Abstract
RNA-guided genome surgery using CRISPR-Cas9 nucleases has shown promise for the treatment of diverse genetic diseases. Yet, the potential of such nucleases for therapeutic applications in nongenetic diseases is largely unexplored. Here, we focus on age-related macular degeneration (AMD), a leading cause of blindness in adults, which is associated with retinal overexpression of, rather than mutations in, the VEGFA gene. Subretinal injection of preassembled, Vegfa gene–specific Cas9 ribonucleoproteins (RNPs) into the adult mouse eye gave rise to mutagenesis at the target site in the retinal pigment epithelium. Furthermore, Cas9 RNPs effectively reduced the area of laser-induced choroidal neovascularization (CNV) in a mouse model of AMD. Genome-wide profiling of Cas9 off-target effects via Digenome-seq showed that off-target mutations were rarely induced in the human genome. Because Cas9 RNPs can function immediately after in vivo delivery and are rapidly degraded by endogenous proteases, their activities are unlikely to be hampered by antibody- and cell-mediated adaptive immune systems. Our results demonstrate that in vivo genome editing with Cas9 RNPs has the potential for the local treatment for nongenetic degenerative diseases, expanding the scope of RNA-guided genome surgery to a new dimension.
Age-related macular degeneration (AMD) is the leading cause of blindness in aged populations in developed countries (Jager et al. 2008). Choroidal neovascularization (CNV) is a major pathologic feature of neovascular AMD and is caused primarily by angiogenic cytokines such as vascular endothelial growth factor A (VEGFA). In fact, VEGFA is a major therapeutic target for the treatment of AMD using monoclonal antibodies or aptamers since it emerged as an important factor in angiogenesis (Leung et al. 1989). Currently, intravitreous anti-VEGF therapy is a mainstay of treatment for neovascular AMD (CATT Research Group et al. 2011; Schmidt-Erfurth et al. 2014). However, these anti-VEGF agents must be injected at least seven times per year, because VEGF is continuously overexpressed in and secreted from diseased retinal pigment epithelium (RPE) cells. In this clinical circumstance, we reasoned that targeted inactivation of the VEGFA gene in RPE cells could reduce the VEGF level below a pathological threshold, leading to a long-term or permanent treatment of AMD, possibly in combination with current anti-VEGF therapy.
The type II CRISPR-Cas9 systems, repurposed from prokaryotic adaptive immune responses, are now widely used for targeted genome modifications in plants, animals, and human cells (Kim et al. 2014; Woo et al. 2015; Zuris et al. 2015). In particular, Cas9 nucleases have shown promise for gene and cell therapy (Maeder and Gersbach 2016). Typically, these nucleases are expressed or delivered in vivo using plasmid DNA or viruses (Yin et al. 2014; Ran et al. 2015). However, plasmid DNA delivery is often inefficient, especially in vivo, and can cause integration of small plasmid fragments degraded by endogenous nucleases at on-target and off-target sites in the genome (Kim et al. 2014). Viral delivery of Cas9 can be highly efficient in vivo (Ran et al. 2015; Long et al. 2016; Nelson et al. 2016; Tabebordbar et al. 2016), but may be hampered by antibodies or T cells induced against the protein (Shankar et al. 2007; Calcedo et al. 2015; Chew et al. 2016). We and others have shown that preassembled Cas9 ribonucleoproteins (RNPs) can be delivered to human primary and stem cells and mice to modify target genes (Kim et al. 2014; Schumann et al. 2015; Zuris et al. 2015). Cas9 RNPs are rapidly turned over in cells, reducing off-target effects. Furthermore, Cas9 RNPs are unlikely to be limited by host immune systems because they function and disappear before the generation of antibodies and T cells directed against them. Currently, despite these advantages of RNPs, the difficult delivery of Cas9 RNPs in vivo limits its utility for therapeutic applications (Zuris et al. 2015). Here, we show that in vivo genome editing of an wild-type gene, whose up-regulation is responsible for pathogenesis, could be a new therapeutic modality for the treatment of nongenetic degenerative diseases. Our ultimate goal is to harness Cas9 RNPs for a clinical application of therapeutic genome surgery in patients with AMD.
Results
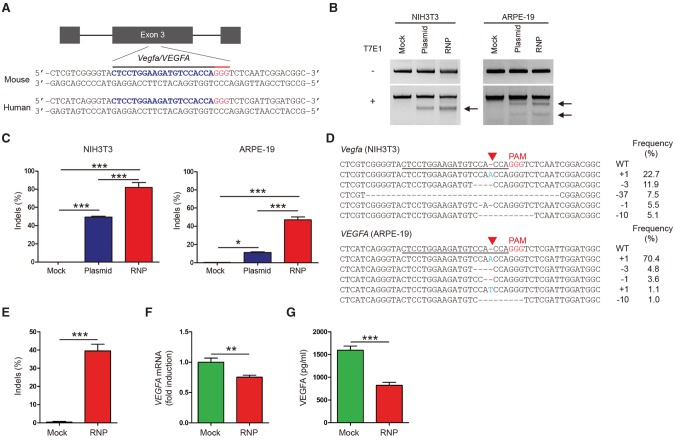
To investigate the potential of Cas9 RNP-mediated in vivo genome surgery for the treatment of AMD, we first identified Cas9 target sites that are conserved in both the human VEGFA gene and the mouse Vegfa gene using Cas-Designer (Park et al. 2015a) and that differ from any other site in the human genome by at least two or three nucleotides using Cas-OFFinder (Bae et al. 2014). We tested four single-chain guide RNAs (sgRNAs) targeting these sites in exons 3 and 4 (labeled as Vegfa-1, 2, 3, and 4), which encode binding sites for VEGF receptors 1 and 2, respectively, in the mouse NIH3T3 cell line (Supplemental Table S1; Supplemental Fig. S1) and the human RPE cell line (ARPE-19). The most active Vegfa-1 sgRNA complexed with the recombinant Cas9 protein and delivered via transfection using cationic lipid induced small insertions and deletions (indels) at the target site with a frequency of 82 ± 5% or 57 ± 3% in NIH3T3 cells or ARPE-19 cells, respectively (Fig. 1A–D). At day 2 post-treatment, Cas9 RNP delivery was much more efficient than plasmid transfection in these cells (Fig. 1C). We also carried out additional experiments using confluent ARPE-19 cells. Cells were harvested at 64 h after transfection and were analyzed using deep sequencing, qPCR, and ELISA to measure indel frequencies, mRNA levels, and protein levels, respectively. Indels were detected at a frequency of 40 ± 8% in the Vegfa-RNP-treated ARPE-19 cells (Fig. 1E). The Cas9 RNP reduced the VEGFA mRNA level by 24 ± 4% and the VEGFA protein level by 52 ± 9% in confluent ARPE-19 cells under post-mitotic conditions (Fig. 1F,G).
Figure 1.
Targeted mutagenesis in the Vegfa/VEGFA gene via Cas9 ribonucleoproteins (RNPs). (A) The target sequence in the Vegfa/VEGFA locus. The PAM sequence and the sgRNA target sequence are shown in red and blue, respectively. (B) Vegfa-specific Cas9 RNP-driven mutations in NIH3T3 and ARPE-19 cells detected by the T7 endonuclease I (T7E1) assay. Arrows indicate the expected positions of DNA bands cleaved by T7E1. (C) Mutation frequencies measured by targeted deep sequencing. Error bars indicate SEM (n = 3). One-way ANOVA and Tukey post-hoc tests: (*) P < 0.05; (***) P < 0.001. (D) Representative mutant DNA sequences at the Vegfa/VEGFA locus. The PAM sequence is shown in red, and inserted nucleotides are shown in blue. The target sequence is underlined. The red triangle indicates the cleavage position. The column on the right indicates the number of inserted or deleted bases and indel frequencies (%). (WT) wild type. (E) Vegfa-specific Cas9 RNP-driven mutation frequencies in confluent ARPE-19 cells at 64 h post-transfection detected by targeted deep sequencing. (F) Relative VEGFA mRNA levels measured by quantitative PCR (qPCR). (G) Secreted VEGFA protein in the supernatant measured by ELISA. Error bars indicate SEM (n = 5). Student's t-test: (**) P < 0.01; (***) P < 0.001.
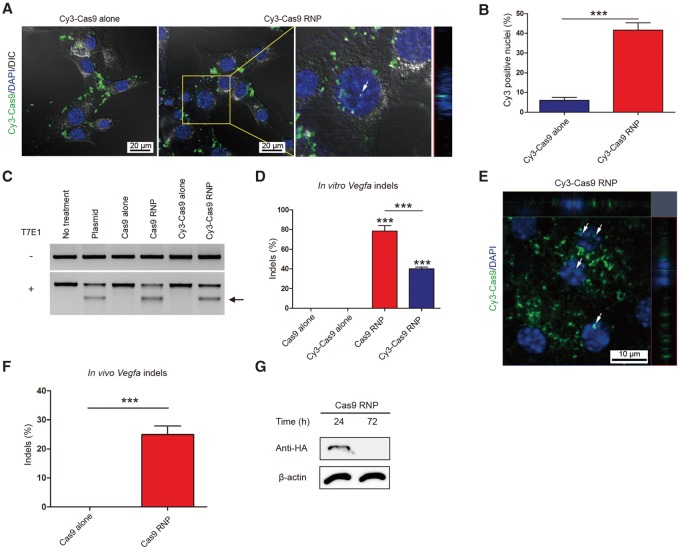
To monitor the localization of Cas9 RNPs in vitro and in vivo, we used Cy3-conjugated Cas9 protein (Deng et al. 2015). Thus, Cy3-Cas9 combined with or without the Vegfa-1 sgRNA was mixed with cationic lipids and delivered into NIH3T3 cells. The Cy3-Cas9 RNP was detected in many nuclei (Fig. 2A,B) and induced indels at the target site (Fig. 2C,D). Interestingly, the proportion of Cy3 positive nuclei (42 ± 6%) (Fig. 2B) was almost equal to the frequency of indels (40 ± 3%) (Fig. 2D) at the target site, suggesting that target sites were almost completely cleaved in cells by nucleus-localized Cas9, and the rate-limiting factor in genome editing was nuclear localization of Cas9. Cy3-Cas9 alone was rarely detected in nuclei and did not induce indels (Fig. 2A,D). Note that Cas9 is a positively charged protein with a pI value of 9.12 and cannot form a complex with cationic lipids in the absence of negatively charged sgRNAs. The Cy3-Cas9 RNP, however, was less active than the unlabeled Cas9 RNP, which induced target-specific mutations at a frequency of 80%.
Figure 2.
In vitro and in vivo delivery of Cy3-labeled Cas9 RNP. (A) Localization of Cy3 dye in NIH3T3 cells transfected with Cy3-labeled Cas9 RNP or Cy3-labeled Cas9 alone (as a control) at 24 h post-transfection. White arrow indicates nuclear colocalization of Cy3 dye. The z-axis image on the right shows that Cy3-Cas9 is localized inside the nucleus. (B) Proportion of Cy3 positive nuclei in total DAPI positive nuclei at 24 h post-transfection. Error bars indicate SEM (n = 3). Student's t-test: (***) P < 0.001. (C) Vegfa-specific Cas9 RNP-mediated mutations in NIH3T3 cells detected by the T7E1 assay. The arrow indicates the expected position of DNA bands cleaved by T7E1. (D) Mutation frequencies were measured using targeted deep sequencing. Error bars are SEM (n = 3). One-way ANOVA and Tukey post-hoc tests: (***) P < 0.001. (E) Representative RPE flat-mount at day 3 post-injection of Cy3-labeled Cas9 RNP into mouse eye. White arrows indicate nuclear colocalization of Cy3 dye. (F) Frequencies of indels induced in vivo determined using genomic DNA isolated from the retinal pigment epithelium (RPE). Indels were analyzed by deep sequencing at day 3 post-injection. Error bars are SEM (n = 5). Student's t-test: (***) P < 0.001. (G) Representative Western blot analysis to measure the level of Cas9 protein in the RPE/choroid/scleral complex 24 and 72 h after injection (n = 4).
Next, we delivered the Vegfa-specific, Cy3-labeled or -unlabeled Cas9 RNP into the adult mouse eye via subretinal injection (Park et al. 2015c). Cy3 dye was observed in the nuclei of the RPE in vivo 3 d after the injection (Fig. 2E). Notably, the subretinal injection of the Cy3-unlabeled Cas9 RNP gave rise to indels with a frequency of 25 ± 3% in RPE cells in the RNP-injected area at day 3 post-injection (n = 5) (Fig. 2F; Supplemental Fig. S2A). Subretinal injection of RNPs did not induce detectable on-target indels in the neural retinal tissue or in the remnant RPE-choroid-sclera complex after RPE removal. Thus, in our experiments, subretinal injection of RNPs can induce indels in RPE cells in the injected area. We also performed Western blot analysis and found that the Cas9 protein was degraded completely at day 3 post-injection (Fig. 2G), showing that Cas9 was rapidly turned over in vivo. Considering that Cas9 expression in vivo using adeno-associated virus (AAV) can evoke host immune responses with distinct cellular and molecular signatures (Chew et al. 2016), rapid degradation of Cas9 after RNP delivery has an advantage for in vivo application.
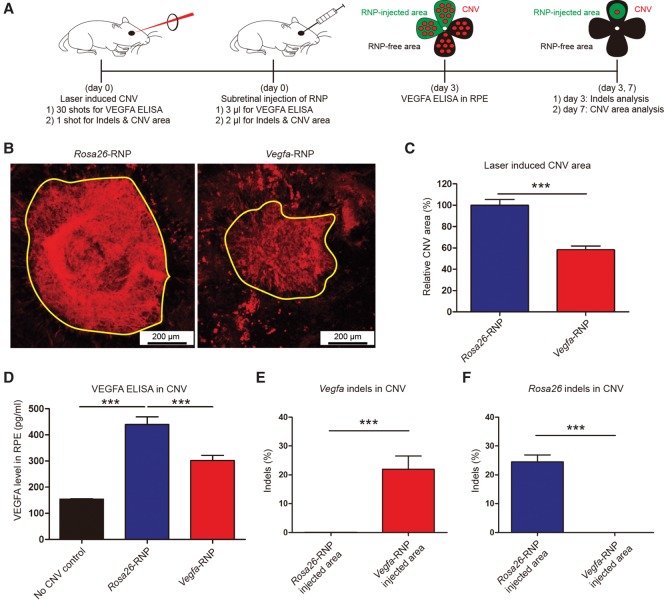
Encouraged by the efficient delivery and high mutation frequency of the Vegfa-specific Cas9 RNP in vitro and in vivo, we investigated whether the Cas9 RNP could be used for the treatment of CNV in a mouse model of AMD (Lambert et al. 2013). Mice with laser-induced CNV were treated by subretinal injection of the Vegfa-specific Cas9 RNP or Rosa26-specific Cas9 RNP (Fig. 3A). The Rosa26-RNP was used as a negative control. At day 3 post-injection, indels were analyzed in RPE cells in the Cas9 RNP-injected CNV area. Cas9-induced indels were detected at a frequency of 22 ± 5% or 24 ± 2% in the Vegfa-RNP-treated or the Rosa26-RNP-treated CNV (n = 5), respectively (Fig. 3E,F; Supplemental Fig. S2B), demonstrating that subretinal injection of the Vegfa-RNP can lead to local treatment in the eye. In addition, the Vegfa-RNP effectively reduced the concentration of the VEGFA protein in the CNV area (300 ± 20 pg/mL, n = 10), compared to the Rosa26-RNP (440 ± 30 pg/mL, n = 10, P < 0.001, One-way ANOVA and Tukey post-hoc tests) (Fig. 3D). At day 7 post-injection, a therapeutic effect was evaluated by assessing the CNV area. In mice treated with the Vegfa-RNP, the CNV area was significantly reduced (58 ± 4%, n = 15, P < 0.001, Student's t-test), compared to that in Rosa26-RNP injected mice (Fig. 3B,C). Our results suggest that targeted inactivation of Vegfa in the RPE using Cas9 RNPs enables therapeutic genome surgery for the local treatment of AMD.
Figure 3.
Subretinal injection of Cas9 RNPs targeting Vegfa reduces the area of laser-induced choroidal neovascularization (CNV) in a mouse model of age-related macular degeneration (AMD). (A) Mice with laser-induced CNV were treated with subretinal injection of the Vegfa-specific preassembled Cas9 RNP (Vegfa-RNP). After the retinal pigment epithelium (RPE) complex in the eye was flat-mounted, the CNV area was analyzed at day 7 post-injection. Genomic DNA isolated from the Cas9 RNP-injected area or from the opposite noninjected area (RNP-free area) was analyzed by deep sequencing. VEGFA ELISA was performed at day 3 post-injection. (B) Representative laser-induced CNV stained with isolectin B4 (IB4) in C57BL/6J mice injected with the Rosa26-specific Cas9 RNP (as a control) or the Vegfa-RNP. The yellow line demarcates the area of CNV. (C) The CNV area. Error bars indicate SEM (n = 15). Student's t-test: (***) P < 0.001. (D) VEGFA level in CNV. Error bars indicate SEM (n = 10). One-way ANOVA and Tukey post-hoc tests: (***) P < 0.001. (E) Indel frequencies at the Vegfa target site in the RPE cells. Error bars indicate SEM (n = 5). Student's t-test: (***) P < 0.001. (F) Indel frequencies at the Rosa26 target site in the RPE cells. Error bars indicate SEM (n = 5). Student's t-test: (***) P < 0.001.
A critical issue in therapeutic genome surgery is the target specificity of CRISPR-Cas9 nucleases. We investigated whether the Vegfa-RNP used in this study caused any off-target mutations in the mouse eye or in human cells. First, using Cas-OFFinder (Bae et al. 2014), we identified 20 potential off-target sites in the mouse genome that are most highly homologous to the Cas9 target site (Supplemental Table S2). Genomic DNA was isolated from the CNV-free, RPE complex in the mouse eye treated with the Cas9 RNP and subjected to targeted deep sequencing. At these 20 sites, no Cas9-induced indels were detected with a frequency greater than 0.1%, demonstrating that no off-target mutations were induced above sequencing error rates (0.1%, on average) (Supplemental Fig. S3).
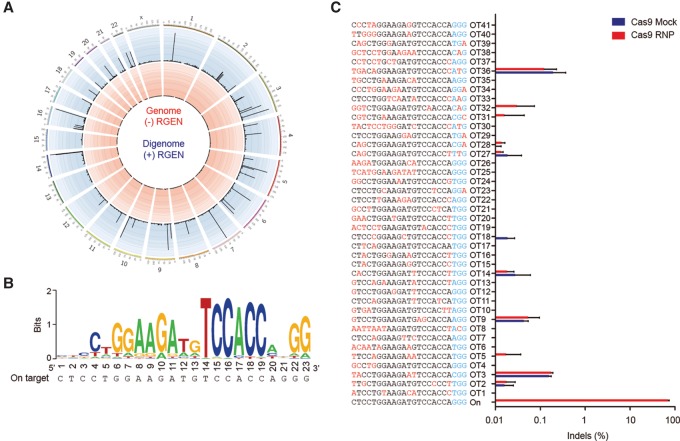
Next, we determined genome-wide off-target effects in the human genome rather than the mouse genome (Fig. 4A,B). Note that the target sequence of this particular Cas9 RNP is conserved in the human VEGFA gene. We assessed the human genome-wide specificity using Digenome-seq (Kim et al. 2015, 2016b), in which cell-free human genomic DNA is digested in vitro using the Vegfa-specific Cas9 RNP and then subjected to whole-genome sequencing. Uniform, rather than random, alignments of sequence reads at in vitro cleavage sites are computationally identified to provide a list of potential off-target sites. Digenome-seq using the Vegfa-RNP revealed 42 in vitro cleavage sites including the on-target site (Supplemental Table S3). To validate or invalidate these sites, we carried out targeted deep sequencing using genomic DNA isolated from Vegfa-RNP-transfected ARPE-19 cells. Although these sites were cleaved efficiently in vitro, off-target indels were not detected at these 41 cleavage sites above sequencing error rates (Fig. 4C). Use of Cpf1 (Zetsche et al. 2015; Kim et al. 2016a), Cas9 variants (Kleinstiver et al. 2016; Slaymaker et al. 2016), or modified gRNAs (Cho et al. 2014; Fu et al. 2014) with improved specificity may avoid this residual off-target effect, if necessary. Taken together, these results show that the Vegfa-RNP is highly specific in both mouse and human cells.
Figure 4.
Genome-wide target specificity of the Vegfa-specific Cas9 RNP revealed by Digenome-seq. (A) Genome-wide Circos plot (Krzywinski et al. 2009) showing in vitro cleavage sites. Human genomic DNA is shown in red, and RGEN-digested genomic DNA is shown in blue. (B) Sequence logo obtained using 42 Digenome-captured sites. (C) Off-target site validated in human ARPE-19 cells by targeted deep sequencing. The mismatched nucleotides are shown in red, and PAM sequences are shown in blue.
Another major concern for mutating the Vegfa gene for the treatment of AMD is its trophic role in the eye. Cone dysfunction is the most significant change and is observed 3 d after conditional deletion of the Vegfa gene in mouse RPE (Kurihara et al. 2012). Note, however, that the Vegfa gene was mutated locally at the site of the RNP injection in our study. We evaluated the integrity of cone opsin at day 7, rather than day 3, post-injection and found that no cone dysfunction had occurred (Supplemental Fig. S4).
Discussion
In this study, we showed that genome surgery using Cas9 RNPs has the potential for local, rather than systemic, treatment for nongenetic diseases such as AMD. Unlike previous studies involving RNA-guided genome editing in vivo to correct genetic defects, this report is focused on inactivating a disease-causing wild-type gene. This approach can broaden the scope of druggable targets from several protein families such as kinases, ion channels, and G protein-coupled receptors to any protein-coding or noncoding genes.
We believe that eye diseases are well-suited for in vivo genome editing using Cas9 RNPs. Because of its compartmented anatomy, subretinal injection is an optimized technique to deliver Cas9 RNPs into RPE cells for the local treatment. The laser-induced CNV model in mice that was used in this study is not suitable for multiple injection of Cas9 RNPs, because CNV formation peaks ∼2 wk after laser treatment and then slightly regressed over time. For multiple injection studies, large animal models such as nonhuman primates should be considered. In human or nonhuman primates, subretinal injection of RNPs in a volume of 100–150 µL would be enough to cover choroidal neovascularization. We envision that genome surgery is not just a metaphor but can be a graphic description of “operations” in the near future by which surgeons or physicians use Cas9 RNPs as microscalpels to cut and paste disease-causing genetic elements in patients.
Methods
Data reporting
No statistical methods were used to predetermine sample size for in vitro and in vivo experiments. The investigators were not blinded to allocation during in vivo experiments, but single-blinded to the outcome assessment, specifically measurement of the CNV area.
Preparation of Cas9 RNPs
Purified Cas9 protein was purchased from ToolGen, Inc. sgRNAs were generated by in vitro transcription using T7 polymerase (New England BioLabs) according to the manufacturer's protocol. Briefly, templates for sgRNA were generated by annealing and extension of two complementary oligonucleotides (Supplemental Table S4). Templates were incubated with T7 RNA polymerase in reaction buffer (40 mM Tris-HCl, 20mM MgCl2, 2 mM spermidine, 1 mM DTT, pH7.9), including NTPs (Jena Bioscience) and RNase inhibitor (New England BioLabs) for 16 h at 37°C. Transcribed sgRNAs were incubated with DNase I (New England BioLabs) for 30 min at 37°C. sgRNAs were purified using RNeasy MinElute Cleanup kit (Qiagen) and quantified using NanoDrop (Thermo Fisher Scientific). Purified sgRNAs (65 µg) were incubated with CIP (1000 units) (Alkaline Phosphatase, New England BioLabs) for the removal of 3-phosphate for 1 h at 37°C. sgRNAs were purified again using the RNeasy MinElute Cleanup kit (Qiagen) and quantified using NanoDrop (Thermo Fisher Scientific). We also tested cell viability and indel efficiency using all Cas9 protein and sgRNA stocks, and high efficiency Cas9 protein and sgRNA stocks were used for in vivo eye injections.
Cy3-labeled Cas9 protein purification
After transformation of the pET28-NLS-Cas9 vector into E. coli strain BL21 (DE3), Cas9 protein expression was induced for 12 h at 18°C with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). The bacterial cells were lysed by sonication; after centrifugation at 20,000g for 30 min, the soluble lysate was mixed with Ni-NTA beads (Qiagen), and Cy3 dye (GE Healthcare) was added at a 1:10 ratio (protein:dye molecules). The mixture was incubated overnight (>12 h) at 4°C in the dark. Cy3-labeled Cas9 was eluted with elution buffer (50 mM Tris-HCl [pH 7.6], 150–500 mM NaCl, 10%–25% glycerol, 0.2 M imidazole) and dialyzed against dialyzing buffer (20 mM HEPES pH 7.5, 150 mM KCl, 1 mM DTT, 10% glycerol). The purified Cy3-labeled Cas9 protein was concentrated using an Ultracel 100K cellulose column (Millipore). The purity of the Cy3-labeled Cas9 protein was determined by SDS-PAGE. The Cy3 labeling efficiency was measured by comparing the absorption spectra of the Cas9 protein (280 nm) and the conjugated Cy3 dye molecule.
Cell culture and transfection
NIH3T3 (ATCC CRL-1658) and ARPE-19 (ATCC CRL-2302) cell lines were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% BCS or FBS at 37°C in a humidified atmosphere containing 5% CO2. NIH3T3 and ARPE-19 cells were not authenticated or tested for mycoplasma contamination. One day before transfection, NIH3T3 and ARPE-19 cells were seeded into 24-well plates at 2 × 104 cells per well, with each well containing 250 µL growth medium lacking antibiotics. For plasmid delivery, cells were transfected with Cas9 (1 µg) and sgRNA (1 µg) expression plasmids using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer's protocol. For RNP delivery, Cas9 protein (4 µg) was incubated with sgRNA (2.25 µg) for 5 min at room temperature, after which 50 µL Opti-MEM (Thermo Fisher Scientific) and 1 µL Lipofectamine 2000 (Thermo Fisher Scientific) were added. After 10 min, the RNP mixture was added to cells in the 24-well plates described above. Cells were harvested 48 h after transfection and analyzed using the T7E1 assay, targeted deep sequencing, and qPCR. For VEGFA expression in confluent RPE cells, ARPE-19 cells were maintained in DMEM/F12 containing 1% FBS after reaching confluency to allow formation of a polarized epithelial layer for the experiments (Park et al. 2015b). ARPE-19 cells in 12-well plates were transfected with 8 µg of Cas9 protein, 4.5 µg of sgRNA, and 3 µL of lipofectamine 2000. Two days after transfection, the transfection growth medium (DMEM + 1% FBS) was gently replaced with 0.5 mL of fresh serum-free medium. After 16 h, cells and media were harvested and analyzed using targeted deep sequencing, qPCR, and ELISA.
Cy3-labeled Cas9 RNP imaging and counting
One day after transfection, cells were fixed in 4% PFA for 10 min at room temperature and then stained with 4′, 6-diamidino-2-phenylindole (DAPI, 1µg/mL; Sigma Aldrich) for 15 min at room temperature. Cells were visualized with a confocal microscope (LSM510, Carl Zeiss) at a magnification of 630×. The scanning parameters were as follows: scaling (x = 0.14 µm/pixel, y = 0.14 µm/pixel, z = 1 µm/pixel), dimensions (x = 1024, y = 1024, z = 6, channels: 3, 12-bit) with objective C-Apochromat 63×/1.20W Korr UV-VIS-IR. Cy3 positive nuclei were counted using ZEN 2 software (black edition, Ver 10.0, Carl Zeiss). To quantify the frequency of Cy3 positive nuclei, we counted the total number of cells and the number of cells with Cy3 staining in the nucleus in a field of view at a magnification of 630× and calculated the average percentage of Cy3 positive nuclei over four fields of view (n = 3).
T7E1 assay
Genomic DNA was isolated from cells and tissues using a DNeasy Tissue Kit (Qiagen) according to the manufacturer's protocol. After target sites were amplified using PCR, the products were denatured and annealed using a thermal cycler. A list of primers used can be found in Supplemental Table S5. Annealed PCR products were incubated with T7 endonuclease I (ToolGen, Inc.) for 25 min at 37°C and analyzed by agarose gel electrophoresis.
Targeted deep sequencing
On-target and potential off-target regions were amplified from genomic DNA using Phusion polymerase (Thermo Fisher Scientific). The PCR amplicons were subjected to paired-end sequencing using an Illumina MiSeq at LAS, Inc. A list of primers used can be found in Supplemental Tables S6, S7, and S8. Indels around the site 3 bp upstream of the PAM sequence were considered to be mutations resulting from Cas9 RNP activity.
RNA extraction and qPCR
Total RNA was isolated from NIH3T3 and ARPE-19 cells using an easy-spinTM Total RNA extraction Kit (iNtRON) according to the manufacturer's protocol. Two hundred fifty nanograms of RNA was then reverse transcribed using SuperScript II (Enzynomics). Quantitative PCR was performed using SYBR Green (KAPA) with the following primers: mouse Vegfa, 5′-ACGTCAGAGAGCAACATCAC-3′ (forward), 5′-CTGTCTTTCTTTGGTCTGCATTC-3′ (reverse); mouse Gapdh, 5′-GCTGAGTATGTCGTGGAGTCTA-3′ (forward), 5′-GTGGTTCACACCCATCACAA-3′ (reverse); human VEGFA-1, 5′-CGAGTACATCTTCAAGCCATCC-3′ (forward), 5′-GGTGAGGTTTGATCCGCATAAT-3′ (reverse); human VEGFA-2, 5′-AGAAGGAGGAGGGCAGAAT-3′ (forward), 5′-CACAGGATGGCTTGAAGATGTA-3′ (reverse); and human GAPDH, 5′-CAATGACCCCTTCATTGACC-3′ (forward), 5′-TTGATTTTGGAGGGATCTCG-3′ (reverse).
VEGFA ELISA using confluent ARPE-19 cells
For human VEGFA ELISA, serum-free supernatants were collected from Vegfa-specific Cas9 RNP-treated confluent ARPE-19 cell cultures after cells were incubated in serum-free medium for 16 h. Secreted VEGFA protein levels were measured using a human VEGF Quantikine ELISA Kit (DVE00, R&D systems) according to the manufacturer's instructions.
In vitro cleavage of genomic DNA and Digenome sequencing
Genomic DNA was isolated from ARPE-19 cells (ATCC) with a DNeasy Tissue Kit (Qiagen). In vitro cleavage of genomic DNA for Digenome sequencing was performed as described previously (Kim et al. 2015, 2016b). Briefly, genomic DNA (20 µg) was incubated with Cas9 protein (16.7 µg) and sgRNA (12.5 µg) in reaction buffer (100 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2, 100 µg/mL BAS, pH 7.9) for 3 h at 37°C. Cleaved genomic DNA was treated with RNase A (50 µg/mL, Sigma Aldrich) for 30 min at 37°C and purified with a DNeasy Tissue Kit (Qiagen). Whole-genome and Digenome sequencing were performed as described previously (Kim et al. 2016b).
Animals
The care, use, and treatment of all animals in this study were in strict agreement with the ARVO “Statement for the Use of Animals in Ophthalmic and Vision Research” and the guidelines established by the Seoul National University Institutional Animal Care and Use Committee. Adult (6 wk old) male SPF C57BL/6J mice were used in the study. Mice were maintained under a 12 h dark–light cycle.
Subretinal injections
Subretinal injection was performed as previously described (Park et al. 2015c). Briefly, RNPs composed of Cas9 protein (8 µg), sgRNA (4.5 µg), and Lipofectamine 2000 (20% v/v) were mixed in 2 µL of injection volume. RNPs (2 or 3 µL) were injected into the subretinal space using a Nanofil syringe with a 33G blunt needle (World Precision Instruments, Inc.) under an operating microscope (Leica Microsystems, Ltd.). Subjects with retinal hemorrhage were excluded from the study.
Laser-induced CNV model
Mice were anesthetized with an intraperitoneal injection of a mixture of tiletamine and zolazepam (1:1, 2.25 mg/kg body weight) and xylazine hydrochloride (0.7 mg/kg body weight). Pupils were dilated with an eye drop containing phenylephrine (0.5%) and tropicamide (0.5%). Laser photocoagulation was performed using an indirect head set delivery system (Iridex) and laser system (Ilooda). The laser wave length was 810 nm. Laser parameters were 200 µm spot size, 1W power, and 100 msec exposure time. Laser burn was induced at the 12 (right eye) or 6 (left eye) o'clock positions around the optic disc with a modification (Lambert et al. 2013). Only burns that produced a bubble without vitreous hemorrhage were included in the study. Subretinal RNP injections were performed in the quadrant of laser burn. RNPs (sgRosa26 or sgVegfa) were randomly allocated to the left or right eye in each mouse. Subretinal injection of Cas9 RNPs produced a bleb. We confirmed that the bleb overlapped with the laser-burn site. Subjects in which the bleb overlapped the laser-burn site were included in further studies. Seven days later, the eyes were fixed in 4% PFA for 1 h at room temperature. RPE complexes (RPE/choroid/sclera) were prepared for immunostaining with isolectin-B4 (Thermo Fisher Scientific, catalog no. I21413, 1:100) overnight at 4°C. The RPE complexes were flat-mounted and viewed with a fluorescent microscope (Eclipse 90i, Nikon) at a magnification of 40×. The CNV area was measured using ImageJ software (1.47v, NIH) by blind observers.
Immunofluorescent staining and imaging
The number of RPE cells in the RPE complex was calculated by counting DAPI stained nuclei in paraffin embedded 4-µm cross-section samples in a high power field area (100 µm × 100 µm, n = 8). Cross-section samples obtained at day 7 post-injection (n = 4) were immunostained with anti-opsin antibody (Millipore, AB5405, 1:1000) and Alexa Fluor 488 antibody (Thermo Fisher Scientific, 1:500). The opsin positive area was measured using ImageJ software (1.47v, NIH) by blind observers. The intracellular distribution of Cy3-Cas9 protein in the RPE flat-mounts was imaged using a confocal microscope (LSM 710, Carl Zeiss). The scanning parameters were as follows: scaling (x = 0.042 µm/pixel, y = 0.042 µm/pixel, z = 0.603 µm/pixel), dimensions (x = 1024, y = 1024, z = 12, channels: 2, 8-bit), and zoom (5.0) with objective C-Apochromat 40×/1.20W Korr M27. ZEN 2 software was used to process the images.
Isolation of RPE sheets and genomic DNA extraction
RPE cell sheets at the RNP delivered site were harvested at day 3 post-injection with or without CNV according to the previously described protocol (Fernandez-Godino et al. 2016). Briefly, enucleated eyes were incubated with 0.1% hyaluronidase (Sigma-Aldrich) for 45 min at 37°C after removal of lens, then transferred in PBS for 30 min on ice. The RNP-injected area was separated from the RNP-free area. Next, neural retina was completely removed from enucleated eyes, and only RPE/choroid/scleral complex was incubated with Trypsin-ETDA solution for 45 min at 37°C. RPE cell sheets were isolated by shaking the eyecup using microforceps, and only monolayer RPE sheets were collected using a glass capillary on the microscopy. Genomic DNA was directly extracted from the collected RPE sheets in the RNP-injected area and subjected to targeted deep sequencing. Briefly, the collected RPE sheets were incubated with lysis buffer (25 mM NaOH, 0.2 mM EDTA) for 20 min at 95°C and pH adjusted to 7.4 using HEPES (50 mM).
Mouse VEGFA ELISA
For mouse VEGFA ELISA, a total of 30 laser burns were induced in the eye, after which RNPs (3 µL) were injected into the subretinal space. At day 3 post-injection, whole RPE complexes were isolated from the retina and frozen for further analysis. Cells were lysed with RIPA buffer, and VEGFA levels were measured using a mouse VEGF Quantikine ELISA Kit (MMV00, R&D systems) according to the manufacturer's instructions.
Western blotting
To analyze RNP levels over time after in vivo RNP delivery, Western blotting of RPE complexes, obtained at 1 and 3 d post-injection, was performed. Samples containing an equal amount of protein (20 µg) were analyzed; Cas9 and beta-actin were detected with an anti-HA high affinity antibody (Roche, 3F10, 1:1000) and an anti-beta-actin antibody (Sigma Aldrich, A2066, 1:1000), respectively. ImageQuant LAS4000 (GE healthcare) was used for digital imaging.
Statistics
Data were analyzed with SPSS software version 18.0 (SPSS, Inc.). P-values were determined by an unpaired, two-sided Student's t-tests or one-way ANOVA and Tukey post-hoc tests for multiple groups. Data are shown as mean with SEM.
Data access
The whole-genome sequencing and deep sequencing data from this study have been submitted to the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession numbers SRX2487860, SRX1801343, and SRX1801344.
Competing interest statement
The authors declare competing financial interests: Details accompany the full-text HTML version of the paper at http://genome.cshlp.org/.
Supplementary Material
Acknowledgments
This work was supported by the Institute for Basic Science (IBS-R021-D1 to J.-S.K.), the Pioneer Research Program of the National Research Foundation of Korea (NRF)/MEST (2012-0009544 to Je.H.K.), the Bio & Medical Technology Development Program of the National Research Foundation, and MSIP (NRF-2015M3A9E6028949 to Je.H.K.). We thank Sunghyun Kim for Digenome-seq analysis.
Author contributions: K.K. performed in vitro experiments, and S.W.P. performed in vivo experiments. Je.H.K. and J.-S.K. supervised the research. All authors discussed the results and commented on the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.219089.116.
Freely available online through the Genome Research Open Access option.
References
- Bae S, Park J, Kim JS. 2014. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30: 1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcedo R, Franco J, Qin Q, Richardson DW, Mason JB, Boyd S, Wilson JM. 2015. Preexisting neutralizing antibodies to adeno-associated virus capsids in large animals other than monkeys may confound in vivo gene therapy studies. Hum Gene Ther Methods 26: 103–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CATT Research Group, Martin DF, Maguire MG, Ying GS, Grunwald JE, Fine SL, Jaffe GJ. 2011. Ranibizumab and bevacizumab for neovascular age-related macular degeneration. N Engl J Med 364: 1897–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew WL, Tabebordbar M, Cheng JK, Mali P, Wu EY, Ng AH, Zhu K, Wagers AJ, Church GM. 2016. A multifunctional AAV–CRISPR–Cas9 and its host response. Nat Methods 13: 868–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. 2014. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 24: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Shi X, Tjian R, Lionnet T, Singer RH. 2015. CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells. Proc Natl Acad Sci 112: 11870–11875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Godino R, Garland DL, Pierce EA. 2016. Isolation, culture and characterization of primary mouse RPE cells. Nat Protoc 11: 1206–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. 2014. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32: 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager RD, Mieler WF, Miller JW. 2008. Age-related macular degeneration. N Engl J Med 358: 2606–2617. [DOI] [PubMed] [Google Scholar]
- Kim S, Kim D, Cho SW, Kim J, Kim JS. 2014. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24: 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, Hwang J, Kim JI, Kim JS. 2015. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods 12: 237–243, 1 p following 243. [DOI] [PubMed] [Google Scholar]
- Kim D, Kim J, Hur JK, Been KW, Yoon SH, Kim JS. 2016a. Genome-wide analysis reveals specificities of Cpf1 endonucleases in human cells. Nat Biotechnol 34: 863–868. [DOI] [PubMed] [Google Scholar]
- Kim D, Kim S, Kim S, Park J, Kim JS. 2016b. Genome-wide target specificities of CRISPR-Cas9 nucleases revealed by multiplex Digenome-seq. Genome Res 26: 406–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, Joung JK. 2016. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529: 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res 19: 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara T, Westenskow PD, Bravo S, Aguilar E, Friedlander M. 2012. Targeted deletion of Vegfa in adult mice induces vision loss. J Clin Invest 122: 4213–4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert V, Lecomte J, Hansen S, Blacher S, Gonzalez ML, Struman I, Sounni NE, Rozet E, de Tullio P, Foidart JM, et al. 2013. Laser-induced choroidal neovascularization model to study age-related macular degeneration in mice. Nature Protoc 8: 2197–2211. [DOI] [PubMed] [Google Scholar]
- Leung DW, Cachianes G, Kuang WJ, Goeddel DV, Ferrara N. 1989. Vascular endothelial growth factor is a secreted angiogenic mitogen. Science 246: 1306–1309. [DOI] [PubMed] [Google Scholar]
- Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN. 2016. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 351: 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Gersbach CA. 2016. Genome-editing technologies for gene and cell therapy. Mol Ther 24: 430–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan WX, et al. 2016. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351: 403–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Bae S, Kim JS. 2015a. Cas-Designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics 31: 4014–4016. [DOI] [PubMed] [Google Scholar]
- Park SW, Kim JH, Park SM, Moon M, Lee KH, Park KH, Park WJ, Kim JH. 2015b. RAGE mediated intracellular Aβ uptake contributes to the breakdown of tight junction in retinal pigment epithelium. Oncotarget 6: 35263–35273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Kim JH, Park WJ, Kim JH. 2015c. Limbal approach-subretinal injection of viral vectors for gene therapy in mice retinal pigment epithelium. J Vis Exp: e53030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al. 2015. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520: 186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt-Erfurth U, Kaiser PK, Korobelnik JF, Brown DM, Chong V, Nguyen QD, Ho AC, Ogura Y, Simader C, Jaffe GJ, et al. 2014. Intravitreal aflibercept injection for neovascular age-related macular degeneration: ninety-six-week results of the VIEW studies. Ophthalmology 121: 193–201. [DOI] [PubMed] [Google Scholar]
- Schumann K, Lin S, Boyer E, Simeonov DR, Subramaniam M, Gate RE, Haliburton GE, Ye CJ, Bluestone JA, Doudna JA, et al. 2015. Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci 112: 10437–10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar G, Pendley C, Stein KE. 2007. A risk-based bioanalytical strategy for the assessment of antibody immune responses against biological drugs. Nat Biotechnol 25: 555–561. [DOI] [PubMed] [Google Scholar]
- Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. 2016. Rationally engineered Cas9 nucleases with improved specificity. Science 351: 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabebordbar M, Zhu K, Cheng JK, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, et al. 2016. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 351: 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo JW, Kim J, Kwon SI, Corvalan C, Cho SW, Kim H, Kim SG, Kim ST, Choe S, Kim JS. 2015. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol 33: 1162–1164. [DOI] [PubMed] [Google Scholar]
- Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. 2014. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol 32: 551–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, et al. 2015. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163: 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuris JA, Thompson DB, Shu Y, Guilinger JP, Bessen JL, Hu JH, Maeder ML, Joung JK, Chen ZY, Liu DR. 2015. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol 33: 73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.