Abstract
MicroRNAs (miRNAs) play a critical role in cancer development and progression. Bioinformatics analyses has identified eukaryotic translation initiation factor 5A2 (eIF5A2) as a target of miR-9. In this study, we attempted to determine whether miR-9 regulates non-small cell lung cancer (NSCLC) cell invasion and migration by targeting eIF5A2 We examined eIF5A2 expression using reverse transcription-quantitative PCR (RT-qPCR) and subsequently transfected A549 and NCI-H1299 NSCLC cells with a miR-9 mimic or miR-9 inhibitor to determine the migration and invasive capability of the cells via wound healing assay and Transwell invasion assay, respectively. E-cadherin and vimentin expression was detected with western blotting. The miR-9 mimic significantly reduced NSCLC cell invasive and metastatic ability, and the miR-9 inhibitor enhanced NSCLC cell migration activity, increasing the number of migrated cells. There was no significant difference between the negative control siRNA and miR-9 mimic groups after knockdown of eIF5A2; western blotting showed that miR-9 regulated E-cadherin and vimentin expression. These data show that miR-9 regulates NSCLC cell invasion and migration through regulating eIF5A2 expression. Taken together, our findings suggest that the mechanism of miR-9-regulated NSCLC cell invasion and migration may be related to epithelial-mesenchymal transition.
Keywords: microRNA-9, eIF5A2, non-small cell lung cancer (NSCLC), invasion and migration
Introduction
Lung cancer is one of the most common malignancies and the leading cause of cancer-related death worldwide [1]. Approximately 80% of patients with lung cancer have non-small cell lung cancer (NSCLC); the rate of mortality is very high, and the 5-year survival rate is less than 15% after initial diagnosis, with metastasis being the primary reason for death [2]. Despite recent improvements in chemotherapy and molecular targeted therapy, the outcome remains poor [3]. The high invasiveness and metastasis remain the major challenges in NSCLC. Therefore, understanding of the factors involved in NSCLC metastasis is necessary for identifying new prognostic biomarkers and for developing new anti-lung cancer strategies.
MicroRNAs (miRNAs) are small, non-coding RNAs that negatively regulate post-transcriptional gene expression by binding to the 3’-untranslated region (3’-UTR) of the target mRNAs, and are involved in regulating essential cellular processes, including proliferation, diversification, metastasis, and apoptosis, particularly in cancer development and progression [4,5]. MiRNAs post-transcriptionally regulate 30% of human genes, suggesting that they may play pivotal roles in physiological and pathological processes [6]. Accumulating evidence describes vital roles for many miRNAs in tumor initiation and metastasis. For example, miR-205 and the miR-200 family influence epithelial-mesenchymal transition (EMT) during cancer metastasis [7]. Additionally, miR-7 inhibits EMT in gastric cancer by targeting IGF1R (insulin-like growth factor 1 receptor) expression [8]. In colorectal carcinoma, miR-30b directly targets the EMT-related gene SIX1 (SIX homeobox 1) to impair metastasis [9]. MiR-185 suppresses cell growth and EMT progression by targeting Six2, and is a novel target for the molecular treatment of liver malignancies [10]. In the present study, we attempted to determine the role of miR-9 in repressing NSCLC cell invasion and metastasis.
MiR-9 levels are downregulated in some cancers, such as ovarian cancer, gastric cancer, and neuroblastoma [11-13]. However, miR-9 expression is upregulated in colorectal cancer, breast cancer, lung cancer, and laryngeal squamous cell carcinomas [14-17]. It has also been indicated that miR-9 could play a major role in tumor progression and tumorigenesis [18]. In esophageal squamous cell carcinoma, miR-9 promotes metastasis by repressing E-cadherin [19].
Eukaryotic translation initiation factor 5A2 (eIF5A2), located on chromosome 3q26, was first discovered in the primary ovarian cancer in 2001 and has been classified as an oncogene [20]. Overexpression of eIF5A2 has been observed in other solid tumors and predicts poor prognosis in colorectal cancer [21], hepatocellular carcinomas (HCC) [22], ovarian cancer [23], and NSCLC [24]. However, whether miR-9 directly targets eIF5A2 to regulate NSCLC cell invasion and metastasis has not been reported. Therefore, in this study, we investigated whether miR-9 regulates NSCLC cell invasion and migration by targeting eIF5A2.
Materials and methods
Cell culture
Human NSCLC cells (A549 and NCI-H1299) were purchased from American Type Culture Collection (Manassas, VA, USA). The cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) and 1% penicillin/streptomycin (P/S; Sigma, St. Louis, MO, USA). The cells were incubated at 37°C in a humidified atmosphere of 5% CO2.
RNA oligoribonucleotides and transfection
The miR-9 mimic, miR-9 inhibitor, control, and negative control siRNA were synthesized by RiboBio (Guangzhou, China). The eIF5A2 siRNA and negative control siRNA were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Western blot
Cells were lysed in protein lysis buffer (Cell Signaling Technology, Danvers, MA, USA) supplemented with protease inhibitor and phosphatase inhibitor (Sigma, St. Louis, MO, USA) following the manufacturer’s protocol. The lysates were centrifuged at 12000×g for 5 min at 4°C and the supernatant was collected. Protein concentrations were determined using a bicinchoninic acid protein assay kit (Sigma, St. Louis, MO, USA). About 20 μg protein from each sample was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). After blocking, the membranes were incubated with primary antibody (Abcam, Cambridge, MA, USA) (anti-eIF5A2, anti-E-cadherin, and anti-vimentin diluted 1:1000 in Tris-buffered saline [TBS] containing 5% bovine serum albumin and 0.1% Tween 20 [TBS/T]) overnight at 4°C. The membranes were washed three times with TBS/T and incubated with a horseradish peroxidase-conjugated secondary antibody (Cell Signaling Technology, Beverly, MA, USA) (1:2000) for 2 h at 37°C. After washing in TBS/T three times, the protein bands were detected using an enhanced chemiluminescence detection system ECL (Biological Industries, Kibbutz Beit Haemek, Israel). Gray value analysis of the protein bands was performed using ImageJ software (National Institutes of Health, Bethesda, MD, USA). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the loading control.
RT-qPCR
Total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. RNA was reverse-transcribed into complementary DNA using a PrimeScript RT Reagent Kit (Takara Dalian, China). The PCR was conducted using a 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) under the following conditions: 95°C for 30 s followed by 40 cycles of 95°C for 5 s, and annealing at 60°C for 34 s. DNA primers specific for miR-9 and U6 were purchased from RiboBio. Relative quantification was performed using the comparative threshold cycle (2-ΔΔCt) method [25]. All reactions were performed in triplicate. The primers used were as follows: eIF5A2: Forward 5’-TATGCAGTGCTCGGCCTTG-3’, Reverse 5’-TTGGAACATCCATGTTGTGAGTAGA-3’; miR-9 mimic: Forward 5’-UCUUUGGUUAUCUAGCUGUAUGA-3’. Reverse 5’-AUACAGCUAGAUAACCAAAGAUU-3’; miR-9 inhibitor: 5’-UCAUACAGCUAGAUAACCAAAGA-3’.
Wound healing assay
After 6-h transfection with miR-9 mimic or inhibitor, negative siRNA, or eIF5A2 siRNA, 2 × 105 cells per well were seeded into 6-well plates and cultured in RPMI 1640 medium containing 10% FBS and 1% P/S at 37°C in a humidified incubator with 5% CO2 for 24 h until 90% confluent, and then the medium was changed to medium containing 0.05% FBS and 1% P/S overnight to synchronize the cells. A wound was then created in the cell monolayer using a 100-μL yellow pipette tip. The wound areas were washed with phosphate-buffered saline and photographed with an inverted light microscope (Olympus IX51, Center Valley, PA, USA) at 0 h and 24 h. The ratio of the remaining wound area relative to the initial wound area was calculated and the wound area was quantified using Image-Pro Plus v. 6.0 software (Media Cybernetics, Bethesda, MD, USA).
Transwell matrigel invasion assay
After 6-h transfection with miR-9 mimic or inhibitor, negative siRNA, or eIF5A2 siRNA (100 nmol/mL), the cells (5 × 104 cells/well) were seeded in the top chamber of a 24-insert Transwell plate containing medium. The inserts in the top chambers were coated with Matrigel (BD Biosciences, San Jose, CA, USA); the bottom chambers contained 10% FBS in the corresponding medium as a chemoattractant. After 24 h, cells on the underside of the inserts were fixed in methanol for 10 min and stained with 0.1% crystal violet. The cells that had invaded to the lower surface were counted under an inverted phase contrast microscope (Olympus; ×40 magnification) and photographed.
Statistical analysis
All experimental data are reported as the mean ± SD. GraphPad Prism 5 (GraphPad, San Diego, CA, USA) was used for statistical analysis. Comparisons of two groups were performed using Student’s t-tests; > 2 independent groups were compared using one-way analysis of variance. P < 0.05 was considered statistically significant.
Results
MiR-9 regulated NSCLC cell invasion and migration
We examined the effect of a miR-9 mimic and miR-9 inhibitor on NSCLC cell invasion and migration by Transwell invasion assay and wound healing assay, respectively. Compared with the control, the miR-9 mimic inhibited NSCLC cell migration capability, while the miR-9 inhibitor enhanced it (Figure 1A, 1B). The Transwell invasion assay showed that, compared with the control, fewer miR-9 mimic-transfected cells crossed the Transwell membrane, and more miR-9 inhibitor-transfected cells crossed the Transwell membranes (Figure 1C). These findings demonstrate that miR-9 regulates NSCLC cell invasion and migration.
Figure 1.
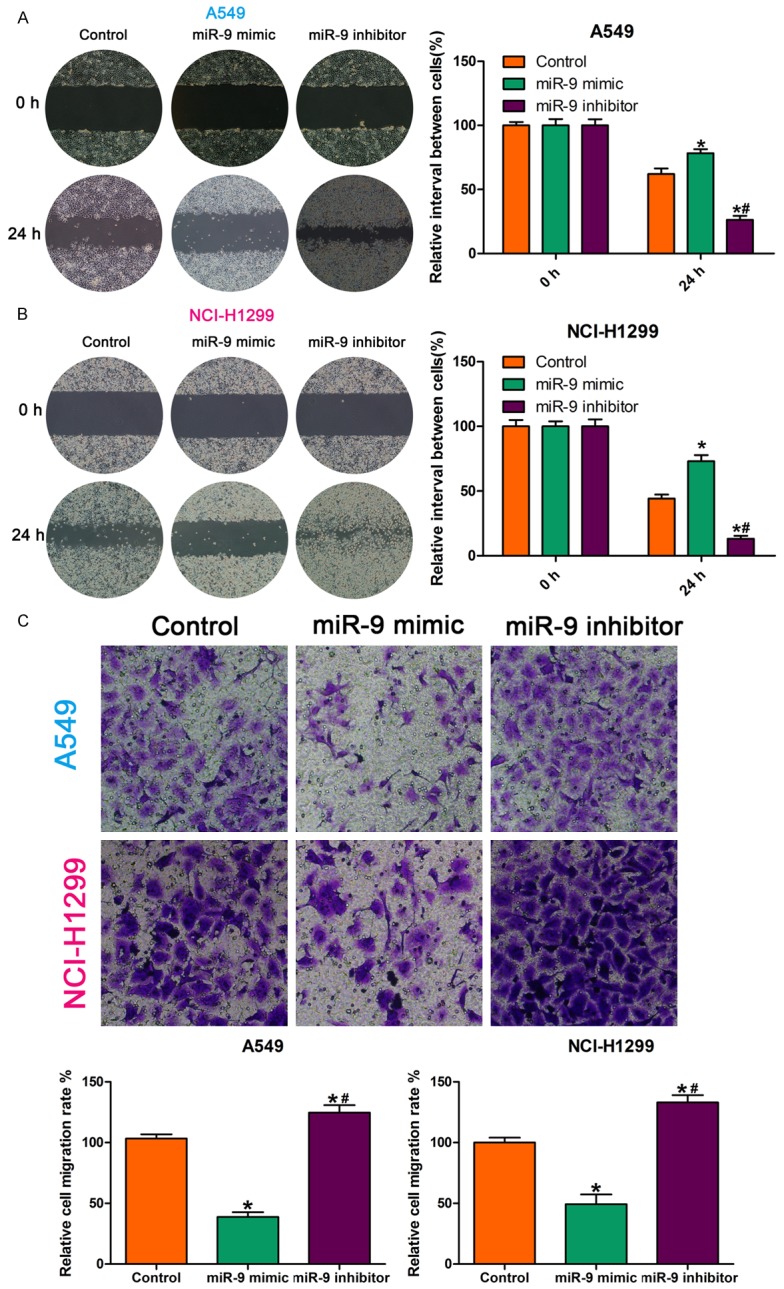
MiR-9 regulated NSCLC cell invasion and migration. A, B. Wound healing assay showing that, compared with the control, the miR-9 mimic reduced cell migration capability and that the miR-9 inhibitor increased it. *P < 0.05 vs. control; #P < 0.05 vs. miR-9 mimic. C. Images and quantification of NSCLC cell migration following treatment with miR-9 mimic, miR-9 inhibitor, or control. Migrated cells were stained with crystal violet and counted. *P < 0.05 vs. control; #P < 0.05 vs. miR-9 mimic. All data are the mean ± SD.
MiR-9 directly targeted and regulated eIF5A2
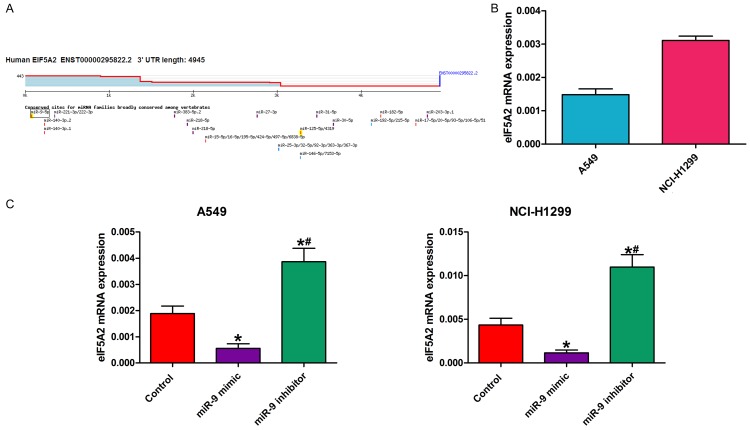
We hypothesized that miR-9 might regulate eIF5A2 expression. We used TargetScan (www.targetscan.org) to predict the eIF5A2-associated miRNAs via bioinformatics analysis (Figure 2A), and detected miR-9 and eIF5A2 mRNA expression by reverse transcription-quantitative PCR (RT-qPCR). Interestingly, miR-9 mRNA expression was negatively associated with eIF5A2 mRNA expression (Figure 2B). We also detected eIF5A2 mRNA expression in NSCLC cells transfected with miR-9 mimic, miR-9 inhibitor, or control. Compared with the control, miR-9 mimic transfection decreased eIF5A2 mRNA expression. However, eIF5A2 mRNA expression was enhanced following transfection with miR-9 inhibitor (Figure 2C).
Figure 2.
MiR-9 regulated EIF5A2 expression in NSCLC cells. A. TargetScan prediction matching miR-9 to the eIF5A2 3’-UTR segment. B. RT-qPCR determination of eIF5A2 mRNA expression in NSCLC cells. C. RT-qPCR detection of eIF5A2 mRNA expression in NSCLC cells in the presence of miR-9 mimic, miR-9 inhibitor, and control. All data are the mean ± SD.
EIF5A2 knockdown suppressed NSCLC cell invasive and migration capability
To determine the role of eIF5A2 in NSCLC cell invasion and migration, we examined the effect of eIF5A2 knockdown on metastasis using Transwell invasion assay and wound healing assay, respectively. The wound healing assay showed that eIF5A2 knockdown increased NSCLC cell motility as compared with cells treated with negative control small interfering RNA (siRNA) (Figure 3A). The Transwell assay showed significantly decreased migrated cells in the eIF5A2 knockdown group as compared with the negative siRNA group (Figure 3B). These findings demonstrate that inhibiting eIF5A2 significantly suppresses the invasive and migration capability in NSCLC cells.
Figure 3.
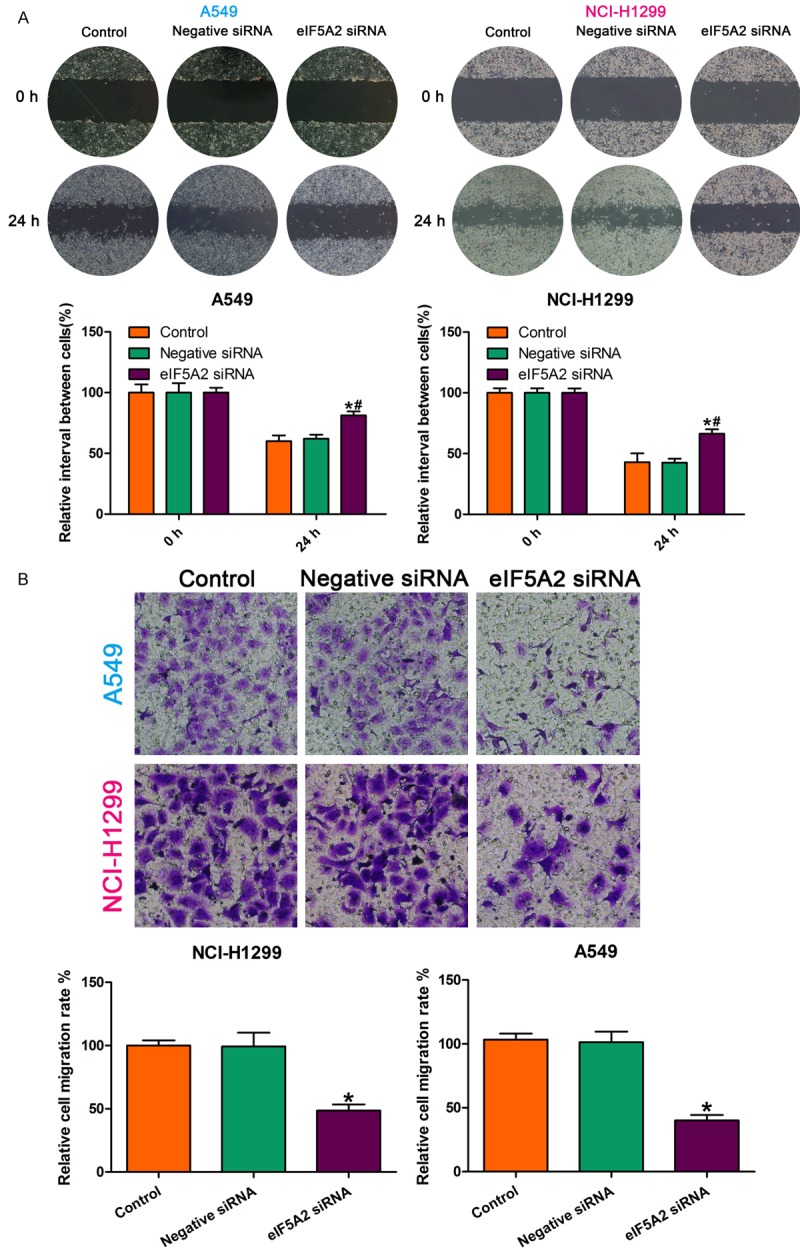
eIF5A2 knockdown inhibited NSCLC cell invasive and migration capability. A. eIF5A2 knockdown enhanced cell migration ability as compared to the effect of the negative control. *P < 0.05 vs. control; #P < 0.05 vs. negative siRNA. B. All data are the mean ± SD.
MiR-9 regulated NSCLC cell invasion and migration by targeting eIF5A2
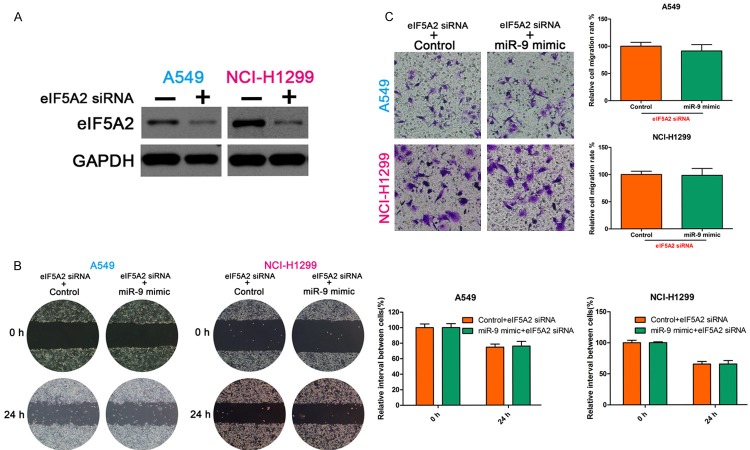
We demonstrated that miR-9 regulates NSCLC cell invasion and migration and that eIF5A2 regulates NSCLC cell invasive and migration capability. Furthermore, we demonstrate that miR-9 regulates the expression of eIF5A2 mRNA. We suspected that miR-9 regulates NSCLC cell invasion and migration by targeting eIF5A2. To confirm this hypothesis, we first silenced eIF5A2 in NSCLC cells using siRNA, and then transfected the cells with miR-9 mimic. Then, we examined the invasive and migration ability of the cells. The wound healing and Transwell assays showed no significant difference between the control and miR-9 mimic groups (Figure 4). Western blotting confirmed the eIF5A2 knockdown (Figure 4A).
Figure 4.
MiR-9 regulated NSCLC cell invasion and migration by targeting eIF5A2. (A) Western blot detection of eIF5A2 expression. GAPDH, loading control. Wound healing (B) and Transwell assays (C) showed no significant difference between control and miR-9 mimic groups after transfection with eIF5A2 siRNA. All data are the mean ± SD.
MiR-9 regulated e-cadherin and vimentin expression
Accumulating evidence has indicated that EMT plays an important role in malignancy and migration and invasive ability in many tumors [26,27]. In the present study, western blotting showed that the miR-9 mimic upregulated E-cadherin expression and downregulated vimentin expression in the NSCLC cells. By contrast, E-cadherin expression was downregulated and vimentin expression was upregulated in NSCLC cells transfected with the miR-9 inhibitor (Figure 5). These data indicate that miR-9 regulates E-cadherin and vimentin expression.
Figure 5.
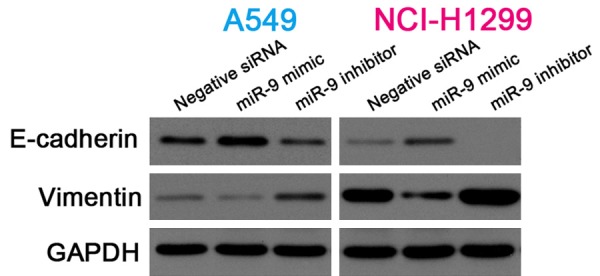
MiR-9 regulated E-cadherin and vimentin expression. Cropped western blots showing E-cadherin and vimentin expression in NSCLC cells transfected with negative siRNA, miR-9 mimic, or miR-9 inhibitor. GAPDH, internal reference.
Discussion
Recent reports have suggested that miRNAs play critical roles in the tumorigenesis and progression of various human cancers [28,29]. Therefore, identifying the tumor-associated miRNAs and their target genes is critical for understanding the role of miRNAs in tumorigenesis and may be important for identifying novel therapeutic targets [30].
MiRNAs, such as oncomiRs or anti-oncomiRs, play important roles in cancer development and progression by acting as activators or inhibitors [31]. More recently, it was indicated that miRNA dysregulation contributes to tumor metastases by targeting genes important to EMT and many other processes in tumor metastases [17]. For example, miR-200c exerts tumor-suppressive effects in NSCLC by suppressing USP25 (ubiquitin-specific peptidase 25) expression, and miR-204 inhibits NSCLC metastasis by suppressing NUAK1 (NUAK family kinase 1) [32,33]. MiR-9, which is selectively expressed in neuronal tissues, was first found to be elevated in primary brain tumors and was described as an essential factor that functions in developing neurons, neural carcinogenesis, or other diseases of the nervous system [34]. Recently, it was reported that miR-9 is a putative metastasis promoter in breast cancer and promotes a metastatic phenotype, including invasion and migration in osteoblasts and osteosarcoma cell lines [35]. EIF5A2 plays critical roles in cell proliferation, metastasis, and apoptosis, and is considered a novel oncogene [36]. eIF5A2 inhibition enhanced NSCLC sensitivity to chemotherapeutics, prevented or reversed EMT, and reduced the migration and invasive capabilities of NSCLC cells [37], and might decrease HCC cell invasion and metastasis [38,39]. In the present study, the miR-9 mimic inhibited NSCLC cell migration capability and reduced the number of membrane-crossing cells, but the miR-9 inhibitor increased cell migration and invasive ability. In addition, we used TargetScan to search for the eIF5A2-associated miRNAs, and found that eIF5A2 is a target gene of miR-9. We also found that miR-9 expression was negatively associated with eIF5A2 expression. To determine whether eIF5A2 regulates the role of miR-9, we knocked down eIF5A2 expression and found that there was no significant difference between the miR-9 mimic and negative control groups. These findings demonstrate that the effect of miR-9 inhibition of NSCLC cell invasion and migration is at least partly mediated through decreased eIF5A2 expression.
EMT is a complex process associated with the loss of E-cadherin expression and the gain of mesenchymal lineage marker expression, and results in increased cell invasion, migration, and proliferation [40]. The activation of EMT triggers tumor cell invasion and dissemination and is therefore considered the initiation step of metastasis [41]. EMT is related to metastasis and poor prognosis in many tumors, including NSCLC [42,43]. It has also previously been shown that eIF5A2 regulates chemoresistance through EMT in many cancers, including NSCLC [37]. In the present study, we confirm that miR-9 is involved in regulating metastasis by targeting E-cadherin and vimentin expression in NSCLC cells. Western blotting demonstrated that compared with the control, the miR-9 mimic increased the expression of the epithelial marker E-cadherin and reduced the expression of the mesenchymal marker vimentin, while the miR-9 inhibitor had the opposite effect, suggesting that EMT may participate in NSCLC cell invasion and metastasis. Together, our findings suggest that miR-9 is a potential drug target for overcoming drug resistance and regulating EMT in NSCLC cells.
To the best of our knowledge, this is the first report on miR-9 directly regulating eIF5A2. We found that miR-9 regulates NSCLC cell invasion and migration by inhibiting eIF5A2, and preventing EMT. This report sheds new light on the role of miR-9 in NSCLC development, and supports the targeting of miR-9, which represents a novel target for therapeutic intervention in NSCLC in the future.
Acknowledgements
This study was supported by Natural Science Fund of Ningbo (No. 2016A610196), Natural Science Fund of Zhejiang Province (No. LQ16H160002) and Zhejiang Provincial Federal Office of Public Health Project (2015KYA222).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Parums DV. Current status of targeted therapy in non-small cell lung cancer. Drugs Today (Barc) 2014;50:503–525. doi: 10.1358/dot.2014.50.7.2185913. [DOI] [PubMed] [Google Scholar]
- 3.Shi L, Zhang B, Sun X, Lu S, Liu Z, Liu Y. MiR-204 inhibits human NSCLC metastasis through suppression of NUAK1. Br J Cancer. 2014;111:2316–2327. doi: 10.1038/bjc.2014.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Selcuklu SD, Donoghue MT, Rehmet K, de Souza Gomes M, Fort A. MicroRNA-9 inhibition of cell proliferation and identification of novel miR-9 targets by transcriptome profiling in breast cancer cells. J Biol Chem. 2012;287:29516–29528. doi: 10.1074/jbc.M111.335943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song Y, Li J, Zhu Y, Dai Y, Zeng T, Liu L. MicroRNA-9 promotes tumor metastasis via repressing e-cadherin in esophageal squamous cell carcinoma. Oncotarget. 2014;5:11669–11680. doi: 10.18632/oncotarget.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 7.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 8.Zhao X, Dou W, He L, Liang S, Tie J, Liu C. MicroRNA-7 functions as an antimetastatic microRNA in gastric cancer by targeting insulin-like growth factor-1 receptor. Oncogene. 2013;32:1363–1372. doi: 10.1038/onc.2012.156. [DOI] [PubMed] [Google Scholar]
- 9.Zhao H, Xu Z, Qin H, Gao Z, Gao L. miR-30b regulates migration and invasion of human colorectal cancer via SIX1. Biochem J. 2014;460:117–125. doi: 10.1042/BJ20131535. [DOI] [PubMed] [Google Scholar]
- 10.Zhu SM, Chen CM, Jiang ZY, Yuan B, Ji M, Wu FH. MicroRNA-185 inhibits cell proliferation and epithelial-mesenchymal transition in hepatocellular carcinoma by targeting Six2. Eur Rev Med Pharmacol Sci. 2016;20:1712–1719. [PubMed] [Google Scholar]
- 11.Guo LM, Pu Y, Han Z, Liu T, Li YX, Liu M. MicroRNA-9 inhibits ovarian cancer cell growth through regulation of NF-kappaB1. FEBS J. 2009;276:5537–5546. doi: 10.1111/j.1742-4658.2009.07237.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, Qi M, Li S, Qi T, Mei H, Huang K. microRNA-9 targets matrix metalloproteinase 14 to inhibit invasion, metastasis, and angiogenesis of neuroblastoma cells. Mol Cancer Ther. 2012;11:1454–1466. doi: 10.1158/1535-7163.MCT-12-0001. [DOI] [PubMed] [Google Scholar]
- 13.Zheng L, Qi T, Yang D, Qi M, Li D, Xiang X. microRNA-9 suppresses the proliferation, invasion and metastasis of gastric cancer cells through targeting cyclin D1 and Ets1. PLoS One. 2013;8:e55719. doi: 10.1371/journal.pone.0055719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muraoka T, Soh J, Toyooka S, Maki Y, Shien K, Furukawa M. Impact of aberrant methylation of microRNA-9 family members on non-small cell lung cancers. Mol Clin Oncol. 2013;1:185–189. doi: 10.3892/mco.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Zhao H, Tang D, Wu J, Yao G, Zhang Q. Overexpressions of microRNA-9 and microRNA-200c in human breast cancers are asso-ciated with lymph node metastasis. Cancer Biother Radiopharm. 2013;28:283–288. doi: 10.1089/cbr.2012.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu S, Jia S, Xu P. MicroRNA-9 as a novel prognostic biomarker in human laryngeal squamous cell carcinoma. Int J Clin Exp Med. 2014;7:5523–5528. [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu L, Chen H, Zhou D, Li D, Bai R, Zheng S. MicroRNA-9 up-regulation is involved in colorectal cancer metastasis via promoting cell motility. Med Oncol. 2012;29:1037–1043. doi: 10.1007/s12032-011-9975-z. [DOI] [PubMed] [Google Scholar]
- 18.Sun J, Fang K, Shen H, Qian Y. MicroRNA-9 is a ponderable index for the prognosis of human hepatocellular carcinoma. Int J Clin Exp Med. 2015;8:17748–17756. [PMC free article] [PubMed] [Google Scholar]
- 19.Song Y, Li J, Zhu Y, Dai Y, Zeng T, Liu L. MicroRNA-9 promotes tumor metastasis via repressing e-cadherin in esophageal squamous cell carcinoma. Oncotarget. 2014;5:11669–11680. doi: 10.18632/oncotarget.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan XY, Sham JS, Tang TC, Fang Y, Huo KK, Yang JM. Isolation of a novel candidate oncogene within a frequently amplified region at 3q26 in ovarian cancer. Cancer Res. 2001;61:3806–3809. [PubMed] [Google Scholar]
- 21.Zhu W, Cai MY, Tong ZT, Dong SS, Mai SJ, Liao YJ. Overexpression of EIF5A2 promotes colorectal carcinoma cell aggressiveness by upregulating MTA1 through C-myc to induce epithelial-mesenchymaltransition. Gut. 2012;61:562–575. doi: 10.1136/gutjnl-2011-300207. [DOI] [PubMed] [Google Scholar]
- 22.Tang DJ, Dong SS, Ma NF, Xie D, Chen LL, Fu L. Overexpression of eukaryotic initiation factor 5A2 enhances cell motility and promotes tumor metastasis in hepatocellular carcinoma. Hepatology. 2010;51:1255–1263. doi: 10.1002/hep.23451. [DOI] [PubMed] [Google Scholar]
- 23.Yang GF, Xie D, Liu JH, Luo JH, Li LJ, Hua WF. Expression and amplification of eIF-5A2 in human epithelial ovarian tumors and overexpression of EIF-5A2 is a new independent predictor of outcome in patients with ovarian carcinoma. Gynecol Oncol. 2009;112:314–318. doi: 10.1016/j.ygyno.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 24.He LR, Zhao HY, Li BK, Liu YH, Liu MZ, Guan XY. Overexpression of eIF5A-2 is an adverse prognostic marker of survival in stage I non-small cell lung cancer patients. Int J Cancer. 2011;129:143–150. doi: 10.1002/ijc.25669. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T))Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Y, DU F, Zhao Q, Jin J, Ma X, Li H. Acquisition of 5-fluorouracil resistance induces epithelial-mesenchymal transitions through the Hedgehog signaling pathway in HCT-8 colon cancer cells. Oncol Lett. 2015;9:2675–2679. doi: 10.3892/ol.2015.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu CG, Calin GA, Meloon B, Gamliel N, Sevignani C, Ferracin M. An oligonucleotide microchip for genome-wide microRNA profiling inhuman and mouse tissues. Proc Natl Acad Sci U S A. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 31.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Tan Q, Yan M, Liu L, Lin H, Zhao F. miRNA-200c inhibits invasion and metastasis of human non-small cell lung cancerby directly targeting ubiquitin specific peptidase 25. Mol Cancer. 2014;13:166. doi: 10.1186/1476-4598-13-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi L, Zhang B, Sun X, Lu S, Liu Z, Liu Y. MiR-204 inhibits human NSCLC metastasis through suppression of NUAK1. Br J Cancer. 2014;111:2316–2327. doi: 10.1038/bjc.2014.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nass D, Rosenwald S, Meiri E, Gilad S, Tabibian-Keissar H. MiR-92b and miR-9/9* are specifically expressed in brain primary tumors and can be used to differentiate primary from metastatic brain tumors. Brain Pathol. 2009;19:375–83. doi: 10.1111/j.1750-3639.2008.00184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fenger JM, Roberts RD, Iwenofu OH, Bear MD, Zhang X, Couto JI. MiR-9 is overexpressed in spontaneous canine osteosarcoma and promotes a metastatic phenotype including invasion and migration in osteoblasts and osteosarcoma cell lines. BMC Cancer. 2016;16:784. doi: 10.1186/s12885-016-2837-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Jiang R, Cui EH, Feng WM, Guo HH, Gu DH. N1-guanyl-1,7-diaminoheptane enhances the chemosensitivity of NSCLC cells tocetuximab through inhibition of eukaryotic translation initiation factor 5A2 activation. Eur Rev Med Pharmacol Sci. 2016;20:1244–1250. [PubMed] [Google Scholar]
- 37.Xu G, Yu H, Shi X, Sun L, Zhou Q, Zheng D. Cisplatin sensitivity is enhanced in non-small cell lung cancer cells by regulatingepithelial-mesenchymal transition through inhibition of eukaryotic translationinitiation factor 5A2. BMC Pulm Med. 2014;14:174. doi: 10.1186/1471-2466-14-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lou B, Fan J, Wang K, Chen W, Zhou X, Zhang J. N1-guanyl-1,7-diaminoheptane (GC7) enhances the therapeutic efficacy of doxorubicin by inhibiting activation of eukaryotic translation initiation factor 5A2 (eIF-5A-2) and preventing the epithelial-mesenchymal transition in hepatocellular carcinoma cells. Exp Cell Res. 2013;319:2708–2717. doi: 10.1016/j.yexcr.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 39.Liu RR, Lv YS, Tang YX, Wang YF, Chen XL, Zheng XX. Eukaryotic translation initiation factor 5A2 regulates the migration and invasion of hepatocellular carcinoma cells via pathways involving reactive oxygen species. Oncotarget. 2016;7:24348–24360. doi: 10.18632/oncotarget.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 41.Tsuji T, Ibaragi S, Hu GF. Epithelial-mesenchymal transition and cell cooperativity in metastasis. Cancer Res. 2009;69:7135–713. doi: 10.1158/0008-5472.CAN-09-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh A, Greninger P, Rhodes D, Koopman L, Violette S, Bardeesy N. A gene expression signature associated with “K-Ras addiction” reveals regulators of EMT and tumor cell survival. Cancer Cell. 2009;15:489–500. doi: 10.1016/j.ccr.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saito RA, Watabe T, Horiguchi K, Kohyama T, Saitoh M, Nagase T. Thyroid transcription factor-1 inhibits transforming growth factor-beta-mediated epithelial-to-mesenchymal transition in lung adenoarcinoma cells. Cancer Res. 2009;69:2783–2791. doi: 10.1158/0008-5472.CAN-08-3490. [DOI] [PubMed] [Google Scholar]