claudin-low cell line (SUM-159PT, Hs 578T, WHIM12) subtypes in isolation. (Excel spreadsheet) Basal-like unique kinases (blue), claudin-low unique (red) or significant kinases common to both subtypes (grey) are indicated along with adjusted p value. Kinases overlapping with patient tumor upregulated kinases (Fig. 2D) are indicated with an asterisk.
Nucleotide variant analysis comparing whole exome sequencing of SUM-229PE subpopulations. (Excel spreadsheet) The “common” tab of the spreadsheet contains nucleotide variants common between the EpCAM+ and EpCAM− SUM-229PE subpopulations, while the “unique” tab of the spreadsheet contains those variants unique to one subpopulation. Q score, Minor Allele Frequency (MAF) and COSMIC database overlap are indicated for each variant.
Cell line methylation fraction (β) values. (Excel spreadsheet) The spreadsheet shows the normalized average methylation fraction β for each Illumina Infinium 450K BeadChip CpG locus for the indicated cell lines upon 24 h DMSO vehicle or 30 nM (SUM-229PE EpCAM+/−, SUM-149PT EpCAM+/−) or 100 nM (SUM-159PT) trametinib treatment. “Methcontrol” column indicates hyper-methylated and “unmethcontrol” column indicates unmethylated genomic DNA controls (see Supplementary Methods).
SUM-159PT Transcriptional correlation with trametinib-induced BRD4 ChIPseq density. (Excel spreadsheet) The spreadsheet contains the merge of SUM-159PT BRD4 stitched ChIPseq peaks and RNAseq data of peak-proximal genes +/− 24 h 100 nM trametinib (see Supplementary Methods). ChIPseq density units are reads per million mapped reads (rpm) and RNAseq data are normalized RSEM transcript abundance estimates. The spreadsheet contains unfiltered ChIPseq data prior to density (rpm) cutoffs and manipulations performed for Fig. 3F.
SUM-159PT ChIPseq data and genic categorization. (Excel spreadsheet) ChIPseq density values for union peaks in reads per million mapped reads (rpm) for the indicated antibodies +/− 24 h trametinib at the indicated doses or +/− 24 h 300 nM JQ1. The “enhancer” spreadsheet classification indicates a putative 5′ enhancer while “3 prime” indicates a putative 3′ enhancer. See Supplementary Methods for union peak, genic category, and “nearby” gene definitions. Spreadsheet contains unfiltered data prior to manipulations for figure construction as described in Supplementary Methods.
HCC1806 ChIPseq data and genic categorization. (Excel spreadsheet) BRD4 ChIPseq density values for union peaks (see Supplementary Methods) in reads per million mapped reads (rpm). The “enhancer” spreadsheet classification indicates a putative 5′ enhancer while “3 prime” indicates a putative 3′ enhancer. See Supplementary Methods for union peak, genic category, and “nearby” gene definitions. Spreadsheet contains unfiltered data prior to manipulations for figure construction as described in Supplementary Methods.