Abstract
Tetraspanin 12 (TSPAN12), as an earliest member of tetraspanin family, has been recently shown to be highly expressed in several malignant tumors, such as lung cancer and breast cancer, which plays an important role in regulating cell proliferation, migration and invasion. However, the functional roles of TSPAN12 in colorectal cancer (CRC) remain largely unclear. In this study, the expression of TSPAN12 was up-regulated compared to that in paracarcinoma tissues. Higher TSPAN12 expression was significantly correlated with TNM stage, tumor size and lymph node metastasis. The vitro functional analysis, including MTT, colony formation, flow cytometry and transwell assays indicated that lentivirus-mediated TSPAN12 knockdown significantly suppressed cell proliferation, migration and invasion, induced cell apoptosis of CRC cells. In addition, knockdown of TSPAN12 remarkably decreased the growth of subcutaneously inoculated tumors in nude mice. Our findings for the first time supported that TSPAN12 might play a positive role in the regulation of CRC cell proliferation, migration and invasion. The inhibition of TSPAN12 may serve as a novel promising therapeutic strategy against human CRC.
Keywords: Colorectal cancer, TSPAN12, shRNA, cell proliferation, invasion
Introduction
Colorectal cancer (CRC) is one of the most leading causes of malignancy-related death worldwide with rapidly increasing incidence in recent years [1,2]. Despite some significant improvements have been achieved in some diagnosis and treatment modalities, the long-term survival rate remains poor for patients in advanced stage, mainly ascribed to local recurrence and distant metastases formation [3,4]. Current studies have demonstrated that a series of complex cascades of molecular mechanisms, including gene mutations, epigenetic modifications and signaling crosstalk [5,6] may affect the treatment outcome in patients with CRC. Therefore, understanding the molecular mechanisms underlying the formation and progression of CRC might contribute to identify novel potential targets for CRC diagnosis and provide more effective methods for CRC prevention and therapy.
Tetraspanins, composed of short N-terminal, C-terminal cytoplasmic tails, small and large extracellular loops and four transmembrane domains, belong to a family of transmembrane proteins [7-9], which are abundantly expressed in nearly all cell and tissue types [10,11] and implicated in various physiological processes, such as cell migration, adhesion and signal-modulation of membrane receptors [7,12]. Recently, emerging evidences have shown that tetraspanins are closely correlated with tumour stage and patients outcome by regulating cell growth, invasion, tumour engraftment and metastasis [13]. As one member of the earliest tetraspanin family, Tetraspanin 12 (TSPAN12) is widely distributed in digestive, urogenital and cardiovascular systems [14]. Interestingly, TSPAN12 has been recently reported to be a p53-regulated gene and its knockdown inhibited cancer cell proliferation and invasion, thereby serving as a potential target for lung cancer therapy [15]. In addition, TSPAN12 has been shown to promote human breast cancer proliferation and tumor xenograft growth, but suppress tumor metastasis [16]. Furthermore, TSPAN12 ablation could cause diminish expressions of multiples SPANX family genes found on various tumors cell types [17]. Based on these evidences, TSPAN12 has been suggested to play a crucial role in cancer progression. However, little is known about the clinical significance and biological function of TSPAN12 in human CRC.
Therefore, the objective of this study was to investigate the exact role of TSPAN12 in CRC by analyzing the correlation between the expression of TSPAN12 and clinicopathological features of patients with CRC. Furthermore, we aimed to investigate its biological function in vitro and vivo CRC cell model based on RNAi-mediated TSPAN12 gene knockdown, as well as the underlying mechanism, in an attempt to gain novel insights into tumorigenesis of CRC.
Materials and methods
Patients and specimens
Total 112 fresh tumor tissues with paired non-cancerous colon tissue were obtained in operation from the Shanghai Changzheng Hospital. All the patients with CRC were histologically and clinically diagnosed and voluntarily signed written informed consents. None of these patients had received radiotherapy or chemotherapy prior to surgical treatment. The clinical data of the CRC patients, including age, tumor size and stage were collected and summarized in Table 1. This study was approved by the Committee on Ethics of Shanghai Changzheng Hospital, China.
Table 1.
Correlations between TSPAN12 expression and clinicopathologic parameters in patients with CRC using qRT-PCR analysis (n = 112)
| Clinical pathologic parameters | Total (n = 112) | Expression of TSPAN12 | P Value (chi-square test) | |
|---|---|---|---|---|
|
| ||||
| Low (n = 66) | High (n = 46) | |||
| Sex | 0.056 | |||
| Male | 66 | 34 | 32 | |
| Female | 46 | 32 | 14 | |
| Age | 0.059 | |||
| < 60 | 49 | 24 | 25 | |
| ≥ 60 | 63 | 42 | 21 | |
| Tumor size (cm) | 0.013* | |||
| < 5 | 78 | 40 | 38 | |
| ≥ 5 | 34 | 26 | 8 | |
| Tumor location | 0.119 | |||
| Rectum | 36 | 25 | 11 | |
| Colon | 76 | 41 | 35 | |
| TNM stage | ||||
| I-II | 81 | 56 | 25 | 0.001** |
| III-IV | 31 | 10 | 21 | |
| Lymph node metastasis | 0.024* | |||
| N0 | 75 | 42 | 33 | |
| N1-N2 | 37 | 24 | 13 | |
For the correlation analysis between tumor size and TSPAN12 expression;
Expression of TSPAN12 was significantly correlated with tumor size. P less than 0.05.
For the correlation analysis between TNM stage and TSPAN12 expression;
Expression of TSPAN12 was significantly correlated with TNM stage. P less than 0.01.
For the correlation analysis between lymph node metastasis and TSPAN12 expression;
Expression of TSPAN12 was significantly correlated with lymph node metastasis. P less than 0.05.
Cell lines and culture
The CRC cell lines HT29, SW1116, RKO, DLD-1 and HCT116, as well as human embryonic kidney cell line (HEK293) were obtained from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). HT29, DLD-1, RKO and HCT116 cells were cultured in RPMI-1640 medium (Hyclone, Biowest) with 10% fetal bovine serum (FBS). SW1116 and HEK293 cells were maintained in DMEM medium (Hyclone, Logan, USA) supplemented with 10% FBS. All these cells were incubated in a humidified atmosphere containing 5% CO2 at 37°C.
Real-time quantitative PCR (RT-qPCR) analysis
Total RNA from human tissues and cells was extracted using Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Single strand cDNA was synthesized from 2 μg total RNA using Superscript II Reverse Transcriptase (Invitrogen, Carlsbad, CA). RT-qPCR was carried out using BioRad Connet Real-Time PCR platform using 20 µl PCR reaction mixtures according to the following cycling conditions: an initial denaturation at 95°C for 1 min, followed by 40 cycles of denaturation at 95°C for 10 s and extension at 60°C for 20 s. The primer sequences were as follows: TSPAN12 (forward): 5’-CCTCTAGCCAGCCAACCTTCC-3’, TSPAN12 (reverse): 5’-GACCCTCCACCCTCCCTCAG-3’, GAPDH (forward): 5’-TGTTGCCATCAATGACCCCTT-3’ and GAPDH (reverse): 5’-CTCCACGACGTACTCAGCG-3’. GAPDH was used as an internal control. All reactions were performed in triplicate. The relative TSPAN12 levels between samples were calculated and statistically compared using 2-ΔΔCt method as previously described [18].
Western blot analysis
The tissue samples and cells were lysed in ice-cold RIPA butter (Beyotime, Shanghai, China) at 4°C for 30 min and then underwent centrifugation at 12000 g for 15 min at 4°C. Subsequently, equal amounts of protein lysates were separated by SDS-PAGE and then transferred onto PVDF membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 1% bovine serum albumin in TBST for 2 h at room temperature and then probed with TSPAN12, Akt, Erk, phospho-Akt, phospho-Erk1/2, caspase-3, E-cadherin, Vimentin (Cell signaling, Danvers, USA) and GAPDH (Proteintech, Chicago, USA) antibodies overnight at 4°C. After washing, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA) for 2 h at room temperature. Finally, the membranes were detected with enhanced chemiluminescence kit (Millipore, Billerica, MA) according to the manufacturer’s instructions. GAPDH was used as the loading control.
Vector construction and transfection
For TSPAN12 silencing, two short hairpin RNAs (shRNAs) for human TSPAN12 (shT-SPAN12-1: 5’-GTTCCGAAGTGTTAACCAGTTTGCAAACTCGGGTTAACACTTCGGAACTTTTTT-3’ and shTSPAN12-2: 5’-CTAGCCCGGGCCGGGACTGTATTCAGTATACTCGAGTATACTGAATACAGTCCCGGCTTTTTTGTTAAT-3’) were synthesized, annealed and validated to be candidate shRNAs. A scrambled shRNA (5’-TTCTCCGAACGTGTCACGT-3’) was used as a negative control. Then stable TSPAN12 shRNA silencing was performed on CRC cell lines HT29 and SW1116 as previously described [19]. Briefly, recombined vector and packaging plasmid were transfected into HEK293T cells to generate lentivirus particles via Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). These generated viral particles were then used to transfect HT29 and SW1116 cells. For evaluation of knockdown efficiency, RT-qPCR and Western blot analysis were performed in transfected cells.
Colony formation assay
To investigate the effects of TSPAN12 on cell growth, a monolayer colony formation assay was performed on HT29 and SW1116 cells. Briefly, lentivirus-infected cells (500 cells/well) were seeded into 6-well plates and washed with PBS after 8 days of incubation. Then cells were fixed with 4% crystal violet, washed with ddH2O and air-dried. The number of colonies with more than 50 cells from different treatments was counted under a light microscope.
MTT assay
MTT assay was used to determine cell viability in HT29 and SW1116 cells. Briefly, cells (2,000 cells per well) were seeded into 96-well plates in triplicates and monitored for 5 consecutive days. Then 20 μL of the MTT solution (5 mg/ml) was added into each well and incubated for 4 h at 37°C. After 4 h incubation, 100 µl acidic isopropanol was added into each well. The OD value in each well was measured with an ELISA reader (Bio-Rad, USA) at a wavelength of 595 nm.
Cell migration and invasion assay
Cell invasion and migration assays were performed on HT29 and SW1116 cells using24-well Transwell cell culture chambers (Corning Incorporated, New York, USA) attached with a membrane filter (8 μm pore size). Approximately 200 μL cell suspension was incubated in the upper chamber either uncoated (for migration assay) or coated (for invasion assay) with 50% Matrigel (BD Biosciences, Bedford, MA). After 24 h incubation, the cells that invaded the Matrigel matrix were fixed with methanol and stained with 0.5% crystal violet. The number of cells that penetrated the membrane was quantified by calculating the mean cell number in five randomly fields per chamber under an inverted light microscope (Nikon, Japan).
Flow cytometry analysis
Flow cytometry combined with Annexin V/7-AAD double stainingwas used to determined cell apoptosis in HT29 and SW1116 following TSPAN12 silencing according to the manufacturer’s instructions. In brief, lentivirus infected HT29 and SW1116 cells were reseeded in 6 cm dishes both at a density of 1 × 105 cells per dish. Then cells were collected and subjected to Annexin V-APC/7-AAD double staining (KeyGEN Biotech, Nanjing, China). Annexin V-APC/7-AAD analysis was performed by FACSCalibur flow cytometer (BD, New Iersey, USA).
Tumorigenesis in nude mice
Total twelve six-week-old nude mice (Shanghai Slac Laboratory Animal Co. Ltd, China) were randomized into two groups with three mice in each group. Equivalent amounts of shTSPAN12-1-infected or shCon-infected CRC cells were prepared and then injected subcutaneously into the mice using a 1 ml needle, respectively. The diameter and size of solid tumor was measured at 7, 14, 21 and 28 days, respectively after inoculation. Then animals were sacrificed 28 days after inoculation and the weight of the tumors was measured and compared among two groups (scramble vs. shTSPAN12-1). This experiment was approved by the ethics committee of Wuhan University.
Statistical analysis
All data were analyzed using SPSS version 19.0 (Chicago, IL) and expressed as mean ± standard deviation (SD) of three independent experiments. All diagrams were drafted by GraphPad Prism 5 software. The Chi-square test was used to analyze TSPAN12 expression and patient characteristics of CRC tissue. Student’s t-test was used to statistically compare the variables of two groups (scramble vs. shTSPAN12-1 or shTSPAN12-2). All statistical differences were accepted as statistically significant at P-value < 0.05.
Results
Correlation between TSPAN12 expression and clinicopathologic parameters of patients with CRC
To investigate whether TSPAN12 is correlated with CRC development and progression, the CRC specimens from patients were analyzed for TSPAN12 expression using RT-qPCR analysis. The results showed the expression level of TSPAN12 was significantly elevated in CRC tissues compared with the adjacent normal tissues (Figure 1A, P < 0.001). To validate this result, Western blotting was performed in representative 8 CRC tissues with paired non-cancerous tissues. As shown in Figure 1B, clearly increased levels of TSPAN12 protein were detected in randomly selected tumor tissues compared with the adjacent normal tissues.
Figure 1.
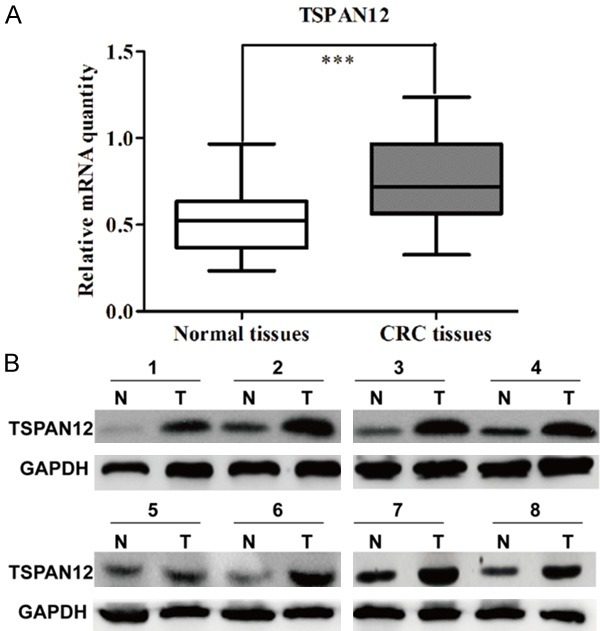
Expression of TSPAN12 was upregulated in CRC tissues. A. The mRNA levels of TSPAN12 in 112 paired CRC tissues by real-time quantitative PCR analysis. B. Western blotting analysis of TSPAN12 protein levels in 8 matched CRC tumor (T) and non-tumor tissues (N). GAPDH was used as internal control. ***P < 0.001.
Subsequently, the median expression level of tissue TSPAN12 (0.72 fold) was used as the cutoff value to divide the 112 CRC patients into two groups (high and low TSPAN12 expression). The association between TSPAN12 expression and the clinicopathologic parameters was shown in Table 1. The TSPAN12 expression was significantly associated with tumor size (P = 0.013), TNM stage (P = 0.001) and lymph node metastasis (P = 0.024). However, no significant association was observed between TSPAN12 expression and patient age (P = 0.056), sex (P = 0.059) and tumor location (P = 0.119). These findings suggest TSPAN12 might play an essential role in CRC cell growth and metastasis.
Lentivirus-mediated efficient TSPAN12 silencing in CRC cells
To further investigate the role of TSPAN12 in CRC at cellular levels, we determined the protein level of TSPAN12 in several CRC cell lines and found it was commonly upregulated in these cells (Figure 2A). Notably, two CRC cell lines, HT29 and SW1116 with relatively higher TSPAN12 expression were chosen to investigate in the following loss-of-function experiments. The expression of TSPAN12 was first knocked down in HT29 and SW1116 cells using lentivirus-mediated shRNA, as confirmed by RT-qPCR and Western blot analysis. As shown in Figure 2B and 2D, the mRNA levels of TSPAN12 in shTSPAN12-1 and shTSPAN12-2 groups of the two cell lines were remarkably reduced as compared with those in scramble groups (P < 0.001). Consistently, the protein levels of TSPAN12 were also obviously downregulated in shTSPAN12-1 and shTSPAN12-2 groups of HT29 (Figure 2C) and SW1116 cells (Figure 2D) as compared with those in scramble groups. Collectively, the endogenous TSPAN12 level in HT29 and SW1116 was stably knocked down by two designed shRNAs. Notably, shTSPAN12-1 had a more obvious knockdown efficiency than shTSPAN12-2 in both cell lines, which was then selected for the subsequent loss-of-function assays.
Figure 2.
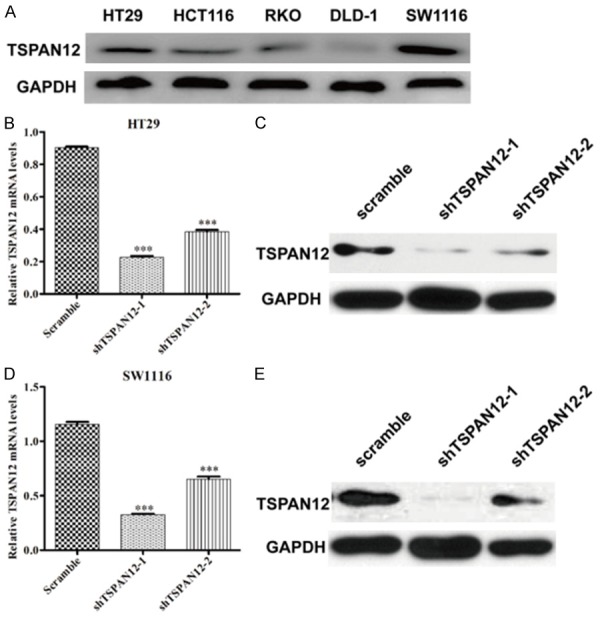
TSPAN12 expression was effectively silenced in CRC cells. (A) Expression analysis of TSPAN12 in five different CRC cell lines by Western blotting. The mRNA levels analysis of TSPAN12 in HT29 (B) and SW1116 (D) cells after shTSPAN12-1 or shTSPAN12-2 infection by RT-qPCR analysis. The protein levels analysis of TSPAN12 in HT29 (C) and SW1116 (E) cells after shTSPAN12-1 or shTSPAN12-2 infection by Western blot analysis. GAPDH was used as internal control. ***P < 0.001 in comparison with non-silencing scrambled control.
Loss-of-function analysis of TSPAN12 by shRNA in CRC cells
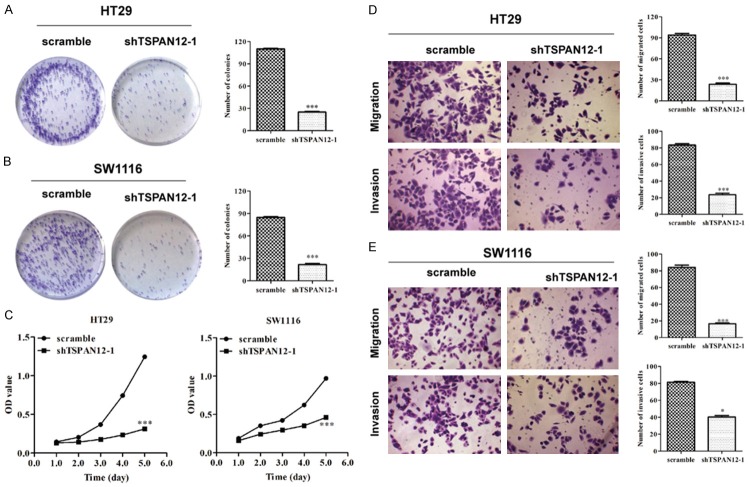
To gain a better insight into the effect of TSPAN12 on cell proliferation, colonyformation and MTT assay were performed on HT29 and SW1116 cells. As shown in Figure 3A and 3B, the smaller and fewer colonies were observed in shTSPAN12-1 groups than in scramble groups of two cell lines (P < 0.001). Moreover, the proliferation rates of shTSPAN12-1-infected cells were relatively slower compared with scrambled groups at the 4th and 5th days in HT29 and SW1116 cells (Figure 3C, P < 0.001). These results indicated that TSPAN12 may play a positive role in CRC cell growth and proliferation.
Figure 3.
Loss-of-function analysis of TSPAN12 in CRC cells, including proliferation and invasion. The number of colonies in HT29 (A) and SW1116 (B) cells after shTSPAN12-1 infection analyzed by colony formation assay. (C) MTT assay was used to determine the proliferative rates of HT29 and SW1116 cells after shTSPAN12-1 infection. Cell migration and invasion assays were conducted on HT29 (D) and SW1116 (E) cells after shTSPAN12-1 infection, as confirmed by statistical analysis. *P < 0.05; ***P < 0.001 in comparison with non-silencing scrambled control.
As our best knowledge, the ability of cancer cells to invade from the primary site into tissues at the site of tumor metastasis is a crucial step in the formation of metastases. Therefore, we determined the migratory and invasive capacity of shTSPAN12-1 in HT29 and SW1116 cells using a matrigel-based transwell. The data indicated that knockdown of TSPAN12 in HT29 (Figure 3D, P < 0.001) and SW1116 (Figure 3E, P < 0.001 and P < 0.05) significantly reduced the migration and invasion potential compared to negative scramble control. These findings suggest that TSPAN12 indeed facilitates CRC cell migration and invasion in vitro.
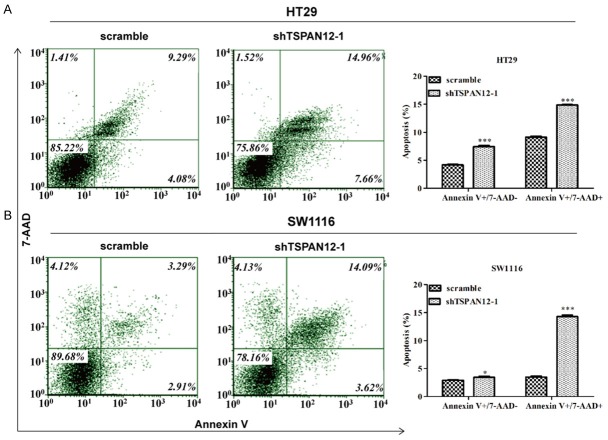
To uncover the mechanism underlying the inhibition of cell proliferation, cell apoptotic rate was detected in HT29 and SW1116 cells following TSPAN12 knockdown. As illustrated in Figure 4A, more HT29 cells were presented early apoptosis (Annexin V+/7-AAD-) and late apoptosis (Annexin V+/7-AAD+) status (P < 0.001) after shTSPAN12-1 infection compared to scrambled shRNA infection. Similar results were also observed in SW1116 cells treated with shTSPAN12-1 (Figure 4B, P < 0.05 and P < 0.001). These results further demonstrated that TSPAN12 knockdown could significantly promote CRC cell apoptosis.
Figure 4.
Knockdown of TSPAN12 significantly promoted cell early and late apoptosis in CRC cells. Flow cytometer combined with Annexin V/7-AAD double staining was used to analyze the percentage of apoptotic cells in HT29 (A) and SW1116 (B) cells following TSPAN12 silencing, as confirmed by statistical analysis. *P < 0.05; ***P < 0.001 in comparison with non-silencing scrambled control.
Knockdown of TSPAN12 impaired tumorigenicity in vivo
To further investigate the role of TSPAN12 in CRC progression, we assessed the effect of TSPAN12 silencing on the growth of CRC xenograft tumors in nude mouse models injected with shTSPAN-1 or scrambled negative control cells. As depicted in Figure 5A and 5D, the size of the tumors was significantly decreased after TSPAN12 knockdown in a time course of 28 days. Moreover, the time-dependent analysis showed that the volume of the tumors was significantly inhibited in mice injected with TSPAN12 silenced HT29 (Figure 5B, P < 0.001) or SW1116 cells (Figure 5E, P < 0.001) compared with the corresponding control cells in a time-dependent manner. Furthermore, the weight of tumors also remarkably decreased in mice inoculated with TSPAN-silenced CRC cells compared with control groups (Figure 5C and 5F). These results were confirmed by analysis of the protein content of TSPAN12 in tissues of a subcutaneous xenograft murine model. The TSPAN12 expression level was completely suppressed in tumor tissues by the infection with shTSPAN12-1 (Figure 5G and 5H). These findings above further demonstrated TSPAN12 silencing could effectively suppress CRC tumorigenesis in vivo.
Figure 5.
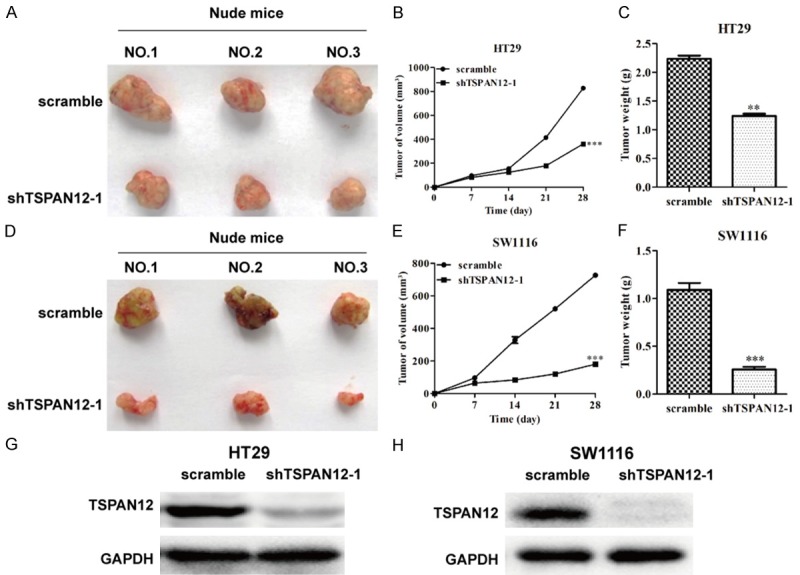
TSPAN12 silencing reduced tumorigenesis of CRC in vivo. (A and D) Representative images of tumors formed in the mice in which shTSPAN12-1 or scrambled control shRNA-infected HT29 and SW1116 cells were injected. Tumor volume altered curve of nude mice injected by HT29 (B) and SW1116 (E) cells transfected with shTSPAN12-1 or scrambled shRNA. Tumor weight alteration of nude mice injected by HT29 (C) and SW1116 (F) cells transfected with shTSPAN12-1 or scrambled shRNA. Expression analysis of TSPAN12 in tumor tissues collected from nude mice injected by HT29/shTSPAN12-1 (G), SW1116/shTSPAN12-1 (H), respectively. **P < 0.01; ***P < 0.001 in comparison with non-silencing scrambled control.
Mechanism study of TSPAN12 silencing was performed in CRC cells
These in vitro and vivo experiments on the role of TSPAN12 in CRC have shown that TSPAN12 play a positive function in CRC proliferation and metastasis. However, the regulatory mechanism of TSPAN12 in the tumorigenesis of CRC remains unclear. Therefore, we detected the expression alteration of some cell proliferation and invasion markers using Western blot analysis. The results indicated that protein levels of p-Akt, p-Erk1/2 and Vimentin in HT29/shTSPAN12-1 (Figure 6A) and SW1116/shTSPAN12-1 (Figure 6B) were significantly downregulated in comparison to both control groups (HT29/scramble and SW1116/scramble), while caspase-3 and E-cadherin were upregulated in HT29 (Figure 6A) and SW1116 (Figure 6B) cells after TSPAN12 knockdown, indicating these proteins play important roles in TSPAN12 induced cell proliferation and metastasis in CRC progression.
Figure 6.
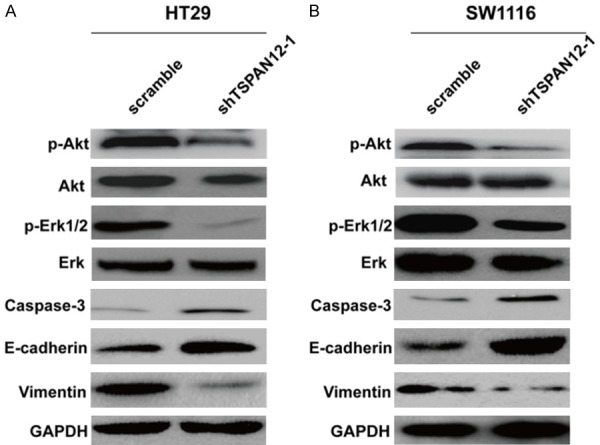
TSPAN12 silencing regulated proteins associated with cell proliferation and metastasis. Western blot analysis of TSPAN12-related molecules, including p-Akt, p-Erk, caspase-3, E-cadherin and Vimentin in HT29 (A) and SW1116 (B) cells, respectively; GAPDH was used as internal control.
Discussion
As an intense interest of current research, aberrant expression of TSPAN12 has been documented to play particularly important roles in human cancers [20], such as breast cancer and lung cancer [15,16]. Additionally, TSPAN12 has been demonstrated to play a critical role in supporting primary tumor xenograft growth in breast cancer and affecting over 400 genes involved in tumor metastasis [16]. In this study, we firstly reported the oncogenicity of TSPAN12 in the pathogenesis and progression of CRC. Through tumor tissues analysis, we found TSPAN12 expression was significantly elevated in tumor tissues of CRC patients. Detailed analysis revealed that increased expression of TSPAN12 was positively correlated with worse clinicopathologic variables, including TNM stage, tumor size and lymph node metastasis. Consistent with this result, higher TSPAN12 expression was also found in cancer-associated stromal tissues [15], lung cancer [21], breast cancer [22,23]. These data strongly suggest TSPAN12 might as an oncogene involved in CRC progression.
RNAi-mediated gene silencing could be effectively used to suppress the proliferation of CRC cells in vitro. In the present study, the expression of TSPAN12 was efficiently knocked down in two CRC cell lines, HT29 and SW1116 to carry out loss-of-function assays. Our data demonstrated that knockdown of TSPAN12 significantly inhibited cell proliferation, cell migration and invasion, and tumor formation in nude mice, which could be explained by some accumulating evidence suggests that altered expression of tetraspanins was associated with tumor progression [24,25]. As a member of the transmembrane 4 superfamily, TSPAN12 has been shown to affect beta-catenin protein expression and Wnt signaling pathway [15,26,27], suggesting its close relationship with cancer cell behavior. Furthermore, we investigated the possible mechanisms of TSPAN12-silencing for decreasing cell proliferation and metastasis. As expected, the findings from the Western blot analysis showed the marked decrease in the p-Akt, p-Erk1/2 and Vimentin expression accompanied by notable increase in the E-cadherin and caspase-3 expression in the CRC cells after transfected with shTSPAN12-1.
Activation of Erk1/2 and Akt is reported to respond to growth factor stimulation that transmits growth and survival signals [28]. Knockdown of TSPAN12 significantly inhibited the phosphorylation of Erk1/2, indicating that TSPAN12 affected cell proliferation possibly might via the blockade of Akt and mitogen-activation protein kinase (MAPK) activation. We also found shTSPAN12-1 mediated TSPAN12-1 knockdown resulted in a significant up-regulation of apoptosis in CRC cells. Caspases, a family of proteases, are shown to govern extrinsic and intrinsic apoptotic pathways, which have been divided into initiators or executioners of apoptosis [29]. Caspase-3 is thought as the primary executioner of apoptosis in the executioner class [30]. Based on these evidences, we could speculate that the inhibited cell growth was partly due to enhanced cell apoptosis induced by TSPAN12-silencing in CRC cells. In addition to cell proliferation, tetraspanins are participated in signal transduction events related with cell motility, including TSPAN8 [31] and TSPAN13 [32]. Similarly, we found knockdown of TSPAN12 could affect cell migration and invasion by altering the expression of E-cadherin and Vimentin. As the hallmark of EMT, E-cadherin plays an important role in the progression of tumor metastasis and its downregulation enhances this procedure [33,34]. Vimentin plays a positive role in mesenchymal cell migration and is required for cytoskeleton elongation remodeling during EMT [35]. Consistent with our results, removal of TSPAN12 leads to decreased cell migration and invasion by diminishing all membrane-type 1-matrix metalloproteinase (MT1-MMP)-dependent function [36]. From the above, we therefore conclude that TSPAN12 might promote migration and invasion through regulating E-cadherin and Vimentin.
In summary, the present study at first time provides new evidence of TSPAN12-silencing could effectively inhibit cell growth, proliferation, migration and invasion during CRC development and also be an effective therapeutic tool for CRC treatment. Even though further studies are needed to uncover the precise mechanism of TSPAN12 in CRC progression, our data still supply a novel molecular basis for understanding and clinical treatment of CRC.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Sung JJ, Ng SC, Chan FK, Chiu HM, Kim HS, Matsuda T, Ng SS, Lau JY, Zheng S, Adler S, Reddy N, Yeoh KG, Tsoi KK, Ching JY, Kuipers EJ, Rabeneck L, Young GP, Steele RJ, Lieberman D, Goh KL Asia Pacific Working Group. An updated Asia pacific consensus recommendations on colorectal cancer screening. Gut. 2015;64:121–132. doi: 10.1136/gutjnl-2013-306503. [DOI] [PubMed] [Google Scholar]
- 3.Chua YJ, Zalcberg JR. Progress and challenges in the adjuvant treatment of stage II and III colon cancers. Expert Rev Anticancer Ther. 2008;8:595–604. doi: 10.1586/14737140.8.4.595. [DOI] [PubMed] [Google Scholar]
- 4.Speetjens FM, Zeestraten EC, Kuppen PJ, Melief CJ, van der Burg SH. Colorectal cancer vaccines in clinical trials. Expert Rev Vaccines. 2011;10:899–921. doi: 10.1586/erv.11.63. [DOI] [PubMed] [Google Scholar]
- 5.de la Chapelle A. Genetic predisposition to colorectal cancer. Nat Rev Cancer. 2004;4:769–780. doi: 10.1038/nrc1453. [DOI] [PubMed] [Google Scholar]
- 6.Wang H, Wang Y, Qian L, Wang X, Gu H, Dong X, Huang S, Jin M, Ge H, Xu C, Zhang Y. RNF216 contributes to proliferation and migration of colorectal cancer via suppressing BECN1-dependent autophagy. Oncotarget. 2016;7:51174–51183. doi: 10.18632/oncotarget.9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beckwith KA, Byrd JC, Muthusamy N. Tetraspanins as therapeutic targets in hematological malignancy: a concise review. Front Physiol. 2015;6:91. doi: 10.3389/fphys.2015.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 9.Levy S, Shoham T. The tetraspanin web modulates immune-signalling complexes. Nat Rev Immunol. 2005;5:136–148. doi: 10.1038/nri1548. [DOI] [PubMed] [Google Scholar]
- 10.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 11.Charrin S, Naour FL, Silvie O, Milhiet PE, Boucheix C, Rubinstein E. Lateral organization of membrane proteins: tetraspanins spin their web. Biochem J. 2009;420:133–154. doi: 10.1042/BJ20082422. [DOI] [PubMed] [Google Scholar]
- 12.Bailey RL, Herbert JM, Khan K, Heath VL, Bicknell R, Tomlinson MG. The emerging role of tetraspanin microdomains on endothelial cells. Biochem Soc Trans. 2011;39:1667–1673. doi: 10.1042/BST20110745. [DOI] [PubMed] [Google Scholar]
- 13.Hemler ME. Tetraspanin proteins promote multiple cancer stages. Nat Rev Cancer. 2014;14:49–60. doi: 10.1038/nrc3640. [DOI] [PubMed] [Google Scholar]
- 14.Garcia-Espana A, Chung PJ, Sarkar IN, Stiner E, Sun TT, Desalle R. Appearance of new tetraspanin genes during vertebrate evolution. Genomics. 2008;91:326–334. doi: 10.1016/j.ygeno.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Otomo R, Otsubo C, Matsushimahibiya Y, Miyazaki M, Tashiro F, Ichikawa H, Kohno T, Ochiya T, Yokota J, Nakagama H. TSPAN12 is a critical factor for cancer-fibroblast cell contact-mediated cancer invasion. Proc Natl Acad Sci U S A. 2014;111:18691–18696. doi: 10.1073/pnas.1412062112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Knoblich K, Wang HX, Sharma C, Fletcher AL, Turley SJ, Hemler ME. Tetraspanin TSPAN12 regulates tumor growth and metastasis and inhibits beta-catenin degradation. Cell Mol Life Sci. 2014;71:1305–1314. doi: 10.1007/s00018-013-1444-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kouprina N, Mullokandov M, Rogozin IB, Collins NK, Solomon G, Otstot J, Risinger JI, Koonin EV, Barrett JC, Larionov V. The SPANX gene family of cancer/testis-specific antigens: rapid evolution and amplification in African great apes and hominids. Proc Natl Acad Sci U S A. 2004;101:3077–3082. doi: 10.1073/pnas.0308532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Xu D, Sharma C, Hemler ME. Tetraspanin12 regulates ADAM10-dependent cleavage of amyloid precursor protein. FASEB J. 2009;23:3674–3681. doi: 10.1096/fj.09-133462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang HX, Li Q, Sharma C, Knoblich K, Hemler ME. Tetraspanin protein contributions to cancer. Biochem Soc Trans. 2011;39:547–552. doi: 10.1042/BST0390547. [DOI] [PubMed] [Google Scholar]
- 21.Wachi S, Yoneda K, Wu R. Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics. 2005;21:4205–4208. doi: 10.1093/bioinformatics/bti688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu R, Wang X, Chen GY, Dalerba P, Gurney A, Hoey T, Sherlock G, Lewicki J, Shedden K, Clarke MF. The prognostic role of a gene signature from tumorigenic breast-cancer cells. N Engl J Med. 2007;356:217–226. doi: 10.1056/NEJMoa063994. [DOI] [PubMed] [Google Scholar]
- 23.Casey T, Bond J, Tighe S, Hunter T, Lintault L, Patel O, Eneman J, Crocker A, White J, Tessitore J, Stanley M, Harlow S, Weaver D, Muss H, Plaut K. Molecular signatures suggest a major role for stromal cells in development of invasive breast cancer. Breast Cancer Res Treat. 2009;114:47–62. doi: 10.1007/s10549-008-9982-8. [DOI] [PubMed] [Google Scholar]
- 24.Hemler ME. Tetraspanin proteins promote multiple cancer stages. Nat Rev Cancer. 2014;14:49–60. doi: 10.1038/nrc3640. [DOI] [PubMed] [Google Scholar]
- 25.Klosek SK, Nakashiro K, Hara S, Goda H, Hasegawa H, Hamakawa H. CD151 regulates HGF-stimulated morphogenesis of human breast cancer cells. Biochem Biophys Res Commun. 2009;379:1097–1100. doi: 10.1016/j.bbrc.2009.01.023. [DOI] [PubMed] [Google Scholar]
- 26.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 27.Valenta T, Hausmann G, Basler K. The many faces and functions of beta-catenin. EMBO J. 2012;31:2714–2736. doi: 10.1038/emboj.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X, Qiao D, Meyer K, Friedl A. Signal transducers and activators of transcription mediate fibroblast growth factor-induced vascular endothelial morphogenesis. Cancer Res. 2009;69:1668–1677. doi: 10.1158/0008-5472.CAN-07-6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bressenot A, Marchal S, Bezdetnaya L, Garrier J, Guillemin F, Plenat F. Assessment of apoptosis by immunohistochemistry to active caspase-3, active caspase-7, or cleaved PARP in monolayer cells and spheroid and subcutaneous xenografts of human carcinoma. J Histochem Cytochem. 2009;57:289–300. doi: 10.1369/jhc.2008.952044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jakob S, Corazza N, Diamantis E, Kappeler A, Brunner T. Detection of apoptosis in vivo using antibodies against caspase-induced neo-epitopes. Methods. 2008;44:255–261. doi: 10.1016/j.ymeth.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 31.Wei L, Li Y, Suo Z. TSPAN8 promotes gastric cancer growth and metastasis via ERK MAPK pathway. Int J Clin Exp Med. 2015;8:8599–8607. [PMC free article] [PubMed] [Google Scholar]
- 32.Arencibia JM, Martin S, Perez-Rodriguez FJ, Bonnin A. Gene expression profiling reveals overexpression of TSPAN13 in prostate cancer. Int J Oncol. 2009;34:457–463. [PubMed] [Google Scholar]
- 33.Christofori G, Semb H. The role of the cell-adhesion molecule E-cadherin as a tumour-suppressor gene. Trends Biochem Sci. 1999;24:73–76. doi: 10.1016/s0968-0004(98)01343-7. [DOI] [PubMed] [Google Scholar]
- 34.Cheng CW, Wu PE, Yu JC, Huang CS, Yue CT, Wu CW, Shen CY. Mechanisms of inactivation of E-cadherin in breast carcinoma: modification of the two-hit hypothesis of tumor suppressor gene. Oncogene. 2001;20:3814–3823. doi: 10.1038/sj.onc.1204505. [DOI] [PubMed] [Google Scholar]
- 35.Schoumacher M, Goldman RD, Louvard D, Vignjevic DM. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J Cell Biol. 2010;189:541–556. doi: 10.1083/jcb.200909113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takino T, Saeki H, Miyamori H, Kudo T, Sato H. Inhibition of membrane-type 1 matrix metalloproteinase at cell-matrix adhesions. Cancer Res. 2007;67:11621–11629. doi: 10.1158/0008-5472.CAN-07-5251. [DOI] [PubMed] [Google Scholar]