Abstract
AIM
To screen clinically relevant microRNAs (miRNAs) silenced by DNA methylation in human hepatocellular carcinoma (HCC).
METHODS
Knockdown of DNA methyltransferases (DNMTs) using siRNAs and miRNA profiling in HCC cell lines were performed to identify DNA hypermethylation-mediated miRNA downregulation. Confirmation using individual quantitative real-time PCR (qRT-PCR) assays was then performed followed by DNA methylation quantification at the promoter of the miRNA genes. Quantification of DNA methylation and miRNA expression was then performed in primary HCC tumor samples and related with clinicopathological variables.
RESULTS
miRNA profiling after DNMT knockdown in HCC cell lines revealed upregulation of miR-23, miR-25 and miR-183. After qRT-PCR confirmation and CpG island methylation quantification of these miRNAs in cell lines, further analysis in primary HCC specimens showed that hsa-miR-183 is hypermethylated in 30% of HCC (n = 40). Expression of mature miR-183 showed an inverse correlation with DNA methylation levels. In HCC cells, DNMT knockdown and 5-aza-2'-deoxycytidine treatment reduced methylation and stimulated expression of miR-183. In HCC patients, hypermethylation at hsa-miR-183 promoter significantly correlates with poor survival (log-rank test P = 0.03). DNA methylation analysis in healthy liver, benign liver tumors (hepatocellular adenoma and focal nodular hyperplasia) and their corresponding adjacent tissues showed absence of hypermethylation supporting the notion that aberrant methylation at hsa-miR-183 is specific for the malignant transformation of hepatocytes.
CONCLUSION
Our data indicate that hypermethylation of hsa-miR-183 is a frequent event in HCC and potentially useful as a novel surrogate diagnostic and prognostic marker.
Keywords: hsa-miR-183, DNA methylation, DNMT knockdown, microRNA microarray
Core tip: A comprehensive screening using microRNA microarray in hepatocellular carcinoma (HCC) cells after DNMT1-, DNMT3A-, and/or DNMT3B-knockdown revealed upregulation of miR-23, miR-25, and miR-183. Using primary HCC tumor tissues, we confirmed frequent DNA hypermethylation at the hsa-mir-183 promoter. Hypermethylation of hsa-miR-183 was not found in benign liver tumors, adjacent tumor tissues as well as healthy livers and significantly correlated with poor prognosis. Therefore it represents a potential novel diagnostic and prognostic marker in HCC.
INTRODUCTION
A class of small non-coding RNAs with the ability to negatively regulate gene expression, named microRNA (miRNA), has been revealed to greatly contribute to tumor development and progression since the last decade[1]. Rather than completely blocking expression of the target genes, miRNAs act specifically in post-transcriptional modulation by fine-tuning of gene expression[2]. Differential expression of miRNAs has been described in almost all tumor types thus also eliciting new opportunities to utilize miRNA as a potential diagnostic or prognostic marker. Depending on their functions, miRNAs are either up- or down-regulated in cancer. After the first report revealing inactivation of miRNA genes by DNA methylation[3], epigenetically mediated silencing of various miRNAs have been reported across different types of human cancers, including human hepatocellular carcinoma[4,5].
DNA methylation aberrations have been shown to contribute to the early steps of malignant transformation[6]. Both miRNA expression and DNA methylation patterns display tissue specificity. Therefore, cancer profiling using those two features have successfully discriminated tumor from healthy tissues and also classified subtypes of tumors with a notable clinical relevance[7].
HCC is among the top five major types of cancer with more than 750000 new cases diagnosed each year and accounts for the third highest cancer mortality rate worldwide[8,9]. However, knowledge about epigenetic aberrations of miRNA genes in HCC is still surprisingly limited. Only few studies have addressed DNA methylation-mediated miRNA silencing in HCC[5,10]. Using in silico and in vitro screening, our previous study has shown frequent DNA methylation aberrations in intergenic miRNA genes and its potential value for specific detection and as a new marker for poor survival in HCC[4]. In the human genome, around 60% miRNAs are intergenic, located far away from any other known genes. The remaining 40% of miRNAs are located within introns of protein-encoding genes. Transcription of these intragenic miRNAs is likely to be regulated in the same direction as the host genes[11]. Some intragenic miRNAs have also been reported to be epigenetically inactivated in HCC[12].
To complement our previous studies, we used a comprehensive experimental screening approach to identify yet unknown hypermethylation mediated miRNA silencing in HCC. Specific DNA methyltransferase (DNMT) knockdown followed by miRNA expression profiling in HCC cell lines was performed. Upregulated miRNAs after DNMT knockdown were selected according to the presence of CpG islands in the 5 kb distance to avoid indirect and unspecific effects of DNMT knockdown. Following this strategy we identified hsa-miR-183 as a new target of aberrant DNA methylation in primary HCC specimens.
MATERIALS AND METHODS
Patient samples and cell lines
Primary tumor specimens were collected at the time of surgery from 40 patients with HCCs, 10 with hepatocellular adenoma (HCA), and 5 with focal nodular hyperplasia (FNH) operated at the Hannover Medical School Germany following a protocol approved by the local ethics committee ("Ethik-Kommission der Medizinischen Hochschule Hannover", head: Prof. Dr. Tröger). After snap froze in liquid nitrogen, the primary tissues were stored at -80 °C until the time of analysis. Clinical specimens were assessed by two independent pathologists following universally accepted criteria for HCC, HCA, and FNH. Morphologic classification of liver tumors and grading of hepatocellular carcinoma were performed according to recommendations as previously described by Lehmann et al[13] and Schlageter et al[14]. Tumor cell content was verified by the pathologists to be at least 70% using HE staining from the reference sections of the snap frozen samples. Basic clinicopathological variables of the patient samples are summarized in Table 1. Seven HCC cell lines (HLE, HLF, HuH7, HepG2, Hep3B, SNU182, and SNU387) and two immortalized hepatocyte lines (THLE-2 and THLE-3) were obtained from the American Tissue Culture Collection (ATCC, Rockville, MD, United States) and grown in conditions recommended by ATCC. Identity of all cell lines was validated using short tandem repeat (STR) profiling following the DSMZ's protocol. All experiments using cell lines were performed at sub-confluent cellular density allowing exponential growth.
Table 1.
Clinopathological variables of hepatocellular carcinoma, hepatocellular adenoma, and focal nodular hyperplasia patients involved in this study
| HCC (n = 40) | HCA (n = 10) | FNH (n = 5) | |
| Age | |||
| < 50 | 12 | 10 | 4 |
| > 50 | 28 | 0 | 1 |
| Sex | |||
| Male | 33 | 1 | 2 |
| Female | 7 | 9 | 3 |
| Etiology | |||
| HBV | 8 | ||
| HCV | 4 | ||
| No infection | 28 | ||
| Tumor differentiation | |||
| Good | 15 | ||
| Moderate | 17 | ||
| Poor | 8 | ||
| Tumor size | |||
| < 5 cm | 20 | 4 | 4 |
| > 5 cm | 20 | 6 | 1 |
| Stage | |||
| I | 5 | ||
| II | 11 | ||
| III | 16 | ||
| IV | 8 | ||
| Number of nodules | |||
| Unilocular | 14 | 9 | 4 |
| Multilocular | 26 | 1 | 1 |
| Cirrhosis | |||
| With cirrhosis | 32 | ||
| Without cirrhosis | 8 | ||
| Survival | |||
| < 3 yr | 18 | ||
| > 3 yr | 17 | ||
| No information | 2 | ||
| Diagnosed < 2 yr ago | 3 |
HCC: Hepatocellular carcinoma; HCA: Hepatocellular adenoma; FNH: Focal nodular hyperplasia; HBV: Hepatitis B virus; HCV: Hepatitis C virus.
DNMT1, DNMT3A, and DNMT3B knockdown
DNMT gene knockdown experiments in HLE cells were performed with pre-designed pools of four siRNA targeting DNMT1, DNMT3A, DNMT3B (ON-TARGET plus and siGENOME SMARTpool siRNAs, Dharmacon/Thermo Scientific, London, United Kingdom) following the manufacturer's protocol. In brief, 2 × 104 cells in 500 μl complete medium were seeded in 24-well plate simultaneously with 100 μl of a previously prepared mixture containing 50 or 100 nmol/L siRNA/well, LipofectamineTM RNAiMAX (Invitrogen, Darmstadt, Germany), and Opti-MEM (Gibco-Invitrogen, Darmstadt, Germany) following recommendations from the manufacture. After 24 h medium containing transfection reagent was replaced by new medium and repeated transfection was performed after 48 h from this point onward. After 3 sequential transfections and re-plating into 12- and 6-well plates, cells were harvested for RNA, DNA, and protein extractions. Two scramble siRNAs (AllStars Negative Control siRNA, Qiagen, Hilden, Germany and Riboxx® control-N1, Riboxx, Dresden, Germany) were included in the experiments as negative controls.
Western blot analysis
Fifteen μg of protein lysate was separated in 10% pre-cast SDS-polyacrylamide gels (Bio-RAD, Munich, Germany) and transferred onto Hybrid-P polyvinylidene difluoride membrane (Amersham Biosciences, Freiburg, Germany). Antibodies used were mouse monoclonal anti-DNMT1 antibody (IMG-261A clone 60B1220.1, Imgenex, San Diego, CA, United States), mouse monoclonal anti-β-actin antibody (ab6276 clone AC-15, Abcam, United Kingdom), and anti-mouse secondary antibody HRP (R1253HRP, Acris, Herford, Germany).
DNA and RNA extraction
Extraction of high molecular weight DNA from the fresh-snap-frozen primary specimens was performed by digestion with proteinase K (Merck, Darmstadt, Germany) followed by phenol/chloroform procedure (ROTI® Carl Roth GmbH, Karlsruhe, Germany) following standard procedures. Total RNA was extracted using TRIZOLTM reagent (Invitrogen, Darmstadt, Germany). For miRNA profiling, RNA was extracted using miRNeasy Mini Kit (Qiagen, Hilden, Germany) following the protocol provided by the manufacturer.
miRNA profiling
Comprehensive miRNA expression profiling after DNMT knockdown was performed with Agilent's high-performance miRNA Microarray Platform (Release 19.0, 8x60K) using its set of optimized reagents and hardware following the supplier's instruction as previously described in detail[15].
Reverse transcription and quantitative real-time PCR
Reverse transcription of mature miRNA species was performed using 10 ng RNA per each reaction using High Capacity cDNA Reverse Transcription Kit and TaqMan® miRNA assays (Applied Biosystem, Darmstadt, Germany) following the manufacturer's recommendation (ID numbers of the TaqMan® assays: 002270 for hsa-miR-183-3p and 002269 for hsa-miR-183-5p. For normalization, RNU48 and U6 were used as reference transcripts. Quantitative real-time PCR was performed using TaqMan Universal PCR Master Mix (Applied Biosystem, Darmstadt, Germany) on ABI Prism 7500 Sequence Detection System (Applied Biosystems, Foster City, CA, United States). For mRNA quantification after DNMTs-knockdowns, RNA (1 μg) was reverse-transcribed using High Capacity cDNA Reverse Transcription Kit (Invitrogen, Darmstadt, Germany) according to the manufacturer's recommendation. Hs00154749_m1 for DNMT1, Hs01027166_m1 for DNMT3A, Hs00171876_m1 for DNMT3B and two references Hs00939627_m1 (GUSB) and Hs02758991_g1 (GAPDH) were used for mRNA expression analysis after DNMTs-knockdown. Expression of miRNAs in primary tumor samples was displayed as fold change after normalized with the paired adjacent liver tissues.
Bisulfite conversion and methylation analysis
For DNA methylation analysis, genomic DNA was treated with sodium bisulfite using EZ DNA Methylation KitTM (Zymo Research, HiSS Diagnostics, Freiburg, Germany) according to the manufacturer's recommended protocol. For each PCR amplification, approximately 25 ng of the bisulfite modified DNA was used. DNA methylation analysis was performed with pyrosequencing as initially described[4] using newly designed primers as available at Table 2. DNA methylation level for a given sample was presented as the mean of all CpG dinucleotide methylation values from two independent pyrosequencing runs. The software Pyro-Q-CpG™ (Qiagen, Hilden, Germany) was used for analyzing DNA methylation levels of each individual CpG dinucleotides. "Hypermethylated" was then defined as methylation value above mean of the adjacent liver tissue plus two times the standard deviation (Meanadj. + 2 × StD).
Table 2.
List of primers used in this study
| Pyrosequencing | Forward | Reverse | Ta ( °C) | MgCl2 (mmol/L) | Sequencing |
| miR-23 cluster | TTTAAGTYGTGTGAAATTATGTGGTAG | ATAAACACCRAAAAACRAATCCA | 60 | 2.5 | TGGTAGTTTATGGTTGTGAG |
| miR-25 cluster | GTGYGGGTTAATYGGATAAGG | AAAAACCCRACRCCTACACTAC | 60 | 2.5 | GYGGGTTTAGAATGAGT |
| miR-183 | GTTATTAATAGGAATGGGGTAG | AAACRACTCTCAACCTCCC | 60 | 1.5 | GAATGGGGTAGTTGAGGG |
DAC treatment in HCC cell lines
DAC treatment in HLF, HuH7, and HepG2 cells in final concentration 100 nmol/L for 5 d was performed as initially described[4].
Statistical analysis
Clustering analysis of the miRNA profiling after DNMT knockdown was performed using Qlucore Omics Explorer v2.2 (Qlucore, Lund, Sweden). For statistical analysis, GraphPad Prism (version 5.01 for Windows, La Jolla, CA, United States) was used. The Mann-Whitney-U test was utilized to compare continuous variables and χ2 test for relationships between categorical variables. To compare survival of HCC patients, Kaplan-Meier curve and long-rank (Mantel-Cox) test were used. For those comparisons, P < 0.05 was considered as statistically significant.
RESULTS
Upregulated miRNAs after DNMT knockdown
Upon DNMT-1, -3A, and -3B knockdowns (Supplementary Figure 1), several miRNAs were shown to be upregulated (Figure 1a). To sort out upregulation of miRNAs directly affected by demethylation due to the DNMT knockdown, we further analyzed presence of CpG island association (within 5 kb) at the miRNA gene promoters. After DNMT knockdown, hsa-miR-23 cluster, hsa-mir-25 cluster, and hsa-mir-183 cluster were shown to be upregulated by using miRNA profiling (Figure 1A). To show direct effects of DNMT knockdowns to alterations of DNA methylation and miRNA expression, we used HLE cells upon DNMT1 knockdown and measured methylation of hsa-miR-183 promoter and expression miR-183. DNMT1 knockdown in HLE cells led to decreased methylation at hsa-miR-183 promoter (P = 0.0015) accompanied by elevated miR-183 expression (P = 0.04) (Figure 1B).
Figure 1.
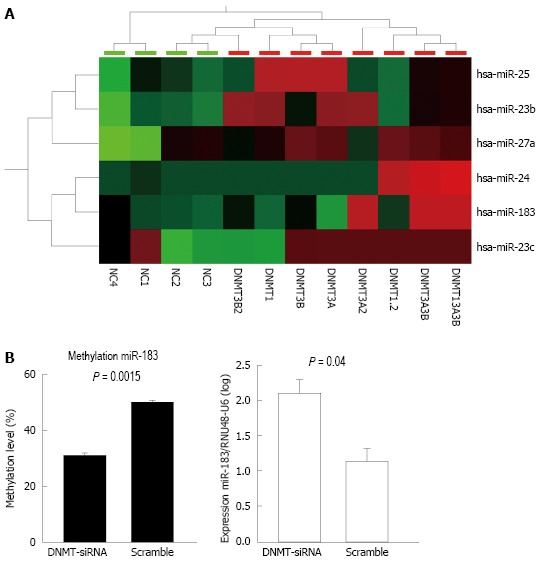
Upregulated microRNAs after DNA methyltransferases knockdown (A); and Methylation and expression changes of hsa-miR-183 after DNA methyltransferases knockdown (B). A: Cluster analysis after single and combined DNMT knockdown using siRNAs (as indicated in the lower panel) shows upregulation of selected microRNAs (right panel). High, intermediate, and low microRNA expression levels in the heatmap are indicated with red, black, and green, respectively. B: Upon DNMT1 knockdown in HLE cell lines, DNA methylation at the promoter of miR-183 decreased significantly (right panel, P = 0.0015) accompanied by elevated miR-183 expression (left panel, P = 0.04). NC = negative control siRNA and DNMT (1, 3A, 3B) = DNA methyl transferase 1, -3A, -3B siRNA treated samples. DNMT: DNA methyltransferase.
Differential DNA methylation in primary HCC specimens
Methylation analysis was performed at promoters of the upregulated miRNAs after DNMT knockdown. First, we performed DNA methylation analysis at the promoter of hsa-miR-23 cluster, hsa-mir-25 cluster, and hsa-mir-183 cluster in 7 HCC cell lines (HLE, HLF, Huh7, HepG2, SNU182, and SNU387) and 2 healthy adult liver epithelial lines (THLE2 and THLE3). Differential DNA methylation was not observed at the promoter of hsa-mir-25 cluster among HCC cell lines and healthy adult liver epithelial lines indicating that the upregulation after DNMT knockdown is likely due to indirect effects. Therefore, further DNA methylation analysis at the promoter of hsa-mir-25 cluster in clinical samples (primary HCC, HCA, FNH tissue samples) was not performed. At the promoter of hsa-miR-23 cluster, although differential methylation was shown in cell lines, we observed constant hypermethylation both in HCC and the adjacent liver tissue (Supplementary Figure 2). We observed differential methylation at hsa-mir-183 transcriptional start site in HCC compared to the corresponding adjacent liver tissues. Hypermethylation was demonstrated in 30% of HCC tumor samples (n = 40) (Figure 2).
Figure 2.
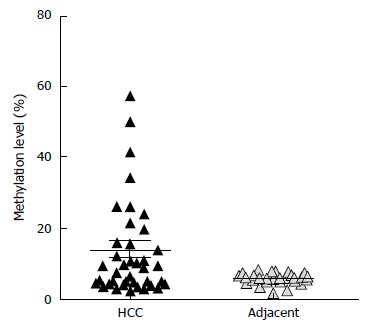
Differential DNA methylation at hsa-mir-183 promoter in primary hepatocellular carcinoma specimens compared to the adjacent liver tissues. High-resolution quantitative DNA methylation analysis using pyrosequencing shows 30% of HCC samples with hypermethylation. "Hypermethylation" was defined as described in Materials and Methods. HCC: Hepatocellular carcinoma.
Deregulated miRNA expression in relation with DNA methylation status
In order to evaluate the functional relevance of differential DNA methylation at hsa-mir-183 locus, we performed expression analysis of mature miR-183 in primary HCC specimens. MiR-183 was frequently downregulated in HCC and its expression negatively correlated with the methylation status (Figure 3).
Figure 3.
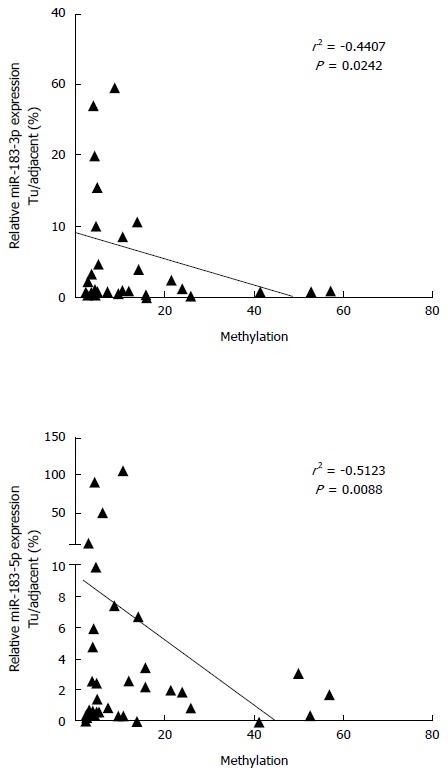
DNA hypermethylation leads to repression of hsas-miR-183 transcription. Shown are scatter plots demonstrating negative correlation between DNA methylation levels and miR-183 expression case by case.
DNA methylation analysis in benign liver tumors and healthy liver tissues
We further analyzed DNA methylation patterns at hsa-mir-183 in benign liver tumor samples. Altogether 10 cases of HCA, 5 cases of FNH, their corresponding adjacent liver tissues, as well as 5 healthy liver tissues were analyzed. Aberrant methylation at hsa-mir-183 was not found in benign liver tumors and healthy liver tissues (Supplementary Figure 3). This indicates that aberrant methylation at hsa-mir-183 might occur only during malignant transformation of hepatocytes.
Impact on HCC survival
HCC patients with hypermethylation at hsa-miR-183 locus had a significant shorter survival (log-rank test, P = 0.03; Figure 4). Age is not significantly different between HCC patients with and without hypermethylation indicating that the gain of methylation at hsa-miR-183 is related to biological course of the disease not merely due to an increase in age. Hypermethylated samples are found more frequently in late HCC although this difference is not statistically significant.
Figure 4.
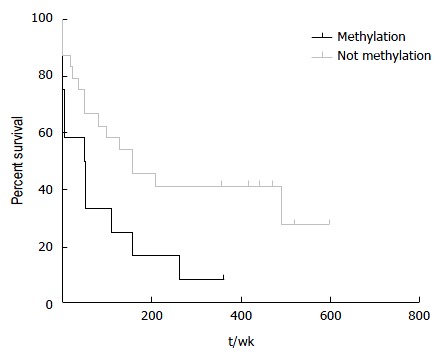
Hypermethylation at hsa-miR-183 significantly correlates with worse survival in hepatocellular carcinoma. The Kaplan-Meier plot shows comparison of HCC patient survival between those with hypermethylated and non-methylated hsa-miR-183 locus (log-rank test P = 0.03, median survivals are 49.5 and 150 wk for patients with hypermethylation and without increased methylation at hsa-miR-183 locus respectively). HCC: Hepatocellular carcinoma.
DISCUSSION
The discovery of miRNA has brought significant improvement in the understanding of regulation of gene expression and highlighted the emerging of a novel class of regulatory molecules in gene transcription[7]. The ability of miRNAs to control gene expression post-transcriptionally reveals a complex and interrelated regulatory network of gene expression. Later, it has been also shown that miRNAs are functionally regulated by epigenetic mechanisms including DNA methylation and histone modifications[3,4,11]. Aberrant DNA methylation and histone modification have been suggested to be an important causal factor for deregulation of miRNA expression, especially in cancer cells.
Liver tumors comprise a range of benign, premalignant, and malignant lesions that differ in histology, etiology, clinical management, and outcome. Aberrations of both DNA methylation and miRNA expression are very specific depending on developmental stage, cell differentiation, and tissue types[5,16,17]. Therefore, using combination of DNA methylation and miRNA expression as clinical relevant markers is potentially of practical relevance in some complex and heterogeneous diseases including liver cancer[5]. Using an in silico screen, our previous study has revealed that aberrant DNA methylation of miRNA genes was a biological event detected only in malignant liver cells and tissues but not in adjacent tumor tissues, benign liver lesions, and in hepatocyte lines[4]. Aberrant DNA methylation at miRNA loci does not occurs at random but appears as highly organized event during the course of hepatocarcinogenesis. Therefore, DNA methylation at miRNA genes has a potential as diagnostic or prognostic marker as well as for guiding alternative adjuvant targeted therapy in HCC because some miRNAs regulated by DNA methylation play a role in modulating therapeutic responses in HCC[5,18,19].
In addition to the in silico screen previously done by us[4], we performed an experimental screen by using siRNA mediated DNMT knockdown to induce re-expression of miRNA expression silenced by DNA methylation. Our experimental screen revealed that hsa-mir-183 is upregulated after DNMT knockdown. Using primary HCC specimens, aberrant DNA methylation is detected in up to 30% of total samples. DNMT1 knockdown leads to significant reduction of DNA methylation at the promoter and increased expression of mature miR-183. Downregulation of miR-183 has been previously described in several cancers such as lung cancer[20], breast cancer[21], or osteosarcoma[22]. Upregulation of miR-183 has also been reported in liver cancer precursors including cirrhotic and pre-malignant lesions[23]. The discrepancy of miR-183 expression in cirrhosis, premalignant liver lesions, and liver cancer might reflect the dynamic evolution during carcinogenesis since miRNAs modulate several mRNAs and transcription factors. Mature miR-183 expression in HCC can be activated by β-catenin through elevated synthesis of polycistronic transcripts of the hsa-miR-182-96-182 cluster[24]. Therefore, in the presence of CTNNB1 mutations, levels of miR-183 expression might not show the expected downregulation in HCC due to complex interactions of this genetic alteration with promoter DNA methylation. In addition, upregulation of miR-183 was not correlated with survival of HCC patients[25] indicating that expression alone is not a useful prognostic marker in HCC.
Our present study showed frequent aberrant DNA methylation at the promoter of hsa-miR-183. Hsa-mir-183 itself is an intergenic miRNA and located at chromosome 7 in a cluster together with hsa-miR-96 and hsa-miR-182. The putative transcriptional start site is predicted at 5207 bp upstream of hsa-mir-183 within a CpG island adjacent to this cluster (CpG 351). The pyrosequencing assay for the detection of DNA methylation used in this study was developed for this CpG island. Aberrant methylation of hsa-miR-183 was absent in benign liver lesions (HCAs, FNHs), adjacent liver tissues (from HCC, HCA, and FNH), as well as healthy liver tissues. This indicates that hypermethylation of hsa-miR-183 is a potential diagnostic marker to differentiate malignant liver tumor from benign lesions especially in cases where relying on histopathology alone might be difficult, for example in distinguishing HCA from well-differentiated HCC. Although involving limited numbers of HCC patients, our present study also shows that aberrant DNA methylation at hsa-miR-183 promoter is associated with poor HCC outcome. Hypermethylation of hsa-miR-183 might not be able to replace other prognostic factors commonly used to predict HCC survival, but offer an additional relevant marker to determine HCC prognosis.
COMMENTS
Background
Hepatocellular carcinoma (HCC) is a leading cause of cancer mortality with limited treatment options, mainly due to late detection of disease. Epigenetic alterations like DNA methylation are now recognized as important contributors to the development and progression of malignancy in humans. However, our knowledge about epigenetic alterations in HCC and their clinical relevance is still limited.
Research frontiers
Determining the clinical relevance of DNA methylation aberrations in microRNA genes and exploring the potential as new biomarker of this epigenetic aberration.
Innovations and breakthroughs
Comprehensive microRNA expression screen after DNA methyltransferase (DNMT) knockdown, avoiding the unspecific side effects of "classical epigenetic screens" using nucleotide analogues like azacytidine or aza-deoxycytidine as DNMT inhibitors.
Applications
hsa-miR-183 gene methylation as new prognostic marker for the clinical management of HCC patients.
Peer-review
In this manuscript, Anwar et al, described the status of miR-183 methylation and related to poor survival in human HCC. In general, this is an interesting manuscript, explaining epigenetic regulation of miR-183 in HCC and its potential role as a novel surrogate and prognostic marker.
Footnotes
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: Germany
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
Institutional review board statement: Primary tumor specimens were collected at the time of surgery as leftover from diagnostics at the Hannover Medical School Germany following protocol approved by the local ethics committee ("Ethik-Kommission der Medizinischen Hochschule Hannover", head: Prof. Dr. Tröger).
Conflict-of-interest statement: all authors declared no conflict of interest.
Data sharing statement: No additional data are available.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: October 10, 2016
First decision: October 20, 2016
Article in press: February 8, 2017
P- Reviewer: Ezzat WM, Ray RB S- Editor: Gong ZM L- Editor: A E- Editor: Liu WX
References
- 1.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kunej T, Godnic I, Ferdin J, Horvat S, Dovc P, Calin GA. Epigenetic regulation of microRNAs in cancer: an integrated review of literature. Mutat Res. 2011;717:77–84. doi: 10.1016/j.mrfmmm.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 4.Anwar SL, Albat C, Krech T, Hasemeier B, Schipper E, Schweitzer N, Vogel A, Kreipe H, Lehmann U. Concordant hypermethylation of intergenic microRNA genes in human hepatocellular carcinoma as new diagnostic and prognostic marker. Int J Cancer. 2013;133:660–670. doi: 10.1002/ijc.28068. [DOI] [PubMed] [Google Scholar]
- 5.Anwar SL, Lehmann U. DNA methylation, microRNAs, and their crosstalk as potential biomarkers in hepatocellular carcinoma. World J Gastroenterol. 2014;20:7894–7913. doi: 10.3748/wjg.v20.i24.7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kunej T, Godnic I, Horvat S, Zorc M, Calin GA. Cross talk between microRNA and coding cancer genes. Cancer J. 2012;18:223–231. doi: 10.1097/PPO.0b013e318258b771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 8.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 9.Gomaa AI, Khan SA, Toledano MB, Waked I, Taylor-Robinson SD. Hepatocellular carcinoma: epidemiology, risk factors and pathogenesis. World J Gastroenterol. 2008;14:4300–4308. doi: 10.3748/wjg.14.4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anwar SL, Lehmann U. MicroRNAs: Emerging Novel Clinical Biomarkers for Hepatocellular Carcinomas. J Clin Med. 2015;4:1631–1650. doi: 10.3390/jcm4081631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kozaki K, Inazawa J. Tumor-suppressive microRNA silenced by tumor-specific DNA hypermethylation in cancer cells. Cancer Sci. 2012;103:837–845. doi: 10.1111/j.1349-7006.2012.02236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen J, Wang S, Zhang YJ, Kappil MA, Chen Wu H, Kibriya MG, Wang Q, Jasmine F, Ahsan H, Lee PH, et al. Genome-wide aberrant DNA methylation of microRNA host genes in hepatocellular carcinoma. Epigenetics. 2012;7:1230–1237. doi: 10.4161/epi.22140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lehmann U, Berg-Ribbe I, Wingen LU, Brakensiek K, Becker T, Klempnauer J, Schlegelberger B, Kreipe H, Flemming P. Distinct methylation patterns of benign and malignant liver tumors revealed by quantitative methylation profiling. Clin Cancer Res. 2005;11:3654–3660. doi: 10.1158/1078-0432.CCR-04-2462. [DOI] [PubMed] [Google Scholar]
- 14.Schlageter M, Terracciano LM, D'Angelo S, Sorrentino P. Histopathology of hepatocellular carcinoma. World J Gastroenterol. 2014;20:15955–15964. doi: 10.3748/wjg.v20.i43.15955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buurman R, Gürlevik E, Schäffer V, Eilers M, Sandbothe M, Kreipe H, Wilkens L, Schlegelberger B, Kühnel F, Skawran B. Histone deacetylases activate hepatocyte growth factor signaling by repressing microRNA-449 in hepatocellular carcinoma cells. Gastroenterology. 2012;143:811–820.e1-15. doi: 10.1053/j.gastro.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 16.Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, Qin L, Wu X, Zheng Y, Yang Y, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232–243. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 17.Perret C. Methylation profile as a new tool for classification of hepatocellular carcinoma. J Hepatol. 2011;54:602–603. doi: 10.1016/j.jhep.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 18.Zhou C, Liu J, Li Y, Liu L, Zhang X, Ma CY, Hua SC, Yang M, Yuan Q. microRNA-1274a, a modulator of sorafenib induced a disintegrin and metalloproteinase 9 (ADAM9) down-regulation in hepatocellular carcinoma. FEBS Lett. 2011;585:1828–1834. doi: 10.1016/j.febslet.2011.04.040. [DOI] [PubMed] [Google Scholar]
- 19.Xu Y, Xia F, Ma L, Shan J, Shen J, Yang Z, Liu J, Cui Y, Bian X, Bie P, et al. MicroRNA-122 sensitizes HCC cancer cells to adriamycin and vincristine through modulating expression of MDR and inducing cell cycle arrest. Cancer Lett. 2011;310:160–169. doi: 10.1016/j.canlet.2011.06.027. [DOI] [PubMed] [Google Scholar]
- 20.Wang G, Mao W, Zheng S. MicroRNA-183 regulates Ezrin expression in lung cancer cells. FEBS Lett. 2008;582:3663–3668. doi: 10.1016/j.febslet.2008.09.051. [DOI] [PubMed] [Google Scholar]
- 21.Lowery AJ, Miller N, Dwyer RM, Kerin MJ. Dysregulated miR-183 inhibits migration in breast cancer cells. BMC Cancer. 2010;10:502. doi: 10.1186/1471-2407-10-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu J, Feng Y, Ke Z, Yang Z, Zhou J, Huang X, Wang L. Down-regulation of miR-183 promotes migration and invasion of osteosarcoma by targeting Ezrin. Am J Pathol. 2012;180:2440–2451. doi: 10.1016/j.ajpath.2012.02.023. [DOI] [PubMed] [Google Scholar]
- 23.Goeppert B, Schmezer P, Dutruel C, Oakes C, Renner M, Breinig M, Warth A, Vogel MN, Mittelbronn M, Mehrabi A, et al. Down-regulation of tumor suppressor A kinase anchor protein 12 in human hepatocarcinogenesis by epigenetic mechanisms. Hepatology. 2010;52:2023–2033. doi: 10.1002/hep.23939. [DOI] [PubMed] [Google Scholar]
- 24.Leung WK, He M, Chan AW, Law PT, Wong N. Wnt/β-Catenin activates MiR-183/96/182 expression in hepatocellular carcinoma that promotes cell invasion. Cancer Lett. 2015;362:97–105. doi: 10.1016/j.canlet.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 25.Liang Z, Gao Y, Shi W, Zhai D, Li S, Jing L, Guo H, Liu T, Wang Y, Du Z. Expression and significance of microRNA-183 in hepatocellular carcinoma. ScientificWorldJournal. 2013;2013:381874. doi: 10.1155/2013/381874. [DOI] [PMC free article] [PubMed] [Google Scholar]


