Abstract
RNA helicase DHX9 is a member of human RNA enzymes. Previous studies have reported that DHX9 is highly expressed in various types of malignant tumor. However, its role in the progression of lung cancer remains to be fully clarified. The present study aims to investigate the oncogenic role of DHX9 in serum, tissues and lung cancer cell lines in vitro. We used RNA interference to downregulate DHX9 expression in A549 cells using a small interfering RNA lentiviral vector. Subsequently, enoxacin was used to inhibit cell proliferation, and this effect was detected using MTT. The results showed that DHX9 was overexpressed in the serum and tissues of lung cancer, especially in small cell lung cancer. Though enoxacin suppressed the proliferation of NSCLC cells, the inhibition effect was diminished when DHX9 was knocked down. In conclusion, the present study provided evidence suggesting that DHX9 was overexpressed in lung cancer and may contribute to the growth of lung cancer, and enoxacin may inhibit the proliferation based on DHX9. Thus DHX9 may be used as a diagnostic marker and a potential therapeutic target for the treatment of NSCLC.
Keywords: DHX9, lung cancer, enoxacin
Introduction
DHX9 (DEAH box protein 9) is also called the RHA (ATP dependent RNA helicase A), LKP (Leukophysin), or NDH II (Nuclear DNA helicase II). DHX spiral enzyme is a branch of spiral human RNA enzymes, consisting of a total of 15 members, mainly involved in the precursor of RNA splicing processing, and the ribosome synthesis, translation and transcription [1]. DHX9 was first discovered in Hela cells. DHX9 gene was located on chromosome 1q25. DHX9 protein has the activity of DNA and RNA unwinding enzyme.
DHX9 is highly expressed in tumor, and is expected to become the new target for cancer treatment [2-6]. In breast cancer, DHX9 blocked the DNA repair function of BRAC-1, resulting in cancer occurrence [7]. Moreover, DHX9 mutations led to an increased incidence of breast cancer [8]. DHX9 was found to regulate the expression of OCT4 through L1TD1-RHA-LIN28 pathway in the embryonic stem cells, and participate in the regulation of stem cells and tumor proliferation [9].
It was found, in prostate cancer, that DHX9 was the downstream genes of SOX4, which could activate the Wnt/β-catenin signaling pathway [10]. SOX4 is a member of the family of SOX, a gene family in stem cells and tumor which plays an important role in the immortalization of cells. The Wnt/β-catenin signaling pathway is closely related to tumor metastasis and resistance [11-13].
DHX9 is closely connected to p53 signaling pathways. In fibroblasts, knocking down DHX9 gene can lead to the aging of fibroblasts. A possible explanation for this mechanism is that DHX9 can inhibit the p53 signaling pathway, while knocking down DHX9 leads to p53 signaling pathway’s activation [14]. DHX9 is also closely connected to NF-κB signaling pathways. As DHX9 is through NF-κB p65 subunit synergy regulates gene transcription [15]. The knocking down of DHX9 by siRNA leads to a reduction of p65, so that of E-selectin, ICAM-1 and IFN-β.
However, studies have found that, in neuroblastoma tumor, suppressor genes KIF 1b is led to tumor inhibition through high expressions of DHX9 in the nucleus; so neuroblastoma was related to the decrease of DHX9 in the nuclei. DHX9 in neuroblastoma may be associated with tumor suppressor [16].
P glycoprotein (P-gp) is one of the ATP combining membrane transport protein family, with functions of energy dependence “pump”. P-gp is one of the important resistance mechanisms of tumor. Intracellular drug can be pumped out extracellular by P-gp. Reducing the intracellular drug concentration makes the cells resistant to chemotherapy drugs. Hence P-gp plays a very important role in cancer drug resistance, especially in acquired drug resistance.
DHX9 can cut down the expression of P-gp. DHX9 is one of the composition of nucleoprotein MEF1. MEF1 combined with MDR1 promoter can raise MDR1 promoter activity, resulting in the over-expression of P glycoprotein [17]. DHX9 gene is found to be over-expressed in cisplatin resistance ovarian cancer cells by cDNA microarray detection [18]. As well, in lymphoma, it was found that patients with Mcl-1 gene had drug resistance to chemotherapy drug AT737. Knocking down DHX9 led lymphoma to become sensitive to AT737, and the p53 gene mediated apoptosis process was activated after silencing DHX9 [19].
There were few studies of DHX9 in lung cancer. Our group identified DHX9 as an over-expressed protein in lung adenocarcinoma by earlier application of iTRAQ combined with mass spectrometry screening. Compared with normal lung tissue, DHX9 was found highly expressed in lung cancer tissue by 1.6 times.
Materials and methods
Participants
Patients (N=64, 22 cases of lung adenocarcinoma, 22 cases of lung squamous carcinoma and 20 cases of small cell lung cancer) were individuals enrolled between 2013 and 2014 with primary lung cancer treated in the department of Respiratory of the second affiliated hospital of Xi’an Jiaotong University (Xi’an, China). The diagnoses were based on clinical and histological examinations of bronchoscopy or lung biopsy. All patients had no history of other cancers and underwent no other prior treatment including chemotherapy or radiotherapy when first diagnosed with lung cancer. 22 controls, age- and gender-matched, were recruited and had no cancer. 22 patients had benign lung diseases including 14 cases of pneumonia, 5 cases of tuberculosis, and 3 cases of lung abscess. All benign lung diseases were also diagnosed based on histology. This study as well as follow-up studies were approved by the Ethics Committee of the second affiliated hospital of Xi’an Jiaotong University; written informed consent had been obtained from all participants.
Tissue and human cancer cells
13 cases of tumor tissues and normal tissues adjacent to carcinoma were collected, including: 6 cases of lung adenocarcinoma, 5 cases of lung squamous carcinoma and 2 cases of small cell lung cancer. These samples served as references for the Western Blot analysis. Additionally, 17 cases of tumor tissues and 7 normal tissues were collected for IHC.
All cell lines (A549, PC9, H446) were obtained from the Chinese Academy of Science. All cell lines were cultured at 37°C in an atmosphere of 5% (v/v) carbon dioxide and in the Roswell Park Memorial Institute medium (RPMI1640).
Lentivirus construction and target screening for RNAi
Four sequences (sites 1, 2, 3, and 4) of the DHX9 gene were selected as the targets for RNA interference (RNAi) (Jiman, Shanghai, China). The sequence of site 1 was 5’-GAAGGATTACTACTCAAGAAACTCGAGTTTCTTGAGTAGTAATCCTTCTTTTTT-3’; the sequence of site 2 was 5’-GGGCTATATCCATCGAAATTTCTCGAGAAATTTCGATGGATATAGCCCTTTTTT-3’; the sequence of site 3 was 5’-ACGACAATGGAAGCGGATATACTCGAGTATATCCGCTTCCATTGTCGTTTTTTT-3’; and the sequence of site 4 was 5’-GTTAAGGAAACCAAGCATATAGCTCGAGCTATATGCTTGGTTTCCTTAATTTTTT-3’. The control non-coding (NC) sequence is 5’-GTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAACTTTTTTACGCGT-3’. Then the pGM-LV-SC5 RNAi vectors containing the DHX9 RNAi sequences were constructed. Co-transduction of the siRNA expression pGMLV-SC5RNAi vectors into the 293 T cells produced alentivirus. Selected lentiviruses containing integrated DHX9 RNAi sequences were used to transfect A549 cells. The PCR method was used to find the most effective gene interference in A549 cells, with non-lentivirus and lentivirus containing NC-shRNA transduction as controls.
Data collection and follow-up
Patient demographic and clinicopathologicdata, tumor location, metastasis and pathological type were collected from medical records at the time of diagnosis. Lung cancer patients received standard therapeutic procedures including neoadjuvant chemotherapy and surgical resection with a wide or radical margin followed by adjuvant chemotherapy.
ELISA for soluble DHX9 in serum
Venous blood samples (~5 ml) were obtained from each participant when first diagnosed with lung cancer. Blood was allowed to coagulate in a serum separator tube. Samples were centrifuged (3,000 × g) for 10 minutes, and serum was transferred into tubes and stored at -80°C until ELISA quantifies soluble DHX9. Serum DHX9 level was measured with a commercial ELISA kit (Elabscience, Wuhan, China) and following the manufacturer’s instructions.
Western blotting
Frozen tumor tissues and adjacent tissues were homogenized on ice with a glass homogenizerand lysed in a RIPA buffer containing protease inhibitor (Sigma, CA, USA) and PMSF. Lysates were sonicated and centrifuged at 13,000 rpm at 4°C for 5 minutes. Then samples were precipitated, and supernatants were collected. A BCA assay kit (Beoytime Biotech, Shanghai, China) was used to measure protein concentration; extracted proteins were subjected to 8% SDS-PAGE, then separated proteins were transferred onto PVDF membranes (Millipore, MA, USA). The PVDF membrane was blocked with TBST (TBS buffer with 0.1% Tween-20) containing 5% skim milk and incubated with human DHX9 antibody (1:500 dilution; Monoclonal Mouse IgG1, Abgent, USA) and GAPDH antibody (1:1000 dilution; Mouse Monoclonal IgG1, Santa Cruz, USA) at room temperature for 2 hours. Subsequently, samples were incubated with HRP-conjugated secondary antibody (1:5000 Dilution, Boster, Wuhan, China). Signals were captured using an HRP Chemiluminescentkit (Boster, Wuhan, China) and CCD camera image system (Bio-Rad, CA, USA).
Immunohistochemistry (IHC)
IHC was performed for 17 cases of lung adenocarcinoma as well as for 7 cases of normal lung tissues. Briefly, 4 μm FFPE tissue sections were deparaffinized in xylene, rehydrated in graded series of ethanol and incubated in a citrate buffer (Dako, Sweden) for 20 minutes at 95-99°C for antigen retrieval. Slides were incubated in 0.5% H2O2 to deactivate endogenous peroxidase. Unspecific binding sites were blocked by 1% bovine serum albumin. Slides were incubated overnight with a primary antibody: monoclonal mouse anti-DHX9 at dilution 1:1,000; A secondary biotinylated antibody was applied, followed by an avidin-biotin-complex (ABC) conjugated to horseradish peroxidase (HRP) according to the manufacturer’s instructions (Vectastain, Vector Burlingame). The result was visualized with diaminobenzidine and counterstained in hematoxylin. Microscopic evaluation of IHC slides was performed using previously described protocols and equipment. A semi-quantitative scoring system was applied to evaluate the intensity of nuclear staining as weak (-), moderate (+), moderate-to-strong (++) or strong (+++). In addition, the proportions of positive tumor cells were estimated for each target protein and presented in percentage.
MTT assay
A549, DHX9-shRNA A549 and NC-shRNA A549 cells were seeded in 96-well plates (5 × 103 cells/well) over-night and then treated with enoxacin (0-128 ug/ml) for 48 hours. 48 hours after the addition of the enoxacin, cell viability was assessed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT Beyotime, Shanghai, China) assay. 4 hours later, dimethylsulphoxide was added into each well. The absorbance at 490 nm (A490) of each well was read on a spectrophotometer. All experiments were performed in quadruplicate.
Statistical analysis
Mann-Whitney U analysis was practiced to determine differences in serum DHX9 level among cancer samples and controls. Non-parametric-received operating characteristic (ROC) curves were generated to assess diagnostic efficiency. All analyses were conducted with SPSS 18.0 (SPSS Inc, Chicago, IL). P<0.05 was considered to be statistically significant.
Results
DHX9 highly expressed in serum of lung cancer patients
Serum DHX9 level was elevated in lung cancer patients compared to controls (4.81±0.36 vs 3.88±0.29, Figure 1 and Table 1). Patients with SCLC had greater serum DHX9 level than patients with NSCLC; meanwhile, patients with lung squamous carcinoma had slightly higher serum DHX9 level than controls, but this was not significant (P=0.36). Serum DHX9 level was not correlated with age or gender (P=0.09, P=0.69; respectively). It varied among patients of different clinical stages: stage III or IV patients had significantly higher DHX9 than those of I or II stages (P<0.05). Similarly, patients with higher smoking index had higher DHX9 than those who never smoked or occasionally smoked (P<0.05, Figure 1C).
Figure 1.
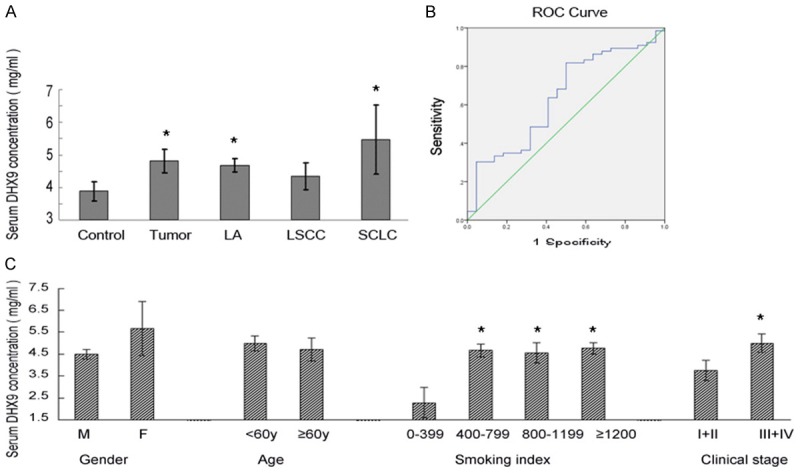
Serum DHX9 level in patients of lung cancer and controls. A. The serum level of DHX9 was highly expressed in tumor patients especially in SCLC patients, *P<0.01. B. ROC analysis for serum DHX9 for distinguishing lung cancer patients from benign lung diseases. C. Stage III or IV patients had significantly higher DHX9 than those with I or II stages (P<0.05). And patients with higher smoking index had higher DHX9 level (P<0.05), *P<0.05.
Table 1.
Patients’ clinical characteristic and serum DHX9 concentration
| Characteristic | N | DHX9 (ng/ml) | P |
|---|---|---|---|
| Gender | |||
| Male | 47 | 4.49±0.22 | |
| Female | 17 | 5.68±1.23 | 0.69 |
| Age (years) | |||
| <60 | 21 | 4.99±0.34 | |
| ≥60 | 43 | 4.72±0.52 | 0.09 |
| Smoking index (Brinkman Index) | |||
| 0-399 | 4 | 2.29±0.70 | |
| 400-799 | 15 | 4.67±0.29 | 0.00** |
| 800-1199 | 18 | 4.56±0.46 | 0.01* |
| ≥1200 | 6 | 4.76±0.27 | 0.01* |
| Clinical stage | |||
| I+II | 10 | 3.76±0.47 | |
| III+IV | 54 | 5.00±0.42 | 0.02* |
P<0.01;
P<0.05.
ROC analysis of serum DHX9 level in lung cancer patients
ROC/AUC analysis revealed the sensitivity and specificity of different serum DHX9 concentrations. ROC curve analysis illustrated that serum DHX9 level was a potential biomarker for screening lung cancer patients from controls (AUC of 0.628; sensitivity 0.803, specificity 0.524, Figure 1B and Table 2).
Table 2.
Serum DHX9 concentration in different pathological and the AUC of ROC curve
| N | DHX9 (ng/ml) | P | AUC | |
|---|---|---|---|---|
| Control | 22 | 3.88±0.29 | ||
| Lung cancer | 64 | 4.81±0.36 | 0.042* | 0.628 |
| Lung adenocarcinoma | 22 | 4.67±0.21 | 0.046* | 0.587 |
| Lung squamous cell carcinoma | 22 | 4.35±0.41 | 0.360 | 0.475 |
| Small cell lung cancer | 20 | 5.46±1.06 | 0.041* | 0.571 |
P<0.05.
DHX9 highly expressed in lung tumor especially in small cell lung cancer
The expression levels of DHX9 were detected in 13 matched clinical tissue samples and 3 lung tumor cell lines by western blotting. The protein expression levels of DHX9 were higher in small cell lung cancer cell line H446 than in the NSCLC lung cell lines A549 and PC9 (Figure 2A). The protein expression levels of DHX9 were increased in the 13 matched adenocarcinoma tissues, as compared to the normal tissues (Figure 2B). In addition, different isoforms of DHX9 exist in the normal tissues and lung tumor tissues. The major band of DHX9 from the normal tissues showed slower mobility than that from tumor tissues (Figure 2B, 2C). DHX9 displayed two isoforms: one of Mr~140,000 in the normal tissues, and one of Mr~128,000 in the tumor tissue or tumor cell lines. We mixed the lysates of normal tissue and H446 cell lines by 1:1, which produced two bands that were similar to those from tumor tissues (Figure 2C). These results suggested that DHX9 protein was probably modified post-translationally in the tumor tissues or cell lines. Immunohistochemistry showed that DHX9 was highly expressed in tumor tissue cells than in normal tissue cells (Figure 2D and Table 3).
Figure 2.
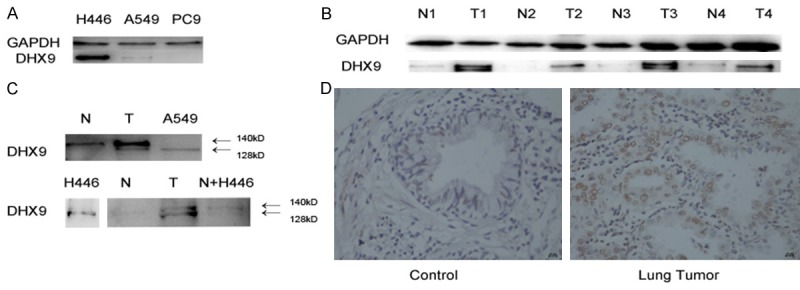
DHX9 level in tissues and cell lines. A. The level of DHX9 was higher in small cell lung cancer cell line H446 than NCSCL cell lines A549 and PC9. B. DHX9 was highly expressed in tumor tissues than compared normal tissues, and there were two bands in tumor tissues. C. DHX9 displayed two isoforms: one isoform of Mr~140,000 in the normal tissues, and one of Mr~128,000 in the tumor tissue or tumor cell lines. In addition, the two bands were similar to those in the tumor tissues when we mixed the lysates of normal tissue and H446 cell lines. D. Immunohistochemistry showed that DHX9 was highly expressed in tumor tissues than in normal controls.
Table 3.
Summary of immunohistochemistry results of lung tumor tissues and control
| IHC of DHX9 | |||||
|---|---|---|---|---|---|
|
| |||||
| - | + | ++ | +++ | P | |
| Control | 4 | 2 | 0 | 1 | |
| Lung Cancer | 1 | 0 | 9 | 7 | 0.005 |
Enoxacin may have suppressed cell proliferation in A549 based on DHX9
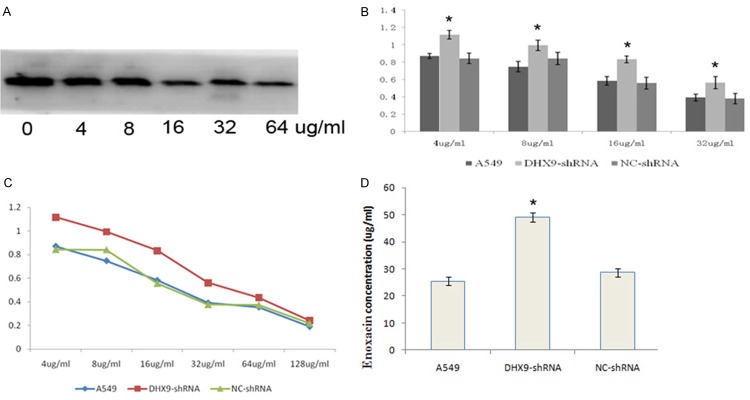
We knocked down the DHX9 in A549 by transfecting the lentivirus expressing DHX9 siRNAs. The DHX9 protein was significantly blocked by the siRNAs, and the silencing effect from the siRNA targeting at site 1 was most significant (Figure 3). DHX9 was decreased as the concentration of enoxacin rose. DHX9 was cut down by enoxacin in a dose-response relationship (Figure 4A). We determined the effect of enoxacin on cell proliferation in A549 by MTT. The results showed that enoxacin significantly inhibited the proliferation of A549 cells (Figure 4B).
Figure 3.
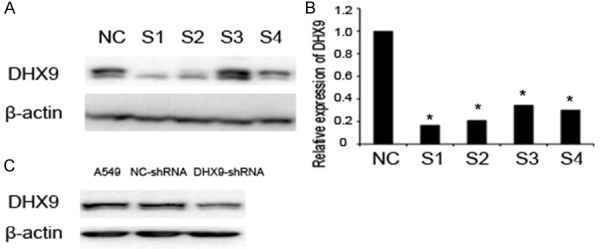
Knockdown of DHX9 in NSCLC cell lines A549. A. Western Blot showed that DHX9 was knocked down by shRNA. B. qPCR showed that the mRNA of DHX9 was knocked down by shRNA. *P<0.01. ShRNA1 was the strongest blocker. C. DHX9 was knocked down by shRNA1.
Figure 4.
DHX9 may be inhibited by enoxacin. A. DHX9 was cut down by enoxacin in a dose-response relationship. B, C. MTT showed that enoxacin significantly inhibited the proliferation of A549 cells. D. The IC50 of enoxacin in DHX9-shRNA-A549 was higher than A549 or NC-shRNA-A549 (IC50: 49.04 ug/ml vs 25.52 ug/ml vs 28.66 ug/ml, P<0.05), *P<0.05.
The effect of enoxacin on cell proliferation in A549 decreased as DHX9 was knocked down. The IC50 of enoxacin in DHX9-shRNA-A549 was higher than A549 or NC-shRNA-A549 (IC50: 49.04 ug/ml vs 25.52 ug/ml vs 28.66 ug/ml, P<0.05) (Figure 4C, 4D).
Discussion
There were few studies of DHX9 in lung cancer. It was discovered that mRNA of DHX9 was over-expressed in lung cancer cell lines and lung cancer tissues by PCR and RT-PCR [20,21]. The tumor suppressor gene p16 can be up-regulated by DHX9 combining with p16 promoter region. P16 and other two Rb tumor suppressor genes, cyclin D and cyclin E, constitutes the Rb pathway. The Rb pathway is common in lung cancer [22]. Wei considered that if there are Rb gene mutations in the lung tumor, the expression of p16 will increase by feedback, and so will DHX9 [21]. In 90% of the patients with small cell lung cancer (SCLC), there are Rb gene mutations. Thus, DHX9 is highly expressed in SCLC than in NSCLC.
In applications with transcriptome sequencing technologies, Sun found 10 mutations (EGFR, TP53, KRAS, RPS6KB2, ATXN2, DHX9, PTPN13, SP1, SPTAN1 and MYOF) in lung adenocarcinoma patients who were non-smoking [23]. 2 out of 27 cases of lung adenocarcinoma patients were found with DHX9 mutations; they also had a poor prognosis. Thus DHX9 can be used as a diagnostic or prognostic marker.
There were two isoforms of DHX9 existent in the normal tissues and lung tumor tissues, but the normal tissues had a higher weight of DHX9. This likely phenomenon has also been found in leukemia cells: the molecular weight of DHX9 in drug-resistant cells was higher than that in the drug-sensitive ones in the SDS-PAGE [24]. These results suggest that some posttranslational modifications of DHX9 occur in cancer or drug-resistant cells. DHX9 may be a therapeutic target in lung cancer.
Quinolones can inhibit the activity of DNA helicases in bacteria. As DHX9 is one of the DNA helicases, we speculated that the quinolones may inhibit the activity of DHX9. DHX9 is an important component of RNA-induced silencing complex (RISC). It had been found that RNA interference could be enhanced by enoxacin: when DHX9 silenced HEK293 cells, the effect of enhancement diminished or disappeared [25]. This suggests that the RNA interference enhancement effect of enoxacin was dependent on DHX9.
Quinolones can inhibit tumor growth and drug resistance. Fluoroquinolone drugs such as enoxacin, norfloxacin, ciprofloxacin and levofloxacin could inhibit the growth of non-small cell lung cancer cell lines H460 [26]. The inhibitory effects were concentration and time dependence. Of all, enoxacin was considered to be the most effective fluoroquinolone, followed by norfloxacin, ciprofloxacin and levofloxacin. In mice leukemia cell line P388/ADR, quinolones strongly increased the intracellular accumulation of adriamycin, while having no effect on P338/ADR without expression of P-gp [27]. Enoxacin could be a candidate drug combined with chemotherapy drugs or other treatments.
Acknowledgements
This study was supported by grants from the Science and Technology Funds of Shaanxi Province (No. 2014KTCL03-02), the Fundamental Research Funds for the Central Universities of China (No. xkjc2015001) and the National Natural Science Foundation of China (No. 81350032). We wish to thank Miaoling Liang for help with the manuscript.
Disclosure of conflict of interest
None.
References
- 1.Abdelhaleem M, Maltais L, Wain H. The human DDX and DHX gene families of putative RNA helicases. Genomics. 2003;81:618–622. doi: 10.1016/s0888-7543(03)00049-1. [DOI] [PubMed] [Google Scholar]
- 2.Lee T, Pelletier J. The biology of DHX9 and its potential as a therapeutic target. Oncotarget. 2016;7:42716–42739. doi: 10.18632/oncotarget.8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mi J, Ray P, Liu J, Kuan CT, Xu J, Hsu D, Sullenger BA, White RR, Clary BM. In vivo selection against human colorectal cancer xenografts identifies an aptamer that targets RNA helicase protein DHX9. Mol Ther Nucleic Acids. 2016;5:e315. doi: 10.1038/mtna.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fidaleo M, Svetoni F, Volpe E, Minana B, Caporossi D, Paronetto MP. Genotoxic stress inhibits Ewing sarcoma cell growth by modulating alternative pre-mRNA processing of the RNA helicase DHX9. Oncotarget. 2015;6:31740–31757. doi: 10.18632/oncotarget.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhattacharya C, Wang X, Becker D. The DEAD/DEAH box helicase, DDX11, is essential for the survival of advanced melanomas. Mol Cancer. 2012;11:82. doi: 10.1186/1476-4598-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee T, Paquet M, Larsson O, Pelletier J. Tumor cell survival dependence on the DHX9 DExH-box helicase. Oncogene. 2016;35:5093–5105. doi: 10.1038/onc.2016.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schlegel BP, Starita LM, Parvin JD. Overexpression of a protein fragment of RNA helicase a causes inhibition of endogenous BRCA1 function and defects in ploidy and cytokinesis in mammary epithelial cells. Oncogene. 2003;22:983–991. doi: 10.1038/sj.onc.1206195. [DOI] [PubMed] [Google Scholar]
- 8.Guenard F, Labrie Y, Ouellette G, Beauparlant CJ, Durocher F, BRCAs I. Genetic sequence variations of BRCA1-interacting genes AURKA, BAP1, BARD1 and DHX9 in French Canadian families with high risk of breast cancer. J Hum Genet. 2009;54:152–161. doi: 10.1038/jhg.2009.6. [DOI] [PubMed] [Google Scholar]
- 9.Narva E, Rahkonen N, Emani MR, Lund R, Pursiheimo JP, Nasti J, Autio R, Rasool O, Denessiouk K, Lahdesmaki H, Rao A, Lahesmaa R. RNA-binding protein L1TD1 interacts with LIN28 via RNA and is required for human embryonic stem cell self-renewal and cancer cell proliferation. Stem Cells. 2012;30:452–460. doi: 10.1002/stem.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai YH, Cheng J, Cheng D, Feasel ME, Beste KD, Peng J, Nusrat A, Moreno CS. SOX4 interacts with plakoglobin in a Wnt3a-dependent manner in prostate cancer cells. BMC Cell Biol. 2011;12:50. doi: 10.1186/1471-2121-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang YG, Luo Y, He DL, Li X, Zhang LL, Peng T, Li MC, Lin YH. Role of Wnt/beta-catenin signaling pathway in epithelial-mesenchymal transition of human prostate cancer induced by hypoxia-inducible factor-1alpha. Int J Urol. 2007;14:1034–1039. doi: 10.1111/j.1442-2042.2007.01866.x. [DOI] [PubMed] [Google Scholar]
- 12.Gupta S, Iljin K, Sara H, Mpindi JP, Mirtti T, Vainio P, Rantala J, Alanen K, Nees M, Kallioniemi O. FZD4 as a mediator of ERG oncogene-induced WNT signaling and epithelial-to-mesenchymal transition in human prostate cancer cells. Cancer Res. 2010;70:6735–6745. doi: 10.1158/0008-5472.CAN-10-0244. [DOI] [PubMed] [Google Scholar]
- 13.Thiago LS, Costa ES, Lopes DV, Otazu IB, Nowill AE, Mendes FA, Portilho DM, Abreu JG, Mermelstein CS, Orfao A, Rossi MI, Borojevic R. The Wnt signaling pathway regulates Nalm-16 b-cell precursor acute lymphoblastic leukemic cell line survival and etoposide resistance. Biomed Pharmacother. 2010;64:63–72. doi: 10.1016/j.biopha.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Lee T, Di Paola D, Malina A, Mills JR, Kreps A, Grosse F, Tang H, Zannis-Hadjopoulos M, Larsson O, Pelletier J. Suppression of the DHX9 helicase induces premature senescence in human diploid fibroblasts in a p53-dependent manner. J Biol Chem. 2014;289:22798–22814. doi: 10.1074/jbc.M114.568535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tetsuka T, Uranishi H, Sanda T, Asamitsu K, Yang JP, Wong-Staal F, Okamoto T. RNA helicase A interacts with nuclear factor kappaB p65 and functions as a transcriptional coactivator. Eur J Biochem. 2004;271:3741–3751. doi: 10.1111/j.1432-1033.2004.04314.x. [DOI] [PubMed] [Google Scholar]
- 16.Chen ZX, Wallis K, Fell SM, Sobrado VR, Hemmer MC, Ramskold D, Hellman U, Sandberg R, Kenchappa RS, Martinson T, Johnsen JI, Kogner P, Schlisio S. RNA helicase A is a downstream mediator of KIF1Bbeta tumor-suppressor function in neuroblastoma. Cancer Discov. 2014;4:434–451. doi: 10.1158/2159-8290.CD-13-0362. [DOI] [PubMed] [Google Scholar]
- 17.Zhong X, Safa AR. Phosphorylation of RNA helicase A by DNA-dependent protein kinase is indispensable for expression of the MDR1 gene product P-glycoprotein in multidrug-resistant human leukemia cells. Biochemistry. 2007;46:5766–5775. doi: 10.1021/bi700063b. [DOI] [PubMed] [Google Scholar]
- 18.Cao MM, Zhang JR, Wang SM, Hu XG, Hu LJ. [Expression of DNA transcription- and repair-related genes in cisplatin-resistant human ovarian carcinoma cell line COC1/DDP] . Di Yi Jun Yi Da Xue Xue Bao. 2005;25:1478–1481. [PubMed] [Google Scholar]
- 19.Mills JR, Malina A, Lee T, Di Paola D, Larsson O, Miething C, Grosse F, Tang H, Zannis-Hadjopoulos M, Lowe SW, Pelletier J. RNAi screening uncovers Dhx9 as a modifier of ABT-737 resistance in an Emu-myc/Bcl-2 mouse model. Blood. 2013;121:3402–3412. doi: 10.1182/blood-2012-06-434365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.An Q, Pacyna-Gengelbach M, Schluns K, Deutschmann N, Guo S, Gao Y, Zhang J, Cheng S, Petersen I. Identification of differentially expressed genes in immortalized human bronchial epithelial cell line as a model for in vitro study of lung carcinogenesis. Int J Cancer. 2003;103:194–204. doi: 10.1002/ijc.10807. [DOI] [PubMed] [Google Scholar]
- 21.Wei X, Pacyna-Gengelbach M, Schluns K, An Q, Gao Y, Cheng S, Petersen I. Analysis of the RNA helicase A gene in human lung cancer. Oncol Rep. 2004;11:253–258. [PubMed] [Google Scholar]
- 22.Myohanen S, Baylin SB. Sequence-specific DNA binding activity of RNA helicase A to the p16INK4a promoter. J Biol Chem. 2001;276:1634–1642. doi: 10.1074/jbc.M004481200. [DOI] [PubMed] [Google Scholar]
- 23.Sun Z, Wang L, Eckloff BW, Deng B, Wang Y, Wampfler JA, Jang J, Wieben ED, Jen J, You M, Yang P. Conserved recurrent gene mutations correlate with pathway deregulation and clinical outcomes of lung adenocarcinoma in never-smokers. BMC Med Genomics. 2014;7:32. doi: 10.1186/1755-8794-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhong X, Safa AR. RNA helicase A in the MEF1 transcription factor complex up-regulates the MDR1 gene in multidrug-resistant cancer cells. J Biol Chem. 2004;279:17134–17141. doi: 10.1074/jbc.M311057200. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Q, Zhang C, Xi Z. Enhancement of RNAi by a small molecule antibiotic enoxacin. Cell Res. 2008;18:1077–1079. doi: 10.1038/cr.2008.287. [DOI] [PubMed] [Google Scholar]
- 26.Mondal ER, Das SK, Mukherjee P. Comparative evaluation of antiproliferative activity and induction of apoptosis by some fluoroquinolones with a human non-small cell lung cancer cell line in culture. Asian Pac J Cancer Prev. 2004;5:196–204. [PubMed] [Google Scholar]
- 27.Zhao YL, Cai SH, Wang L, Kitaichi K, Tatsumi Y, Nadai M, Yoshizumi H, Takagi K, Takagi K, Hasegawa T. Possible involvement of P-glycoprotein in the biliary excretion of grepafloxacin. Clin Exp Pharmacol Physiol. 2002;29:167–172. doi: 10.1046/j.1440-1681.2002.03627.x. [DOI] [PubMed] [Google Scholar]