This study elucidates how a cyanobacterial primary producer acclimates to heterotrophic partnership by modulating the expression levels of key metabolic genes. Heterotrophic bacteria can indirectly regulate the physiology of the photoautotrophic primary producers, resulting in physiological changes identified here, such as increased intracellular ROS. Some of the interactions inferred from this model system represent putative principles of metabolic coupling in phototrophic-heterotrophic partnerships.
KEYWORDS: consortia, cyanobacteria, heterotroph, microbial interactions, transcriptome
ABSTRACT
The mechanisms by which microbes interact in communities remain poorly understood. Here, we interrogated specific interactions between photoautotrophic and heterotrophic members of a model consortium to infer mechanisms that mediate metabolic coupling and acclimation to partnership. This binary consortium was composed of a cyanobacterium, Thermosynechococcus elongatus BP-1, which supported growth of an obligate aerobic heterotroph, Meiothermus ruber strain A, by providing organic carbon, O2, and reduced nitrogen. Species-resolved transcriptomic analyses were used in combination with growth and photosynthesis kinetics to infer interactions and the environmental context under which they occur. We found that the efficiency of biomass production and resistance to stress induced by high levels of dissolved O2 increased, beyond axenic performance, as a result of heterotrophic partnership. Coordinated transcriptional responses transcending both species were observed and used to infer specific interactions resulting from the synthesis and exchange of resources. The cyanobacterium responded to heterotrophic partnership by altering expression of core genes involved with photosynthesis, carbon uptake/fixation, vitamin synthesis, and scavenging of reactive oxygen species (ROS).
IMPORTANCE This study elucidates how a cyanobacterial primary producer acclimates to heterotrophic partnership by modulating the expression levels of key metabolic genes. Heterotrophic bacteria can indirectly regulate the physiology of the photoautotrophic primary producers, resulting in physiological changes identified here, such as increased intracellular ROS. Some of the interactions inferred from this model system represent putative principles of metabolic coupling in phototrophic-heterotrophic partnerships.
INTRODUCTION
Interspecies microbial interactions are controlled by the genome-encoded functions belonging to individual organisms and from their responses to environmental cues. Community-level responses are a function of all species, including those in low abundance, and comprehensive analyses require species-level resolution (1). Most microbial communities in nature are structurally and functionally complex, and it is technically challenging to make species-specific observations of behavior. Hence, model microbial consortia, maintained under controlled environments, are attractive for interrogating the principles by which multispecies interactions mediate the exchange of nutrients, vitamins/cofactors, and energy under different growth conditions and environmental constraints (2–6).
Phototroph-heterotroph partnerships are essentially ubiquitous in photic environments and mediate key biogeochemical and ecological processes on a global scale (7). In this study, we employed a bottom-up approach to infer and test specific interactions occurring within a constructed binary consortium containing a unicellular cyanobacterium, Thermosynechococcus elongatus BP-1, and an obligate aerobic heterotroph, Meiothermus ruber strain A (here T. elongatus and M. ruber, respectively). T. elongatus is a thermophilic, unicellular cyanobacterium previously investigated in numerous ecophysiological and biotechnological studies (8–13). Its genome is well characterized (14) and was first isolated from a hot spring cyanobacterial mat environment near Beppu, Japan (15). M. ruber strain A is an aerobic, heterotrophic thermophile isolated from an enrichment culture of Synechococcus sp. strain JA-3-3-Ab, which was sampled from a cyanobacterial mat inhabiting the outflow of Octopus Spring in Yellowstone National Park (WY, USA) (16, 17), and shares 98.6% nucleotide identity to the 16S rRNA gene of a closely related strain, Meiothermus ruber DSM 1279 (18). At the genome level, both M. ruber strains display substantial functional relatedness with regard to carbohydrate and energy metabolism, including genes for glycolysis, tricarboxylic acid cycle, oxidative pentose phosphate, Entner-Doudoroff, and aerobic respiratory electron transfer pathways (16). Similarly to M. ruber DSM 1279, strain A lacks an assimilatory nitrate reduction pathway; hence, it depends upon reduced N sources produced by T. elongatus when cocultured in minimal medium containing only nitrate. This consortium was constructed specifically to identify interactions underlying acclimation responses to heterotrophic partnership. Simplified consortia, such as the system presented here, are useful tools for studying microbial interactions at the species level. It is much more difficult to interrogate species-resolved responses to partnership in natural systems, in which membership cannot be controlled. Typically, such studies have invoked only bulk measurements of physiological or biochemical activities derived from the entire community or have examined subsets, focusing on only the dominant members (19).
In this study, analyses were performed on both member species by interrogating transcriptional and physiological data associated with a commensal “producer-consumer” interaction (20). These data were collected across tightly controlled steady states maintained via discrete incident irradiance (Ii) and dissolved O2 tension (pO2) treatments. The strains were chosen because of an obligate dependency of M. ruber on cyanobacterium-derived carbon and nitrogen, when the consortium was grown in autotrophic minimal medium containing nitrate. These are representative thermophiles derived from hot spring cyanobacterial mats that are hallmark habitats for high solar Ii and pO2 that significantly influence microbial interactions and the ecosystem properties (21–25). We designed controlled cultivation experiments that compared axenic growth of T. elongatus to growth in the consortium and concluded that M. ruber acted as a commensal partner supported by direct metabolite exchange. We then hypothesized that T. elongatus sensed and acclimated to its partner and that this behavior represented indirect interspecies regulation where each species coordinated some transcriptional events in response to the other. We conclude that some of these behaviors that relate to the most foundational functions of life, such as carbon and energy acquisition, may represent generalizable principles for phototroph-heterotroph interactions that can occur in habitats subjected to a dynamic range of light and oxygen tensions.
RESULTS
Photoautotrophically supported binary culture.
T. elongatus supported heterotrophic growth of M. ruber under photoautotrophic growth conditions (i.e., CO2/HCO3− as the sole carbon sources) in the defined medium containing nitrate with no vitamin amendments (see Fig. S1 in the supplemental material). Stable compositional and metabolic steady states were maintained under turbidostat control. The relative abundances of T. elongatus and M. ruber remained stable across the treatments and maintained average values of 90.2% ± 2.0% and 9.1% ± 1.5% cell counts, respectively. Because M. ruber lacks the ability to assimilate nitrate (16) (Fig. S2), its capacity to maintain a stable population indicates that a consistent flux of both organic carbon and organic/assimilated nitrogen emanates from the cyanobacterium. Relative extracellular metabolite measurements (i.e., peak area of quantifiable metabolites) confirmed that organic carbon and nitrogen were available for heterotrophic growth and present in the form of citric acid and several amino acids, respectively, although compounds in lower abundance were not accounted for (Fig. S3).
(A) A representative confocal micrograph (1 of 40) of cells maintained as a stable, photosynthetically supported binary culture of T. elongatus (red; autofluorescence) supporting M. ruber (green; SYBR gold). Bar, 10 µm. (B) Relative abundances measured in percentage of cells counted via fluorescence-activated cell sorting. Values represent the means from three independent steady states held under different oxygen tensions (pO2 = 0, 0.3, and 0.6 atm-O2). Error bars represent ±1 standard deviation. Download FIG S1, PDF file, 0.1 MB (104.6KB, pdf) .
Copyright © 2017 Bernstein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Batch growth and nitrate assay. Each data point represents the mean from three biological replicates. The optical densities (OD600) of M. ruber cells are matched to the M. ruber nitrate samples. Error bars represent ±1 standard deviation. The abiotic control experiment revealed no change in nitrate concentration, which remained constant at 106 ± 3 mg liter−1 over 48 h. Download FIG S2, PDF file, 0.2 MB (184.2KB, pdf) .
Copyright © 2017 Bernstein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Extracellular metabolite measurements of axenic T. elongatus (Ax) and the binary culture (Bi). Specific turbidostat steady-state conditions are designated with the following abbreviations: HL, high light (1,995 µmol photons m−2 s−1); ML, medium light (1,190 µmol photons m−2 s−1); LL, low light (197 µmol photons m−2 s−1); HO, high O2 (pO2 = 0.6 atm); MO, medium O2 (pO2 = 0.3 atm); LO, low O2 (pO2 = 0.0 atm). Measurements are the means from two biological replicates with standard errors. Download FIG S3, PDF file, 0.3 MB (304.2KB, pdf) .
Copyright © 2017 Bernstein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Concordance of replication shown as the ranges between biological duplicate measurements of mRNA abundances (RPKM) values for 2,479 genes under each steady-state condition for which T. elongatus gene expression was analyzed. Specific turbidostat steady-state conditions are designated with the following abbreviations: AX, axenic T. elongatus; BC, binary culture; HL, high light (1,995 µmol photons m–2 s–1); ML, medium light (1,190 µmol photons m–2 s–1); LL, low light (197 µmol photons m–2 s–1); HO, high O2 (pO2 = 0.6 atm); MO, medium O2 (pO2 = 0.3 atm); LO, low O2 (pO2 = 0.0 atm). Download FIG S4, PDF file, 0.2 MB (166KB, pdf) .
Copyright © 2017 Bernstein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Concordance of replication shown as the ranges between biological replicate measurements of mRNA abundances (RPKM) values for 4,492 genes under each steady-state condition for which species-resolved gene expression was analyzed; binary cultivation of T. elongatus and M. ruber. Specific turbidostat steady-state conditions are designated with the following abbreviations: HL, high light (1,995 µmol photons m–2 s–1); ML, medium light (1,190 µmol photons m–2 s–1); LL, low light (197 µmol photons m–2 s–1); HO, high O2 (pO2 = 0.6 atm); MO, medium O2 (pO2 = 0.3 atm); LO, low O2 (pO2 = 0.0 atm). Replication of transcriptomic analyses was performed in duplicate with the exception of condition HL-LO, for which replication was performed in quadruplicate. Download FIG S5, PDF file, 0.1 MB (155.9KB, pdf) .
Copyright © 2017 Bernstein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
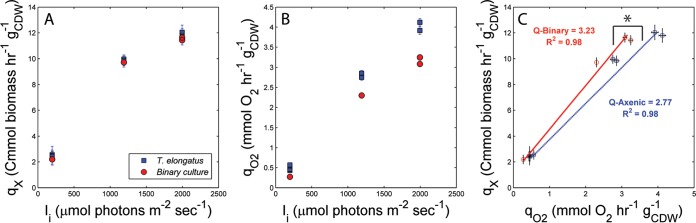
Turbidostat steady states, controlled by incident irradiance (Ii), yield the respective maximum specific growth rates (µ) for each condition. Axenic T. elongatus and the binary culture µ values increased with Ii values that were between 350 and 2,000 µmol photons m−2 s−1. Cell growth did not saturate or became inhibited within the bounds of experimentally imposed Ii treatments. The µ values measured for the binary consortium were nearly identical to those for the T. elongatus axenic control under each Ii treatment. The cultures (binary and axenic) approached 2.4-h doubling times at the highest Ii treatments (2,000 µmol photons m−2 s−1). As expected, analysis of the specific growth rate, on a carbon-mole basis (Cmmol biomass h−1 g−1cell dry weight), indicated that heterotrophic partnership had a negligible effect on the net conversion of inorganic carbon to biomass compared to the T. elongatus axenic control (Fig. 1A). The specific rates of O2 production measured in the binary consortium were lower than those in the T. elongatus axenic control (Fig. 1B). Hence, the photosynthetic quotients, approximated here as the Cmol fixed into biomass by moles O2 produced, were greater in the binary culture than in the T. elongatus axenic control (Fig. 1C). The binary culture captured more of its reductant in biomass than did axenic T. elongatus as a result of concurrent heterotrophic growth, inferred to be supported by cyanobacterium-derived reduced carbon and nitrogen sources.
FIG 1 .
Growth and photosynthesis kinetics of the T. elongatus axenic (blue) and binary (red) cultures held under variable irradiance-controlled turbidostat steady states. (A) Specific growth rates on a Cmol basis. (B) Specific rate of net O2 production (net photosynthesis). (C) Growth rate plotted against net photosynthesis. The slope of these curves represents the photosynthetic quotients or yields of carbon uptake into biomass per net photosynthetic output. Each data point represents the average of measurements taken every minute over a minimum of three reactor residence times held under each respective steady state. Error bars represent ±1 standard deviation. *, significant differences with >99% certainty as determined by t test of unequal sample sizes, assuming equal variance; CDW, cell dry weight.
Interaction leads to reduced oxygen sensitivity.
The binary consortium exhibited a decreased sensitivity to O2 stress compared to the T. elongatus axenic control (Fig. 2). A linear decrease in µ was observed, for both the axenic and binary cultures, as the partial pressure of O2 (pO2; corresponding to inlet gas composition) was raised from 0 through 0.59. This decrease was ameliorated in the binary culture. The maximum pO2 treatment provided a dissolved O2 concentration of 366 µM or 281% of air saturation (at 52°C). Oxygen sensitivities were measured as the absolute slope(s) 0.127 ± 0.004 and 0.086 ± 0.005 h atm-O2−1 (± standard error) for the T. elongatus axenic control and binary consortium, respectively.
FIG 2 .
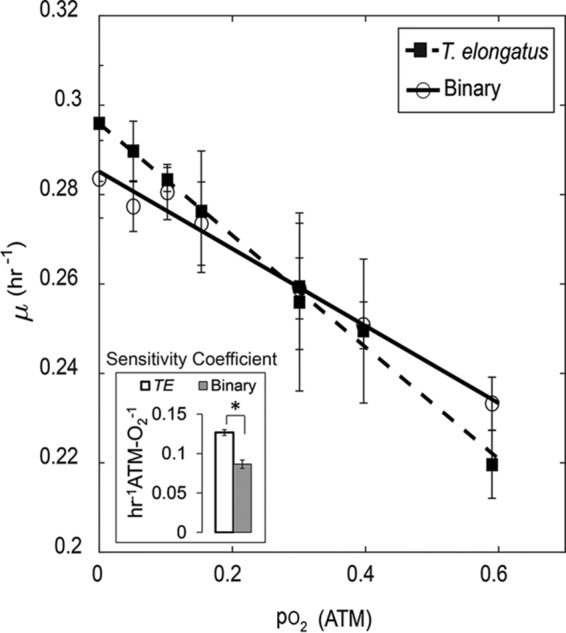
Oxygen inhibition profiles of steady-state specific growth rates controlled under increasing oxygen tensions (pO2 of in-gas). The absolute value of the slopes represents the sensitivity coefficients which are inversely proportional to the growth resistance to O2 stress for each culture. The sensitivity coefficients were determined to be 0.127 ± 0.004 and 0.086 ± 0.005 (± standard error) for the T. elongatus axenic (TE) and binary (Binary) cultures, respectively. Each data point represents the average of measurements taken every minute over a minimum of three reactor residence times held under each respective steady state. Error bars represent ±1 standard deviation. *, significant differences with >99% certainty as determined by t test of unequal sample sizes, assuming equal variance.
Cyanobacterial responses to heterotrophic partnership.
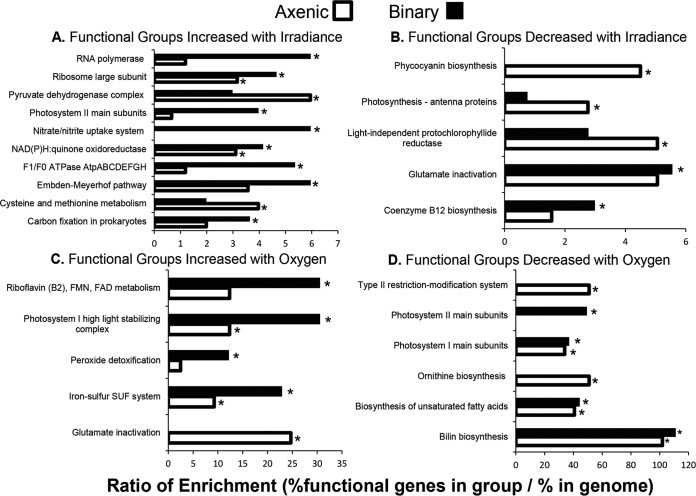
T. elongatus showed a different transcriptional response in the presence of M. ruber than the axenic control. Light- and O2-responsive genes were defined as those showing ≥2-fold change between the lower and maximum experimental bounds with respect to Ii and pO2. Of the 2,476 protein-encoding genes in T. elongatus, 354 and 339 were determined to be light responsive under axenic and binary steady-state conditions (pO2 = 0 atm-O2), respectively. Similarly, 105 and 60 genes were determined to be O2 responsive under axenic and binary steady-state conditions (Ii = 1,995 µmol photons m−2 s−1), respectively. Different gene function categories were significantly enriched (P < 0.05) from groups determined to be either light or oxygen regulated (Fig. 3).
FIG 3 .
Enriched gene functions responding to irradiance and pO2. Ratios on the x axis depict the percentage of genes of a particular function (shown on the y axis) in a given category (e.g., RNA polymerase genes increasing their expression with irradiance under binary conditions, top black bar of the upper left figure) divided by the percentage of genes of the same function in the genome as a whole. For comparison, the ratios of functional enrichment are shown for both axenic (white) and binary (black) cultivation with at least one of the two being significantly enriched with a P value of <0.05 as determined by Fisher’s exact test (indicated by *). (A) Functional groups that increased with increasing irradiance. (B) Functional groups that decreased with increasing irradiance. (C) Functional groups that increased with increasing O2 tension (pO2); SUF, sulphur assimilation. (D) Functional groups that decreased with increasing O2 tension. FMN, flavin mononucleotide; FAD, flavin adenine dinucleotide.
Genes whose transcripts were shown to be responsive by increasing with irradiance included those associated with photosystem II (PS II) (psbV, psbX, and psbV2; tll1285, tsr2013, and tll1284) and β-carboxysome (ccmK1 and ccmL; tll0946 and tll0945) functions. In addition to their response to Ii, a greater number of PS II genes showed elevated transcript abundance during heterotrophic partnership (Fig. 3A). T. elongatus genes encoding significantly enriched functions related to pyruvate metabolism (pdhB and pdhA; tll0204 and tlr1169) and metabolism of the amino acids cysteine and methionine (cysE and metE; tlr0851 and tlr1190; coaX and panC-cmk; tll1149 and tll2450) were light responsive with Ii, but a smaller number of these genes exhibited such changes in expression as a result of M. ruber partnership compared to axenic growth (Fig. 3B).
The majority of functions involving O2-responsive genes showed higher enrichment ratios under binary than under axenic T. elongatus growth. These include PS I-stabilizing products, peroxide detoxification, and assembly of iron-sulfur clusters. The specific genes in these categories which showed increased expression with pO2 include photosystem-stabilizing genes hliC and hliA (tsr0446 and tsl2208), iron sulfur genes sufD and iscU (tlr1905 and tll1093), and peroxide detoxification genes grxD (tll0874) and tll1454 (Fig. 3C). Among genes showing decreased expression with increasing O2, both type II restriction modification system gene tll1944 and ornithine biosynthesis gene argJ (tll1911) were enriched only under axenic conditions.
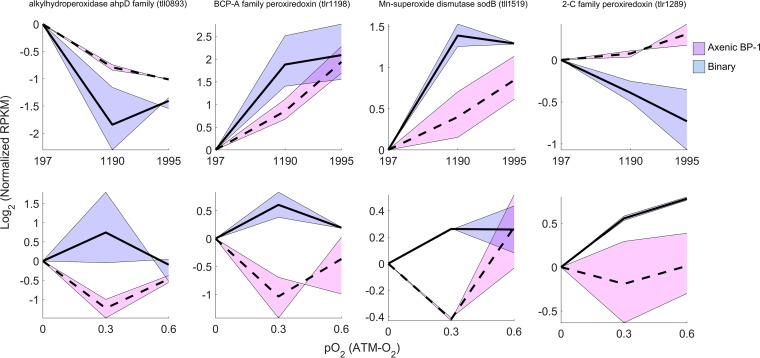
Another acclimation response to partnership was observed as increased relative intracellular abundances of reactive oxygen and nitrogen species (ROS and RNS, respectively) during binary cultivation compared to the T. elongatus axenic controls (Fig. 4). The fluorescence of an oxidized ROS-RNS reporter dye [5-(6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFHAD)] increased under binary cultivation but, unexpectedly, was insensitive to the increasing pO2 treatments. T. elongatus showed differential expression of ROS detoxification transcripts in the presence of M. ruber across a broad range of Ii- and pO2-controlled steady states. In many cases, the expression patterns of these T. elongatus transcripts showed opposite trends between axenic and binary cultivation across the Ii and pO2 treatments (Fig. 5). Generally, an increased relative abundance of these transcripts was observed under binary cultivation compared to the T. elongatus axenic control. These results show that T. elongatus acclimates to partnership with M. ruber by changing the expression of ROS detoxification genes, which is likely driven by a heterotrophically mediated increase in ROS-RNS.
FIG 4 .
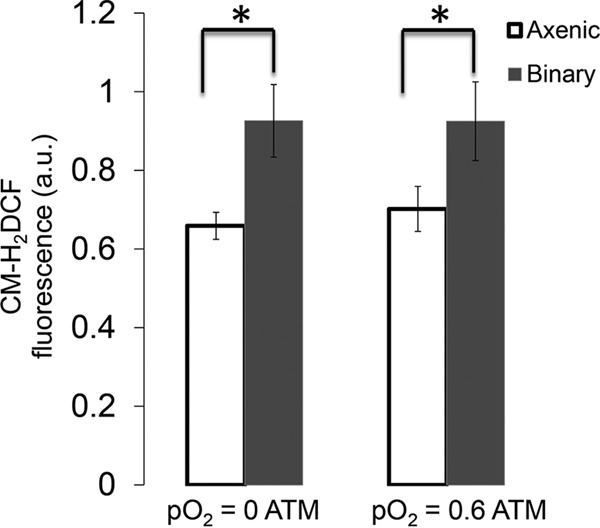
Intracellular reactive oxygen/nitrogen (ROS-RNS) increased under binary cultivation compared to axenic T. elongatus controls. Average intensity (n = 3) of the 525-nm fluorescence emitted from cells excited at 488 nm, corresponding to the abundance of oxidized ROS-RNS reporter dye (CM-H2DCFHDA) for the T. elongatus axenic and binary cultures maintained under low and high oxygen tensions. Error bars represent ±1 standard deviation. *, significant differences with >99% certainty as determined by Tukey’s test.
FIG 5 .
Relative abundance profiles for T. elongatus transcripts of ROS-RNS detoxification genes that showed at least a 1.5-fold change over either the irradiance- or pO2-controlled steady states. Solid black lines and shaded blue regions represent the means and ranges of T. elongatus genes expressed during binary cultivation. Similarly, dashed black lines and shaded red regions represent the means and ranges of T. elongatus genes expressed during axenic cultivation. The transcripts identified include a putative alkylhydroperoxidase ahpD family gene (tll0893), a BCP-A family peroxiredoxin (tlr1198), an Mn-superoxide dismutase (sodB; tll1519), and a 2-Cys family peroxiredoxin (tlr1289). “Normalized RPKM” is the average of duplicate RPKM values measured under each steady state divided by the average RPKM corresponding to the lowest incident irradiance (197 µmol photons m−2 s−1) or pO2 (0 atm-O2).
Coordinated transcriptional patterns as a result of metabolic coupling.
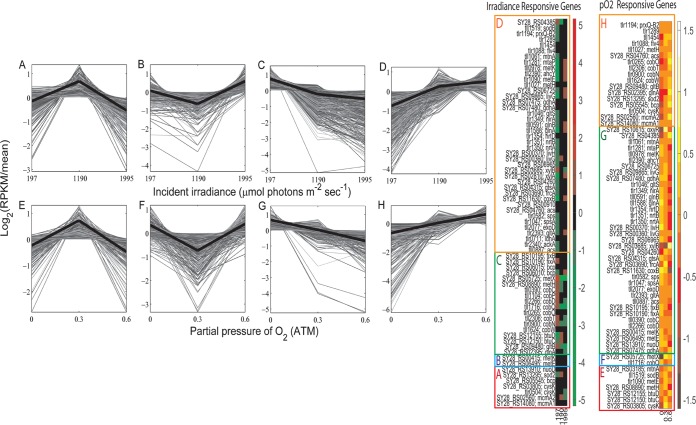
A guiding hypothesis was that since T. elongatus supported M. ruber via exchange of essential resources (i.e., C, N, and O2), M. ruber must coordinate its gene expression to accommodate the variable physiology of the cyanobacterium across the gradients of Ii and pO2 conditions. Examination of individual transcript profiles was performed by considering clusters around four main patterns specified by the centroid or “eigen-genes” corresponding to the Ii and pO2 treatments used to establish steady state (Fig. 6). Clusters A to D were calculated to examine responses to Ii treatments, and clusters E to H were calculated for examining the transcriptional effects from increasing O2 tension. All clusters contained both T. elongatus and M. ruber genes. Cluster A exhibits a tent-shaped eigen-gene with maximum relative mRNA abundances at the midpoint Ii (1,190 µmol photons m−2 s−1). The midpoint was chosen as a sampling condition because it is the theoretical saturating irradiance (26). Genes in cluster B responded inversely to those in cluster A. Clusters C and D contain genes that show a relative decrease or increase with Ii, respectively. Note that the changes observed for the relative abundance of transcripts with increasing Ii also correspond to increasing specific growth and photosynthesis rates (Fig. 1). Clusters E to H contain transcripts that trend with increasing pO2 treatments (0 to 0.59 atm-O2) and show profiles that are analogous to clusters A to D. Changes observed for the relative abundance of transcripts with increasing pO2 correspond to a linear decrease in the specific growth rate (Fig. 2). Statistically enriched gene functions were identified for both T. elongatus and M. ruber within each cluster (Tables S1 and S2) to infer which processes may have been involved in metabolite exchange and acclimation to partnership.
FIG 6 .
K-means clusters of T. elongatus and M. ruber relative mRNA abundance profiles that share correlated expression patterns over irradiance- and pO2-controlled steady states. Heavy black lines represent the centroid (i.e., eigen-gene) calculated within each cluster. Cluster A, 201 T. elongatus plus 259 M. ruber genes; cluster B, 255 T. elongatus plus 188 M. ruber genes; cluster C, 971 T. elongatus plus 444 M. ruber genes; cluster D, 914 T. elongatus plus 324 M. ruber genes; cluster E, 211 T. elongatus plus 328 M. ruber genes; cluster F, 187 T. elongatus plus 309 M. ruber genes; cluster G, 913 T. elongatus plus 468 M. ruber genes; cluster H, 888 T. elongatus plus 387 M. ruber genes. Heat maps show the expression patterns [log2(RPKM/mean)] for specific functional genes from each organism.
Statistical/functional enrichment of T. elongatus gene functions within clusters of mRNA profiles. Download TABLE S1, PDF file, 0.2 MB (166.5KB, pdf) .
Copyright © 2017 Bernstein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Statistical/functional enrichment of M. ruber gene functions within clusters of mRNA profiles. Download TABLE S2, PDF file, 0.2 MB (247.9KB, pdf) .
Copyright © 2017 Bernstein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primary productivity and carbon exchange.
T. elongatus supported M. ruber growth via the production and transfer of reduced carbon. Cyanobacterial processes involved in biosynthesis and transport of organic carbon shared common transcriptional patterns with M. ruber genes encoding enzymes used in carbon uptake and metabolism of the same (or similar) gene products. Principal genes involved in organic acid synthesis of T. elongatus grouped into clusters that increased with Ii (cluster D) and/or decreased with pO2 (cluster G). These included acetyl coenzyme A (CoA) synthetase (acs; tll0887), acetate kinase (ackA; tlr2340), lactate dehydrogenase (ldhA; tlr0711), and citrate synthase (gltA; tlr2393). Functions involved in synthesis and export of larger biomolecules (i.e., sugars, peptides, and extracellular polymeric substance [EPS]) also grouped into clusters D and G. These included a putative exopolysaccharide synthesis gene (exoD; tll2077), sucrose synthase (spsA; tlr1047), and a sucrose degradation enzyme (sps; tlr0582). In conjunction with T. elongatus, M. ruber genes involved in the uptake and metabolism of compounds related to export and synthesis of T. elongatus-derived organic carbon were also found in clusters D and/or G. Notable examples included genes encoding putative acetyl-CoA synthetase (acs; SY28_RS04760 and SY28_RS00910), cytochrome c oxidase (coxB; SY28_RS11630), monosaccharide uptake systems (frcA and gtsAB; SY28_RS03690, SY28_RS04315, and SY28_RS04260), xylose isomerase (xylA; SY28_RS02810), xylulokinase (xylB; SY28_RS03685), an ABC-type multisugar uptake system (SY28_RS06965), and branched-chain amino acid uptake (livGH; SY28_RS00360 and SY28_RS00370).
Nitrogen and amino acid exchange.
Since M. ruber lacks an assimilatory nitrate reductase, T. elongatus was assumed to provide reduced N during binary cultivation. The relative abundances of transcripts encoding the T. elongatus nitrate uptake system from nrtABD (tlr1350, tlr1351, and tlr1354) increased with Ii, decreased with pO2, and grouped appropriately into clusters D and G. Other T. elongatus nitrogen metabolism genes found in these clusters include glutamine synthetase gene glnA (tll1588), a nitrogen regulatory protein gene (glnB; tll0591), an assimilatory ferredoxin-nitrate reductase gene (nirA; tlr1349), and a glutamate symporter gene (gltS; tlr1046). Several M. ruber nitrogen-associated genes also grouped into clusters D and/or G, including a glutamate dehydrogenase gene (gdhA; SY28_RS07480 and SY28_RS07475) and amino acid uptake system genes (SY28_RS09865 and SY28_RS06725). However, some key genes required for N acquisition by M. ruber grouped into clusters C and/or H, showed opposite expression patterns with respect to Ii and pO2, and effectively increased with the specific growth and photosynthesis rates. These include glnA (SY28_RS02395) and the large subunit of glutamate synthase (gltB; SY28_RS09480) and suggest the potential for direct exchange of glutamine and glutamate from T. elongatus as growth requirements increase with the specific growth and photosynthesis rates.
Methionine and vitamin B12 exchange.
Cobalamin (B12) auxotrophy in M. ruber is indicated by the absence of a complete B12 synthesis pathway and a B12-independent methylcitrate pathway (prpBCDF) required for conversion of propionyl-CoA to succinyl-CoA. M. ruber contains the vitamin B12-dependent methylmalonyl-CoA mutase subunits (mcmA1 and -A2; SY28_RS14080 and SY28_RS02560) which grouped into clusters A and H. T. elongatus is a B12 prototroph, as confirmed by the capacity for axenic growth in the absence of vitamin B12 and by the presence of a complete vitamin B12 biosynthesis pathway in the genome. The relative abundances of transcripts encoding vitamin B12 uptake/scavenging gene products in M. ruber (btuCD; SY28_RS12150 and SY28_RS12155) decreased with Ii in concurrence with decreased T. elongatus transcripts (all within cluster C) encoding vitamin B12 biosynthesis, including those required for the insertion of cobalt (cobWNT; tll1624, tlr0900, and tll2306) and for the conversion of cobyrinic acid diamine to the vitamin B12 coenzyme from cobOQDPC (tlr0265, tll1716, tll2266, tll1104, and tll0390). Both T. elongatus and M. ruber expressed transcripts encoding the vitamin B12-dependent homocysteine methyltransferase (metH) but were negatively correlated and grouped across Ii and pO2 treatments into opposing clusters D (tll1027) and C (SY28_RS08890), indicating that methionine may be directly exchanged from T. elongatus as growth requirements increase with the specific growth rate. Both strains also expressed vitamin B12-independent homocysteine methyltransferase (metE) transcripts which grouped into clusters D and E compared to clusters B and G for tlr1090 and SY28_RS06495, respectively.
The relative abundances of T. elongatus transcripts encoding the degradation and salvage of methionine (ahcY, metK, mtaP, and mtnA; tll2390, tll0978, tlr1281, and tll1061) increased with Ii but decreased with pO2 treatments and grouped within clusters D and G, respectively. Uniform grouping of M. ruber transcripts encoding methionine degradation and salvage was not observed as they were spread across multiple clusters grouped by the Ii and pO2 treatments: metK (SY28_RS00415; clusters B and G) and mtnA (SY28_RS03185; cluster E). In contrast, the relative abundance of M. ruber transcripts encoding methionine biosynthesis proteins decreased with Ii, and they were grouped into cluster C, including metHX (SY28_RS08890 and SY28_RS05725). Genes involved in cysteine biosynthesis shared common transcriptional patterning between species, indicating that while M. ruber may have salvaged cyanobacterium-derived methionine, it likely synthesized its own cysteine via the vitamin B12-independent pathway as growth requirements increased. These include cysteine synthases (cysK; tlr0504 and SY28_RS03805) and serine O-acetyltransferases (cysE; tlr0851 and SY28_RS05065), which cogrouped into clusters D and A, respectively.
Oxidative stress responses.
The relative abundance of T. elongatus transcripts encoding enzymes involved with ROS detoxification generally increased with increasing Ii and pO2, and the transcripts grouped into the appropriate clusters D and/or H. Notable examples include an NAD(P)H-oxygen oxidoreductase (flv4; tlr1088), 2-Cys family peroxiredoxins (tll1454 and tlr1289), a periplasmic peroxiredoxin (prxQ-B2; tlr1194), and an Mn-superoxide dismutase (sodB; tll1519). M. ruber contains an H2O2-responsive transcriptional regulator (oxyR; SY28_RS10615), which grouped into clusters D and G and is located adjacent to a putative manganese catalase (SY28_RS10610). However, M. ruber peroxidase (bcp; SY28_RS05545, SY28_RS06010, and SY28_RS06015) and superoxide dismutase (sod2; SY28_RS13295) genes responded differently to Ii treatments than did oxyR and related cyanobacterial profiles and were grouped into clusters A and C. These genes generally increased with pO2 and grouped with the T. elongatus genes into cluster H (increased with pO2 and decreasing µ). M. ruber genes associated with electron transfer processes that are potentiators of ROS (27, 28) grouped into cluster G, which decreased with increasing pO2 treatments and increased with specific growth and photosynthesis rates. These include subunits for an electron transfer flavoprotein (fixAB; SY28_RS10190 and SY28_RS10195), NADH-dehydrogenase (SY28_RS04385), and principal components of the NADH-quinone oxidoreductase (nuoDFGHIJKN).
DISCUSSION
The cyanobacterium T. elongatus responded to heterotrophic partnership with M. ruber by altering the expression of key functional genes. This primary result is evidence of indirect interspecies regulation. It is important to note that the turbidostat culturing platform provided a constant, optically thin, nutrient-replete environment. Hence, the addition of M. ruber to T. elongatus cultures did not alter the growth environment by reducing availability of actinic light or nutrients needed to support T. elongatus. The net specific rate of O2 production decreased during binary cultivation compared to the T. elongatus axenic controls. These rates are functionally equivalent to net photosynthesis rates and account for the gross rate of oxygenic photosynthesis minus all oxygen-consuming reactions, including photorespiration (RuBisCO oxygenase activity) and heterotrophic respiration (29, 30). The net photosynthesis rates are conservative interpretations for the lower bound of oxygenic photosynthesis and relatable as proxy measurements for the minimum consortium-wide energy acquisition rates, assuming 0.125 quanta absorbed and 2 NADPH produced per mole O2 formed via PS II-mediated charge separation (31). Hence, the energy efficiency for biomass production is greater in the binary consortium than in the T. elongatus axenic control. These increases likely result from heterotrophic capture of reductant that would otherwise be lost from the system as either photosynthate or necromass. The addition of M. ruber partnership also resulted in an observed decrease in O2 sensitivity compared to T. elongatus axenic controls (Fig. 2). A decrease in sensitivity is equivalent to an increase in O2 stress resistance. We note that this effect was observed under identical pO2 treatments (binary versus axenic), which supported O2 tensions sufficient to render the bulk effects resulting from heterotrophic O2 removal as negligible. However, attachment or close cellular proximity of M. ruber to T. elongatus cells may create localized gradients and microheterogeneities in Ii and pO2 experienced by the T. elongatus population. Interactions that occur when cells from each species are in close contact could account for the observed differences in O2 sensitivity and the photosynthetic quotients which were based on measurements taken from the well-mixed (bulk) volume.
The M. ruber-induced decrease of O2 sensitivity (Fig. 2) originally led us to hypothesize that heterotrophic partnership reduced cyanobacterial oxidative stress and that this effect could be observed by comparing intracellular ROS-RNS between axenic and binary conditions. Interestingly, we found that heterotrophic partnership had the opposite effect and that increased pO2 had no effect on intracellular ROS-RNS (Fig. 4). However, the binary culture’s observed decrease in O2 sensitivity did correspond to enrichment of genes within the functional category photosystem I high-light-stabilizing complex (Fig. 3). The T. elongatus genes in this category include hliACD (tsl2208, tsr0446, and tsr1916) and tsl0063, annotated as a member of the CAB/ELIP/HLIP protein family, which are known to be stress-induced genes that help cyanobacteria cope with free radicals and excess excitation energy (32, 33). The increases in ROS-RNS observed as a direct result of heterotrophic partnership corresponded with increased relative transcript abundances of T. elongatus genes required for mitigating ROS, showing that the cyanobacterium acclimates to heterotrophic partnership by increasing protection from oxidative stress (Fig. 4 and 5). This is contrary to previous observations made in consortia constructed from oligotrophic cyanobacteria and heterotrophs, in which Prochlorococcus species have been reported to benefit and/or depend upon heterotrophic bacteria, such as Alteromonas-like species, to reduce oxidative stress (34, 35). In contrast, evidence of cyanobacterium-mediated ROS mitigation was reported within the binary consortium of Synechococcus sp. strain PCC 7002 coupled with Shewanella putrefaciens W3-18-1, cultured under very different conditions than those employed in the current study (2). Both T. elongatus and Synechococcus sp. PCC 7002 were isolated from eutrophic environments (mat and marine sediment, respectively), which can presumably support higher levels of heterotrophic growth than oligotrophic marine environments. Cyanobacteria are equipped to detoxify intracellular ROS-RNS and mitigate oxidative stress, because ROS production is an inherent by-product of oxygenic photosynthesis and photosynthetic electron transfer (36, 37), while heterotrophic bacteria are recognized for production of extracellular ROS (27). The principle that is inferred from these collective results is that some cyanobacteria, such as Synechococcus species adapted to eutrophic environments exposed to high irradiance, specialize in ROS-RNS mitigation (38, 39).
The metabolic dependency of M. ruber upon T. elongatus for reduced carbon, nitrogen, and certain vitamins (i.e., B12, biotin, and niacin) was corroborated through the coexpression of genes encoding enzymes that metabolize and transport shared metabolites. These inferences are made through the basic assumption that the coexpressed genes, i.e., correlated mRNA abundances between the two species, correspond to protein frequency and activity. Clustering the relative mRNA abundance profiles of the two organisms together provided the means to infer which specific metabolic exchanges are coordinated and how these interactions respond to Ii and pO2. Specifically, the results suggest that multiple carbon compounds derived from the cyanobacterium could be exchanged and taken up by M. ruber. While evidence for the exchange of organic acids (e.g., acetate) was observed, polysaccharides, peptides, and EPS may have also supplied the carbon and reductant required to support stable M. ruber populations. Ample evidence also supports the likelihood that amino acids can serve as a source of reduced N (and possibly carbon) required by M. ruber. For example, the expression of genes involved in the synthesis, transport, and salvage of specific amino acids showed coordinated patterns shared by each species. Specifically, T. elongatus transcripts of methionine and glutamate biosynthesis genes clustered into groups that increased with µ (clusters D and G) while M. ruber homologs showed opposite patterns (clusters C and H), suggesting the specific exchange of these amino acids. The relative abundance profiles for M. ruber genes encoding vitamin B12-dependent methionine synthesis showed expression patterns in common with vitamin B12 salvage and T. elongatus vitamin B12 synthesis genes, indicating that one exchanged resource (e.g., vitamin B12) may affect requirement of another exchangeable resource (e.g., methionine) that has a closely linked pathway.
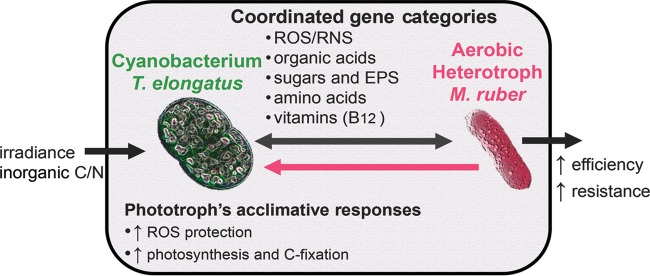
The collection of experimental results presented here clearly shows that the cyanobacterium responded and acclimated to heterotrophic partnership. In this experimental system, T. elongatus is either indirectly regulated by environmental changes instigated by M. ruber or directly regulated via molecular signals. The integrated kinetics- and global transcriptomics-based inferences were not targeted enough to capture a mechanism for direct interspecies regulation. Although direct interspecies regulation cannot be ruled out, our conclusion is that this heterotrophic partnership establishes an indirect interspecies regulation of gene expression resulting in measurable changes of growth and photosynthesis kinetics of the binary culture compared to the cyanobacterial axenic control. The outcomes described here can be generalized to understand microbial cyanobacterium-heterotroph interactions better (Fig. 7). For instance, the cyanobacterium was inferred to sense the presence of its commensal heterotrophic partner and respond by altering its gene expression. Interspecies modulation of gene expression, via indirect regulation, is supported by results reported in previous studies that investigated very different cyanobacterium-heterotroph platforms (2, 4), and a number of notable commonalities emerge. A previous study, investigating a consortium composed of cyanobacterium Synechococcus sp. PCC 7002 coupled with Shewanella putrefaciens W3-18-1 (2), reported a >2-fold change in mRNA abundances of cyanobacterial genes (compared to axenic) that encode enzymes belonging to the same functional groups that were statistically enriched from T. elongatus in the current study (Fig. 3). These include some of the most notable examples, such as carbon concentration, carboxysome synthesis, PS II, oxidative phosphorylation, methionine metabolism, and Fe-S cluster biogenesis. Similarly to the results from this study, Synechococcus sp. PCC 7002 was also reported to show >2-fold changes in B12 salvage processes when partnered with S. putrefaciens W3-18-1, although the current study investigated a cyanobacterial B12 prototroph. Another study, which investigated the transcriptional responses of Synechococcus sp. WH8102 cocultured with Vibrio parahaemolyticus, also reported changes in expression of genes sharing the functions reported here. These include amino acid biosynthesis, cofactor biosynthesis, PS II, NAD(P)H-dehydrogenase, and ATPase. The functional similarities of cyanobacterial genes being influenced and potentially indirectly regulated by the activity of a heterotrophic partner are remarkable considering the difference in species, their origins, and the treatments and culturing platforms compared. Interspecies modulation of gene expression likely serves as a fundamental principle enabling microbial communities to coordinate their metabolism and coacclimate to each other and environmental cues.
FIG 7 .
Schematic outline of responses to heterotrophic partnership observed and inferred as indirect interspecies regulation events and metabolic coupling.
MATERIALS AND METHODS
Bacterial strains and culturing media.
Both Thermosynechococcus elongatus strain BP-1 and Meiothermus ruber strain A were grown axenically and as a binary consortium in a modified BG-11 (mBG-11) medium (BP-1 medium) containing 17.6 mM NaNO3, 0.304 mM MgSO4⋅7H2O, 0.175 mM KH2PO4, 0.245 mM CaCl2⋅2H2O, 0.0028 mM Na2EDTA, and 0.0144 mM FeCl3. The mBG-11 was supplemented with 1 ml liter−1 trace mix: 46.2544 mM H3BO3, 9.1458 mM MnCl2⋅4H2O, 0.772 mM ZnSO4⋅7H2O, 1.611 mM Na2MoO4⋅2H2O, 0.316 mM CuSO4⋅5H2O, and 0.170 mM Co(NO3)2⋅6H2O. During binary culture, both species were supported by the autotrophic growth of T. elongatus (i.e., no organic substrate was added for the growth of M. ruber). Axenic starter cultures of M. ruber were cultured in mBG-11 supplemented with 1% yeast extract. Starter cultures were generated in 150-ml sealed serum bottles charged with 50 ml mBG-11 plus 15 mM NaHCO3 (adjusted to pH 7.5) under 10% CO2 in N2 headspace. Photobioreactors were inoculated to an optical density at 730 nm (OD730) of 0.01 with exponentially growing starter culture(s).
Turbidostat photobioreactor operation.
This study modified a previously reported continuous stirred tank reactor operated with feedback control as a turbidostat (40–43). Briefly, a “white-light” photobioreactor (7.5-liter vessel) was constructed with evenly spaced sets of fluorescent lights (Sylvania; model FP14/835/ECO, Pentron 3500 K, 14 W) at each quadrant of the cylindrical bioreactor vessel (both 8- and 12-bulb configurations). A custom dimmer was controlled with a 4- to 20-mA signal commanded from a New Brunswick Scientific BioFlo 310 bioreactor controller. The reactor and light bank apparatus were isolated from external ambient light. The working volume of the reactor (5.5 liters) was held at a constant temperature of 52°C and pH 7.5. A Clark-type oxygen sensor coupled with two independent off-gas sensors (Blue Sense; Cell and Ferm models) was used to measure dO2 and off-gas O2 and CO2, respectively. Incident scalar irradiance and the transmitted irradiance used to control turbidostat operation were maintained using custom photovoltaic cells coupled with a digital pA/mV multimeter (Keithley 2700 Integra Series; Tektronix, Beaverton, OR). The dilution rate of the culture was determined gravimetrically from the effluent medium. Steady states were defined by stable (<10% variation from the mean) measurements of dilution rate, OD730, and dissolved O2 for a minimum of 3 residence times. These measurements, as well as off-gas CO2, pH, temperature, and acid/base addition, were data logged at 1-min intervals; hence, the replication of values used to calculate the specific rates (presented in Fig. 1 and 2) is means and standard deviations from a minimum of 604 measurements (for the fastest residence time of 3.36 h; dilution). Additional details for bioreactor-enabled measurements are available in Text S1 in the supplemental material.
Supplemental text describing additional details pertaining to the methodology and results presented in the manuscript and supplemental files. Download TEXT S1, DOCX file, 0.04 MB (41.6KB, docx) .
Copyright © 2017 Bernstein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Flow cytometry.
The flow cytometry data were obtained using a BD Influx fluorescence-activated cell sorter (FACS; BD Biosciences, San Jose, CA). Using the 488-nm excitation from a Sapphire LP laser (Coherent Inc., Santa Clara, CA) at 100 mW, samples were analyzed using a 70-μm nozzle. Optimization and calibration of the FACS were performed before each analysis using 3-μm Ultra Rainbow fluorescent particles (Spherotech, Lake Forest, IL). The ratio of the two distinct populations of cells within a mixed microbial community was identified from 50,000 recorded cells using size and complexity gates with FCS Express (Los Angeles, CA) flow cytometry software. Cell counts are presented as the percentage of total counting events.
RNA isolation and sequencing.
Cells for total RNA isolation were harvested from steady-state growth conditions by previously reported methods (2, 43). Transcriptomic samples were sampled in a minimum of two biological duplicates sampled from steady states in a minimum of 1 residence time (1/dilution rate) apart from each other. Briefly, cells were collected via centrifugation at 7,000 rpm for 5 min at 4°C, frozen in liquid nitrogen, and stored at −80°C. All RNA analyses were repeated in biological duplicates. To analyze the response of T. elongatus alone to the addition of a heterotrophic partner, RNA was extracted with TRIzol and sequenced using SOLiD (sequencing by oligonucleotide ligation and detection) technology with no depletion of rRNA (see the supplemental material for details). However, as binary culture samples contained <10% M. ruber by cell count, the extraction and sequencing methodology was modified when collecting data used to specifically examine the correlation of transcriptional responses between T. elongatus and M. ruber. For this, RNA was extracted using the SV total RNA isolation system (Promega, Madison, WI) followed by genomic DNA removal and purification using the Turbo DNA-free kit (Life Technologies, Carlsbad, CA). An Agilent 2100 Bioanalyzer was used to assess the integrity of the RNA samples. Only RNA samples having an RNA integrity number between 8 and 10 were used. cDNA libraries were constructed using the Ovation Universal transcriptome sequencing (RNA-seq) system (NuGEN, San Carlos, CA). This kit was used to incorporate specific InDA-C probes used to deplete rRNA from the binary culture experiments. Probes were designed to deplete both 16S and 23S rRNA from both T. elongatus and M. ruber. cDNA libraries were examined using the Agilent 2100 Bioanalyzer to confirm proper size and construction and were sequenced on an Illumina NextSeq 500 sequencer. Reads from all samples were aligned to the genomes of T. elongatus BP-1 (NCBI accession no. NC_004113) and the RAST gene model of Meiothermus ruber strain A (NCBI accession no. NZ_JXOP01000000) using the Burrows-Wheeler Aligner (BWA) with the default settings (44). Gene counts were determined using HTSeq (45) and were normalized first with DESeq2 (46) followed by normalization to the gene length in kilobases. Additional details on RNA sequencing and transcriptomic data analyses are available in the supplemental material.
Transcriptomic analyses.
mRNA abundance profiles (given in reads per kilobase per million [RPKM]) for each steady-state condition (measured in biological duplicate) were filtered to remove any gene with an average count of zero in any condition and any gene with an average RPKM of <15. Binary culture expression profiles (genes from both organisms) were grouped via correlation-based K-means clustering (see the supplemental material). Prior to clustering, filtering was employed to mask the bottom 30% of genes with the smallest variance across each respective profile (i.e., flat profiles). Genes were determined to be significantly different in their expression when comparing two conditions if they showed a >2.0-fold change in their expression with an adjusted P value of <0.05. Significant enrichment (or functional enrichment) was performed as previously described (43). Briefly, enrichment was defined as the percentage of genes of a given function in the profile depicted (for example, increased expression with increased irradiance under binary conditions) divided by the percentage of genes in that same function in the genome as a whole. Enrichment ratios were then determined to be statistically significant using Fisher’s exact test with any ratio with a P value of <0.05 classed as significant.
ROS-RNS measurements.
Relative abundances of reactive oxygen and nitrogen species (ROS-RNS) were measured using a previously reported method (47). Briefly, we used the cell-permeant fluorescent dye 5-(6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFHDA; Cayman Chemicals, Ann Arbor, MI), for which increased dichlorofluorescein (DCF) fluorescence is a proxy measurement of reactive oxygen/nitrogen species; increased fluorescence is proportional to increased intracellular ROS-RNS. Cell suspensions were collected from a steady-state culture and analyzed in triplicate (3 subfractions of cells collected from each steady state). For each replicate, a 1-ml aliquot was treated with 10 mM CM-H2DCFHDA freshly dissolved in 100% Me2SO (Sigma-Aldrich, St. Louis, MO) to a final concentration of 10 μM. Concurrently, a 1-ml aliquot of control cells was treated with 1 µl of Me2SO. Cells were transferred to opaque black 96-well plates (Nunc A/S, Roskilde, Denmark) and incubated in the dark with intermittent shaking for 30 min at room temperature. Fluorescence was measured at 525 nm after excitation at 488 nm on a SpectraMax Gemini XS spectrofluorometer. The control measurements were subtracted from the CM-H2DCFHDA-treated samples to correct for nonspecific fluorescence.
Axenic T. elongatus transcript abundances and gene annotations. Download DATA SET S1, XLSX file, 0.8 MB (822.7KB, xlsx) .
Copyright © 2017 Bernstein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Binary culture, T. elongatus and M. ruber, transcript abundances, gene annotations, and clustering assignments. Download DATA SET S2, XLSX file, 4.4 MB (4.5MB, xlsx) .
Copyright © 2017 Bernstein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Accession number(s).
Raw sequencing files are available at the NCBI-GEO repository under accession numbers GSE94125 and GSE93859.
ACKNOWLEDGMENTS
This research was supported by the U.S. DOE Office of Biological and Environmental Research (BER) Genomic Science Program and is a contribution of the Fundamental Scientific Focus Area. A portion of the RNA sequencing was performed by the Environmental Molecular Sciences Laboratory (user project 49356), a national scientific user facility sponsored by DOE BER and located at PNNL. The corresponding author, H.C.B., is grateful for support given by the Linus Pauling Distinguished Postdoctoral Fellowship, a Laboratory Directed Research program at PNNL. PNNL is operated for the DOE by Battelle Memorial Institute under contract DE-AC05-76RLO 1830.
We acknowledge Leo Kucek, Lye Meng Markillie, Charles Resch, Colin J. Brislawn, Moiz Charania, Christopher Overall, Erika Zink, Tom Metz, and Wayne Curtis for their technical assistance and helpful discussions.
REFERENCES
- 1.Hooper DU, Chapin FS, Ewel JJ, Hector A, Inchausti P, Lavorel S, Lawton JH, Lodge DM, Loreau M, Naeem S, Schmid B, Setälä H, Symstad AJ, Vandermeer J, Wardle DA. 2005. Effects of biodiversity on ecosystem functioning: a consensus of current knowledge. Ecol Monogr 75:3–35. doi: 10.1890/04-0922. [DOI] [Google Scholar]
- 2.Beliaev AS, Romine MF, Serres M, Bernstein HC, Linggi BE, Markillie LM, Isern NG, Chrisler WB, Kucek LA, Hill EA, Pinchuk GE, Bryant DA, Wiley HS, Fredrickson JK, Konopka A. 2014. Inference of interactions in cyanobacterial-heterotrophic co-cultures via transcriptome sequencing. ISME J 8:2243–2255. doi: 10.1038/ismej.2014.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein HC, Paulson SD, Carlson RP. 2012. Synthetic Escherichia coli consortia engineered for syntrophy demonstrate enhanced biomass productivity. J Biotechnol 157:159–166. doi: 10.1016/j.jbiotec.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tai V, Paulsen IT, Phillippy K, Johnson DA, Palenik B. 2009. Whole-genome microarray analyses of Synechococcus-Vibrio interactions. Environ Microbiol 11:2698–2709. doi: 10.1111/j.1462-2920.2009.01997.x. [DOI] [PubMed] [Google Scholar]
- 5.Aharonovich D, Sher D. 2016. Transcriptional response of Prochlorococcus to co-culture with a marine Alteromonas: differences between strains and the involvement of putative infochemicals. ISME J 10:2892–2906. doi: 10.1038/ismej.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biller SJ, Coe A, Chisholm SW. 2016. Torn apart and reunited: impact of a heterotroph on the transcriptome of Prochlorococcus. ISME J 10:2831–2843. doi: 10.1038/ismej.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falkowski PG, Barber RT, Smetacek V. 1998. Biogeochemical controls and feedbacks on ocean primary production. Science 281:200–207. doi: 10.1126/science.281.5374.200. [DOI] [PubMed] [Google Scholar]
- 8.Arai T, Kino K. 2008. A cyanophycin synthetase from Thermosynechococcus elongatus BP-1 catalyzes primer-independent cyanophycin synthesis. Appl Microbiol Biotechnol 81:69–78. doi: 10.1007/s00253-008-1623-y. [DOI] [PubMed] [Google Scholar]
- 9.Eberly JO, Ely RL. 2012. Photosynthetic accumulation of carbon storage compounds under CO2 enrichment by the thermophilic cyanobacterium Thermosynechococcus elongatus. J Ind Microbiol Biotechnol 39:843–850. doi: 10.1007/s10295-012-1092-2. [DOI] [PubMed] [Google Scholar]
- 10.Onai K, Morishita M, Itoh S, Okamoto K, Ishiura M. 2004. Circadian rhythms in the thermophilic cyanobacterium Thermosynechococcus elongatus: compensation of period length over a wide temperature range. J Bacteriol 186:4972–4977. doi: 10.1128/JB.186.15.4972-4977.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang P, Battchikova N, Paakkarinen V, Katoh H, Iwai M, Ikeuchi M, Pakrasi HB, Ogawa T, Aro EM. 2005. Isolation, subunit composition and interaction of the NDH-1 complexes from Thermosynechococcus elongatus BP-1. Biochem J 390:513–520. doi: 10.1042/BJ20050390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abed RM, Dobretsov S, Sudesh K. 2009. Applications of cyanobacteria in biotechnology. J Appl Microbiol 106:1–12. doi: 10.1111/j.1365-2672.2008.03918.x. [DOI] [PubMed] [Google Scholar]
- 13.Henry CS, Bernstein HC, Weisenhorn P, Taylor RC, Lee JY, Zucker J, Song HS. 2016. Microbial community metabolic modeling: a community data-driven network reconstruction. J Cell Physiol 231:2339–2345. doi: 10.1002/jcp.25428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakamura Y, Kaneko T, Sato S, Ikeuchi M, Katoh H, Sasamoto S, Watanabe A, Iriguchi M, Kawashima K, Kimura T, Kishida Y, Kiyokawa C, Kohara M, Matsumoto M, Matsuno A, Nakazaki N, Shimpo S, Sugimoto M, Takeuchi C, Yamada M, Tabata S. 2002. Complete genome structure of the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. DNA Res 9:123–130. doi: 10.1093/dnares/9.4.123. [DOI] [PubMed] [Google Scholar]
- 15.Yamaoka T, Satoh K, Katoh S. 1978. Photosynthetic activities of a thermophilic blue-green-alga. Plant Cell Physiol 19:943–954. [Google Scholar]
- 16.Thiel V, Tomsho LP, Burhans R, Gay SE, Schuster SC, Ward DM, Bryant DA. 2015. Draft genome sequence of the Deinococcus-Thermus bacterium Meiothermus ruber strain A. Genome Announc 3:e00202-15. doi: 10.1128/genomeA.00202-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thiel V, Wood JM, Olsen MT, Tank M, Klatt CG, Ward DM, Bryant DA. 2016. The dark side of the Mushroom Spring microbial mat: life in the shadow of chlorophototrophs. I. Microbial diversity based on 16S rRNA gene amplicons and metagenomic sequencing. Front Microbiol 7:919. doi: 10.3389/fmicb.2016.00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loginova LG, Egorova LA, Golovacheva RS, Seregina LM. 1984. Thermus ruber sp. nov., nom. rev. Int J Syst Bacteriol 34:498–499. doi: 10.1099/00207713-34-4-498. [DOI] [Google Scholar]
- 19.Davies J-M, Nowlin WH, Matthews B, Mazumder A. 2010. Temporal discontinuity of nutrient limitation in plankton communities. Aquat Sci 72:393–402. doi: 10.1007/s00027-010-0143-x. [DOI] [Google Scholar]
- 20.Bernstein HC, Carlson RP. 2012. Microbial consortia engineering for cellular factories: in vitro to in silico systems. Comput Struct Biotechnol J 3:e201210017. doi: 10.5936/csbj.201210017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward DM, Bateson MM, Ferris MJ, Kühl M, Wieland A, Koeppel A, Cohan FM. 2006. Cyanobacterial ecotypes in the microbial mat community of Mushroom Spring (Yellowstone National Park, Wyoming) as species-like units linking microbial community composition, structure and function. Philos Trans R Soc Lond B Biol Sci 361:1997–2008. doi: 10.1098/rstb.2006.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allewalt JP, Bateson MM, Revsbech NP, Slack K, Ward DM. 2006. Effect of temperature and light on growth of and photosynthesis by Synechococcus isolates typical of those predominating in the Octopus Spring microbial mat community of Yellowstone National Park. Appl Environ Microbiol 72:544–550. doi: 10.1128/AEM.72.1.544-550.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kühl M, Glud RN, Ploug H, Ramsing NB. 1996. Microenvironmental control of photosynthesis and photosynthesis-coupled respiration in an epilithic cyanobacterial biofilm. J Phycol 32:799–812. doi: 10.1111/j.0022-3646.1996.00799.x. [DOI] [Google Scholar]
- 24.Kühl M, Lassen C, Jørgensen BB. 1994. Optical properties of microbial mats: light measurements with fiber-optic microprobes, p 149–166. In Stal LJ, Caumette P (ed), Microbial mats. Springer, New York, NY. [Google Scholar]
- 25.Nowack S, Olsen MT, Schaible GA, Becraft ED, Shen G, Klapper I, Bryant DA, Ward DM. 2015. The molecular dimension of microbial species: 2. Synechococcus strains representative of putative ecotypes inhabiting different depths in the Mushroom Spring microbial mat exhibit different adaptive and acclimative responses to light. Front Microbiol 6:626. doi: 10.3389/fmicb.2015.00626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakshaug E, Bricaud A, Dandonneau Y, Falkowski PG, Kiefer DA, Legendre L, Morel A, Parslow J, Takahashi M. 1997. Parameters of photosynthesis: definitions, theory and interpretation of results. J Plankton Res 19:1637–1670. doi: 10.1093/plankt/19.11.1637. [DOI] [Google Scholar]
- 27.Diaz JM, Hansel CM, Voelker BM, Mendes CM, Andeer PF, Zhang T. 2013. Widespread production of extracellular superoxide by heterotrophic bacteria. Science 340:1223–1226. doi: 10.1126/science.1237331. [DOI] [PubMed] [Google Scholar]
- 28.González-Flecha B, Demple B. 1995. Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli. J Biol Chem 270:13681–13687. doi: 10.1074/jbc.270.23.13681. [DOI] [PubMed] [Google Scholar]
- 29.Bernstein HC, Kesaano M, Moll K, Smith T, Gerlach R, Carlson RP, Miller CD, Peyton BM, Cooksey KE, Gardner RD, Sims RC. 2014. Direct measurement and characterization of active photosynthesis zones inside wastewater remediating and biofuel producing microalgal biofilms. Bioresour Technol 156:206–215. doi: 10.1016/j.biortech.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Bernstein HC, Brislawn C, Renslow RS, Dana K, Morton B, Lindemann SR, Song HS, Atci E, Beyenal H, Fredrickson JK, Jansson JK, Moran JJ. 2017. Trade-offs between microbiome diversity and productivity in a stratified microbial mat. ISME J 11:405–414. doi: 10.1038/ismej.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Falkowski PG, Raven JA. 2013. Aquatic photosynthesis. Princeton University Press, Princeton, NJ. [Google Scholar]
- 32.Adamska I. 2001. The Elip family of stress proteins in the thylakoid membranes of pro- and eukaryota, p 487–505. In Aro E-M, Andersson B (ed), Regulation of photosynthesis. Springer, New York, NY. [Google Scholar]
- 33.He Q, Dolganov N, Björkman O, Grossman AR. 2001. The high light-inducible polypeptides in Synechocystis PCC6803 expression and function in high light. J Biol Chem 276:306–314. doi: 10.1074/jbc.M008686200. [DOI] [PubMed] [Google Scholar]
- 34.Morris JJ, Johnson ZI, Szul MJ, Keller M, Zinser ER. 2011. Dependence of the cyanobacterium Prochlorococcus on hydrogen peroxide scavenging microbes for growth at the ocean’s surface. PLoS One 6:e16805. doi: 10.1371/journal.pone.0016805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris JJ, Kirkegaard R, Szul MJ, Johnson ZI, Zinser ER. 2008. Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by “helper” heterotrophic bacteria. Appl Environ Microbiol 74:4530–4534. doi: 10.1128/AEM.02479-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dietz KJ. 2011. Peroxiredoxins in plants and cyanobacteria. Antioxid Redox Signal 15:1129–1159. doi: 10.1089/ars.2010.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raven JA. 2011. The cost of photoinhibition. Physiol Plant 142:87–104. doi: 10.1111/j.1399-3054.2011.01465.x. [DOI] [PubMed] [Google Scholar]
- 38.Zhu Y, Graham JE, Ludwig M, Xiong W, Alvey RM, Shen G, Bryant DA. 2010. Roles of xanthophyll carotenoids in protection against photoinhibition and oxidative stress in the cyanobacterium Synechococcus sp. strain PCC 7002. Arch Biochem Biophys 504:86–99. doi: 10.1016/j.abb.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 39.Scott NL, Xu Y, Shen G, Vuletich DA, Falzone CJ, Li Z, Ludwig M, Pond MP, Preimesberger MR, Bryant DA, Lecomte JT. 2010. Functional and structural characterization of the 2/2 hemoglobin from Synechococcus sp. PCC 7002. Biochemistry 49:7000–7011. doi: 10.1021/bi100463d. [DOI] [PubMed] [Google Scholar]
- 40.Melnicki MR, Pinchuk GE, Hill EA, Kucek LA, Stolyar SM, Fredrickson JK, Konopka AE, Beliaev AS. 2013. Feedback-controlled led photobioreactor for photophysiological studies of cyanobacteria. Bioresour Technol 134:127–133. doi: 10.1016/j.biortech.2013.01.079. [DOI] [PubMed] [Google Scholar]
- 41.Bernstein HC, Konopka A, Melnicki MR, Hill EA, Kucek LA, Zhang S, Shen G, Bryant DA, Beliaev AS. 2014. Effect of mono- and dichromatic light quality on growth rates and photosynthetic performance of Synechococcus sp. PCC 7002. Front Microbiol 5:488. doi: 10.3389/fmicb.2014.00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernstein HC, Charania MA, McClure RS, Sadler NC, Melnicki MR, Hill EA, Markillie LM, Nicora CD, Wright AT, Romine MF. 2015. Multi-omic dynamics associate oxygenic photosynthesis with nitrogenase-mediated H2 production in Cyanothece sp. ATCC 51142. Sci Rep 5:16004. doi: 10.1038/srep16004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernstein HC, McClure RS, Hill EA, Markillie LM, Chrisler WB, Romine MF, McDermott JE, Posewitz MC, Bryant DA, Konopka AE, Fredrickson JK, Beliaev AS. 2016. Unlocking the constraints of cyanobacterial productivity: acclimations enabling ultrafast growth. mBio 7:e00949-16. doi: 10.1128/mBio.00949-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sander J, Nowaczyk M, Buchta J, Dau H, Vass I, Deák Z, Dorogi M, Iwai M, Rögner M. 2010. Functional characterization and quantification of the alternative PsbA copies in Thermosynechococcus elongatus and their role in photoprotection. J Biol Chem 285:29851–29856. doi: 10.1074/jbc.M110.127142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baroli I, Gutman BL, Ledford HK, Shin JW, Chin BL, Havaux M, Niyogi KK. 2004. Photo-oxidative stress in a xanthophyll-deficient mutant of Chlamydomonas. J Biol Chem 279:6337–6344. doi: 10.1074/jbc.M312919200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) A representative confocal micrograph (1 of 40) of cells maintained as a stable, photosynthetically supported binary culture of T. elongatus (red; autofluorescence) supporting M. ruber (green; SYBR gold). Bar, 10 µm. (B) Relative abundances measured in percentage of cells counted via fluorescence-activated cell sorting. Values represent the means from three independent steady states held under different oxygen tensions (pO2 = 0, 0.3, and 0.6 atm-O2). Error bars represent ±1 standard deviation. Download FIG S1, PDF file, 0.1 MB (104.6KB, pdf) .
Copyright © 2017 Bernstein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Batch growth and nitrate assay. Each data point represents the mean from three biological replicates. The optical densities (OD600) of M. ruber cells are matched to the M. ruber nitrate samples. Error bars represent ±1 standard deviation. The abiotic control experiment revealed no change in nitrate concentration, which remained constant at 106 ± 3 mg liter−1 over 48 h. Download FIG S2, PDF file, 0.2 MB (184.2KB, pdf) .
Copyright © 2017 Bernstein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Extracellular metabolite measurements of axenic T. elongatus (Ax) and the binary culture (Bi). Specific turbidostat steady-state conditions are designated with the following abbreviations: HL, high light (1,995 µmol photons m−2 s−1); ML, medium light (1,190 µmol photons m−2 s−1); LL, low light (197 µmol photons m−2 s−1); HO, high O2 (pO2 = 0.6 atm); MO, medium O2 (pO2 = 0.3 atm); LO, low O2 (pO2 = 0.0 atm). Measurements are the means from two biological replicates with standard errors. Download FIG S3, PDF file, 0.3 MB (304.2KB, pdf) .
Copyright © 2017 Bernstein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Concordance of replication shown as the ranges between biological duplicate measurements of mRNA abundances (RPKM) values for 2,479 genes under each steady-state condition for which T. elongatus gene expression was analyzed. Specific turbidostat steady-state conditions are designated with the following abbreviations: AX, axenic T. elongatus; BC, binary culture; HL, high light (1,995 µmol photons m–2 s–1); ML, medium light (1,190 µmol photons m–2 s–1); LL, low light (197 µmol photons m–2 s–1); HO, high O2 (pO2 = 0.6 atm); MO, medium O2 (pO2 = 0.3 atm); LO, low O2 (pO2 = 0.0 atm). Download FIG S4, PDF file, 0.2 MB (166KB, pdf) .
Copyright © 2017 Bernstein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Concordance of replication shown as the ranges between biological replicate measurements of mRNA abundances (RPKM) values for 4,492 genes under each steady-state condition for which species-resolved gene expression was analyzed; binary cultivation of T. elongatus and M. ruber. Specific turbidostat steady-state conditions are designated with the following abbreviations: HL, high light (1,995 µmol photons m–2 s–1); ML, medium light (1,190 µmol photons m–2 s–1); LL, low light (197 µmol photons m–2 s–1); HO, high O2 (pO2 = 0.6 atm); MO, medium O2 (pO2 = 0.3 atm); LO, low O2 (pO2 = 0.0 atm). Replication of transcriptomic analyses was performed in duplicate with the exception of condition HL-LO, for which replication was performed in quadruplicate. Download FIG S5, PDF file, 0.1 MB (155.9KB, pdf) .
Copyright © 2017 Bernstein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Statistical/functional enrichment of T. elongatus gene functions within clusters of mRNA profiles. Download TABLE S1, PDF file, 0.2 MB (166.5KB, pdf) .
Copyright © 2017 Bernstein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Statistical/functional enrichment of M. ruber gene functions within clusters of mRNA profiles. Download TABLE S2, PDF file, 0.2 MB (247.9KB, pdf) .
Copyright © 2017 Bernstein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental text describing additional details pertaining to the methodology and results presented in the manuscript and supplemental files. Download TEXT S1, DOCX file, 0.04 MB (41.6KB, docx) .
Copyright © 2017 Bernstein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Axenic T. elongatus transcript abundances and gene annotations. Download DATA SET S1, XLSX file, 0.8 MB (822.7KB, xlsx) .
Copyright © 2017 Bernstein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Binary culture, T. elongatus and M. ruber, transcript abundances, gene annotations, and clustering assignments. Download DATA SET S2, XLSX file, 4.4 MB (4.5MB, xlsx) .
Copyright © 2017 Bernstein et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.