Abstract
Extensive research has been performed regarding the integration sites of murine leukemia retrovirus (MLV) for the identification of proto-oncogenes. To date, the overlap of mutations within specific oligonucleotides across different tumor genomes has been regarded as a rare event; however, a recent study of MLV integration into the oncogene Zfp521 suggested the existence of a hotspot oligonucleotide for MLV integration. In the current review, we discuss the hotspots of MLV integration into several genes: c-Myc, Stat5a and N-myc, as well as ZFP521, as examined in tumor genomes. From this, MLV integration convergence within specific oligonucleotides is not necessarily a rare event. This short review aims to promote re-consideration of MLV integration within the tumor genome, which involves both well-known and potentially newly identified and novel mechanisms and specifications.
Introduction
Retroviruses such as avian leukosis retrovirus,1 murine leukemia retrovirus (MLV)2 and feline leukemic retrovirus3 are involved in the development of a wide variety of diseases including malignant neoplasms, most particularly soft tissue tumors or hematopoietic tumors including lymphoma and leukemia. Retroviral genomes are integrated into the host cellular genome, becoming proviral genomes that contribute to tumorigenesis; this step is one of the essential steps in the retroviral life cycle. The inserted retroviral genome (the provirus) is thus replicated stably in the course of host cell division.4 By becoming a part of host genome, the provirus promoter and enhancer elements can instead promote downstream gene expression.2 When the provirus is inserted into the vicinity of cellular oncogenes controlling host cell growth and expansion, the affected cells will occupy the lymph node tissues and form lymphoma tissues.5, 6 To date, many researchers have analyzed the retrovirus integration sites, in order to identify neighboring oncogenes that are transcriptionally activated in the tumor genome.5
MLV integration represents an old but still relevant unsolved problem in the field of oncology. To identify the selectivity of genomic virus integration sites, the vast majority of previously reported studies have used methods involving the infection of cell culture systems with retroviruses, followed by the subsequent identification and comparison of respective integration sites in the transfected cell genomes.7 Previous studies have shown that γ-retroviruses, including MLV, exhibit strong bias for integrating near active gene promoters and associated CpG islands.8, 9, 10, 11 On the other hand, there are reports that transcriptionally active genes are favored by integration,8 or that enhancers are major target of MLV vector integration.10 As discussed later in this review, the bromodomain and extraterminal domain (BET) family of proteins are noted as the key factor of MLV integration near transcription start sites.12 In particular, whole genome analysis using second-generation sequencing has facilitated the recent discovery that MLV integration site selection is driven by strong enhancers and active promoters in an integration assay using cultured cells.9, 10 DNA secondary structural fluctuation around an integration hotspot might contribute to the open chromatin structure in the transcriptionally active site that would likely promote proviral integration after the physical breakage of the double-stranded DNA (dsDNA).13, 14
In association with MLV integration bias, there have been extensive discussions regarding the clinical application of MLV vectors. For gene therapy, an MLV-based vector was found to have integrated into the LIM domain only 2 (LMO2) gene in human T cells in a gene therapy trial15, 16 and thereby induced T-cell leukemia in the two patients.17, 18, 19, 20 However, the majority opinion was that the selection of LMO2 as an insertion site in the patients was accidental. In addition, the tumorigenesis caused by MLV insertion has become an issue of concern with respect to the clinical application of induced pluripotent stem (iPS) cells, whereas the present protocol for iPS cell preparation uses a plasmid vector that does not have tumorigenesis potential.21
A model mouse strain of MLV-induced lymphomagenesis
As an experimental model for studies of MLV-induced tumorigenesis, an inbred strain of mice such as AKR can be used that is susceptible to spontaneous tumors by the integration of MLV;22 this susceptibility arises from endogenous (that is, previously genetically acquired in the germ line) MLV provirus genomes.23 BXH224 and AKXD25 are well-known mouse strains that have been used to study the process of leukemogenesis. Using these strains and other strains, the identification of integration sites has been a powerful tool for the study of oncogenes. In fact, many integration sites have been registered in the retroviral-tagged cancer gene database (http://variation.osu.edu/rtcgd/), which has in turn been used for the study of retroviral tumorigenesis, wherein a numbering system based on the ecotropic viral integration site (EviN; N=1, 2, 3,..) was applied by Copeland and colleagues,26, 27 to summarize the MLV integration site. Notably, many integration sites within identical genes are registered in the database; however, integration site overlap at the oligonucleotide level is rare in this database with the exception of c-Myc.
SL/Kh strain
SL/Kh represents an inbred mouse strain derived from AKR that we have developed previously and wherein we have achieved MLV-mediated induction of lymphoma.28, 29, 30, 31 One of the advantages of using this strain for the study of integration is the extremely biased integration into specified genes that occurs in these mice, as observed in tumors. This strain possesses pathogenic endogenous ecotropic murine virus 11 (Emv11) that has been mapped onto chromosome 7 and is shared with the AKR mouse strain.23, 32 In the SL/Kh strain, the pro- to pre-B cells in the bone marrow expand28, 33 and newly acquire more than one copy of the proviral genome during the process of lymphomagenesis;34 >90% of these mice spontaneously develop sIgM- pro- or pre-B lymphomas that are positive for BP-1 by the age of 6 months.28, 32 Notably, the phenotypic stability and the tumor incidence are significantly higher than those in other models, BXH224 and AKXD.25 We have thus used the SL/Kh mouse strain to investigate MLV integration because of the high incidence and reproducibility of the phenotype.29
Phenotypes of MLV-induced lymphomagenesis
In the reported cases of spontaneous hematopoietic tumors, granulocytes, B cells, T cells and their immature lineages serve as host cells and can develop tumors such as leukemia or lymphoma following MLV infection. The tumor phenotype in tumor-susceptible strains might be attributed to the host genetic background of the mice,22, 24, 35 as well as viral tropism and the function of target genes.36 Tropism of the viruses, that is, which type of cells are infected by viruses, is determined by the presence of the retroviral receptor on the host cells and the Env protein on the viral particle;37 both myeloid and lymphoid cell lineages express retroviral receptor. In addition, the capsid domain of Gag polyprotein, which is the major structural protein among the MLV-encoded proteins, mediates the assembly of many retroviral proteins. Capsid confers tropism that determines the susceptibility to the intracellular restriction factors Friend virus susceptibility1 (Fv1) on chromosome 4 that inhibits the development of MLV by interaction with capsid.38 Tumor phenotype might therefore directly depend on the gene structure of proviral genome. Moloney MLV induces almost exclusively lymphoid disease; however, when replacing the long terminal repeat39 with that of retrovirus 4070A, the MLV causes myeloid leukemias.39
With respect to the genetic background of mice, we previously identified a locus responsible for pre-B-cell expansion, Bomb1 near Mit319 on chromosome 3, via the genetic study of SL/Kh. A congenic mouse on a NFS/N mouse background carrying the Bomb1 locus of SL/Kh mice demonstrated expansion of pre-B-cell lymphocytes28, 33 and microsatellite instability near Bank1 gene locus, an adaptor molecule of pre-B receptors in pre-B-cell lymphomas in SL/Kh mice.40 Therefore, the microsatellite instability of the responsible locus as well as other background factors might cooperatively determine the phenotype of the tumor.
Retroviral genome integration into the host genome
Molecular integration mechanism
The molecular biology of MLV integration is fairly well understood. Following retroviral infection via viral receptor molecules on host cell membrane, viral dsDNA synthesis by reverse transcriptase begins in the cytoplasm and is completed before or after virus entry into the nucleus. A viral enzyme integrase (IN) that is encoded in the retroviral genome cleaves the termini of the viral DNA; the viral dsDNA then forms a pre-integration complex and enters the nucleus. IN forms a tetramer or oligomer at the terminus of the pre-integration complex DNA; these catalyze the host DNA cleavage reaction in a staggered manner. It has been suggested that the terminal ends of the dsDNA might be critical for integration, and that the presence of a dinucleotide capsid at the terminal 3′-ends is particularly essential.13, 41 Subsequently, the free energy change released from the broken phosphodiester bonds in the host dsDNA promotes the formation of new bonds joining the viral DNA ends to the ends of cleaved host DNA. DNA synthesis extends from the host DNA flanking the host-viral DNA junctions and fills in the gaps adjacent to the viral DNA, displacing the viral DNA ends. Consequently, four bases are duplicated on either side of the MLV proviral genome; during HIV-1 integration, five nucleotides are duplicated.42 These duplicated nucleotides have been analyzed from the perspective of integration selectivity; however, no significant tendency of the sequence motifs has been identified. For this reason, integration has been believed to be random as for the target nucleotides.
Hotspots of MLV integration
Using the tumor DNAs from spontaneous tumor model mice such as BXH2, AKXD and SL/Kh, inverse PCR was performed,26, 42, 43 which involves digestion of the host cell DNA with a restriction enzyme such as SacII, which recognizes CpG islands near gene-coding regions, followed by a ligation reaction using T4-ligase.36 The development of inverse PCR-dependent cloning has allowed the identification of numerous integration sites that have in turn been used for the identification of novel oncogenes26 such as Stat5a,43 Hipk2, Fiz144 and Zfp521,45 which are frequent integration loci. Before this technical development, the distribution of integration sites within a given gene were not sufficiently investigated and there were few studies providing detailed data regarding the integration within a short segment in a single gene. Here, the term ‘hotspot of integration' is used to imply that the integration site is shared by more than one different tumor, despite the fact that the probability is prospected to be substantially lower for integration into such a narrow range.
Hotspots in c-Myc and N-myc
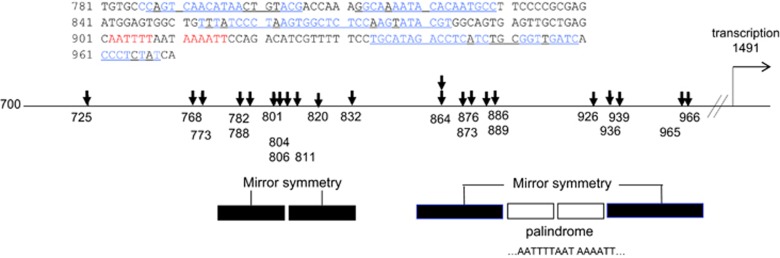
Systematic analysis of integration site has been limited to the study of integration into the c-myc proto-oncogene.46, 47, 48 Here we reviewed MLV integration sites within the c-myc promoter area in particular. As mentioned previously, the systematic analysis did not elucidate a definite trend. However, several integration sites were shared by tumors in various strains of mice, suggesting that certain sites might be preferred by MLV integration.49 In a systematic analysis of SL3-3 MLV integration sites, many nucleotides were shared by different tumors. For instance, nucleotide numbers 773, 832, 864 and 889 (M1234.5 in GENBANK) were actually shared by different tumors in different strains (Figure 1 and Table 1). We noted that c-Myc promoter consists of palindromic and mirror symmetry sequence motifs around the integration sites (Figure 1).
Figure 1.
MLV integration sites within c-myc (M12345). Sequence motifs in the c-myc promoter. The shown sequence represents the mirror symmetry in blue and palindrome in red. The white and black boxes with lines represent the pairs of palindromic and mirror symmetry motifs, respectively. The downward arrows indicate the previously reported integration sites shown by the numbers.
Table 1. Each host gene locus is indicated via its registered number in GenBank.
| Host gene | Location | Previously reported sites | SL/Kh | Tumor phenotype | Reference |
|---|---|---|---|---|---|
| c-myc | Promoter | ||||
| M12345 | |||||
| 725 | T cell | 47 | |||
| 768 | T cell | 71 | |||
| 773 | 773 | T cell | 72 | ||
| 782, 788 | Lymphoma | AF52467873 | |||
| 801 | 804 | B cell | AF52431573 | ||
| 811 | T cell | 72 | |||
| 820 | B cell | 47 | |||
| 832 | 832 | Unknown | AF52432473 | ||
| 853 | Pre-B cell | ||||
| 864 | 864 | T cell | Z6982574 | ||
| 876, 886, 889 | 889 | B cell | BH85954726 | ||
| 926 | Unknown | AF1931425 | |||
| 932 | |||||
| 939 | 939 | T, B mixed | 5 | ||
| 966 | 966 | B cell | 75 | ||
| Pim-1 | 3′ of the last | 6980 | 6980 | B cell | 77 |
| M13945 | Eleventh exon | 6982 | B cell | 77 | |
| 6991 | T cell | 78 | |||
| 7000, 7005 | 7005 | T cell | 78 | ||
| 7064 | 7063 | T cell | 78 | ||
| Sox4 | |||||
| AX695422 | First exon | ||||
| 12 168 | B cell, pre-B cell | 26 | |||
| 12 173 | B cell | 75 | |||
| 12 175 | B cell | 75 | |||
| 12 176 | T cell | 26 | |||
| 12 182 | Pre-B cell myeloid | 26 | |||
| N-myc | 3′ | 6307 | ND | 72 | |
| M12731 | 6311 | T cell | 5 | ||
| 6313, 6314, 6315 | 76 | ||||
| 6328 | 6328 | T, B mix | 5 | ||
| 6329 | 6336 | Lymphoma | 73 | ||
| 6344 | 6338 | Pre-B cell | 50 | ||
| 6345 | 6344 | Pre-B cell | 26 | ||
| Zfp521 | First intron | ||||
| AC142257 | 83 098, 83 101 | 83 098 | B cell, pre-B cell | 45, 55 | |
| 83 117 | 83 117 | B cell, pre-B cell | 45, 55 | ||
| Meis 1 | |||||
| AL603984 | Upstream of the first exon | 199 385 | |||
| 199 389 | 199 389 | B cell | BH86008126 | ||
| 199 389 | Myeloid | BH86052126 | |||
| AL645570 | Eleventh intron | 77 862 | Pre-B cell | ||
| 77 872 | Pre-B cell | ||||
| 77 878 | Myeloid | BH86007726 | |||
| Cyclin D2 | Promoter | ||||
| AC163747 | 130 896 | B cell | 77 | ||
| 130 903 | T cell | 75 | |||
SL/Kh indicates the integration sites in pre-B-cell lymphoma in SL/Kh mice. The superscripted numbers indicate the reference number in the text. Previous reported sites indicate the nucleotide number based on the GenBank sequence for the individual accession numbers listed. In the c-myc gene, No. 1491 represents the transcriptional start nucleotide. ND, no determined.
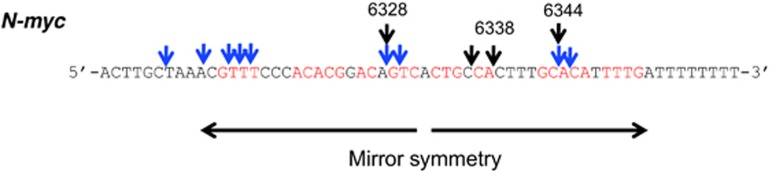
Similarly, MLV integration sites were shared by lymphoid tumors in the third exon and the 3′-flanking sequence in N-myc.50, 51 Numbers 6328 and 6344 (M12731) were shared by different tumors in different strains and mirror symmetry sequence motifs around the integration sites (Figure 2).
Figure 2.
MLV integration within N-myc (number 6300-, M12731) (Table 1). The red letters indicate the mirror symmetry motif in the N-myc gene. The two vertical arrows indicate the paired regions of mirror symmetry. The black and blue downward arrows represent the integration sites in SL/Kh mice and other previously reported sites.
Hotspots in Stat5a
Next, we consider the MLV integration sites in Stat5a. The encoded STAT5A protein is a member of the signal inducer and activator of transcription (STAT) family. These proteins form a dimer that translocates into the nucleus and exerts transcriptional activity by binding to the γ-interferon activation site palindromic element in the promoters of target anti-apoptotic genes.52, 53 The products of these genes, such as c-Myc, Pim-1, Bcl-xL and Cyclin D1,54 regulate proliferation and anti-apoptosis in hematopoietic cells; STAT5A in particular contributes to IL-7-induced B-cell precursor expansion.43, 49
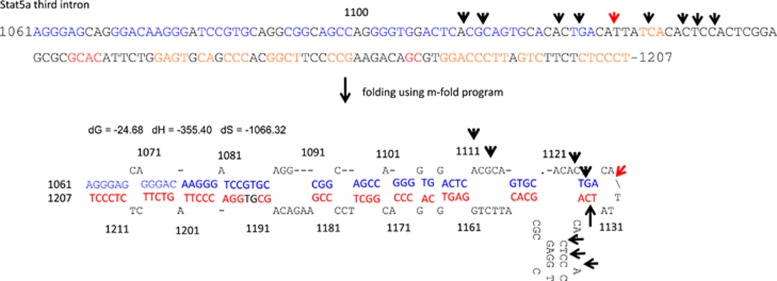
In pre-B-cell lymphoma in SL/Kh mice, MLV integration within the 400 bp second intron of the Stat5a gene was observed.43 Notably, several integration sites were shared by different tumors of this strain.32, 41, 43 In particular, a 170 bp segment within the second intron was found to be the hotspot for MLV integration (Figure 3). Furthermore, within this region, several nucleotides were shared by different pre-B-cell tumors.
Figure 3.
MLV integration sites within Stat5a. The sequence represents the whole sequence of AF049104 from (5′-)1061th–1207th nt (-3′) (Stat5a gene sequence in GenBank, renumbered according to the nucleotide number form the start of the gene, BLAST: https://blast.ncbi.nlm.nih.gov). The colored letters in blue and red represent the pair of palindromic sequences in the above sequence of which stem of the hairpin structure that is anticipated by m-fold analysis. dG, dH and dS represent Gibbs free energy, enthalpy, and entropy change (kcal/mol) in folding. [Na+]=1.0 mM. Folding temperature is equal to 37°C. The most frequent site (N0.1130, shown by a red arrow) that is shared by different pre-B-cell tumor is shown.41
Hotspots in Zfp521
The Zfp521 (zinc finger protein 521) gene was identified as a common integration site 3 (Evi3) in the genomes of B-cell lymphomas in the AKXD mouse strain.45, 55 We also reported that this gene is the most frequent integration site as well in pre-B-cell lymphoma in SL/Kh mice.32 The encoded ZFP521 regulates and activates pre-B-cell receptor signal pathways.45
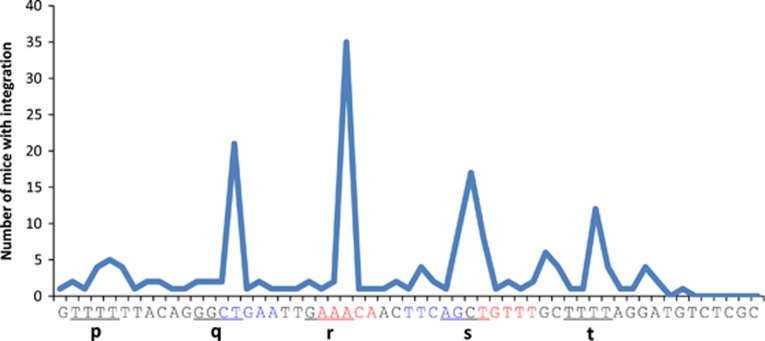
In more than 140 male SL/Kh mice, >80% pe-B-lymphoma genomes acquired the integrated proviral genome.32 Specifically, the integration sites in the genome of pre-B-cell tumors in SL/Kh mice (p, q, r, s, t) are shown in Figure 4. The integration occurred once or twice within a 50 bp segment located in the region of the second to the third exon of Zfp521 in the pre-B-cell lymphoma genome during the lifetime of the mouse. Two independent integrations into the identical gene in the genome of a single mouse are undoubtedly a rare occurrence. Therefore, we can state that the second to the third exon is one of the hotspots of MLV integration.
Figure 4.
MLV integration within Zfp521. The numbers of mice with MLV integration are presented. In the vertical sequence of Zfp521, underlines represent the four oligonucleotides duplicated by the integration in the host cell genome when the integration occurs at p, q, r, s, or t. There were mice that have more than one integration sites in the lymphoma tissue genome. The r and s positions are shared by hematopoietic tumor of SL/Kh and AKXD strains. Four integration site data are added to our previously data. A hotspot ‘s' consists of two nucleotide nucleotides G and C.45, 55
Common features of hotspot sequences
As discussed in above, the sharing at the nucleotide level within the integration tissues in the hematopoietic tumors are not rare events as would be expected, as shown by SL3-3 integration in c-Myc and Emv11 in Zfp521, in spite of the fact that it is statistically almost impossible that MLV targets the specified short oligonucleotide as the integration sites. Therefore, the common features of these hotspots within the host genes should be considered. To date, reports have indicated that the promoter region and enhancer comprise the targets of the integration,9, 10 or transcriptional active site are favored for intefration.8, 56 With respect to the latter and as discussed in further detail later, BET mediates the association of MLV IN and the transcriptional start site in the open chromatin of the host cell.10
With respect to DNA structure, the integrations within Zfp521 frequently occurred at ~10 bp intervals and were symmetrically distributed on either side of the most frequent integration site (p, q, r, s, t in Figure 4).45, 55 The 10 bp intervals suggest that MLV integration sites are distributed on the nucleosome surface of the 10 bp periodical outward-facing DNA major grooves in chromatin. To our knowledge, no distinct data has been published regarding 10 bp periodical integration of MLV into the host genome; however, it has been demonstrated that HIV-1 integration sites are periodically distributed on the nucleosome surface by statistical analysis.46 Our data of 10 bp interval between integration in Zfp521 provides the first direct evidence of periodical retroviral integration.
The transcriptional factor-binding motifs within the promoter and enhancer might also be involved in the choice of integration site.57 In addition, the junction of DNA between the viral and host genome might generate novel motifs required for binding transcription factors and other adaptor proteins, to effect higher transcription of the target gene.32, 41 For example, the MLV integration site induces the formation of transcription factor complexes on palindromic sequences during the development of pre-B lymphomagenesis and alters the transcriptional activity of Stat5a. Conversely, the transcription factor preference for particular DNA sequence motifs might determine the selection of the integration site. MLV integration induces the formation of transcription factor complexes consisting of GATA, CREB and C/EBPβ on palindromic sequences and on the TATA-box in the Stat5a gene. The presumed secondary structure in the open chromatin such as the hairpin structure in the 170 bp segment in Stat5a might assist the binding of transcription factors that enhance host gene transcription (Figure 2).38 Furthermore, the palindromic motifs might contribute to posttranscriptional stabilization of MLV-host gene chimera RNA through the generation of secondary structure after transcription or of open chromatin structure that controls the transcriptional activity of the host gene when the double helix DNA is rewound. In actuality, such DNA structures are anticipated using m-fold analysis (http://mfold.rna.albany.edu/?q=mfold/DNA-Folding-Form) (Figure 3).
Previously, Holman et al.58 also suggested that palindromes are statistically favored by MLV integration. Palindromic motifs were also observed in the hotspots in c-Myc, Stat5a, and Zfp521 In Zfp521, the integration occurred at the site indicated as ‘:' in the sequence 5′-CTGAATTG AAAC:AACTTCAGCTGTTT-3′ with 10 bp periodicity at p, q, r, s and t (Figure 4).45 Such alternating palindrome sequences are definitely rare motifs. It is noticed that the integration occurs at the middle position, r, of the above sequence, suggesting that the sequence may confer the selectivity of the integration into the position surrounding the palindrome. This plot represents a probability density function of MLV insertional mutation. Pim1 and Sox4 are also targets of MLV integration in hematopoietic tumor genomes.12, 27, 41 In Pim1 (M13945), the hotspots of integration are positioned in the palindromic sequences at the site indicated by a colon (‘:') as follows: 5′-CCCTGCG:TGAC:GACGCAGGG-3′ and 5′-CCAGGTCCCTGGAGGAGCCTC:CCAC:AAGGGAAAGAGACTACTTCACTGGTCCTGG-3′, where the pair of underlined sequences and dotted-underlined sequences represent palindromic sequences. In Sox4 (AX695422), the integration sites are positioned near the middle of the palindrome as well in 5′-GGAGCGCGGGGGCGTTAGTGGA:CCCGCG:CT:CC-3′ (Table 1). The preference for the palindrome requires a rigorous statistical study. However, as the sharing of integration sites by different tumors has been found to not be a rare event as had been expected, the DNA structure factor may contribute to the integration site selection.
To better understand the contribution of the host gene to MLV integration, we need to review recent research advances in the control of open chromatin regions in association with IN interaction with histones and its binding to the host gene by recruitment of BET.
Recent advance of molecular basis of MLV integration
BET proteins and MLV integration
Recent studies have yielded great advancements in our understanding of MLV integration.59 BET proteins (Brd2, Brd3 and Brd4) are the cellular-binding partners of MLV IN and preferentially engage open chromatin regions that are enriched for transcription start sites, CpG islands, DNaseI-hypersensitive sites and proto-oncogenes.11, 56, 60, 61, 62, 63 BET family proteins behave as a scaffold on chromatin to recruit E2F proteins, histone deacetylases, histone H4-specific acetyltransferases including GCN5, and chromatin remodeling proteins.64 The BET proteins are characterized by an extraterminal domain and two N-terminal bromodomains that recognize and bind acetylated lysines on histone H3 and H4 tails on chromatin.65 In vitro interaction analysis and co-immunoprecipitation of MLV IN in human cells revealed that different MLV IN domains including the C-terminal domain, the catalytic core domain and the IN C-terminus interacts with the BET family of proteins.66 It has also been shown that purified recombinant Brd4 (1–720) bound with high affinity to MLV IN and stimulated MLV concerted integration in vitro.59 Through the recognition of open chromatin structure, BET proteins have been suggested to contribute to the tethering of the MLV pre-integration complex to the host DNA.11, 67 Furthermore, BET protein knockdown and treatment with the cell-permeable small molecule JQ-1, a BET bromodomain inhibitor, were shown to reduce MLV integration frequencies at transcription start sites.60 These observations suggest that BET proteins navigate the MLV genome and promote efficient MLV integration around transcription start sites associated with an open chromatin structure.56
In addition, BRD4 has a critical role in germinal center response by regulating Bcl-6 and nuclear factor-κB activation.68 The double bromodomain protein Brd2 promotes B-cell expansion and mitogenesis.69 Accordingly, these proteins might broadly contribute to tumorigenesis in cooperation with MLV integration in the development of spontaneous lymphoma.70 Recent advances suggest that integration selection needs to be understood in the context of the epigenetic modification of histones or nucleosomes. Such control mechanisms of transcriptional activity will likely provide novel standpoints of oncogene function and a better understanding of MLV insertional mutagenesis as well.
Conclusion
Detailed analysis of MLV integration between tumors within and between mouse strains indicates that the convergence of integration sites within a narrow oligonucleotide range is not necessarily a rare event, and that the integration mutagenesis process might be unique among tumor mutagenesis mechanisms. Furthermore, specific DNA structural factors may contribute to the integration site selection, facilitating the generation of ‘hotspot' motifs in tumor genomes.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research on Innovative Areas, MEXT, Japan (project title ‘Synergy of Fluctuation and Structure' and number 2502). This funding agency had no role in the study design, data collection, analysis, decision to publish or preparation of the manuscript.
Footnotes
The authors declare no conflict of interest.
References
- Withers-Ward ES, Kitamura Y, Barnes JP, Coffin JM. Distribution of targets for avian retrovirus DNA integration in vivo. Genes Dev 1994; 8: 1473–1487. [DOI] [PubMed] [Google Scholar]
- Baumbach WR, Colston EM, Cole MD. Integration of the BALB/c ecotropic provirus into the colony-stimulating factor-1 growth factor locus in a myc retrovirus-induced murine monocyte tumor. J Virol 1988; 62: 3151–3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura T, Brost H, Kabisch A, Lampert F, Hadwiger-Fangmeier A, Niemann H. Detection of fms-oncogene-specific tyrosine kinase activity in human leukemia cells. J Cancer Res Clin Oncol 1989; 115: 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beveren C, Goddard JG, Berns A, Verma IM. Structure of Moloney murine leukemia viral DNA: nucleotide sequence of the 5' long terminal repeat and adjacent cellular sequences. Proc Natl Acad Sci USA 1980; 77: 3307–3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen GM, Skapura D, Justice MJ. Genetic profile of insertion mutations in mouse leukemias and lymphomas. Genome Res 2000; 10: 237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell 1991; 65: 737–752. [DOI] [PubMed] [Google Scholar]
- Kurdi-Haidar B, Levine F, Roemer K, LaPorte P, Friedmann T. Provirus-anchored long-range (PAL) mapping of mammalian genomes. Genomics 1993; 15: 305–310. [DOI] [PubMed] [Google Scholar]
- Wu XL, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science 2003; 300: 1749–1751. [DOI] [PubMed] [Google Scholar]
- LaFave MC, Varshney GK, Gildea DE, Wolfsberg TG, Baxevanis AD, Burgess SM. MLV integration site selection is driven by strong enhancers and active promoters. Nucleic Acids Res 2014; 42: 4257–4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Ravin SS, Su L, Theobald N, Choi U, Macpherson JL, Poidinger M et al. Enhancers are major targets for murine leukemia virus vector integration. J Virol 2014; 88: 4504–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvaratskhelia M, Sharma A, Larue RC, Serrao E, Engelman A. Molecular mechanisms of retroviral integration site selection. Nucleic Acids Res 2014; 42: 10209–10225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rijck J, de Kogel C, Demeulemeester J, Vets S, El Ashkar S, Malani N et al. The BET family of proteins targets moloney murine leukemia virus integration near transcription start sites. Cell Rep 2013; 5: 886–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruyama T, Liu W, Yoshikawa K. In vitro murine leukemia retroviral integration and structure fluctuation of target DNA. PLoS One 2012; 7: e31533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz RA, Gravuer K, Skalka AM. A preferred target DNA structure for retroviral integrase in vitro. J Biol Chem 1998; 273: 24190–24195. [DOI] [PubMed] [Google Scholar]
- Davé UP, Akagi K, Tripathi R, Cleveland SM, Thompson MA, Yi M et al. Murine leukemias with retroviral insertions at Lmo2 are predictive of the leukemias induced in SCID-X1 patients following retroviral gene therapy. PLoS Genet 2009; 5: e1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCcormack MP, Wulffraat N, Leboulch P et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science 2003; 302: 415–419. [DOI] [PubMed] [Google Scholar]
- Pike-Overzet K, van der Burg M, Wagemaker G, van Dongen JJ, Staal FJ. New insights and unresolved issues regarding insertional mutagenesis in X-linked SCID gene therapy. Mol Ther 2007; 15: 1910–1916. [DOI] [PubMed] [Google Scholar]
- Swazo NK. Calculating risk/benefit in X-linked severe combined immune deficiency disorder (X-SCID) gene therapy trials: the task of ethical evaluation. J Med Philos 2006; 31: 533–564. [DOI] [PubMed] [Google Scholar]
- Ginn SL, Curtin JA, Kramer B, Smyth CM, Wong M, Kakakios A et al. Treatment of an infant with X-linked severe combined immunodeficiency (SCID-X1) by gene therapy in Australia. Med J Aust 2005; 182: 458–463. [DOI] [PubMed] [Google Scholar]
- Kohn DB, Sadelain M, Glorioso JC. Occurrence of leukaemia following gene therapy of X-linked SCID. Nat Rev Cancer 2003; 3: 477–488. [DOI] [PubMed] [Google Scholar]
- Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science 2008; 322: 949–953. [DOI] [PubMed] [Google Scholar]
- Lenz J, Crowther R, Klimenko S, Haseltine W. Molecular cloning of a highly leukemogenic, ecotropic retrovirus from an AKR mouse. J Virol 1982; 43: 943–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haran-Ghera N, Peled A, Wu L, Shortman K, Brightman BK, Fan H. The effects of passive antiviral immunotherapy in AKR mice: I. The susceptibility of AKR mice to spontaneous and induced T cell lymphomagenesis. Leukemia 1995; 9: 1199–1206. [PubMed] [Google Scholar]
- Shaughnessy Jr JD, Largaespada DA, Tian E, Fletcher CF, Cho BC, Vyas P et al. Mrvi1, a common MRV integration site in BXH2 myeloid leukemias, encodes a protein with homology to a lymphoid-restricted membrane protein Jaw1. Oncogene 1999; 18: 2069–2084. [DOI] [PubMed] [Google Scholar]
- Liao X, Buchberg AM, Jenkins NA, Copeland NG. Evi-5, a common site of retroviral integration in AKXD T-cell lymphomas, maps near Gfi-1 on mouse chromosome 5. J Virol 1995; 69: 7132–7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Shen H, Akagi K, Morse HC, Malley JD, Naiman DQ et al. New genes involved in cancer identified by retroviral tagging. Nat Genet 2002; 32: 166–174. [DOI] [PubMed] [Google Scholar]
- Akagi K, Suzuki T, Stephens RM, Jenkins NA, Copeland NG. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res 2004; 32: D523–D527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka T, Tsuruyama T, Kaszynski R, Kometani K, Minato N, Nakamura T et al. Bone marrow pre-B expansion by SL/Kh-Bomb1 locus: not sufficient for lymphomagenesis. Leuk Res 2008; 32: 309–314. [DOI] [PubMed] [Google Scholar]
- Abujiang P, Yamada Y, Haller O, Kobayashi H, Kamoto T, Lu LM et al. The origin of SL family mice. Lab Anim Sci 1996; 46: 410–417. [PubMed] [Google Scholar]
- Yamada Y, Matsushiro H, Ogawa MS, Okamoto K, Nakakuki Y, Toyokuni S et al. Genetic predisposition to pre-B lymphomas in SL/Kh strain mice. Cancer Res 1994; 54: 403–407. [PubMed] [Google Scholar]
- Shimada MO, Yamada Y, Nakakuki Y, Okamoto K, Fukumoto M, Honjo T et al. SL/KH strain of mice: a model of spontaneous pre-B-lymphomas. Leuk Res 1993; 17: 573–578. [DOI] [PubMed] [Google Scholar]
- Hiai H, Tsuruyama T, Yamada Y. Pre-B lymphomas in SL/Kh mice: a multifactorial disease model. Cancer Sci 2003; 94: 847–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LM, Shimada R, Higashi S, Zeng Z, Hiai H. Bone marrow pre-B-1 (Bomb1): a quantitative trait locus inducing bone marrow pre-B-cell expansion in lymphoma-prone SL/Kh mice. Cancer Res 1999; 59: 2593–2595. [PubMed] [Google Scholar]
- Hiai H, Yamada Y, Abujiang P, Lu L, Kamoto T, Tsuruyama T. Genetic and epigenetic susceptibility to endogenous retrovirus-induced lymphomas in SL mice. Prog Exp Tumor Res 1999; 35: 64–77. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Largaespada DA, Lee MP, Johnson LA, Ohyashiki K, Toyama K et al. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat Genet 1996; 12: 154–158. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Largaespada DA, Shaughnessy Jr JD, Jenkins NA, Copeland NG. Cooperative activation of Hoxa and Pbx1-related genes in murine myeloid leukaemias. Nat Genet 1996; 12: 149–153. [DOI] [PubMed] [Google Scholar]
- Katane M, Fujita R, Takao E, Kubo Y, Aoki Y, Amanuma H. An essential role for the His-8 residue of the SDF-1alpha-chimeric, tropism-redirected Env protein of the Moloney murine leukemia virus in regulating postbinding fusion events. J Gene Med 2004; 6: 260–267. [DOI] [PubMed] [Google Scholar]
- Mortuza GB, Dodding MP, Goldstone DC, Haire LF, Stoye JP, Taylor IA. Structure of B-MLV capsid amino-terminal domain reveals key features of viral tropism, gag assembly and core formation. J Mol Biol 2008; 376: 1493–1508. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herguido C, Guiu J, D'Altri T, Ingles-Esteve J, Dzierzak E, Espinosa L et al. Hematopoietic stem cell development requires transient Wnt/beta-catenin activity. J Exp Med 2012; 209: 1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaszynski RH, Akatsuka S, Hiratsuka T, Jin G, Ozeki M, Okuno T et al. A quantitative trait locus responsible for inducing B-cell lymphoblastic lymphoma is a hotspot for microsatellite instability. Cancer Sci 2010; 101: 800–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruyama T, Hiratsuka T, Jin G, Imai Y, Takeuchi H, Maruyama Y et al. Murine leukemia retrovirus integration induces the formation of transcription factor complexes on palindromic sequences in the signal transducer and activator of transcription factor 5a gene during the development of pre-B lymphomagenesis. Am J Pathol 2011; 178: 1374–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldgur Y, Craigie R, Cohen GH, Fujiwara T, Yoshinaga T, Fujishita T et al. Structure of the HIV-1 integrase catalytic domain complexed with an inhibitor: a platform for antiviral drug design. Proc Natl Acad Sci USA 1999; 96: 13040–13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruyama T, Nakamura T, Jin G, Ozeki M, Yamada Y, Hiai H. Constitutive activation of Stat5a by retrovirus integration in early pre-B lymphomas of SL/Kh strain mice. Proc Natl Acad Sci USA 2002; 99: 8253–8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruyama T, Imai Y, Takeuchi H, Hiratsuka T, Maruyama Y, Kanaya K et al. Dual retrovirus integration tagging: identification of new signaling molecules Fiz1 and Hipk2 that are involved in the IL-7 signaling pathway in B lymphoblastic lymphomas. J Leukoc Biol 2010; 88: 107–116. [DOI] [PubMed] [Google Scholar]
- Hiratsuka T, Takei Y, Ohmori R, Imai Y, Ozeki M, Tamaki K et al. ZFP521 contributes to pre-B-cell lymphomagenesis through modulation of the pre-B-cell receptor signaling pathway. Oncogene 2015; 35: 3227–3238. [DOI] [PubMed] [Google Scholar]
- Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res 2007; 17: 1186–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AA, Sorensen AB, Schmidt J, Pedersen FS. Analysis of wild-type and mutant SL3-3 murine leukemia virus insertions in the c-myc promoter during lymphomagenesis reveals target site hot spots, virus-dependent patterns, and frequent error-prone gap repair. J Virol 2005; 79: 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin G, Tsuruyama T, Yamada Y, Hiai H. Svi3: a provirus common integration site in c-myc in SL/Kh pre-B lymphomas. Cancer Sci 2003; 94: 791–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran LM, Cory S, Adams JM. Transposition of the immunoglobulin heavy chain enhancer to the myc oncogene in a murine plasmacytoma. Cell 1985; 40: 71–79. [DOI] [PubMed] [Google Scholar]
- Setoguchi M, Higuchi Y, Yoshida S, Nasu N, Miyazaki Y, Akizuki S et al. Insertional activation of N-myc by endogenous Moloney-like murine retrovirus sequences in macrophage cell lines derived from myeloma cell line-macrophage hybrids. Mol Cell Biol 1989; 9: 4515–4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePinho RA, Legouy E, Feldman LB, Kohl NE, Yancopoulos GD, Alt FW. Structure and expression of the murine N-myc gene. Proc Natl Acad Sci USA 1986; 83: 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle JN, Schwaller J, Parganas E, Wang DM, Cain D, Aster JC et al. Stat5 is essential for the myelo- and lymphoproliferative disease induced by TEL/JAK2. Mol Cell 2000; 6: 693–704. [DOI] [PubMed] [Google Scholar]
- Nosaka T, Kawashima T, Misawa K, Ikuta K, Mui ALF, Kitamura T. STAT5 as a molecular regulator of proliferation, differentiation and apoptosis in hematopoietic cells. EMBO J 1999; 18: 4754–4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanakura Y, Matsumura I, Kitamura T, Wakao H, Tanaka H, Hashimoto K et al. Transcriptional regulation of the cyclin D1 promoter by STAT5: its involvement in cytokine-dependent growth of hematopoietic cells. EMBO J 1999; 18: 1367–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland NG, Warming S, Liu P, Suzuki T, Akagi K, Lindtner S et al. Evi3, a common retroviral integration site in murine B-cell lymphoma, encodes an EBFAZ-related Kruppel-like zinc finger protein. Blood 2003; 101: 1934–1940. [DOI] [PubMed] [Google Scholar]
- Sharma A, Larue RC, Plumb MR, Malani N, Male F, Slaughter A et al. BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc Natl Acad Sci USA 2013; 110: 12036–12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felice B, Cattoglio C, Cittaro D, Testa A, Miccio A, Ferrari G et al. Transcription factor binding sites are genetic determinants of retroviral integration in the human genome. PLoS ONE 2009; 4: e4571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman AG, Coffin JM. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc Natl Acad Sci USA 2005; 102: 6103–6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elis E, Ehrlich M, Prizan-Ravid A, Laham-Karam N, Bacharach E. p12 tethers the murine leukemia virus pre-integration complex to mitotic chromosomes. PLoS Pathog 2012; 8: e1003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci USA 2003; 100: 8758–8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmuth F, Blankenfeldt W, Geyer M. Structures of the dual bromodomains of the P-TEFb-activating protein Brd4 at atomic resolution. J Biol Chem 2009; 284: 36547–36556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara T, Nakamura Y, Jang MK, Nakano K, Tanaka A, Ozato K et al. Structural basis for acetylated histone H4 recognition by the human BRD2 bromodomain. J Biol Chem 2010; 285: 7610–7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Prakash C, Sum C, Gong Y, Li Y, Kwok JJ et al. Bromodomain-containing protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. J Biol Chem 2012; 287: 43137–43155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta SS, Maetzig T, Maertens GN, Sharif A, Rothe M, Weidner-Glunde M et al. Bromo- and extraterminal domain chromatin regulators serve as cofactors for murine leukemia virus integration. J Virol 2013; 87: 12721–12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Knapp S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat Rev Drug Discov 2014; 13: 337–356. [DOI] [PubMed] [Google Scholar]
- Aiyer S, Swapna GV, Malani N, Aramini JM, Schneider WM, Plumb MR et al. Altering murine leukemia virus integration through disruption of the integrase and BET protein family interaction. Nucleic Acids Res 2014; 42: 5917–5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe BL, Larue RC, Yuan C, Hess S, Kvaratskhelia M, Foster MP. Structure of the Brd4 ET domain bound to a C-terminal motif from gamma-retroviral integrases reveals a conserved mechanism of interaction. Proc Natl Acad Sci USA 2016; 113: 2086–2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Yang Y, Wang Z, Gao X, Zheng B. BRAD4 plays a critical role in germinal center response by regulating Bcl-6 and NF-kappaB activation. Cell Immunol 2015; 294: 1–8. [DOI] [PubMed] [Google Scholar]
- Belkina AC, Blanton WP, Nikolajczyk BS, Denis GV. The double bromodomain protein Brd2 promotes B cell expansion and mitogenesis. J Leukoc Biol 2014; 95: 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan SV, Bhadury J, Nilsson LM, Green LC, McLure KG, Nilsson JA. BET bromodomain inhibitors synergize with ATR inhibitors to induce DNA damage, apoptosis, senescence-associated secretory pathway and ER stress in Myc-induced lymphoma cells. Oncogene 2016, e-pub ahead of print 25 January 2016 doi:10.1038/onc.2015.521. [DOI] [PubMed]
- Rassart E, Shang M, Boie Y, Jolicoeur P. Studies on emerging radiation leukemia virus variants in C57BL/Ka mice. J Virol 1986; 58: 96–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R, Trubetskoy A, Suzuki T, Jenkins NA, Copeland NG, Lenz J. Genome-based identification of cancer genes by proviral tagging in mouse retrovirus-induced T-cell lymphomas. J Virol 2003; 77: 2056–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund AH, Turner G, Trubetskoy A, Verhoeven E, Wientjens E, Hulsman D et al. Genome-wide retroviral insertional tagging of genes involved in cancer in Cdkn2a-deficient mice. Nat Genet 2002; 32: 160–165. [DOI] [PubMed] [Google Scholar]
- Sorensen AB, Duch M, Amtoft HW, Jorgensen P, Pedersen FS. Sequence tags of provirus integration sites in DNAs of tumors induced by the murine retrovirus SL3-3. J Virol 1996; 70: 4063–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Minehata K, Akagi K, Jenkins NA, Copeland NG. Tumor suppressor gene identification using retroviral insertional mutagenesis in Blm-deficient mice. EMBO J 2006; 25: 3422–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HC, Martins CP, Bronkhorst Y, Randel E, Berns A, Fero M et al. Identification of oncogenes collaborating with p27Kip1 loss by insertional mutagenesis and high-throughput insertion site analysis. Proc Natl Acad Sci USA 2002; 99: 11293–11298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkers H, Allen J, Knipscheer P, Romeijn L, Hart A, Vink E et al. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Genet 2002; 32: 153–159. [DOI] [PubMed] [Google Scholar]
- Selten G, Cuypers HT, Boelens W, Robanus-Maandag E, Verbeek J, Domen J et al. The primary structure of the putative oncogene pim-1 shows extensive homology with protein kinases. Cell 1986; 46: 603–611. [DOI] [PubMed] [Google Scholar]