Abstract
Cancer treatment is becoming more and more individually based as a result of the large inter-individual differences that exist in treatment outcome and toxicity when patients are treated using population-based drug doses. Polymorphisms in genes encoding drug-metabolizing enzymes and transporters can significantly influence uptake, metabolism, and elimination of anticancer drugs. As a result, the altered pharmacokinetics can greatly influence drug efficacy and toxicity. Pharmacogenetic screening and/or drug-specific phenotyping of cancer patients eligible for treatment with chemotherapeutic drugs, prior to the start of anticancer treatment, can identify patients with tumors that are likely to be responsive or resistant to the proposed drugs. Similarly, the identification of patients with an increased risk of developing toxicity would allow either dose adaptation or the application of other targeted therapies. This review focuses on the role of genetic polymorphisms significantly altering the pharmacokinetics of anticancer drugs. Polymorphisms in DPYD, TPMT, and UGT1A1 have been described that have a major impact on the pharmacokinetics of 5-fluorouracil, mercaptopurine, and irinotecan, respectively. For other drugs, however, the association of polymorphisms with pharmacokinetics is less clear. To date, the influence of genetic variations on the pharmacokinetics of the increasingly used monoclonal antibodies has hardly been investigated. Some studies indicate that genes encoding the Fcγ-receptor family are of interest, but more research is needed to establish if screening before the start of therapy is beneficial. Considering the profound impact of polymorphisms in drug transporters and drug-metabolizing enzymes on the pharmacokinetics of chemotherapeutic drugs and hence, their toxicity and efficacy, pharmacogenetic and pharmacokinetic profiling should become the standard of care.
Key Points
| Genetic mutations in genes can affect the pharmacokinetics of drugs. |
| Altered metabolism of drugs can result in a decreased therapeutic response and increased toxicity. |
| Personalized medicine requires detailed analyses of the patient’s genome and phenotypic consequences. |
Introduction
Cancer treatment is becoming more and more individually based as a result of the large inter-individual differences in treatment outcome and toxicity. Factors responsible for inter-individual variability in pharmacokinetics and pharmacodynamics include drug–drug interactions, ethnicity, age, renal and liver function, comorbidities, nutritional status, smoking, and alcohol consumption. However, genetic factors may have an even greater impact on drug efficacy and toxicity [1]. In oncology, genetic variations can be found either in the tumor genome as somatic mutations, influencing the choice of chemotherapeutic treatment or as germline mutations, potentially altering individual drug pharmacology [2].
Pharmacogenetics is the study of the inherited basis of inter-individual differences in the efficacy and toxicity of drugs. Pharmacogenetic screening and/or drug-specific phenotyping of cancer patients eligible for treatment with chemotherapeutic drugs, prior to the start of anticancer treatment, can identify patients with tumors that are likely to be responsive or resistant to the proposed drugs. Patients with an unfavorable clinical or genetic make-up would be candidates for alternative treatment modalities. Similarly, the identification of patients with an increased risk of developing toxicity would allow either dose adaptation or the application of other targeted therapies. Polymorphisms in the human genome, affecting either expression or functionality of enzymes and transporters involved in the distribution and metabolism of anticancer drugs, can influence drug efficacy and toxicity and thereby the treatment outcome of patients.
The metabolism of xenobiotics is often divided into three phases: modification (phase I), conjugation (phase II), and elimination (most often in urine or bile). Phase I drug-metabolizing enzymes, especially members of the cytochrome P450 (CYP) family, are responsible for oxidation, reduction, and hydrolysis of drugs [3]. Phase II drug-metabolizing enzymes, such as glutathione S-transferases (GSTs) and uridine diphosphate glucuronosyltransferases (UGTs), mainly inactivate or activate drugs by conjugation reactions [4]. Polymorphisms in these enzymes have frequently been described to influence the pharmacokinetics of several anticancer drugs. Genes encoding key enzymes in the anabolic and catabolic pathway of purine and pyrimidine analogs, such as thiopurine S-methyltransferase (TPMT) and dihydropyrimidine dehydrogenase (DPD), are known to contain many polymorphisms and functional mutations affecting the enzymatic activity [5, 6]. Polymorphisms in genes encoding drug efflux transporters, such as P-glycoprotein (P-gp) and breast cancer resistance protein (BCRP), can greatly influence gastrointestinal uptake and excretion of anticancer drugs [7]. Monoclonal antibodies (mAbs) are increasingly being used in the treatment of cancer. However, limited information is known about the influence of genetic variations on the pharmacokinetics of mAbs [8].
This review focuses on the role of genetic polymorphisms in altering the pharmacokinetics of anticancer drugs. Table 1 provides an overview of the currently used anticancer drugs, their metabolic pathways, and if a genetic polymorphism significantly alters its pharmacokinetics.Anaplastic lymphoma kinase
Table 1.
Overview of the currently used anticancer drugs, their metabolic pathways, and if genetic polymorphisms significantly alter their pharmacokinetics
| Anti-cancer drug | Target | Metabolizing enzymes | Transporters/receptors | Polymorphism-altering pharmacokinetics |
|---|---|---|---|---|
| Alkylating agents | ||||
| Bendamustine | DNA | CYP1A2 | No, mainly non-enzymatic | |
| Busulfan | DNA | CYP2C9, CYP2B6, GSTs | Yes [159, 160] | |
| Carmustine | DNA | Unknown | ||
| Chlorambucil | DNA | GSTs | Yes [161] | |
| Cyclophosphamide | DNA | CYP3A4, CYP3A5, CYP2B6, CYP2C19, GSTs | Yes [32, 162] | |
| Dacarbazine | DNA | CYP1A1, CYP1A2, CYP2E1 | Unknown | |
| Estramustine | DNA | Unknown | ||
| Hydroxycarbamide | DNA | Unknown | ||
| Ifosfamide | DNA | CYP2A6, CYP2B1, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP3A4, CYP3A5, GSTs | Yes [9, 162] | |
| Lomustine | DNA | Unknown | ||
| Mechlorethamine | DNA | Unknown | ||
| Melphalan | DNA | LAT1, LAT2 | No [163] | |
| Temozolomide | DNA | No, mainly non-enzymatic | ||
| Procarbazine | DNA | CYP2B6, CYP1A4, CYP3A5 | Unknown | |
| Thiotepa | DNA | CYP3A4,CYP3A5 CYP2B6 | Yes [9] | |
| Treosulfan | DNA | No, mainly non-enzymatic [160] | ||
| Antimetabolites | ||||
| Azacitidine | DNA/RNA | CDA | Yes [164] | |
| Capecitabine | DNA/RNA | DPD | Yes [70] | |
| Cladribine | DNA/RNA | dCK | Unknown | |
| Clofarabine | DNA/RNA | dCK | Yes [92, 93] | |
| Cytarabine | DNA/RNA | dCK | MDR1 | Yes [86] |
| Decitabine | DNA/RNA | CDA, dCK | Yes [90, 95] | |
| Fludarabine | DNA/RNA | dCK | Yes [91] | |
| Fluorouracil | DNA/RNA | DPD, GSTs | Yes [49] | |
| Gemcitabine | DNA/RNA | CDA, dCK | Yes [87–89] | |
| Mercaptopurine | DNA/RNA | TPMT | Yes [72, 73] | |
| Methotrexate | DNA/RNA | MTHFR | SLC, MDR1 | Yes [71, 81–84] |
| Nelarabine | DNA/RNA | Unknown | ||
| Pemetrexed | DNA/RNA | Unknown | ||
| Tegafur | DNA/RNA | DPD, CYP2A6, CYP2C8, CYP1A2 | Yes [57] | |
| Tioguanine | DNA/RNA | TPMT | Yes [165] | |
| Anti-mitotic cytostatics | ||||
| Cabazitaxel | Microtubule | CYP3A4 | Unknown | |
| Docetaxel | Microtubule | CYP1B1, CYP2B6, CYP3A4, CYP3A5 | MDR1, BCRP | Yes [98, 105] |
| Paclitaxel | Microtubule | CYP2C8, CYP3A4, CYP3A5 | MDR1, BCRP | Yes [98, 105] |
| Vinblastine | Microtubule | CYP3A4,CYP3A5, GSTs | Unknown | |
| Vincristine | Microtubule | CYP3A4,CYP3A5, GSTs | MDR1 | No [166] |
| Vinorelbine | Microtubule | CYP2D6, CYP2E1, CYP3A4,CYP3A5, GSTs | Unknown | |
| Anti-tumor antibacterials | ||||
| Bleomycin | DNA/RNA | BLMH, GSTs | Yes [167] | |
| Dactinomycin | DNA/RNA | GSTs | MDR1 | No [168] |
| Daunorubicin | DNA | GSTs | MDR1 | No [108] |
| Doxorubicin | DNA | CYP2B6, CYP3A4, CYP3A5, CYP2D6, GSTs, UGTs | MDR1, BCRP | Yes [106] |
| Epirubicin | DNA | UGTs | MDR1, SLC | No [107] |
| Idarubicin | DNA | CYP2D6, CYP2C9, GSTs | MDR1 | Unknown |
| Mitomycin | DNA | GSTs | Unknown | |
| Mitoxantrone | DNA | CYP1B1, CYP3A4, CYP3A5, GSTs | MDR1 | Unknown |
| Topoisomerase inhibitors | ||||
| Etoposide | Topoisomerase | CYP1A2, CYP2E1, CYP3A4, CYP3A5, GSTs, UGTs | MDR1 | Yes [98] |
| Irinotecan | Topoisomerase | CYP3A4, CYP3A5, UGTs | MDR1, BCRP | Yes [169] |
| Teniposide | Topoisomerase | CYP3A4, CYP3A5, UGTs | Unknown | |
| Topotecan | Topoisomerase | CYP3A4, CYP3A5, UGTs | BCRP | No [170] |
| Anti-hormones | ||||
| Abiraterone | Androgen receptor | Unknown | ||
| Anastrozole | Aromatase | CYP3A4, CYP3A5, CYP2C8, CYP19A1, UGTs | Yes [171] | |
| Bicalutamide | Androgen receptor UGTs | MDR1, BCRP | Yes [113] | |
| Enzalutamide | Androgen receptor | CYP2C8, CYP3A4, CYP3A5 | Unknown | |
| Exemestane | Aromatase | CYP3A4, CYP3A5, CYP4A11, CYP1A2, CYP19A1, UGTs | Yes [172] | |
| Flutamide | Aromatase | CYP1A2 | No [9] | |
| Letrozole | Aromatase | CYP3A4, CYP3A5, CYP2A6, CYP19A1 | Yes [172] | |
| Megestrol | Estrogen receptor | Unknown | ||
| Nilutamide | Androgen receptor | Unknown | ||
| Tamoxifen | Estrogen receptor | CYP2D6, CYP3A5, CYP3A4, CYP2C9, CYP2C19, CYP1B1, UGTs | Yes [173] | |
| Fulvestrant | Estrogen receptor | CYP3A4, CYP3A5 | No, mainly non-enzymatic | |
| Tyrosine kinase inhibitors | ||||
| Afatinib | EGFR | No, mainly non-enzymatic | ||
| Axitinib | VEGF-R 1-3 | CYP3A4, CYP1A2, CYP2C19, UGTs | MDR1 | No [125] |
| Bosutinib | BCR-ABL/SRc | CYP3A4 | MDR1 | No [115] |
| Crizotinib | ALK | CYP3A4, CYP3A5 | Unknown | |
| Dabrafenib | BRAF | CYP2C8, CYP3A4 | Unknown | |
| Dasatinib | BCR-ABL | CYP3A4 | MDR1, BCRP | No [115] |
| Erlotinib | EGFR | |||
| Gefetinib | EGFR | CYP3A4, CYP3A5, CYP2D6 | MDR1, BCRP | No [28] |
| Imatinib | BCR-ABL | CYP3A4, CYP3A5, CYP2C8 | MDR1, BCRP, SLC | Yes [118, 119] |
| Lapatinib | HER-2 | CYP3A4, CYP3A5, CYP2C19, CYP2C8 | Unknown | |
| Nilotinib | BCR-ABL | CYP3A4, CYP2C8 | BCRP, SLC | No [115] |
| Olaparib | PARP | CYP3A4 | Unknown | |
| Pazopanib | Multi | CYP3A4, CYP1A2, CYP2C8 | Unknown | |
| Ponatinib | BCR-ABL | CYP3A4 | No [115] | |
| Regorafenib | Multi | CYP3A4, UGTs | Unknown | |
| Ruxolitinib | JAK | CYP3A4, CYP2C9 | Unknown | |
| Sorafenib | Multi | CYP3A4, UGTs | BCRP | Yes [126] |
| Sunitinib | Multi | CYP3A4, CYP3A5 | MDR1, BCRP | Yes [174] |
| Vandetanib | Multi | CYP3A4 | Unknown | |
| Vemurafenib | BRAF | CYP3A4 | No, mainly non-enzymatic | |
| Biologicals | ||||
| Bevacizumab | VEGF | Unknown | ||
| Brentuximab | CD30 | CYP3A4, CYP2D6 | Unknown | |
| Cetuximab | EGFR | FCGRT | Yes [145–148] | |
| Ipilimumab | CTLA-4 | Unknown | ||
| Nivolumab | PD-1 | Unknown | ||
| Ofatumumab | CD20 | Unknown | ||
| Panitumumab | EGFR | Unknown | ||
| Pembrolizumab | PD-1 | Unknown | ||
| Pertuzumab | HER-2 | Unknown | ||
| Rituximab | CD20 | FCGRT | Yes [138–142] | |
| Trastuzumab | HER-2 | FCGRT | Yes [143, 144] | |
| Immunomodulants | ||||
| Lenalidomide | Bone marrow | No, mainly non-enzymatic | ||
| Pomalidomide | Bone marrow | CYP1A2, CYP3A4, CYP2C19, CYP2D6 | Unknown | |
| Thalidomide | Bone marrow | CYP2C19 | Yes [175] | |
| Non-categorized | ||||
| Asparaginase (PEG) | l-Asparagine | No [71] | ||
| Bortezomib | Proteasome | CYP3A4, CYP2C19, CYP1A2 | No [176] | |
| Carboplatin | DNA | GSTs | Yes [162] | |
| Cisplatin | DNA | CYP2E1, CYP3A4, CYP3A5, GSTs | Yes [177] | |
| Oxaliplatin | DNA | GSTs | Yes [98] | |
| Temsirolimus | mTOR | CYP3A4 | Unknown | |
| Trabectedin | DNA | CYP3A4, (CYP2C19, CYP2C9, CYP2D6, CYP2E1) | Unknown | |
The enzymes and transporters for which genetic polymorphisms are known to significantly alter the pharmacokinetics are indicated in bold
ALK anaplastic lymphoma kinase, BCR-ABL/SRc breakpoint cluster region protein-Abelson murine leukemia viral oncogene homolog/proto-oncogene tyrosine-protein kinase src, BRAF serine/threonine-protein kinase B-Raf, CD20 cluster of differentiation 20, CD30 cluster of differentiation 30, CTLA-4 cytotoxic T-lymphocyte-associated protein 4, CYP cytochrome P450, EGFR epidermal growth factor receptor, GSTs glutathione, HER-2 human epidermal growth factor receptor 2, JAK janus kinase, mTOR mammalian target of rapamycin, multi various tyrosine kinases, PARP Poly (ADP-ribose) polymerase, PD-1 Programmed cell death protein, S-transferases, UGT uridine diphosphate glucuronosyltransferase, VEGF vascular endothelial growth factor, VEGFR 1-3 vascular endothelial growth factor subtypes 1-3
Cytochrome P450 (CYP)-Mediated Phase I-Metabolizing Enzymes
Phase I reactions are catalyzed by CYP enzymes, a large superfamily of membrane-bound proteins, located predominantly in the endoplasmatic reticulum. The CYP1, CYP2, and CYP3 families are most frequently involved in drug metabolism (Table 2). Several factors may cause inter-individual variations in CYP450 activity: genetic polymorphisms, changes in physiological conditions such as age, sex, and disease, or environmental factors such as smoking, drugs, and certain foods.
Table 2.
Polymorphisms in phase I and phase II metabolic enzymes affecting pharmacokinetics of anticancer drugs
| Drugs | Gene | Mutations | dbSNP ID | ESP MAF | ExAC MAF | PK parameters | |
|---|---|---|---|---|---|---|---|
| African American | European American | ||||||
| Erlotinib | CYP1A2 | c.1042+43G>A | rs2472304 | 0.13 | 0.56 | 0.46 | Plasma concentrations [11] |
| Tegafur | CYP2A6 | CYP2A6*1A | Increased 5-FU formation [178] | ||||
| CYP2A6*4A | del | Decreased 5-FU formation [178] | |||||
| CYP2A6*4A-H | del | CL, AUC [13, 14] | |||||
| CYP2A6*7 (c.1412T>C) | rs5031016 | nr | 9 × 10−4 | 1.1 × 10−2 | CL, AUC [13, 14] | ||
| CYP2A6*9 (c.-48T>G) | rs28399433 | 0.08 | 0.06 | 0.10 | CL, AUC [13, 14, 178] | ||
| Cyclofosfamide | CYP2B6 | CYP2B6*6 | CL [15–18, 32] | ||||
| (c.516G>T) | rs3745274 | 0.37 | 0.25 | 0.27 | |||
| (c.785A>G) | rs2279343 | nr | nr | 0.06 | |||
| Busulfan | CYP2C9 | CYP2C9*2 (c.430C>T) | rs1799853 | 0.03 | 0.13 | 0.09 | Decreased CL [19] |
| Cyclophosphamide | CYP2C19 | CYP2C19*2 (c.681G>A) | rs4244285 | 0.17 | 0.15 | 0.19 | Reduced CL [16, 20, 179, 180] |
| CYP2C19*3 (c.636G>A) | rs4986893 | 5 × 10−4 | 2 × 10−4 | 6 × 10−3 | |||
| Gefitinib Tamoxifen |
CYP2D6 | CYP2D6*2xN | duplication | Endoxifen plasma concentrations [21–23, 28] | |||
| CYP2D6*3 (c.775delA) | rs35742686 | 4.5 × 10−3 | 1.7 × 10−2 | 1.3 × 10−2 | Gefitinib plasma concentrations [29, 30] | ||
| CYP2D6*4 (c.506-1G>A) | rs3892097 | 0.07 | 0.19 | 0.17 | |||
| CYP2D6*5 | del | ||||||
| CYP2D6*6 (c.454delT) | rs5030655 | 2 × 10−3 | 9 × 10−3 | 7.9 × 10−3 | |||
| CYP2D6*9 (c.841_843del) | rs5030656 | 5.9 × 10−3 | 2.7 × 10−2 | 1.9 × 10−2 | |||
| CYP2D6*10 (c.100C>T) | rs1065852 | 0.12 | 0.22 | 0.25 | |||
| CYP2D6*17 (c.320C>T) | rs28371706 | 0.17 | 1.8 × 10−2 | 1.8 × 10−2 | |||
| (c.886C>T) | rs16947 | 0.49 | 0.34 | 0.34 | |||
| Busulfan Chloorambucil Melphalan |
GSTA1 | GSTA1*B (c.-135T>C) | rs3957357 | nr | nr | nr | Reduced CL busulfan [32, 33] |
| Thiotepa | GSTP1 | GSTP1 (c.341C>T) | rs1138272 | 0.02 | 0.08 | 0.06 | Reduced CL thiotepa and tepa [34] |
| Irinotecan | UGT1A1 | UGT1A1*6 (c.211G>A) | rs4148323 | 0.01 | 0.01 | 0.02 | Reduced CL SN-38 [36–38, 42] |
| UGT1A1*28a | rs8175347 | ||||||
AUC area under the curve, CL clearance, ExAc Exome Aggregation Consortium, ESP Exome Sequencing Project, MAF minor allele frequency, nr not reported, PK pharmacokinetic, 5-FU 5-fluorouracil
aUGT1A1*28 occurs with a frequency of 0.26–0.31 in Caucasians, 0.42–0.56 in African Americans, and only 0.09–0.16 in Asian populations [181]
The phase I, polymorphic xenobiotic-metabolizing CYP enzymes can be mainly divided into two classes: Class I, composed of CYP1A1, CYP1A2, CYP2E1, and CYP3A4, which are well conserved, do not have many clinically important functional polymorphisms, and are active in the metabolism of precarcinogens and drugs. Class II, composed of CYP2B6, CYP2C9, CYP2C19, and CYP2D6, which are highly polymorphic and active in the metabolism of drugs, but not of precarcinogens [9]. In this review, we discuss all three CYP families, with a special focus on Class II enzymes and their polymorphisms.
CYP1
In the CYP1 family, only one member, i.e., CYP1A2, has been associated with altered cancer drug metabolism. The CYP1A2 enzyme is involved in the metabolism of more than 20 clinically used drugs and the enzyme accounts for approximately 15 % of the total CYP450 amount in the human liver [10]. In lung cancer patients, the CYP1A2*1M variant has been associated with higher maximum plasma concentration values after the intake of 150 mg of erlotinib, suggesting reduced enzyme activity. The impact on drug efficacy and toxicity is so far unknown [11].
CYP2
The most important polymorphic enzymes in cancer drug metabolism are members of the CYP2 family, i.e., CYP2A6, CYP2B6, CYP2C9, CYP2C19, and CYP2D6. The CYP2A6 enzyme is involved in the activation of the 5-fluorouracil (5-FU) prodrug tegafur. In a set of 45 Chinese livers with 20 polymorphic variants, the CYP2A6*4 allele was mainly responsible for decreased in vitro microsomal formation of 5-FU from tegafur, whereas the CYP2A6*1B variant was associated with increased in vitro 5-FU formation [12]. In 23 Asian patients treated with irinotecan, oxaliplatin, and tegafur for metastatic gastrointestinal cancer, the CYP2A6*4, *7, and *9 variants were associated with a lower metabolic ratio of tegafur (area under the curve [AUC] ratio of 5-FU to tegafur) [13].
The impact of CYP2A6 polymorphisms (*4A, *7, and *9) on tegafur pharmacokinetics was studied in 58 Japanese patients. Although the CYP2A6 genotype did not affect the AUC of 5-FU, the clearance of tegafur was 58 % lower in patients with two variant alleles of CYP2A6 than in patients with the wild-type or 1 variant allele [14].
The CYP2B6 enzyme converts cyclophosphamide to its active form 4-hydroxycyclophosphamide. The most common functionally deficient allele is CYP2B6*6. A total of 644 plasma samples collected over a 5-year period, from 49 B-cell non-Hodgkin lymphoma (NHL) patients aged ≤18 years receiving cyclophosphamide (250 mg/m2), were used to characterize a population pharmacokinetic model. Polymorphisms in genes including CYP2B6 and CYP2C19 were analyzed. The presence of at least one CYP2B6*6 variant allele was associated with a lower cyclophosphamide clearance, as compared with homozygous wild-type patients, but there was no impact on clinical outcome [15]. However, several other reports have shown that the *6 allele is associated with a higher rate of cyclophosphamide 4-hydroxylation [16–18]. The overall effect of CYP2B6*6 expression on the pharmacokinetics and therapeutic efficacy/toxicity of cyclophosphamide seems difficult to predict and would depend on whether the dominant effect is reduced enzyme expression or increased specific enzyme activity.
Patients with the CYP2C9 *1/*2 or *2/*2 genotype undergoing hemopoietic stem cell transplantation may have decreased metabolism of busulfan as compared with patients with the wild-type genotype. However, other genetic and clinical factors may also influence the metabolism of busulfan [19]. The CYP2C19 enzyme plays a role in the metabolism of cyclophosphamide, ifosfamide, tamoxifen, and thalidomide. A splice site mutation in exon 5 (CYP2C19*2) and a premature stopcodon in exon 4 (CYP2C19*3) represent the most predominant null alleles [10]. With regard to cyclophosphamide and CYP2C19 activity, poor metabolizers are theoretically expected to have a reduced response and low toxicity upon therapy with cyclophosphamide, as a result of decreased CYP2C19-mediated activation. However, for CYP2C19*2 and CYP2C19*3, no effect on the pharmacokinetics of cyclophosphamide was observed in two larger trials conducted in Japanese and European patients [16, 20]. This might be owing to the fact that cyclophosphamide is activated via multiple CYP enzyme pathways.
The CYP2D6 gene is the best-studied member of the CYP family, with over 40 variant alleles [10]. In breast cancer patients, CYP2D6 plays an important role in the activation of tamoxifen into endoxifen. In several studies, clear associations were found between CYP2D6 status and plasma endoxifen concentrations [21–23]. However, a clear exposure-response effect remains controversial. In two of the largest prospective-retrospective studies from BIG 1-98 and ATAC, no association was found between the CYP2D6 genotype and breast cancer recurrence, although genotyping was performed in tumor DNA and massive departures from the Hardy–Weinberg equilibrium have been noted [24–26]. These controversial findings and the partial contribution of the genotype in explaining inter-individual variability in plasma concentrations of endoxifen imply that tailored tamoxifen treatment may not be fully realized through pharmacogenetics of metabolizing enzymes alone [27].
Lung cancer patients designated as CYP2D6 poor metabolizers might theoretically have increased concentrations of gefitinib as compared with individuals designated as CYP2D6 extensive metabolizers. However, other genetic and clinical factors may also influence concentrations of gefitinib. The pharmacokinetics and pharmacogenomics were not associated with significantly different toxicities, response rates, or survival times with gefitinib [28, 29].
CYP3
The CYP3A subfamily is involved in the metabolism of more than 50 % of clinically used drugs, including several anticancer drugs such as cyclophosphamide, ifosfamide, thiotepa, etoposide, teniposide, docetaxel, paclitaxel, irinotecan, toremifene, vinblastine, vincristine, vinorelbine, gefitinib, imatinib, and erlotinib. The enzyme activity of CYP3A ranges widely among subjects, and its activity is largely affected by non-genetic factors such as age, endogenous hormone levels, health status, and environmental stimuli. Although approximately 40 allelic variants have been described for CYP3A4, it has been found that genetic variability in CYP3A alone is insufficient to explain its widely ranging enzyme activity and therefore is not indicated in clinical practice [30]. Recently, it was demonstrated that the CYP3A4*22 (rs35599367:C>T) and the CYP3A5*3 (rs776746:A>G) polymorphisms have a small but clinical insignificant impact on the pharmacokinetics of sunitinib, but so far no other studies have demonstrated an impact on the pharmacokinetics of anticancer drugs [31].
Non-CYP Phase II-Metabolizing Enzymes
Several clinical relevant gene polymorphisms associated with phase II drug metabolism and pharmacokinetics of anticancer drugs have been reported in the literature. The GST enzyme family and UGT enzymes have been most intensively studied (Table 2).
Glutathione S-Transferase
Four subfamilies of GSTs exist, namely, GSTA, GSTM, GSTP, and GSTT. GSTA1 plays an important role in the detoxification of, busulfan, melphalan, and chlorambucil. GSTA1*B might be of clinical relevance in busulfan treatment because two studies in children demonstrated that the presence of GSTA1*B reduced the clearance of busulfan up to 30 % [32, 33]. In clinical practice, this might require dose adjustments of busulfan, at least in children, based on the GSTA1*B genotype.
In 124 Caucasian patients treated with high-dose chemotherapy for metastatic breast, ovarian, and testicular tumors, the clearance of thiotepa and tepa was predominantly affected by the GSTP1 C341T polymorphism, which had a frequency of 9.3 %. This allele variant increased non-inducible thiotepa clearance by 52 % and decreased tepa clearance by 32 % in heterozygous patients, which resulted in an increase in combined exposure to thiotepa and tepa of 45 % in homozygous patients [34].
Uridine Diphosphate Glucuronosyltransferases
The UGT enzymes are a superfamily of enzymes responsible for the glucuronidation of target substrates. The transfer of glucuronic acid renders xenobiotics and other endogenous compounds water soluble, allowing for their biliary or renal elimination. Unconjugated hyperbilirubinemias, such as Gilbert’s syndrome and Crigler–Najjar syndrome, have been found to be associated with polymorphic variants of UGT1A1, especially with UGT1A1*28 [35]. Currently, over 113 different UGT1A1 variants have been described throughout the gene. These variants can confer reduced or increased activities, as well as inactive or normal enzymatic phenotypes.
Irinotecan is converted to its active metabolite SN-38. SN-38 is further metabolized to SN-38-glucuronide by various hepatic and extrahepatic UGT1A isozymes, mainly UGT1A1. Impaired glucuronidation activity of the UGT1A1 enzyme has been linked with elevated levels of SN-38, leading to toxicities. UGT1A1*28 involves an extra TA repeat in the UGT1A1 promoter region and is the variant most frequently contributing to interpatient variability in irinotecan pharmacokinetics and toxicities. This information led to the revision of the irinotecan label by the US Food and Drug Administration. Both the *28 and *6 alleles have been well studied in regard to pharmaceutical toxicities. In particular, both alleles have shown associations with the development of irinotecan toxicities [36–38]. UGT1A1*28 occurs with a frequency of 0.26–0.31 in Caucasians, and 0.42–0.56 in African Americans, and only 0.09–0.16 in Asian populations [39, 40]. UGT1A1*6 has allele frequencies in Japanese, Korean, and Chinese populations of 0.13, 0.23, and 0.23, respectively [41].
Several studies hypothesized that patients with the *1/*1 genotype would tolerate a higher dose than the standard recommended dose of 180 mg/m2, while patients with the *28/*28 genotype would require dose reduction. A prospective genotype-guided phase I study in colorectal cancer patients receiving irinotecan monotherapy indeed demonstrated a maximum tolerated dose (MTD) of 850 mg, 700 mg, and 400 mg in patients with the *1/*1 genotype, *1/*28 genotype, and *28/*28 genotype, respectively. Interestingly, although the irinotecan AUC increased according to the different MTDs in each genotype group, the mean SN-38 AUC levels were comparable across the different MTDs in each genotype group [42].
A similar, genotype-guided dose escalation study in colorectal patients receiving FOLFIRI (5-FU, folinic acid, irinotecan) identified the MTD of irinotecan to be 370 mg/m2 and 310 mg/m2 in patients with the *1/*1 genotype and *1/*28 genotype, respectively. Patients with the *28/*28 genotype were excluded [43]. Recently, it was demonstrated that high-dose irinotecan (260 mg/m2) FOLFIRI combined with bevacizumab did not improve the overall response rate in metastatic colorectal cancer patients with the *1/*1 or *1/*28 genotype [44]. Whether irinotecan dose escalation in wild-type UGT1A1 patients contributes to improved clinical outcome is therefore questionable.
Enzymes of Purine and Pyrimidine Metabolism
Structural analogs of nucleobases and nucleosides are used in the treatment of cancer, viral infections, and inflammatory diseases. These nucleobase and nucleoside analogs are inactive produgs that are taken up by the cell via specific nucleobase or nucleoside transporters and subsequently phosphorylated intracellularly to their pharmacologically active triphosphate form [45]. The incorporation of nucleoside triphosphate analogs into DNA causes termination of DNA elongation and often also resistance to proofreading exonucleases. Some of these analogs also inhibit key enzymes (e.g., ribonucleotide reductase, thymidylate synthase, or dCMP deaminase) involved in the generation of purine and pyrimidine nucleotides for RNA and DNA synthesis. Opposing the activation of these purine and pyrimidine analogs are enzymes that inactivate or degrade the parent compounds or one of its anabolic products. Thus, a deficiency of a key enzyme in the anabolic or catabolic pathway of these purine and pyrimidine analogs will not only affect the clinical efficacy and toxicity of the drug but is also likely to alter the pharmacokinetics of the drug (Table 3).
Table 3.
Genotypes affecting pharmacokinetics of drugs targeting purine and pyrimidine metabolism
| Drugs | Gene | Mutations | dbSNP ID | ESP MAF | ExAC MAF | PK parameters | |
|---|---|---|---|---|---|---|---|
| African American | European American | ||||||
| 5-FU Capecitabine Tegafur |
DPYD | c.1905+1G>A | rs3918290 | 9.0 × 10−4 | 5.8 × 10−3 | 5.2 × 10−3 | V max, t 1/2, AUC, CL [58, 59, 61, 182] |
| c.1679T>G | rs55886062 | 0 | 6 × 10−4 | 3.5 × 10−4 | CL [61] | ||
| c.2846A>T | rs67376798 | 9.0 × 10−4 | 5.5 × 10−3 | 2.6 × 10−3 | CL [61] | ||
| c.2579delA | rs746991079 | nr | nr | 3.3 × 10−5 | V max (uracil) [62] | ||
| c.1129-5923C>G | rs75017182 | nr | nr | nr | V max (uracil) [62] | ||
| DPYS | c.1506delC | rs147965145 | 2.6 × 10−3 | 0 | 2.4 × 10−4 | T max, C max, AUC [66] | |
| UPB1 | c.254C>A | rs34035085 | 1.5 × 10−2 | 0 | 1.4 × 10−3 | Altered uracil flux [68] | |
| Capecitabine | MTHFR | c.655C>T (C677T) | rs1801133 | 0.12 | 0.35 | 0.30 | t 1/2 [70] |
| Thiopurines | TPMT | TMPT*2 (c.238G>C) | rs1800462 | 0 | 2.3 × 10−3 | 1.4 × 10−3 | TGN [77, 78] |
| TPMT*3B (c.460G>A) | rs1800460 | 1.0 × 10−2 | 3.7 × 10−2 | 2.7 × 10−2 | |||
| TMPT*3C (c.719A>G) | rs1142345 | 5.3 × 10−2 | 4.2 × 10−2 | 3.7 × 10−2 | |||
| Methotrexate | MTHFR | c.665C>T (C677T) c.1286A>C |
rs1801133 | 0.12 | 0.35 | 0.30 | CL, serum concentrations MTX [183, 184] |
| rs1801131 | 0.16 | 0.31 | 0.30 | Serum concentrations MTX [184] | |||
| ARID5B | c.1200-6044T>C | rs4948502 | nr | nr | nr | Serum concentrations MTX [185] | |
| c.734-5030T>C | rs4948496 | nr | nr | nr | Serum concentrations MTX [185] | ||
| c.276+7693C>A | rs4948487 | nr | nr | nr | Serum concentrations MTX [185] | ||
| ABCC2 | c.1234A>G Knockout model |
rs765027508 | nr | nr | 8.2 × 10−6 | t 1/2 [186] | |
| AUC [187] | |||||||
| SLCO1B1 | c.521T>C | rs4149056 | 3.6 × 10−2 | 0.16 | 0.13 | Serum concentrations MTX, AUC, CL [185, 188] | |
| c.388A>G | rs2306283 | 0.23 | 0.40 | 0.48 | Serum concentrations MTX, AUC, CL [188] | ||
| c.1865+248G>A | rs4149081 | nr | nr | nr | CL [189] | ||
| c.1865+4846T>C | rs11045879 | nr | nr | nr | CL[189] | ||
| Hap*5 | CL[190] | ||||||
| Hap*15 | CL [190] | ||||||
| Hap*23 | CL [190] | ||||||
| Hap*31 | CL [190] | ||||||
| Gemcitabine | CDA | Hap*3 (c.208G>A) | rs60369023 | nr | nr | 2.9 × 10−4 | AUC, CL, C max [88] |
| THU induceda | AUC, CL [191] | ||||||
| CNTN4 | c.2398+70G>T | rs4685596 | nr | nr | nr | AUC, C max [87] | |
| ALOX5AP | c.495-204A>G | rs4769060 | nr | nr | nr | AUC, Vss, C max [87] | |
| c.495-523T>C | rs3935645 | nr | nr | nr | Vss, C max [87] | ||
| c.341+12C>A | rs3803277 | 0.44 | 0.44 | 0.49 | Vss, C max [87] | ||
| DMD | c.1331+127G>A | rs5928065 | nr | nr | nr | AUC, Vss [87] | |
| HEXDC | c.15T>G | rs1141463 | 0.38 | 0.30 | 0.35 | AUC, Vss [87] | |
| Decitabine | CDA | mRNA expression activity | Plasma concentrations decitabine [95] | ||||
AUC area under the curve, ExAc Exome Aggregation Consortium, CDA cytidine deaminase, CL clearance, C max maximum plasma concentration, ESP Exome Sequencing Project, MAF minor allele frequency, nr not reported, PK pharmacokinetic, TGN thioguanine nucleotides, t 1/2 elimination half-life, V max maximum enzymatic conversion capacity, Vss volume of distribution at steady state, 5-FU 5-fluorouracil
aCDA deficiency was achieved in mice by treatment with tetrahydrouridine [191]
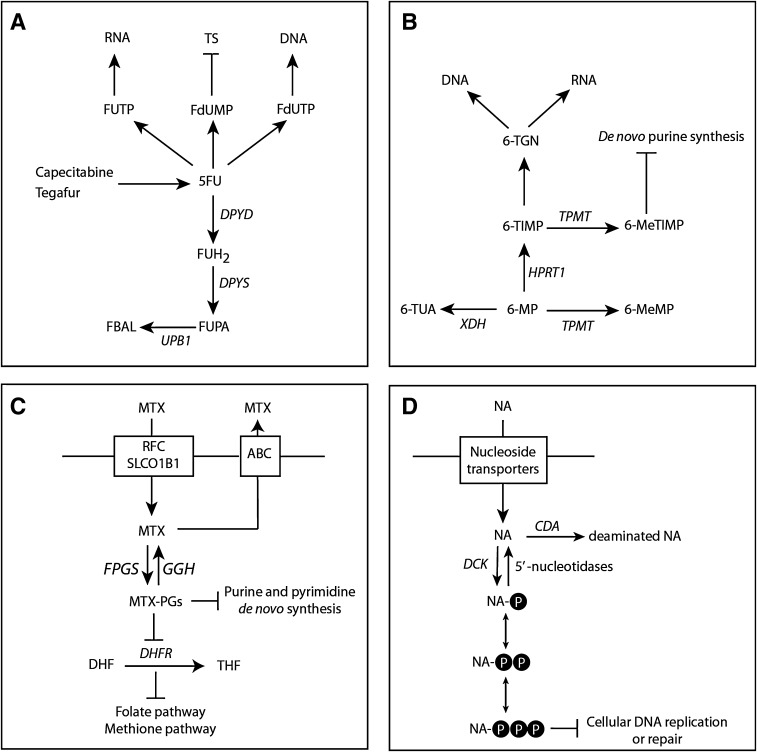
5-FU and its oral prodrug capecitabine (Xeloda®, F. Hoffmann-La Roche AG, Basel, Switzerland) are two of the most frequently prescribed chemotherapeutic drugs for the adjuvant and palliative treatment of patients with cancers of the gastrointestinal tract, breast, and head and neck [46, 47]. Both 5-FU and capecitabine need to undergo enzymatic activation to fluoropyrimidine nucleotides to exert their cytotoxic effects (Fig. 1a). However, the degradation of 5-FU plays a significant role as more than 80 % of 5-FU is catabolized by DPD [48]. 5-FU has a narrow therapeutic index and an increased exposure to 5-FU, owing to a reduced activity of DPD, can thus result in severe or even lethal toxicity [49]. For DPD activities within the normal range, conflicting results have been published as to whether a correlation exists between the DPD activity and the clearance of 5-FU [50–52]. Compelling results, however, have shown that patients with a partial or complete DPD deficiency have a reduced capacity to degrade 5-FU and are at risk of developing severe 5-FU-associated toxicity [53]. To date, many mutations and polymorphisms have been described in the gene encoding DPD (DPYD) and ample evidence has been provided that carriers of the c.1905+1G>A, c.1679T>C, c.2846A>T, and c.1129-5923C>G/hapB3 variant have a strongly increased risk of developing toxicity [6, 54–57].
Fig. 1.
Metabolism of drugs interfering with purine and pyrimidine synthesis. a Metabolism of fluoropyrimidine-containing drugs. b Thiopurine metabolism. c Methotrexate metabolism. d Nucleoside metabolism. ABC adenosine triphosphate-binding cassette family of transporters, CDA cytidine deaminase, DCK deoxycytidine kinase, DHF dihydrofolate, DHFR dihydrofolate reductase, DPYD dihydropyrimidine dehydrogenase, DPYS dihydropyrimidinase, FBAL fluoro-β-alanine, FGPS folylpolyglutamate synthase, FUH 2 5-fluoro-dihydrouracil, FUPA fluoro-β-ureidopropionate, GGH γ-glutamyl hydrolase, HPRT1 hypoxanthine-guanine phosphoribosyltransferase, MTX methotrexate, MTX-PGs MTX-polyglutamate, NA (deoxy)nucleoside analogs, RFC reduced folate carrier, SLCO1B1 solute carrier organic anion transporter B, THF tetrahydrofolate, dehydrogenase/oxidase, TPMT thiopurine-S-methyltransferase, UPB1 β-ureidopropionase, XDH xanthine, 5-FU 5-fluorouracil, 6-MeMP 6-methylmercaptopurine, 6-MeTIMP 6-methylthioinosine monophosphate, 6-MP 6-mercaptopurine, 6-TGN 6-thioguanine nucleotides, 6-TIMP 6-thioinosine monophosphate, 6-TUA 6-thiouric acid
In a two-compartment model with Michaelis–Menten elimination, the mean Vmax value was 40 % lower in patients heterozygous for the c.1905+1G>A mutation in DPYD compared with controls [58]. Non-compartmental analysis showed that the mean AUC was 1.5-fold and 1.3-fold higher in carriers of the c.1905+1G>A mutation treated with 300 mg/m2 and 450 mg/m2, respectively, when compared with controls. The mean terminal half-life of 5-FU was 2.1-fold and 1.7-fold longer at 300 mg/m2 and 450 mg/m2, respectively, compared with controls [58]. Furthermore, a clinical pharmacological study of a patient with a complete deficiency owing to homozygosity for the c.1905+1G>A mutation in DPYD demonstrated minimal catabolism of 5-FU, with a tenfold longer half-life of 5-FU compared with patients with a normal DPD activity [59, 60]. A decreased clearance of plasma 5-FU concentrations was also noticed for carriers of the c.2846A>T and c.1679T>G mutations [61]. In addition, an oral uracil loading test to identify DPD-deficient patients showed altered pharmacokinetics of uracil in patients who were carriers for the c.1905+1G>A, c.2846A>T, c.2579delA, c.1679 T>G, or c.1129-5923C>G mutation [62].
Patients with a complete dihydropyrimidinase (DPYS) deficiency, the second enzyme of the pyrimidine degradation pathway, present with strongly elevated levels of dihydropyrimidines and moderately elevated levels of uracil and thymine [63]. Patients with a partial DHP deficiency also show an impaired flux through the pyrimidine degradation pathway and are prone to the development of severe toxicity after the administration of 5-FU [64–67]. The identification of a healthy individual showing altered catabolism of uracil due to heterozygosity for a mutation in UPB1 suggests that also patients with a β-ureidopropionase deficiency, the third enzyme of the pyrimidine degradation pathway, might be at risk of developing 5-FU toxicity [68, 69]. Although methylenetetrahydrofolate reductase (MTHFR) is not involved in the degradation of capecitabine, a borderline decrease in the elimination-half-life of capecitabine was observed for the c.677C>T mutation in MTHFR [70]. The c.677C>T mutation in MTHFR reduces the enzyme activity and presumably increases the level of 5,10-methyleneterahydrofolate, a substrate of thymidylate synthase. Thus, a direct causal relationship between MTHFR genotype and apparent elimination half-life of capecitabine is not likely.
6-Mercaptopurine (6-MP) is an analog of guanine and hypoxanthine, which is widely used in the treatment of patients with inflammatory bowel disease and patients with acute lymphoblastic leukemia [71, 72]. The principal cytotoxic and immunosuppressive effects of thiopurine drugs are caused by incorporation of thioguanine nucleotides into DNA or RNA (Fig. 1b). Opposing the principal enzyme of the anabolic pathway, hypoxanthine-guanine phosphoribosyl transferase, are the two catabolic enzymes xanthine oxidase and TPMT. Xanthine oxidase is responsible for oxidation of 6-MP into the inactive metabolite 6-thiouric acid, whereas TPMT methylates 6-MP to form the inactive metabolite 6-MP. Therefore, TPMT plays a pivotal role in the production of active thiopurine metabolites by diverting a proportion of available substrates away from the anabolic pathway of thiopurines to generate methylated metabolites. To date, more than 35 variants in the gene encoding TPMT have been associated with decreased TPMT activity [72]. Three variants TPMT*2, TMPT*3A, and TMPT*3C account for 80–85 % of intermediate or low enzyme activity in the Caucasian population [5]. Individuals who are heterozygous carriers or homozygous for an inherited functional mutation in TPMT have an increased risk of developing life-threatening myelosuppressive effects of thiopurines. Patients who are heterozygous for a TPMT deficiency require a lower dose of thiopurines (30–50 % of the regular dose) and substantial reduced doses (>tenfold) or the use of alternative agents is recommended in patients homozygous for a TPMT deficiency [72, 73]. Upfront screening of patients for variants in TPMT, followed by a dose reduction in heterozygous or homozygous carriers of a variant, reduced hematological events during thiopurine treatment of inflammatory bowel disease [74]. Furthermore, a similar treatment efficacy was obtained in carriers treated with a reduced thiopurine dose as compared with that observed in controls [74].
The oral bioavailability of 6-MP is very low owing to extensive intestinal and hepatic metabolism by xanthine oxidase [75]. The maximum concentration of 6-MP in plasma is observed approximately 1.3 h after oral administration of 6-MP and the elimination half-life is approximately 1.8 h [76]. One approach to ensure optimal dosing of thiopurines is to monitor the thiopurine metabolites in erythrocytes [72]. A population pharmacokinetic model has been developed to predict the concentrations of thioguanine nucleotides in erythrocytes in pediatric patients with acute lymphoblastic leukemia and the most influential covariate examined proved to be the TMPT genotype [77]. In a physiologically based pharmacokinetic model, the predicted thioguanine nucleotides in erythrocytes in patients with heterozygous or homozygous variants in the TPMT gene were twofold and tenfold higher, respectively, compared with those observed in patients with wild-type TMPT [78].
Methotrexate (MTX) is most frequently used for the treatment of patients with rheumatoid arthritis as well as patients with acute lymphoblastic leukemia. Nevertheless, MTX can cause severe dose-limiting adverse events and organ toxicities [79]. MTX is a structural analog of folic acid and it enters the cell via the reduced folate carrier (solute carrier family 19 member 1B1, SLC19A1) or the solute carrier organic anion transporter B1 (SLCO1B1) (Fig. 1c). In the cytoplasm, MTX is polyglutamated by folylpolyglutamate synthase, which enhances its retention in the cell. This process can be reversed by the enzyme γ-glutamyl hydrolase. MTX and MTX-polyglutamate inhibit dihydrofolate reductase, which catalyses the conversion of dihydrofolate into tetrahydrofolate. Because reduced folates are required for both the de novo thymidylate and purine synthesis, inhibition of dihydrofolate reductase results in direct inhibition of both pathways (Fig. 1). In addition, methotrexate polyglutamate metabolites also bind directly and inhibit thymidylate synthase and aminoimidazolecarboximide ribonucelotide formyltransferase (purine de novo pathway) [80]. Efflux of MTX from the cell occurs via members of the adenosine-5′-triphosphate-binding cassette (ABC) family of transporters, including ABCB1 [81]. Genetic variations in pharmacokinetic genes involved in MTX metabolism can be major determinants of clinical response and toxicity (Table 3) [71, 81–84].
Deoxycytidine kinase (dCK) is responsible for the initial activation of a number of clinically important anticancer drugs such as cytarabine, gemcitabine, decitabine, fludarabine, and clofarabine (Fig. 1d). Impaired dCK expression or activity in cells results in resistance to these drugs, whereas overexpression of dCK in dCK-deficient cell lines increased the sensitivity of dCK-activated deoxynucleoside analogs, indicating that dCK plays a key role in their metabolism and pharmacological activities [45]. Cytidine deaminase is an important determinant of the efficacy and cytotoxicity of cytarabine, gemcitabine, and decitabine because these deoxynucleoside analogs are readily deaminated and thereby inactivated by cytidine deaminase [85]. The pharmacokinetics of cytarabine [86], gemcitabine [87–89], decitabine [90], fludarabine [91], and clofarabine [92, 93] have been thoroughly investigated in cancer patients. To date, limited information is only available regarding the potential impact of altered levels of the cytidine deaminase gene (CDA) on the pharmacokinetics of gemcitabine [87–89, 94] and decitabine [95].
Drug Transporters
Polymorphisms in genes encoding drug efflux transporters, such as P-gp and BCRP, can influence uptake and excretion of anticancer drugs (Table 4). This contributes to the inter-individual variability in pharmacokinetics and, as a consequence, to large differences in treatment response between cancer patients [7, 96].
Table 4.
Polymorphisms in drug transporter genes affecting pharmacokinetics of anticancer drugs
| Drugs | Gene | Mutations | dbSNP ID | ESP MAF | ExAC MAF | PK parameters | |
|---|---|---|---|---|---|---|---|
| African American | European American | ||||||
| Docetaxel | ABCB1 | c.1236T>C | rs1128203 | 0.22 | 0.43 | 0.54 | CL [1] |
| c.3435T>C | rs1045642 | 0.23 | 0.48 | 0.50 | CL [192] | ||
| Paclitaxel | ABCB1 | c.1236T>C | rs1128203 | 0.22 | 0.43 | 0.54 | AUC [193] |
| c.2677T>A/G | rs2032582 | nr | nr | 0.54 | AUC [193] | ||
| Etoposide | ABCB1 | c.3435T>C | rs1045642 | 0.77 | 0.47 | 0.50 | CL [194] |
| Doxorubicin | ABCB1 | c.1236T>C | rs1128203 | 0.22 | 0.43 | 0.54 | C max [106] |
| c.2677T>A/G | rs2032582 | nr | nr | 0.54 | CL [106] | ||
| SLC22A16 | c.146A>G | rs714368 | 0.36 | 0.22 | 0.25 | AUC [195] | |
| c.312T>C | rs6907567 | 0.36 | 0.22 | 0.25 | AUC [195] | ||
| Irinotecan | ABCB1 | c.1236T>C | rs1128203 | 0.22 | 0.43 | 0.54 | AUC, CL [110] |
| Hap*2 | CL [111] | ||||||
| Bicalutamide | ABCG2 | c.421C>A | rs2231142 | 0.03 | 0.11 | 0.12 | AUC, T max, C max, t 1/2, CL plasma concentrations [113] |
| Topotecan | ABCG2 | c.421C>A | rs2231142 | 0.03 | 0.11 | 0.12 | F [196] |
| Imatinib | ABCB1 | c.1236T>C | rs1128203 | 0.22 | 0.43 | 0.54 | C min, CL, F [116, 117] |
| c.2677T>A/G | rs2032582 | nr | nr | 0.54 | CL, F [117] | ||
| c.3435T>C | rs1045642 | 0.23 | 0.48 | 0.50 | CL, F [117] | ||
| Hap*4 | C min [116] | ||||||
| ABCG2 | c.421C>A | rs2231142 | 0.03 | 0.11 | 0.12 | C min, CL [118, 119] | |
| SLC22A1 | c.480C>G | rs683369 | 0.05 | 0.22 | 0.17 | CL, C min [128] | |
| Gefitinib | ABCG2 | c.421C>A | rs2231142 | 0.03 | 0.11 | 0.12 | C ss,min/C 1,min [123] |
| Sunitinib | ABCB1 | c.2677T>A/G | rs2032582 | nr | nr | 0.54 | CL [31] |
The drug accumulation at the steady-state was assessed as the ratio of C ss,min to C 1,min, where C ss,min was the average pretreatment concentration on days 8, 15, 22 and 28, and C 1,min was the pretreatment concentration before the second dose
AUC area under the curve, ExAc Exome Aggregation Consortium, ESP Exome Sequencing Project, CL clearance, C max maximum plasma concentration, C min trough plasma concentration, C ss,min /C 1,min, F oral bioavailability, MAF minor allele frequency, nr not reported, PK pharmacokinetic, T max time to maximum plasma concentration, t 1/2 elimination half-life
P-gp is a member of the ABC superfamily of membrane transporters and is involved in the active transport of lipophilic and amphipathic molecules through lipid membranes [97]. P-gp is encoded by the multidrug resistance 1 (MDR1) gene (ABCB1), located at chromosome 7q21. A number of polymorphisms described in this gene significantly influence the pharmacokinetics of several anticancer drugs. There are three main polymorphisms influencing the activity of P-gp; the c.2677G>T/A single nucleotide polymorphism (SNP) in exon 21 leads to a change in the amino acid sequence from Ala (G) to Ser (T) or Thr (A), possibly resulting in increased P-gp function [98, 99]. The second polymorphism is in exon 26 at wobble position c.3435C>T, resulting in a more than twofold lower P-gp expression in the duodenum [100]. The third one is also a synonymous SNP, at c.1236T in exon 12, which does not directly affect expression of P-gp but may have an indirect effect such as altering RNA stability for P-gp [101].
BCRP, also called mitoxantrone resistant protein (MXR) or placenta-specific ABC transporter, is another member of the ABC transporter superfamily. BCRP is encoded by the ABCG2 gene located at chromosome 4q22 [102]. A functional SNP (c.421C>A) in exon 5 has been identified, resulting in a Gln (C) to Lys (A) amino acid substitution, which proved to be associated with decreased BCRP expression levels and altered substrate specificity [103].
Docetaxel and paclitaxel are cytotoxic taxanes inhibiting mitosis leading to cancer cell death, which are mainly used in the treatment of breast, ovarian, and lung cancer [104]. For taxanes, the ABCB1 gene is considered one of the best candidates to become a biomarker underlying variations in clinical responses and toxicity owing to pharmacokinetic differences [105].
Doxorubicin, an anthracycline widely used as mono- or combination therapy in the treatment of solid tumors including breast cancer, is also the substrate of P-gp and BCRP [106]. Significantly altered clearance and lower plasma concentration of doxorubicin was observed in patients harboring any of the three above-described polymorphisms in ABCB1. For c.421C>A in ABCG2, no significant influence on doxorubicin pharmacokinetics was observed [106]. Pharmacokinetics of other anthracyclines, such as epirubicin and daunorubicin, were not altered by polymorphisms in drug transporter genes [107, 108].
Irinotecan, a topoisomerase I inhibitor, plays a major role in the treatment of colorectal cancer as monotherapy or in combination with 5-FU [109]. Elimination pathways of irinotecan are partially mediated by P-gp and BCRP. A study investigating polymorphisms in genes encoding these transporters showed that only the polymorphism c.1236C>T in ABCB1 was associated with significantly increased exposure to irinotecan and its active metabolite SN-38 in individuals homozygous for the T allele [110]. In addition, a significant association has been observed for ABCB1 haloptype*2 containing both c.1236C>T, c.2677G>T/A, and c. 3435C>T, with reduced renal clearance of irinotecan [111].
Bicalutamide, a non-steroidal pure anti-androgen that competitively blocks the growth-stimulating effects of androgens, is used in the treatment of prostate cancer as monotherapy or in combination with a luteinizing hormone-releasing hormone analog [112]. P-gp and BCRP are involved in the disposition of bicalutamide. The pharmacokinetic parameters of bicalutamide did not show any significant differences between ABCB1 genotype groups for the three main polymorphisms previously described [113]. However, for ABCG2 it was shown that the c.421C>A polymorphism influenced plasma concentrations of bicalutamide with subjects homozygous for the c.421AA genotype exhibiting significantly higher plasma concentrations than those with the c.421CC or c.421CA genotype [113].
Tyrosine kinase inhibitors (TKIs) are a relatively new class of oral targeted anticancer therapy. TKIs are designed to compete with adenosine-5′-triphosphate in the tyrosine kinase receptor mutated and/or over-expressed in cancer tissues, thereby blocking the signaling important for tumor growth [114]. Most TKIs are transported by P-gp and BCRP, thus polymorphisms in genes encoding these transporters are likely to influence the pharmacokinetics of TKIs. Most studied in this respect is the first approved TKI; imatinib, used in the treatment of chronic myeloid leukemia and gastrointestinal stromal tumors [115]. However, conflicting results have been reported as to whether ABCB1/ABCG2 polymorphisms affect the pharmacokinetics of imatinib [116–122]. For ABCG2, the results are more consisted, with the c.421C>A SNP resulting in significant lower plasma concentrations and changes in the clearance of imatinib [118, 119]. Another first-generation TKI gefitinib, a selective epidermal growth factor receptor inhibitor used in the treatment of non-small-cell lung cancer, showed higher drug accumulation in patients with c.421C>A SNP in ABCG2 [123]. No relationship with gefitinib AUC was found with polymorphisms in either ABCB1 or ABCG2 [28]. For the newer second- and third-generation TKIs, axitinib, bosutinib, nilotinib, dasatinib, sorafenib, and ponatinib, the substrate affinity for both efflux transporters is lower than measured for imatinib. Therefore, their efficacy is not significantly affected by polymorphisms in genes encoding these transporters [115, 124–126]. Increased clearance of sunitinib, a multi-targeted TKI used in the treatment of renal cell carcinoma, has been shown for homozygote genotypes of c.2677G>A/T SNP in ABCB1 [31].
Another increasingly recognized group of transporters involved in the pharmacokinetics of anticancer drugs are the influx transporters of the solute carrier family, also known as the human organic cation transporter 1 (hOCT1), encoded by the SLC22A1 gene [127]. hOCT1 is expressed in several tissues and organs where its activity contributes to the uptake and elimination of endogenous small organic toxic by-products and drugs. The c.480C>G polymorphism in this gene is the most studied one in relation to pharmacodynamic effects, but only for imatinib has an association been found between genotype and clearance [128].
Immunoglobulin-Metabolizing Enzymes
Monoclonal antibodies (mAbs) are increasingly being used in the treatment of cancer, owing to their high specificity and activity, combined with the expanding knowledge on specific tumor targets [129]. mAbs are immunoglobulins produced with recombinant DNA technology and can be fully human, humanized chimeric (human/murine), or murine [130].
The response to mAbs may be difficult to predict owing to several sources of variability, partly explained by inter-individual variability in pharmacokinetics [131]. mAbs are hydrophilic high-molecular-weight proteins and their pharmacokinetic properties are therefore different from conventional chemical agents. mAbs used in cancer treatment are, or derive from, human immunoglobulin G (IgG). Therefore, the pharmacokinetic properties of mAbs are similar to those of IgG [131]. The IgG structure can be divided into two identical binding portions (Fab) and a crystallizable portion (Fc). The Fc portion binds to the neonatal Fc receptor (FcRn) expressed on phagocytic cells of the reticuloendothelial system, which is involved in IgG protection from intracellular catabolism [132]. Intracellular catabolism is the main route for elimination of IgGs and mAbs with a Fc portion [133]. Knock-out mice that do not produce FcRn have a much higher IgG elimination (lower half-life) than wild-type mice [134].
FcRn is encoded by FCGRT, a gene located on chromosome 19 [135]. To date, little is known about potential polymorphisms of this gene influencing the pharmacokinetics of mAbs [8]. A variable number of tandem repeats (VNTR) in the FCGRT promotor region has been described and immunoglobulin therapy proved to be more efficient in VNTR3/VNTR3 homozygous patients than in VNTR2/VNTR3 patients [136]. For cetuximab, a significant lower distribution clearance was shown in VNTR3/VNTR3 patients compared with VNTR2/VNTR3 [8].
In addition, cells of the reticuloendothelial system express various types of Fcγ-receptors, which are also expected to play a role in the elimination of mAbs, through internalization and degradation by lysosomes in these cells after binding of the mAb to the Fcγ-receptors [137]. Two of these Fcγ-receptors are FcγRIIA and FcγRIIIA and several studies described the influence of a polymorphisms in FCGR2A or FCGR3A, the genes encoding the FcRIIA and FcRIIIA receptors, respectively, on therapy outcomes for rituximab [138–142], trastuzumab [143, 144], and cetuximab [145–148]. The G to A point mutation described in the FCGR2A gene generates two FcγRIIA allotypes, with either a histidine (H) or arginine (R) at amino acid position 131. The T to G substitution described in FCGR3A generates two FcγRIIIA allotypes, with either a phenylalanine (F) or valine (V) at amino acid position 158 in the membrane-proximal Ig-like loop. Human IgG binds more strongly to cells homozygous for FcγRIIA-131H and FcγRIIIA-158V than to cells homozygous for FcγRIIA-131R and FcγRIIIA-158F [149, 150].
Cetuximab, a chimeric immunoglobulin monoclonal antibody that targets the epidermal growth factor receptor, is used in the treatment of metastatic colorectal cancer in combination with chemotherapy or as monotherapy [151]. Several studies explored the role of FCGR polymorphisms in the treatment outcome of cetuximab, but have conflicting results. In some studies, FcγRIIIA-158F/F was correlated with response and a longer progression-free survival [146], while in other studies FcγRIIIA-158V/V was associated with longer progression-free survival [147] or no difference on progression-free survival at all was observed [145, 148]. For FcγRIIA-131 H/H, in two out of three studies, a better disease control rate and progression-free survival was observed than those for FcγRIIA-131 R/R [147, 148, 152].
Rituximab, a chimeric immunoglobulin monoclonal antibody that targets the B-cell-surface antigen CD20, is used in the treatment of diffuse large B-cell lymphoma in combination with chemotherapy or as monotherapy [153]. For rituximab, several studies showed that FcγRIIIA-158V/V patients had a longer progression-free survival than F carriers [138–142].
Trastuzumab, a humanized immunoglobulin monoclonal antibody that targets the human epidermal growth factor receptor (HER2), is a major therapeutic agent in the treatment of HER2-positive breast cancer in combination with chemotherapy or as monotherapy [154]. Some studies on trastuzumab show a similar effect as observed with rituximab, with a better clinical response for FcγRIIIA-158V/V patients [143, 144], while others could not confirm these results [155].
For cetuximab, rituximab, and trastuzumab, the underlying mechanism of FCGR polymorphisms was speculated to be pharmacodynamic, owing to a more efficient FcγRIIa/FcγRIIIa-dependent cytotoxicity. However, because these receptors are also involved in the elimination of mAbs, it can be hypothesized that these polymorphisms also impact mAb clearance. For infliximab, a mAb that is not used in cancer treatment but instead is frequently used to control inflammatory diseases, a higher infliximab elimination rate constant in FcγRIIIA-158V/V patients was observed than in F carriers, leading to a faster infliximab underexposure and relapse of disease [156]. These findings can also explain why in vitro studies on rituximab showed a much stronger correlation, with the concentration leading to 50 % of maximal lysis about fourfold lower for FcγRIIIA-158V/V patients than for F carriers, than found in vivo [157].
Conclusions
Cancer treatment is becoming more and more individually based to increase drug efficacy and reduce adverse responses to therapy. Pharmacogenetic screening and/or drug-specific phenotyping of cancer patients eligible for treatment with chemotherapeutic drugs, prior to the start of anticancer treatment, can not only identify patients with tumors that are likely to be responsive or resistant to the proposed drugs but also patients prone to develop severe toxicity. Ample evidence is now available that polymorphisms in DPYD, TPMT, and UGT can profoundly affect the pharmacokinetics of 5-FU, mercaptopurine, and irinotecan, respectively [158]. Considering the common use of these three drugs in the treatment of cancer patients, the severe toxicity in patients carrying functional polymorphisms in these genes, it would be preferable to screen these patients prior to the start of the therapy. For most other chemotherapeutic drugs, however, the association of gene mutations and pharmacokinetics is less clear, which may be because of a minor impact of genetics compared with non-genetic factors such as diet, co-medication, health status, and renal and liver function. These agents may be candidates for dose individualization by a phenotype-based approach such as therapeutic drug monitoring.
In the past decades, huge progress has been made in the rapid characterization of SNPs, enabling the clinical application of pretreatment pharmacogenetic screening. However, the scarcity of information on functional characteristics of many SNPs indicates the need for future research, allowing pharmacogenetic and pharmacokinetic screenings to become the standard of care.
Compliance with Ethical Standards
Funding
No sources of funding were used to assist with the preparation of this review.
Conflict of interest
Daphne Bertholee, Dr. Jan Gerard Maring, and Dr. André B.P. van Kuilenburg have no conflicts of interest that are relevant to the content of this review.
References
- 1.Bosch TM, Huitema AD, Doodeman VD, et al. Pharmacogenetic screening of CYP3A and ABCB1 in relation to population pharmacokinetics of docetaxel. Clin Cancer Res. 2006;12(19):5786–5793. doi: 10.1158/1078-0432.CCR-05-2649. [DOI] [PubMed] [Google Scholar]
- 2.Weng L, Zhang L, Peng Y, Huang RS. Pharmacogenetics and pharmacogenomics: a bridge to individualized cancer therapy. Pharmacogenomics. 2013;14(3):315–324. doi: 10.2217/pgs.12.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamb DC, Waterman MR, Kelly SL, Guengerich FP. Cytochromes P450 and drug discovery. Curr Opin Biotechnol. 2007;18(6):504–512. doi: 10.1016/j.copbio.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Jancova P, Anzenbacher P, Anzenbacherova E. Phase II drug metabolizing enzymes. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2010;154(2):103–116. doi: 10.5507/bp.2010.017. [DOI] [PubMed] [Google Scholar]
- 5.McLeod HL, Krynetski EY, Relling MV, Evans WE. Genetic polymorphism of thiopurine methyltransferase and its clinical relevance for childhood acute lymphoblastic leukemia. Leukemia. 2000;14:567–572. doi: 10.1038/sj.leu.2401723. [DOI] [PubMed] [Google Scholar]
- 6.Rosmarin D, Palles C, Church D, et al. Genetic markers of toxicity from capecitabine and other fluorouracil-based regimens: investigation in the QUASAR2 study, systematic review, and meta-analysis. J Clin Oncol. 2014;32(10):1031–1039. doi: 10.1200/JCO.2013.51.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ieiri I. Functional significance of genetic polymorphisms in P-glycoprotein (MDR1, ABCB1) and breast cancer resistance protein (BCRP, ABCG2) Drug Metab Pharmacokinet. 2012;27(1):85–105. doi: 10.2133/dmpk.DMPK-11-RV-098. [DOI] [PubMed] [Google Scholar]
- 8.Passot C, Azzopardi N, Renault S, et al. Influence of FCGRT gene polymorphisms on pharmacokinetics of therapeutic antibodies. mAbs. 2013;5(4):614–9. doi:10.4161/mabs.24815. [DOI] [PMC free article] [PubMed]
- 9.Rodriguez-Antona C, Ingelman-Sundberg M. Cytochrome P450 pharmacogenetics and cancer. Oncogene. 2006;25(11):1679–1691. doi: 10.1038/sj.onc.1209377. [DOI] [PubMed] [Google Scholar]
- 10.van Schaik RH. Cancer treatment and pharmacogenetics of cytochrome P450 enzymes. Invest New Drugs. 2005;23(6):513–522. doi: 10.1007/s10637-005-4019-1. [DOI] [PubMed] [Google Scholar]
- 11.Choi HG, Jeon JY, Im YJ, et al. Pharmacokinetic properties of two erlotinib 150 mg formulations with a genetic effect evaluation in healthy Korean subjects. Clin Drug Investig. 2015;35(1):31–43. doi: 10.1007/s40261-014-0248-4. [DOI] [PubMed] [Google Scholar]
- 12.Wang H, Bian T, Liu D, et al. Association analysis of CYP2A6 genotypes and haplotypes with 5-fluorouracil formation from tegafur in human liver microsomes. Pharmacogenomics. 2011;12(4):481–492. doi: 10.2217/pgs.10.202. [DOI] [PubMed] [Google Scholar]
- 13.Park SR, Hong YS, Lim HS, et al. Phase I clinical and pharmacokinetic/pharmacogenetic study of a triplet regimen of S-1/irinotecan/oxaliplatin in patients with metastatic colorectal or gastric cancer. Cancer Chemother Pharmacol. 2013;72(5):953–964. doi: 10.1007/s00280-013-2272-0. [DOI] [PubMed] [Google Scholar]
- 14.Hirose T, Fujita K, Nishimura K, et al. Pharmacokinetics of S-1 and CYP2A6 genotype in Japanese patients with advanced cancer. Oncol Rep. 2010;24(2):529–536. doi: 10.3892/or_00000889. [DOI] [PubMed] [Google Scholar]
- 15.Veal GJ, Cole M, Chinnaswamy G, et al. Cyclophosphamide pharmacokinetics and pharmacogenetics in children with B-cell non-Hodgkin’s lymphoma. Eur J Cancer. 2016;55:56–64. doi: 10.1016/j.ejca.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakajima M, Komagata S, Fujiki Y, et al. Genetic polymorphisms of CYP2B6 affect the pharmacokinetics/pharmacodynamics of cyclophosphamide in Japanese cancer patients. Pharmacogenet Genom. 2007;17(6):431–445. doi: 10.1097/FPC.0b013e328045c4fb. [DOI] [PubMed] [Google Scholar]
- 17.Xie H, Griskevicius L, Stahle L, et al. Pharmacogenetics of cyclophosphamide in patients with hematological malignancies. Eur J Pharm Sci. 2006;27(1):54–61. doi: 10.1016/j.ejps.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 18.Ariyoshi N, Ohara M, Kaneko M, et al. Q172H replacement overcomes effects on the metabolism of cyclophosphamide and efavirenz caused by CYP2B6 variant with Arg262. Drug Metab Dispos. 2011;39(11):2045–2048. doi: 10.1124/dmd.111.039586. [DOI] [PubMed] [Google Scholar]
- 19.Uppugunduri CR, Rezgui MA, Diaz PH, et al. The association of cytochrome P450 genetic polymorphisms with sulfolane formation and the efficacy of a busulfan-based conditioning regimen in pediatric patients undergoing hematopoietic stem cell transplantation. Pharmacogenomics J. 2014;14(3):263–271. doi: 10.1038/tpj.2013.38. [DOI] [PubMed] [Google Scholar]
- 20.Ekhart C, Doodeman VD, Rodenhuis S, et al. Influence of polymorphisms of drug metabolizing enzymes (CYP2B6, CYP2C9, CYP2C19, CYP3A4, CYP3A5, GSTA1, GSTP1, ALDH1A1 and ALDH3A1) on the pharmacokinetics of cyclophosphamide and 4-hydroxycyclophosphamide. Pharmacogenet Genom. 2008;18(6):515–523. doi: 10.1097/FPC.0b013e3282fc9766. [DOI] [PubMed] [Google Scholar]
- 21.Jin Y, Desta Z, Stearns V, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;97(1):30–39. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- 22.Gjerde J, Hauglid M, Breilid H, et al. Effects of CYP2D6 and SULT1A1 genotypes including SULT1A1 gene copy number on tamoxifen metabolism. Ann Oncol. 2008;19(1):56–61. doi: 10.1093/annonc/mdm434. [DOI] [PubMed] [Google Scholar]
- 23.Borges S, Desta Z, Li L, et al. Quantitative effect of CYP2D6 genotype and inhibitors on tamoxifen metabolism: implication for optimization of breast cancer treatment. Clin Pharmacol Ther. 2006;80(1):61–74. doi: 10.1016/j.clpt.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 24.Regan MM, Leyland-Jones B, Bouzyk M, et al. CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the Breast International Group 1-98 Trial. J Natl Cancer Inst. 2012;104(6):441–451. doi: 10.1093/jnci/djs125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rae JM, Drury S, Hayes DF, et al. ATAC trialists. CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. J Natl Cancer Inst. 2012;104(6):452–460. doi: 10.1093/jnci/djs126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakamura Y, Ratain MJ, Cox NJ, et al. Re: CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the Breast International Group 1-98 Trial. J Natl Cancer Inst. 2012;104(16):1264. doi:10.1093/jnci/djs304(author reply 1266–8). [DOI] [PubMed]
- 27.de Vries Schultink AH, Zwart W, Linn SC, et al. Effects of pharmacogenetics on the pharmacokinetics and pharmacodynamics of tamoxifen. Clin Pharmacokinet. 2015;54(8):797–810. doi: 10.1007/s40262-015-0273-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi H, Sato K, Niioka T, et al. Relationship among gefitinib exposure, polymorphisms of its metabolizing enzymes and transporters, and side effects in Japanese patients with non-small-cell lung cancer. Clin Lung Cancer. 2015;16(4):274–28. doi:10.1016/j.cllc.2014.12.004. [DOI] [PubMed]
- 29.Hirose T, Fujita K, Kusumoto S, et al. Association of pharmacokinetics and pharmacogenomics with safety and efficacy of gefitinib in patients with EGFR mutation positive advanced non-small cell lung cancer. Lung Cancer. 2016;93:69–76. doi: 10.1016/j.lungcan.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 30.Deenen MJ, Cats A, Beijnen JH, Schellens JH. Part 2: pharmacogenetic variability in drug transport and phase I anticancer drug metabolism. Oncologist. 2011;16(6):820–834. doi: 10.1634/theoncologist.2010-0259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diekstra MH, Klumpen HJ, Lolkema MP, et al. Association analysis of genetic polymorphisms in genes related to sunitinib pharmacokinetics, specifically clearance of sunitinib and SU12662. Clin Pharmacol Ther. 2014;96(1):81–89. doi: 10.1038/clpt.2014.47. [DOI] [PubMed] [Google Scholar]
- 32.Johnson GG, Lin K, Cox TF, et al. CYP2B6*6 is an independent determinant of inferior response to fludarabine plus cyclophosphamide in chronic lymphocytic leukemia. Blood. 2013;122(26):4253–4258. doi: 10.1182/blood-2013-07-516666. [DOI] [PubMed] [Google Scholar]
- 33.Gaziev J, Nguyen L, Puozzo C, et al. Novel pharmacokinetic behavior of intravenous busulfan in children with thalassemia undergoing hematopoietic stem cell transplantation: a prospective evaluation of pharmacokinetic and pharmacodynamic profile with therapeutic drug monitoring. Blood. 2010;115(22):4597–4604. doi: 10.1182/blood-2010-01-265405. [DOI] [PubMed] [Google Scholar]
- 34.Ekhart C, Doodeman VD, Rodenhuis S, et al. Polymorphisms of drug-metabolizing enzymes (GST, CYP2B6 and CYP3A) affect the pharmacokinetics of thiotepa and tepa. Br J Clin Pharmacol. 2009;67(1):50–60. doi: 10.1111/j.1365-2125.2008.03321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strassburg CP. Pharmacogenetics of Gilbert’s syndrome. Pharmacogenomics. 2008;9(6):703–715. doi: 10.2217/14622416.9.6.703. [DOI] [PubMed] [Google Scholar]
- 36.Hu ZY, Yu Q, Pei Q, Guo C. Dose-dependent association between UGT1A1*28 genotype and irinotecan-induced neutropenia: low doses also increase risk. Clin Cancer Res. 2010;16(15):3832–3842. doi: 10.1158/1078-0432.CCR-10-1122. [DOI] [PubMed] [Google Scholar]
- 37.Hoskins JM, Goldberg RM, Qu P, et al. UGT1A1*28 genotype and irinotecan-induced neutropenia: dose matters. J Natl Cancer Inst. 2007;99(17):1290–1295. doi: 10.1093/jnci/djm115. [DOI] [PubMed] [Google Scholar]
- 38.Onoue M, Terada T, Kobayashi M, et al. UGT1A1*6 polymorphism is most predictive of severe neutropenia induced by irinotecan in Japanese cancer patients. Int J Clin Oncol. 2009;14(2):136–142. doi: 10.1007/s10147-008-0821-z. [DOI] [PubMed] [Google Scholar]
- 39.Beutler E, Gelbart T, Demina A. Racial variability in the UDP-glucuronosyltransferase 1 (UGT1A1) promoter: a balanced polymorphism for regulation of bilirubin metabolism? Proc Natl Acad Sci. 1998;95(14):8170–8174. doi: 10.1073/pnas.95.14.8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall D, Ybazeta G, Destro-Bisol G, et al. Variability at the uridine diphosphate glucuronosyltransferase 1A1 promoter in human populations and primates. Pharmacogenetics. 1999;9(5):591–599. doi: 10.1097/00008571-199910000-00006. [DOI] [PubMed] [Google Scholar]
- 41.Akaba K, Kimura T, Sasaki A, et al. Neonatal hyperbilirubinemia and mutation of the bilirubin uridine diphosphate-glucuronosyltransferase gene: a common missense mutation among Japanese, Koreans and Chinese. Biochem Mol Biol Int. 1998;46(1):21–26. doi: 10.1080/15216549800203512. [DOI] [PubMed] [Google Scholar]
- 42.Innocenti F, Schilsky RL, Ramirez J, et al. Dose-finding and pharmacokinetic study to optimize the dosing of irinotecan according to the UGT1A1 genotype of patients with cancer. J Clin Oncol. 2014;32(22):2328–2334. doi: 10.1200/JCO.2014.55.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toffoli G, Cecchin E, Gasparini G, et al. Genotype-driven phase I study of irinotecan administered in combination with fluorouracil/leucovorin in patients with metastatic colorectal cancer. J Clin Oncol. 2010;28(5):866–871. doi: 10.1200/JCO.2009.23.6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manfredi S, Bouche O, Rougier P, et al. High-dose FOLFIRI plus bevacizumab in the treatment of metastatic colorectal cancer patients with two different UGT1A1 genotypes: FFCD 0504 Study. Mol Cancer Ther. 2015;14(12):2782–2788. doi: 10.1158/1535-7163.MCT-15-0293. [DOI] [PubMed] [Google Scholar]
- 45.Van Rompay AR, Johansson M, Karlsson A. Substrate specificity and phosphorylation of antiviral and anticancer nucleoside analogues by human deoxyribonucleoside kinases and ribonucleoside kinases. Pharmacol Ther. 2003;100:119–139. doi: 10.1016/j.pharmthera.2003.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meyerhardt JA, Mayer RJ. Systemic therapy for colorectal cancer. N Engl J Med. 2005;352:476–487. doi: 10.1056/NEJMra040958. [DOI] [PubMed] [Google Scholar]
- 47.Twelves C, Wong A, Nowacki MP, et al. Capecitabine as adjuvant treatment for stage III colon cancer. N Engl J Med. 2005;352:2696–2704. doi: 10.1056/NEJMoa043116. [DOI] [PubMed] [Google Scholar]
- 48.Heggie GD, Sommadossi JP, Cross DS, et al. Clinical pharmacokinetics of 5-fluorouracil and its metabolites in plasma, urine, and bile. Cancer Res. 1987;47:2203–2206. [PubMed] [Google Scholar]
- 49.van Kuilenburg ABP. Dihydropyrimidine dehydrogenase and the efficacy and toxicity of 5-fluorouracil. Eur J Cancer. 2004;40:939–950. doi: 10.1016/j.ejca.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 50.Etienne MC, Chatelut E, Pivot X, et al. Co-variables influencing 5-fluorouracil clearance during continuous venous infusion: a NONMEM analysis. Eur J Cancer. 1998;34:92–97. doi: 10.1016/S0959-8049(97)00345-6. [DOI] [PubMed] [Google Scholar]
- 51.Etienne MC, Lagrange JL, Dassonville O, et al. Population study of dihydropyrimidine dehydrogenase in cancer patients. J Clin Oncol. 1994;12:2248–2253. doi: 10.1200/JCO.1994.12.11.2248. [DOI] [PubMed] [Google Scholar]
- 52.Di Paolo A, Danesi R, Falcone A, et al. Relationship between 5-fluorouracil disposition, toxicity and dihydropyrimidine dehydrogenase activity in cancer patients. Ann Oncol. 2001;12:1301–1306. doi: 10.1023/A:1012294617392. [DOI] [PubMed] [Google Scholar]
- 53.Maring JG, van Kuilenburg ABP, Haasjes J, et al. Reduced 5-FU clearance in a patient with low DPD activity due to heterozygosity for a mutant allele of the DPYD gene. Br J Cancer. 2002;86:1028–1033. doi: 10.1038/sj.bjc.6600208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Froehlich TK, Amstutz U, Aebi S, et al. Clinical importance of risk variants in the dihydropyrimidine dehydrogenase gene for the prediction of early-onset fluoropyrimidine toxicity. Int J Cancer. 2015;136:730–739. doi: 10.1002/ijc.29025. [DOI] [PubMed] [Google Scholar]
- 55.Terrazzino S, Cargnin S, Del Re M, et al. DPYD IVS14+1G>A and 2846A>T genotyping for the prediction of severe fluoropyrimidine-related toxicity: a meta-analysis. Pharmacogenomics. 2013;14:1255–1272. doi: 10.2217/pgs.13.116. [DOI] [PubMed] [Google Scholar]
- 56.Lee AM, Shi Q, Pavey E, et al. Pmc4271081; DPYD variants as predictors of 5-fluorouracil toxicity in adjuvant colon cancer treatment (NCCTG N0147). J Natl Cancer Inst. 2014;106(12). doi:10.1093/jnci/dju298. [DOI] [PMC free article] [PubMed]
- 57.Meulendijks D, Henricks LM, Sonke GS, et al. Clinical relevance of DPYD variants c.1679T>G, c.1236G>A/HapB3, and c.1601G>A as predictors of severe fluoropyrimidine-associated toxicity: a systematic review and meta-analysis of individual patient data. Lancet Oncol. 2015;16:1639–1650. doi: 10.1016/S1470-2045(15)00286-7. [DOI] [PubMed] [Google Scholar]
- 58.van Kuilenburg ABP, Hausler P, Schalhorn A, et al. Evaluation of 5-fluorouracil pharmacokinetics in cancer patients with a c.1905+1G>A mutation in DPYD by means of a Bayesian limited sampling strategy. Clin Pharmacokinet. 2012;51:163–174. doi: 10.1007/BF03257473. [DOI] [PubMed] [Google Scholar]
- 59.Diasio RB, Beavers TL, Carpenter JT. Familial deficiency of dihydropyrimidine dehydrogenase: biochemical basis for familial pyrimidinemia and severe 5-fluorouracil-induced toxicity. J Clin Invest. 1988;81:47–51. doi: 10.1172/JCI113308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson MR, Wang K, Diasio RB. Profound dihydropyrimidine dehydrogenase deficiency resulting from a novel compound heterozygote genotype. Clin Cancer Res. 2002;8:768–774. [PubMed] [Google Scholar]
- 61.Morel A, Boisdron-Celle M, Fey L, et al. Clinical relevance of different dihydropyrimidine dehydrogenase gene single nucleotide polymorphisms on 5-fluorouracil tolerance. Mol Cancer Ther. 2006;5:2895–2904. doi: 10.1158/1535-7163.MCT-06-0327. [DOI] [PubMed] [Google Scholar]
- 62.van Staveren MC, van Kuilenburg AB, Guchelaar HJ, et al. Pmc4767209; evaluation of an oral uracil loading test to identify DPD-deficient patients using a limited sampling strategy. Br J Clin Pharmacol. 2016;81:553–561. doi: 10.1111/bcp.12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Kuilenburg ABP, Dobritzsch D, Meijer J, et al. Dihydropyrimidinase deficiency: phenotype, genotype and structural consequences in 17 patients. Biochim Biophys Acta. 2010;1802:639–648. doi: 10.1016/j.bbadis.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 64.Sumi S, Imaeda M, Kidouchi K, et al. Population and family studies of dihydropyrimidinuria: prevalence, inheritance mode, and risk of fluorouracil toxicity. Am J Med Genet. 1998;78:336–340. doi: 10.1002/(SICI)1096-8628(19980724)78:4<336::AID-AJMG6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 65.van Kuilenburg ABP, Meinsma JR, Zonnenberg BA, et al. Dihydropyrimidinase deficiency and severe 5-fluorouracil toxicity. Clin Cancer Res. 2003;9:4363–4367. [PubMed] [Google Scholar]
- 66.Thomas HR, Ezzeldin HH, Guarcello V, et al. Genetic regulation of dihydropyrimidinase and its possible implication in altered uracil catabolism. Pharmacogenet Genom. 2007;17:973–987. doi: 10.1097/FPC.0b013e3282f01788. [DOI] [PubMed] [Google Scholar]
- 67.Kummer D, Froehlich TK, Joerger M, et al. Dihydropyrimidinase and beta-ureidopropionase gene variation and severe fluoropyrimidine-related toxicity. Pharmacogenomics. 2015;16:1367–1377. doi: 10.2217/pgs.15.81. [DOI] [PubMed] [Google Scholar]
- 68.Thomas HR, Ezzeldin HH, Guarcello V, et al. Genetic regulation of beta-ureidopropionase and its possible implication in altered uracil catabolism. Pharmacogenet Genom. 2008;18:25–35. doi: 10.1097/FPC.0b013e3282f2f134. [DOI] [PubMed] [Google Scholar]
- 69.van Kuilenburg AB, Dobritzsch D, Meijer J, et al. β-ureidopropionase deficiency: phenotype, genotype and protein structural consequences in 16 patients. Biochim Biophys Acta. 2012;1822:1096–1108. doi: 10.1016/j.bbadis.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 70.Queckenberg C, Erlinghagen V, Baken BC, et al. Pharmacokinetics and pharmacogenetics of capecitabine and its metabolites following replicate administration of two 500 mg tablet formulations. Cancer Chemother Pharmacol. 2015;76:1081–1091. doi: 10.1007/s00280-015-2840-6. [DOI] [PubMed] [Google Scholar]
- 71.Mei L, Ontiveros EP, Griffiths EA, et al. Pharmacogenetics predictive of response and toxicity in acute lymphoblastic leukemia therapy. Blood Rev. 2015;29(4):243–249. doi: 10.1016/j.blre.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Coskun M, Steenholdt C, de Boer NK, Nielsen OH. Pharmacology and optimization of thiopurines and methotrexate in inflammatory bowel disease. Clin Pharmacokinet. 2016;55:257–274. doi: 10.1007/s40262-015-0316-9. [DOI] [PubMed] [Google Scholar]
- 73.Adam dB, Jacqz-Aigrain E. Pharmacogenetic determinants of mercaptopurine disposition in children with acute lymphoblastic leukemia. Eur J Clin Pharmacol. 2012;68:1233–42. doi:10.1007/s00228-012-1251-4. [DOI] [PubMed]
- 74.Coenen MJ, de Jong DJ, van Marrewijk CJ, et al. Identification of patients with variants in TPMT and dose reduction reduces hematologic events during thiopurine treatment of inflammatory bowel disease. Gastroenterology. 2015;149:907–17.e7; doi:10.1053/j.gastro.2015.06.002. [DOI] [PubMed]
- 75.Zimm S, Collins JM, Riccardi R, et al. Variable bioavailability of oral mercaptopurine: is maintenance chemotherapy in acute lymphoblastic leukemia being optimally delivered? N Engl J Med. 1983;308:1005–1009. doi: 10.1056/NEJM198304283081705. [DOI] [PubMed] [Google Scholar]
- 76.Lonnerholm G, Kreuger A, Lindstrom B, et al. Plasma and erythrocyte concentrations of mercaptopurine after oral administration in children. Pediatr Hematol Oncol. 1986;3:27–35. doi: 10.3109/08880018609031198. [DOI] [PubMed] [Google Scholar]
- 77.Hawwa AF, Collier PS, Millership JS, et al. Pmc2675766; population pharmacokinetic and pharmacogenetic analysis of 6-mercaptopurine in paediatric patients with acute lymphoblastic leukaemia. Br J Clin Pharmacol. 2008;66:826–837. doi: 10.1111/j.1365-2125.2008.03281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ogungbenro K, Aarons L. Pmc4500328; physiologically based pharmacokinetic model for 6-mercpatopurine: exploring the role of genetic polymorphism in TPMT enzyme activity. Br J Clin Pharmacol. 2015;80:86–100. doi: 10.1111/bcp.12588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kapoor G, Sinha R, Abedin S. Experience with high dose methotrexate therapy in childhood acute lymphoblastic leukemia in a tertiary care cancer centre of a developing country. Pediatr Blood Cancer. 2012;59:448–453. doi: 10.1002/pbc.24081. [DOI] [PubMed] [Google Scholar]
- 80.Lehman NL. Future potential of thymidylate synthase inhibitors in cancer therapy. Expert Opin Investig Drugs. 2002;11:1775–1787. doi: 10.1517/13543784.11.12.1775. [DOI] [PubMed] [Google Scholar]
- 81.Lima A, Sousa H, Monteiro J, et al. Genetic polymorphisms in low-dose methotrexate transporters: current relevance as methotrexate therapeutic outcome biomarkers. Pharmacogenomics. 2014;15:1611–1635. doi: 10.2217/pgs.14.116. [DOI] [PubMed] [Google Scholar]
- 82.Inoue K, Yuasa H. Molecular basis for pharmacokinetics and pharmacodynamics of methotrexate in rheumatoid arthritis therapy. Drug Metab Pharmacokinet. 2014;29:12–19. doi: 10.2133/dmpk.DMPK-13-RV-119. [DOI] [PubMed] [Google Scholar]
- 83.Moya P, Salazar J, Arranz MJ, et al. Methotrexate pharmacokinetic genetic variants are associated with outcome in rheumatoid arthritis patients. Pharmacogenomics. 2016;17:25–29. doi: 10.2217/pgs.15.150. [DOI] [PubMed] [Google Scholar]
- 84.Hider SL, Bruce IN, Thomson W. The pharmacogenetics of methotrexate. Rheumatology (Oxford) 2007;46:1520–1524. doi: 10.1093/rheumatology/kem147. [DOI] [PubMed] [Google Scholar]
- 85.Serdjebi C, Milano G, Ciccolini J. Role of cytidine deaminase in toxicity and efficacy of nucleosidic analogs. Expert Opin Drug Metab Toxicol. 2015;11:665–672. doi: 10.1517/17425255.2015.985648. [DOI] [PubMed] [Google Scholar]
- 86.Krogh-Madsen M, Bender B, Jensen MK, et al. Population pharmacokinetics of cytarabine, etoposide, and daunorubicin in the treatment for acute myeloid leukemia. Cancer Chemother Pharmacol. 2012;69:1155–1163. doi: 10.1007/s00280-011-1800-z. [DOI] [PubMed] [Google Scholar]
- 87.Knights J, Sato Y, Kaniwa N, et al. Genetic factors associated with gemcitabine pharmacokinetics, disposition, and toxicity. Pharmacogenet Genom. 2014;24:15–25. doi: 10.1097/FPC.0000000000000016. [DOI] [PubMed] [Google Scholar]
- 88.Sugiyama E, Kaniwa N, Kim SR, et al. Pharmacokinetics of gemcitabine in Japanese cancer patients: the impact of a cytidine deaminase polymorphism. J Clin Oncol. 2007;25:32–42. doi: 10.1200/JCO.2006.06.7405. [DOI] [PubMed] [Google Scholar]
- 89.Maring JG, Wachters FM, Slijfer M, et al. Pmc2868996; pharmacokinetics of gemcitabine in non-small-cell lung cancer patients: impact of the 79A>C cytidine deaminase polymorphism. Eur J Clin Pharmacol. 2010;66:611–617. doi: 10.1007/s00228-010-0799-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Karahoca M, Momparler RL. Pmc3570332; pharmacokinetic and pharmacodynamic analysis of 5-aza-2′-deoxycytidine (decitabine) in the design of its dose-schedule for cancer therapy. Clin Epigenetics. 2013;5:3. doi: 10.1186/1868-7083-5-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.McCune JS, Vicini P, Salinger DH, et al. Pmc4282607; population pharmacokinetic/dynamic model of lymphosuppression after fludarabine administration. Cancer Chemother Pharmacol. 2015;75:67–75. doi: 10.1007/s00280-014-2618-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bonate PL, Cunningham CC, Gaynon P, et al. Population pharmacokinetics of clofarabine and its metabolite 6-ketoclofarabine in adult and pediatric patients with cancer. Cancer Chemother Pharmacol. 2011;67:875–890. doi: 10.1007/s00280-010-1376-z. [DOI] [PubMed] [Google Scholar]
- 93.Long-Boyle J, Huang J, Rydholm N, et al. Pharmacokinetics of clofarabine in patients with high-risk inherited metabolic disorders undergoing brain-sparing hematopoietic cell transplantation. J Clin Pharmacol. 2011;51:679–686. doi: 10.1177/0091270010372519. [DOI] [PubMed] [Google Scholar]
- 94.Wong A, Soo RA, Yong WP, Innocenti F. Clinical pharmacology and pharmacogenetics of gemcitabine. Drug Metab Rev. 2009;41:77–88. doi: 10.1080/03602530902741828. [DOI] [PubMed] [Google Scholar]
- 95.Mahfouz RZ, Jankowska A, Ebrahem Q, et al. Pmc3577958; increased CDA expression/activity in males contributes to decreased cytidine analog half-life and likely contributes to worse outcomes with 5-azacytidine or decitabine therapy. Clin Cancer Res. 2013;19:938–948. doi: 10.1158/1078-0432.CCR-12-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nagasubramanian R, Innocenti F, Ratain MJ. Pharmacogenetics in cancer treatment. Annu Rev Med. 2003;54:437–452. doi: 10.1146/annurev.med.54.101601.152352. [DOI] [PubMed] [Google Scholar]
- 97.Schwab M, Eichelbaum M, Fromm MF. Genetic polymorphisms of the human MDR1 drug transporter. Annu Rev Pharmacol Toxicol. 2003;43:285–307. doi: 10.1146/annurev.pharmtox.43.100901.140233. [DOI] [PubMed] [Google Scholar]
- 98.Ekhart C, Rodenhuis S, Smits PHM, et al. An overview of the relations between polymorphisms in drug metabolising enzymes and drug transporters and survival after cancer drug treatment. Cancer Treat Rev. 2009;35(1):18–31. doi:10.1016/j.ctrv.2008.07.003. [DOI] [PubMed]
- 99.Cascorbi I, Gerloff T, Johne A, et al. Frequency of single nucleotide polymorphisms in the P-glycoprotein drug transporter MDR1 gene in white subjects. Clin Pharmacol Ther. 2001;69(3):169–174. doi: 10.1067/mcp.2001.114164. [DOI] [PubMed] [Google Scholar]
- 100.Balram C, Sharma A, Sivathasan C, Lee EJD. Frequency of C3435T single nucleotide MDR1 genetic polymorphism in an Asian population: phenotypic–genotypic correlates. Br J Clin Pharmacol. 2002;56(1):78–83. doi: 10.1046/j.1365-2125.2003.01820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hoffmeyer S, Burk O, von Richter O, et al. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci. 2000;97(7):3473–3478. doi: 10.1073/pnas.97.7.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Allen JD, Schinkel AH. Multidrug resistance and pharmacological protection mediated by the breast cancer resistance protein (BCRP/ABCG2) Mol Cancer Ther. 2002;1(6):427–434. [PubMed] [Google Scholar]
- 103.Imai Y, Nakane M, Kage K, et al. C421A polymorphism in the human breast cancer resistance protein gene is associated with low expression of Q141K protein and low-level drug resistance. Mol Cancer Ther. 2002;1(8):611–616. [PubMed] [Google Scholar]
- 104.Oshiro C, Marsh S, McLeod H. Taxane pathway. Pharmacogenet Genom. 2009;19(12):979–983. doi: 10.1097/FPC.0b013e3283335277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Iuliis F, Salerno G, Taglieri L, Scarpa S. Are pharmacogenomic biomarkers an effective tool to predict taxane toxicity and outcome in breast cancer patients? Literature review. Cancer Chemother Pharmacol. 2015;76(4):679–690. doi: 10.1007/s00280-015-2818-4. [DOI] [PubMed] [Google Scholar]
- 106.Lal S, Wong ZW, Sandanaraj E, et al. Influence of ABCB1 and ABCG2 polymorphisms on doxorubicin disposition in Asian breast cancer patients. Cancer Sci. 2008;99(4):816–823. doi: 10.1111/j.1349-7006.2008.00744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jamieson D, Lee J, Cresti N, Jackson R. Pharmacogenetics of adjuvant breast cancer treatment with cyclophosphamide, epirubicin and 5-fluorouracil. Cancer Chemother Pharmacol. 2014;74(4):667–674. doi: 10.1007/s00280-014-2541-6. [DOI] [PubMed] [Google Scholar]
- 108.Thompson P, Wheeler HE, Delaney SM, Lorier R, Broeckel U, Devidas M, et al. Pharmacokinetics and pharmacogenomics of daunorubicin in children: a report from the Children’s Oncology Group. Cancer Chemother Pharmacol. 2014;74(4):831–838. doi: 10.1007/s00280-014-2535-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Saltz L. Irinotecan-based combinations for the adjuvant treatment of stage III colon cancer. Oncology (Williston Park). 2000;14(12 Suppl 14):47–50. [PubMed] [Google Scholar]
- 110.Mathijssen RH, Marsh S, Karlsson MO, et al. Irinotecan pathway genotype analysis to predict pharmacokinetics. Clin Cancer Res. 2003;9(9):3246–3253. [PubMed] [Google Scholar]
- 111.Sai K, Kaniwa N, Itoda M, et al. Haplotype analysis of ABCB1/MDR1 blocks in a Japanese population reveals genotype-dependent renal clearance of irinotecan. Pharmacogenetics. 2003;13(12):741–757. doi: 10.1097/00008571-200312000-00005. [DOI] [PubMed] [Google Scholar]
- 112.Cockshott ID. Bicalutamide: clinical pharmacokinetics and metabolism. Clin Pharmacokinet. 2004;43(13):855–878. doi: 10.2165/00003088-200443130-00003. [DOI] [PubMed] [Google Scholar]
- 113.Kim K, Cha Y, Lee H, et al. Effects of the ABCG2 and ABCB1 drug transporter polymorphisms on the pharmacokinetics of bicalutamide in humans. Clin Chim Acta. 2015;438:7–11. doi:10.1016/j.cca.2014.08.006. [DOI] [PubMed]
- 114.van Erp NP, Gelderblom H, Guchelaar HJ. Clinical pharmacokinetics of tyrosine kinase inhibitors. Cancer Treat Rev. 2009;35(8):692–706. doi: 10.1016/j.ctrv.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 115.Polillo M, Galimberti S, Baratè C, et al. Pharmacogenetics of BCR/ABL inhibitors in chronic myeloid leukemia. Int J Mol Sci. 2015;16(9):22811–22829. doi: 10.3390/ijms160922811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dulucq S, Bouchet S, Turcq B, et al. Multidrug resistance gene (MDR1) polymorphisms are associated with major molecular responses to standard-dose imatinib in chronic myeloid leukemia. Blood. 2008;112(5):2024–2027. doi: 10.1182/blood-2008-03-147744. [DOI] [PubMed] [Google Scholar]
- 117.Gurney H, Wong M, Balleine RL, et al. Imatinib disposition and ABCB1 (MDR1, P-glycoprotein) genotype. Clin Pharmacol Ther. 2007;82(1):33–40. doi: 10.1038/sj.clpt.6100201. [DOI] [PubMed] [Google Scholar]
- 118.Petain A, Kattygnarath D, Azard J, Innovative Therapies with Children with Cancer European Consortium et al. Population pharmacokinetics and pharmacogenetics of imatinib in children and adults. Clin Cancer Res. 2008;14(21):7102–7109. doi: 10.1158/1078-0432.CCR-08-0950. [DOI] [PubMed] [Google Scholar]
- 119.Takahashi N, Miura M, Scott SA, et al. Influence of CYP3A5 and drug transporter polymorphisms on imatinib trough concentration and clinical response among patients with chronic phase chronic myeloid leukemia. J Hum Genet. 2010;55(11):731–737. doi: 10.1038/jhg.2010.98. [DOI] [PubMed] [Google Scholar]
- 120.Seong SJ, Lim M, Sohn SK, et al. Influence of enzyme and transporter polymorphisms on trough imatinib concentration and clinical response in chronic myeloid leukemia patients. Ann Oncol. 2013;24(3):756–760. doi: 10.1093/annonc/mds532. [DOI] [PubMed] [Google Scholar]
- 121.Vivona D, Lima LT, Rodrigues AC, et al. ABCB1 haplotypes are associated with P-gp activity and affect a major molecular response in chronic myeloid leukemia patients treated with a standard dose of imatinib. Oncol Lett. 2014;7(4):1313–1319. doi: 10.3892/ol.2014.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vine J, Cohen SB, Ruchlemer R, et al. Polymorphisms in the human organic cation transporter and the multidrug resistance gene: correlation with imatinib levels and clinical course in patients with chronic myeloid leukemia. Leuk Lymphoma. 2014;55(11):2525–2531. doi: 10.3109/10428194.2014.893307. [DOI] [PubMed] [Google Scholar]
- 123.Li J, Cusatis G, Brahmer J, et al. Association of variant ABCG2 and the pharmacokinetics of epidermal growth factor receptor tyrosine kinase inhibitors in cancer patients. Cancer Biol Ther. 2007;6(3):432–438. doi: 10.4161/cbt.6.3.3763. [DOI] [PubMed] [Google Scholar]
- 124.Lu L, Saunders VA, Leclercq TM, et al. Ponatinib is not transported by ABCB1, ABCG2 or OCT-1 in CML cells. Leukemia. 2015;29(8):1792–1794. doi: 10.1038/leu.2015.35. [DOI] [PubMed] [Google Scholar]
- 125.Brennan M, Williams JA, Chen Y, et al. Meta-analysis of contribution of genetic polymorphisms in drug-metabolizing enzymes or transporters to axitinib pharmacokinetics. Eur J Clin Pharmacol. 2011;68(5):645–655. doi: 10.1007/s00228-011-1171-8. [DOI] [PubMed] [Google Scholar]
- 126.Boudou-Rouquette P, Narjoz C, Golmard JL, et al. Early sorafenib-induced toxicity is associated with drug exposure and UGTIA9 genetic polymorphism in patients with solid tumors: a preliminary study. PLoS One. 2012;7(8):e42875. doi: 10.1371/journal.pone.0042875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kerb R, Brinkmann U, Chatskaia N, et al. Identification of genetic variations of the human organic cation transporter hOCT1 and their functional consequences. Pharmacogenetics. 2002;12(8):591–595. doi: 10.1097/00008571-200211000-00002. [DOI] [PubMed] [Google Scholar]
- 128.Di Paolo A, Polillo M, Capecchi M, et al. The c.480C>G polymorphism of hOCT1 influences imatinib clearance in patients affected by chronic myeloid leukemia. Pharmacogenomics J. 2014;14(4):328–335. doi: 10.1038/tpj.2014.7. [DOI] [PubMed] [Google Scholar]
- 129.Wold ED, Smider V, Felding BH. Antibody therapeutics in oncology. Immunother Open Acc. 2016;2(108). doi:10.4172/imt.1000108. [PMC free article] [PubMed]
- 130.Breedveld F. Therapeutic monoclonal antibodies. Lancet. 2000;355(9205):735–40. doi:10.1016/S0140-6736(00)01034-5. [DOI] [PubMed]
- 131.Ternant D, Paintaud G. Pharmacokinetics and concentration–effect relationships of therapeutic monoclonal antibodies and fusion proteins. Exp Opin Biol Ther. 2005;5:S37–S47. doi: 10.1517/14712598.5.1.S37. [DOI] [PubMed] [Google Scholar]
- 132.Keizer RJ, Huitema AD, Schellens JH, Beijnen JH. Clinical pharmacokinetics of therapeutic monoclonal antibodies. Clin Pharmacokinet. 2010;49(8):493–507. doi: 10.2165/11531280-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 133.Ternant D, Bejan-Angoulvant T, Passot C, et al. Clinical pharmacokinetics and pharmacodynamics of monoclonal antibodies approved to treat rheumatoid arthritis. Clin Pharmacokinet. 2015;54(11):1107–1123. doi: 10.1007/s40262-015-0296-9. [DOI] [PubMed] [Google Scholar]
- 134.Borvak J, Richardson J, Medesan C, et al. Functional expression of the MHC class I-related receptor, FcRn, in endothelial cells of mice. Int Immunol. 1998;10(9):1289–1298. doi: 10.1093/intimm/10.9.1289. [DOI] [PubMed] [Google Scholar]
- 135.Mikulska JE, Pablo L, Canel J, Simister NE. Cloning and analysis of the gene encoding the human neonatal Fc receptor. Eur J Immunogenet. 2000;27(4):231–240. doi: 10.1046/j.1365-2370.2000.00225.x. [DOI] [PubMed] [Google Scholar]
- 136.Gouilleux-Gruart V, Chapel H, Chevret S, et al. DEFI Study Group. Efficiency of immunoglobulin G replacement therapy in common variable immunodeficiency: correlations with clinical phenotype and polymorphism of the neonatal Fc receptor. Clin Exp Immunol. 2013;171(2):186–194. doi: 10.1111/cei.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tabrizi MA, Tseng CM, Roskos LK. Elimination mechanisms of therapeutic monoclonal antibodies. Drug Discov Today. 2006;11(1–2):81–88. doi: 10.1016/S1359-6446(05)03638-X. [DOI] [PubMed] [Google Scholar]
- 138.Kim DH, Jung HD, Kim JG, et al. FCGR3A gene polymorphisms may correlate with response to frontline R-CHOP therapy for diffuse large B-cell lymphoma. Blood. 2006;108(8):2720–2725. doi: 10.1182/blood-2006-01-009480. [DOI] [PubMed] [Google Scholar]
- 139.Weng WK, Negrin RS, Lavori P, Horning SJ. Immunoglobulin G Fc receptor FcgammaRIIIa 158 V/F polymorphism correlates with rituximab-induced neutropenia after autologous transplantation in patients with non-Hodgkin’s lymphoma. J Clin Oncol. 2010;28(2):279–284. doi: 10.1200/JCO.2009.25.0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Persky DO, Dornan D, Goldman BH, et al. Fc gamma receptor 3a genotype predicts overall survival in follicular lymphoma patients treated on SWOG trials with combined monoclonal antibody plus chemotherapy but not chemotherapy alone. Haematologica. 2012;97(6):937–942. doi: 10.3324/haematol.2011.050419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ahlgrimm M, Pfreundschuh M, Kreuz M, et al. The impact of Fc-gamma receptor polymorphisms in elderly patients with diffuse large B-cell lymphoma treated with CHOP with or without rituximab. Blood. 2011;118(17):4657–4662. doi: 10.1182/blood-2011-04-346411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cartron G, Dacheux L, Salles G, et al. Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcγRIIIa gene. Blood. 2002;99(3):754–758. doi: 10.1182/blood.V99.3.754. [DOI] [PubMed] [Google Scholar]
- 143.Musolino A, Naldi N, Bortesi B, et al. Immunoglobulin G fragment C receptor polymorphisms and clinical efficacy of trastuzumab-based therapy in patients with HER-2/neu-positive metastatic breast cancer. J Clin Oncol. 2008;26(11):1789–1796. doi: 10.1200/JCO.2007.14.8957. [DOI] [PubMed] [Google Scholar]
- 144.Tamura K, Shimizu C, Hojo T, et al. FcgammaR2A and 3A polymorphisms predict clinical outcome of trastuzumab in both neoadjuvant and metastatic settings in patients with HER2-positive breast cancer. Ann Oncol. 2011;22(6):1302–1307. doi: 10.1093/annonc/mdq585. [DOI] [PubMed] [Google Scholar]
- 145.Kjersem JB, Skovlund E, Ikdahl T, et al. FCGR2A and FCGR3A polymorphisms and clinical outcome in metastatic colorectal cancer patients treated with first-line 5-fluorouracil/folinic acid and oxaliplatin +/− cetuximab. BMC Cancer. 2014;14(1):1–9. doi: 10.1186/1471-2407-14-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhang W, Gordon M, Schultheis AM, et al. FCGR2A and FCGR3A polymorphisms associated with clinical outcome of epidermal growth factor receptor expressing metastatic colorectal cancer patients treated with single-agent cetuximab. J Clin Oncol. 2007;25(24):3712–3718. doi: 10.1200/JCO.2006.08.8021. [DOI] [PubMed] [Google Scholar]
- 147.Bibeau F, Lopez-Crapez E, Di Fiore F, et al. Impact of FcγRIIa-FcγRIIIa polymorphisms and KRAS mutations on the clinical outcome of patients with metastatic colorectal cancer treated with cetuximab plus irinotecan. J Clin Oncol. 2009;27(7):1122–1129. doi: 10.1200/JCO.2008.18.0463. [DOI] [PubMed] [Google Scholar]
- 148.Rodriguez J, Zarate R, Bandres E, et al. Fc gamma receptor polymorphisms as predictive markers of cetuximab efficacy in epidermal growth factor receptor downstream-mutated metastatic colorectal cancer. Eur J Cancer. 2012;48(12):1774–1780. doi: 10.1016/j.ejca.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 149.Koene HR, Kleijer M, Algra J, et al. FcγRIIIa-158V/F polymorphism influences the binding of IgG by natural killer cell FcγRIIIa, independently of the FcγRIIIa-48L/R/H phenotype. Blood. 1997;90(3):1109–1114. [PubMed] [Google Scholar]
- 150.van Sorge NM, van der Pol WL, van de Winkel JG. FcgammaR polymorphisms: implications for function, disease susceptibility and immunotherapy. Tissue Antigens. 2003;61(3):189–202. doi: 10.1034/j.1399-0039.2003.00037.x. [DOI] [PubMed] [Google Scholar]
- 151.Van Cutsem E, Köhne C, Hitre E, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360(14):1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 152.Geva R, Vecchione L, Kalogeras KT, et al. FCGR polymorphisms and cetuximab efficacy in chemorefractory metastatic colorectal cancer: an international consortium study. Gut. 2015;64(6):921–928. doi: 10.1136/gutjnl-2014-307234. [DOI] [PubMed] [Google Scholar]
- 153.Feugier P, Van Hoof A, Sebban C, et al. Long-term results of the R-CHOP study in the treatment of elderly patients with diffuse large B-cell lymphoma: a study by the Groupe d’Etude des Lymphomes de l’Adulte. J Clin Oncol. 2005;23(18):4117–4126. doi: 10.1200/JCO.2005.09.131. [DOI] [PubMed] [Google Scholar]
- 154.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344(11):783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 155.Hurvitz SA, Betting DJ, Stern HM, et al. Analysis of Fcgamma receptor IIIa and IIa polymorphisms: lack of correlation with outcome in trastuzumab-treated breast cancer patients. Clin Cancer Res. 2012;18(12):3478–3486. doi: 10.1158/1078-0432.CCR-11-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ternant D, Berkane Z, Picon L, et al. Assessment of the influence of inflammation and FCGR3A genotype on infliximab pharmacokinetics and time to relapse in patients with Crohn’s disease. Clin Pharmacokinet. 2014;54(5):551–562. doi: 10.1007/s40262-014-0225-3. [DOI] [PubMed] [Google Scholar]
- 157.Dall’Ozzo S, Tartas S, Paintaud G, et al. Rituximab-dependent cytotoxicity by natural killer cells: influence of FCGR3A polymorphism on the concentration-effect relationship. Cancer Res. 2004;64(13):4664–9. [DOI] [PubMed]
- 158.Patel JN, Papachristos A. Personalizing chemotherapy dosing using pharmacological methods. Cancer Chemother Pharmacol. 2015;76(5):879–896. doi: 10.1007/s00280-015-2849-x. [DOI] [PubMed] [Google Scholar]
- 159.Huezo-Diaz P, Uppugunduri CR, Tyagi AK, et al. Pharmacogenetic aspects of drug metabolizing enzymes in busulfan based conditioning prior to allogenic hematopoietic stem cell transplantation in children. Curr Drug Metab. 2014;15(3):251–264. doi: 10.2174/1389200215666140202214012. [DOI] [PubMed] [Google Scholar]
- 160.ten Brink MH, Zwaveling J, Swen JJ, et al. Personalized busulfan and treosulfan conditioning for pediatric stem cell transplantation: the role of pharmacogenetics and pharmacokinetics. Drug Discov Today. 2014;19(10):1572–86. doi:10.1016/j.drudis.2014.04.005. [DOI] [PubMed]
- 161.Pandya U, Srivastava SK, Singhal SS, et al. Activity of allelic variants of Pi class human glutathione S-transferase toward chlorambucil. Biochem Biophys Res Commun. 2000;278(1):258–262. doi:10.1006/bbrc.2000.3787. [DOI] [PubMed]
- 162.Ekhart C, Rodenhuis S, Smits PHM, et al. Relations between polymorphisms in drug-metabolising enzymes and toxicity of chemotherapy with cyclophosphamide, thiotepa and carboplatin. Pharmacogenet Genom. 2008;18(11):1009–1015. doi: 10.1097/FPC.0b013e328313aaa4. [DOI] [PubMed] [Google Scholar]
- 163.Kuhne A, Kaiser R, Schirmer M, et al. Genetic polymorphisms in the amino acid transporters LAT1 and LAT2 in relation to the pharmacokinetics and side effects of melphalan. Pharmacogenet Genom. 2007;17(7):505–517. doi: 10.1097/FPC.0b013e3280ea77cd. [DOI] [PubMed] [Google Scholar]
- 164.Fanciullino R, Mercier C, Serdjebi C, et al. Yin and yang of cytidine deaminase roles in clinical response to azacitidine in the elderly: a pharmacogenetics tale. Pharmacogenomics. 2015;16(17):1907–1912. doi: 10.2217/pgs.15.135. [DOI] [PubMed] [Google Scholar]
- 165.Lennard L, Cartwright CS, Wade R, Vora A. Thiopurine dose intensity and treatment outcome in childhood lymphoblastic leukaemia: the influence of thiopurine methyltransferase pharmacogenetics. Br J Haematol. 2015;169(2):228–240. doi: 10.1111/bjh.13240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Guilhaumou R, Simon N, Quaranta S, et al. Population pharmacokinetics and pharmacogenetics of vincristine in paediatric patients treated for solid tumour diseases. Cancer Chemother Pharmacol. 2011;68(5):1191–1198. doi: 10.1007/s00280-010-1541-4. [DOI] [PubMed] [Google Scholar]
- 167.de Haas EC, Zwart N, Meijer C, et al. Variation in bleomycin hydrolase gene is associated with reduced survival after chemotherapy for testicular germ cell cancer. J Clin Oncol. 2008;26(11):1817–1823. doi: 10.1200/JCO.2007.14.1606. [DOI] [PubMed] [Google Scholar]
- 168.Hill CR, Cole M, Errington J, Malik G, Boddy AV, Veal GJ. Characterisation of the clinical pharmacokinetics of actinomycin D and the influence of ABCB1 pharmacogenetic variation on actinomycin D disposition in children with cancer. Clin Pharmacokinet. 2014;53(8):741–751. doi: 10.1007/s40262-014-0153-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kim TW, Innocenti F. Insights, challenges, and future directions in irinogenetics. Ther Drug Monit. 2007;29(3):265–270. doi: 10.1097/FTD.0b013e318068623b. [DOI] [PubMed] [Google Scholar]
- 170.Stewart CF, Tagen M, Schwartzberg LS, et al. Phase I dosage finding and pharmacokinetic study of intravenous topotecan and oral erlotinib in adults with refractory solid tumors. Cancer Chemother Pharmacol. 2014;73(3):561–568. doi: 10.1007/s00280-014-2385-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Abubakar MB, Wei K, Gan SH. The influence of genetic polymorphisms on the efficacy and side effects of anastrozole in postmenopausal breast cancer patients. Pharmacogenet Genom. 2014;24(12):575–581. doi: 10.1097/FPC.0000000000000092. [DOI] [PubMed] [Google Scholar]
- 172.Turkistani A, Marsh S. Pharmacogenomics of third-generation aromatase inhibitors. Expert Opin Pharmacother. 2012;13(9):1299–1307. doi: 10.1517/14656566.2012.687721. [DOI] [PubMed] [Google Scholar]
- 173.Mwinyi J, Vokinger K, Jetter A, et al. Impact of variable CYP genotypes on breast cancer relapse in patients undergoing adjuvant tamoxifen therapy. Cancer Chemother Pharmacol. 2014;73(6):1181–1188. doi: 10.1007/s00280-014-2453-5. [DOI] [PubMed] [Google Scholar]
- 174.Diekstra MHM, Swen JJ, Boven E, et al. CYP3A5 and ABCB1 polymorphisms as predictors for sunitinib outcome in metastatic renal cell carcinoma. Eur Urol. 2015;68(4):621–9. doi:10.1016/j.eururo.2015.04.018. [DOI] [PubMed]
- 175.Matsuzawa N, Nakamura K, Matsuda M, et al. Influence of cytochrome P450 2C19 gene variations on pharmacokinetic parameters of thalidomide in Japanese patients. Biol Pharm Bull. 2012;35(3):317–320. doi: 10.1248/bpb.35.317. [DOI] [PubMed] [Google Scholar]
- 176.Quinn DI, Nemunaitis J, Fuloria J, et al. Effect of the cytochrome P450 2C19 inhibitor omeprazole on the pharmacokinetics and safety profile of bortezomib in patients with advanced solid tumours, non-Hodgkin’s lymphoma or multiple myeloma. Clin Pharmacokinet. 2012;48(3):199–209. doi: 10.2165/00003088-200948030-00006. [DOI] [PubMed] [Google Scholar]
- 177.Roco A, Cayún J, Contreras S, Stojanova J, Quiñones L. Can pharmacogenetics explain efficacy and safety of cisplatin pharmacotherapy? Front Genet. 2014;5:391. doi: 10.3389/fgene.2014.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yamamiya I, Yoshisue K, Ishii Y, et al. Effect of CYP2A6 genetic polymorphism on the metabolic conversion of tegafur to 5-fluorouracil and its enantioselectivity. Drug Metab Dispos. 2014;42(9):1485–1492. doi: 10.1124/dmd.114.058008. [DOI] [PubMed] [Google Scholar]
- 179.Vangsted AJ, Søeby K, Klausen TW, et al. No influence of the polymorphisms CYP2C19 and CYP2D6 on the efficacy of cyclophosphamide, thalidomide, and bortezomib in patients with multiple myeloma. BMC Cancer. 2010;10(1):1–8. doi: 10.1186/1471-2407-10-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Li Y, Hou J, Jiang H, et al. Polymorphisms of CYP2C19 gene are associated with the efficacy of thalidomide based regimens in multiple myeloma. Haematologica. 2007;92(9):1246–1249. doi: 10.3324/haematol.11319. [DOI] [PubMed] [Google Scholar]
- 181.Barbarino JM, Haidar CE, Klein TE, Altman RB. PharmGKB summary: very important pharmacogene information for UGT1A1. Pharmacogenet Genom. 2014;24(3):177–183. doi: 10.1097/FPC.0000000000000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.van Kuilenburg ABP, Maring JG, Schalhorn A, et al. Pharmacokinetics of 5-fluorouracil in patients heterozygous for the IVS14+1G>A mutation in the dihydropyrimidine dehydrogenase gene. Nucleosides Nucleotides Nucleic Acids. 2008;27:692–698. doi: 10.1080/15257770802145009. [DOI] [PubMed] [Google Scholar]
- 183.Faganel Kotnik B, Grabnar I, Bohanec Grabar P, et al. Association of genetic polymorphism in the folate metabolic pathway with methotrexate pharmacokinetics and toxicity in childhood acute lymphoblastic leukaemia and malignant lymphoma. Eur J Clin Pharmacol. 2011;67:993–1006. doi:10.1007/s00228-011-1046-z. [DOI] [PubMed]
- 184.Kantar M, Kosova B, Cetingul N, et al. Methylenetetrahydrofolate reductase C677T and A1298C gene polymorphisms and therapy-related toxicity in children treated for acute lymphoblastic leukemia and non-Hodgkin lymphoma. Leuk Lymphoma. 2009;50:912–917. doi: 10.1080/10428190902893819. [DOI] [PubMed] [Google Scholar]
- 185.Csordas K, Lautner-Csorba O, Semsei AF, et al. Associations of novel genetic variations in the folate-related and ARID5B genes with the pharmacokinetics and toxicity of high-dose methotrexate in paediatric acute lymphoblastic leukaemia. Br J Haematol. 2014;166:410–420. doi: 10.1111/bjh.12886. [DOI] [PubMed] [Google Scholar]
- 186.Hulot JS, Villard E, Maguy A, et al. A mutation in the drug transporter gene ABCC2 associated with impaired methotrexate elimination. Pharmacogenet Genom. 2005;15:277–285. doi: 10.1097/01213011-200505000-00002. [DOI] [PubMed] [Google Scholar]
- 187.Vlaming ML, van Esch A, van de Steeg S, et al. Impact of abcc2 multidrug resistance-associated protein (MRP) 2], abcc3 (MRP3), and abcg2 (breast cancer resistance protein) on the oral pharmacokinetics of methotrexate and its main metabolite 7-hydroxymethotrexate. Drug Metab Dispos. 2011;39:1338–1344. doi: 10.1124/dmd.111.038794. [DOI] [PubMed] [Google Scholar]
- 188.Radtke S, Zolk O, Renner B, et al. Germline genetic variations in methotrexate candidate genes are associated with pharmacokinetics, toxicity, and outcome in childhood acute lymphoblastic leukemia. Blood. 2013;121:5145–5153. doi: 10.1182/blood-2013-01-480335. [DOI] [PubMed] [Google Scholar]
- 189.Trevino LR, Shimasaki N, Yang W, et al. Pmc2793040; germline genetic variation in an organic anion transporter polypeptide associated with methotrexate pharmacokinetics and clinical effects. J Clin Oncol. 2009;27:5972–5978. doi: 10.1200/JCO.2008.20.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ramsey LB, Bruun GH, Yang W, et al. Pmc3246196; rare versus common variants in pharmacogenetics: SLCO1B1 variation and methotrexate disposition. Genome Res. 2012;22:1–8. doi: 10.1101/gr.129668.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ciccolini J, Dahan L, Andre N, et al. Cytidine deaminase residual activity in serum is a predictive marker of early severe toxicities in adults after gemcitabine-based chemotherapies. J Clin Oncol. 2010;28:160–165. doi: 10.1200/JCO.2009.24.4491. [DOI] [PubMed] [Google Scholar]
- 192.Tran A, Jullien V, Alexandre J, et al. Pharmacokinetics and toxicity of docetaxel: role of CYP3A, MDR1, and GST polymorphisms. Clin Pharmacol Ther. 2006;79(6):570–580. doi: 10.1016/j.clpt.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 193.Yamaguchi H, Hishinuma T, Endo N, et al. Genetic variation in ABCB1 influences paclitaxel pharmacokinetics in Japanese patients with ovarian cancer. Int J Gynecol Cancer. 2006;16(3):979–985. doi: 10.1111/j.1525-1438.2006.00593.x. [DOI] [PubMed] [Google Scholar]
- 194.Kishi S, Yang W, Boureau B, et al. Effects of prednisone and genetic polymorphisms on etoposide disposition in children with acute lymphoblastic leukemia. Blood. 2004;103(1):67–72. doi: 10.1182/blood-2003-06-2105. [DOI] [PubMed] [Google Scholar]
- 195.Lal S, Wong ZW, Jada SR, et al. Novel SLC22A16 polymorphisms and influence on doxorubicin pharmacokinetics in Asian breast cancer patients. Pharmacogenomics. 2007;8(6):567–575. doi: 10.2217/14622416.8.6.567. [DOI] [PubMed] [Google Scholar]
- 196.Sparreboom A, Loos WJ, Burger H, et al. Effect of ABCG2 genotype on the oral bioavailability of topotecan. Cancer Biol Ther. 2005;4(6):650–658. doi: 10.4161/cbt.4.6.1731. [DOI] [PubMed] [Google Scholar]