Abstract
Up-frameshift protein 1 (UPF1) is an ATP-dependent RNA helicase that has essential roles in RNA surveillance and in post-transcriptional gene regulation by promoting the degradation of mRNAs. Previous studies revealed that UPF1 is associated with the 3′ untranslated region (UTR) of target mRNAs via as-yet-unknown sequence features. Herein, we aimed to identify characteristic sequence features of UPF1 targets. We identified 246 UPF1 targets by measuring RNA stabilization upon UPF1 depletion and by identifying mRNAs that associate with UPF1. By analyzing RNA footprint data of phosphorylated UPF1 and two CLIP-seq data of UPF1, we found that 3′ UTR but not 5′ UTRs or open reading frames of UPF1 targets have GC-rich motifs embedded in high GC-content regions. Reporter gene experiments revealed that GC-rich motifs in UPF1 targets were indispensable for UPF1-mediated mRNA decay. These findings highlight the important features of UPF1 target 3′ UTRs.
RNA degradation plays a central role in the RNA surveillance machinery for aberrant mRNAs and the post-transcriptional regulation of gene expression for normal mRNAs. Cooperation among RNA helicases, RNA-binding proteins, and microRNAs triggers mRNA degradation by recognizing specific structural or sequence features of target mRNAs (Balagopal et al. 2012; Wu and Brewer 2012).
One of the best characterized RNA surveillance machineries is nonsense-mediated mRNA decay (NMD), which eliminates premature termination codon (PTC)-containing aberrant mRNAs that produce potentially harmful truncated proteins (Schweingruber et al. 2013; Lykke-Andersen and Jensen 2015; Kurosaki and Maquat 2016). In human cells, spliced mRNAs harbor exon junction complexes (EJCs), comprising EIF4A3, Y14 (RBM8A), MAGOH, and CASC3, positioned at 20–24 nt upstream of the exon–exon boundary, as a consequence of pre-mRNA splicing. Most human genes encode the termination codon in the last exon. Thus, on the mRNAs of these genes, all EJCs are located upstream of the termination codon. If an EJC is present at >50–55 nt downstream from the termination codon, such termination codon is generally recognized as the PTC that triggers NMD. According to a current model, the UPF1-SMG1 complex associates with a PTC through the eukaryotic release factors, eRF1 (encoded by ETF1) and eRF3 (encoded by GSPT1), which are the components of the translation termination complex (Kashima et al. 2006; Schoenberg and Maquat 2012). The NMD factors UPF2 and UPF3A or UPF3B (UPF3X) are recruited to the EJC and bridge UPF1 to the EJC. Consequently, SMG1 phosphorylates UPF1 (Isken et al. 2008; Lykke-Andersen and Jensen 2015). Phosphorylated UPF1 (p-UPF1) recruits SMG6 and SMG5-SMG7, resulting in SMG6-mediated endonucleolytic cleavage (Huntzinger et al. 2008; Eberle et al. 2009) and SMG5-SMG7-mediated exonucleolytic degradation (Unterholzner and Izaurralde 2004), respectively. Thus, UPF1 plays a central role in the NMD pathway.
Recent studies revealed that UPF1 is also involved in the normal mRNA decay process, such as STAU1-mediated mRNA decay (SMD). UPF1 directly interacts with STAU1 to promote mRNA degradation (Kim et al. 2005). The NMD factors UPF2 and UPF3X are not involved in SMD. STAU1 recognizes normal mRNAs harboring a STAU1-binding site (SBS) in the 3′ UTR. The SBS is formed by intra-molecular base-pairing within the 3′ UTR, by inter-molecular base-pairing between two different 3′ UTRs (Gong et al. 2013) or between an mRNA 3′ UTR and a long noncoding RNA (lncRNA) (Gong and Maquat 2011). In contrast, another study reported that STAU1 binds preferentially to intra-molecular duplexes in 3′ UTRs, but hardly binds to inter-molecular RNA structures (Sugimoto et al. 2015).
UPF1 is highly conserved among eukaryotes, from yeast to humans, indicating its important biological activities (Culbertson and Leeds 2003). Upf1 knockout mice show embryonic lethality (Medghalchi et al. 2001), suggesting that not only the accumulation of aberrant mRNAs, but also the dysregulation of physiological gene expression might cause this phenotype. UPF1-dependent mRNA decay contributes to the post-transcriptional regulation of a considerable proportion of normal mRNAs (Tani et al. 2012b). UPF1 preferentially associates with the 3′ UTR (Hurt et al. 2013; Zünd et al. 2013; Gregersen et al. 2014; Kurosaki et al. 2014; Lee et al. 2015). The binding of UPF1 to the 3′ UTR is associated with RNA repression in mESC (Hurt et al. 2013). Thus, the sequence features of 3′ UTRs are thought to play an important role in UPF1-dependent mRNA decay. However, the context features in the 3′ UTRs of UPF1 targets (normal mRNAs) remain poorly understood.
In the present study, we aimed to identify UPF1 targets to uncover the context features of mRNAs in UPF1-dependent mRNA decay using combinatorial analysis to measure RNA stability and identify mRNAs that associate with UPF1. Previous studies have determined UPF1 target genes based on the changes in expression level following depletion of UPF1 (Mendell et al. 2004; Viegas et al. 2007). However, in that strategy, it was difficult to assess whether these changes reflected the suppression effect of RNA degradation. Indeed, our previous study revealed that the expression levels of specific transcription factors were regulated via RNA degradation; therefore, the depletion of UPF1 would also affect transcription, resulting in changes in RNA levels (Maekawa et al. 2015). Thus, the depletion of RNA degradation factors would cause the up-regulation of indirect targets via an increased transcription rate. To overcome the erroneous detection in previous studies, we measured the changes of RNA decay rates directly instead of the RNA expression level during depletion of UPF1 (Tani et al. 2012b). To determine RNA decay rates for each transcript, we developed a new method using high-throughput sequencing, called 5′-bromouridine (BrU) immunoprecipitation chase-deep sequencing analysis (BRIC-seq) (Tani et al. 2012a; Imamachi et al. 2014). In BRIC-seq, the newly synthesized RNAs are labeled metabolically with BrU under transcriptionally undisturbed conditions. The decrease of BrU-labeled RNAs is measured chronologically to determine RNA half-lives by high-throughput sequencing. We also tested whether UPF1 was associated with transcripts using ribonucleoprotein (RNP) immunoprecipitation followed by high-throughput sequencing (RIP-seq). We then discovered sequence features in the 3′ UTRs of UPF1 targets by analyzing previously reported RNA footprint data for phosphorylated UPF1 (Kurosaki et al. 2014) and two cross-linking immunoprecipitation sequencing (CLIP-seq) data for UPF1 (Lee et al. 2015; Van Nostrand et al. 2016).
Results
BRIC-approach discriminates PTC-containing UPF1 targets from normal mRNAs
To assess whether our approach using BRIC-seq could detect UPF1-mediated RNA degradation targets, we measured the RNA degradation kinetics of PTC-containing UPF1 targets such as TUBA1B, PRMT1, and RPS9 mRNAs. The short RNA half-lives of these mRNAs were prolonged upon UPF1 depletion (Supplemental Fig. S1A).
A large number of genes express several isoforms during post-transcriptional processing, and the PTC-containing isoforms are recognized by NMD machinery. The GENCODE project has identified many PTC-containing mRNAs, based on high-throughput sequencing (Harrow et al. 2012) and PTC-containing mRNAs are also annotated in the RefSeq database. To further assess whether our approach using BRIC-seq could discriminate UPF1 target mRNAs on a genome-wide scale, we compared the RNA decay rates of PTC-containing isoforms (isoform level) registered in RefSeq and GENCODE v19 with the sum of all isoforms (gene level) (Supplemental Table S2). The RNA half-lives of PTC-containing isoforms (median of T1/2 = ∼3.9 h) were shorter than those of all isoforms (median of T1/2 = ∼7.2 h) in control cells. However, the distribution of RNA half-lives of PTC-containing isoforms was shifted toward longer RNA half-lives upon depletion of UPF1 (see the medians of T1/2 in Supplemental Fig. S1B). We also analyzed the changes in mRNA abundance of PTC-containing isoforms and the sum of all isoforms after depletion of the NMD factors UPF1, UPF2, and SMG1. PTC-containing isoforms were up-regulated after depletion of these NMD factors. However, the sum of expression levels of all isoforms did not change (Supplemental Fig. S2A–E). These results indicated that the BRIC-seq approach could discriminate UPF1-dependent RNA decay targets (PTC-containing mRNAs) from non-UPF1 targets.
Identification of bona fide UPF1 target mRNAs
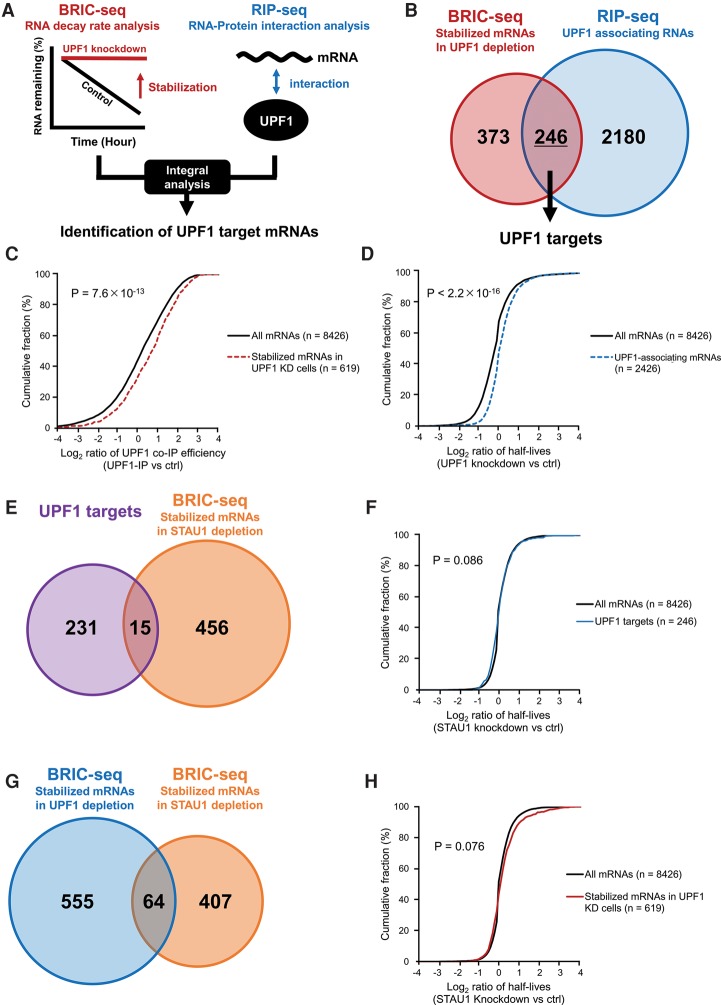
To identify bona fide endogenous UPF1 targets, we measured the changes in mRNA decay rates following depletion of the UPF1 protein in HeLa tet-off cells using previously reported BRIC-seq data with modified half-life calculation (Tani et al. 2012b). Briefly, BRIC-seq data collected at 0, 4, 8, and 12 h were aligned to the human genome (hg19) and transcriptome (RefSeq). To calculate mRNA decay rates for each transcript, the relative mRNA remaining after 0 h was measured from the number of the aligned BrU-labeled mRNAs at each time point. As a result, the mRNA decay rates of 8426 transcripts were determined. By comparing the half-lives between the control and UPF1 knockdown, the half-lives of 619 transcripts were identified as being prolonged by at least twofold following depletion of UPF1 (Fig. 1B).
Figure 1.
Identification of UPF1 and SMD target mRNAs, including UPF1 targets partially regulated by the STAU1 protein. (A) Schematic of the method to identify UPF1 targets based on the combinatorial analysis of BRIC-seq and RIP-seq. (B) Venn diagram illustrating the overlap of the two types of mRNAs: those with more than twofold prolonged half-lives upon UPF1-knockdown in HeLa tet-off cells (red: BRIC-seq, 619 mRNAs) and those enriched by more than twofold in the HA-UPF1 immunoprecipitation fraction relative to input fraction (blue: RIP-seq, 2426 mRNAs). The intersection of stabilized mRNAs in UPF1-knockdown cells (BRIC-seq) and UPF1-associating mRNAs (RIP-seq) were judged as bona fide UPF1 target mRNAs (246 mRNAs). (C) Cumulative distribution of changes in UPF1 association of 619 transcripts stabilized upon UPF1 depletion (red dashed line; median: 0.66) and 8426 whole transcripts (black solid line; median: 0.17). Changes in UPF1 association were calculated as the number of mapped tags of RNA-seq immunoprecipitated with HA-UPF1 by reads per kilobase of transcript per million mapped reads [RPKM]/the number of mapped tags of input RNA-seq [RPKM] ≥2. P-values were calculated using the Wilcoxon rank-sum test. (D) Cumulative distribution of changes in RNA stability of 2426 UPF1 associating transcripts (blue dashed line; median: 0.02) and 8426 whole transcripts (black solid line; median: −0.20). Changes in RNA stability were determined by the following calculation: the half-life of each transcript in UPF1 depletion/the half-life of each transcript in control. P-values were calculated using the Wilcoxon rank-sum test. (E) Venn diagram illustrating the overlap of two types of genes: UPF1 target mRNAs (purple: 246 genes) and mRNAs with more than twofold prolonged half-lives upon STAU1-knockdown in HeLa tet-off cells (orange: BRIC-seq, 471 genes). The intersection of the UPF1 target mRNAs and stabilized mRNAs in STAU1-knockdown cells (BRIC-seq) indicates STAU1-mediated mRNA decay (SMD) target mRNAs (15 genes). (F) Cumulative distribution of changes in RNA stability of 246 UPF1 target transcripts (blue line; median: 0.032) and 8426 whole transcripts (black line; median: 0.000). Changes in RNA stability were determined by the following calculation: the half-life of each transcript in STAU1 depletion/the half-life of each transcript in control. P-values were calculated using the Wilcoxon rank-sum test. (G) Venn diagram illustrating the overlap of two types of mRNAs: those with more than twofold prolonged half-lives upon UPF1-knockdown (blue: BRIC-seq, 619 genes containing 246 UPF1 targets and 373 transcripts stabilized by UPF1 depletion but not associated with UPF1; see B) and STAU1-knockdown in HeLa tet-off cells (orange: BRIC-seq, 471 genes). The number of the stabilized mRNAs upon UPF1-knockdown (BRIC-seq) and those upon STAU1-knockdown (BRIC-seq) contains 64 mRNAs. (H) Cumulative distribution of changes in RNA stability of mRNAs with more than twofold prolonged half-lives upon UPF1-knockdown (red line; median: 0.008) and 8426 whole transcripts (black line; median: 0.000). Changes in RNA stability were determined by the following calculation: the half-life of each transcript in STAU1 depletion/the half-life of each transcript in control. P-values were calculated using the Wilcoxon rank-sum test.
Next, we identified UPF1-associating mRNAs using RIP-seq, in which transcripts coimmunoprecipitating with UPF1 were identified by high-throughput sequencing analysis. As a result, 2426 transcripts were at least twofold enriched in the UPF1-immunoprecipitated sample compared with the input sample. A CLIP experiment for endogenous UPF1 proteins was performed to validate whether UPF1-associating mRNAs were directly bound to UPF1. We confirmed the association of the UPF1 protein and its target mRNAs (Supplemental Fig. S4D). This result suggests that we successfully detected UPF1-associating transcripts using RIP-seq (Fig. 1B).
The cumulative curve of UPF1-mediated immunoprecipitation efficiency (UPF1 co-IP efficiency) of 619 transcripts stabilized by UPF1 depletion were compared with that of the 8426 transcripts comprising the parent population (all mRNAs containing both stabilized and nonstabilized mRNAs upon UPF1 depletion). In this analysis, UPF1 co-IP efficiency indicated the ratio of the RPKM of transcripts in the UPF1-immunoprecipitated sample to the RPKM of transcripts in the input sample from RIP-seq data. As a result of this comparison, these 619 stabilized transcripts upon UPF1 depletion tended to be coimmunoprecipitated more efficiently with the UPF1 protein compared with all mRNAs (Fig. 1C; Supplemental Fig. S3A), corresponding to the association of UPF1 to its target mRNAs. In this work, we mentioned the mRNAs identified by RIP-seq or similar experiments as UPF1-associating transcripts. Moreover, the cumulative RNA stability changes of 2426 UPF1-associating transcripts upon UPF1 depletion were compared with the 8426 transcripts of the parent population. In this analysis, “RNA stability” indicated the relative RNA decay half-life under UPF1 knockdown conditions compared with the control conditions. The result showed that these UPF1-associating transcripts tended to be more stabilized in UPF1 knockdown cells, corresponding to UPF1 targets being stabilized upon depletion of UPF1 (Fig. 1D; Supplemental Fig. S3B).
Finally, we compared the BRIC-seq and RIP-seq results. As a result, we identified 246 transcripts (40% of the transcripts selected by BRIC-seq data and 10% of the transcripts selected by RIP-seq data) that had at least twofold prolonged RNA half-life in UPF1-knockdown cells and were enriched in the UPF1-immunoprecipitated sample. We used these 246 transcripts as bona-fide UPF1 targets in the subsequent analyses (Supplemental Tables S3, S4).
UPF1 target mRNAs are enriched for transcription factors and cell-cycle regulators
To assess the biological functions of the UPF1 target mRNAs, GO term enrichment analysis was performed. Transcription factors (GO: 0006350, transcription P = 6.7 × 10−6, Benjamini-adjusted P = 8.5 × 10−3) were enriched among these targets, which agreed with previous studies (McIlwain et al. 2010; Hurt et al. 2013). In a previous study, UPF1 depletion induced the arrest at the early S phase via an ATR-dependent DNA damage response (Azzalin and Lingner 2006). Correspondingly, cell-cycle regulators (GO: 00511726, regulation of cell cycle P = 3.4 × 10−5, Benjamini-adjusted P = 2.1 × 10−2) were also enriched among UPF1 targets (Supplemental Table S5).
Certain UPF1 target mRNAs are regulated partially by the STAU1 protein
Previous studies revealed that STAU1 is involved in UPF1-dependent normal mRNA decay (Kim et al. 2005). However, to what extent UPF1-regulated normal mRNAs are regulated by STAU1 is not fully understood. We next sought to assess the contribution of the STAU1 protein to UPF1-dependent mRNA decay by measuring the changes in RNA decay rates of the 246 UPF1 targets after depletion of STAU1, using previously reported BRIC-seq data (Maekawa et al. 2015). We determined that the RNA decay rates of 471 transcripts were at least twofold prolonged after depletion of STAU1. A comparison of the 471 STAU1 targets with the 246 UPF1 targets showed that only 15 transcripts (6%) of UPF1 targets were stabilized in STAU1-knockdown cells (Fig. 1E). Sixty-four of 619 stabilized transcripts upon UPF1 depletion (15%) were stabilized in STAU1-knockdown cells (Fig. 1G). Moreover, the cumulative changes in stability of the UPF1 targets upon STAU1 depletion were compared with that of all 8426 transcripts upon STAU1 depletion. The changes of RNA decay of the UPF1 targets upon STAU1 depletion were not significantly changed compared with those of all 8426 transcripts upon STAU1 depletion (Fig. 1F). In addition, there was no significant difference between the cumulative changes of stability of whole stabilized mRNAs by UPF1 depletion and that of all mRNAs upon UPF1 depletion (Fig. 1H). These results indicate that a small proportion of UPF1 targets are regulated by STAU1. Thus, the majority of UPF1 targets are regulated independently of STAU1.
UPF1 target mRNAs were up-regulated by the knockdown of UPF1, UPF2, and SMG1
To assess whether these NMD factors are required for UPF1-dependent normal mRNA decay, we measured the changes in RNA abundances of six UPF1 targets after depletion of these NMD factors. All tested UPF1 targets were up-regulated after depletion of UPF1, UPF2, and SMG1 (Supplemental Fig. S4A–D). These results suggested that UPF2 and SMG1 are also involved in UPF1-dependent normal mRNA decay.
Prediction of phosphorylated UPF1-binding sites on the 3′ UTRs of UPF1 target mRNAs
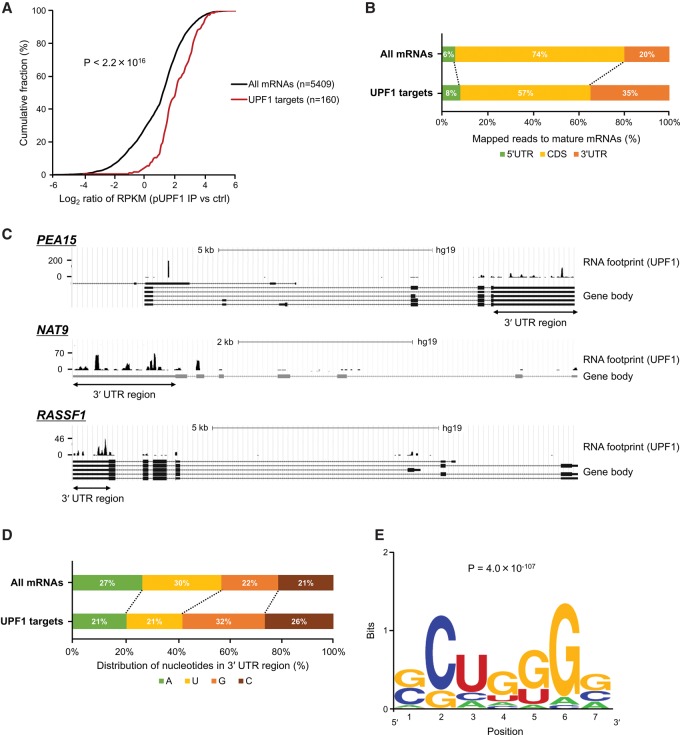
Phosphorylation of UPF1 is a necessary step to degrade PTC-containing mRNAs (Schweingruber et al. 2013). Phospho-UPF1 (p-UPF1) is enriched at the 3′ UTRs of PTC-containing mRNAs (Kurosaki et al. 2014) and causes the recruitment of SMG6 and SMG5-SMG7 to degrade mRNAs. Thus, p-UPF1 on mRNAs is an effective marker for identification of mRNA regulated by UPF1. To examine whether p-UPF1 binding sites are enriched in the mRNAs of our UPF1 targets, we analyzed genome-wide RNA footprint data for p-UPF1 (Kurosaki et al. 2014). In this analysis, we excluded transcripts that could not be detected in the RNA footprint data from among all 8426 transcripts. As a result, 5414 and 160 transcripts were selected as all transcripts and UPF1 targets, respectively, and these data sets were used in the subsequent analysis. We measured the sequence reads derived from RNAs associated with p-UPF1 along 160 UPF1 targets and all 5414 mRNAs. The 160 UPF1 targets tended to bind to p-UPF1 more frequently (Fig. 2A). In addition, the preferential p-UPF1 binding to the 3′ UTRs of UPF1 targets was observed compared with the 5′ UTRs and coding DNA sequence (CDS) (Fig. 2B), as previously reported (Kurosaki et al. 2014). Sequencing reads derived from RNAs associated with p-UPF1 were enriched in the 3′ UTRs of specific UPF1 targets (Fig. 2C). We found that p-UPF1 residence in the 3′ UTRs of UPF1 targets was biased toward high G and C nucleotide content (Fig. 2D).
Figure 2.
Prediction of phosphorylated UPF1-binding sites on 3′ UTRs of UPF1 target RNAs. (A) Cumulative distribution of all mRNAs (black) and UPF1 targets (red) in association with phospho-UPF1. P-values were calculated using the Wilcoxon rank-sum test. (B) The distribution of mapped reads derived from mRNAs associated with p-UPF1 in the indicated RNA regions. (C) Examples of p-UPF1 association with bona fide UPF1 target mRNAs. The y-axis indicates numbers of sequencing reads derived from mRNAs associated with p-UPF1. mRNAs transcribed from these genes are UPF1 targets. (D) Relative ratio of nucleotides in p-UPF1-binding sites in 3′ UTRs of mRNAs. (E) p-UPF1-binding motifs in 3′ UTRs of UPF1 target mRNAs were predicted using MEME software. The motif length was restricted to 5–8 bp. The motif with the lowest P-value (P = 4.0 × 10−107) is shown.
We next sought to predict p-UPF1 binding motifs in 3′ UTR of UPF1 targets using the MEME software (Bailey et al. 2006). GC-rich motifs, which contain the CUG sequence, were identified in the 3′ UTRs of UPF1 targets, but not in the 5′ UTRs and CDSs (Fig. 2E; Supplemental Table S6). This result suggested that GC-rich motifs in the 3′ UTR are target motifs in UPF1-dependent mRNA decay.
In addition, we analyzed the publicly available CLIP-seq data for UPF1 target mRNAs (Lee et al. 2015; Van Nostrand et al. 2016). In accordance with our conclusion obtained from the RNA footprint data for p-UPF1, UPF1 is enriched on GC-rich sequence in the 3′ UTR in other cell contexts (HEK293 cells and K562 cells) (Supplemental Fig. S5).
Characterization of context features in 3′ UTRs of UPF1 targets
AU-rich elements (ARE) are well known as degradative motifs in the 3′ UTRs of mRNAs. Specific RNA-binding proteins bind to AREs, resulting in the regulation of stability of mRNAs containing AREs. The presence of a high AU-content surrounding an ARE is important for ARE-mediated mRNA decay (Spasic et al. 2011). A previous study found that UPF1 preferentially binds G-rich sequences in 3′ UTRs in mESCs (Hurt et al. 2013). Inspired by these observations, we analyzed whether the predicted GC-rich motifs in bona-fide 246 UPF1 target mRNAs are embedded in regions with higher GC-content. As expected, the GC-content of 3′ UTRs of UPF1 target mRNAs were 8%–12% higher than that of the other mRNAs (Supplemental Fig. S6A). This result suggested that the high GC-content of the 3′ UTRs of UPF1 targets are important in UPF1-dependent mRNA decay. Indeed, mRNAs with high GC content in their 3′ UTRs were preferentially stabilized by UPF1 depletion (Supplemental Fig. S7A) and were associated with UPF1 (Supplemental Fig. S7B,C). We next sought to assess whether the high GC-content in the UPF1 targets is conserved between humans and mice. We prepared the orthologous data sets for humans and mice from the RefSeq database to compare GC-content of 3′ UTRs between the two species (Fong et al. 2013). The GC-content of mouse UPF1 target orthologs were significantly higher in 3′ UTRs. This analysis also demonstrated that the high GC-content of the UPF1 targets are conserved between humans and mice (Supplemental Fig. S6B). Therefore, high GC-content in the 3′ UTRs are one of the important features to identify UPF1 targets.
To further characterize the sequence features of UPF1 targets, we measured mononucleotide content and dinucleotide content in the 3′ UTRs of UPF1 targets and the other genes with >55% GC-content (median GC-content of UPF1 targets). The G content and several dinucleotide (GA and AG) content in UPF1 targets were higher than those in the other genes. On the other hand, the C content and a dinucleotide (CC) content in UPF1 targets were lower than in the other genes (Supplemental Fig. S8A,B). These results suggested that the presence of G content and several dinucleotide (GA and AG) content surrounding GC-rich motifs in the 3′ UTR are also important for UPF1-mediated mRNA decay.
Transposable elements (SINE, LINE, and LTR) are not significantly contained in the 3′ UTRs of UPF1 targets
Transposable elements (TEs) such as the long terminal repeat (LTR), long interspersed element (LINE), and short interspersed element (SINE) in the 3′ UTRs of mRNAs play an important role in gene expression (Feschotte 2008). Alu elements, classified as SINEs, are inserted into the 3′ UTRs of several SMD target mRNAs. mRNA–lncRNA or mRNA–mRNA hybrids are targeted via base-pairing with complementary Alu elements. These hybridizations via complementary Alu elements are recognized by the STAU1 protein and are degraded in the SMD pathway (Gong and Maquat 2011; Gong et al. 2013). In addition, it was reported that UPF1 interacts with LINEs to negatively regulate LINE constituents (Taylor et al. 2013). To assess whether 3′ UTRs containing TEs are preferentially degraded by UPF1, we analyzed TE-containing mRNAs. The transcripts with LTRs and mammalian-wide interspersed repeat (MIR) elements, classified as SINEs, in the 3′ UTRs were slightly enriched in UPF1 targets (Supplemental Fig. S9A,B). TE-containing mRNAs were immunoprecipitated with UPF1 (Supplemental Fig. S9D,E) but not with p-UPF1 (Supplemental Fig. S9F,G). In addition, no significant changes in RNA stability after depletion of UPF1 were observed for these mRNAs (Supplemental Fig. S9B,C). Thus, we did not observe significant degradation of TE-containing mRNAs mediated by UPF1.
Phosphorylated UPF1-binding motifs are related to RNA decay in a UPF1-dependent manner
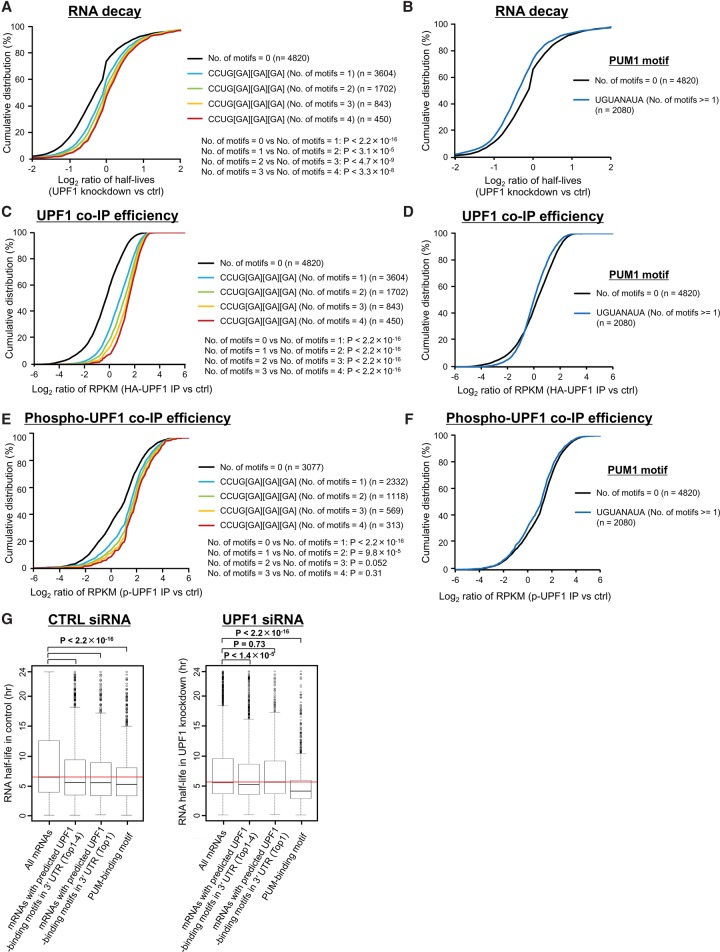
To assess whether transcripts with predicted GC-rich motifs in their 3′ UTRs (Fig. 2E) are recognized and degraded in a UPF1-dependent manner, we measured the UPF1-binding affinity and UPF1-dependent RNA degradation for transcripts with and without the predicted motifs by analyzing BRIC-seq, RIP-seq, and RNA footprint data for p-UPF1, respectively. The top four GC-rich motifs that are significantly found in the 3′ UTRs of UPF1 targets were used in this analysis. mRNAs containing these top four motifs were significantly stabilized after depletion of UPF1 and significantly bound to UPF1 and p-UPF1, depending on the increased numbers of motifs in the 3′ UTR (Fig. 3A,C,E). In contrast, mRNAs with a binding motif for PUM1, a well-known mRNA decay factor (Morris et al. 2008), were not significantly stabilized after the depletion of UPF1 and bound to neither UPF1 nor p-UPF1 (Fig. 3B,D,F).
Figure 3.
Phosphorylated UPF1-binding motifs are related to RNA decay in a UPF1-dependent manner. (A,B) Cumulative distribution of relative half-lives (UPF1 knockdown/control) of mRNAs harboring the indicated number of any four UPF1-binding motifs (CCUGGGG, CCUGGGA, CCUGGAA, CCUGAGA; these are compressed as CCUG[GA][GA][GA] in the caption) (A), or PUM1-binding motif (UGUANAUA) (B) in their 3′ UTRs. Data were constructed from BRIC-seq data. The numbers shown to the right indicate the number of mRNAs included in the group. P-values were calculated using the Wilcoxon rank-sum test. (C,D) Cumulative distribution of the relative enrichment (IP of UPF1/normal IgG control) of mRNAs harboring the indicated number of any four UPF1-binding motifs (C), or PUM1-binding motif (D) in their 3′ UTRs in a RIP-seq experiment. (E,F) Cumulative distribution of the relative enrichment (IP of p-UPF1/normal IgG control) of mRNAs harboring the indicated number of any four UPF1-binding motifs (E), or PUM1-binding motif (F) in their 3′ UTRs in a genome-wide RNA footprinting experiment. (G) The RNA half-life distribution for all mRNAs (leftmost box); mRNAs with any predicted UPF1-binding motifs (top 1–4) (CCUGGGG, CCUGGGA, CCUGGAA, or CCUGAGA) (second box from the left); mRNAs with predicted UPF1-binding motifs (top 1) (CCUGGGG) (third box from the left); and mRNAs with PUM1-binding motifs (UGUANAUA) (rightmost box) in the control (left panel) and UPF1 siRNA transfected cells (right panel). Red horizontal lines indicate the median value of the half-life distribution for all mRNAs.
In general, transcripts with degradative motifs have shorter RNA half-lives. Indeed, the RNA half-lives of transcripts with binding motifs for PUM1 were significantly shorter than the other mRNAs (Fig. 3G). The short half-lives of transcripts with p-UPF1 binding motifs were prolonged by depletion of UPF1. In contrast, transcripts with binding motifs for PUM1 were not stabilized upon UPF1 depletion (Fig. 3G). In conclusion, we believe that the predicted GC-rich sequences are degradative motifs in UPF1-dependent mRNA decay.
Phosphorylated UPF1-binding motifs are responsible for UPF1-mediated mRNA decay
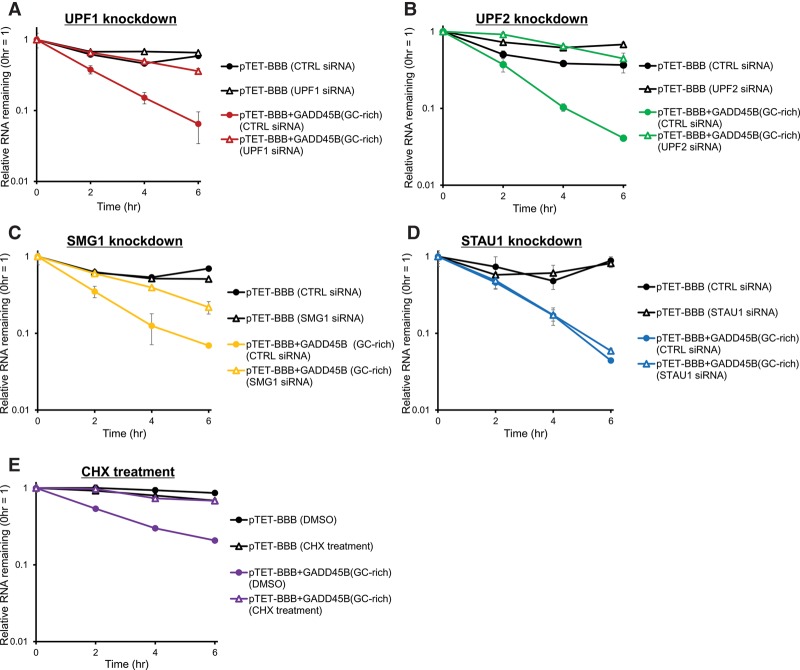
To test whether the mRNAs harboring p-UPF1-binding motifs in 3′ UTRs are degraded in a UPF1-dependent manner in vivo, the tet-off reporter gene expression system was used. We constructed a plasmid harboring rabbit beta globin gene fused with the GADD45B 3′ UTR (containing a GC-rich region) under Tet-mediated transcriptional regulation (Fig. 4A). In HeLa tet-off cells, the rabbit beta globin transcript fused to the GC-rich region of GADD45B 3′ UTR was more unstable than the parental rabbit beta globin transcript. The stability of the GC-rich sequence-fused beta globin mRNA increased after depletion of UPF1, UPF2, and SMG1 (Fig. 4A–C). Because UPF1-dependent NMD is a translation-dependent process, we next tested whether GC-rich sequence-fused transcripts were also degraded dependent on translation. The stability of the GC-rich sequence-fused transcript was also increased by inhibition of translation using Cycloheximide (CHX), a translational inhibitor (Fig. 4E). In contrast, the RNA decay rates of rabbit beta globin transcript fused to GC-rich region of the GADD45B 3′ UTR did not change after STAU1 depletion (Fig. 4D). These results indicate that the GADD45B 3′ UTR containing the GC-rich sequence specifically triggers UPF1-dependent mRNA decay.
Figure 4.
Phosphorylated UPF1-binding motifs trigger RNA decay in a UPF1-dependent manner. (A–E) HeLa tet-off cells were transfected with pTET-BBB vector or pTET-BBB+GADD45B 3′ UTR vector. pGL4.13 luciferase (Luc) expression vector was cotransfected as an internal control. Doxycycline was added to the medium to stop transcription from the tet-responsive promoter, and total RNA was collected at 0, 2, 4, and 6 h after addition of doxycycline. qRT-PCR was performed to monitor Luc and beta globin mRNA levels and determine the relative RNA remaining compared to time 0 h. For each point, the beta globin mRNA level was normalized to the Luc mRNA levels. The decay rates of rabbit beta globin mRNA derived from pTET-BBB or pTET-BBB+GADD45B 3′ UTR (GC-rich region) were measured in UPF1- (A), UPF2- (B), SMG1- (C), or STAU1- (D) depleted cells; or cycloheximide- (E) treated cells and compared with the control condition. Values represent the mean ± SD from triplicate experiments (biological replicates).
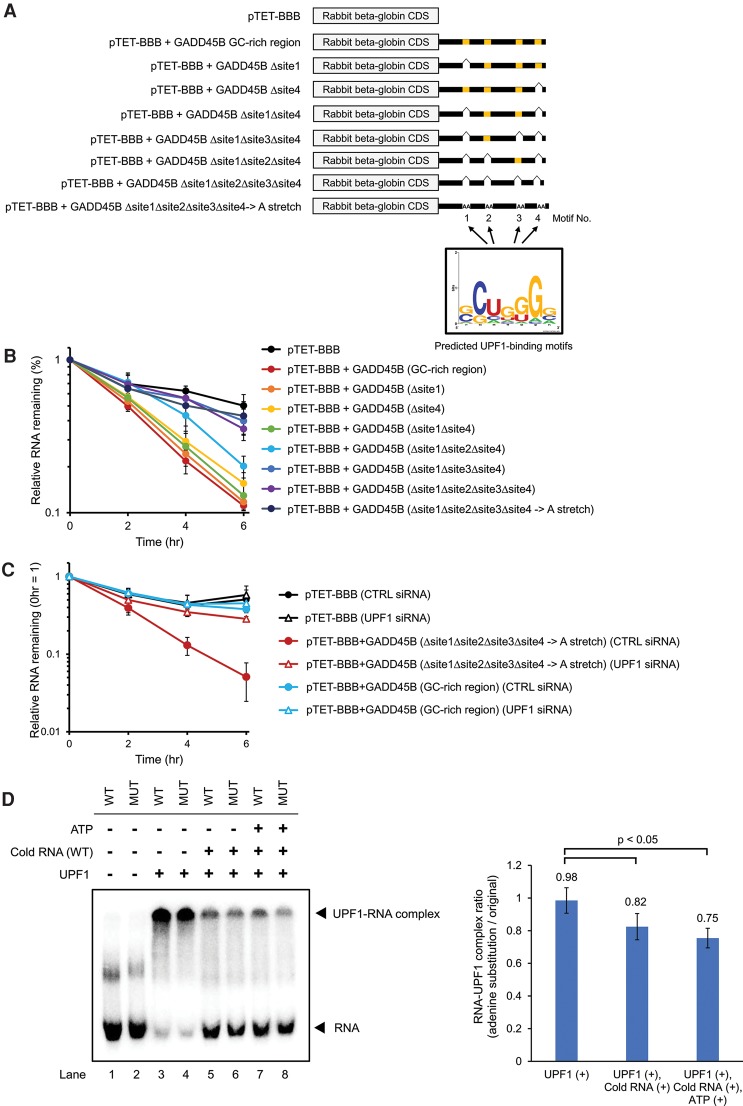
To test the functional significance of the p-UPF1-binding motifs in mRNA decay, we performed mutational analysis of the p-UPF1 binding motifs in the GADD45B 3′ UTR, (Fig. 5A; Supplemental Fig. S10). Deletion of one or two p-UPF1 binding motifs from the GC-rich region had weak effects on the mRNA decay rates (Fig. 5B). In contrast, the deletion of three or four motifs or the substitution of four motifs to adenine sequence cancelled the accelerated mRNA decay (Fig. 5B). In addition, the RNA decay rate of the adenine substitution mutant did not change on UPF1 depletion. It was also almost the same as the RNA decay rate of rabbit beta globin transcript fused to GADD45B 3′ UTR on UPF1 depletion (Fig. 5C). Taken together, these p-UPF1 binding motifs (site 1–4) are related functionally to UPF1-mediated mRNA decay.
Figure 5.
Phosphorylated UPF1-binding motifs trigger RNA decay in a UPF1-dependent manner. (A) The 3′ UTR of GADD45B mRNA contains four p-UPF1 binding motifs. (B,C) The decay of rabbit beta globin mRNA derived from pTET-BBB, pTET-BBB fused with GADD45B 3′ UTR, pTET-BBB fused with mutant GADD45B 3′ UTR deleted with indicated p-UPF1-binding motif, and pTET-BBB fused with mutant GADD45B 3′ UTR with substitution of contiguous adenylic acid sequence (A, stretch) were measured in the cells without siRNA transfection (B) or in the cells transfected with indicated siRNA (C). HeLa tet-off cells were transfected with a pGL4.13 luciferase (Luc) expression vector and indicated pTET-BBB rabbit beta globin reporter vectors. Doxycycline was then added to the medium to stop transcription from the tet-responsive promoter, and total RNA was collected at 0, 2, 4, and 6 h. qRT-PCR was performed to monitor Luc and beta globin levels and determine the relative RNA remaining. For each point, the beta globin mRNA level was normalized to the Luc RNA levels. Values represent mean ± SD from triplicate experiments. (D) RNA electrophoretic gel mobility shift assay was performed to determine the binding and release of UPF1 from the GADD45B 3′ UTR RNA (WT) or its adenine substitution mutant RNA (MUT). Five-tenths picomoles 32P-labeled RNAs were incubated with 500 ng UPF1 (lanes 3–8) or without UPF1 (lanes 1,2) for 15 min at 10°C. In lanes 5–8, 1 pmol nonlabeled WT GADD45B 3′ UTR RNA was added to the reactions after initial incubation and then incubated for another 30 min with ATP (lanes 7,8) or without ATP (lanes 5,6). Reaction solutions were then electrophoresed in a 4.5% native PAGE gel. The positions of the RNA probe and the UPF1-RNA complex are indicated on the left. The bar plot indicates the quantification of relative RNA release rate from UPF1 protein (adenine substitution mutant/WT 3′ UTR). Values represent the mean ± SD from triplicate experiments (biological replicates).
Delayed dissociation of UPF1 from p-UPF1 binding motifs
It was proposed that UPF1 target specificity is achieved by preferential release of UPF1 from nontargets (Lee et al. 2015). We hypothesized that the preferential release of UPF1 from nontargets over UPF1 targets harboring GC-rich sequences results in UPF1 stalling on its target mRNAs. If this is the case, we would observe delayed dissociation of UPF1 from RNAs containing p-UPF1 binding motifs. To test this hypothesis, we compared the dissociation of UPF1 from the wild-type (WT) GADD45B 3′ UTR with that from the adenine substitution mutant of the GADD45B 3′ UTR. A gel mobility shift assay showed that UPF1 was indiscriminately bound to 32P-labeled GADD45B 3′ UTR (WT) and its adenine substitution mutant (MUT) (Fig. 5D, lanes 3,4). After incubation of the 32P-RNA:UPF1 complex in the presence of cold WT RNA, dissociation of UPF1 from WT was faster than that from MUT (Fig. 5D, lanes 5,6, left panel). In addition, this tendency was enhanced by the addition of ATP (Fig. 5D, lanes 7,8, left panel). These results indicated the delayed dissociation of UPF1 from GC-rich motifs.
Discussion
An effective method to determine UPF1 targets
The mRNAs for transcription factors have comparatively shorter RNA half-lives (Maekawa et al. 2015), suggesting that the expression of transcription factors is influenced by post-transcriptional regulation. Consistent with this, transcription factors were enriched among the UPF1 targets in this study (Supplemental Table S5). This result suggested that several genes regulated by these transcription factors may be also up-regulated indirectly by an increased transcription rate, rather than by repression of mRNA decay after depletion of UPF1. We think that the indirect effect complicates the determination of UPF1 targets. In a previous study, Kurosaki et al. (2014) noted that the putative UPF1 targets identified by screening up-regulated mRNAs upon UPF1 depletion (Viegas et al. 2007) were not enriched for binding to p-UPF1. This is because these putative UPF1 targets included many indirect targets that were up-regulated by transcription factors whose mRNAs are regulated by UPF1. In contrast, the UPF1 targets identified in this study were clearly enriched for binding to p-UPF1 (Fig. 2A). Therefore, our combinatorial strategy based on BRIC-seq and RIP-seq is effective for identification of bona-fide UPF1 targets (Fig. 1A,B). Moreover, cell-cycle regulating genes were enriched among the UPF1 targets identified in this study (Supplemental Table S5), which was consistent with the function of UPF1 in cell-cycle regulation (Azzalin and Lingner 2006). This result also showed that we correctly identified UPF1 targets associated with the biological function of the UPF1 protein.
Context features in the 3′ UTRs of UPF1 targets
We first described the context features for UPF1 targets, based on the analysis of p-UPF1 binding sites in UPF1 targets using genome-wide RNA footprint data for p-UPF1. The p-UPF1 binding locations were more frequently detected in the 3′ UTRs of UPF1 targets compared with the other mRNAs (Fig. 2B,C). We predicted p-UPF1 binding motifs including the CUG sequence in the 3′ UTRs of UPF1 targets (Fig. 2E). Moreover, rabbit beta globin reporter analysis supported the idea that p-UPF1 binding motifs are responsible for UPF1-mediated mRNA degradation (Figs. 4, 5). We tested whether p-UPF1 binding motifs were sufficient to trigger UPF1-mediated decay by introducing these motifs in the 3′ UTRs of non-UPF1 targets; however, insertion of p-UPF1 binding motifs did not destabilize non-UPF1 targets (Supplemental Fig. S11). This result suggested that the presence of p-UPF1 binding motifs are necessary but not sufficient to trigger accelerated mRNA decay. Therefore, we investigated whether additional factors were present to accelerate UPF1-mediated mRNA decay. As a result, we discovered high GC-contents in the 3′ UTRs of UPF1 targets, which was consistent with a previous report (Hurt et al. 2013). In addition, sequence preference analysis showed that the 3′ UTRs of UPF1 targets contain significant G contents and several dinucleotide (GA and AG) contents (Supplemental Fig. S8). This suggested that a G-rich environment around the p-UPF1 binding motifs in the 3′ UTR is also important to trigger UPF1-mediated mRNA decay.
Model for the mechanism of UPF1-dependent mRNA decay via the 3′ UTR
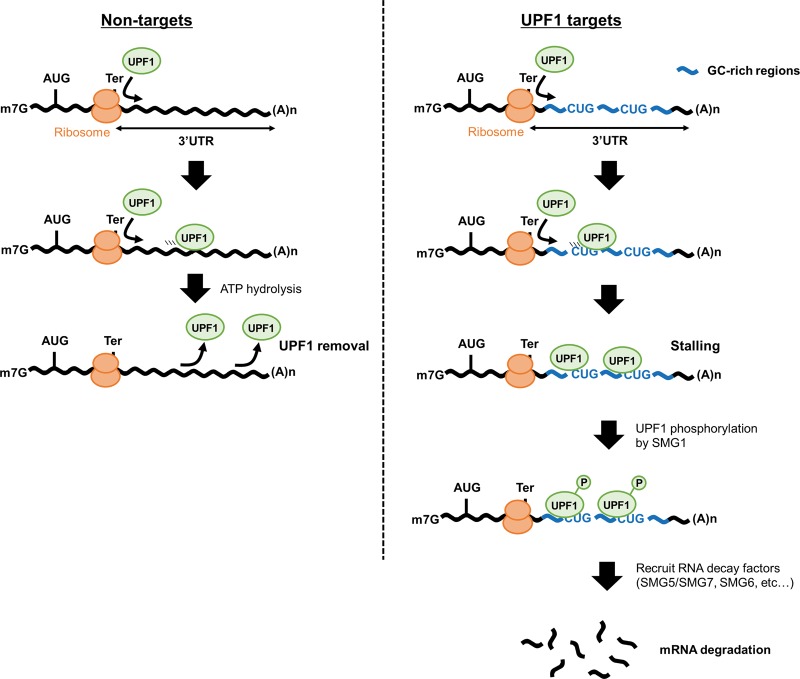
Single-molecule analysis for the UPF1 protein revealed that UPF1 effectively translocates along single-stranded RNA (Fiorini et al. 2015). The ATPase and helicase activities of UPF1 play an important role in the dissociation of UPF1 from RNAs (Franks et al. 2010), and this ATPase activity becomes weakened by association with poly(rG) and poly(rG:rC) (Bhattacharya et al. 2000). Recent studies also suggested that a reduction in the ATPase activity of UPF1 causes UPF1 accumulation on RNAs (Lee et al. 2015). These results suggest that UPF1 translocation and dissociation from RNA is slower over GC-rich sequences and UPF1 is retained over the GC-rich sequence. Corresponding to this idea, the dissociation of UPF1 from the wild-type GADD45B 3′ UTR was slower than that from the substitution mutant GADD45B 3′ UTR lacking p-UPF1 binding motifs (site 1–4) (Fig. 5D). The stalled UPF1 on mRNA is progressively phosphorylated, and the phosphorylation of UPF1 activates its mRNA decay (Kurosaki et al. 2014; Durand et al. 2016). Recent studies revealed that UPF1 accumulation on the 3′ UTR occurs in a translation-dependent manner (Hurt et al. 2013; Zünd et al. 2013; Gregersen et al. 2014; Kurosaki et al. 2014). Moreover, UPF1-associated mRNAs were up-regulated following translation inhibition (Hurt et al. 2013). In agreement with previous work, UPF1-mediated mRNA decay via the 3′ UTR is dependent on the translation process (Fig. 4E). Taken together, the following model is proposed (Fig. 6): When UPF1 is moving along the mRNA, its ATPase activity becomes weakened at high GC-content regions, causing the UPF1 protein to stall at GC-rich sequences in the 3′ UTRs of UPF1 targets. UPF1 stalling at RNAs increases the chance of phosphorylation via SMG1, which results in promotion of RNA degradation by recruiting RNA degradation factors, such as SMG5/SMG7 or SMG6. On the other hand, UPF1 is easily released from the 3′ UTR without GC-rich sequences (Fig. 5D), even when UPF1 is loaded onto the 3′ UTR; therefore, mRNAs lacking a GC-rich sequence apparently tolerate UPF1 (Fig. 5B).
Figure 6.
Model for the mechanism of UPF1-dependent mRNA decay. UPF1 protein is loaded onto target mRNAs via translation and then translocates over the 3′ UTR. In non-GC-rich regions (left), UPF1 is removed from mRNAs. In contrast, in GC-rich regions (right), UPF1 stalls at CUG-centered GC-rich motifs, which promotes mRNA decay.
In conclusion, our study provides bona fide UPF1 targets and context features in 3′ UTR of UPF1 targets for a better understanding of the mechanism of UPF1-dependent mRNA decay. Furthermore, our study highlights the practical importance of direct measurement of RNA stability to identify RNA targets of RNA decay factors such as UPF1.
Methods
RNA analysis
RIP-seq, qRT-PCR, mRNA decay assay, and RNA electrophoretic mobility shift assay protocols are detailed in the Supplemental Material, Supplemental Table S1.
5′-bromo-uridine immunoprecipitation chase-deep sequencing (BRIC-seq) analysis
BRIC-seq data for UPF1 or STAU1 knockdown were downloaded from the DDBJ Sequence Read Archive (DRA) under accession numbers DRA000591 and DRA001215, respectively. BRIC-seq data sets were processed, and the half-lives of transcripts derived from each gene were calculated as previously described (Imamachi et al. 2014).
As described by a previous study (Trcek et al. 2013), the PTC-containing mRNAs that escaped from NMD were degraded with half-lives similar to those of normal PTC-free mRNAs. As a result, biphasic decay of PTC-containing mRNAs was observed. Therefore, we assumed that the “RNA half-life” in our study meant the half-life derived from the first phase of the biphasic decay, because in almost all cases, the inflection point between the first phase and the second phase comes at a later time from the time point of 50% RNA remaining (compared with 0 h).
Data analysis of phosphorylated UPF1-binding sites
RNA footprint data for p-UPF1, HITS-CLIP, and eCLIP data for UPF1 were downloaded from the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE60045, GSE69586, and GSE80039, respectively. Adapter sequences were removed, and low-quality reads were discarded. The filtered reads were mapped to the reference human genome (hg19) using Bowtie 2 (version 2.2.4) (Langmead and Salzberg 2012) in local mode (--local). The abundance of RefSeq transcripts was measured using RPKM with Cufflinks. We also detected the peaks of p-UPF1-binding sites using Pyicoclip (version 2.0.6b) (Althammer et al. 2011). The artifact peaks, which resembled blocks resulting from duplicates, were discarded from the detected peaks. To predict p-UPF1-binding motifs, p-UPF1 peaks within the 3′ UTRs of 246 UPF1 targets were selected, and these sequences were used for MEME analysis (Bailey et al. 2006). The 3′ UTR sequences of all 8426 transcripts with one or more RPKM were used as the background model for MEME. 3′ UTRs defined in the RefSeq annotation data were used for the aforementioned analysis. To estimate the distribution of sequence reads derived from p-UPF1-binding sites, the reads mapping to 5′ UTRs, CDSs, and 3′ UTRs were counted using htseq-count (Anders et al. 2015), and the GTF file of each region was derived from RefSeq annotation data, respectively. Computational analyses were performed using custom scripts in Perl, Python, and R (R Core Team 2016).
Data access
High throughput sequencing data generated in this study have been submitted to the DDBJ Sequence Read Archive (DRA; http://trace.ddbj.nig.ac.jp/dra/) under accession number DRA003675. Source code for the data analysis is available from Supplemental Custom Scripts and at https://github.com/Imamachi-n/UPF1_study_script_collection. All lists of GC-content, motifs in 3′ UTRs, high-throughput sequencing data, and repetitive elements for all mRNAs are listed in Supplemental Table S3.
Supplementary Material
Acknowledgments
This work was financially supported by MEXT KAKENHI (Grant Numbers 221S0002 and 21115001), the Funding Program for World-Leading Innovative R&D on Science and Technology of the Japan Society for the Promotion of Science and Research Fellowship of the Japan Society for the Promotion of Science.
Author contributions: N.I. and N.A. conceived the project. N.I. performed most of the experiments and bioinformatic analysis. K.A.S. performed the RIP experiments. Y.S. performed the sequence analyses and initial bioinformatics. N.I., Y.S., and N.A. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.206060.116.
References
- Althammer S, González-Vallinas J, Ballaré C, Beato M, Eyras E. 2011. Pyicos: a versatile toolkit for the analysis of high-throughput sequencing data. Bioinformatics 27: 3333–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzalin CM, Lingner J. 2006. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol 16: 433–439. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. 2006. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34: W369–W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopal V, Fluch L, Nissan T. 2012. Ways and means of eukaryotic mRNA decay. Biochim Biophys Acta 1819: 593–603. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. 2000. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6: 1226–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culbertson MR, Leeds PF. 2003. Looking at mRNA decay pathways through the window of molecular evolution. Curr Opin Genet Dev 13: 207–214. [DOI] [PubMed] [Google Scholar]
- Durand S, Franks TM, Lykke-Andersen J. 2016. Hyperphosphorylation amplifies UPF1 activity to resolve stalls in nonsense-mediated mRNA decay. Nat Commun 7: 12434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle AB, Lykke-Andersen S, Mühlemann O, Jensen TH. 2009. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol 16: 49–55. [DOI] [PubMed] [Google Scholar]
- Feschotte C. 2008. Transposable elements and the evolution of regulatory networks. Nat Rev Genet 9: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorini F, Bagchi D, Le Hir H, Croquette V. 2015. Human Upf1 is a highly processive RNA helicase and translocase with RNP remodelling activities. Nat Commun 6: 7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong JH, Murphy TD, Pruitt KD. 2013. Comparison of RefSeq protein-coding regions in human and vertebrate genomes. BMC Genomics 14: 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TM, Singh G, Lykke-Andersen J. 2010. Upf1 ATPase-dependent mRNP disassembly is required for completion of nonsense-mediated mRNA decay. Cell 143: 938–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Maquat LE. 2011. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 470: 284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong C, Tang Y, Maquat LE. 2013. mRNA–mRNA duplexes that autoelicit Staufen1-mediated mRNA decay. Nat Struct Mol Biol 20: 1214–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregersen LH, Schueler M, Munschauer M, Mastrobuoni G, Chen W, Kempa S, Dieterich C, Landthaler M. 2014. MOV10 is a 5′ to 3′ RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3′ UTRs. Mol Cell 54: 573–585. [DOI] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. 2012. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22: 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Kashima I, Fauser M, Saulière J, Izaurralde E. 2008. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA 14: 2609–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt JA, Robertson AD, Burge CB. 2013. Global analyses of UPF1 binding and function reveal expanded scope of nonsense-mediated mRNA decay. Genome Res 23: 1636–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamachi N, Tani H, Mizutani R, Imamura K, Irie T, Suzuki Y, Akimitsu N. 2014. BRIC-seq: a genome-wide approach for determining RNA stability in mammalian cells. Methods 67: 55–63. [DOI] [PubMed] [Google Scholar]
- Isken O, Kim YK, Hosoda N, Mayeur GL, Hershey JW, Maquat LE. 2008. Upf1 phosphorylation triggers translational repression during nonsense-mediated mRNA decay. Cell 133: 314–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. 2006. Binding of a novel SMG-1–Upf1–eRF1–eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev 20: 355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Furic L, Desgroseillers L, Maquat LE. 2005. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120: 195–208. [DOI] [PubMed] [Google Scholar]
- Kurosaki T, Maquat LE. 2016. Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci 129: 461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Li W, Hoque M, Popp MW, Ermolenko DN, Tian B, Maquat LE. 2014. A post-translational regulatory switch on UPF1 controls targeted mRNA degradation. Genes Dev 28: 1900–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Pratt GA, Martinez FJ, Yeo GW, Lykke-Andersen J. 2015. Target discrimination in nonsense-mediated mRNA decay requires Upf1 ATPase activity. Mol Cell 59: 413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S, Jensen TH. 2015. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol 16: 665–677. [DOI] [PubMed] [Google Scholar]
- Maekawa S, Imamachi N, Irie T, Tani H, Matsumoto K, Mizutani R, Imamura K, Kakeda M, Yada T, Sugano S, et al. 2015. Analysis of RNA decay factor mediated RNA stability contributions on RNA abundance. BMC Genomics 16: 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwain DR, Pan Q, Reilly PT, Elia AJ, McCracken S, Wakeham AC, Itie-Youten A, Blencowe BJ, Mak TW. 2010. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc Natl Acad Sci 107: 12186–12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. 2001. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet 10: 99–105. [DOI] [PubMed] [Google Scholar]
- Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. 2004. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet 36: 1073–1078. [DOI] [PubMed] [Google Scholar]
- Morris AR, Mukherjee N, Keene JD. 2008. Ribonomic analysis of human Pum1 reveals cis-trans conservation across species despite evolution of diverse mRNA target sets. Mol Cell Biol 28: 4093–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2016. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
- Schoenberg DR, Maquat LE. 2012. Regulation of cytoplasmic mRNA decay. Nat Rev Genet 13: 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweingruber C, Rufener SC, Zünd D, Yamashita A, Mühlemann O. 2013. Nonsense-mediated mRNA decay—mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta 1829: 612–623. [DOI] [PubMed] [Google Scholar]
- Spasic M, Friedel CC, Schott J, Kreth J, Leppek K, Hofmann S, Ozgur S, Stoecklin G. 2011. Genome-wide assessment of AU-rich elements by the AREScore algorithm. PLoS Genet 8: e1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, Vigilante A, Darbo E, Zirra A, Militti C, D'Ambrogio A, Luscombe NM, Ule J. 2015. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature 519: 491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Mizutani R, Salam KA, Tano K, Ijiri K, Wakamatsu A, Isogai T, Suzuki Y, Akimitsu N. 2012a. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res 22: 947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H, Imamachi N, Salam KA, Mizutani R, Ijiri K, Irie T, Yada T, Suzuki Y, Akimitsu N. 2012b. Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biol 9: 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MS, LaCava J, Mita P, Molloy KR, Huang CR, Li D, Adney EM, Jiang H, Burns KH, Chait BT, et al. 2013. Affinity proteomics reveals human host factors implicated in discrete stages of LINE-1 retrotransposition. Cell 155: 1034–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trcek T, Sato H, Singer RH, Maquat LE. 2013. Temporal and spatial characterization of nonsense-mediated mRNA decay. Genes Dev 27: 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L, Izaurralde E. 2004. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell 16: 587–596. [DOI] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. 2016. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13: 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas MH, Gehring NH, Breit S, Hentze MW, Kulozik AE. 2007. The abundance of RNPS1, a protein component of the exon junction complex, can determine the variability in efficiency of the Nonsense Mediated Decay pathway. Nucleic Acids Res 35: 4542–4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Brewer G. 2012. The regulation of mRNA stability in mammalian cells: 2.0. Gene 500: 10–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zünd D, Gruber AR, Zavolan M, Mühlemann O. 2013. Translation-dependent displacement of UPF1 from coding sequences causes its enrichment in 3′ UTRs. Nat Struct Mol Biol 20: 936–943. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.