Abstract
The T-box gene family refers to a group of transcription factors that share a highly conserved, sequence-specific DNA-binding domain (T-box) containing around 180-amino acids. According to HUGO gene nomenclature committee (HGNC), there are 18 T-box family members. These T-box genes have been implicated essential roles during embryogenesis and cardiac development, given their specific expression pattern in developing mammalian heart for several T-box genes, including TBX5. TBX5 is consisted of three transcriptional variants which cover 9 exons and encode two distinct isoforms that differ in N-terminus. TBX5 is probably the most frequently studied T-box gene over the past decade due to the typical cardiac defects observed in Holt-Oram syndrome (HOS), which is caused by TBX5 mutation. Most of the mutations are within exons 3-7 where locate sequence coding for the T-box domain. Notably, a variety of cardiac defects, as well as abnormalities in limb and other organs have been seen in HOS syndrome with different kinds of TBX5 mutations, suggesting a heterogeneous disease mechanism. We have performed a meta-analysis of TBX5 and found a significant correlation between its single nucleotide polymorphism (SNP) rs3825214 (A to G), and risk of atrial fibrillation and its subtypes, supporting TBX5 as a master transcription factor for cardiac development. In addition, bioinformatics analysis of this SNP identified several TFs that may be affected for their binding affinity with TBX5. Identification and characterization of more TBX5 mutations and SNPs hold promise for therapeutic strategy targeting TBX5 associated developmental abnormalities and diseases.
Keywords: T-box gene TBX5 mutation, Holt-Oram syndrome, SNP rs3825214, atrial fibrillation, meta-analysis
Introduction
The T-box gene family is an ancient gene family as indicated by phylogenetic analysis. T-box genes are believed to have arisen from a common metazoan ancestor and from a genome wide duplication that occurred over 600 million years ago during the early evolution of vertebrates [1]. These T-box family members, which share a highly conserved 180 amino acid T-box DNA-binding domain, exist in a wide range of organisms, including nematodes, frog, chick, mouse, and human [2-3]. No apparent sequence similarity was observed between T-box and any other DNA-binding motif of known transcription factors (TFs) [4]. Therefore, the T-box genes are unique and have been implicated in early embryonic cell fate decision, regulation of the development of extraembryonic structures, embryonic patterning, as well as many aspects of organogenesis [5,6]. Among these gene family members, TBX5 has been extensively studied over the past decade due to its mutation and correlation with the typical cardiac defects observed in Holt-Oram syndrome (HOS) [7,8]. However, there are multiple mutations of TBX5 which mostly occur within the T-box coding region. The resultant clinical symptoms and cardiac or limb defects are not unanimous, suggesting a diverse genotype/phenotype correlation and a heterogeneous disease mechanism [9].
In this manuscript, we first summarized the T-box family of TFs by focusing on the group of TBX-2, one of the five subgroups of T-box family members. The close subfamily member of TBX5, gene TBX4, was briefly reviewed with focus on illustration and comparison of the mRNA transcripts of human and mouse T-box 5. We then summarized the mutations of TBX5 and their association with HOS. The pathological manifestations and the mechanisms that cause the diversity of HOS were as described. A meta-analysis of TBX5 single nucleotide polymorphism (SNP) indicates that rs3825214 (A to G) is highly associated with decreased risk of atrial fibrillation (AF) and its subtypes.
Materials and methods
Gene structure of human and mouse T-box 5 transcription factor
The NCBI (The National Center for Biotechnology Information) gene database were searched for human and mouse T-box 5 transcription factor using terms TBX-5/homo sapiens and Tbx-5/mus musculus respectively. The structure of mRNA transcripts of human and mouse T-box 5, including their untranslated region, coding sequence (exons) and the regions where locate the binding and other functional motifs, was compared and illustrated based on data most recently updated in the NCBI database in December 2016 (https://www.ncbi.nlm.nih.gov/gene/).
Meta-analysis of TBX5 SNP rs3825214
The MEDLINE and NCBI databases were searched for eligible articles until the end of 2016 using keywords “Tbx5”, “rs3825214”, and “atrial fibrillation”. Google academic searching was also performed to obtain additional information that may be relevant for above searching terms. The inclusion and exclusion criteria was set for eligible studies and due to limited sample size of the studies selected, the statistical efficacy of the meta-analysis was calculated using web-based software Power and Precision Version 4 (available at http://www.power-analysis.com). This is a stand-alone statistical power analysis software that is used for the calculation of a sample size for a planned study. The power to detect an association between SNP rs3825214 and lone or total AFs was conducted to obtain the odds ratio (OR) with acceptable type I error probability of 0.05.
Bioinformatics analysis of TBX5 SNP rs3825214
Potential transcription factor binding sites that may be affected by SNP rs3825314 and its normal allele were searched using the Transcription Factor Affinity Prediction software sTRAP (available at http://trap.molgen.mpg.de/cgi-bin/trap_two_seq_form.cgi). This is a web-based tool that was developed and maintained at the Computational Molecular Biology Department at the Max Plank Institute for Molecular Genetics in Berlin, Germany. Candidate factors that show a binding difference with a p-value less than 0.05 were selected and subjected to function analysis based on literature review.
Results and discussion
T-box family of transcription factors
Based on HUGO gene nomenclature committee, there are 18 T-box gene family members (http://www.genenames.org/cgi-bin/genefamilies/set/766), including one pseudogene T-box 23 (TBX-23). These genes are divided into five subgroups, including T (Brachyury), TBX1, TBX2, TBX6, and TBR1 (Figure 1). The T-box family members are generally known as essential transcription factors for heart and limb development, as at least seven members of the T-box gene family are expressed in the embryonic heart in humans and vertebrate models. These genes include TBX1-5, TBX18 and TBX20, most of which belong to the TBX-2 subfamily. TBX-2 subfamily members have recently been extensively studied given their important function during multiple biological processes. Among these subfamily members, TBX2 has been implicated in several developmental processes such as patterning and morphogenesis of a wide range of tissues, coordinating cell fate and organs of limb, kidney, lung, mammary gland, heart, as well as craniofacial structure.TBX2 is also overexpressed in several cancers including carcinoma, melanoma, pancreatic, small cell lung cancer, breast, bladder, and liver cancers and can suppress senescence, a cellular process also relates to cancer development [3]. TBX3 is an important transcriptional repressor which involves multiple tissue development. TBX3 is also overexpressed in multiple cancer cells including bladder carcinoma, melanoma, malignant primary breast cancer, and immortalized breast cancer cell lines [4,5,10,11]. Other functions of TBX3 include potential roles in bone development [12]. These studies suggest the importance of TBX3 in the proliferation and specification of cells and in relevant tissue development.
Figure 1.
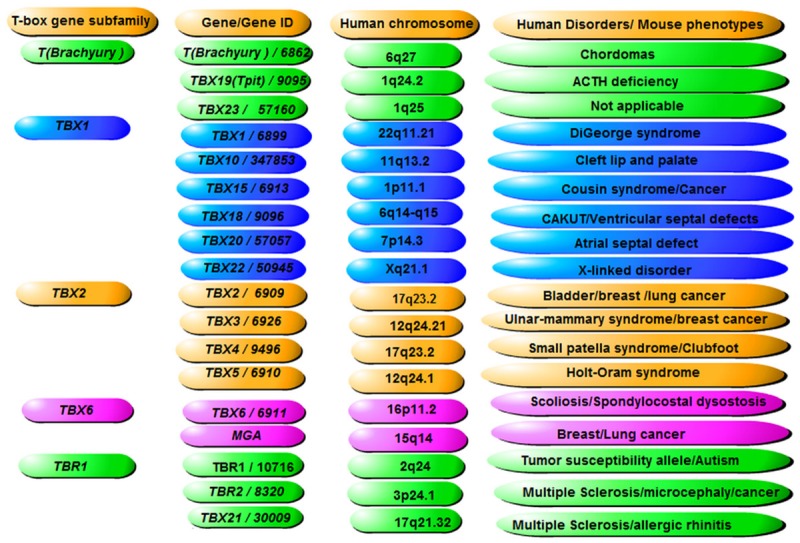
Human T-box family of transcription factors. Based on HUGO gene nomenclature committee, there are 18 T-box gene family members (http://www.genenames.org/cgi-bin/genefamilies/set/766), including one pseudogene T-box 23 (TBX-23). These genes are divided into 5 subgroups, including T (Brachyury), TBX1, TBX2, TBX6, and TBR1. Listed are gene and gene IDs, their chromosomal localizations and the corresponding human diseases or mouse phenotypes that relate to the listed genes respectively.
T-box family of transcription factor-TBX4
It has been generally accepted that TBX3 is the closest family member of TBX5, as both TBX3 and TBX5 have been demonstrated to play essential roles in heart development, while mutations of TBX3 and TBX5 have been associated with HOS with distinct cardiac defect [13]. However, Tbx4 is also an important member of the T-box transcription factor family, which plays important roles during embryonic development through modulating expression of target genes [5]. Murine Tbx4 is known to be essential for the formation of the umbilicus, skeletal muscular and hindlimb development. Tbx4 is associated primarily with the hindlimb, which may involve regulation of Tbx2, but forelimb expression of Tbx4 has also been observed [14,15]. In addition, Tbx4 was identified as a novel transcriptional activator of Shox2 in both fore- and hind-limbs, which strongly suggests that Shox2 acts as a feedback modulator of Tbx4 during limb development [16]. Haploinsufficiency of human TBX4 causes small patella syndrome (SPS), an autosomal-dominant skeletal dysplasia characterized by patellar aplasia or hypoplasia and by anomalies of the knee, pelvis, and foot, including disrupted ossification of the ischia and inferior pubicrami [17]. TBX4 mutations might contribute to childhood-onset pulmonary arterial hypertension (PAH) through a decreased activation of the BMP pathway [18]. Hindlimb specific Tbx4 expression may have evolved concomitantly with the evolution of pelvic fins in fish, which is the origin of the posterior limb pairing [19].
In mice, Tbx4 null mutants lack chorio-allantoic fusion, which prevents formation of the umbilical vessels and results in early lethal at embryonic day 10.5 [15]. In addition, Tbx4-null mouse embryos have abnormal hindlimb development and severe defects in growth of the allantois and vasculogenesis. Such embryos are early lethal due to their inability to establish an umbilical connection and failure to form vessels in the allantois by endothelial cells [15].Previous mouse genetics studies also indicated an important role for Tbx4 and Tbx5 in lung growth and branching through interaction with fibroblast growth factor-10 (Fgf10) but not during tracheal/bronchial cartilage development [20-22]. Notably, the T-box genes Tbx5 and Tbx4 are the earliest factors required to initiate forelimb and hind limb outgrowth respectively.Tbx5 and Tbx4 directly regulate the expression of Fgf10 and may establish an FGF signaling loop that drives successful limb outgrowth [15].
T-box family of transcription factor-TBX5
As a critical member of the T-box family of transcription factors, TBX5 is essential for early cellular commitment, differentiation, and, especially as an evolutionarily conserved dosage-sensitive regulator for heart and limb development [23,24]. Based on latest NCBI database, human TBX5 gene is consisted of three transcriptional variants, namely 1, 3, and 4 (Figure 2A). Variant 1 is the longest TBX5 transcript encoding isoform I, while variant 4 also encode the same isoform I, but lack some 5’-UTR sequence compared to variant 1. Variant 3, which encodes isoform II, lacks one exon at the 5’-end and thus, leads to shorter protein at the N-terminus. Mouse Tbx5 gene is consisted of five transcriptional variants that encode five isoforms a-e (Figure 2B). All five variants encode proteins that contain two nuclear localization segments. The first segment (NLS1) is located within the DNA binding domain and the second segment (NLS2) is located at the C-terminal region [25]. The nuclear availability of Tbx5 is a measure of its transcriptional activity. The T-box is required for specific DNA binding and protein-protein interactions, and previous experiments have demonstrated that Tbx5 transcriptionally activates multiple target genes expressed during cardiac development. These genes include ANF, CX40 and SRF, which may singly or synergistically work with their cooperative partners NKX2-5, GATA4 and TBX20 [26]. A normal structure and dosage of TBX5 is essential for upper limb and cardiac development; and mutations in this gene are associated with Holt-Oram syndrome, in which about 85% of the affected individuals have a structural heart defect and/or abnormalities in the cardiac conduction system [27].
Figure 2.
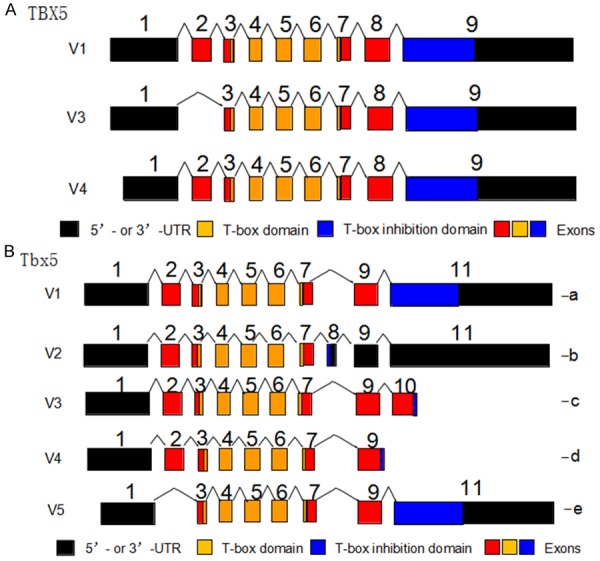
TBX5/Tbx5 gene structure and its functional domains. A. Based on latest NCBI (National Center for Biotechnology Information) database, the human TBX5 gene is consisted of three transcriptional variants, namely 1, 3, and 4. Variant 1 is the longest TBX5 transcript encoding isoform I, while variant 4 also encode the same isoform I, but lack some 5’-UTR sequence compared with variant 1. Variant 3, which encodes isoform II, lacks one exon at the 5’-end and thus, leads to shorter protein at the N-terminus. B. The Tbx5 gene contains five variants (V1, V2, V3, V4, and V5) that are able to encode five Tbx5 protein isoforms a, b, c, d, and e. Variant 1 is consisted of 11 exons and encodes isoform a. Variant 2 is the only transcript that has exon 8 and encodes isoform b. Variant 3 is consisted of 10 exons and encodes isoform c. Variant 4 is consisted of 9 exons, represents the shortest transcript and encodes isoform d. Variant 5 lacks exon 2 and encodes isoform e. All the variants encode two functional domains: the T-box domain (DNA binding domain) and the T-box inhibition domain (Mediate protein-protein interaction). ATG: start codon; TGA: stop codon.
TBX5 and Holt-Oram syndrome pathogenesis
Holt-Oram syndrome (HOS) is an autosomal dominant disorder and a rare syndrome (1 in 100000 live births) characterized by forelimb and cardiac congenital abnormalities. It was first reported by Holt and Oram in 1960 in family members over 4 generations [28]. Clinically, there are variations for Holt-Oram syndrome with the cardiac and skeletal defects vary widely ranging from mild to severe [29]. The most common cardiac anomalies associated with Holt-Oram syndrome include atrial and ventricular septal defects and conduction disease or atrial fibrillation. These classic cardiac defects have been attributed to abnormal interactions of the T-box domain with NKX2-5 and GATA4 [30]. It has been shown that interactions between TBX5 and NKX2-5 are essential for development of the conduction system. Furthermore, TBX5 and NKX2-5 both act synergistically to upregulate CX40 expression which is involved in the normal development of atrioventricular conduction [30]. It has also been shown that there is a physical interaction between TBX5 and members of myocyte enhancer factor 2 C (MEF2C). This interaction leads to a synergistic activation of the α-cardiac myosin heavy chain MYH6, a structural protein of cardiomyocytes [31]. TBX5 is preferentially expressed in the left side of the developing heart where major cardiac anomalies occur. The interactions between TBX5 and the FGF10-FGF8 loop and the SALL4 pathway may explain the pathogenesis of the classic radial ray deficiency of HOS, while the non-classic upper limb phenotypes of HOS may be attributed to the complex interactions between TBX5 and other mesodermal and ectodermal factors [32,33].
The majority of TBX5 mutations introduce premature stop codons and thus truncated proteins that are unable to bind DNA and cause haplo-insufficiency of T-box activity. These mutations are usually accompanied with severe upper limb and cardiac malformations. Some of previous missense mutations of TBX5 and their associated clinical features were summarized in Table 1. It has been reported that mu-tations causing alteration near the amino-terminal end of the T-box will interfere with binding of the major groove of target DNA and cause significant cardiac malformations, while mutations causing amino acid changes at the carboxyl end of the T-box will impact the interaction between TBX5 and the minor groove of target DNA and thus severe limb abnormalities [27]. Reduced Tbx5 dosage may contribute to the risk of heart defects via transcriptional regulation of its target genes [34], while Tbx5 loss-of-function mutation has been associated with lone atrial fibrillation and familial dilated cardiomyopathy [9,35-37]. Notably, correlation between TBX5 mutations and clinical phenotypes of Holt-Oram syndrome have previously been established. For instance, Gly80Arg caused significant cardiac malformations but minor skeletal abnormalities, while Arg237Gln and Arg237Trp caused extensive upper limb malformations but less significant cardiac abnormalities [27]. The Asp61Tyr mutation was seen in a patient with severe aortic and mitral valve prolapse. Meanwhile, the Gly125Arg missense mutation and the intragenic duplication have all been associated with unique limb and cardiac phenotypes [33]. As to the transcriptional regulation of TBX5 and its potential working mechanism in relation to cardiac anomalies, it was previously shown that there is an interaction between Tbx5 and LMP4, a PDZ-LIM domain protein [38]. LMP4 acts as a repressor of Tbx5 activity and is associated with its nucleus and cytoplasm subcellular localization, a cellular activity that is also mediated by a nuclear export signal (NES), the CRM1 export protein [39]. TBX5, as well as Tbx20 and Nkx2-5, may interact with the BAF (Brg1/Brm-associated factor) chromatin remodeling complex and play essential roles during heart development [40]. More recently, TBX5 has been associated with the nucleosome remodeling and deacetylase (NuRD) complex and related to the phenotypic consequence of heart development or congenital heart disease [41,42]. TBX5 mutations may affect its interaction with above complexes, thereby constitute a putative molecular mechanism of limb and heart defects (Figure 3).
Table 1.
Summary of some of the TBX5 mutations and human diseases
| TBX5 mutation | The upper limb anomaly | The severity of cardiac anomaly |
|---|---|---|
| Asp61tyr | Mild RRD | Severe: aortic and mitral valve prolapse with agenesis of the left pericardium |
| Gly80Arg | Mild RRD | Combined defects (severe) |
| Ile106Val | Phocomelia Mild RRD | No cardiac anomalies |
| Gly125Arg | Radial head dislocation | No cardiac anomalies |
| Carpal synostosis | PAF, ASD | |
| Scapular dysplasia | VSD | |
| iRBBB | ||
| Arg237Gln | RRD of variable severity ± triphalangeal thumb | No cardiac anomaly |
| Phocomelia | Mild: isolated ASD or VSD PFO | |
| Severe: not specified | ||
| Arg237Trp | Mild RRD | No cardiac anomaly |
| Severe RRD | Mild: ASD | |
| Severe: AV canal, VSD |
RRD = radial ray deficiency, ASD = atrial septal defect, VSD = ventricular septal defect, PFO = Patent Foramen Ovale, PAF = paroxysmal atrial fibrillation, iRBBB = incomplete right bundle branch block.
Figure 3.
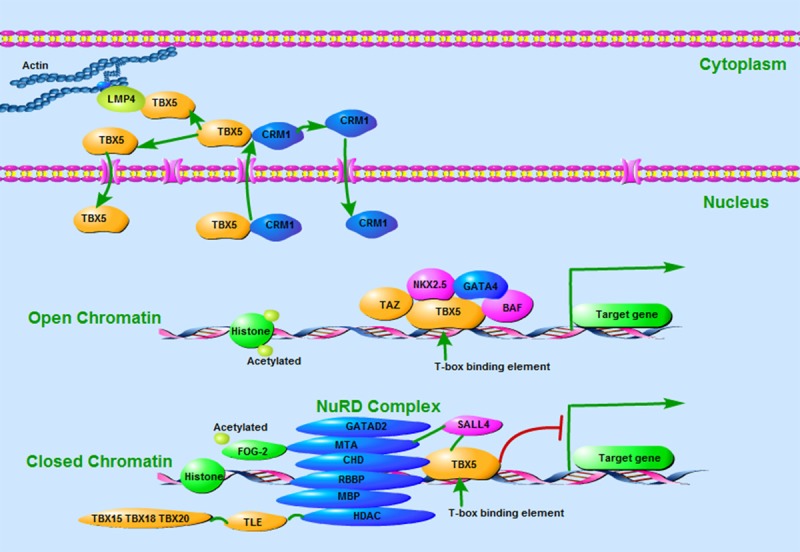
Protein interactions in regulation of Tbx5 transcriptional activity. (1) TBX5 interaction with the PDZ-LIM domain protein (LMP4) or the CRM1 export protein has been associated with its subcellular localization between nucleus and cytoplasm. (2) TBX5 interacts with the BAF chromatin remodeling complex and other transcription factors to activate expression of its downstream target genes. (3) TBX5 binds the NuRD complex via interaction with CHD4 and recruits it to regulatory regions containing T-box binding elements. The NuRD complex deacetylates histones and remodels chromatin to a transcriptionally inactive state, and thus represses target gene expression.
TBX5 SNP rs3825214 is associated with decreased risk of atrial fibrillation
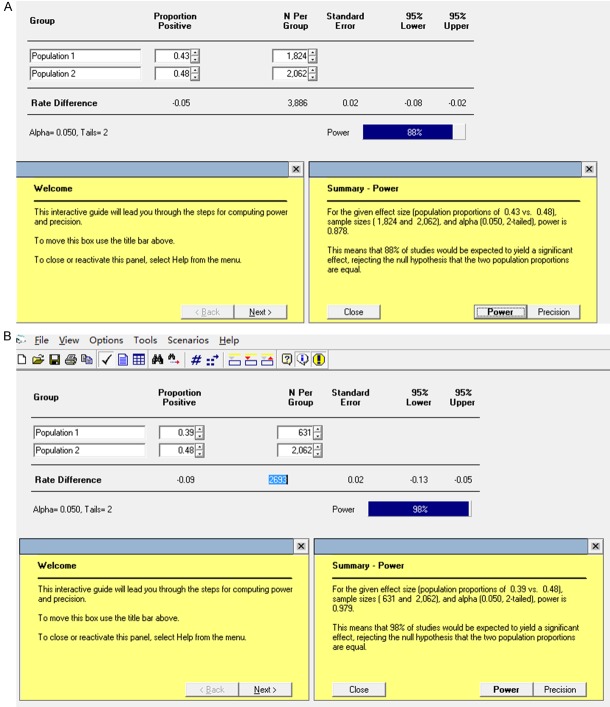
TBX5 SNPs have been associated with several diseases or with increased risk of the disease, such as rs2701108 and barrett’s esophagus which will increase the risk of esophageal adenocarcinoma [43,44]. Previous studies have established the correlation between TBX5 mutations and atrial fibrillation or cardiac defects [9,35-37]. Interestingly, as a master transcription factor for cardiac development, there are only a few reports that show a disease correlation of TBX5 SNPs with cardiac defects, including rs3825214 [45,46]. A previous case-control study with the Chinese Han population has shown a moderate association between rs3825214 and atrial fibrillation but a highly significant association between the G allele of rs3825214 and lone atrial fibrillation [45]. This observation was further confirmed in similar study with larger sample size [46]. We have performed a meta-analysis to specifically evaluated the correlation between TBX5 SNP rs3825214 and atrial fibrillation. The results suggested that rs3825214 showed strong correlation with total and lone atrial fibrillation, atrial fibrillation with hypertension and with both genders (Figure 4). As only limited sample size from two eligible studies were included, we have evaluated the statistical efficacy of the meta-analysis using Power and Precision Version 4 software. The results showed that the statistical power for total atrial fibrillation is close to 88% (Figure 5A), while the efficacy for lone atrial fibrillation is approximately 98% (Figure 5B). These results demonstrate that about 88% and 98% of the studies were expected to yield a significant effect with unequal two populations, and therefore, support a significant correlation between TBX5 SNP rs3825314 and the decreased risk of cardiac anomalies.
Figure 4.
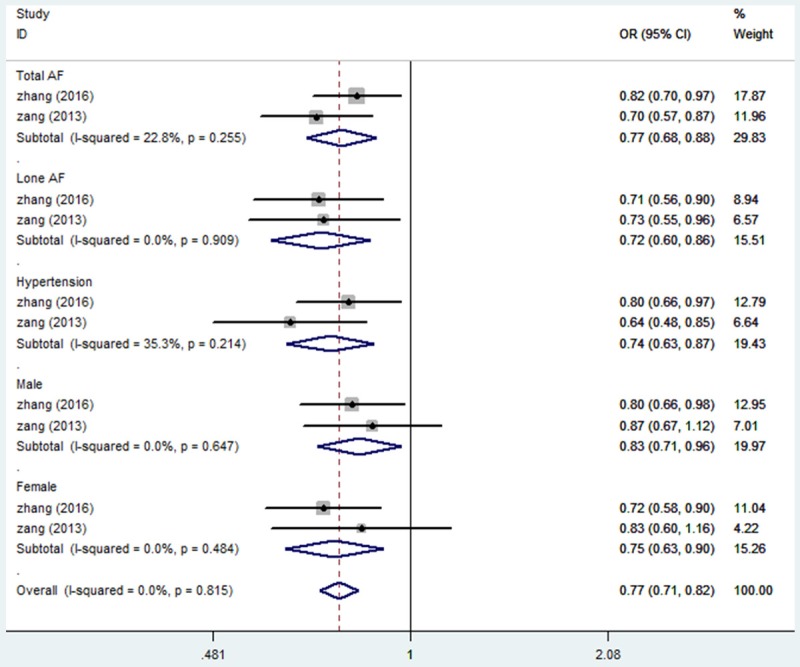
Forest plot of allele comparison of SNP rs3825214 of TBX5. For overall comparison (G vs A, association of rs3825214 and AF), rs3825214 is associated with a significantly decreased risk of AF in allelic comparison (G vs A: OR: 0.72, 95% CI 0.6-0.86), and decreased risk of Lone AF (G vs A: OR: 0.77, 95% CI 0.68-0.88). rs3825214 was associated with AF in either male or female groups.
Figure 5.
Results of the efficacy of meta-analysis of AF and Lone AF. Illustrated are sample/population size and statistical powers for total AF (A) and Lone AF (B). The power to detect an association of rs3825214 with total AF was 88%, while the power of Lone AF was 98%.
Candidate factors interacting with rs3825214 and its normal allele
We have performed in silico sequence analysis of SNP rs3825214 and its normal allele using sTRAP software. This sequence analysis software, available at strap.molgen.mpg.de, is able to analyze variations in the DNA sequence and predicts quantitative changes to the binding strength of any TFs [47]. We successfully identified 7 candidate TFs, including FOXM1, SF1, ERR1, ALPHACP1, ATF3, PAX3, and CP2. The interactions between TBX5 and these factors may potentially be affected due to the sequence variation of this SNP (p<0.05, Figure 6). Based on literature review, these candidate TFs have previously been implicated important biological functions, including cardiac development, which, to some extent, might be attributed to its correlation with TBX5 SNP rs3825214 [48-55]. The SNP variation of rs3825214 may affect the affinity of TFs with TBX5, as SNP rs3825214 has been shown to be adjacent to the enhancer of TBX5 [56]. It is, therefore, predictable that the interaction between TBX5 and NKX2-5 and, possibly GATA4, may be affected, leading to altered expression of downstream target genes and corresponding biological effects.
Figure 6.
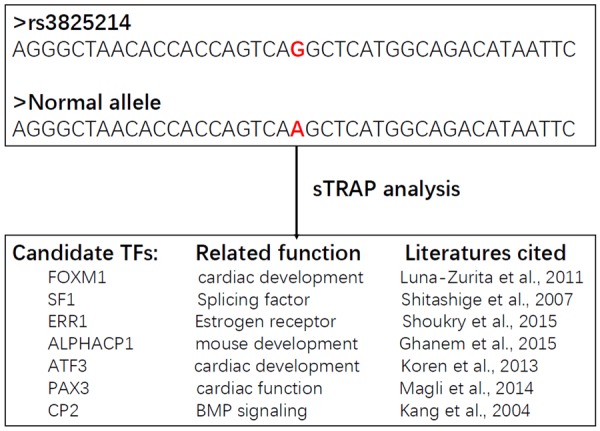
Results of sTRAP analysis of rs3825214 and its normal allele. The sequence of SNP rs3825214 (G allele) and its normal allele (A allele) was shown (top panel). After sTRAP analysis, seven candidate TFs, including FOXM1, SF1, ERR1, ALPHACP1, ATF3, PAX3, and CP2 that potentially show a binding strength change with a p-value less than 0.05 were selected (bottom panel). Based on literature review, these candidate TFs have been implicated essential roles in cardiac development, as well as other important biological functions.
In summary
TBX5 has been established as a master transcription factor for cardiac development. It has also been shown that TBX5, as well as TBX4 are essential for limb development. TBX5 mutations and SNPs may cause the cardiac and limb diseases and/or disease risk for corresponding defects under a variety of biological bases. Identification of these mutations and SNPs and their underlying disease-causing mechanisms will help with targeted therapeutic strategy against TBX5 associated disorders, majorly Holt-Oram syndrome.
Acknowledgements
This study was supported by the innovation program of Jiangsu province, China (Q.Z.), and the National Science Foundation China grants, China (Nos. 31271399, 81472047, and 81672229, Q.Z., J.G., and Y.L).
Disclosure of conflict of interest
None.
References
- 1.Agulnik SI, Garvey N, Hancock S, Ruvinsky I, Chapman DL, Agulnik I, Bollag R, Papaioannou VE, Silver LM. Evolution of mouse T-box genes by tandem duplication and cluster dispersion. Genetics. 1996;144:249–254. doi: 10.1093/genetics/144.1.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Minguillon C, Logan M. The comparative genomics of T-box genes. Brief Funct Genomic Proteomic. 2003;2:224–233. doi: 10.1093/bfgp/2.3.224. [DOI] [PubMed] [Google Scholar]
- 3.Abrahams A, Parker MI, Prince S. The T-box transcription factor Tbx2: its role in development and possible implication in cancer. IUBMB Life. 2010;62:92–102. doi: 10.1002/iub.275. [DOI] [PubMed] [Google Scholar]
- 4.Wilson V, Conlon FL. The T-box family. Genome Biol. 2002;3:3008. doi: 10.1186/gb-2002-3-6-reviews3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE. T-Box genes in vertebrate development. Annu Rev Genet. 2005;39:219–239. doi: 10.1146/annurev.genet.39.073003.105925. [DOI] [PubMed] [Google Scholar]
- 6.DeBenedittis P, Jiao K. Alternative splicing of T-box transcription factor genes. Biochem Biophys Res Commun. 2011;412:513–517. doi: 10.1016/j.bbrc.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lichiardopol C, Militaru C, Popescu B, Hila G, Mixich F. Holt-Oram syndrome. Rom J Morphol Embryol. 2007;48:67–70. [PubMed] [Google Scholar]
- 8.Mori AD, Bruneau BG. TBX5 mutations and congenital heart disease: Holt-Oram syndrome revealed. Curr Opin Cardiol. 2004;19:211–215. doi: 10.1097/00001573-200405000-00004. [DOI] [PubMed] [Google Scholar]
- 9.Ma JF, Yang F, Mahida SN, Zhao L, Chen X, Zhang ML, Sun Z, Yao Y, Zhang YX, Zheng GY, Dong J, Feng MJ, Zhang R, Sun J, Li S, Wang QS, Cao H, Benjamin EJ, Ellinor PT, Li YG, Tian XL. TBX5 mutations contribute to early-onset atrial fibrillation in Chinese and Caucasians. Cardiovasc Res. 2006;109:442–450. doi: 10.1093/cvr/cvw003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan W, Huang X, Chen C, Gray J, Huang T. TBX3 and its isoform TBX3+2a are functionally distinctive in inhibition of senescence and are overexpressed in a subset of breast cancer cell lines. Cancer Res. 2004;64:5132–5139. doi: 10.1158/0008-5472.CAN-04-0615. [DOI] [PubMed] [Google Scholar]
- 11.Hoogaars W, Barnett P, Rodriguez M, Clout D, Moorman A, Goding C, Christoffels V. TBX3 and its splice variant TBX3 + exon 2a are functionally similar. Pigment Cell Melanoma Res. 2008;21:379–387. doi: 10.1111/j.1755-148X.2008.00461.x. [DOI] [PubMed] [Google Scholar]
- 12.Govoni KE, Lee SK, Chadwick RB, Yu H, Kasukawa Y, Baylink DJ, Mohan S. Whole genome microarray analysis of growth hormone-induced gene expression in bone: T-box3, a novel transcription factor, regulates osteoblast proliferation. Am J Physiol Endocrinol Metab. 2006;291:E128–E136. doi: 10.1152/ajpendo.00592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alby C, Bessieres B, Bieth E, Attie-Bitach T, Fermont L, Citony I, Razavi F, Vekemans M, Escande F, Manouvrier S, Malan V, Amiel J. Contiguous gene deletion of TBX5 and TBX3 leads to a varible phenotype with combined features of Holt-Oram and ulnar-mammary syndromes. Am J Med Genet A. 2013;161A:1797–1802. doi: 10.1002/ajmg.a.36054. [DOI] [PubMed] [Google Scholar]
- 14.Naiche LA, Arora R, Kania A, Lewandoski M, Papaioannou VE. Identity and fate of Tbx4-expressing cells reveal developmental cell fate decisions in the allantois, limb, and external genitalia. Dev Dyn. 2011;240:2290–2300. doi: 10.1002/dvdy.22731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naiche LA, Papaioannou VE. Loss of Tbx4 blocks hindlimb development and affects vascularization and fusion of the allantois. Development. 2003;130:2681–2693. doi: 10.1242/dev.00504. [DOI] [PubMed] [Google Scholar]
- 16.Glaser A, Arora R, Hoffmann S, Li L, Gretz N, Papaioannou VE, Rappold GA. Tbx4 interacts with the short stature homeobox gene Shox2 in limb development. Dev Dyn. 2014;243:629–639. doi: 10.1002/dvdy.24104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bongers EM, Duijf PH, van Beersum SE, Schoots J, Van Kampen A, Burckhardt A, Hamel BC, Losan F, Hoefsloot LH, Yntema HG, Knoers NV, van Bokhoven H. Mutations in the human TBX4 gene cause small patella syndrome. Am J Hum Genet. 2004;74:1239–1248. doi: 10.1086/421331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerstjens-Frederikse WS, Bongers EM, Roofthooft MT, Leter EM, Douwes JM, Van Dijk A, Vonk-Noordegraaf A, Dijk-Bos KK, Hoefsloot LH, Hoendermis ES, Gille JJ, Sikkema-Raddatz B, Hofstra RM, Berger RM. TBX4 mutations (small patella syndrome) are associated with childhood-onset pulmonary arterial hypertension. J Med Genet. 2013;50:500–506. doi: 10.1136/jmedgenet-2012-101152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horton AC, Mahadevan NR, Minguillon C, Osoegawa K, Rokhsar DS, Ruvinsky I, de Jong PJ, Logan MP, Gibson-Brown JJ. Conservation of linkage and evolution of developmental function within the Tbx2/3/4/5 subfamily of T-box genes: implications for the origin of vertebrate limbs. Dev Genes Evol. 2008;218:613–628. doi: 10.1007/s00427-008-0249-5. [DOI] [PubMed] [Google Scholar]
- 20.Cebra-Thomas JA, Bromer J, Gardner R, Lam GK, Sheipe H, Gilbert SF. T-box gene products are required for mesenchymal induction of epithelial branching in the embryonic mouse lung. Dev Dyn. 2003;22:82–90. doi: 10.1002/dvdy.10208. [DOI] [PubMed] [Google Scholar]
- 21.Arora R, Metzger RJ, Papaioannou VE. Multiple roles and interactions of Tbx4 and Tbx5 in development of the respiratory system. PLoS Genet. 2012;8:1002866. doi: 10.1371/journal.pgen.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sountoulidis A, Stavropoulos A, Giaglis S, Apostolou E, Monteiro R, Chuva de Sousa Lopes SM, Chen H, Stripp BR, Mummery C, Andreakos E, Sideras P. Activation of the canonical bone morphogenetic protein (BMP) pathway during lung morphogenesis and adult lung tissue repair. PLoS One. 2012;7:41460. doi: 10.1371/journal.pone.0041460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hariri F, Nemer M, Nemer G. T-box factors: insights into the evolutionary emergence of the complex heart. Ann Med. 2012;44:680–693. doi: 10.3109/07853890.2011.607468. [DOI] [PubMed] [Google Scholar]
- 24.Yamak A, Georges RO, Sheikh-Hassani M, Morin M, Komati H, Nemer M. Novel exons in the tbx5 gene locus generate protein isoforms with distinct expression domains and function. J Biol Chem. 2015;290:6844–6856. doi: 10.1074/jbc.M114.634451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Collavoli A, Hatcher CJ, He J, Okin D, Deo R, Basson CT. TBX5 nuclear localization is mediated by dual cooperative intramolecular signals. J Mol Cell Cardiol. 2003;35:1191–1195. doi: 10.1016/s0022-2828(03)00231-1. [DOI] [PubMed] [Google Scholar]
- 26.Greulich F, Rudat C, Kispert A. Mechanisms of T-box gene function in the developing heart. Cardiovasc Res. 2011;91:212–222. doi: 10.1093/cvr/cvr112. [DOI] [PubMed] [Google Scholar]
- 27.Basson CT, Huang T, Lin RC, Bachinsky DR, Weremowicz S, Vaglio A, Bruzzone R, Quadrelli R, Lerone M, Romeo G, Silengo M, Pereira A, Krieger J, Mesquita SF, Kamisago M, Morton CC, Pierpont ME, Müller CW, Seidman JG, Seidman CE. Different TBX5 interactions in heart and limb defined by Holt-Oram syndrome mutations. Proc Natl Acad Sci U S A. 1999;96:2919–2924. doi: 10.1073/pnas.96.6.2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basson CT, Cowley GS, Solomon SD. The clinical and genetic spectrum of the Holt-Oram Syndrome (hand-heart syndrome) N Engl J Med. 1994;330:885–891. doi: 10.1056/NEJM199403313301302. [DOI] [PubMed] [Google Scholar]
- 29.Postma AV, van de Meerakker JB, Mathijssen IB, Barnett P, Christoffels VM, Ilgun A, Lam J, Wilde AA, Lekanne Deprez RH, Moorman AF. A gain-of-function TBX5 mutation is associated with atypical Holt-Oram syndrome and paroxysmal atrial fibrillation. Circ Res. 2008;102:1433–1442. doi: 10.1161/CIRCRESAHA.107.168294. [DOI] [PubMed] [Google Scholar]
- 30.Linhares VL, Almeida NA, Menezes DC, Elliott DA, Lai D, Beyer EC, Campos de Carvalho AC, Costa MW. Transcriptional regulation of the murine Connexin40 promoter by cardiac factors Nkx2-5, GATA4 and Tbx5. Cardiovasc Res. 2004;64:402–411. doi: 10.1016/j.cardiores.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh TK, Song FF, Packham EA, Buxton S, Robinson TE, Ronksley J, Self T, Bonser AJ, Brook JD. Physical interaction between TBX5 and MEF2C is required for early heart development. Mol Cell Biol. 2009;29:2205–2218. doi: 10.1128/MCB.01923-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koshiba-Takeuchi K, Takeuchi JK, Arruda EP, Kathiriya IS, Mo R, Hui CC, Srivastava D, Bruneau BG. Cooperative and antagonistic interactions between Sall4 and Tbx5 pattern the mouse limb and heart. Nat Genet. 2006;38:175–183. doi: 10.1038/ng1707. [DOI] [PubMed] [Google Scholar]
- 33.Al-Qattan MM, Abou Al-Shaar H. Molecular basis of the clinical features of Holt-Oram syndrome resulting from missense and extended protein mutations of the TBX5 gene as well as TBX5 intragenic duplications. Gene. 2015;560:129–136. doi: 10.1016/j.gene.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 34.Polk RC, Gergics P, Steimle JD, Li H, Moskowitz IP, Camper SA, Reeves RH. The pattern of congenital heart defects arising from reduced Tbx5 expression is altered in a down syndrome mouse model. BMC Dev Biol. 2015;15:30. doi: 10.1186/s12861-015-0080-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDermott DA, Hatcher CJ, Basson CT. Atrial fibrillation and other clinical manifestations of altered TBX5 dosage in typical Holt-Oram syndrome. Circ Res. 2008;103:e96. doi: 10.1161/CIRCRESAHA.108.181834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang ZC, Ji WH, Ruan CW, Liu XY, Qiu XB, Yuan F, Li RG, Xu YJ, Liu X, Huang RT, Xue S, Yang YQ. Prevalence and spectrum of TBX5 mutation in patients with lone atrial fibrillation. Int J Med Sci. 2016;13:60–67. doi: 10.7150/ijms.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang XL, Qiu XB, Yuan F, Wang J, Zhao CM, Li RG, Xu L, Xu YJ, Shi HY, Hou XM, Qu XK, Xu YW, Yang YQ. TBX5 loss-of-function mutation contributes to familial dilated cardiomyopathy. Biochem Biophys Res Commun. 2015;459:166–171. doi: 10.1016/j.bbrc.2015.02.094. [DOI] [PubMed] [Google Scholar]
- 38.Camarata T, Bimber B, Kulisz A, Chew TL, Yeung J, Simon HG. LMP4 regulates Tbx5 protein subcellular localization and activity. J Cell Biol. 2006;174:339–348. doi: 10.1083/jcb.200511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kulisz A, Simon HG. An evolutionarily conserved nuclear export signal facilitates cytoplasmic localization of the Tbx5 transcription factor. Mol Cell Biol. 2008;28:1553–1564. doi: 10.1128/MCB.00935-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeuchi JK, Lou X, Alexander JM, Sugizaki H, Delgado-Olguín P, Holloway AK, Mori AD, Wylie JN, Munson C, Zhu Y, Zhou YQ, Yeh RF, Henkelman RM, Harvey RP, Metzger D, Chambon P, Stainier DY, Pollard KS, Scott IC, Bruneau BG. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat Commun. 2011;2:187. doi: 10.1038/ncomms1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Waldron L, Steimle JD, Greco TM, Gomez NC, Dorr KM, Kweon J, Temple B, Yang XH, Wilczewski CM, Davis IJ, Cristea IM, Moskowitz IP, Conlon FL. The cardiac TBX5 interactome reveals a chromatin remodeling network essential for cardiac septation. Dev Cell. 2016;36:262–275. doi: 10.1016/j.devcel.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boogerd CJ, Evans SM. TBX5 and NuRD divide the heart. Dev Cell. 2016;36:242–244. doi: 10.1016/j.devcel.2016.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palles C, Chegwidden L, Li X, Findlay JM, Farnham G, Castro Giner F, Peppelenbosch MP, Kovac M, Adams CL, Prenen H, Briggs S, Harrison R, Sanders S, MacDonald D, Haigh C, Tucker A, Love S, Nanji M, deCaestecker J, Ferry D, Rathbone B, Hapeshi J, Barr H, Moayyedi P, Watson P, Zietek B, Maroo N, Gay L, Underwood T, Boulter L, McMurtry H, Monk D, Patel P, Ragunath K, Al Dulaimi D, Murray I, Koss K, Veitch A, Trudgill N, Nwokolo C, Rembacken B, Atherfold P, Green E, Ang Y, Kuipers EJ, Chow W, Paterson S, Kadri S, Beales I, Grimley C, Mullins P, Beckett C, Farrant M, Dixon A, Kelly S, Johnson M, Wajed S, Dhar A, Sawyer E, Roylance R, Onstad L, Gammon MD, Corley DA, Shaheen NJ, Bird NC, Hardie LJ, Reid BJ, Ye W, Liu G, Romero Y, Bernstein L, Wu AH, Casson AG, Fitzgerald R, Whiteman DC, Risch HA, Levine DM, Vaughan TL, Verhaar AP, van den Brande J, Toxopeus EL, Spaander MC, Wijnhoven BP, van der Laan LJ, Krishnadath K, Wijmenga C, Trynka G, McManus R, Reynolds JV, O’Sullivan J, MacMathuna P, McGarrigle SA, Kelleher D, Vermeire S, Cleynen I, Bisschops R, Tomlinson I, Jankowski J. Polymorphisms near TBX5 and GDF7 are associated with increased risk for barrett’s esophagus. Gastroenterology. 2015;148:367–378. doi: 10.1053/j.gastro.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Becker J, May A, Gerges C, Anders M, Schmidt C, Veits L, Noder T, Mayershofer R, Kreuser N, Manner H, Venerito M, Hofer JH, Lyros O, Ahlbrand CJ, Arras M, Hofer S, Heinrichs SK, Weise K, Hess T, Böhmer AC, Kosiol N, Kiesslich R, Izbicki JR, Hölscher AH, Bollschweiler E, Malfertheiner P, Lang H, Moehler M, Lorenz D, Ott K, Schmidt T, Nöthen MM, Hackelsberger A, Schumacher B, Pech O, Vashist Y, Vieth M, Weismüller J, Knapp M, Neuhaus H, Rösch T, Ell C, Gockel I, Schumacher J. The Barrett-associated variants at GDF7 and TBX5 also increase esophageal adenocarcinoma risk. Cancer Med. 2016;5:888–891. doi: 10.1002/cam4.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zang X, Zhang S, Xia Y, Li S, Fu F, Li X, Wang F, Zhang R, Tian X, Gao L, Zhang J, Yang Y, Tu X, Wang Q. SNP rs3825214 in TBX5 is associated with lone atrial fibrillation in Chinese Han population. PLoS One. 2013;8:e64966. doi: 10.1371/journal.pone.0064966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang R, Tian X, Gao L, Li H, Yin X, Dong Y, Yang Y, Xia Y. Common variants in the TBX5 gene associated with atrial fibrillation in a Chinese Han population. PLoS One. 2016;11:e0160467. doi: 10.1371/journal.pone.0160467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Manke T, Heinig M, Vingron M. Quantifying the effect of sequence variation on regulatory interactions. Human Mutation. 2010;31:477–483. doi: 10.1002/humu.21209. [DOI] [PubMed] [Google Scholar]
- 48.Luna-Zurita L, Stirnimann CU, Glatt S, Kaynak BL, Thomas S, Baudin F, Samee MA, He D, Small EM, Mileikovsky M, Nagy A, Holloway AK, Pollard KS, Müller CW, Bruneau BG. Complex interdependence regulates heterotypic transcription factor distribution and coordinates cardiogenesis. Cell. 2016;164:999–1014. doi: 10.1016/j.cell.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luna-Zurita L, Stirnimann CU, Glatt S, Kaynak BL, Thomas S, Baudin F, Samee MA, He D, Small EM, Mileikovsky M, Nagy A, Holloway AK, Pollard KS, Müller CW, Bruneau BG. Expression of Foxm1 transcription factor in cardiomyocytes is required for myocardial development. PLoS One. 2011;6:e22217. doi: 10.1371/journal.pone.0022217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shitashige M, Naishiro Y, Idogawa M, Honda K, Ono M, Hirohashi S, Yamada T. Involvement of splicing factor-1 in beta-catenin/T-cell factor-4-mediated gene transactivation and pre-mRNA splicing. Gastroenterology. 2007;132:1039–1054. doi: 10.1053/j.gastro.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 51.Shoukry A, Shalaby SM, Etewa RL, Ahmed HS, Abdelrahman HM. Association of estrogen receptor β and estrogen-related receptor α gene polymorphisms with bone mineral density in postmenopausal women. Mol Cell Biochem. 2015;405:23–31. doi: 10.1007/s11010-015-2391-5. [DOI] [PubMed] [Google Scholar]
- 52.Ghanem LR, Kromer A, Silverman IM, Chatterji P, Traxler E, Penzo-Mendez A, Weiss MJ, Stanger BZ, Liebhaber SA. The Poly(C) binding protein Pcbp2 and its retrotransposed derivative Pcbp1 are independently essential to mouse development. Mol Cell Biol. 2015;36:304–319. doi: 10.1128/MCB.00936-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Koren L, Elhanani O, Kehat I, Hai T, Aronheim A. Adult cardiac expression of the activating transcription factor 3, ATF3, promotes ventricular hypertrophy. PLoS One. 2013;8:e68396. doi: 10.1371/journal.pone.0068396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Magli A, Schnettler E, Swanson SA, Borges L, Hoffman K, Stewart R, Thomson JA, Keirstead SA, Perlingeiro RC. Pax3 and Tbx5 specify whether PDGFRα+ cells assume skeletal or cardiac muscle fate in differentiating embryonic stem cells. Stem Cells. 2014;32:2072–2083. doi: 10.1002/stem.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kang HC, Chae JH, Kim BS, Han SY, Kim SH, Auh CK, Yang SI, Kim CG. Transcription factor CP2 is involved in activating mBMP4 in mouse mesenchymal stem cells. Mol Cells. 2004;17:454–461. [PubMed] [Google Scholar]
- 56.Smemo S, Campos LC, Moskowitz IP, Krieger JE, Pereira AC, Nobrega MA. Regulatory variation in a TBX5 enhancer leads to isolated congenital heart disease. Hum Mol Genet. 2012;21:3255–3263. doi: 10.1093/hmg/dds165. [DOI] [PMC free article] [PubMed] [Google Scholar]