Abstract
Pulmonary surfactant (PS), which is synthesized by type II alveolar epithelial cells (AECIIs), maintains alveolar integrity by reducing surface tension. Many premature neonates who lack adequate PS are predisposed to developing respiratory distress syndrome (RDS), one of the leading causes of neonatal morbidity and mortality. PS synthesis is influenced and regulated by various factors, including microRNAs. Previous in vitro studies have shown that PS synthesis is regulated by miR-26a in fetal rat AECIIs. This study aimed to investigate the role of miR-26a in PS synthesis in vivo. To obtain a miR-26a-1/miR-26a-2 double knockout mouse model, we used the clustered regularly interspaced short palindromic repeat/CRISPR-associated protein 9 (CRISPR/Cas9) system, an important genome editing technology. Real-time PCR was performed to determine the miR-26a levels in various organs, as well as the mRNA levels of surfactant-associated proteins. Moreover, AECIIs and surfactant-associated proteins in lung tissues were analyzed by hematoxylin-eosin staining and immunohistochemistry. Homozygous offspring of miR-26a-1/miR-26a-2 double knockout mice generated using the CRISPR/Cas9 system were successfully obtained, and PS synthesis and the number of AECIIs were significantly increased in the miR-26a knockout mice. These results indicate that miR-26a plays an important role in PS synthesis in AECIIs.
Keywords: miR-26a, AECII, PS, CRISPR/Cas9, knockout mice
Introduction
Pulmonary surfactant (PS) is a complex fluid that consists of phospholipids, neutral lipids, and proteins and is synthesized and secreted by type II alveolar epithelial cells (AECIIs) [1]. It maintains alveolar integrity by reducing surface tension and increases lung compliance. Premature neonates, who have immature lungs and lack adequate PS, are predisposed to developing respiratory distress syndrome (RDS), one of the leading causes of neonatal morbidity and mortality [2]. Lung maturation and PS synthesis in AECIIs are influenced and regulated by a variety of factors, such as circulating hormones, cell-cell interactions, and local paracrine mediators [3-5]. In addition to these factors, microRNAs (miRNAs) have been increasingly recognized to be involved in the regulation of fetal lung maturation [6].
miRNAs are a class of small, endogenous, non-coding RNAs (approximately 22-24 nucleotides in length) that regulate eukaryotic gene expression by binding to the 5’-untranslated regions (UTRs) of target messenger RNAs (mRNAs), typically leading to the repression of protein translation or mRNA degradation [7,8]. It has been speculated that miRNAs are involved in the regulation of almost every aspect of cell physiology [9,10]. Many studies have highlighted roles of miRNAs in the regulation of fetal lung maturation and PS metabolism [6,11,12]. For example, transgenic overexpression of miR17-92 has been reported to promote the proliferation and to inhibit the differentiation of epithelial progenitor cells in developing lungs, whereas deletion of this miRNA has been shown to result in lung hypoplasia [13,14]. Additionally, overexpression of miR-127 has been demonstrated to significantly decrease the terminal bud count, increase the terminal and internal bud sizes, and cause unevenness in bud sizes [15]. Furthermore, miR-150 [16] and miR-375 [17] have been reported to be involved in the regulation of PS secretion. In our previous study, miR-26a was one of seven miRNAs that showed significant changes in expression, as determined by miRNA profiling, at three time points in the developing rat lung [18]. Zhang et al. [11] have shown that PS synthesis is regulated by miR-26a in fetal rat AECIIs. These reports have indicated potential roles of miR-26a in lung development and PS synthesis and metabolism.
Mature miR-26a includes both miR-26a-1 and miR-26a-2, which have the same sequence but are derived from different chromosomes [19]. In mice, mmu-miR-26a-1 is located on chromosome 9 (119031796-119031885 [+]), and mmu-miR-26a-2 is located on chromosome 10 (126995530-126995613 [+]). In this study, to further investigate the biological functions of miR-26a in vivo, we sought to generate miR-26a-1/miR-26a-2 double knockout (miR-26a-1-/-/miR-26a-2-/-) mice using the clustered regularly interspaced short palindromic repeat/CRISPR-associated protein 9 (CRISPR/Cas9) system.
In recent years, various genome editing techniques have been developed, including the use of zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the CRISPR/Cas9 system. However, the CRISPR/Cas9 system has significant advantages over ZFNs or TALENs, including increased target accuracy, the ability to simultaneously knock out multiple target genes, easier and less time-consuming experimental procedures, and a lack of species limitations. In addition, because the Cas9 gene is consistently used this system, only two short guide RNAs (sgRNAs) targeting murine miR-26a-1 and miR-26a-2 need to be synthesized. The gene targeting system, with CRISPR RNA-guided Cas9 nuclease, has been extensively utilized to edit the genomes of several organisms [20-24]. The CRISPR/Cas9 system has been demonstrated to be very highly efficient in live mice. However, reports of double knockout mice, especially for miRNAs, are rare. In the Cas9/gRNA system, when the RNA-guided Cas9 protein is combined with an sgRNA, it can target and cleave DNA sequences [25-28]. Subsequently, genetically modified animals can be generated through the introduction of double-strand breaks (DSBs) and non-homologous end joining (NHEJ)-mediated repair [29,30], leading to the deletion of a DNA fragment at the target locus. To dissect the functions of gene family members and to analyze relationships among signaling pathways, mice with two or more mutated genes are required [21]. For such purposes, the CRISPR/Cas9 system is a useful and highly efficient genetic tool, provided that the double knockout mice (miR-26a-1/miR-26a-2 knockout mice in this study) are viable and that they can be produced from a single embryo [21]. In this study, homozygous offspring of miR-26a-1-/-/miR-26a-2-/- mice generated using the CRISPR/Cas9 system were successfully obtained and represented a suitable model for studying the role of miR-26a in vivo.
Materials and methods
Animals
C57BL/6J and FVB mice were purchased from Nanjing Biomedical Research Institute (NBRI) of Nanjing University, China, and were maintained on a normal 12 h light/dark schedule at a constant temperature and humidity. The housing conditions met the specific pathogen-free (SPF) standards. All procedures were performed in accordance with protocols approved by the Nanjing Medical University Animal Care Committee.
Microinjection
A Cas9 expression plasmid containing the SP6 promoter was used as a template for in vitro transcription (IVT) using an mMESSAGE mMACHINE SP6 Kit (Invitrogen, CA, US) after linearization and purification. As previously described [22], we constructed fusion crRNA and tracrRNA expression vectors with a customizable synthetic (sgRNA) template using miR-26a-1F/R and miR-26a-2F/R primers (Table 1A). The T7-sgRNA PCR product was purified and used as a template for IVT, performed using a T7 kit (TaKaRa, Japan). Female FVB mice (7-8 weeks old) were superovulated using pregnant mare serum gonadotropin (PMSG) and were injected with human chorionic gonadotropin (HCG) after 48 h. The superovulated female mice (FVB mice) were mated with C57BL/6J stud males, and fertilized embryos were collected from their oviducts. miR-26a-1 and miR-26a-2 sgRNAs (12.5 ng/uL each) were mixed with Cas9 mRNA (25 ng/μL), and the mixture was microinjected into the cytoplasm of the female mouse embryos at the pronuclei stage. The injected zygotes were cultured in KSOM with amino acids at 37°C and 5% CO2 in air until the blastocyst stage (at 3.5 days). Subsequently, approximately 15-25 blastocysts were transferred into the uterus of each pseudopregnant FVB female.
Table 1A.
sgRNA primer sequences
| mmu-mir-26a-1 | F: 5’-GGGCTCTTTCCTTAGACTTGG-3’ |
| R: 5’-GACCTGCTTTGCTCATAACACTC-3’ | |
| mmu-mir-26a-2 | F: 5’-GTTGGTGCTGATGTGGGCTAG-3’ |
| R: 5’-CTGGGAGACAGAGTGGATTGC-3’ |
Genotyping and breeding
After approximately 19 days, DNA was extracted from the pups’ tails (founder mice), and the mice were genotyped by PCR and sequencing. Then, male (7-week-old) and female (4-week-old) founder mice were mated with wild-type mice. The next generation of mice (F1 mice) was obtained and genotyped until mice positive for either or both mutations in miR-26a-1 and/or miR-26a-2 were identified, indicating the successful generation of gene knockout mouse strains. Positive mice (> 3 females and> 3 males) were mated with wild-type mice (SPF standard), and the PCR products from the tail DNA samples were identified after restriction digestion with T7 endonuclease I (T7EI) and resolution in a 1% agarose gel. The 20 μL PCR mixture contained 10 μL master mix (Vazyme Biotech, Nanjing, China), 8.2 μL ddH2O, 1 μL DNA, and 0.4 μL each of the miR-26a-1 forward and reverse primers or 0.4 μL each of the miR-26a-2 forward and reverse primers. The PCR amplification program was as follows: 94°C for 5 minutes; 35 cycles at 94°C for 30 seconds, 55°C for 30 seconds, and 72°C for 60 seconds; and 72°C for 7 minutes. The amplicons were separated by 1.0% agarose gel electrophoresis.
RNA extraction and real-time quantitative polymerase chain reaction
Total RNA was extracted from mouse organs using Trizol reagent (Invitrogen, CA, US) according to the manufacturer’s instructions. The ratio of absorbance at 260 to that at 280 nm (A260/280) was used to assess RNA purity and quantity. First-strand cDNA was generated using a TaqMan MicroRNA Reverse Transcriptase Kit (Applied Biosystems, US) and oligo(dT) primers, which were designed and produced by Sangon Biotech (Shanghai, China) according to the manufacturer’s instructions. The probes were designed to target miR-26a-5p and U6 for reverse transcription (5× dilution) and real-time PCR (20× dilution). Moreover, RNA samples from mouse lungs were subjected to reverse transcription with a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, US). Real-time PCR was performed using SYBR Green (Roche, Shanghai, China), and specific primers for SP-A (Sftpa1) and SP-B (Sftpb) were synthesized according to published cDNA sequences (Table 1B). PCR was performed with an ABI 7500 thermal cycler (Applied Biosystems) using the following two-step cycling program: 95°C for 10 minutes; 50°C for 2 minutes and 40 cycles at 60°C for 1 minute; and 95°C for 15 seconds. Dissociation curves were generated for both genes under the following conditions: 95°C for 15 seconds, 60°C for 60 seconds, and 95°C for 15 seconds. The expression levels of the miR-26a-5p genes were normalized to that of an internal control, U6, to obtain relative threshold cycle (ΔCt) values. Relative quantification of gene expression in multiple samples was achieved by normalization against an endogenous control, GAPDH. Then, the relative expression levels were compared between the wild-type mice and knockout mice using the comparative Ct (ΔΔCt) method (ΔΔCt = ΔCt of the wild-type mice -ΔCt of the knockout mice) or the 2-ΔΔCT method. A P < 0.05 was considered to indicate significance.
Table 1B.
Real-time PCR primer sequences
| mmu-SP-A | F: 5’-GCCTTCACCCTCTTCTTGACT-3 |
| R: 5’-ACCATCTCTCCCATCTCTGC-3 | |
| mmu-SP-B | F: 5’-CTGCTTCCTACCCTCTGCTG-3’ |
| R: 5’-ATCCTCACACTCTTGGCACA-3’ | |
| mmu-GAPDH | F: 5’-GGTGAAGGTCGGTGTGAACG-3’ |
| R: 5’-CTCGCTCCTGGAAGATGGTG-3’ |
Immunohistochemistry and hematoxylin-eosin staining
The mice were anesthetized with 2% chloral hydrate (0.2 mL/10 g) and transcardially perfused with 4% paraformaldehyde (PFA) in PBS. Dissected lungs were post-fixed overnight with 4% PFA in PBS. To obtain coronal sections, the lungs were cryoprotected by overnight immersion in 30% sucrose in PBS and embedded in OCT compound. Sections of 50 µm thickness were prepared using a cryostat, permeabilized with 0.3% Triton X-100 in PBS and blocked with 2% BSA and 0.3% Triton X-100 in PBS. The sections were incubated overnight with primary antibodies, rinsed with PBS, incubated with secondary antibodies, and developed using DAB. The primary antibodies used included a rabbit anti-podoplanin/gp36 antibody (Abcam, Shanghai, China) and rabbit anti-prosurfactant protein C antibody (Millipore, Shanghai, China) for AECIs and AECIIs and rabbit anti-prosurfactant protein A and B antibodies for PS. As described above, the mouse lungs were pretreated for hematoxylin-eosin staining, and the obtained sections were dewaxed with xylene and incubated in a series of ethanol/water solutions. Then, the sections were stained with hematoxylin, differentiated with ethanol hydrochloride, transferred to eosin solution. Subsequently, the sections were dehydrated and mounted. Finally, microscopic analysis was performed.
Statistical analysis
The data are presented as the mean ± SEM. All of the in vitro experiments were repeated three times (n = 6). The data were analyzed using the unpaired t-test with GraphPad Prism 5.0, and a P < 0.05 was considered to indicate statistical significance.
Results
Construction of CRISPR/Cas9 plasmids targeting miR-26a-1 and miR-26a-2
To select Cas9 target sites in the miR-26a-1 and miR-26a-2 genes, we searched for 20nucleotide sequences that were followed by the protospacer-adjacent motif (PAM) sequence (NGG) (Figure 1). To examine the effectiveness of gene knockout, we sequenced the PCR fragments and ensured that all of the heterozygous mice had deletions of at least 4 base pairs. Additionally, more than three mice harbored biallelic miR-26a-1 and miR-26a-2 double mutations, as determined by sequencing.
Figure 1.
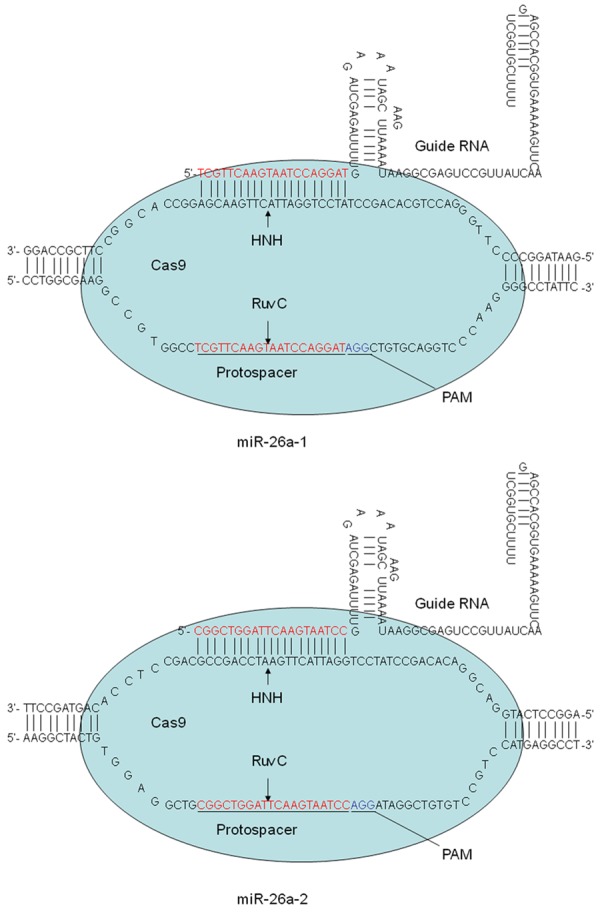
Map of Cas9 protein bound to sgRNAs. The 20-nucleotide sequences in miR-26a-1 and miR-26a-2 followed by the protospacer-adjacent motif (PAM) sequence (NGG) can be used to target and cleave DNA sequences.
One-step generation of double mutant mice by zygote injection
To determine whether miR-26a-1/miR-26a-2 double mutant mice could be produced from single embryos, we co-microinjected Cas9 mRNA (25 ng/μL) and sgRNA (12.5 g/μL) into the pronuclei of fertilized eggs. The blastocysts derived from the injected embryos were transplanted into foster mothers, and the resulting newborn pups were genotyped. A total of 18 pups were born from 90 embryos injected with in vitro-synthesized RNA and transferred into foster mothers (20% live birth rate) (Table 2). In total, 15 mice carried targeted mutations in all four alleles of the miR-26a-1 and miR-26a-2 genes, and the numbers of mice carrying mutations in either or both genes are shown in Table 2. More than 30% of the pups were biallelic miR-26a-1 and miR-26a-2 double mutants (Table 2). These results demonstrate that postnatal mice carrying biallelic mutations in two different genes can be generated with high efficiency.
Table 2.
Generation of knockout mice using the CRISPR-Cas system
| Gene | Strain | Cas9/sgRNA doses (ng/ul) | Numbers injected/transferred (%) | Total number of newborns (%) | Mutant number |
|---|---|---|---|---|---|
| mir26a-1 | C57BL/6J | 7 | |||
| mir26a-2 | C57BL/6J | 25/12.5+12.5 | 107/74 (69) | 18 (20) | 3 |
| 1&2 | C57BL/6J | 5 |
Breeding strategy and genotyping
Once the F1 mice were obtained, breeding cages were established as described in Table 3A and Table 3B. A total of 3 female and 3 male knockout mice were selected and mated with wild-type mice, as shown in Table 3A. Embryos from pregnant mice were transplanted into the uteri of surrogate mice under SPF conditions, as shown in Table 3B. The mouse genotypes were identified by 1% agarose gel electrophoresis (Figure 2A and 2B) and sequencing (Figure 2C). No difference in miR-26a expression was detected in the double-heterozygous or single-homozygous mice compared with the wild-type mice. Phenotypic analysis of miR-26a-1-/-/miR-26a-2-/- mice revealed that some of the mice were blind and had white abdominal hair, but the reason for the observed phenotype was unknown. However, most of the double knockout mice did not phenotypically differ from the wild-type mice. The homozygous mice that were subsequently obtained did not show any differences in body weight, birth rate, pregnancy rate, or growth rate compared with the wild-type mice.
Table 3A.
Breeding strategy
| Number | Sex | Strain | mir-26a-1 | mir-26a-2 | Gen |
|---|---|---|---|---|---|
| 5# | ♂ | C57BL/6J | HET | HET | F1 |
| 19# | ♂ | C57BL/6J | HET | HET | F1 |
| 24# | ♂ | C57BL/6J | HET | HET | F1 |
| 2# | ♀ | C57BL/6J | HET | HET | F1 |
| 21# | ♀ | C57BL/6J | HET | HET | F1 |
| 22# | ♀ | C57BL/6J | WT | HET | F1 |
Method of purification: HET (mir-26a-1+/-/mir-26a-2+/-) × WT (mir-26a-1+/+/ mir-26a-2+/+).
Table 3B.
Purification strategy
| Transplant recipient | Embryo type | Number of transplantations | Number of recipients |
|---|---|---|---|
| C57BL/6J♀ | 2-cell | 9 | 1 |
| C57BL/6J♀ | 2-cell | 23 | 2 |
| C57BL/6J♀ | 4-cell | 16 | 1 |
| C57BL/6J♀ | 4-cell | 32 | 2 |
Figure 2.
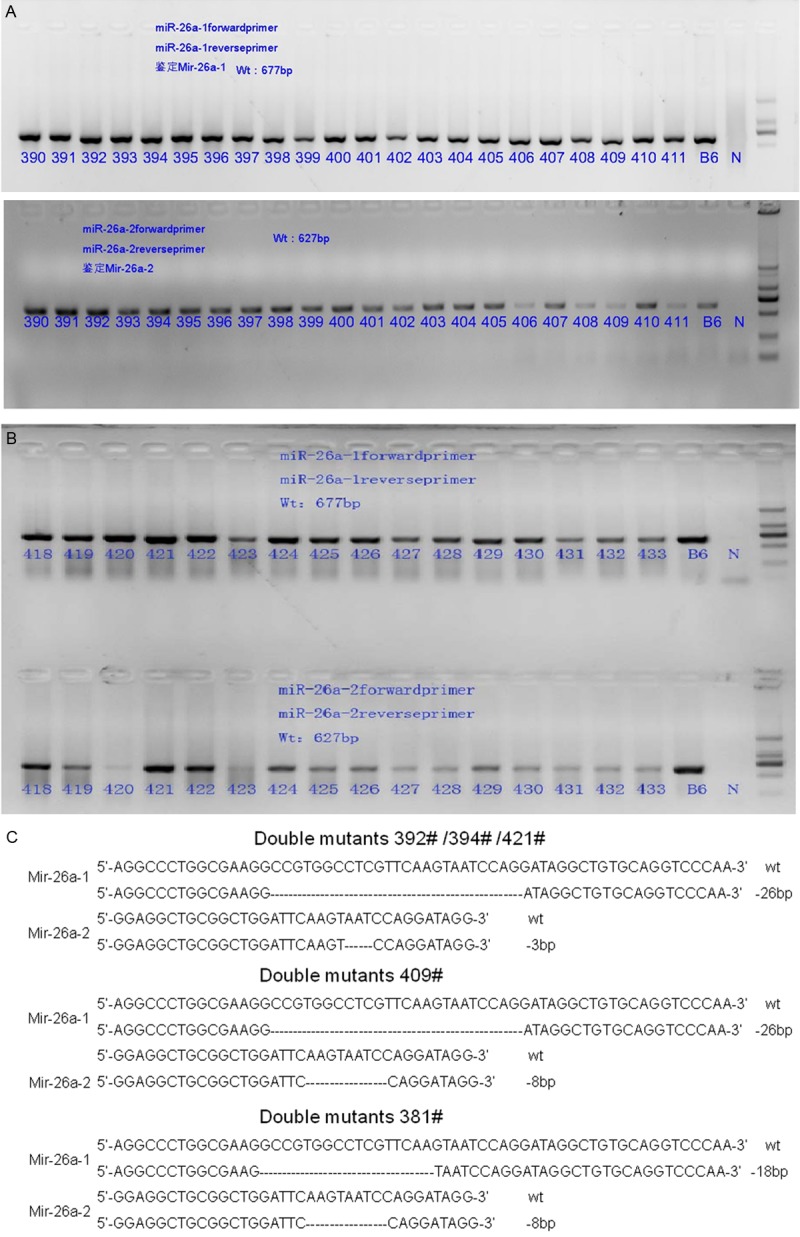
A, B. The PCR products were separated by 1% agarose gel electrophoresis. B6-blank control; and N-negative control. C. Genotyping was performed by sequencing; the different deletion locations and fragments of the three types of mutants are shown. The numbers #392, #394, #421, #381, and #409 are representative numbers of all homozygous mice.
Expression of miR-26a, SP-A and SP-B
Various organs were excised from homozygous mutant mice (n> 6) and wild-type mice (n> 6), and the miR-26a expression levels were measured by real-time PCR. miR-26a expression was downregulated in various organs in the knockout mice compared with the wild-type mice, with the greatest decrease in expression observed in the lungs (Figure 3). To determine whether lung surfactant synthesis in knockout mice is affected by miR-26a, the SP-A and SP-B levels were measured by real-time quantitative PCR. The results revealed that the SP-A and SP-B mRNA levels were significantly increased in the lungs of the miR-26a knockout mice (Figure 6) compared with those of the wild-type mice.
Figure 3.
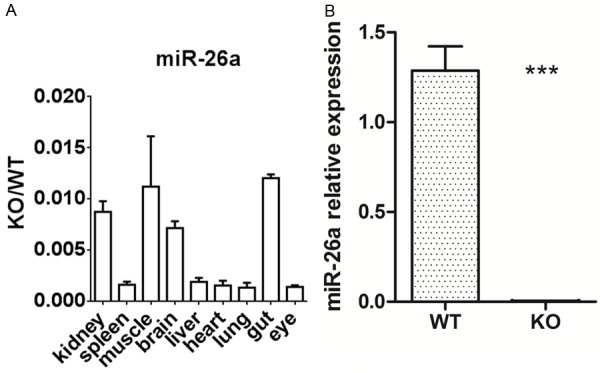
A. miR-26a expression in the lung showed the greatest change among various organs. B. miR-26a expression was significantly downregulated in the lungs of the KO mice compared with those of the WT mice (P < 0.05). KO: knockout mice; WT: wild-type mice.
Figure 6.
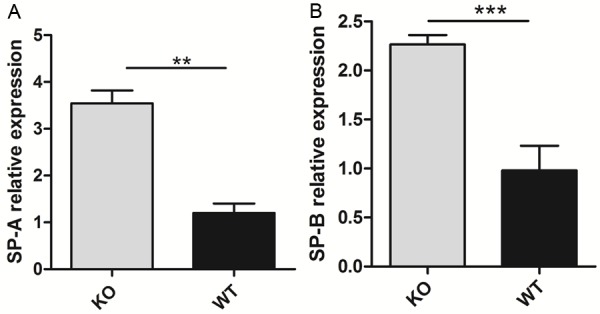
Real-time PCR was performed to determine the SP-A and SP-B mRNA levels, which were significantly increased in the KO mice compared with the WT mice (P < 0.05).
Hematoxylin-eosin staining and immunohistochemistry
To observe differences in lung tissue structure between the miR-26a double knockout and wild-type (5-week-old) mice, hematoxylin-eosin staining and immunohistochemistry (IHC) were performed. The number of AECIIs was significantly increased and that of AECIs was significantly decreased in the knockout mice compared with the wild-type mice (Figures 4 and 5), indicating the increased maturity of the lung tissues of the double knockout mice. To determine whether surfactant synthesis in the knockout mice was affected by miR-26a, the SP-A and SP-B levels were detected by IHC (Figure 7). The results indicated that the lungs of the miR-26a knockout mice contained more mature cells and were thus more conducive to the synthesis and secretion of PS.
Figure 4.
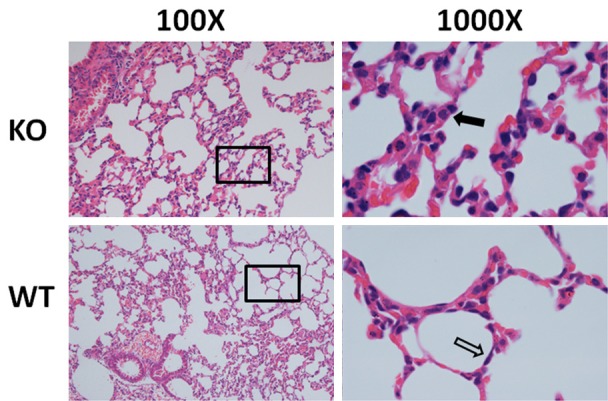
Hematoxylin-eosin staining of lung tissues. More type II alveolar epithelial cells (black arrow) and fewer type I alveolar epithelial cells were detected in the KO mice compared with the WT mice. Representative images (n> 4 per genotype) are shown.
Figure 5.
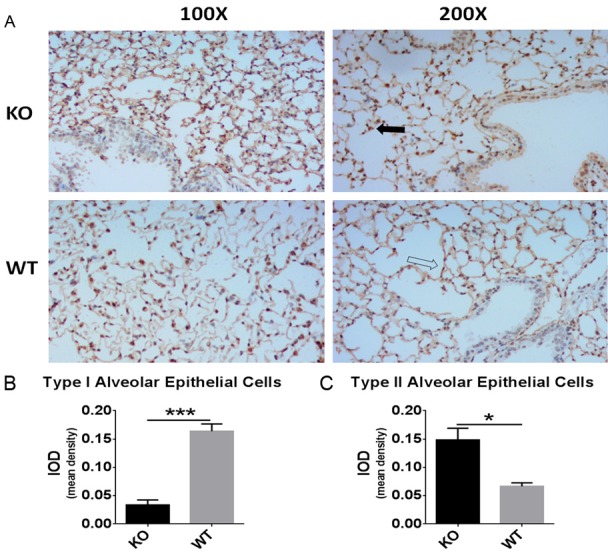
Significant lung morphological changes were observed in IHC analysis. A. Type I alveolar epithelial cells were identified with a rabbit anti-podoplanin/gp36 antibody (indicated by the black arrow), and type II alveolar epithelial cells were identified with a rabbit anti-prosurfactant protein C antibody (indicated by the white arrow). B, C. Analysis of the mean IODs revealed that significantly more type II alveolar epithelial cells and significantly fewer type I alveolar epithelial cells were present in the KO mice compared with the WT mice. The data are presented as the mean ± standard deviation (*P < 0.05).
Figure 7.
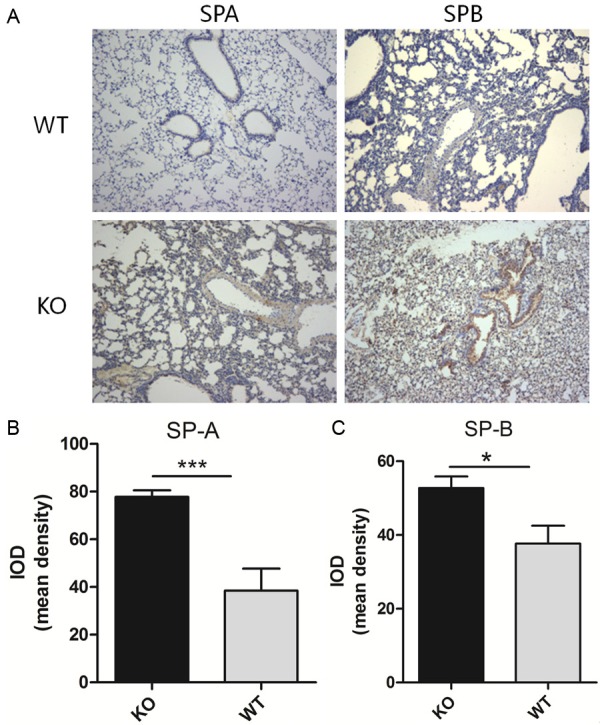
A. SP-A and SP-B were identified by IHC with rabbit anti-prosurfactant protein A and rabbit anti-prosurfactant protein B, respectively. Representative images (n> 4 per genotype) are shown. B, C. Analysis of the mean IODs revealed that the SP-A and SP-B levels were significantly increased in the KO mice compared with the WT mice. The data are presented as the mean ± standard deviation (*P < 0.05).
Discussion
With advances in perinatal medicine and neonatal intensive care technologies, the birth and survival rates of premature infants with structurally immature lungs have increased, and the incidence of RDS has also increased. Inadequate PS synthesis in premature infants can result in RDS, which is the leading cause of neonatal morbidity and mortality. PS is synthesized and secreted by AECIIs but not by AECIs. It maintains alveolar integrity by reducing surface tension and participates in host defense and the regulation of inflammation in the lungs [1,31]. In this study, we aimed to investigate the role of miR-26a in PS synthesis using knockout mice.
Studies have suggested that lung maturation and PS synthesis in AECIIs are influenced and regulated by a number of hormones and factors, including glucocorticoids, prolactin, insulin, and growth factors [3-5]. In addition, miRNAs, including miR-26a, have been increasingly recognized to be involved in the regulation of fetal lung maturation [6]. Links between miR-26a and circulating hormones, such as lactogenic hormones, have been reported in the literature [32]. However, it is unclear whether miR-26a impacts lung development by regulating circulating hormones. It is well known that miR-26a exerts multiple biological effects in lung-related diseases; it plays a key role in various types of carcinoma, including lung adenocarcinoma [33], and participates in inflammation, including pulmonary inflammation [34]. Furthermore, serum miR-26a has been used as a potential biomarker for the diagnosis of various diseases [35]. Jiang et al. [34]. have shown that circulating miR-26a in wheezing children and AIPI rats is associated with recurrent childhood wheezing with asthmatic risk, and Liu et al. [36]. have demonstrated that miR-26a enhances the metastatic potential of lung cancer cells via the AKT pathway by targeting PTEN. Additionally, Cinegaglia et al. [33] have reported that miR-26a is significantly under-expressed in lung adenocarcinoma and that serum miR-26a might be a useful biomarker for the diagnosis of several diseases [35]. Our previous study has shown that miR-26a is differentially expressed in fetal rat lung tissues during various stages of pregnancy [11]. In addition, we have demonstrated that PS synthesis is reduced upon miR-26a overexpression in vitro [11]. In this study, we sought to generate miR-26a-1/miR-26a-2 double knockout mice using the CRISPR/Cas9 system to further study the biological functions of miR-26a in vivo.
The CRISPR/Cas9 system is a useful gene knockout technique for investigating gene function in vivo; since its development, this technique has been utilized to generate knockout cell lines [25,26] and animal models [22,23]. In this study, we constructed CRISPR/Cas9 plasmids targeting miR-26a-1 and miR-26a-2. Different NGG PAM sequences for S. pyogenes Cas9 in the mouse genome were selected to induce targeted DNA cleavage, as shown in Figure 1. Mice harboring mutations in both genes were produced by coinjection of Cas9 with miR-26a-1 and miR-26a-2 sgRNAs into zygotes. The efficiency of obtaining mice carrying mutations in both targeted genes reached 28%, and approximately 56% of the mice harbored a biallelic mutation in one of the targeted genes. Thus, double knockout mice could be generated within 32 days. As mentioned above, the crossing of mice with the appropriate genotypes and the further breeding required to obtain homozygous offspring for studying miR-26a gene function were time consuming.
In this study, the homozygous double knockout mice represented an ideal experimental model to analyze the function of miR-26a in vivo. Our results obtained in vivo were more credible and convincing than those obtained in vitro. The number of mature cells, AECIIs, and PS synthesis were increased in the lungs of the miR-26a-1/miR-26a-2 double knockout mice, indicating the close involvement of miR-26a in lung development and its influence on PS synthesis. However, whether miR-26a directly affects surfactant synthesis or whether these findings are due to some undefined molecular mechanism(s) affected by the miRNA knockout is unclear, and further research is needed.
There are potential limitations to our study. First, miR-26a is selectively expressed in bronchial and alveolar epithelial cells of the murine lung and is upregulated in the adult lung following the postnatal period of lung development [37,38]. Although the miR-26a deficiency of the strain used in this study was not specific to the lungs, it is still an important model for studying lung disease. In addition, considering that miR-26a overexpression may cause lethality, we created a gene knockout mouse model using the CRISPR/Cas9 system. Second, there may be off-target effects of the CRISPR/Cas9 system, but they can be resolved by conducting a more thorough sequencing analysis, as previously reported [21]. Finally, the mechanism by which miR-26a regulates PS synthesis in lung AECIIs in vivo is unclear and needs to be further explored. Therefore, the miR-26a knockout mouse model that we generated will enable us to study the biological functions of miR-26a and to elucidate its roles in lung development and pathogenesis. These findings may allow for increased understanding and the development of novel approaches for the prevention and cure of RDS.
Acknowledgements
This work was supported by funding from the National Natural Science Foundation of China (No. 81270725).
Disclosure of conflict of interest
None.
References
- 1.Andreeva AV, Kutuzov MA, Voyno-Yasenetskaya TA. Regulation of surfactant secretion in alveolar type II cells. Am J Physiol Lung Cell Mol Physiol. 2007;293:L259–L271. doi: 10.1152/ajplung.00112.2007. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez RJ. Management of respiratory distress syndrome: an update. Respir Care. 2003;48:279–287. [PubMed] [Google Scholar]
- 3.Mulugeta S, Beers MF. Surfactant protein C: its unique properties and emerging immunomodulatory role in the lung. Microbes Infect. 2006;8:2317–2323. doi: 10.1016/j.micinf.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Ballard PL. Hormonal regulation of pulmonary surfactant. Endocr Rev. 1989;10:165–181. doi: 10.1210/edrv-10-2-165. [DOI] [PubMed] [Google Scholar]
- 5.Gross I. Regulation of fetal lung maturation. Am J Physiol. 1990;259:L337–L344. doi: 10.1152/ajplung.1990.259.6.L337. [DOI] [PubMed] [Google Scholar]
- 6.Nana-Sinkam SP, Karsies T, Riscili B, Ezzie M, Piper M. Lung microRNA: from development to disease. Expert Rev Respir Med. 2009;3:373–385. doi: 10.1586/ers.09.30. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 9.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 10.Sonkoly E, Pivarcsi A. Advances in microRNAs: implications for immunity and inflammatory diseases. J Cell Mol Med. 2009;13:24–38. doi: 10.1111/j.1582-4934.2008.00534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang XQ, Zhang P, Yang Y, Qiu J, Kan Q, Liang HL, Zhou XY, Zhou XG. Regulation of pulmonary surfactant synthesis in fetal rat type II alveolar epithelial cells by microRNA-26a. Pediatr Pulmonol. 2014;49:863–872. doi: 10.1002/ppul.22975. [DOI] [PubMed] [Google Scholar]
- 12.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 13.Lu Y, Thomson JM, Wong HY, Hammond SM, Hogan BL. Transgenic over-expression of the microRNA miR-17-92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol. 2007;310:442–453. doi: 10.1016/j.ydbio.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp PA, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhaskaran M, Wang Y, Zhang H, Weng T, Baviskar P, Guo Y, Gou D, Liu L. MicroRNA-127 modulates fetal lung development. Physiol Genomics. 2009;37:268–278. doi: 10.1152/physiolgenomics.90268.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weng T, Mishra A, Guo Y, Wang Y, Su L, Huang C, Zhao C, Xiao X, Liu L. Regulation of lung surfactant secretion by microRNA-150. Biochem Biophys Res Commun. 2012;422:586–589. doi: 10.1016/j.bbrc.2012.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Mishra A, Chintagari NR, Gou D, Liu L. Micro-RNA-375 inhibits lung surfactant secretion by altering cytoskeleton reorganization. IUBMB Life. 2010;62:78–83. doi: 10.1002/iub.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y, Kai G, Pu XD, Qing K, Guo XR, Zhou XY. Expression profile of microRNAs in fetal lung development of Sprague-Dawley rats. Int J Mol Med. 2012;29:393–402. doi: 10.3892/ijmm.2011.855. [DOI] [PubMed] [Google Scholar]
- 19.Icli B, Dorbala P, Feinberg MW. An emerging role for the miR-26 family in cardiovascular disease. Trends Cardiovasc Med. 2014;24:241–248. doi: 10.1016/j.tcm.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li D, Qiu Z, Shao Y, Chen Y, Guan Y, Liu M, Li Y, Gao N, Wang L, Lu X, Zhao Y, Liu M. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol. 2013;31:681–683. doi: 10.1038/nbt.2661. [DOI] [PubMed] [Google Scholar]
- 23.Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, Xiang AP, Zhou J, Guo X, Bi Y, Si C, Hu B, Dong G, Wang H, Zhou Z, Li T, Tan T, Pu X, Wang F, Ji S, Zhou Q, Huang X, Ji W, Sha J. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 24.Hwang WY, Fu Y, Reyon D, Maeder ML, Kaini P, Sander JD, Joung JK, Peterson RT, Yeh JR. Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS One. 2013;8:e68708. doi: 10.1371/journal.pone.0068708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 27.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jinek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, Anders C, Hauer M, Zhou K, Lin S, Kaplan M, Iavarone AT, Charpentier E, Nogales E, Doudna JA. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science. 2014;343:1247997. doi: 10.1126/science.1247997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song Y, Yuan L, Wang Y, Chen M, Deng J, Lv Q, Sui T, Li Z, Lai L. Efficient dual sgRNA-directed large gene deletion in rabbit with CRISPR/Cas9 system. Cell Mol Life Sci. 2016;73:2959–2968. doi: 10.1007/s00018-016-2143-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crouch E, Wright JR. Surfactant proteins A and D and pulmonary host defense. Annu Rev Physiol. 2001;63:521–554. doi: 10.1146/annurev.physiol.63.1.521. [DOI] [PubMed] [Google Scholar]
- 32.Muroya S, Hagi T, Kimura A, Aso H, Matsuzaki M, Nomura M. Lactogenic hormones alter cellular and extracellular microRNA expression in bovine mammary epithelial cell culture. J Anim Sci Biotechnol. 2016;7:8. doi: 10.1186/s40104-016-0068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cinegaglia NC, Andrade SC, Tokar T, Pinheiro M, Severino FE, Oliveira RA, Hasimoto EN, Cataneo DC, Cataneo AJ, Defaveri J, Souza CP, Marques MM, Carvalho RF, Coutinho LL, Gross JL, Rogatto SR, Lam WL, Jurisica I, Reis PP. Integrative transcriptome analysis identifies deregulated microRNA-transcription factor networks in lung adenocarcinoma. Oncotarget. 2016;7:28920–28934. doi: 10.18632/oncotarget.8713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang C, Yu H, Sun Q, Zhu W, Xu J, Gao N, Zhang R, Liu L, Wu X, Yang X, Meng L, Lu S. Extracellular microRNA-21 and microRNA-26a increase in body fluids from rats with antigen induced pulmonary inflammation and children with recurrent wheezing. BMC Pulm Med. 2016;16:50. doi: 10.1186/s12890-016-0216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leidinger P, Brefort T, Backes C, Krapp M, Galata V, Beier M, Kohlhaas J, Huwer H, Meese E, Keller A. High-throughput qRT-PCR validation of blood microRNAs in non-small cell lung cancer. Oncotarget. 2016;7:4611–4623. doi: 10.18632/oncotarget.6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu B, Wu X, Liu B, Wang C, Liu Y, Zhou Q, Xu K. MiR-26a enhances metastasis potential of lung cancer cells via AKT pathway by targeting PTEN. Biochim Biophys Acta. 2012;1822:1692–1704. doi: 10.1016/j.bbadis.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 37.Moschos SA, Williams AE, Perry MM, Birrell MA, Belvisi MG, Lindsay MA. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genomics. 2007;8:240. doi: 10.1186/1471-2164-8-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Williams AE, Moschos SA, Perry MM, Barnes PJ, Lindsay MA. Maternally imprinted microRNAs are differentially expressed during mouse and human lung development. Dev Dyn. 2007;236:572–580. doi: 10.1002/dvdy.21047. [DOI] [PMC free article] [PubMed] [Google Scholar]


