Abstract
Triple-negative breast cancer (TNBC) is one of the most aggressive subtypes of breast cancer, with a significantly higher recurrence and mortality rate. There is an urgent need to uncover the mechanism underlying TNBC and establish therapeutic targets. Long non-coding RNAs (lncRNAs) are involved in a series of biological functions and provide novel insights into the molecular mechanism of cancer. Based on their expression specificity and large number, lncRNAs are likely to serve as the basis for clinical applications in oncology. In our previous study, we utilized RNA sequencing (RNA-seq) to explore the lncRNAs expression profiles in TNBC and identified that small nucleolar RNA host gene 12 (SNHG12) was remarkably increased in TNBC. However, the role of SNHG12 in TNBC has not been clarified. Herein, we determine that SNHG12 is upregulated in TNBC, and its high expression is significantly correlated with tumor size and lymph node metastasis. Mechanistic investigations show that SNHG12 is a direct transcriptional target of c-MYC. Silencing SNHG12 expression inhibits TNBC cells proliferation and apoptosis promotion, whereas SNHG12 overexpression has the opposite effect. In addition, we reveal that SNHG12 may promote cells migration by regulating MMP13 expression. To the best of our knowledge, it is the first report indicating that SNHG12 is involved in breast cancer. Taken together, our findings suggest that SNHG12 contributes to the oncogenic potential of TNBC and may be a promising therapeutic target.
Keywords: Long non-coding RNA, triple-negative breast cancer, SNHG12, c-MYC
Introduction
Breast cancer is the most common malignancy in women worldwide [1]. Triple-negative breast cancer (TNBC), which accounts for 12 to 17% of breast cancer cases, is characterized by lacking expression of hormone receptor (HR) and human epidermal growth factor receptor-2 (HER-2) [2]. Although it comprises a small percentage of all breast cancer cases, TNBC represents a major clinical problem and is under intense investigation due to its aggressive nature. Patients diagnosed with TNBC are usually younger, more likely to have larger tumors, and have an increased possibility of distant metastasis and mortality [3,4]. Thus, it is urgent to elucidate the molecular mechanisms underlying TNBC and identify available therapeutic targets.
Long non-coding RNAs (lncRNAs) are defined as RNAs with lengths greater than 200 nt and lacking protein-coding potential [5]. LncRNAs were initially viewed as the products generated from the background noise of transcription [6]. However, it has become increasingly clear that lncRNAs are involved in a series of biological functions [7]. To date, thousands of lncRNAs have been reported to drive many important cancer phenotypes through their interactions with other cellular macromolecules [8-10]. A deeper understanding of lncRNAs will provide novel insights into the molecular mechanism of cancer. Based on their expression specificity and large number, lncRNAs are likely to serve as the basis for many clinical applications in oncology [10].
In our previous study, we utilized RNA sequencing (RNA-seq) to explore the lncRNAs expression profiles in TNBC [11]. We found numerous differentially expressed lncRNAs between TNBC and non-tumorous breast tissues, among which a lncRNA, small nucleolar RNA host gene 12 (SNHG12), was remarkably increased in TNBC. However, the function of SNHG12 in TNBC has not been clarified. Herein, we confirmed the expression of SNHG12 in TNBC tissues and its clinical significance. Furthermore, we explored the mechanism of the high expression of SNHG12 in TNBC. Gain- and loss-of-function approaches were adopted to investigate the biological effect of SNHG12 in TNBC cells. Finally, the potential mechanism of SNHG12 was explored. Thus, our study shed light on the utilization of SNHG12 as a potential novel therapeutic target for TNBC.
Materials and methods
Tissue specimens and ethics statement
Tissue specimens were collected from patients who underwent modified radical mastectomy at the First Affiliated Hospital of Wenzhou Medical University. Postoperative histopathological examination and immunohistochemistry were performed to identify TNBC. A total of 102 TNBC tumor tissues and 95 noncancerous breast tissues were included in this study. The Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University approved this study, and written informed consents were acquired from all patients. Expression levels of SNHG12 and c-MYC in an independent TNBC cohort were also downloaded from The Cancer Genome Atlas (TCGA) database [12].
RNA extraction and quantitative real-time PCR (qRT-PCR)
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract total RNA from tissues or cells according to the manufacturer’s instructions. Cytoplasmic & Nuclear RNA Purification Kit (Norgen, Belmont, CA, USA) was used to isolate and purifie cytoplasmic and nuclear RNA. RNA reverse transcription to cDNA was performed with a reverse transcription kit (Toyobo, Osaka, Japan). QRT-PCR was performed using Thunderbird SYBR qPCR Mix (Toyobo, Osaka, Japan) to detect genes expression levels. The results were normalized to the expression of β-actin. Primers sequences are listed in Table 1.
Table 1.
The primer sequences used for qRT-PCR
| Gene name | Forward primer sequences (5’-3’) | Reverse primer sequences (5’-3’) |
|---|---|---|
| SNHG12 | GTGATACTGAGGAGGTGAG | CCTTCTGCTTCCCATAGAG |
| CD164 | AGAACAGTGCCTTAGAAGAC | GCTCACATCAAGTTACATCC |
| KRAS | GAGTACAGTGCAATGAGGGAC | CCTGAGCCTGTTTTGTGTCTAC |
| MMP13 | TCCTGATGTGGGTGAATACAATG | GCCATCGTGAAGTCTGGTAAAAT |
| E-cadherin | CGAGAGCTACACGTTCACGG | GGGTGTCGAGGGAAAAATAGG |
| Slug | AGGACACATTAGAACTCACA | CTACACAGCAGCCAGATT |
| FOXO1 | GGGCCCTAATTCGGTCATGT | CGCCCGTTAACTGCAGATGT |
| Notch1 | AGGACCTCATCAACTCACA | CGTTCTTCAGGAGCACAA |
| β-actin | GGGAAATCGTGCGTGACATTAAGG | CAGGAAGGAAGGCTGGAAGAGTG |
| GAPDH | ATGGGTGTGAACCATGAGAA | GTGCTAAGCAGTTGGTGGTG |
Cell culture and transfection
Human TNBC cell lines MDA-MB-231 and BT-549 were purchased from the Chinese Academy of Sciences (Shanghai, China). The MDA-MB-231 cells were cultured in L15 medium (Invitrogen, USA), while the BT-549 cells were cultured in RPMI 1640 medium (Invitrogen, USA). All media were supplemented with 10% fetal bovine serum (10% FBS, Gibco), and cell lines were maintained at 37°C in a humidified atmosphere of 5% CO2. Small interfering RNA (siRNA) and non-specific negative control siRNA (NC) were synthesized by GenePharma (Shanghai, China). The siRNA oligonucleotide sequences targeting SNHG12 were as follows: siRNA-1, sense 5’-UGUGAUACUGAGGAGGUGATT-3’, and antisense 5’-UCACCUCCUCAGUAUCACATT-3’; siRNA-2, sense 5’-GCAGUGUGCUACUGAACUUTT-3’, and antisense 5’-AAGUUCAGUAGCACACUGCTT-3’. To overexpress c-MYC, the full-length coding sequence was amplified and subcloned into the GV230 vector. Lipofectamine-iMAX (Invitrogen, USA) was used to transfect siRNA and Lipofectamine-2000 (Invitrogen, USA) was used to transfect c-MYC-expressing plasmid into cell lines.
Generation of SNHG12-overexpressing cell lines
Recombinant lentiviruses packing genomes encoding human full-length SNHG12 (Lv-SNHG12) and a negative control sequence (Lv-vector) were purchased from GenePharma (Shanghai, China). Infection efficiency was examined by qRT-PCR, and the cells were selected with 1 μg/ml puromycin to create stable overexpression cell lines.
Cell proliferation assay
Cell Counting Kit-8 (CCK-8, Dojindo, Japan) was employed to determine cell proliferation. Briefly, cells were seeded into 96-well plates. An aliquot of 10 μl CCK-8 solution was added to each well, and the plate was incubated at 37°C for 4 hours. At the indicated time points, the absorbance at 450 nm was measured using a spectrophotometer. For the colony formation assay, 500 cancer cells per well were seeded into six-well plates. After being cultured for two weeks, the plates were washed with phosphate-buffered saline (PBS), and then, the cells were fixed with 4% paraformaldehyde for 15 min and stained with crystal violet for 10 min before the staining solution was washed away with PBS.
Apoptosis assay
Harvested cells were double stained with fluorescein isothiocyanate-Annexin V and propidium iodide using an APC Annexin V Apoptosis Detection Kit (BioLegend, San Diego, CA, USA). The cells were then analyzed using a flow cytometer (FACScan, BD Biosciences) equipped with CellQuest software (BD Biosciences). The cells were separated into viable cells, early apoptotic cells, advanced apoptotic cells, and dead cells, and the relative ratio of early apoptotic cells was compared in each experiment.
Transwell assay
Transwell plates (Costar, New York, NY) were used for the Transwell assay. Cells were seeded onto Transwell inserts, and growth medium containing 20% FBS was used as a chemoattractant in the bottom chamber. After incubation at 37°C for 24 h, cells that did not migrate through the Transwell inserts were manually removed with a cotton swab. Cells that had migrated through the filter pores were fixed with methanol, stained with 0.1% crystal violet, and observed under a microscope. The number of migration tumor cells was counted from five randomly selected 20 fold fields for each experiment and averaged.
Luciferase reporter assay
The promoter region of SNHG12 was synthesized and then inserted into the pGL3 vector. The reporter constructs were generated by subsequent PCR-based cloning, and they contained the wild type promoter or mutated c-MYC binding sites. C-Myc plasmid or vector and the luciferase reporter construct were co-transfected into TNBC cells. Luciferase activity was measured by the Dual Luciferase Assay System (Promega, USA) according to the manufacturer’s instructions.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed using the Pierce Agarose ChIP Kit according to the manufacturer’s protocol (Invitrogen, USA). C-MYC antibody was obtained from Abcam. PCR was employed to analyze the specific sequences from immunoprecipitation. The primers spanning c-MYC-bound motif 1 (B1) were as follows: sense 5’-CCTTGGCCTCCCAAAGTGCTG-3’ and antisense 5’-CTCCAGCCTGGGTGATACAGC-3’. The primers spanning c-MYC-bound motif 2 (B2) were as follows: sense 5’-CCCCCTCGGACTCCCAAAGTGC-3’ and antisense 5’-TGGGATTACAGGCGCCCGCCA-3’.
Statistical analysis
Student’s t-test was used to evaluate significant differences between the two groups in gene expression levels and cellular experiments. Clinicopathological characteristics were evaluated using the Chi-square test or Wilcoxon test as appropriate. Correlation analyses were assessed using the Pearson correlation analysis. Data was presented as the mean ± standard deviation from three independent experiments. P values less than 0.05 were considered statistically significant. All statistical analyses were performed using SPSS version 22.0 (Chicago, IL).
Results
SNHG12 was a bona fide lncRNA and upregulated in TNBC
Firstly, to validate whether SNHG12 was a lncRNA, the Coding-Potential Assessment Tool (CPAT) was employed to predict its protein-coding potential, as it had a prediction accuracy of 96% [13]. The result showed that SNHG12 was a bona fide lncRNA with a low coding probability (Figure 1A). Classic lncRNA HOTAIR and mRNA GAPDH were used as negative and positive controls, respectively. Further, we confirmed the increased expression of SNHG12 in 102 TNBC tumor tissues compared to 95 noncancerous breast tissues by qRT-PCR. The result showed that SNHG12 was significantly upregulated in TNBC (P < 0.001, Figure 1B). Interestingly, the MiTranscriptome database revealed that SNHG12 was also upregulated in multiple cancers, such as thyroid, lung, and bladder cancer (Figure 1C) [14]. Despite the fact that most lncRNAs displayed modest sequence conservation due to less evolution pressure, lncRNA SNHG12 owned a highly conserved region across mammals (Figure 1D). Collectively, the conservation of SNHG12 and its aberrant expression across multiple cancers suggested a functionally important role for SNHG12.
Figure 1.
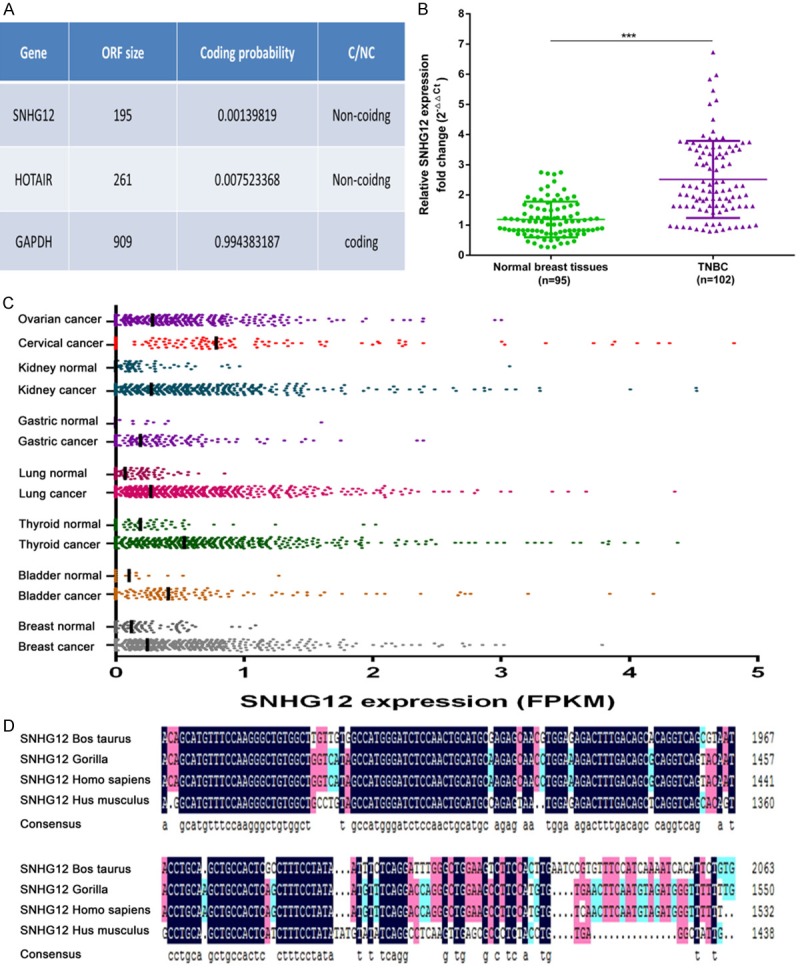
SNHG12 was a bona fide lncRNA and upregulated in TNBC. A. The RNA sequences of SNHG12, HOTAIR, and GAPDH were put into the Coding-Potential Assessment Tool, and both SNHG12 and HOTAIR were predicted to be non-coding RNAs, while GAPDH was identified to code for protein. B. SNHG12 expression was examined by qRT-PCR in 102 TNBC tissues and 95 noncancerous breast tissues. The expression levels of SNHG12 were normalized to ACTIN. C. MiTranscriptome expression data for SNHG12 across multiple cancer and normal tissue type cohorts. The RNA expression level was calculated by the fragments per kilobase of exon per million fragments mapped (FPKM) algorithm, and the median was labeled. D. Alignment of SNHG12 with lncRNA sequence from the indicated species showed SNHG12 owned a highly conserved region across mammals. ***P < 0.001.
SNHG12 was associated with tumor size and lymph node metastasis
To annotate SNHG12 expression to clinically relevant parameters, SNHG12 expression was assessed across the qRT-PCR cohort. Patients were classified into low or high expression groups depending on the median expression value of SNHG12. As shown in Table 2, the expression level of SNHG12 was significantly correlated with tumor size (P = 0.012) and lymph node metastasis (P = 0.041). Patients with higher SNHG12 expression were inclined to have larger tumors and metastatic lymph nodes. However, there was no correlation between the expression level of SNHG12 and age, distant metastasis, stage, or tumor grade.
Table 2.
Relationship between SNHG12 expression and clinicopathological characteristics
| Characteristics | Patients frequency (%) | SNHG12 | P value | |
|---|---|---|---|---|
|
| ||||
| Low | High | |||
| Total | 102 | 51 | 51 | |
| Age | ||||
| < 60 | 52 (51.0) | 27 | 25 | 0.692 |
| > 60 | 50 (49.0) | 24 | 26 | |
| Tumor size | 0.012*,a | |||
| < 2 cm | 29 (28.4) | 20 | 9 | |
| 2-5 cm | 69 (67.6) | 30 | 39 | |
| > 5 cm | 4 (3.9) | 1 | 3 | |
| Lymph node metastasis | 0.041* | |||
| No | 38 (37.3) | 24 | 14 | |
| Yes | 64 (62.7) | 27 | 37 | |
| Distant metastasis | 1.000 | |||
| No | 93 (91.2) | 46 | 47 | |
| Yes | 9 (8.8) | 5 | 4 | |
| Stage | 0.053 | |||
| I-II | 71 (69.6) | 40 | 31 | |
| III-IV | 31 (30.4) | 11 | 20 | |
| Tumor grade | 0.739a | |||
| I | 8 (7.8) | 3 | 5 | |
| II | 62 (60.8) | 33 | 29 | |
| III | 32 (31.4) | 15 | 17 | |
Wilcoxon test;
P < 0.05.
SNHG12 was activated by c-MYC
To explore the mechanism of the high expression of SNHG12, the MEME software program (http://meme-suite.org) was used for searching possible transcription factor-binding sites. The result showed that there were two putative c-MYC-bound motifs at 1564 bp and 780 bp upstream of SNHG12 (Figure 2E). Next, we analyzed the data from the TCGA database, which included 116 TNBC patients. The result showed that transcription factor c-MYC mRNA level was positively correlated with the SNHG12 transcript level in the TNBC tissues (P < 0.001, Figure 2A). Meanwhile, a tighter correlation could be observed between c-MYC protein level and SNHG12 transcript level (P < 0.001, Figure 2B). We addressed whether the upregulation of SNHG12 in TNBC was mediated by c-MYC. The expression of c-MYC was downregulated by targeted siRNA and upregulated by an overexpression plasmid in TNBC cell lines MDA-MB-231 and BT-549 (data not shown). Our results indicated that the depletion of c-MYC by siRNA in both cell lines significantly reduced SNHG12 transcript levels (P < 0.05, Figure 2C). In addition, SNHG12 levels were markedly increased in MDA-MB-231 and BT-549 cells transfected with c-MYC overexpression plasmid (P < 0.05, Figure 2D). To determine whether SNHG12 is a direct transcriptional target of c-MYC, the promoter region of SNHG12 was cloned into a luciferase reporter plasmid. These two core binding motifs were then mutated, either individually or combined, and tested in luciferase promoter reporter assays (Figure 2E). Upon c-MYC overexpression, cells transfected with a reporter containing the wild type SNHG12 promoter regions exhibited dramatically increased expression, while mutations in the c-MYC binding motif abolished the effects of c-Myc on the promoter activity of SNHG12 (P < 0.001, Figure 2F). In addition, ChIP assays showed that c-MYC directly bounds to the SNHG12 promoter regions (Figure 2G). Together, these results suggest that c-MYC promoted SNHG12 expression transcriptionally through direct interaction with its promoter region.
Figure 2.
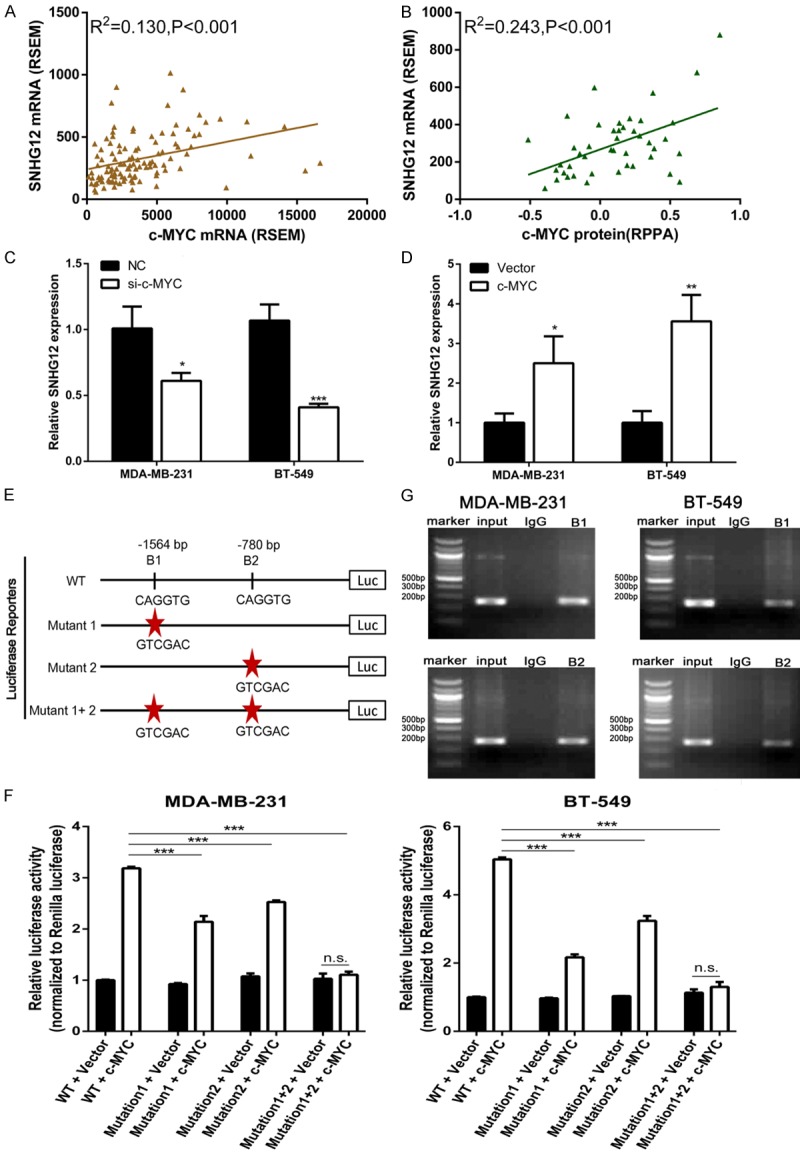
SNHG12 was activated by c-MYC. A, B. The TNBC cohort in the TCGA database was accessed and analyzed. The correlation between c-MYC mRNA level as well as c-MYC protein level and lncRNA SNHG12 transcript level was analyzed. The RNA expression level was calculated by the RNASeq by the Expectation Maximization (RSEM) algorithm, and protein expression level was represented for Reverse Phase Protein microArray (RPPA). C, D. The expression level of SNHG12 was determined using qRT-PCR in TNBC cells following treatment of cells with c-MYC siRNA or overexpression plasmid, respectively. E. A schematic diagram depicting the promoter region of SNHG12 containing two c-MYC-bound motifs at 1564 bp (B1) and 780 bp (B2) upstream of SNHG12. Sequences having mutations in (B1) and (B2) were also shown. F. Dual luciferase assays on MDA-MB-231 and BT-549 cells cotransfected with firefly luciferase constructs containing the SNHG12 promoters and vector or c-MYC-expressing plasmid. G. Chromatin from MDA-MB-231 and BT-549 cells was immunoprecipitated with the indicated antibodies. PCR was performed on immunoprecipitated DNAs or soluble chromatin using the specific primer pair for the SNHG12 promoter. Input was used as a positive control and IgG as a negative control. *P < 0.05, **P < 0.01, ***P < 0.001.
Attenuated expression of SNHG12 suppressed TNBC cells proliferation and migration
To investigate the biological functions of SNHG12 in TNBC, we designed two siRNA oligonucleotides to suppress the endogenous expression of SNHG12 in MDA-MB-231 and BT-549 cells. First, qRT-PCR assays revealed that the expression of SNHG12 was significantly reduced by specific siRNA-2 (P < 0.001, Figure 3A). CCK-8 assays showed that the repression of SNHG12 significantly decreased the proliferation of MDA-MB-231 and BT-549 cells (P < 0.001, Figure 3B). Colony formation assays also revealed that SNHG12 downregulation greatly attenuated the colony numbers of TNBC cells (P < 0.01, Figure 3C). To further investigate whether the proliferative effects of SNHG12 on TNBC cells resulted from alteration of the cell cycle or apoptosis, we performed flow cytometric analysis which showed that the fraction of apoptotic cells was significantly increased among the siRNA2-treated cells compared with the NC-treated cells (P < 0.001, Figure 3D). However, knockdown of SNHG12 had a limited impact on the TNBC cell cycle. Further, the effects of SNHG12 on cell migration were evaluated. Transwell assays showed that the inhibition of SNHG12 markedly impaired TNBC cells migration ability when compared with control cells (P < 0.001, Figure 3E).
Figure 3.
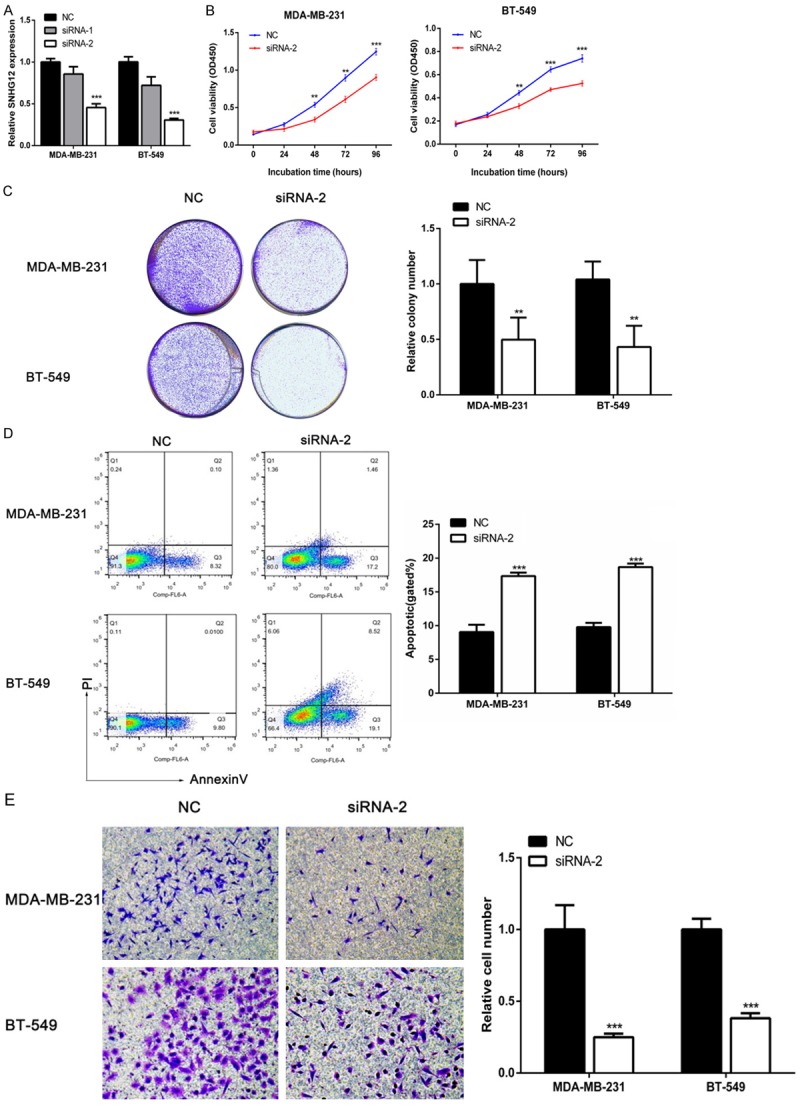
Attenuated expression of SNHG12 suppressed TNBC cells proliferation, migration and induced apoptosis. A. The SNHG12 expression level was determined by qRT-PCR when MDA-MB-231 and BT-549 cells were transfected with siRNA. B, C. The relative cell proliferation in MDA-MB-231 and BT-549 cells was measured after the cells were transfected with siRNA-2 or negative control (NC) using CCK-8 assays and colony-forming assays. D. The apoptosis rate of the two TNBC cell lines transfected with siRNA-2 or NC were measured using flow cytometry assays. E. Transwell assays were used to determine the migration ability of the cells that were transfected with siRNA-2 or NC. **P < 0.01, ***P < 0.001.
Enforced expression of SNHG12 promoted TNBC cells proliferation and migration
To further verify the biological function of SNHG12 in TNBC cells, MDA-MB-231 and BT-549 cells that stably overexpressed SNHG12 were established. QRT-PCR showed that the expression level of SNHG12 was dramatically increased in MDA-MB-231 and BT-549 cells (P < 0.01, Figure 4A). Furthermore, CCK-8 assays and colony formation assays showed that SNHG12-overexpressing TNBC cells showed an increased ability for cell proliferation compared with control cells (P < 0.001, Figure 4B and 4C). Flow cytometry analysis showed that enforced expression of SNHG12 remarkably decreased the apoptotic rate of TNBC cells (P < 0.05, Figure 4D). In addition, Transwell assays demonstrated that SNHG12 enhanced the migration of TNBC cells (P < 0.001, Figure 4E). Taken together, these results suggested that SNHG12 was crucial for the proliferation, apoptosis, and migration of TNBC.
Figure 4.
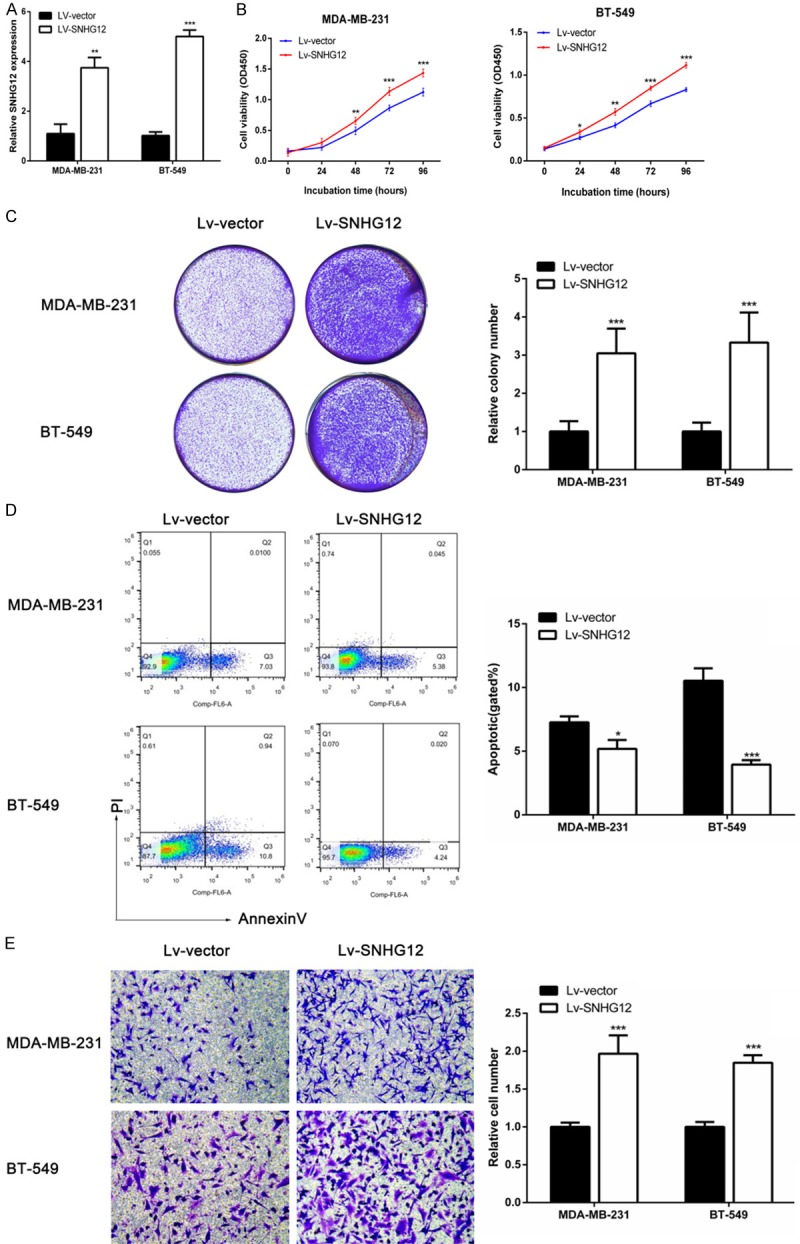
Enforced expression of SNHG12 promoted TNBC cells proliferation, migration and decreased apoptosis. A. Stable overexpression of SNHG12 was detected in MDA-MB-231 and BT-549 cells using qRT-PCR after transfection of a lentivirus harboring the full-length human SNHG12 sequence. B, C. CCK-8 assays and colony-forming assays were used to determine the cells’ proliferation. D. Flow cytometry assays were performed to analyze the cells apoptosis when TNBC cells stably overexpressed SNHG12. E. The cells migration in MDA-MB-231 and BT-549 cells were measured after the cells were overexpressed SNHG12 or vector using Transwell assays. *P < 0.05, **P < 0.01, ***P < 0.001.
SNHG12 was mainly located in cytoplasm and regulated MMP13
To explore the molecular mechanisms by which SNHG12 contributes to the phenotypes of TNBC cells, potential targets were investigated. By analyzing the RNA expression profile in our previous study, we established a co-expression network based on SNHG12 and screened seven genes as candidate SNHG12-regulated genes. Next, we further verified the regulating effect of SNHG12 on these candidates. The qRT-PCR assays showed that knockdown of SNHG12 could downregulate MMP13, while overexpression of SNHG12 could upregulate MMP13 (P < 0.01, Figure 5A and 5B). These results suggested that MMP13 was a potential target of SNHG12. Generally, the mechanism of lncRNA in part began with its cellular localization [9]. The cytoplasmic lncRNAs could modulate mRNA stability or translation and influence cellular signaling cascades, while nuclear lncRNAs were enriched for functionality involving chromatin interactions, transcriptional regulation, and RNA processing. Therefore, we fractionated TNBC cell lines into nuclear and cytoplasmic fractions and detected the subcellular localization of SNHG12. The result revealed that SNHG12 molecules were mainly located within the cytoplasm rather than the nucleus (P < 0.001, Figure 5C). Collectively, these results implied that SNHG12 exerting its biological function depends on regulating MMP13 expression at the post-transcriptional level.
Figure 5.
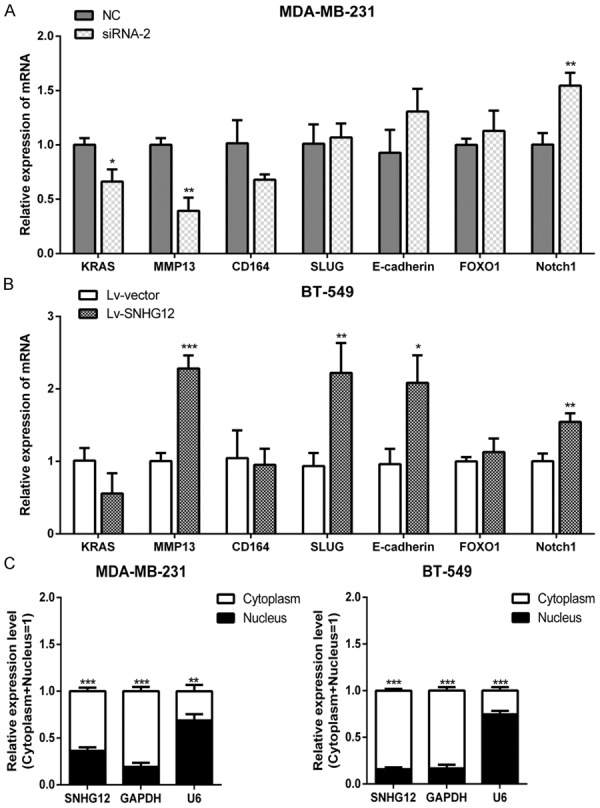
SNHG12 was mainly located in the cytoplasm and regulated MMP13. A, B. The expression levels of seven genes were determined using qRT-PCR in TNBC cells when SNHG12 was decreased or upregulated, respectively. C. SNHG12 levels in the nucleus and cytoplasmic compartments of MDA-MB-231 and BT-549 cells were determined using qRT-PCR. GAPDH was used as the cytoplasmic control, and U6 snoRNA was used as the nuclear control. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
TNBC is one of the most aggressive subtypes of breast cancer with a significantly higher recurrence and mortality rate [2]. Despite the fact that hormone therapy has led to great clinical success in luminal subtype breast cancer, as well as trastuzumab therapy in the HER2 subtype, TNBC still lacks effective therapeutic targets [15]. At present, there is an urgent need to uncover the mechanism underlying TNBC and establish therapeutic targets.
During the post-genome period, lncRNAs have become a focus of research to elucidate the complex mechanisms of cancers [9]. As a category of lncRNA, small nucleolar RNA host genes (SNHGs), which carry the snoRNA-encoding sequences in their introns, have been reported to contribute to the progression of cancers. For example, SNHG2 (also known as GAS5) was found to regulate apoptosis and proliferation in breast cancer [16,17]. SNHG5 has been reported to suppress gastric cancer progression by trapping protein in the cytosol [18]. Cao et al. found that SNHG6 functions as a competing endogenous RNA (ceRNA) to promote the progression of hepatocellular carcinoma [19]. Besides, it was demonstrated that the upregulation of SNHG20 predicts poor prognosis in hepatocellular cancer and colorectal cancer [20,21]. In sum, SNHGs are widely involved in tumor progression.
In 2015, Zhai et al. became the first to find that a novel lncRNA SNHG12 is significantly upregulated in endometrial cancer. Expression inhibition of SNHG12 caused repression of proliferation, increased cell apoptosis, and G1 phase arrest in endometrial cancer cells [22]. Subsequently, SNHG12 was reported to promote cell proliferation and migration in osteosarcoma [23]. In this study, we found that SNHG12 is significantly upregulated in TNBC tissues compared with normal tissues. Moreover, in vitro experiments confirmed that SNHG12 acts as an onco-lncRNA in TNBC, which promoted cell proliferation and migration as well as inhibited apoptosis. To the best of our knowledge, it is the first report indicating that SNHG12 is involved in breast cancer.
To explore the mechanism of the high expression of SNHG12 in TNBC, we revealed that c-MYC could bind to the promoter and activated transcription of SNHG12 directly. C-MYC is considered an “amplifier” of transcription on a global scale, and is always overexpressed or amplified in breast cancer [24-26]. However, previous studies on c-MYC-mediated transcriptional regulation have focused mainly on coding transcripts. Specific relations between c-MYC and lncRNAs have only begun to be investigated. Recently, Hart and his colleagues revealed that 534 lncRNAs were dysregulated in response to c-MYC overexpression in human B-cells including SNHG15 and SNHG16 [27]. The regulatory effect of c-MYC on a broad segment of lncRNAs opens up a new area of c-MYC activity and tumorigenesis mechanisms.
MMP13 is part of a cluster of matrix metalloproteinases (MMPs), and it was originally identified in breast cancer [28]. By degrading the extracellular matrix, MMP13 is known to promote tumor invasion and metastasis [29]. A higher expression level of MMP13 is always associated with more aggressive behaviors and poorer outcome in cancers [28]. Our study revealed that the mRNA expression levels of MMP13 decreased with decreasing SNHG12 levels and increased with the upregulation of SNHG12, which indicated that MMP13 was a potential target of SNHG12. In addition, the NPInter database indicated that SNHG12 was able to bind several RNA-binding proteins such as IGF2BP3, EWSR1 and hnRNP as well as a lot of miRNAs. Therefore, we hypothesized that SNHG12 may act a scaffold mediating RNA-binding proteins or act a ceRNA to stabilize MMP13 expression at the post-transcriptional level [30]. Further research needs to be performed to confirm our hypothesis.
It is worth noting that our study still has several limitations. First, the biological role of SNHG12 has not yet been determined by in vivo experiments. Second, the mechanism by which SNHG12 regulates proliferation and apoptosis needs to be elucidated. High-throughput sequencing technology and microarray technology provide an efficient solution to screen out potential cellular signaling cascades regulated by SNHG12.
In summary, we determined that SNHG12 is strongly upregulated in TNBC, and its high expression is significantly correlated with tumor size and lymph node metastasis. Moreover, we revealed that SNHG12 overexpression is induced by a transcription factor c-MYC. As documented in the MDA-MB-231 and BT-549 cell lines, silencing SNHG12 expression inhibited cells proliferation and apoptosis promotion, whereas SNHG12 overexpression had the opposite effect. In addition, we revealed that SNHG12 may promote cells migration by regulating MMP13 expression. Taken together, our findings suggest that SNHG12 could contribute to the oncogenic potential of TNBC and be a promising therapeutic target.
Acknowledgements
This study was funded by the Key Project of Science and Technology Innovation Team of Zhejiang Province (2013TD10), National Natural Science Foundation of China (No. 81372380), and Zhejiang Province Natural Science Foundation of China (LY17H160053).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 3.Podo F, Buydens LM, Degani H, Hilhorst R, Klipp E, Gribbestad IS, Van Huffel S, van Laarhoven HW, Luts J, Monleon D, Postma GJ, Schneiderhan-Marra N, Santoro F, Wouters H, Russnes HG, Sorlie T, Tagliabue E, Borresen-Dale AL. Triple-negative breast cancer: present challenges and new perspectives. Mol Oncol. 2010;4:209–229. doi: 10.1016/j.molonc.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol. 2010;7:683–692. doi: 10.1038/nrclinonc.2010.154. [DOI] [PubMed] [Google Scholar]
- 5.St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet. 2015;31:239–251. doi: 10.1016/j.tig.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hudson WH, Ortlund EA. The structure, function and evolution of proteins that bind DNA and RNA. Nat Rev Mol Cell Biol. 2014;15:749–760. doi: 10.1038/nrm3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 9.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest. 2016;126:2775–2782. doi: 10.1172/JCI84421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang F, Liu YH, Dong SY, Yao ZH, Lv L, Ma RM, Dai XX, Wang J, Zhang XH, Wang OC. Co-expression networks revealed potential core lncRNAs in the triple-negative breast cancer. Gene. 2016;591:471–477. doi: 10.1016/j.gene.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The cancer genome Atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Park HJ, Dasari S, Wang S, Kocher JP, Li W. CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iyer MK, Niknafs YS, Malik R, Singhal U, Sahu A, Hosono Y, Barrette TR, Prensner JR, Evans JR, Zhao S, Poliakov A, Cao X, Dhanasekaran SM, Wu YM, Robinson DR, Beer DG, Feng FY, Iyer HK, Chinnaiyan AM. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mourtada-Maarabouni M, Pickard MR, Hedge VL, Farzaneh F, Williams GT. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene. 2009;28:195–208. doi: 10.1038/onc.2008.373. [DOI] [PubMed] [Google Scholar]
- 17.Williams GT, Hughes JP, Stoneman V, Anderson CL, McCarthy NJ, Mourtada-Maarabouni M, Pickard M, Hedge VL, Trayner I, Farzaneh F. Isolation of genes controlling apoptosis through their effects on cell survival. Gene Ther Mol Biol. 2006;10:255–262. [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao L, Guo H, Zhou B, Feng J, Li Y, Han T, Liu L, Li L, Zhang S, Liu Y, Shi J, Zheng D. Long non-coding RNA SNHG5 suppresses gastric cancer progression by trapping MTA2 in the cytosol. Oncogene. 2016;35:5770–5780. doi: 10.1038/onc.2016.110. [DOI] [PubMed] [Google Scholar]
- 19.Cao C, Zhang T, Zhang D, Xie L, Zou X, Lei L, Wu D, Liu L. The long non-coding RNA, SNHG6-003, functions as a competing endogenous RNA to promote the progression of hepatocellular carcinoma. Oncogene. 2016 doi: 10.1038/onc.2016.278. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 20.Li C, Zhou L, He J, Fang XQ, Zhu SW, Xiong MM. Increased long noncoding RNA SNHG20 predicts poor prognosis in colorectal cancer. BMC Cancer. 2016;16:655. doi: 10.1186/s12885-016-2719-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang D, Cao C, Liu L, Wu D. Up-regulation of LncRNA SNHG20 predicts poor prognosis in hepatocellular carcinoma. J Cancer. 2016;7:608–617. doi: 10.7150/jca.13822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhai W, Li X, Wu S, Zhang Y, Pang H, Chen W. Microarray expression profile of lncRNAs and the upregulated ASLNC04080 lncRNA in human endometrial carcinoma. Int J Oncol. 2015;46:2125–2137. doi: 10.3892/ijo.2015.2897. [DOI] [PubMed] [Google Scholar]
- 23.Ruan W, Wang P, Feng S, Xue Y, Li Y. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes cell proliferation and migration by upregulating angiomotin gene expression in human osteosarcoma cells. Tumour Biol. 2016;37:4065–4073. doi: 10.1007/s13277-015-4256-7. [DOI] [PubMed] [Google Scholar]
- 24.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hart JR, Roberts TC, Weinberg MS, Morris KV, Vogt PK. MYC regulates the non-coding transcriptome. Oncotarget. 2014;5:12543–12554. doi: 10.18632/oncotarget.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leeman MF, Curran S, Murray GI. The structure, regulation, and function of human matrix metalloproteinase-13. Crit Rev Biochem Mol Biol. 2002;37:149–166. doi: 10.1080/10409230290771483. [DOI] [PubMed] [Google Scholar]
- 29.Sendon-Lago J, Seoane S, Eiro N, Bermudez MA, Macia M, Garcia-Caballero T, Vizoso FJ, Perez-Fernandez R. Cancer progression by breast tumors with Pit-1-overexpression is blocked by inhibition of metalloproteinase (MMP)-13. Breast Cancer Res. 2014;16:505. doi: 10.1186/s13058-014-0505-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hao Y, Wu W, Li H, Yuan J, Luo J, Zhao Y, Chen R. NPInter v3.0: an upgraded database of noncoding RNA-associated interactions. Database (Oxford) 2016:2016. doi: 10.1093/database/baw057. [DOI] [PMC free article] [PubMed] [Google Scholar]


