Abstract
Phenotypic differences within populations and between closely related species are often driven by variation and evolution of gene expression. However, most analyses have focused on the effects of genomic variation at cis-regulatory elements such as promoters and enhancers that control transcriptional activity, and little is understood about the influence of post-transcriptional processes on transcript evolution. Post-transcriptional modification of RNA by N6-methyladenosine (m6A) has been shown to be widespread throughout the transcriptome, and this reversible mark can affect transcript stability and translation dynamics. Here we analyze m6A mRNA modifications in lymphoblastoid cell lines (LCLs) from human, chimpanzee and rhesus, and we identify patterns of m6A evolution among species. We find that m6A evolution occurs in parallel with evolution of consensus RNA sequence motifs known to be associated with the enzymatic complexes that regulate m6A dynamics, and expression evolution of m6A-modified genes occurs in parallel with m6A evolution.
Gene expression differences tuned by regulatory changes are considered to be a major contributor to phenotypic differences between closely related species (King and Wilson 1975; Romero et al. 2012; Villar et al. 2014). The majority of effort in deciphering the basis for gene expression evolution has focused on identification of cis-acting genomic regulatory elements and trans-acting regulatory factors that drive differences between species and populations (Schmidt et al. 2010; Zheng et al. 2010; He et al. 2011; Ni et al. 2012; McVicker et al. 2013; Arnold et al. 2014; Villar et al. 2015). Largely unexplored is whether gene regulation has also differentiated at the post-transcriptional level during evolution and what impact if any that may have on gene expression. As the most prevalent post-transcriptional mRNA modification, the biochemistry and molecular genetics of N6-methyladenosine (m6A) RNA transcript modification have been heavily investigated because of its ubiquity among eukaryotes (Desrosiers et al. 1974; Dominissini et al. 2012; Meyer et al. 2012; Batista et al. 2014; Luo et al. 2014), its reversible nature (Jia et al. 2011; Zheng et al. 2013; Liu et al. 2014), its effect on mRNA lifetime and translation efficiency (Wang et al. 2014, 2015), and its role in the regulation of key regulators of biological pathways and human disease (Fustin et al. 2013; Schwartz et al. 2013; Batista et al. 2014). Previous studies also investigated the conservation of the m6A methylome between human and mouse (Dominissini et al. 2012; Chen et al. 2015), and within Arabidopsis (Luo et al. 2014). Yet virtually nothing is known about the evolutionary or population dynamics of this important post-transcriptional modification.
To explore whether evolutionary forces shape gene regulation through m6A at the post-transcriptional level, we mapped the landscape of m6A modifications across the transcriptome of lymphoblastoid cell lines (LCLs) derived from human, chimpanzee, and rhesus.
Results
Comparisons on human, chimpanzee, and rhesus m6A modification
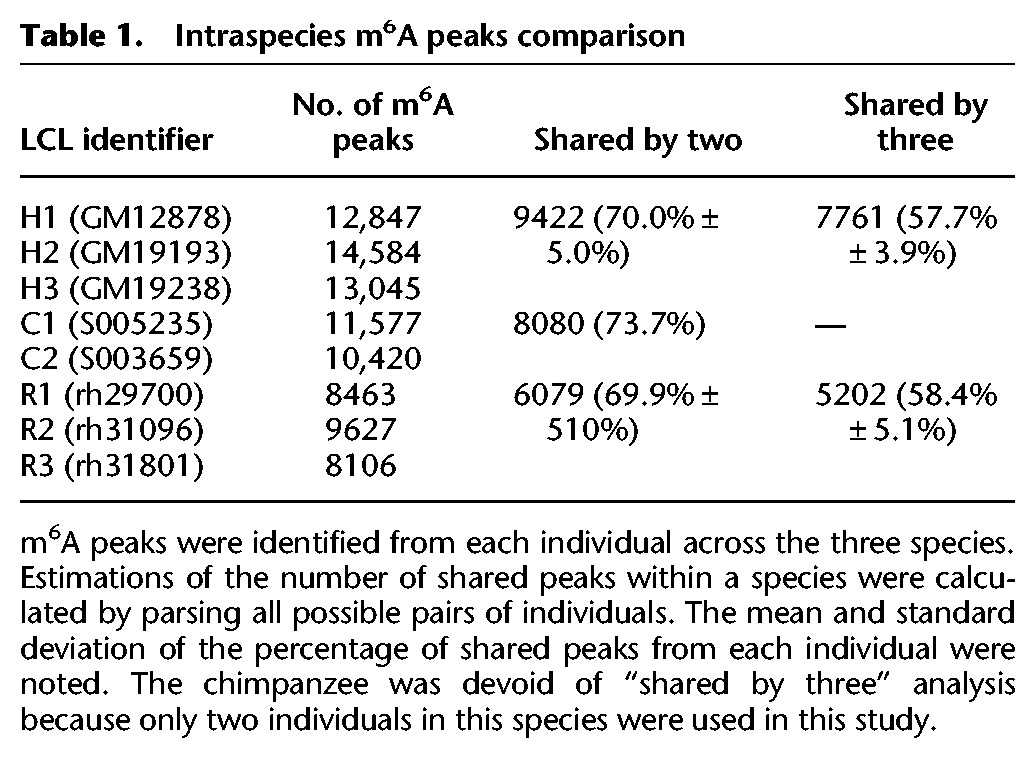
We performed m6A-seq (Dominissini et al. 2012) on LCLs from three human individuals, two chimpanzee individuals, and three rhesus individuals. For each cell line, we performed at least five biological replicates in order to generate accurate estimates of m6A modification levels for transcripts from each genomic locus (Supplemental Fig. S1). m6A peaks were identified in each individual using a peak calling approach modified from Dominissini et al. (2012) (see Methods). For each individual, all biological replicates of m6A immunoprecipitation (IP) and the matched input were fit using a negative binomial model to identify significantly enriched windows (see Methods). On average, we found 13,492 m6A peaks (FDR < 1%) in human LCLs (Supplemental Tables S1–S3), with 9422 (70.0% ± 5.0%) peaks shared between at least two human LCLs and 7761 (57.7% ± 3.9%) peaks identified across all three human LCLs (Table 1). As a control, we examined the variation within replicate experiments from the same human individual and found that 95.3% ± 3.3% of m6A peaks were shared between replicates. At the gene transcript level, 6584 human genes expressed m6A-modified transcripts, accounting for an average of approximately two m6A peaks within transcription units from each gene (Supplemental Fig. S2). Among these transcription units, 4621 (70.2%) were m6A-modified in all three human LCLs. About 10,999 chimpanzee (Supplemental Tables S4, S5) and 8732 rhesus (Supplemental Tables S6–S8) m6A peaks were identified (Table 1). The reduced number of m6A peaks in rhesus appears to result from incomplete annotation of gene structure, especially in untranslated regions (UTRs) (Supplemental Fig. S3).
Table 1.
Intraspecies m6A peaks comparison
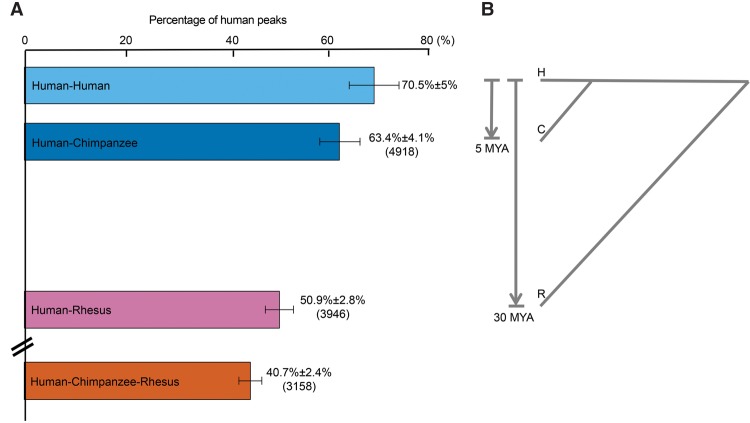
To enable interspecies comparison, orthologous regions expressed from the three genomes were analyzed for m6A peaks (see Methods). In total, 7763 human peaks and their counterparts in chimpanzee and rhesus were used in the interspecies comparison. The number of shared m6A peaks corresponds with the divergence time between human and the other two species (Fig. 1). We found that 4918 (63.4% ± 4.1%) of human m6A peaks were shared with chimpanzee, 3946 (50.9% ± 2.8%) were shared with rhesus, and 3158 (40.7% ± 2.4%) were shared in all three species (Fig. 1). In summary, these data indicate that a considerable fraction of m6A-modified peaks are conserved within and between species and thus are amenable to quantitative analysis for evolutionary patterns of change.
Figure 1.
Interspecies m6A peaks comparison. (A) We compared the m6A peaks and plotted the percentage of the shared peaks between individuals (H–H, H–C, H–R, and H–C–R) by parsing all possible combinations. The mean was plotted as the bars; error bar, SD. The “Human–Human” bar was plotted as a reference of zero diverged time to human. From the top to the bottom: pairwise comparison between two human individuals, pairwise comparison between human and chimpanzee (4918 peaks), pairwise comparison between human and rhesus (3946 peaks), and three-way comparison across human, chimpanzee, and rhesus (3158 peaks). (B) Divergence time between species.
The evolution of m6A modification in primates
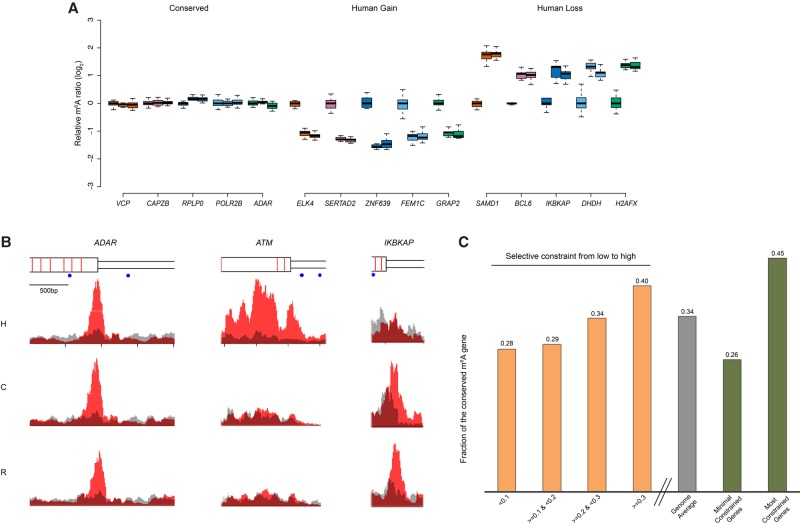
To investigate evolutionary modes of m6A modification, we classified m6A peaks according to patterns of their intra- and interspecies variation. In the absence of a background model for neutral patterns of m6A evolution, it is challenging to define which m6A modifications are under natural selection. However, an empirical model-free statistical test allows the identification of m6A modifications that follow patterns consistent with stabilizing selection or with directional selection (Gilad et al. 2006; Romero et al. 2012). We reasoned that m6A modifications under stabilizing selection should have constant m6A levels both within and between species, while those under directional selection in human should have significant lineage-specific elevated or reduced m6A levels in human but constant levels between chimpanzee and rhesus. For each m6A peak, one-way ANOVA was applied to estimate intraspecies variation between individuals (see Methods). To perform interspecies comparison, enrichment scores of m6A peaks were fitted by a mixed effects model in which species was considered as the fixed effect and individuals within species were considered as random effects. Maximum likelihood (ML) was used to estimate parameters, and hypothesis testing was performed to infer differences between species (see Methods). In total, 2861 m6A peaks (Supplemental Table S9) were conserved at orthologous positions within and between species, displaying stability of m6A modification for at least ∼30 Myr of evolution (Table 2). By using rhesus as an out-group, we also identified 320 instances of m6A gain (Supplemental Table S10) and 30 instances of m6A loss in the human lineage (see discussion below) (Table 2; Supplemental Table S11). Examples of m6A conservation among species, as well as m6A gain or loss in the human lineage, are shown in Figure 2, A and B.
Table 2.
The number of m6A peaks with different evolutionary patterns
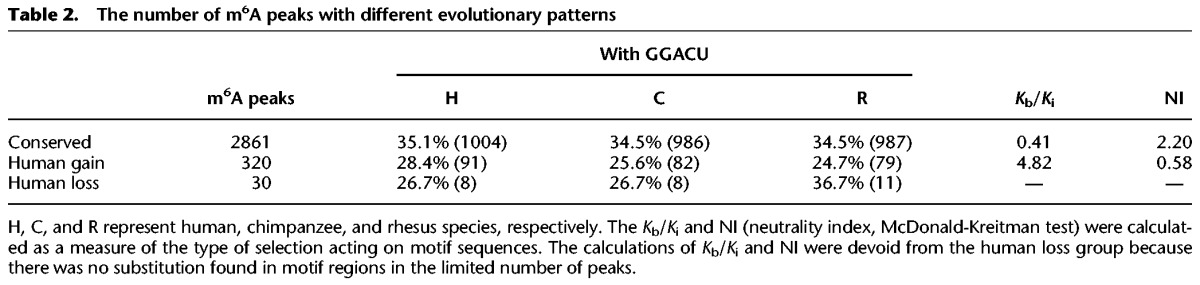
Figure 2.
The evolution of m6A modification. (A) The top five genes from the groups of “conserved,” “human gain,” and “human loss” are presented as examples. The m6A signals in the “conserved” group show low variation both within and between species, following the pattern of stabilizing selection. The “human gain” and “human loss” represent m6A modifications specifically changed in human lineage, which follow the pattern of directional selection. Data points from all biological replicates were included. The m6A signals of each biological replicate from chimpanzee and rhesus were normalized to the mean of enrichment score of all human experiments, and the log2 ratio was plotted in the y-axis to represent differential m6A signals between species. For each gene, from left to right represented human, chimpanzee, and rhesus. (B) m6A-IP and input signals were plotted. We chose one example from each of the three categories. Signals on the transcripts of ADAR (adenosine deaminase, RNA-specific), ATM (ATM serine/threonine kinase), and IKBKAP (inhibitor of kappa light polypeptide gene enhancer in B cells, kinase complex-associated protein) represent “conserved,” “human gain,” and “human loss,” respectively. Red represents m6A-IP signal; gray, input. From the top to the bottom: gene structure (wider rectangle indicates exon; narrow rectangle, UTR; red bar, exon boundary; blue dot, GGACT motif), human signal, chimpanzee signal, and rhesus signal. (C) Human m6A-modified genes were split into different groups according to the selective constraint defined in the ExAC (Exome Aggregation Consortium) data (see Methods). With the increase of selective constraint, we observed the increased percentage of conserved m6A modifications. (Four yellow bars) Q1, Q2, and Q3 represented the quartiles of Z-scores defined by ExAC; the lower the Z-scores, the less the selective constraint. (Gray bar) Genome average (N = 5940). (Two green bars) The minimal constrained genes (N = 296) and the most constrained genes (N = 780).
The selective constraints of DNA sequences correspond to m6A conservation
The conservation of m6A modifications across species does not necessarily mean that the modifications are under purifying selection, as shared ancestry could also lead to conservation. However, we are able to determine whether m6A evolution mirrors the patterns of gene-level sequence evolutionary constraints. To determine whether conserved m6A modifications are associated with genes under selective constraints at the sequence level, we incorporated the data from ExAC (Exome Aggregation Consortium), in which they designed a sophisticated statistical framework to infer selective constraint of genes (see Methods) (Samocha et al. 2014). We split the human m6A-modified genes from our study into four groups according to the sequence selective constraints defined by ExAC (from low to high) (Fig. 2C, four yellow bars) and observed an increase in the percentage of transcripts bearing conserved m6A modifications. Genome-wide, 34.0% of human genes with m6A-modified transcripts (and with chimp and rhesus orthologs) show m6A conservation. When we considered a set of genes defined by ExAC as under minimal constraints, we found that 25.8% show such conservation of m6A modifications on their corresponding transcripts. Conversely, in the most highly constrained gene set defined by ExAC, 45.4% show m6A conservation on their corresponding transcripts. These results indicate that conservation or divergence of exon sequences broadly correspond, respectively, to conservation or divergence of m6A.
However, this relationship is likely to be driven by specific sequence elements and not broad patterns of exon variation and evolution. Indeed, specific RNA sequence motifs are known to be associated with m6A in the human transcriptome (Dominissini et al. 2012; Meyer et al. 2012), and these motifs have been shown to be responsible for binding of proteins associated with m6A modification (Jia et al. 2011; Wang et al. 2014, 2015). Motif analysis of our experimental results demonstrated a consistent set of m6A consensus sequences in all three species, including the previously reported RRACH motif (particularly GGACU) (Dominissini et al. 2012; Meyer et al. 2012) that emerged as the most statistically significant motif for all three species. This motif was present in 48.5%, 65.6%, and 60.5% m6A peaks in human, chimpanzee, and rhesus, respectively (Supplemental Fig. S4). The representation and significance level of the top motif are far better than the secondary and tertiary motifs, although these motifs may be biologically significant as well.
Given the strong conservation of this consensus motif across species in this study and in previous studies (Dominissini et al. 2012; Meyer et al. 2012), we considered whether the evolution of m6A modifications are associated with changes in this motif at the nucleotide sequence level. We compared different evolutionary modes for the occurrence of GGACU (encoded as GGACT in the genomic DNA sequence). For the 2861 evolutionarily stable peaks, the frequency of motif occurrences in orthologous sequences from all three species was ∼35% (Table 2). However, for the 320 m6A peaks gained in human, the motif occurrence in associated orthologous sequences in all three species was reduced to 25%–28%. There were only 30 cases of human m6A loss, which involved similar absolute numbers (between eight and 11) of motifs found in orthologous sequences from the three species. Thus, at least for the previously identified and most significant m6A-associated motif, we observed a higher level of conservation for the motif when m6A marks were conserved among species and a lower level of conservation for the motif when m6A marks had evolved.
To test whether evolution of m6A corresponds to patterns of selection at the motif sequences, we devised an analogy of the Ka/Ks test for positive selection on m6A peaks at the DNA sequence level (see Methods) (Nei and Gojobori 1986; Hahn et al. 2004). Positions in GGACT consensus sequences in the genome were considered equivalent to nonsynonymous sites, and positions in flanking nonmotif regions (from the nonmotif base to the peak boundary) were considered equivalent to synonymous sites. We measured the ratio of substitution per site within the motif (Kb) to the substitution per site within nonmotif regions (Ki) and used the rhesus as an out-group to test the fixed differences within motifs relative to nonmotifs (Hahn et al. 2004) (Table 2). m6A peaks gained in humans showed a Kb/Ki ratio of 4.82 (P = 0.012, Fisher's exact test), indicating that positive selection may have acted on m6A modifications newly acquired in the human lineage. The Kb/Ki ratio is less than one in the conserved peaks (Kb/Ki = 0.41, P = 0.072, Fisher's exact test).
As a control, we considered the Kb/Ki ratio of the human-gained GGACT motifs and surrounding sequences in nontranscribed regions where GGACT motifs presumably have no role in facilitating m6A modifications, and we found that Kb/Ki = 1.02 (see Methods), consistent with the theoretical ratio of 1.0 for neutrality. Thus, the ratio of the human-gained m6A peaks in transcribed portions of the genome significantly deviated from the neutral expectation. These results were confirmed by the McDonald–Kreitman test using the human polymorphism data from the dbSNP database (see Methods) (McDonald and Kreitman 1991; Sherry et al. 2001). The neutrality indexes (NIs) in “human gain” and “conserved” m6A peaks were 0.58 and 2.2, respectively, indicating positive selection on “human gain” peaks and purifying selection on “conserved” peaks. Thus, selection on GGACT motifs in m6A modification peaks appears to be associated with the evolution of m6A modifications between species.
The evolution of m6A modification is reflected in the evolution of mRNA abundance
Because m6A has recently been recognized as an important modulator of mRNA post-transcriptional regulation (Dominissini et al. 2012; Meyer et al. 2012; Wang et al. 2014, 2015), we investigated the extent to which evolution of m6A modification is reflected in the evolution of mRNA abundance among species. Indeed, we find that m6A divergence is correlated with expression divergence of mRNA transcripts. Transcripts with evolved m6A modification show significant expression divergence compared with transcripts with conserved m6A modifications (Fig. 3A). To investigate how m6A modification evolution relates to gene expression evolution, we compared the expression levels of orthologous transcripts with either conserved or diverged m6A modification. Notably, we observed that transcripts that have gained m6A modification in the human lineage are generally more highly expressed in humans compared with the other two species, while transcripts that have lost m6A modifications show mostly reduced expression in human (Fig. 3B; Supplemental Fig. S5). Additionally, orthologous genes with conserved m6A modifications show comparatively less expression divergence and similar numbers of slightly up- and down-regulated genes. These trends also hold true in the chimpanzee-centric analysis (Supplemental Fig. S6). Consistent with this result, when intraspecies expression variation in human was examined, individual-specific m6A modifications were more likely to associate with the elevated mRNA expression level (Supplemental Fig. S7). To verify that the positive correlation between mRNA change and m6A change does not result from an artifact of detecting m6A modification from genes with different expression levels, we devised a control analysis for the ability to detect m6A at different expression levels. We examined genes that carry both shared and individual-specific m6A between human individuals, and we also used a FPKM filter to exclude lower expressed genes (Supplemental Fig. S8). In this case, the positive correlation is still observed. Taken together, these results indicate a positive correlation between the evolution of m6A modification and the evolution of m6A-modified mRNA abundance. m6A modification and mRNA abundance appear to evolve in the same direction, with evolutionary gain of m6A corresponding to the evolution of higher mRNA levels.
Figure 3.
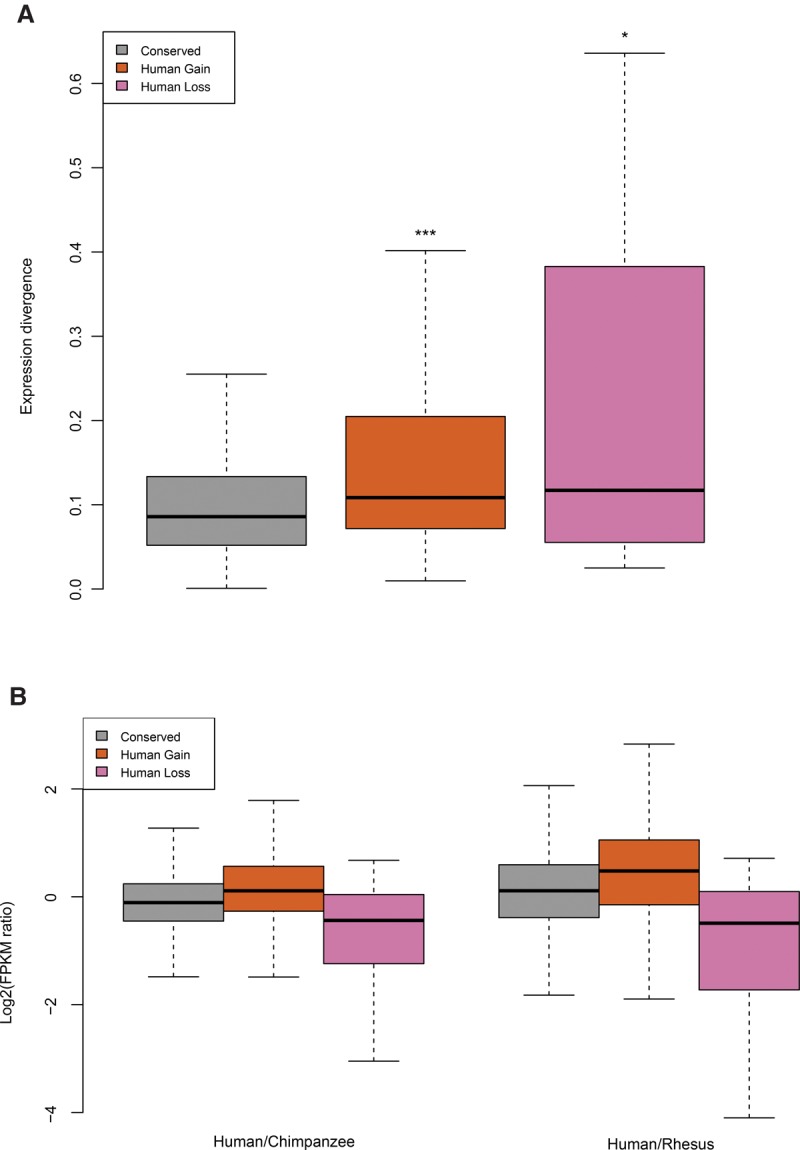
m6A evolution and gene expression divergence. (A) The expression divergence of m6A-modified orthologous genes were plotted along the y-axis. These values were calculated from the coefficient of variation of log2 transformed individual FPKMs. The Wilcoxon test was performed to calculate significant level of statistical differences between “conserved” and the other groups: (***) P < 10−8, (*) P < 10−2. (B) The expression change of orthologous genes was plotted against groups of m6A-modified genes. The “conserved” group has a similar number of up- and down-regulated genes. Human gain m6A-modified genes demonstrated more genes up-regulated compared with chimpanzee and rhesus orthologs, while human loss genes showed more genes were down-regulated. (Left) Human compared to chimpanzee; (right) human compared to rhesus. Conserved, N = 2118; human gain, N = 250; and human loss, N = 30.
Discussion
Comparison of m6A modification in multiple species allowed us to examine the evolutionary mode of post-transcriptional regulation in primates. We identified sets of m6A modification that are consistent with patterns of stabilizing or directional selection. Signals of selection were also identified at the DNA sequence level for m6A-modified peaks, particularly in the GGACT consensus sequences. When we directly compared the change of m6A levels to the change of mRNA expression between species, we found that elevated gene expression evolution is correlated with evolutionary gains of m6A signal. Although the mark itself mediates accelerated translation and decay of mRNA in specific systems where gain of m6A is associated with increased mRNA turnover (Wang et al. 2014, 2015), m6A modification evolution and gene expression evolution appear to track one another. This positive correlation also holds true between the transcripts from two different individuals within a species (Supplemental Fig. S7). When testing this hypothesis using an independent public data set performed in HeLa cells (Liu et al. 2014), we found that upon METTL3 and WTAP knockdown, there is a slight but statistically significant down-regulation of the targeted genes, which is consistent with the pattern observed in our study (siMETTL3 P = 0.00107; siWTAP P = 0.0185; siMETTL14 P = 0.992; one-sided Wilcoxon rank-sum test). However, we did not observe a global correlation when directly comparing the m6A enrichment score to the mRNA levels transcriptome-wide (the correlation between the m6A enrichment score and mRNA abundance in human transcripts carrying m6A modification is −0.09). Considering that transcript levels are impacted by multiple regulatory mechanisms, perhaps this is not surprising. We propose that the evolutionary patterns we observe—increased modification level along with the up-regulation of the transcripts—provide a wider dynamic range for the control of mRNA turnover and protein production. Additional functional studies to determine the cell biological and biochemical role of m6A modification will be needed to gain additional insights regarding the effect the evolution of these post-transcriptional modifications have on the relationship between genotype and phenotype.
We found that the gain of m6A peaks in the human lineage showed a strong enrichment in protein coding sequences compared with “conserved” m6A peaks and the well-known m6A topology (Supplemental Fig. S9a); we also noticed an enrichment of m6A peaks in 5′ UTR and start codons when plotting peak density along the human transcript (Supplemental Figs. S9b, S10). These observations imply that m6A deposited in those regions may have functional significance, which has not been widely appreciated previously. Consistent with this idea, a recent study reported that the 5′ UTR m6A modification could promote cap-independent translation initiation, which served as a selective mRNA translation mechanism (Meyer et al. 2015; Zhou et al. 2015). Together, the results associated with the evolutionary pattern of m6A modifications we have observed in this study add a post-transcriptional dimension to the hypothesis that regulatory changes largely contributed to phenotypic differences between closely related species.
Methods
LCL cell culture and m6A-seq library preparation
The human and chimpanzee LCLs were ordered from Coriell (https://www.coriell.org), and three rhesus LCLs were kindly provided by Dr. Eric Vallender from the New England Primate Research Center. In total, three human LCLs (GM12878, GM19193, GM19238), two chimpanzee LCLs (S005235, S003659), and three rhesus LCLs (rh29700, rh31096, rh31801) were used in this study. All LCLs were derived from adult females. LCLs were cultured in RPMI1640 media (Life Technologies no. 11875-085) with 15% FBS (Life Technologies no. 16000-044) and penicillin–streptomycin (Life Technologies no. 15140-122). Fifty million cells were harvested for each m6A-seq experiment, and six biological replicates were performed for each individual.
The m6A IP experiment was performed following the method of Dominissini et al. (2012, 2013). Total RNA was extracted from about 50 million cells (5PRIME no. 2302340) and followed by poly(A) mRNA purification (Life no. K1580-01). mRNA was fragmented into ∼100 nt by heating to 70°C for 10 min in fragmentation buffer (100 mM Tris-HCl at pH 7.4, 100 mM ZnCl2). The fragmented mRNA was purified by EtOH precipitation. For each sample, an equal amount of mRNA was retained from each biological replicate and an aliquot of pooled replicates served as input. m6A antibody was purchased from Synaptic Systems (no. 202003, lot 42). Each IP reaction is composed of 2–5 µg mRNA, 200 U RNasin (Promega no. N2516), 1× IP buffer (50 mM Tris-HCl at pH 7.4, 750 mM NaCl, 0.5% NP-40 [Thermo Scientific no. 28324]), and 10 µg m6A antibody in 500 µL final volume. The IP product was eluted with 1× elution buffer (1× IP Buffer, 6.7 mM m6A [Sigma no. M2780], and RNasin), and recovered by EtOH precipitation. About 50 ng input mRNA and 50–100 ng IP-ed mRNA were subjected to NGS library preparation using the TueSeq mRNA stranded kit (Illumina no. RS-122-2101/2), with 11 PCR cycles to enrich fragments while minimizing PCR duplicates. The libraries were sequenced on the Illumina HiSeq2000/2500 platform (50-bp single-end) at the Institute of Genomics and Systems Biology's (IGSB) high-throughput genome analysis core at the University of Chicago.
RNA-seq data processing and m6A peak calling
Sequencing reads were aligned to the reference genome (human: hg38; chimpanzee: panTro4; rhesus: rheMac2) using TopHat (v2.0.14) (Kim et al. 2013). If one read could be mapped to multiple locations, a random location will be chosen. Gene structure annotations were downloaded from UCSC hg38 RefSeq (human) and Ensembl release 76 (chimpanzee and rhesus). In total, 25 million mappable reads were sampled from each IP and input experiment. The longest isoform was used if multiple isoforms were detected. Aligned reads were extended to 100 bp (average fragments size) and converted from genome-based coordinates to isoform-based coordinates in order to eliminate the interference from intron in peak calling.
The peak calling method was modified from Dominissini et al. (2012). To call m6A peaks, the longest isoform of each human gene was scanned using a 100-bp sliding window with 10-bp steps. To reduce bias from potential inaccurate gene structure annotation and the arbitrary usage of the longest isoform, windows with reads counts less than 1/20 of the top window in both m6A-IP and input sample were excluded. For each gene, the reads count in each window was normalized by the median count of all windows of that gene. A negative binomial model was used to identify the differential windows between IP and input samples by using the edgeR package (Robinson et al. 2010), combining information from all replicates in the two groups. The window was called as positive if the FDR < 1% and log2(enrichment score) ≥ 1. Overlapping positive windows were merged. The following four numbers were calculated to obtain the enrichment score of each peak (or window): reads count of the IP sample in the current peak/window (a), median read count of the IP sample in all 100-bp windows on the current mRNA (b), read count of the input sample in the current peak/window (c), and median read count of the input sample in all 100-bp windows on the current mRNA (d). The enrichment score of each window was calculated as (a × d)/(b × c).
Orthologous peak mapping
We downloaded the orthologous gene table from Ensembl (release 76). BLAT (Kent 2002) was used to align the homologous regions in orthologous genes. An m6A peak was mapped from one species to another if more than half the length of the peak in the original genome can be mapped to the new genome; this ensured sequence similarity. If one peak has more than one orthologous peak, the counterpart with the highest enrichment score was chosen. Only peaks successfully mapped to other species were considered in interspecies analysis.
Estimating m6A divergence within and between species
Interspecies m6A divergences were estimated both qualitatively and quantitatively.
Qualitative comparison was based on m6A peak overlap. Pair-wise comparison using all possible pairs between individuals from two species was used to calculate mean and standard deviation.
To perform quantitative estimation, we applied a linear mixed model with a fixed factor for species and a random factor for individuals within species:
where yijk is the log2(enrichment score) of one m6A peak measured in species i for replicate k of individual j. The term µ represents the overall mean, αi is the effect of species i, βj(i) is the random effect of individual j in species i and , ε is the residual component, and . The parameters were estimated via ML using R package nlme (http://cran.r-project.org/web/packages/nlme/index.html).
The null hypothesis is
where H, C, and R represent human, chimpanzee, and rhesus, respectively.
An interspecies differential m6A peak will be called if (1) the null hypothesis is rejected (P < 0.05); (2) the difference of enrichment scores between samples was >50% (1.5-fold).
Intraspecies m6A peak divergence was estimated using a similar approach. Qualitative estimation was based on peak overlap between possible pairs of individuals. One-way ANOVA was applied to estimate a quantitative difference in interspecies variation between individuals that pass an equal variance test (P > 0.05). Otherwise, the nonparametric Kruskal-Wallis test was used. Quantitatively different peaks were called when the null hypothesis was rejected (P < 0.05) and the difference of enrichment scores between samples was >50% (1.5-fold).
Identification of m6A evolutionary mode
By using rhesus as an out-group, m6A peaks were categorized into “conserved,” “human gain,” and “human loss,” which represent candidates of m6A that are under stabilizing selection or directional selection. We combined intra- and interspecies divergence estimations and set criteria as described below:
An evolutionarily conserved m6A peak must meet the following criteria: (1) Orthologous peaks were called in all three species; (2) the peaks are not quantitatively different among the species (human, chimpanzee, and rhesus); (3) the peaks are not quantitatively different among individuals in each species. All peaks pass the above criteria were ranked by the standard deviation of all samples, from small to large.
A human-specific m6A peak (human gain) must meet the following criteria: (1) No orthologous peaks were identified in either chimpanzee or rhesus; (2) the peaks are quantitatively different in human versus chimpanzee and in human versus rhesus, but not different in chimpanzees versus rhesus; and (3) the peaks are not quantitatively different among human individuals. Similarly, a human loss m6A peak meets the following criteria: (1) Orthologous peaks were called in both chimpanzee and rhesus, but not in human; (2) the peaks are quantitatively different in human versus chimpanzee and in human versus rhesus, but not in chimpanzee versus rhesus; and (3) the peaks are not quantitatively different among chimpanzee individuals and among rhesus individuals. All peaks that passed the above criteria were ranked by the ratio of interspecies-to-intraspecies mean squared, from large to small. The chimpanzee and rhesus were considered as one species during the mean square calculation, since the m6A in those two nonhuman species is considered as conserved in this case. Peaks we call “chimpanzee gain” and “chimpanzee loss” were defined in the same way.
Analysis of negative selection on m6A modification
Gene constraint data was downloaded from ExAC (release 0.3) (Samocha et al. 2014). They used the 1000 Genomes Project data and the chimpanzee genome to infer intraspecies genetic variation and designed a sophisticated statistical framework to infer selective constraint according to the observed and expected variants in a gene-based analysis. A Z-score was assigned to each gene to represent the χ2 deviation of observation from expectation. Genes were split into four groups using the 25th percentile, 50th percentile, and 75th percentile of the Z-score and were further refined by (1) chimpanzee and rhesus orthologs and (2) the transcript carrying m6A modification. Then the percentage of the genes with conserved m6A modification on their transcripts was calculated. The same percentage was also calculated for the three groups of genes filtered by the same two criteria described above: the minimal constrained genes (−0.1 < Z < 0.1; N = 296), genome average (all genes except the “minimal constrained genes”; N = 5940), and the most constrained genes (Z > 3.09; N = 780).
Motif analysis and selection on motif sequences
HOMER (Heinz et al. 2010) was used to search for motifs in each set of m6A peaks. The longest isoform of all genes was used as background. In order to reveal the natural selection acting on sequences of m6A peaks, the substitutions inside the GGACT motif were considered as nonsynonymous mutations (Kb) and the substitutions in the nonmotif region of that peak (Ki) were considered as synonymous mutations. The concept of this analysis is an analog to the calculation of nonsynonymous substitution rate (Ka) and synonymous substitution rate (Ks) in the coding region. Multiple sequence alignments were performed, and the sequences of rhesus were used as an out-group to infer DNA substitutions in the human lineage. The substitution rates in the GGACT motif (Kb) and the nonmotif region (Ki) were calculated. The Fisher's exact test was used to test for an excess of differences in motif region relative to nonmotif region or vice versa. Since many m6A peaks were located around the boundary of a stop codon, we performed the same analysis on all peaks and peaks that did not overlap with CDS to avoid potential effects from CDS regions (sequences in CDS regions are usually under more purifying selection). Both analyses demonstrated the same conclusion, and the results of excluding CDS-peaks were presented in the main text and Table 2. To establish a neutral background, we did the same Kb/Ki calculation for the “human gain” GGACT in noncoding and nontranscribed regions and its surrounding sequences. Surrounding sequences were set as ±50 bp from the GGACT motif sequence. The McDonald–Kreitman test was also applied to detect selection on m6A peaks and verify results from the Kb/Ki test. Common SNPs (minor allele frequency >1%) were downloaded from the NCBI dbSNP database (Sherry et al. 2001) to estimate the polymorphism in these peaks. Similar to the way we estimated the divergence between human and chimpanzee, the polymorphisms in motif regions were considered as nonsynonymous and in nonmotif regions were considered as synonymous. The NI (NI = (Pn × Ds)/(Ps × Ds)) was calculated for both conserved and diverged m6A peaks.
Gene expression analysis
Cufflinks (v2.2.1) was used to calculate the FPKM of each gene to represent their mRNA expression level (Trapnell et al. 2010). Expression divergence was calculated as the coefficient of variation of log2 transformed FPKMs between orthologous genes.
Data access
The m6A-seq data from this study have been submitted in the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE70299.
Supplementary Material
Acknowledgments
We thank Dr. Eric Vallender from the New England Primate Research Center for providing three rhesus macaque LCLs. We also thank Dr. Martin Kreitman, Dr. Barbara Stranger, Dr. Robert Arthur, Alec Victorsen, Dr. Aashisha Jha, Dr. Bin He, and other members in the White laboratories for comments and discussions. We thank Dr. M.H. Wright for helping editing and discussing the manuscript. Work by K.P.W. and L.M. was supported by the National Institutes of Health (NIH) U54HG006996. The experiments performed in C.H.’s laboratory are supported by NIH R01HG006827. C.H. is an investigator of the Howard Hughes Medical Institute. B.Z. is supported by the Howard Hughes Medical Institute International Student Research Fellowship.
Author contributions: K.P.W. and L.M. planned and designed the project in consultation with C.H.; K.C. and B.Z. advised m6A IP. L.M. and X.H. designed the peak calling methods and quantitative model. L.M. performed m6A-seq experiments with help from A.T. and J.H.T. and carried out computational analyses. L.M. and K.P.W. wrote the paper with the contributions from all authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.212563.116.
References
- Arnold CD, Gerlach D, Spies D, Matts JA, Sytnikova YA, Pagani M, Lau NC, Stark A. 2014. Quantitative genome-wide enhancer activity maps for five Drosophila species show functional enhancer conservation and turnover during cis-regulatory evolution. Nat Genet 46: 685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista PJ, Molinie B, Wang J, Qu K, Zhang J, Li L, Bouley DM, Lujan E, Haddad B, Daneshvar K, et al. 2014. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell 15: 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Hao YJ, Zhang Y, Li MM, Wang M, Han W, Wu Y, Lv Y, Hao J, Wang L, et al. 2015. m6A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell 16: 289–301. [DOI] [PubMed] [Google Scholar]
- Desrosiers R, Friderici K, Rottman F. 1974. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci 71: 3971–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. 2012. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485: 201–206. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, Rechavi G. 2013. Transcriptome-wide mapping of N6-methyladenosine by m6A-seq based on immunocapturing and massively parallel sequencing. Nat Protoc 8: 176–189. [DOI] [PubMed] [Google Scholar]
- Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, et al. 2013. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155: 793–806. [DOI] [PubMed] [Google Scholar]
- Gilad Y, Oshlack A, Smyth GK, Speed TP, White KP. 2006. Expression profiling in primates reveals a rapid evolution of human transcription factors. Nature 440: 242–245. [DOI] [PubMed] [Google Scholar]
- Hahn MW, Rockman MV, Soranzo N, Goldstein DB, Wray GA. 2004. Population genetic and phylogenetic evidence for positive selection on regulatory mutations at the factor VII locus in humans. Genetics 167: 867–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Bardet AF, Patton B, Purvis J, Johnston J, Paulson A, Gogol M, Stark A, Zeitlinger J. 2011. High conservation of transcription factor binding and evidence for combinatorial regulation across six Drosophila species. Nat Genet 43: 414–420. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. 2011. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 7: 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC. 1975. Evolution at two levels in humans and chimpanzees. Science 188: 107–116. [DOI] [PubMed] [Google Scholar]
- Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, Jia G, Yu M, Lu Z, Deng X, et al. 2014. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10: 93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo GZ, MacQueen A, Zheng G, Duan H, Dore LC, Lu Z, Liu J, Chen K, Jia G, Bergelson J, et al. 2014. Unique features of the m6A methylome in Arabidopsis thaliana. Nat Commun 5: 5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351: 652–654. [DOI] [PubMed] [Google Scholar]
- McVicker G, van de Geijn B, Degner JF, Cain CE, Banovich NE, Raj A, Lewellen N, Myrthil M, Gilad Y, Pritchard JK. 2013. Identification of genetic variants that affect histone modifications in human cells. Science 342: 747–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. 2012. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 149: 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoview A, Skabkin MA, Elemento O, Pestova T, Qian SB, Jaffrey SR. 2015. 5′ UTR m6A promotes cap-independent translation. Cell 163: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Gojobori T. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3: 418–426. [DOI] [PubMed] [Google Scholar]
- Ni X, Zhang YE, Negre N, Chen S, Long M, White KP. 2012. Adaptive evolution and the birth of CTCF binding sites in the Drosophila genome. PLoS Biol 10: e1001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero IG, Ruvinsky I, Gilad Y. 2012. Comparative studies of gene expression and the evolution of gene regulation. Nat Rev Genet 13: 505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, Kosmicki JA, Rehnström K, Mallick S, Kirby A, et al. 2014. A framework for the interpretation of de novo mutation in human disease. Nat Genet 46: 944–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. 2010. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328: 1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Agarwala SD, Mumbach MR, Jovanovic M, Mertins P, Shishkin A, Tabach Y, Mikkelsen TS, Satija R, Ruvkun G, et al. 2013. High-resolution mapping reveals a conserved, widespread, dynamic mRNA methylation program in yeast meiosis. Cell 155: 1409–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. 2001. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29: 308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28: 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D, Flicek P, Odom DT. 2014. Evolution of transcription factor binding in metazoans: mechanisms and functional implications. Nat Rev Genet 15: 221–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villar D, Berthelot C, Aldridge S, Rayner TF, Lukk M, Pignatelli M, Park TJ, Deaville R, Erichsen JT, Jasinska AJ, et al. 2015. Enhancer evolution across 20 mammalian species. Cell 160: 554–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. 2014. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505: 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. 2015. N6-methyladenosine modulates messenger RNA translation efficiency. Cell 161: 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Zhao H, Mancera E, Steinmetz LM, Snyder M. 2010. Genetic analysis of variation in transcription factor binding in yeast. Nature 464: 1187–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al. 2013. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 49: 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. 2015. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 526: 591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.