Abstract
This study was aimed to investigate the functional role of miR-15a in breast cancer cells in response to DNA damage and to illustrate the possible potential underlying molecular mechanism(s). Human breast cancer cell lines MCF-7 cells and/or MDA-MB-231 cells were pre-treated with or without bleomycin. Cells were transfected with corresponding vectors. qRT-PCR was used to detect the expression of mRNA or miRNA, and immunoprecipitation and immunoblot analysis were performed to explore the status of protein association. Cell apoptosis was analyzed with flow cytometry. The results showed that neuronal apoptosis inhibitory protein (NAIP) was negatively regulated by p53 in MCF-7 cells, and NAIP expression was still high in bleomycin-treated MCF-7 cells. In addition, we observed that miR-15a expression was regulated by p53, and the effects of miR-15a on DNA damage was also mediated by p53. Furthermore, the results revealed that the cell apoptosis was mediated by miR-15a. Taken together, this study reveals that p53 negatively regulates NAIP expression by targeting miR-15a processing from primary into precursor miRNA in breast cancer.
Keywords: Breast cancer, neuronal apoptosis inhibitory protein (NAIP), DNA damage, miR-15a, tumor suppressor p53
Introduction
Breast cancer remains to be one of the most common malignancies among females worldwide, which is characterized by a high morbidity [1]. To date, the mechanism of multi-drug resistance (MDR) in breast cancer is complicated [2]. Therefore, this would be of great significance to explore the possible potential underlying molecular mechanism and novel molecular targets for breast cancer.
Neuronal apoptosis inhibitory protein (NAIP) is a protein encoded by NAIP gene in humans. The gene is a part of a 500 kb inverted duplication on chromosome 5q13 [3]. It has been reported that NAIP is abnormally expressed in different cancers and is involved in some significant signaling pathways [4]. Recently, research on NAIP has become one of the hot spots in the anti-tumor therapy method [5,6]. In addition, cell apoptosis is an active and programmed death process that is regulated by the endogenous genes [7]. Cell apoptosis has been well acknowledged to be closely correlated with the progress of breast cancer [7,8]. Previous study reported that NAIP was highly expressed in breast cancer, and it may play an important role in the mechanisms of MDR in tumor cells to various chemotherapeutic agents [9,10].
It has been reported that tumor suppressor p53 expression is altered in a variety of cancers [11,12]. p53 plays pivotal roles in various cell functions including cell cycle arrest, DNA damage, and apoptosis [13,14]. Previous studies have reported that p53 regulates several microRNAs (miRs), such as miR-34 and miR-15 at both transcriptional level and post-transcriptional level [15,16]. The maturation of miRNA consists of two steps, from primary miRNA (pri-miR) into precursor miRNA (pre-miR), and further interacts with DICER to become mature miRNA [17]. Kazuya et al. has reported that abnormal expression of p53 inhibits the Bcell lymphoma-2 (Bcl-2) in response to DNA damage in non-small cell lung carcinoma (NSCLC) cell lines by regulation of the miR-1915 processing [18]. However, little information is available regarding the potential roles of p53 expression in the miR-15a processing in regulating the process of NAIP in response to DNA damage in breast cancer.
Therefore, in the current study, we investigated how p53 regulated anti-apoptotic NAIP in the breast cancer cells. Our study revealed that miR-15a regulated the cell apoptosis in breast cancer cells by negative mediation of NAIP expression, and this process was mediated by tumor suppressor p53 at the post-transcriptional level. Our study might provide a novel mechanism by which p53 negatively modulates NAIP expression by controlling miR-15a processing.
Materials and methods
Cell culture
MCF7 cells and MDA-MB-231 cells were obtained from the Chinese Academy of Sciences’ Type Culture Collection (Shanghai, China). Cells were maintained in high-glucose Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (v/v) fetal bovine serum (FBS; Sigma-Aldrich, St Louis, MO, USA), 100 μg of streptomycin/ml (Sigma-Aldrich), and 100 U of penicillin/ml (Sigma-Aldrich). Cultures were incubated at 37°C in 5% CO2. Small interfering RNA (siRNA) and miRNA inhibitors were transfected by using of Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol.
Immunoprecipitation and immunoblot analysis
Cells were suspended in 1% Nonidet P-40 lysis buffer (50 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% Nonidet P-40, 10 mM NaF, 1 mM Na3VO3, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 10 µg/mL leupeptin, and 10 µg/mL pepstatin), and then were incubated on ice for 30 min. After centrifugation, cell lysates were immunoprecipitated with anti-NAIP or anti-p53 antibodies (Abcam, Cambridge, UK). The immunoprecipitates were washed three times with 0.1% Nonidet P-40 lysis buffer and immunoblot analysis was carried out. Cell lysates or immunoprecipitated proteins were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gels and then were transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA, USA). Thereafter, the membranes were incubated with anti-NAIP, anti-p53, or anti-α-tubulin antibodies. Immunoreactive protein bands were visualized using chemiluminescence (PerkinElmer, Wellesley, MA, USA).
siRNA transfection
Complementary DNA (cDNA) of p53 was amplified by PCR from a human fetal brain cDNA library, and then cloned into the pcDNA3-Flagvector. Gene-specific siRNA for p53 (Qiagen, Hilden, Germany), and control siRNA (Qiagen) were used. Cell transfection was performed using the Liperfection 2000 based on manufacture’s protocol.
Real-time RT-PCR
Total RNAs were isolated from cells with TRIsure (Bioline, London, UK) according to the manufacturer’s protocol. cDNA was synthesized with the PrimeScript 1st strand cDNA Syntheses kit (Takarabio, Shiga, Japan) according to the supplied protocol. Real-time RT-PCR was performed with the KAPA SYBRFAST ABI Prism qPCR kit (Nippon Genetics, Tokyo, Japan) according to manufacturer’s instructions. The following conditions were used: incubation for 10 min at 95°C, denaturation for 15 s at 95°C, annealing for 60 s at 60°C, and extension for 30 s at 72°C.
Cell apoptosis assay
Cells apoptosis was quantified with a flow cytometry using Annexin V-FITC cell apoptosis kit (Invitrogen, USA) according to manufacturer’s protocol. Briefly, after transfection, the cell medium was replaced with serum-free RPMI 1640 medium (Invitrogen, Carlsbad, CA). Cells were harvested, washed 3 times with phosphate buffer saline (PBS) buffer (PH 7.4), and then were resuspended in the staining buffer. Thereafter, 5 μL of annexin-V-FITC and 5 μL of propidium iodide (PI) were mixed with the cells. After incubation at room temperature for 10 min, the mixtures were analyzed using the FACS can flow cytometry system (Scalibur-Becton Dickinson, SanJose, CA). Annexin V-positive and propidium iodide-negative cells were considered as apoptotic cells.
Statistical analysis
All data were expressed as mean ± standard error of mean (SEM). Independent sample t-test was used to calculate the difference between two groups using the graph prism 5.0 software (GraphPad Prism, San Diego, CA). Post-hoc Tukey-test was used to calculate the difference among groups. A value of P<0.05 was considered as statistically significant.
Results
NAIP protein expression was negatively regulated by p53
To analyze the correlation between NAIP expression and p53, MCF-7 cells were treated with or without bleomycin and then were transfected with the si-p53. The expression levels of NAIP were determined by immunoblot analysis. As shown in Figure 1A, the results showed that the expression level of p53 was significantly increased and while NAIP level was statistically decreased in response to bleomycin administration. Interestingly, we found that the effects of bleomycinon the expression of NAIP were reversed by silencing of p53. In addition, the results revealed that the expression of NAIP was upregulated following administration of bleomycin (Figure 1B). These results indicated that p53 negatively regulated NAIP expression in MCF-7 cells.
Figure 1.
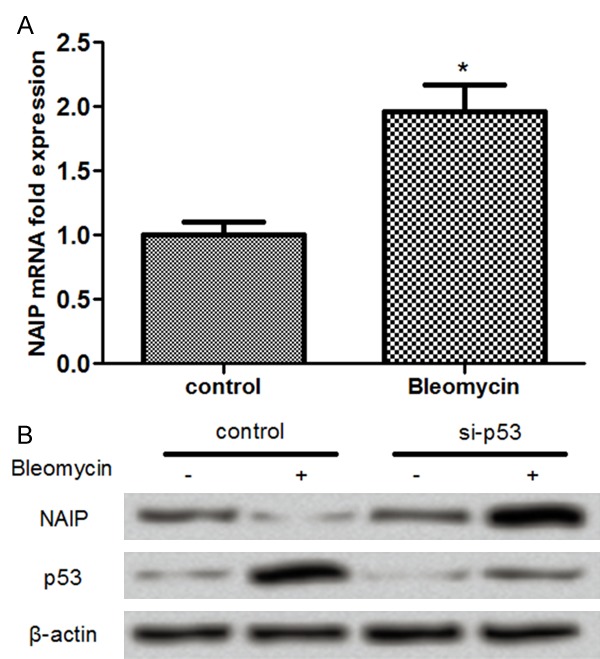
Neuronal apoptosis inhibitory protein (NAIP) was negatively regulated by p53 in MCF-7 cells. A: NAIP expression was increased in MCF-7 cells treated with silenced p53; B: Bleomycin treatment produced high level of NAIP in MCF-7 cells. *: P<0.05 compared to the control.
P53 regulated miR-15a in response to DNA damage
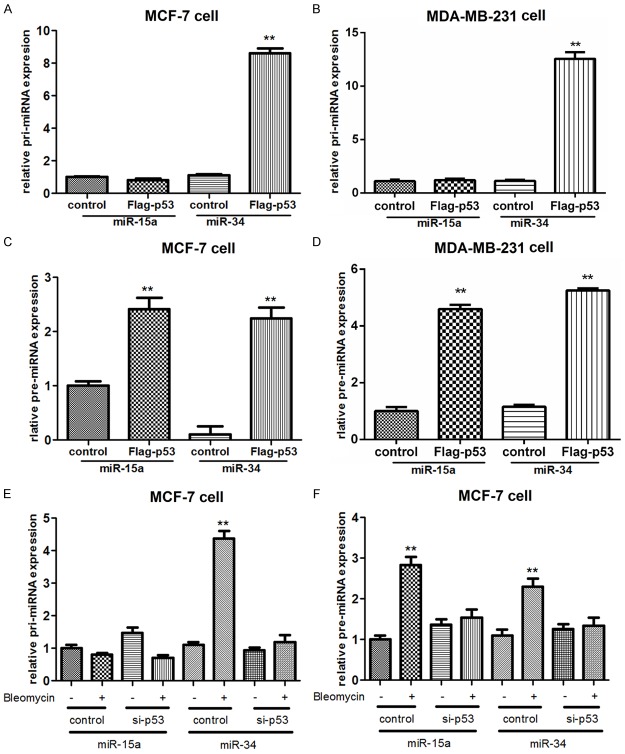
To address whether p53 regulated the expression of NAIP via mediating miR-15a, we first investigated whether miR-15a was a target of p53 in two breast cancer cell lines, MCF-7 cells and MDA-MB-231 cells (Figure 2). The results showed that miR-15a was highly expressed in the two cell lines transfected with the Flag-p53 (both P<0.01) (Figure 2A and 2B), and while NAIP expression was markedly decreased by Flag-p53 (Figure 2C and 2D). In addition, miR-15a expression was significantly decreased in the two kinds of cells treated with silenced p53, but was highly expressed in cells treated with Bleomycin (both P<0.01) (Figure 2E and 2F). These results suggested that the inductive effect of p53 on NAIP expression was mediated by miR-15a in breast cancer cells, namely, p53 controlled the expression of miR-15a in response to bleomycin exposure.
Figure 2.

Expression of miR-15a in two breast cancer cell lines MCF-7 cells and MDA-MB-231 cells treated with Flag-p53 or si-p53. A, B: miR-15a expression was mediated by p53 in the two kinds of breast cancer cells; C, D: NAIP expression was negatively regulated by p-53 in the two kinds of breast cancer cells; E, F: Further results showed that miR-15a expression was decreased by the silenced p53, but was increased in cells treated with bleomycin. **: P<0.01 and #: P<0.05 compared to the control.
The processing of miR-15a was mediated by p53
To further investigate the correlation between p53 and miR-15a processing, we therefore transfected the vectors of Flag-p53 and control vector into two breast cancer cell lines, MCF-7 cells and MDA-MB-231 cells (Figure 3). The above results showed that the expression of miR-15a was regulated by p53, we further analyzed whether p53 regulated miR-15a in a transcriptional dependent manner. Compared with the control group (pri-miR-34 in cells), no significant differences were found in pri-miR-15a expression between Flag-p53 group and control group in both MCF-7 cells and MDA-MB-231 cells (Figure 3A and 3B), but the expression of pre-miR-15a was significantly increased in the Flag-p53 treated MCF-7 cells and MDA-MB-231 cells compared to the control group (both P<0.01) (Figure 3C and 3D). Besides, the expression of pri-miR-15a was not significantly changed in the MCF-7 cells treated with silenced p53 and bleomycin compared to that in the control group (miR-34) (Figure 3E), whereas the pre-miR-15a expression was significantly increased in bleomycin group in the MCF-7 cells (P<0.01; Figure 3F). These results indicated that p53 regulated miR-15a expression in a transcriptional dependent manner.
Figure 3.
miR-15a expression was regulated by p53 in a transcriptional dependent manner in breast cancer cells. A, B: No significant difference was found in the pri-miR-15a expression in cells treated with Flag-p53 compared to the miR-34 group in both MCF-7 cells and MDA-MB-231 cells; C, D: Pre-miR-15a was significantly increased in the Flag-p53 cells compared to the control group in both MCF-7 cells and MDA-MB-231 cells; E: Pri-miR-15a expression was not apparently changed in MCF-7 cells among different groups compared to the miR-34 group; F: Pre-miR-15a expression was significantly increased in the bleomycin group. **: P<0.01 compared to the control.
miR-15a reduced the NAIP expression and induced cell apoptosis
To further investigate the correlation between miR-15a and NAIP and the effects of miR-15a abnormal expression on cell apoptosis, we analyzed the expression of NAIP in miR-15a inhibited cells (Figure 4). After MCF-7 cells were treated with bleomycin, the NAIP protein expression was markedly decreased compared to its control, however, NAIP expression was also notably decreased in the miR-15a inhibited cells compared to its control (Figure 4A), suggesting that inhibition of miR-15a could reduce the NAIP expression. In addition, we analyzed the influence of miR-15a abnormal expression on cell apoptosis in both MCF-7 cells and MDA-MB-231 cells. The data showed that the cell apoptosis was significantly induced bleomycin in bothMCF-7 cells and MDA-MB-231 cells treated with bleomycin (both P<0.01) (Figure 4B and 4C). When the cells were treated with miR-15a inhibitor, the apoptotic cells were also significantly increased compared to its control (both P<0.05). These data revealed that inhibition of miR-15a could induce cell apoptosis in breast cancer cells.
Figure 4.
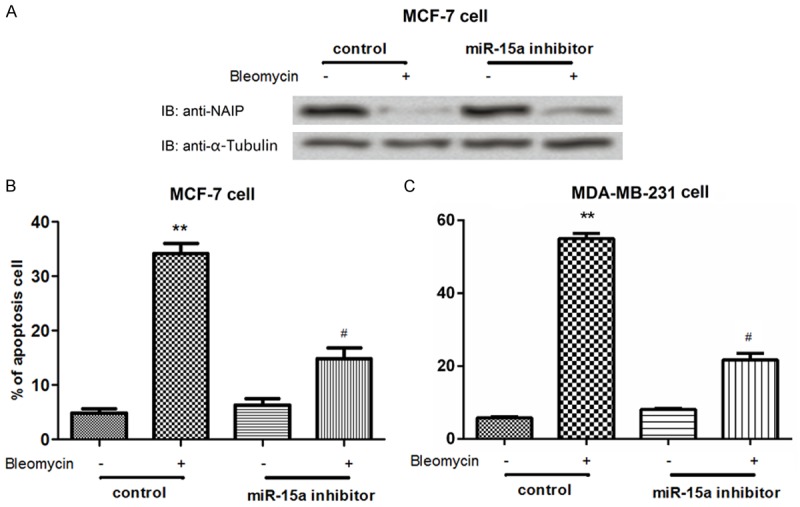
miR-15a expression mediated the expression of NAIP at post-transcriptional level and induced cell apoptosis. A: Protein level of NAIP in miR-15a inhibited MCF-7 cells; B, C: The percentage of apoptotic cells was increased by administration of bleomycin, but was decreased by the inhibited miR-15a in both MCF-7 cells and MDA-MB-231 cells. **: P<0.01 and #: P<0.05 compared the control.
Discussion
Accumulating studies have demonstrated that miRNAs play significant roles in the pathogenesis of breast cancer, including the cell proliferation, apoptosis, and/or metastasis [19,20]. It has been well demonstrated that cell apoptosis is responsible for the progress and development of breast cancer [21,22]. The tumor suppressor p53 was reported to be abnormally down-expressed in breast cancer [23,24]. In this study, we analyzed the significant roles of miR-15a, a cancer-related suppressor protein, in the induction of breast cancer cell apoptosis and tried to illustrate its potential underlying molecular mechanism.
Breast cancer cells were pre-treated with or without bleomycin, an anti-tumor drug that is often used to induce the cell apoptosis [25]. In agreement with a previous study [26], our results showed that the cell apoptosis inhibitor protein NAIP was highly expressed in breast cancer cells treated with the anti-tumor drug bleomycin. Then, the cells were treated with or without bleomycin and silenced p53 to further investigate the effects of administration of bleomycin and silencing of p53 on expression of NAIP. The data showed that NAIP expression was negatively correlated with the p53 expression. These data suggested that the tumor suppressor p53 was down-regulated while NAIP was highly expressed in the bleomycin-treated breast cancer cells, and we therefore speculated that the abnormal expression of NAIP might be correlated with breast cancer.
Subsequently, we tested the expression of miR-15a in breast cancer cells. Previous evidence had shown that miR-15a was abnormally down-expressed in various kinds of cancers, such as lung cancer, prostate cancer and ovarian cancer [27-29]. Recently, lorio et al. has proved that miR-15a is down-regulated in breast cancer cells [30]. However, few studies have been conducted to examine the functional role of miR-15a in the pathogenesis of breast cancer. Our data showed that miR-15a was significantly up-regulated in the p53 overexpressed cells, indicating that miR-15a expression was regulated by p53. Further results showed that miR-15a expression was increased in bleomycin treated cells, but was significantly down-regulated in the bleomycin treated and silenced p53 cells, suggesting that miR-15a expression was regulated by p53 in response to DNA damage caused by bleomycin. Meanwhile, we tested the effects of miR-15a expression on cell apoptosis and NAIP expression. It has been proved that NAIP in bleomycin treated cancer cells was often up-regulated. NAIP is a direct inhibitor of the cell apoptosis executor caspase-3 and caspase-7 [31]. Our results showed that NAIP expression was induced by the silenced miR-15a, suggesting that miR-15a was negatively correlated with NAIP expression in breast cancer. Besides, Cimmino et al. proved that miR-15 could induce cell apoptosis by targeting Bcl-2 in cancers [32], and Bonci et al. demonstrated that miR-15a controlled the apoptosis in prostate cancer [28]. The data presented in our study showed that cell apoptosis was reduced by silenced miR-15a in the bleomycin treated cells, suggesting that miR-15a is a promoter of breast cancer cell apoptosis but was an inhibitor of NAIP expression.
Furthermore, it has been well-known that p53 is involved in various biological processes by regulating the miRNAs at the transcriptional or post-transcriptional level [33]. Kazuya et al. proved that miR-1915 was involved in DNA damage, and this process was mediated by p53 at the post-transcriptional level [18]. Similar evidence was also found in the lung cancer in the study of Rahman et al. [34]. Our study showed that the pri-miR-15a expression was not apparently changed in the Flag-p53 treated cells, but the expression of pre-miR-15a was significantly increased. Based on our results, we speculated that p53 might regulate the miR-15a processing at the post-transcriptional level.
To sum up, the data presented that down-regulation of miR-15a reduced the NAIP expression and induced the cell apoptosis in response to DNA damage in breast cancer cells, and this process was mediated by the tumor suppressor p53 at the post-transcriptional level. This study may provide theoretical basis for illustrating the significant roles of miR-15a in regulating cell apoptosis in breast cancer, and may provide a new insight for the anti-tumor drug application in breast cancer. However, further studies should be performed to confirm the significant roles of miR-15a in breast cancer.
Disclosure of conflict of interest
None.
References
- 1.Autier P, Boniol M, Smans M, Sullivan R, Boyle P. Statistical analyses in Swedish randomised trials on mammography screening and in other randomised trials on cancer screening: a systematic review. J R Soc Med. 2015;108:440–50. doi: 10.1177/0141076815593403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mao Q, Unadkat JD. Role of the breast cancer resistance protein (BCRP/ABCG2) in drug transport-An update. AAPS J. 2015;17:65–82. doi: 10.1208/s12248-014-9668-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ting JP, Lovering RC, Alnemri ES, Bertin J, Boss JM, Davis BK, Flavell RA, Girardin SE, Godzik A, Harton JA, Hoffman HM, Hugot JP, Inohara N, Mackenzie A, Maltais LJ, Nunez G, Ogura Y, Otten LA, Philpott D, Reed JC, Reith W, Schreiber S, Steimle V, Ward PA. The NLR gene family: an official nomenclature. Immunity. 2008;28:285–7. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Almagro M, Vucic D. The inhibitor of apoptosis (IAP) proteins are critical regulators of signaling pathways and targets for anti-cancer therapy. Exp Oncol. 2012;34:200–211. [PubMed] [Google Scholar]
- 5.Saleem M, Qadir MI, Perveen N, Ahmad B, Saleem U, Irshad T, Ahmad B. Inhibitors of apoptotic proteins: new targets for anticancer therapy. Chem Biol Drug Des. 2013;82:243–251. doi: 10.1111/cbdd.12176. [DOI] [PubMed] [Google Scholar]
- 6.Dynek JN, Vucic D. Antagonists of IAP proteins as cancer therapeutics. Cancer Lett. 2013;332:206–214. doi: 10.1016/j.canlet.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Xiao Y, Nimmer P, Sheppard GS, Bruncko M, Hessler P, Lu X, Roberts-Rapp L, Pappano WN, Elmore SW, Souers AJ, Leverson JD, Phillips DC. MCL-1 is a key determinant of breast cancer cell survival: Validation of MCL-1 dependency utilizing a highly selective small molecule inhibitor. Mol Cancer Ther. 2015;14:1837–1847. doi: 10.1158/1535-7163.MCT-14-0928. [DOI] [PubMed] [Google Scholar]
- 8.Merchant S, Aboagye EO, Lim A, Kozlowski K, Patel N, Steel J, Cleator S, Shousha S, Varghese V, Coombes RC. Abstract P5-01-02: Evaluation of apoptosis in breast cancer using the novel PET probe [18F] ICMT-11 in patients treated with neoadjuvant FEC chemotherapy: Initial assessment of optimum imaging time and relation to caspase-3 immunostaining. Cancer Research. 2015;75 P5-01-02-P05-01-02. [Google Scholar]
- 9.Shaw JA, Page K, Blighe K, Hava N, Guttery D, Ward B, Brown J, Ruangpratheep C, Stebbing J, Payne R. Genomic analysis of circulating cell-free DNA infers breast cancer dormancy. Genome Res. 2012;22:220–231. doi: 10.1101/gr.123497.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi J, Hwang YK, Choi YJ, Yoo KE, Kim JH, Nam SJ, Yang JH, Lee SJ, Yoo KH, Sung KW, Koo HH, Im YH. Neuronal apoptosis inhibitory protein is overexpressed in patients with unfavorable prognostic factors in breast cancer. J Korean Med Sci. 2007;22(Suppl):S17–23. doi: 10.3346/jkms.2007.22.S.S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Song H, Hollstein M, Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nature Cell Biol. 2007;9:573–580. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- 12.Muller PA, Vousden KH. p53 mutations in cancer. Nature Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 13.Nicol SM, Bray SE, Black HD, Lorimore SA, Wright EG, Lane DP, Meek DW, Coates PJ, Fuller-Pace FV. The RNA helicase p68 (DDX5) is selectively required for the induction of p53-dependent p21 expression and cell-cycle arrest after DNA damage. Oncogene. 2013;32:3461–3469. doi: 10.1038/onc.2012.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, Baer R, Gu W. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell. 2012;149:1269–1283. doi: 10.1016/j.cell.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rokavec M, Li H, Jiang L, Hermeking H. The p53/miR-34 axis in development and disease. J Mol Cell Biol. 2014;6:214–230. doi: 10.1093/jmcb/mju003. [DOI] [PubMed] [Google Scholar]
- 16.Shi L, Jackstadt R, Siemens H, Li H, Kirchner T, Hermeking H. p53-induced miR-15a/16-1 and AP4 form a double-negative feedback loop to regulate epithelial-mesenchymal transition and metastasis in colorectal cancer. Cancer Res. 2014;74:532–542. doi: 10.1158/0008-5472.CAN-13-2203. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki HI, Miyazono K. Emerging complexity of microRNA generation cascades. J Biochem. 2011;149:15–25. doi: 10.1093/jb/mvq113. [DOI] [PubMed] [Google Scholar]
- 18.Nakazawa K, Dashzeveg N, Yoshida K. Tumor suppressor p53 induces miR-1915 processing to inhibit Bcl-2 in the apoptotic response to DNA damage. FEBS J. 2014;281:2937–2944. doi: 10.1111/febs.12831. [DOI] [PubMed] [Google Scholar]
- 19.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 20.Song SJ, Poliseno L, Song MS, Ala U, Webster K, Ng C, Beringer G, Brikbak NJ, Yuan X, Cantley LC. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013;154:311–324. doi: 10.1016/j.cell.2013.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu J, Du W, Yan F, Wang Y, Li H, Cao S, Yu W, Shen C, Liu J, Ren X. Myeloid-derived suppressor cells suppress antitumor immune responses through IDO expression and correlate with lymph node metastasis in patients with breast cancer. J Immunol. 2013;190:3783–3797. doi: 10.4049/jimmunol.1201449. [DOI] [PubMed] [Google Scholar]
- 22.Brosseau C, Pirianov G, Colston K. Role of insulin-like growth factor binding protein-3 in 1, 25-dihydroxyvitamin-D3-induced breast cancer cell apoptosis. Int J Cell Biol. 2013;2013:960378. doi: 10.1155/2013/960378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosoda M, Yamamoto M, Nakano K, Hatanaka KC, Takakuwa E, Hatanaka Y, Matsuno Y, Yamashita H. Differential expression of progesterone receptor, FOXA1, GATA3, and p53 between pre-and postmenopausal women with estrogen receptor-positive breast cancer. Breast Cancer Res Treat. 2014;144:249–261. doi: 10.1007/s10549-014-2867-0. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto M, Hosoda M, Nakano K, Jia S, Hatanaka KC, Takakuwa E, Hatanaka Y, Matsuno Y, Yamashita H. p53 accumulation is a strong predictor of recurrence in estrogen receptor-positive breast cancer patients treated with aromatase inhibitors. Cancer Sci. 2014;105:81–88. doi: 10.1111/cas.12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ozben T. Oxidative stress and apoptosis: impact on cancer therapy. J Pharm Sci. 2007;96:2181–2196. doi: 10.1002/jps.20874. [DOI] [PubMed] [Google Scholar]
- 26.Hunter AM, LaCasse EC, Korneluk RG. The inhibitors of apoptosis (IAPs) as cancer targets. Apoptosis. 2007;12:1543–1568. doi: 10.1007/s10495-007-0087-3. [DOI] [PubMed] [Google Scholar]
- 27.Bhattacharya R, Nicoloso M, Arvizo R, Wang E, Cortez A, Rossi S, Calin GA, Mukherjee P. MiR-15a and MiR-16 control Bmi-1 expression in ovarian cancer. Cancer Res. 2009;69:9090–9095. doi: 10.1158/0008-5472.CAN-09-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bonci D, Coppola V, Musumeci M, Addario A, Giuffrida R, Memeo L, D’Urso L, Pagliuca A, Biffoni M, Labbaye C. The miR-15a-miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat Med. 2008;14:1271–1277. doi: 10.1038/nm.1880. [DOI] [PubMed] [Google Scholar]
- 29.Aqeilan R, Calin GA, Croce CM. miR-15a and miR-16-1 in cancer: discovery, function and future perspectives. Cell Death Differ. 2010;17:215–220. doi: 10.1038/cdd.2009.69. [DOI] [PubMed] [Google Scholar]
- 30.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Ménard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM. MicroRNA gene expression deregulation in human breast cancer. Cancer Research. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 31.Maier JK, Lahoua Z, Gendron NH, Fetni R, Johnston A, Davoodi J, Rasper D, Roy S, Slack RS, Nicholson DW. The neuronal apoptosis inhibitory protein is a direct inhibitor of caspases 3 and 7. J Neurosci. 2002;22:2035–2043. doi: 10.1523/JNEUROSCI.22-06-02035.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng Y, Yao Z, Klionsky DJ. How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol. 2015;25:354–363. doi: 10.1016/j.tcb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rahman M, Lovat F, Romano G, Calore F, Acunzo M, Bell EH, Nana-Sinkam P. miR-15b/16-2 regulates factors that promote p53 phosphorylation and augments the DNA damage response following radiation in the lung. J Biol Chem. 2014;289:26406–26416. doi: 10.1074/jbc.M114.573592. [DOI] [PMC free article] [PubMed] [Google Scholar]