Abstract
Prostate cancer (PCa) is a leading cause of cancer-related deaths in elder men. This disease has limited therapeutic options and poor prognosis as the underlying molecular mechanisms are not clearly understood. LncRNA UCA1 functions as an oncogene in many types of cancers. However, the role of UCA1 in PCa remains unclear. In the present study, we showed that UCA1 was significantly up-regulated in PCa cell lines and tissue samples. High UCA1 expression was positively associated with high gleason score, advanced TNM stage and shorter overall survival of PCa patients. Inhibition of UCA1 suppressed PCa cells proliferation, migration and invasion in vitro. Moreover, UCA1 depletion inhibited the growth of PCa cells in vivo. In addition, we found that ATF2 was a direct target gene of UCA1. UCA1 regulated ATF2 expression through functioning as a competing endogenous RNA (ceRNA). UCA1 directly interacted with miR-204 and decreased the binding of miR-204 to ATF2 3’UTR, which suppressed the degradation of ATF2 mRNA by miR-204. In summary, we unveil a branch of the UCA1-miR-204-ATF2 pathway that regulates the progression of PCa. The inhibition of UCA1 expression may be a promising strategy for PCa therapy.
Keywords: Prostate cancer, lncRNA UCA1, competing endogenous RNA, miR-204, ATF2
Introduction
Prostate cancer (PCa) is the most commonly diagnosed malignancy and the second leading cause of cancer death among American men [1]. Androgen receptor (AR) plays a critical role in the progression of prostate cancer, and androgen deprivation therapy (ADT) is the first line therapy for most first time diagnostic prostate cancer patients [2,3]. However, despite initial response rates of 80-90%, patients will progress to castration-resistant prostate cancer (CRPC) and even metastatic prostate cancer [4]. CRPC has limited therapeutic options and poor prognosis, and is currently the leading cause of death among male cancer patients [5]. Thus, a better understanding of the mechanisms involved in the pathogenesis of PCa and more effective therapeutic approaches are instantly required.
Recent studies showed that more than 90% of the transcripts from the human genome do not code for proteins [6]. These non-coding transcripts which contain more than 200 nucleotides are called long non-coding RNAs (lncRNAs) [7]. Recently, many studies showed that lncRNAs were frequently dysregulation in various tumors and have multiple functions in a wide range of biological processes, such as carcinogenesis, metastasis and angiogenesis [8,9]. Dysregulated expression of lncRNAs were found in various cancers. For example, Liu et al showed that lncRNA GAS5 was down-regulated in stomach cancer tissues and enhanced G1 cell cycle arrest via binding to YBX1 to regulate p21 expression in stomach cancer [10]. Qiao et al found that lncRNA FER1L4 suppressed cell proliferation and cell cycle by regulating PTEN expression in endometrial carcinoma [11].
Human urothelial carcinoma associated 1 (UCA1) gene is located in chromosome 19p13.12, which was first identified in human bladder carcinoma [12]. Li et al showed that overexpression of lncRNA UCA1 promoted osteosarcoma progression and correlated with poor prognosis [13]. Zhang et al suggested that lncRNA UCA1 overexpression predicted a poor clinical outcome in ovarian cancer patients receiving adjuvant chemotherapy [14]. Wang et al revealed that lncRNA UCA1 inhibited esophageal squamous cell carcinoma growth by regulating the Wnt signaling pathway [15]. However, whether the aberrant expression of UCA1 in PCa is associated with malignancy remains unknown, and the mechanism through which UCA1 exerts its oncogenic activity needs to be illustrated.
In the present study, we showed that UCA1 was significantly increased in PCa and associated with high gleason score, advanced TNM stage and shorter overall survival. Inhibition of UCA1 suppressed PCa cells proliferation, migration and invasion both in vitro and in vivo. Furthermore, we found that ATF2 was a direct target gene of UCA1. UCA1 regulated ATF2 expression by acting as a competing endogenous RNA (ceRNA). UCA1 directly interacted with miR-204 and decreased the binding of miR-204 to ATF2 3’UTR, which suppressed the degradation of ATF2 mRNA by miR-204. This study provided the first evidence that UCA1 promoted PCa progression through ATF2, suggesting that UCA1 and ATF2 may be potential therapeutic targets for PCa treatment.
Material and methods
Tissue samples
A total of 47 paired PCa tissues and adjacent non-tumor tissues were obtained from patients who underwent radical prostatectomies at the Department of Urology, People’s Hospital of Zhengzhou University, between January 2010 and December 2011. All samples were confirmed by pathological examination and stored in liquid nitrogen for later total RNA extraction. All patients recruited in this study did not receive any preoperative treatments. The ethics committees at Zhengzhou University approved this protocol, and all samples were collected following a written informed consent from the patients.
Cell culture
The human prostate cancer cell lines LNCaP, PC-3, DU145 and normal prostate epithelial cell line RWPE-1 were obtained from the American Type Culture Collection (ATCC). The four cell lines are all cultured in RPMI1640 medium supplemented with 10% fetal bovine serum (FBS, Gibco), 100 U/ml penicillin, and 100 mg/ml streptomycin (Gibco) at 37°C in humidified air containing 5% of CO2.
Plasmid construction and transfection
To construct UCA1 knockdown cells, PCa cells were infected with lentivirus particles expressing shRNA against UCA1 (sh-UCA1) and selected by puromycin (1 ug/ml) for one week. The target sequence is as follows: GCCATATGAAGACACCCTA. SiRNA sequences for ATF2 (si-ATF2), miR-204 mimics and negative controls (Con) were purchased from GenePharma. Transfections were carried out using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
RNA isolation and quantitative real-time PCR (qRT-PCR)
Total RNA was isolated using Trizol reagent (Invitrogen). First strand cDNA was generated using the Reverse EasyScript One Step gDNA Removal and cDNA Synthesis SuperMix (Trangene). QRT-PCR was performed by using SYBR Green Master Mixture (Roche) reagent in ABI7500 real-time PCR instrument. GAPDH was used as an internal control. The relative levels of gene expression were calculated by the 2-ΔΔCt method. qRT-PCR primers are as follows: UCA1 forward: 5’-TTTATGCTTGAGCCTTGA-3’, reverse 5’-CTTGCCTGAAATACTTGC-3’; ATF2 forward: 5’-TGCCTGTTGCTATTCCTGC-3’, reverse 5’-GCTCTTCTCCGACGACCACT-3’; GAPDH, forward 5’-GAAGGTCGGAGTCAACGGATT-3’, reverse 5’-CGCTCCTGGAAGATGGTGAT-3’. All experiments were performed in triplicate.
Cell proliferation assay
The cell proliferation was assessed using the Cell Counting Kit-8 (Dojindo) according to the manufacturer’s protocol. Briefly, 1×105 cells were seeded into each well of 24-well plates. Before proliferation assessment, 30 μl of CCK-8 reagent with 300 μl phenol-free RPMI1640 medium was added to each well, and incubated at 37°C for 2 h. Viable cells were evaluated by absorbance measurements at 450 nm at each time point. Each experiment was performed in triplicate and repeated three times.
Cell migration and invasion assays
For transwell migration assay, 1×105 transfected PCa cells were plated in the top chamber with the non-coated membrane (24-well insert; pore size, 8 μm; BD Biosciences). For invasion assay, matrigel (BD biosciences) was polymerized in transwell inserts for 45 min at 37°C. In both assays, cells were plated in the top chamber in medium without serum; the lower chamber was filled with 10% FBS and EGF (25 ng/ml) (Sigma) was used as a chemoattractant. Cells were incubated for 24 h and the cells that did not migrate or invade through the pores were removed by a cotton swab. Cells on the lower surface of the membrane were stained with crystal violet and counted. Each experiment was performed in triplicate.
Western blot analysis
Cells were washed once with cold PBS and lysis in RIPA buffer containing protease inhibitors. Approximately 30 μg of protein was separated with 10% SDS-PAGE gel and blotted onto nitrocellulose membranes. Then membranes were blocked with 5% skim milk at room temperature for 1 h and then incubated with primary antibodies (Abcam) at 4°C overnight, followed by TBST wash and 1 h incubation with horseradish peroxidase conjugated secondary antibodies at room temperature. Protein bands were visualized by a Molecular Imager ChemiDoc XRS System (Bio-Rad Laboratories).
Luciferase assay
Cells grown in the 96-well plate were co-transfected with either empty vector (pmirGLO) or miR-204 and luciferase reporter comprising 3’UTR of ATF2, wild type (Wt-UCA1) or mutant UCA1 (Mut-UCA1) fragment, using Lipofectamie 2000 (Invitrogen). Cells were harvested 48 h after transfection and luciferase activity was measured as chemiluminescence in a luminometer (PerkinElmer Life Sciences) using the Dual-Luciferase reporter assay system (Promega) according to the manufacturer’s protocol.
Xenograft mouse model
PC-3 cells (1×106) stably expressing sh-UCA1 were subcutaneously injected into either side of the flank area of 4-week-old male athymic nude mice. Tumor size was measured (0.5× length × width2) in mice on a weekly basis. After 6 weeks, the nude mice were sacrificed and the tumor tissues were excised and fixed in 4% paraformaldehyde solution for further study. All animal studies were performed under the supervision and guidelines of the Institutional Animal Care and Use Committee of Zhengzhou University.
RNA immunoprecipitation (RIP)
Anti-MS2 RIP and anti-AGO2 RIP was performed as previously describe by using the EZ4magna RNA Immunoprecipitation Kit (Millipore) [16].
Statistical analysis
All the statistical analyses were performed using SPSS 17.0 software. Data was presented as mean ± SD from at least three separate experiments. Overall survival rates were calculated according to the Kaplan-Meier method and survival curves were plotted. For comparisons, Student’s t test was used when two groups comparison. A P value less than 0.05 was considered significant.
Results
Expression of UCA1 was increased in PCa and associated with poor prognosis
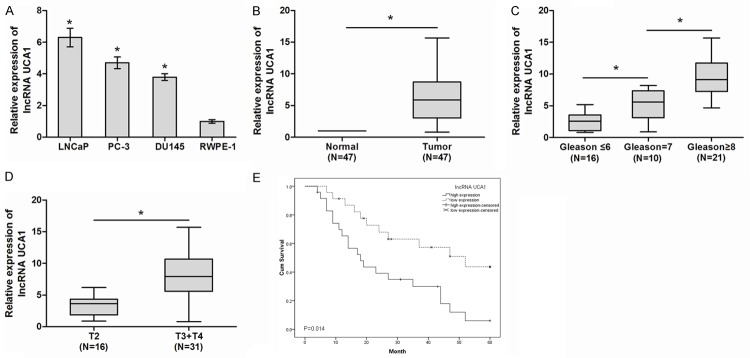
To determine whether UCA1 was involved in the tumorigenesis of PCa, qRT-PCR was used to detect the expression of UCA1 in PCa cell lines and tissue samples. Our results showed that UCA1 expression was significantly increased in all three PCa cell lines (LNCaP, PC-3 and DU145) compared with that in normal prostate epithelial cell line RWPE-1 (Figure 1A, P<0.05). In addition, we explored the expression levels of UCA1 in 47 paired PCa tissues and adjacent non-tumor tissues. QRT-PCR showed that UCA1 expression was significantly up-regulated in PCa tissues compared with adjacent non-tumor tissues (Figure 1B, P<0.05). Examination of the association between UCA1 expression and clinical pathological features showed that UCA1 up-regulation was correlated with high gleason score and advanced TNM stage (Figure 1C, 1D, P<0.05). Furthermore, Kaplan-Meier analysis showed that overall survival of PCa patients with high UCA1 expression had a significantly poorer prognosis than those with low UCA1 expression (Figure 1E, P<0.05). These results suggested that UCA1 is involved in PCa aggressiveness and progression.
Figure 1.
LncRNA UCA1 expression and its clinical significance in PCa. (A) Relative expression of UCA1 in PCa cells (LNCaP, PC-3 and DU145) and normal prostate epithelial cell line RWPE-1. (B) Relative expression of UCA in pair 47 PCa tissue samples and adjacent non-tumor tissue samples. (C, D) UCA1 expression correlated positively with high gleason score (C) and advanced TNM stage (D). (E) The Kaplan-Meier overall survival curves by UCA1 levels. Patients with elevated UCA1 expression showed reduced survival times compared with patients with low levels of UCA1 expression (log-rank test, P<0.05). *P<0.05.
Knockdown of UCA1 suppressed cell proliferation and metastasis in vitro and in vivo
To investigated the role of UCA1 in PCa progression, we stably inhibited UCA1 in two PCa cell lines (LNCaP and PC-3) with lentiviruses carrying shRNA for UCA1 (sh-UCA1) and a control nonspecific shRNA (Con) (Figure 2A, P<0.05). CCK8 assay showed that inhibition of UCA1 suppressed PCa cell proliferation (Figure 2B, P<0.05). Transwell assay showed that knockdown UCA1 inhibited PCa cell migration and invasion ability (Figure 2C, 2D, P<0.05). Furthermore, effects of PCa cells treatment with sh-UCA1 on the growth were also evaluated in vivo. During the whole tumor growth period, tumors from the sh-UCA1 transfected PCa cells grew lower than that of control nonspecific shRNA (Con) transfected groups (Figure 3A, P<0.05). After a 6-week inoculation, the average weight of tumors developed from sh-UCA1 transfected cells was obviously larger than those of Con treated group (Figure 3B, P<0.05). These results reported that UCA1 could promote the proliferation and metastasis of PCa cells both in vitro and in vivo.
Figure 2.
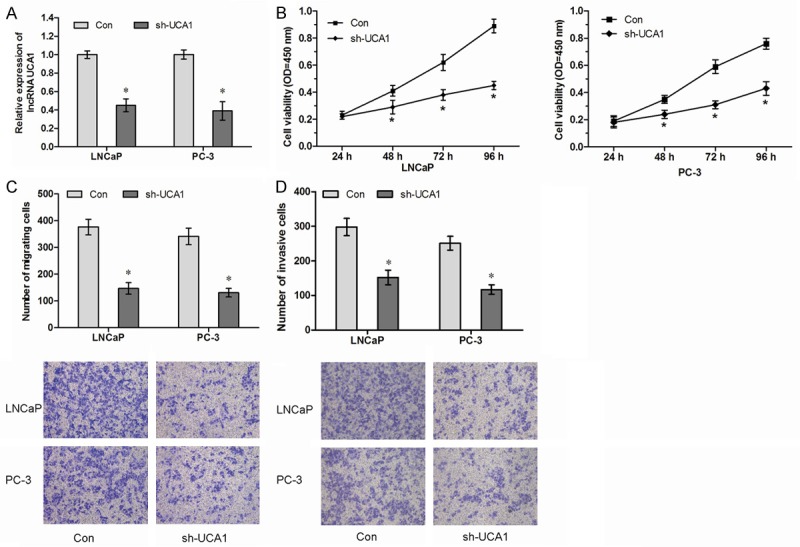
Knockdown of lncRNA UCA1 inhibits PCa cell proliferation and metastasis in vitro. A. LNCaP and PC-3 were transfected with sh-UCA1. The relative expression of UCA1 was examined by qRT-PCR and normalized to GAPDH expression. B. Knockdown of UCA1 decreased proliferation of LNCaP and PC-3 cells, as detected by CCK8 assay. C. Knockdown of UCA1 decreased migration of LNCaP and PC-3 cells, as detected by Transwell migration assay. D. Knockdown of UCA1 suppressed invasion of LNCaP and PC-3 cells, as detected by Transwell invasion assay. *P<0.05.
Figure 3.
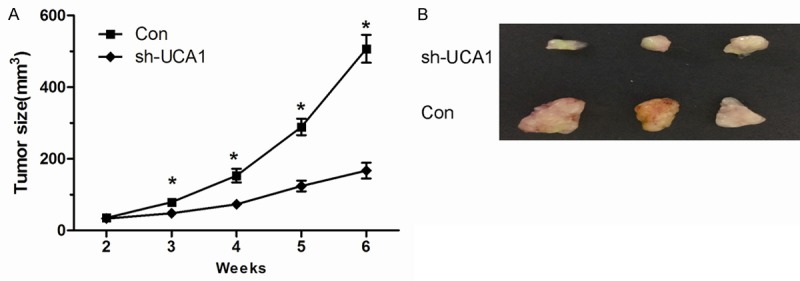
Knockdown of lncRNA UCA1 inhibits tumor growth in vivo. A. Tumor growth curves measured after injection of PC-3 cells stably transfected with sh-UCA1 or Con. The tumor size was calculated every 7 days from 2 to 6 weeks. B. Photographs of tumors excised 6 weeks after inoculation of stably sh-UCA1 transfected PC-3 cells into nude mice. *P<0.05.
MiR-204 bind to and suppressed UCA1 expression
Recently, mounting evidence showed that lncRNAs contain motif with sequence complementary to miRNAs and have an inhibition effect on miRNAs expression and activity [17]. A recent study reported that UCA1 could sponge endogenous miR-204 and inhibited its activity in colorectal cancer [18]. However, the direct interaction of UCA1 and miR-204 in PCa remains unclear. To confirm the direct binding between UCA1 and miR-204, we performed RIP assay to pull-down miRNAs interacted with UCA1. The results showed that UCA1 was significantly enriched for miR-204 compared to the empty vector (MS2) and UCA1 with mutations in miR-204 targeting sites (mut-UCA1) (Figure 4A, P<0.05). In addition, we constructed luciferase reporter plasmid containing the wild-type UCA1 (Wt-UCA1) or mutant UCA1 (Mut-UCA1) with mutated miR-204 binding sites. We found that miR-204 overexpression decreased the luciferase activities of the Wt-UCA1 reporter vector but not empty vector or Mut-UCA1 reporter vector (Figure 4B, P<0.05). Finally, we performed anti-AGO2 RIP to detect whether UCA1 was regulated by miR-204 in an AGO2 dependent manner. Endogenous UCA1 pull-down by AGO2 was significantly enriched in miR-204 overexpressed cells (Figure 4C, P<0.05), indicating that miR-204 is a bonafide UCA1 targeting miRNA.
Figure 4.
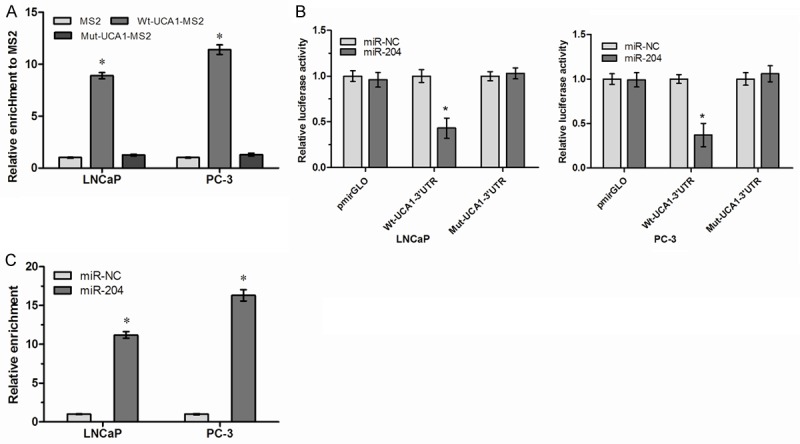
lncRNA UCA1 is physically associated with miR-204. A. MS2-RIP followed by qRT-PCR to detect endogenous miR-204 associated with UCA1. B. Luciferase activity in LNCaP and PC3 cells co-transfected with miR-204 and luciferase reporters containing nothing (pmirGLO), Wt-UCA1 or Mut-UCA1. C. Anti-AGO2 RIP was performed in LNCaP and PC3 cells transfected with miR-204, followed by qRT-PCR to detect UCA1 associated with AGO2. *P<0.05.
UCA1 functioned as a ceRNA of ATF2
Recent studies showed that ATF2 is a target gene of miR-204 [19]. Thus, we suspected that UCA1 may regulate ATF2 expression through functioning as a ceRNA. To confirm this hypothesis, we detected the mRNA and protein level of ATF2 in UCA1 overexpression PCa cells. Surprisedly, we found that up-regulated expression of UCA1 promoted ATF2 expression both in mRNA and protein levels (Figure 5A, 5B, P<0.05). For rescue experiment, we transiently transfected with miR-204 in Wt-UCA1 or Mut-UCA1 overexpressed cells. Overexpression of Wt-UCA1, but not the Mut-UCA1, increased ATF2 expression. Furthermore, Overexpression of miR-204 abolished the increase of ATF2 induced by UCA1 (Figure 5C, 5D, P<0.05).
Figure 5.
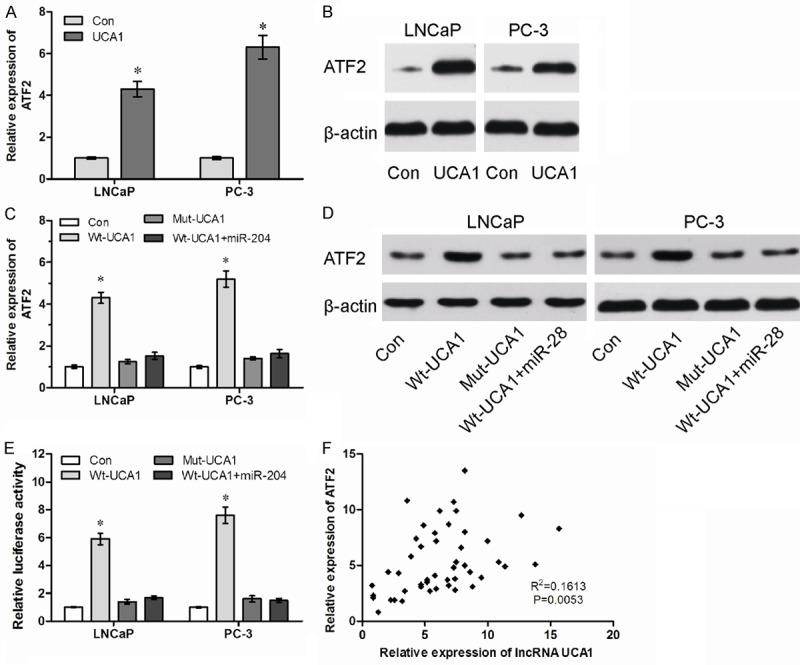
LncRNA UCA1 functions as a ceRNA of ATF2. A. The mRNA levels of ATF2 in control (Con) and UCA1 overexpression (UCA1) PCa cells were determined by qRT-PCR. B. The protein levels of ATF2 in Con and UCA1 PCa cells were determined by western blot. C. The relative mRNA levels of ATF2 in Wt or Mut UCA1 overexpressed cells with or without overexpression of miR-204. D. The relative protein levels of ATF2 in Wt or Mut UCA1 overexpressed cells with or without overexpression of miR-204. E. The relative luciferase activity of ATF2 3’UTR in Wt or Mut UCA1 overexpressed cells with or without overexpression of miR-204. F. The correlation analysis between UCA1 and ATF2 expression in 47 paired PCa tissue samples. *P<0.05.
To further confirm whether UCA1 influenced ATF2 expression depends on the regulation of ATF2 3’UTR, we constructed luciferase reporters containing ATF2 3’UTR (pmirGLO-ATF2). The luciferase plasmid (pmirGLO-ATF2) was transfected into the Wt-UCA1 or Mut-UCA1 overexpressed cells with or without miR-204. We found that Overexpression of Wt-UCA1, but not the Mut-UCA1, increased the luciferase activity of ATF2 3’UTR, while miR-204 abolished this effect (Figure 5E, P<0.05). We next detected the expression levels of ATF2 in 47 PCa tissues. We found that lncRNA UCA1 expression was significantly correlated with ATF2 mRNA expression (Figure 5F, P<0.05). All these results suggested an important role of UCA1 in regulating ATF2 expression by competitively binding miR-204.
LncRNA UCA1 promoted cell proliferation and metastasis through regulation of ATF2
Finally, we detected whether UCA1 promoted cell proliferation and metastasis through regulation with ATF2 expression. We transfected si-ATF2 in UCA1 overexpression PCa cells (Figure 6A, P<0.05). We found that decreased expression of ATF2 abolished the promotion of cell proliferation mediated by UCA1 overexpression (Figure 6B, P<0.05). Furthermore, our data showed that inhibition of ATF2 abolished the promotion of cell migration and invasion mediated by UCA1 overexpression (Figure 6C, 6D, P<0.05). These results suggested that UCA1 promotes cell progression through regulation of ATF2 (Figure 7).
Figure 6.
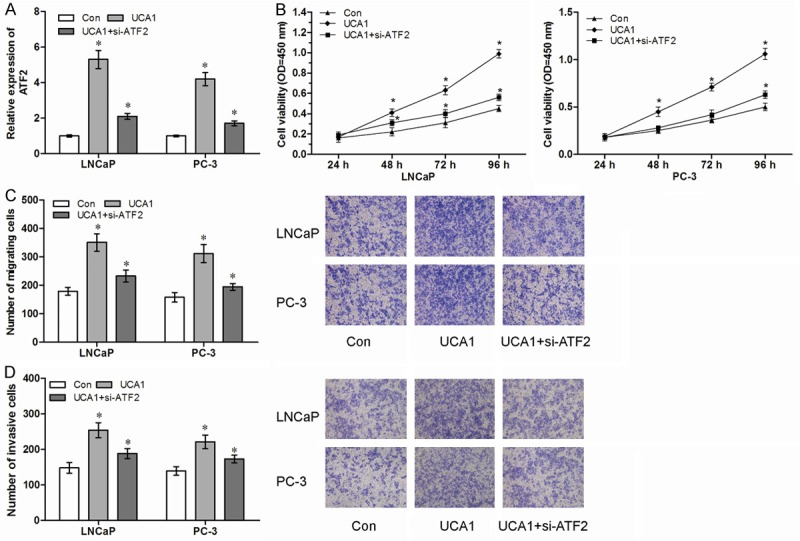
LncRNA UCA1 promotes cell proliferation and metastasis through regulation of ATF2. (A) The relative expression of ATF2 in UCA1 overexpression cells with or without siRNA against ATF2 (si-ATF2). (B-D) Decreased expression of ATF2 abolished the promotion of proliferation (B), migration (C) and invasion (D) by UCA1 overexpression. *P<0.05.
Figure 7.
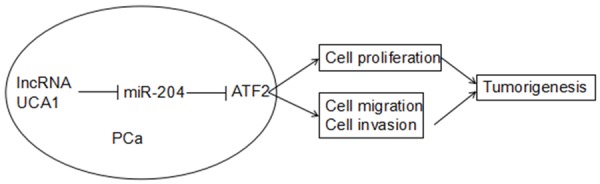
Schematic model of lncRNA UCA1 functions as a ceRNA to regulate ATF2 expression by sponging miR-204 in PCa.
Discussion
LncRNAs are transcribed RNA molecules >200 nucleotides in length that lack significant protein-coding potential, however, they can regulate protein-coding genes at epigenetic, transcriptional and post-transcriptional levels and play central roles in physiological processes [2]. Increasing evidence suggests that a variety of lncRNAs are frequently aberrantly expressed in cancers, exhibiting spatially and temporally regulated expression patterns. These differential expression lncRNAs are closely related to tumorigenesis, metastasis, prognosis or diagnosis, serving as roles of oncogenes or tumor suppressor genes [20]. Thus, more efforts should be made to deeply clarify the biological and molecular mechanisms of lncRNAs in cancer.
In this study, we detected the expression of lncRNA UCA1 in PCa. Our data showed that lncRNA UCA1 was significantly increased in PCa cell lines and tissues, high expression of UCA1 was closely associated with high gleason score and advanced TNM stage. Moreover, we demonstrated that high lncRNA UCA1 expression was correlated with poorer overall survival of PCa patients. In function assay, we showed that inhibition lncRNA UCA1 expression suppressed prostate cell proliferation and metastasis both in vitro and in vivo. Therefore, our findings suggested that lncRNA UCA1 could act as an oncogene and a potential therapeutic target for PCa.
Increasing evidences demonstrated that lncRNAs play critical roles in PCa progression. For example, Jia et al showed that lncRNA DANCR promoted prostate cancer invasion and metastasis through repressing the expression of TIMP2/3, and suggested that lncRNA DANCR could be a potential target for preventing prostate cancer metastasis [21]. Wang et al showed that lncRNA MALAT1 bind to EZH2 and enhanced oncogenic activities of EZH2 in castration-resistant prostate cancer [22]. Misawa et al suggested that androgen-induced lncRNA SOCS2-AS1 promoted cell growth and inhibited apoptosis by regulating tumor necrosis factor superfamily 10 (TNFSF10) in prostate cancer cells [23].
However, to our knowledge, the mechanism related to ceRNA in PCa is still poorly. In this study, we demonstrated that lncRNA UCA1 promoted cell proliferation and metastasis by functioning as a ceRNA of ATF2. UCA1 completed with ATF2 for common miR-204 which was an important tumor suppressor miRNA in various cancers. UCA1 directly interacted with miR-204 and decreased the binding of miR-204 to ATF2 3’UTR, which suppressed the degradation of ATF2 mRNA by miR-204. Previous studies showed that lncRNA UCA1 could function as ceRNAs of other mRNAs in other cancers. For example, Nie et al showed that lncRNA UCA1 functioned as an oncogene in NSCLC, acting mechanistically by up-regulating ERBB4 in part through ‘sponging’ miR-193a-3p [24]. Wang indicated that up-regulated lncRNA UCA1 contributed to the progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway [25]. Our study enriched the ceRNA network regulated by UCA1.
Activating transcription factor 2 (ATF2) is a member of the basic helix-loop-helix (b-ZIP) transcription factor family, whose transcriptional activities are mediated by stress-activated kinases JNK/p38 and activated by phosphorylation on threonine residues [26,27]. Recent studies indicated that ATF2 play an important role in tumor progression. For example, Wu et al showed that ATF2 exerted an oncogenic role in renal cell progression and correlated with aggressive clinico-pathological characteristics and poor prognosis of renal cancer patients [28]. Zhang et al showed that miR-622 suppressed proliferation, invasion and migration by directly targeting ATF2 in glioma cells [29]. Lacono et al suggested that ATF2 contributed to cisplatin resistance in non-small cell lung cancer and celastrol induceds cisplatin resensitization through inhibition of JNK/ATF2 pathway [30]. However, whether ATF2 expression is regulated by lncRNA remains unclear. In the present study, we showed that ATF2 was regulated by lncRNA UCA1 via a ceRNA mechanism. Our data indicated that overexpression of lncRNA UCA1 promoted ATF2 expression. Moreover, lncRNA UCA1 expression was positively correlated with ATF2 expression in PCa tissues. These data strongly demonstrated that ATF2 is a direct target gene of lncRNA UCA1.
In summary, our study demonstrated that lncRNA UCA1 could function as a ceRNA to promote ATF2 expression by sponging miR-204. Therefore, lncRNA UCA1 might serve as a therapeutic target as well as prognostic biomarker in PCa.
Acknowledgements
This project was supported by The Foundation and Frontier Technology Research Program of Henan Health Department (No.A-001-0068).
Disclosure of conflict of interest
None.
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Sharifi N, Gulley JL, Dahut WL. Androgen deprivation therapy for prostate cancer. JAMA. 2005;294:238–244. doi: 10.1001/jama.294.2.238. [DOI] [PubMed] [Google Scholar]
- 3.Karantanos T, Corn PG, Thompson TC. Prostate cancer progression after androgen deprivation therapy: mechanisms of castrate resistance and novel therapeutic approaches. Oncogene. 2013;32:5501–5511. doi: 10.1038/onc.2013.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Lorenzo G, Buonerba C, Autorino R, De Placido S, Sternberg CN. Castration-resistant prostate cancer. Drugs. 2010;70:983–1000. doi: 10.2165/10898600-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 5.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, Asangani IA, Ateeq B, Chun SY, Siddiqui J, Sam L, Anstett M, Mehra R, Prensner JR, Palanisamy N, Ryslik GA, Vandin F, Raphael BJ, Kunju LP, Rhodes DR, Pienta KJ, Chinnaiyan AM, Tomlins SA. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 7.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 8.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Zhao J, Zhang W, Gan J, Hu C, Huang G, Zhang Y. lncRNA GAS5 enhances G1 cell cycle arrest via binding to YBX1 to regulate p21 expression in stomach cancer. Sci Rep. 2015;5:10159. doi: 10.1038/srep10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiao Q, Li H. LncRNA FER1L4 suppresses cancer cell proliferation and cycle by regulating PTEN expression in endometrial carcinoma. Biochem Biophys Res Commun. 2016;478:507–512. doi: 10.1016/j.bbrc.2016.06.160. [DOI] [PubMed] [Google Scholar]
- 12.Nortier JL, Martinez MC, Schmeiser HH, Arlt VM, Bieler CA, Petein M, Depierreux MF, De Pauw L, Abramowicz D, Vereerstraeten P, Vanherweghem JL. Urothelial carcinoma associated with the use of a Chinese herb (aristolochia fangchi) N Engl J Med. 2000;342:1686–1692. doi: 10.1056/NEJM200006083422301. [DOI] [PubMed] [Google Scholar]
- 13.Li W, Xie P, Ruan WH. Overexpression of lncRNA UCA1 promotes osteosarcoma progression and correlates with poor prognosis. J Bone Oncol. 2016;5:80–85. doi: 10.1016/j.jbo.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Cao X, Zhang X, Sheng H, Tao K. UCA1 overexpression predicts clinical outcome of patients with ovarian cancer receiving adjuvant chemotherapy. Cancer Chemother Pharmacol. 2016;77:629–634. doi: 10.1007/s00280-016-2963-4. [DOI] [PubMed] [Google Scholar]
- 15.Wang X, Gao Z, Liao J, Shang M, Li X, Yin L, Pu Y, Liu R. lncRNA UCA1 inhibits esophageal squamous-cell carcinoma growth by regulating the Wnt signaling pathway. J Toxicol Environ Health A. 2016;79:407–418. doi: 10.1080/15287394.2016.1176617. [DOI] [PubMed] [Google Scholar]
- 16.Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ, Tao QF, Liu F, Pan W, Wang TT, Zhou CC, Wang SB, Wang YZ, Yang Y, Yang N, Zhou WP, Yang GS, Sun SH. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell. 2014;25:666–681. doi: 10.1016/j.ccr.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 17.Jalali S, Bhartiya D, Lalwani MK, Sivasubbu S, Scaria V. Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS One. 2013;8:e53823. doi: 10.1371/journal.pone.0053823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bian Z, Jin L, Zhang J, Yin Y, Quan C, Hu Y, Feng Y, Liu H, Fei B, Mao Y, Zhou L, Qi X, Huang S, Hua D, Xing C, Huang Z. LncRNA-UCA1 enhances cell proliferation and 5-fluorouracil resistance in colorectal cancer by inhibiting miR-204-5p. Sci Rep. 2016;6:23892. doi: 10.1038/srep23892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang S, Gao L, Thakur A, Shi P, Liu F, Feng J, Wang T, Liang Y, Liu JJ, Chen M, Ren H. miRNA-204 suppresses human non-small cell lung cancer by targeting ATF2. Tumour Biol. 2016;37:11177–11186. doi: 10.1007/s13277-016-4906-4. [DOI] [PubMed] [Google Scholar]
- 20.Gibb EA, Vucic EA, Enfield KS, Stewart GL, Lonergan KM, Kennett JY, Becker-Santos DD, MacAulay CE, Lam S, Brown CJ. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011;6:e25915. doi: 10.1371/journal.pone.0025915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia J, Li F, Tang XS, Xu S, Gao Y, Shi Q, Guo W, Wang X, He D, Guo P. Long noncoding RNA DANCR promotes invasion of prostate cancer through epigenetically silencing expression of TIMP2/3. Oncotarget. 2016;7:37868–37881. doi: 10.18632/oncotarget.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang D, Ding L, Wang L, Zhao Y, Sun Z, Karnes RJ, Zhang J, Huang H. LncRNA MALAT1 enhances oncogenic activities of EZH2 in castration-resistant prostate cancer. Oncotarget. 2015;6:41045–41055. doi: 10.18632/oncotarget.5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Misawa A, Takayama K, Urano T, Inoue S. Androgen-induced long noncoding RNA (lncRNA) SOCS2-AS1 promotes cell growth and inhibits apoptosis in prostate cancer cells. J Biol Chem. 2016;291:17861–17880. doi: 10.1074/jbc.M116.718536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nie W, Ge HJ, Yang XQ, Sun X, Huang H, Tao X, Chen WS, Li B. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371:99–106. doi: 10.1016/j.canlet.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 25.Wang F, Ying HQ, He BS, Pan YQ, Deng QW, Sun HL, Chen J, Liu X, Wang SK. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6:7899–917. doi: 10.18632/oncotarget.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodríguez M, Domingo E, Alonso S, Frade JG, Eiros J, Crespo MS, Fernández N. The unfolded protein response and the phosphorylations of activating transcription factor 2 in the trans-activation of il23a promoter produced by β-glucans. J Biol Chem. 2014;289:22942–22957. doi: 10.1074/jbc.M113.522656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruhat A, Jousse C, Carraro V, Reimold AM, Ferrara M, Fafournoux P. Amino acids control mammalian gene transcription: activating transcription factor 2 is essential for the amino acid responsiveness of the CHOP promoter. Mol Cell Biol. 2000;20:7192–7204. doi: 10.1128/mcb.20.19.7192-7204.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu DS, Chen C, Wu ZJ, Liu B, Gao L, Yang Q, Chen W, Chen JM, Bao Y, Qu L, Wang LH. ATF2 predicts poor prognosis and promotes malignant phenotypes in renal cell carcinoma. J Exp Clin Cancer Res. 2016;35:108. doi: 10.1186/s13046-016-0383-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang R, Luo H, Wang S, Chen Z, Hua L, Wang HW, Chen W, Yuan Y, Zhou X, Li D, Shen S, Jiang T, You Y, Liu N, Wang H. MiR-622 suppresses proliferation, invasion and migration by directly targeting activating transcription factor 2 in glioma cells. J Neurooncol. 2015;121:63–72. doi: 10.1007/s11060-014-1607-y. [DOI] [PubMed] [Google Scholar]
- 30.Iacono M, Monica V, Vavalà T, Gisabella M, Saviozzi S, Bracco E, Novello S, Papotti M, Scagliotti GV. ATF2 contributes to cisplatin resistance in nonsmall cell lung cancer and celastrol induces cisplatin resensitization-through inhibition of JNK/ATF2 pathway. Int J Cancer. 2015;136:2598–2609. doi: 10.1002/ijc.29302. [DOI] [PubMed] [Google Scholar]