Abstract
TRIM24, originally known as intermediary factor 1-alpha, is involved in the development of several cancers. This study aimed to evaluate the expression level and prognostic value of TRIM24 in cervical cancer. In the present study, we showed that the expression of TRIM24 was markedly upregulated in cervical cancer cell lines and cancerous specimens at both transcriptional and translational levels. TRIM24 expression was analyzed in 147 archived cervical cancer specimens using immunohistochemistry, and the correlation between TRIM24 expression and clinicopathological parameters was evaluated. Statistical analysis suggested that TRIM24 expression was significantly correlated with clinical stage and (P=0.007) and lymphatic metastasis (P=0.001). Patients with higher TRIM24 expression had shorter overall (P=0.005) and recurrence-free (P=0.011) survival time. Moreover, we found that silencing TRIM24 by short hairpin RNAi caused an inhibition of cell migration and invasion. Further study indicated that TRIM24 induced cervical cancer cell migration and invasion was through the NF-κB and AKT signaling pathways. In conclusion, TRIM24 is overexpressed in cervical cancer and regulates malignant cell metastasis, which makes TRIM24 a candidate therapeutic target for cervical cancer.
Keywords: TRIM24, cervical cancer, progression, NF-κB, AKT, epithelial-mesenchymal transition
Introduction
Cervical carcinoma is one of the most common gynecological malignancies, with more than 520000 new cases reported annually [1]. In spite of recent advances in surgery and chemo-radiotherapy, the long-term outcome for patients with metastatic or recurrent diseases still remains poor, with a 5-year survival rate of <30% [2]. Therefore, it would be beneficial to clinically discover biomarkers that predict tumor development and progression, identify their molecular mechanisms in cervical cancer patients with high potential for tumor metastasis, and then design more effective therapeutic strategies and novel therapeutic targets.
TRIM24 belongs to the TRIM family of structurally related proteins that has an N-terminal TRIM domain with potential self-assembly properties, a C-terminal region containing a PHD finger and a bromodomain, and a nuclear receptor interaction box [3-5]. It was originally known as transcription intermediary factor 1-alpha (TIF1α), a co-regulator that interact with retinoic acid receptor (RAR) in a ligand-dependent fashion [6]. TRIM24 associates with chromatin, which is mediated by bromodomain-DNA and bromodomain-nucleosome interactions [7]. Existing research showed that TRIM24 is involved in modulating gene expression during mouse embryonic development [8]. More recently, TRIM24 has been identified as a partner of the p53 tumor suppressor, as it interacts with phosphorylated p53 to target it for degradation and termination of the DNA damage response [9]. Accumulating evidence showed that TRIM24 is a crucial regulator in cellular proliferation and tumor progression [10,11]. In vitro studies suggested that knockdown of TRIM24 suppresses cell proliferation, cell cycle progression, and in vivo tumor development, whereas overexpression of TRIM24 promotes cell growth [12]. Likewise, elevated expression of TRIM24 could promote gastric cancer cell growth and chemoresistance [13]. These studies suggested that TRIM24 may serve as an important factor during cancer progression. However, the clinical and biological roles of TRIM24 in human cervical cancer still need to be addressed.
In the present study, we examined the expression of TRIM24 in 147 cervical cancer tissue specimens using immunohistochemistry and analyzed the correlation between its expression and clinicopathological factors. We also investigated the effect of TRIM24 on the biological behavior of cervical cancer cell lines, revealing the potential mechanisms of TRIM24 on cervical cancer metastasis.
Materials and methods
Patients and tissue specimens
The present study included 147 patients with stage Ib-IIb cervical cancer who were treated at the First Affiliated Hospital of Shenzhen University between 2004 and 2009. None of the patients had been given chemo-radiotherapy before surgery. All patients were staged according to the International Federation of Gynecology and Obstetrics (FIGO) criteria. The histological subtype was assigned according to the criteria of the World Health Organization classification. The clinical and pathologic parameters were reviewed from impatient medical records and presented in Table 1. Seven freshly collected paired cervical cancerous/normal tissues were frozen and stored in liquid nitrogen until further use. The Institutional Review Board approved the present study and informed consent was obtained from all patients for the use of their tissue samples.
Table 1.
Correlation between TRIM24 expression levels and clinicopathological characters in cervical cancer patients
| Clinical characters | No. of Patients | TRIM24 | P | |
|---|---|---|---|---|
|
| ||||
| High | Low | |||
| Age (y) | ||||
| ≤50 | 85 | 40 | 45 | |
| >50 | 62 | 27 | 35 | .673 |
| FIGO Stage | ||||
| Ib | 106 | 41 | 65 | |
| IIa + IIb | 41 | 26 | 15 | .007 |
| Differentiation | ||||
| Well + Moderately | 58 | 25 | 33 | |
| Poorly | 89 | 42 | 47 | .627 |
| Timor Size | ||||
| ≤4 cm | 110 | 46 | 64 | |
| >4 cm | 37 | 21 | 16 | .115 |
| Histological Type | ||||
| SCC | 101 | 45 | 56 | |
| AC | 46 | 22 | 24 | .712 |
| Nodal Metastasis | ||||
| + | 25 | 19 | 6 | |
| - | 122 | 48 | 74 | .001 |
| Total No. of Patients | 147 | 67 | 80 | |
SCC: squamous cell cancer; AC: Adenocarcinoma.
Cell lines
Primary normal cervical epithelial cells (NCECs) were collected from non-cancerous cervical tissue according to the previous report [14]. Cervical cancer cell lines, including Hela, Hela229, Siha, ME-180, C33A, CaSKi, and MS7501, were obtained from the Chinese Academy of Sciences (Shanghai, China) and cultured in DMEM medium (Invitrogen). All cells were supplemented with 10% fetal bovine serum (HyClone, Logan, UT, USA) and 1% penicillin/streptomycin (Invitrogen).
Immunohistochemistry (IHC)
Paraffin-embedded samples were obtained from 147 patients for immunohistochemical analysis. In brief, 4-μm paraffin-embedded sections were deparaffinized and rehydrated. Endogenous peroxidase activity was blocked by 3% hydrogen peroxide for 15 min. After antigen retrieval, sections were incubated with 5% serum to avoid the non-specific binding. The sections were incubated overnight at 4°C with anti-TRIM24 antibody (1:500 dilution; Proteintech, Chicago, IL). After the primary antibody was washed off, the sections were incubated with prediluted secondary antibody (Santa Cruz Biotechnology). The sections were then stained with DAB (3,3-diaminobenzidine) and then counterstained with hematoxylin. For blank control, the primary antibody was omitted. For negative control, the primary antibody was replaced by nonimmune serum.
Two independent investigators examined all slides randomly. Five views were examined per slide at 400× magnification. Immunostaining of TRIM24 was scored following a semi-quantitative scale by evaluating, in representative tumor areas, the intensity and percentage of cells showing higher immunostaining than the control cells. Nuclear staining of the tumor cells was considered as positive immunostaining. The intensity of TRIM24 nuclear staining was also scored as 0 (no staining), 1 (weak), 2 (strong). Percentage scores were assigned as: 1, 1-25%; 2, 26-50%; 3, 51-75%; 4, 76-100%. The scores of each tumor sample were multiplied to give a final score of 0 to 8 and the total expression of TRIM24 was determined as low with score ≤4, and as high with score >4.
Small RNA transfection
Small interfering RNAs targeting two splice variants of TRIM24 and FAM-labeled negative control siRNA were purchased from Guangzhou Ruibo Company, Ltd. (Guangzhou, China). In vitro transient transfection was performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) as recommended by the manufacturer.
RNA extraction and reverse transcription-PCR
Total RNA from cervical cancer and normal tissues was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The extracted RNA was pretreated with RNase-free DNase, and 2 μg RNA from each sample was used for cDNA synthesis with random hexamers. Real-time PCR was performed using the Applied Biosystems 7500 Sequence Detection system. Primers were designed as follows: TRIM24 forward 5’-GTTTACCAAACCCTAGAATGCAGG-3’, and reverse 5’-AAGTTTATCAAACGTGGAGGCG-3’; a-catenin forward 5’-CAACCCTTGTAAACACCAAT-3’ and reverse 5’-CCTTCTCCAAGAAATTCTCA-3’; Vimentin forward 5’-AGGAAATGGCTCGTCACCTTCGTGAATA-3’ and reverse 5’-GGAGTGTCGGTTGTTAAGAACTAGAGCT-3’; Fibronectin forword 5’-TTATGACGACGGGAAGACCT-3’ and reverse, 5’-GCTGGATGGAAAGATTACTC-3’; Snail forword 5’-ACCACTATGCCGCGCTCTT-3’ and reverse, 5’-GGTCGTAGGGCTGCTGGAA-3’; Twist forword 5’-CGGGAGTCCGCAGTCTTA-3’ and reverse, 5’-TGAATCTTGCTCAGCTTGTC-3’; GAPDH forward primer 5’-GACCCCTTCATTGACCTCAAC-3’, reverse primer 5’-CTTCTCCATGGTGGTGAAGA-3’. Real-time PCR was then employed to determine the fold increase of TRIM24 mRNA in each of the cervical tumors relative to the paired normal cervical tissues, with each pair taken from the same patient. Expression data were normalized to the geometric mean of the housekeeping GAPDH gene to control the variability in expression levels.
Western blot analysis
Total proteins from cells and tissues were extracted in cell lysis buffer (Pierce, Rockford, IL) and quantified. Samples of 40 μg of protein were separated by SDS-PAGE. Proteins were transferred to PVDF membranes (Millipore, Billerica, MA, USA) and incubated overnight at 4°C with antibody against TRIM24 (1:1000, Proteintech, USA), p-IκBα, IκBα, p-AKT, AKT (1:1000; Cell signaling, Boston, MA, USA) and GAPDH (1:2000; Santa Cruz, USA). After washing, the membrane was incubated with Horseradish Peroxidase conjugating secondary antibodies. Finally, membrane was detected with anenhanced chemiluminescence kit (Amersham Bioscience, Piscataway, NJ) according to the manufacturer’s instructions.
Migration and invasion assay
Cell migration and invasion assay was performed using a 24-well transwell chamber with a pore size of 8 μm (Costar, Cambridge, MA) (In invasion assays, the inserts were coated with 20 μl Matrigel). Forty-eight hours after the transfection, cells were trypsinized, and 1×105 cells in 100 μl of serum-free medium were transferred to the upper chamber and incubated for 24 h. Medium supplemented with 20% FBS was added to the lower chamber. After incubation, the non-invaded cells on the upper membrane surface were removed with a cotton tip, and the cells that passed through the filter were fixed with 4% paraformaldehyde and stained with hematoxylin. Three independent experiments were performed and the data are presented as the mean ± SD.
Statistical analysis
SPSS (Chicago, IL) statistical software version 16.0 was used for all statistical analyses. A χ2 test was used to examine possible correlations between TRIM24 expression and clinicopathologic factors. Survival curves were generated using the Kaplan-Meier method and compared using the log-rank test. All p values are based on a two-sided statistical analysis, and P<0.05 was considered to indicate statistical significance.
Results
Overexpression of TRIM24 in cervical cancer
In order to investigate TRIM24 expression pattern in cervical cancer, Western blotting and real-time PCR analyses were performed using 7 pairs of cervical cancer and adjacent non-cancerous tissues, NCEC and cervical cancer cell lines. The expressions of both the TRIM24 protein and mRNA were markedly upregulated in multiple cervical cancer cell lines, in comparison with those in NCEC (Figure 1A). In addition, comparative analysis showed that TRIM24 mRNA and protein levels were differentially upregulated in all 7 cervical cancer samples compared with matched adjacent non-cancerous tissues (Figure 1B).
Figure 1.
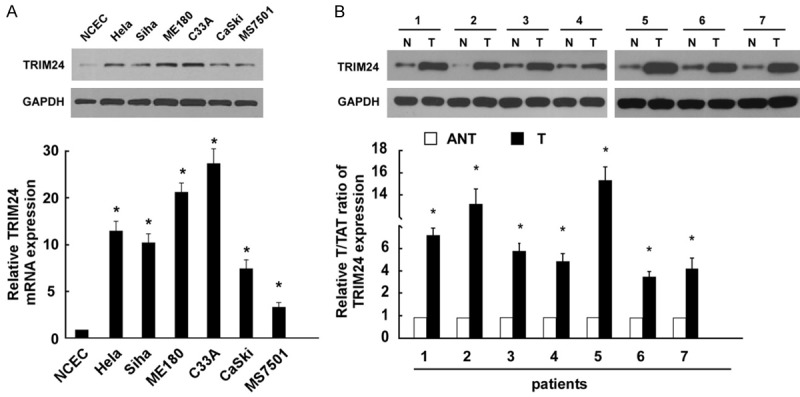
TRIM24 is over-expressed in cervical cancer. A. Western blot and real-time PCR analysis of TRIM24 expression in normal cervical epithelial cells (NCECs) and 6 cervical cancer cell lines (Hela, Hela229, Siha, ME-180, C33A, CaSKi, and MS7501). B. Western blot and real-time PCR analysis of TRIM24 expression in matched primary cancer tissues (T) and the adjacent non-tumour tissues (ANT); GAPDH was used as a loading control. Bars represent the means of three independent experiments. *P<0.05.
TRIM24 expression is associated with clinical characteristics and prognosis
The relationship between TRIM24 expression and clinicopathological characters was evaluated (Table 1). IHC staining showed the expression levels of TRIM24 were significantly increased in patients with advanced stages (Figure 2A and 2B; Table 1, P=0.007) and lymphatic metastasis (Figure 2C and 2D; Table 1, P=0.001). These results suggest that TRIM24 is associated to the aggressiveness of cervical cancer patients. The overall survival rate and recurrence-free survival rate of the cervical cancer following surgical treatment were analyzed using the Kaplan-Meier method. The results showed that the patients with high levels of TRIM24 had significant worse overall survival (Figure 3A, P=0.005) and recurrence-free survival (Figure 3B, P=0.011) than those with low levels of TRIM24 in cervical cancer. Multivariable Cox regression analysis revealed that high expression of TRIM24 could be an independent prognostic factor in cervical cancer (Table 2).
Figure 2.
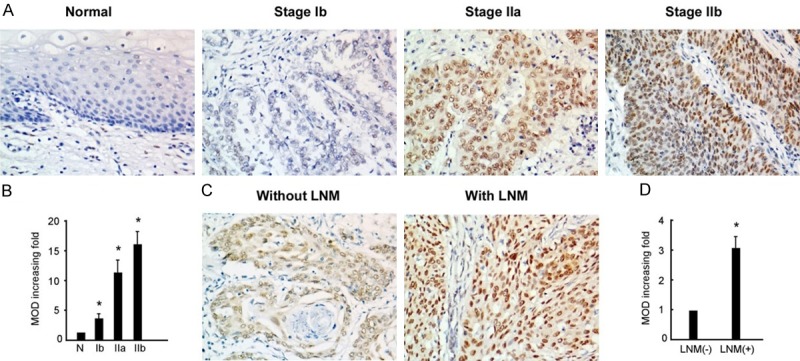
Expression levels of TRIM24 in cervical cancer. A. Representative IHC staining images of TRIM24 expression levels in normal cervical epithelium and stage Ib-IIb tumor tissues. B. Statistical analysis of average MODs in normal cervical tissues and patients with different FIGO stage. C. Representative IHC staining images of TRIM24 expression in patients with or without lymph node metastasis. D. Statistical analysis of average MODs in patients without LNM compared with those with LNM. *P<0.05.
Figure 3.
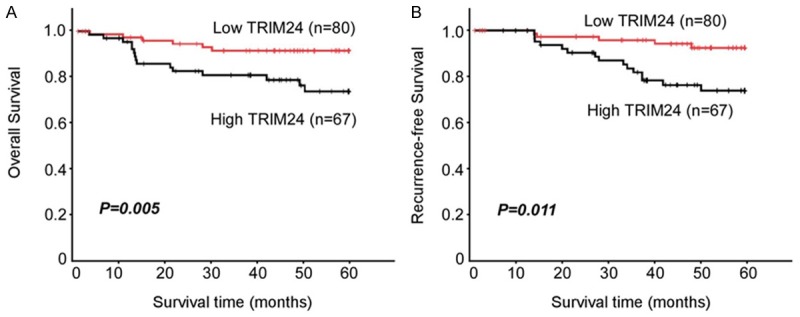
Kaplan-Meier survival curves for overall (A) and recurrence-free (B) survivals of cervical cancer patients with low TRIM24-expressing versus high TRIM24-expressing tumours.
Table 2.
Univariable and Multivariable analysis for overall survival in cervical cancer patients
| Prognostic variables | Univariable | Multivariable | ||
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | P | HR (95% CI) | P | |
| Age (>50 vs ≤50) | 1. 258 (0.365-4.353) | 0.526 | ||
| FIGO Stage (II vs Ib) | 2. 964 (0.504-6.626) | 0.026 | ||
| Differentiation (Grade 3 vs 1/2) | 1.944 (0.185-7.243) | 0.304 | ||
| Tumor size (>4 cm vs ≤4 cm) | 2.764 (1.035-5.097) | 0.037 | ||
| Histological type (SCC vs AC) | 1.043 (0.056-4.953) | 0.690 | ||
| LN Metastasis (+ vs -) | 4.155 (0.267-12.648) | 0.015 | 4.004 (1.126-14.277) | 0.034 |
| TRIM24 expression (high vs low) | 3.754 (0.205-9.590) | 0.012 | 3.383 (0.246-9.489) | 0.026 |
SCC: squamous cell cancer; AC: Adenocarcinoma.
Depletion of TRIM24 inhibits cervical cancer cell invasion
In light of the obvious connection between TRIM24 expression and pelvic lymph node metastasis, we further investigated the effect of TRIM24 on invasion and migration in Siha and CaSki cell lines. As shown in Figure 4A, TRIM24 protein levels were decreased in the TRIM24-silenced cells. Moreover, the migration and invasion ability of Siha and CaSki cells was reduced after being transfected with siTRIM24 (P<0.05, compared with controls) (Figure 4B). These results suggested that TRIM24 may play an important role in tumor metastasis in cervical cancer.
Figure 4.
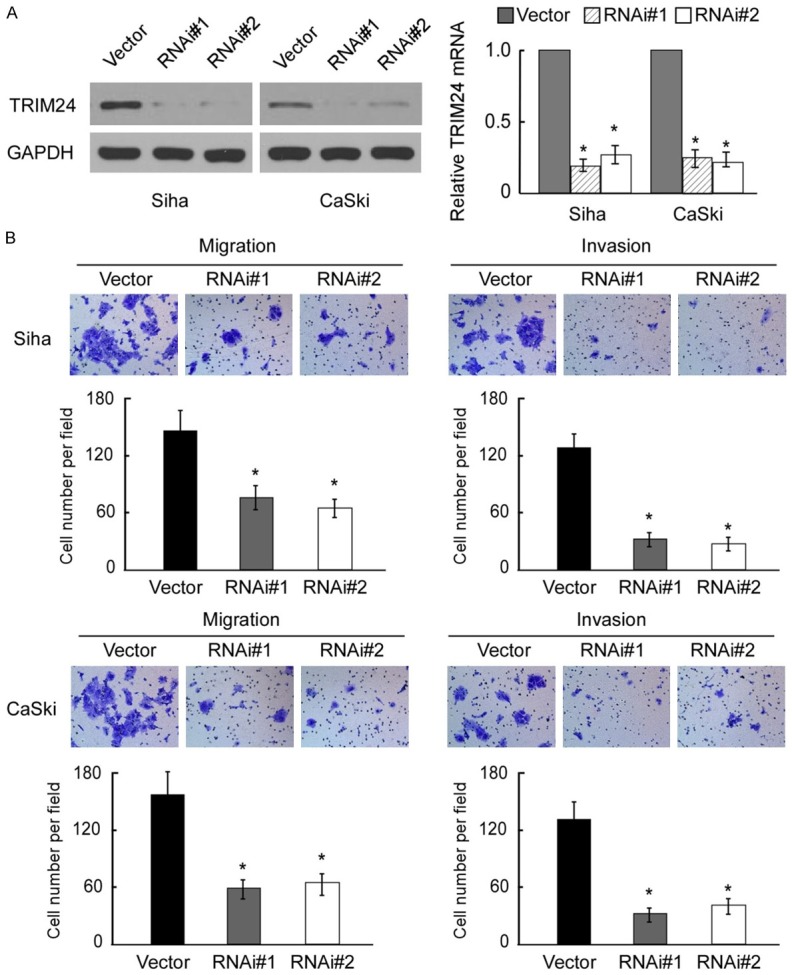
Depletion of TRIM24 inhibits cancer cell migration and invasion. A. Western blotting analysis of of TRIM24 expression in vector and TRIM24-transduced Siha and CaSki cells; GAPDH was used as a loading control. B. Reduced migration and invasion ability of Siha and CaSki cells post-transfection with siTRIM24 (P<0.05, compared with controls).
TRIM24 induces EMT (epithelial mesenchymal transition) by regulating the NF-κB and AKT signaling in cervical cancer cells
To investigate the potential mechanisms of TRIM24 modulation in tumor cell metastasis, we tested the effect of TRIM24 knockdown on the levels of EMT related genes and several signaling pathways. As expected, depletion of TRIM24 downregulated the level of p-IκBα and p-AKT in both Siha and CaSKi cells (Figure 5A). Moreover, the expression of EMT-related genes such as α-cadherin, Vimentin, Fibronectin, Snail, and Twist, was analyzed by real-time PCR. After knockdown of TRIM24, the expression of α-cadherin was up-regulated, while Vimentin, Fibronectin, Snail, and Twist were down-regulated in each siTRIM24 group compared with the negative control group (Figure 5B). Furthermore, we examined the expression of TRIM24, p-IκBα, p-AKT, and Snail in 10 freshly prepared cervical cancer tissues. Real-time PCR and correlation analyses showed that expression of TRIM24 was positively correlated with expression of p-IκBα (r=0.721; P=0.019), p-AKT (r=0.700; P=0.024), and Snail (r=0.749; P=0.013) (Figure 5C). These results further demonstrated that TRIM24 mediated EMT and metastasis of cervical cancer cells via the NF-κB and AKT signaling cascade.
Figure 5.
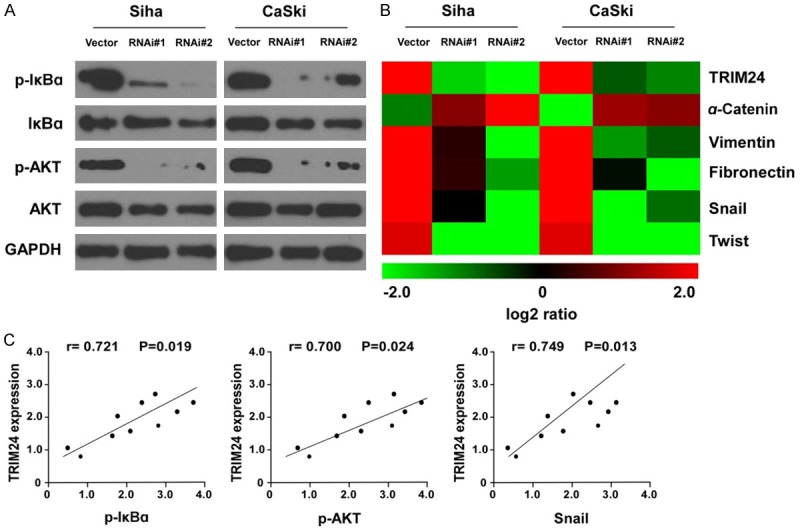
Knockdown of TRIM24 decreased NF-κB and AKT signaling in cervical cancer cells. A. Western blotting analysis revealed that TRIM24 knockdown downregulated the protein levels of p-IκB and p-AKT without significant changes of IκB and AKT in cervical cancer cell lines. B. Real-time PCR of the expressions of TRIM24 and EMT-relevant markers a-Catenin, fibronectin and vimentin, as well as EMT regulators Snail and Twist in RNAi vector and TRIM24 shRNA 1- and 2-transduced Siha and CaSki cells. C. Correlation analyses of TRIM24, p-IκB, p-AKT, and Snail in 10 freshly prepared cervical cancer tissues.
Discussion
Despite the improvement in diagnostic and therapeutic methods, invasive cervical cancer remains one of the most devastating diseases, and it has a dismal prognosis. Therefore, the screening of predictive biomarkers is extremely important in improving patients’ survival time. In the present study, we found upregulated TRIM24 expression in cervical cancer tissues in compared with that in normal cervical tissues, and positive correlation of TRIM24 overexpression with clinical features and outcomes, suggesting TRIM24 may participate in cervical cancer progression and could be used as a diagnostic indicator in cervical cancer.
Recently, tripartite motif (TRIM) protein family members, characterized by conserved amino (N)-terminal zinc-finger domains of a RING-type E3 ubiquitin ligase, B-boxes and coiled coil, have been shown to be crucial regulators in tumorigenesis [15]. As a histone reader that activates estrogen-dependent genes, TRIM24 is associated with cellular proliferation in embryonic stem cells and tumor cells [11,16]. Of note, TRIM24 has been found to be upregulated in several human malignancies including acute promyelocytic leukaemia, papillary thyroid carcinoma, breast cancer, and non-small cell lung cancer. However, the expression pattern of TRIM24 as well as its correlation with clinical and pathological features in cervical cancer has not yet been investigated. In the present study, we found that the expression of TRIM24 in cervical cancer tissues was higher than in adjacent normal cervical tissues. There was a close correlation between TRIM24 overexpression and tumor stage and lymph lode metastasis. In addition, TRIM24 overexpression was correlated with poor prognosis in cervical cancer patients, which was consistent with previous reports in that higher TRIM24 expression is correlates with tumor aggressiveness and poor clinical outcomes [17]. These studies indicate that TRIM24 may play an important role in cervical cancer progression.
To explore the relationship between TRIM24 and the biological behavior of cervical cancer cells, we blocked TRIM24 function by using siRNA in the Siha and CaSki cell lines. We found that TRIM24 depletion caused an obvious decrease in the migration and invasion potentials of both cell lines, which was in accord with precious reports concerning its role in other cancers. Liu et al. demonstrated that small interfering RNA induced down-regulation of TRIM24 promoted apoptosis, decreased EMT related proteins expression, and reduced migration and invasion ability of hepatocellular carcinoma cell line HepG2 [18]. Li et al. reported that TRIM24 siRNA treatment have a measurable blocking effect on cell invasion in non-small cell lung cancer cell lines [19]. Moreover, this study showed that the expression level of TRIM24 was higher in cervical cancer patients with lymph nodes metastasis. Thus, these studies together revealed that TRIM24 played an important role in cell metastasis of cervical cancer.
Activation of NF-κB pathway plays an important role in the tumor development in many types of cancers [20]. In cervical cancer, NF-κB signaling is involved in the progression of cervical cancer [21], probably by mediating a negative feedback loop to regulate HPV replication and that this feedback loop could be associated with control of the viral copy numbers [22]. The human papillomavirus 16 (HPV16) E1 and E2 could both mediate NF-κB activation [23]. Interestingly, NF-κB has been reported to be constitutively activated in human cervical cancer to promote EMT [24]. In recent functional study, Xue et al. investigated the role of TRIM24 in NF-κB activation. They showed that overexpression of TRIM24 increased cyclin D1, cyclin E and upregulated NF-κB and AKT signaling in bladder cancer cells, suggesting that TRIM24 might regulate cyclin D1 expression and cell proliferation by activating NF-κB and AKT signaling pathways [25]. During our study, we demonstrated that TRIM24 might contribute to invasion, metastasis and progression of cervical cancer, and silencing TRIM24 inhibited the EMT of human cervical cancer cells through NF-κB and AKT pathways. Our results provide the preliminary molecular mechanisms underlying the role of TRIM24 in cervical cancer EMT and metastasis.
In conclusion, we have revealed that TRIM24 expression is increased in cervical cancer and correlated with clinicopathological features and prognosis. TRIM24 contributed to the cancer cell metastasis through NF-κB and AKT signaling pathways.
Disclosure of conflict of interest
None.
References
- 1.Arbyn M, Castellsague X, de Sanjose S, Bruni L, Saraiya M, Bray F, Ferlay J. Worldwide burden of cervical cancer in 2008. Ann Oncol. 2011;22:2675–2686. doi: 10.1093/annonc/mdr015. [DOI] [PubMed] [Google Scholar]
- 2.Yeung TL, Leung CS, Yip KP, Au Yeung CL, Wong ST, Mok SC. Cellular and molecular processes in ovarian cancer metastasis. A Review in the theme: cell and molecular processes in cancer metastasis. Am J Physiol Cell Physiol. 2015;309:C444–456. doi: 10.1152/ajpcell.00188.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borden KL. RING domains: master builders of molecular scaffolds? J Mol Biol. 2000;295:1103–1112. doi: 10.1006/jmbi.1999.3429. [DOI] [PubMed] [Google Scholar]
- 4.Dyson MH, Rose S, Mahadevan LC. Acetyllysine-binding and function of bromodomain-containing proteins in chromatin. Front Biosci. 2001;6:D853–865. doi: 10.2741/dyson. [DOI] [PubMed] [Google Scholar]
- 5.Le Douarin B, Zechel C, Garnier JM, Lutz Y, Tora L, Pierrat P, Heery D, Gronemeyer H, Chambon P, Losson R. The N-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le Douarin B, Nielsen AL, You J, Chambon P, Losson R. TIF1 alpha: a chromatin-specific mediator for the ligand-dependent activation function AF-2 of nuclear receptors? Biochem Soc Trans. 1997;25:605–612. doi: 10.1042/bst0250605. [DOI] [PubMed] [Google Scholar]
- 7.Remboutsika E, Yamamoto K, Harbers M, Schmutz M. The bromodomain mediates transcriptional intermediary factor 1alpha -nucleosome interactions. J Biol Chem. 2002;277:50318–50325. doi: 10.1074/jbc.M203759200. [DOI] [PubMed] [Google Scholar]
- 8.Torres-Padilla ME, Zernicka-Goetz M. Role of TIF1alpha as a modulator of embryonic transcription in the mouse zygote. J Cell Biol. 2006;174:329–338. doi: 10.1083/jcb.200603146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain AK, Allton K, Duncan AD, Barton MC. TRIM24 is a p53-induced E3-ubiquitin ligase that undergoes ATM-mediated phosphorylation and autodegradation during DNA damage. Mol Cell Biol. 2014;34:2695–2709. doi: 10.1128/MCB.01705-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katzenellenbogen BS. Estrogen receptors: bioactivities and interactions with cell signaling pathways. Biol Reprod. 1996;54:287–293. doi: 10.1095/biolreprod54.2.287. [DOI] [PubMed] [Google Scholar]
- 11.Tsai WW, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S, Tsai CY, Shi X, Schwarzer D, Plunkett W, Aronow B, Gozani O, Fischle W, Hung MC, Patel DJ, Barton MC. TRIM24 links a non-canonical histone signature to breast cancer. Nature. 2010;468:927–932. doi: 10.1038/nature09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang LH, Yin AA, Cheng JX, Huang HY, Li XM, Zhang YQ, Han N, Zhang X. TRIM24 promotes glioma progression and enhances chemoresistance through activation of the PI3K/Akt signaling pathway. Oncogene. 2015;34:600–610. doi: 10.1038/onc.2013.593. [DOI] [PubMed] [Google Scholar]
- 13.Miao ZF, Wang ZN, Zhao TT, Xu YY, Wu JH, Liu XY, Xu H, You Y, Xu HM. TRIM24 is upregulated in human gastric cancer and promotes gastric cancer cell growth and chemoresistance. Virchows Arch. 2015;466:525–532. doi: 10.1007/s00428-015-1737-4. [DOI] [PubMed] [Google Scholar]
- 14.Hou T, Ou J, Zhao X, Huang X, Huang Y, Zhang Y. MicroRNA-196a promotes cervical cancer proliferation through the regulation of FOXO1 and p27Kip1. Br J Cancer. 2014;110:1260–1268. doi: 10.1038/bjc.2013.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11:792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- 16.Allton K, Jain AK, Herz HM, Tsai WW, Jung SY, Qin J, Bergmann A, Johnson RL, Barton MC. Trim24 targets endogenous p53 for degradation. Proc Natl Acad Sci U S A. 2009;106:11612–11616. doi: 10.1073/pnas.0813177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui Z, Cao W, Li J, Song X, Mao L, Chen W. TRIM24 overexpression is common in locally advanced head and neck squamous cell carcinoma and correlates with aggressive malignant phenotypes. PLoS One. 2013;8:e63887. doi: 10.1371/journal.pone.0063887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X, Huang Y, Yang D, Li X, Liang J, Lin L, Zhang M, Zhong K, Liang B, Li J. Overexpression of TRIM24 is associated with the onset and progress of human hepatocellular carcinoma. PLoS One. 2014;9:e85462. doi: 10.1371/journal.pone.0085462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Sun L, Tang Z, Fu L, Xu Y, Li Z, Luo W, Qiu X, Wang E. Overexpression of TRIM24 correlates with tumor progression in non-small cell lung cancer. PLoS One. 2012;7:e37657. doi: 10.1371/journal.pone.0037657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Man X, He J, Kong C, Zhu Y, Zhang Z. Clinical significance and biological roles of CARMA3 in human bladder carcinoma. Tumour Biol. 2014;35:4131–4136. doi: 10.1007/s13277-013-1540-2. [DOI] [PubMed] [Google Scholar]
- 21.Pallavi S, Anoop K, Showket H, Alo N, Mausumi B. NFKB1/NFKBIa polymorphisms are associated with the progression of cervical carcinoma in HPV-infected postmenopausal women from rural area. Tumour Biol. 2015;36:6265–6276. doi: 10.1007/s13277-015-3312-7. [DOI] [PubMed] [Google Scholar]
- 22.Nakahara T, Tanaka K, Ohno S, Egawa N, Yugawa T, Kiyono T. Activation of NF-kappaB by human papillomavirus 16 E1 limits E1-dependent viral replication through degradation of E1. J Virol. 2015;89:5040–5059. doi: 10.1128/JVI.00389-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prabhavathy D, Subramanian CK, Karunagaran D. Re-expression of HPV16 E2 in SiHa (human cervical cancer) cells potentiates NF-kappaB activation induced by TNF-alpha concurrently increasing senescence and survival. Biosci Rep. 2015:35. doi: 10.1042/BSR20140160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang AC, Hsu SC, Kuo CL, Liao CL, Lai KC, Lin TP, Wu SH, Lu HF, Tang NY, Yang JS, Chung JG. Involvement of matrix metalloproteinases in the inhibition of cell invasion and migration through the inhibition of NF-[kappa] B by the new synthesized ethyl 2-[N-p-chlorobenzyl-(2’-methyl)] anilino-4-oxo-4,5-dihydrofuran-3-carboxylate (JOTO1007) in human cervical cancer Ca ski cells. In Vivo. 2009;23:613–619. [PubMed] [Google Scholar]
- 25.Xue D, Zhang X, Zhang X, Liu J, Li N, Liu C, Liu Y, Wang P. Clinical significance and biological roles of TRIM24 in human bladder carcinoma. Tumour Biol. 2015;36:6849–6855. doi: 10.1007/s13277-015-3393-3. [DOI] [PubMed] [Google Scholar]


