Abstract
A-to-I RNA editing is a conserved widespread phenomenon in which adenosine (A) is converted to inosine (I) by adenosine deaminases (ADARs) in double-stranded RNA regions, mainly noncoding. Mutations in ADAR enzymes in Caenorhabditis elegans cause defects in normal development but are not lethal as in human and mouse. Previous studies in C. elegans indicated competition between RNA interference (RNAi) and RNA editing mechanisms, based on the observation that worms that lack both mechanisms do not exhibit defects, in contrast to the developmental defects observed when only RNA editing is absent. To study the effects of RNA editing on gene expression and function, we established a novel screen that enabled us to identify thousands of RNA editing sites in nonrepetitive regions in the genome. These include dozens of genes that are edited at their 3′ UTR region. We found that these genes are mainly germline and neuronal genes, and that they are down-regulated in the absence of ADAR enzymes. Moreover, we discovered that almost half of these genes are edited in a developmental-specific manner, indicating that RNA editing is a highly regulated process. We found that many pseudogenes and other lncRNAs are also extensively down-regulated in the absence of ADARs in the embryo but not in the fourth larval (L4) stage. This down-regulation is not observed upon additional knockout of RNAi. Furthermore, levels of siRNAs aligned to pseudogenes in ADAR mutants are enhanced. Taken together, our results suggest a role for RNA editing in normal growth and development by regulating silencing via RNAi.
Adenosine deaminases that act on RNA (ADARs) are enzymes that convert adenosine to inosine, a process commonly referred to as RNA editing (Bass 2006). ADARs act on completely, or largely, double-stranded RNA (dsRNA) (Bass 2006). A-to-I RNA editing is widespread, with modifications occurring in more than half of the human transcriptome (Bazak et al. 2014). As inosine (I) and guanosine (G) base pair similarly, RNA editing in coding regions of genes can alter the amino acid sequence of proteins. In humans, these changes affect many proteins in the brain, including the glutamate-gated channels, the serotonin 2C receptor and the voltage-gated potassium channel (Burns et al. 1997; Werry et al. 2008; Nicholas et al. 2010; Pullirsch and Jantsch 2010). Despite these examples, most known editing sites in humans reside in noncoding regions, mainly in repetitive genomic structures (like Alu repetitive elements), and in the 3′ UTRs of genes (Athanasiadis et al. 2004; Blow et al. 2004; Kim et al. 2004; Levanon et al. 2004, 2005; Barak et al. 2009; Li et al. 2009b; Kleinberger and Eisenberg 2010; Osenberg et al. 2010; Paz-Yaacov et al. 2010; Wu et al. 2011). The A-to-I edits in 3′ UTRs can affect base-pairing with small regulatory RNAs, including microRNAs and siRNAs, thus altering gene expression (Kawahara et al. 2007; Hundley and Bass 2010; Wang et al. 2013). Therefore, A-to-I editing in both coding and noncoding regions of mRNA can be biologically significant.
RNA editing is essential and conserved across metazoans: Altered editing patterns in humans and mice have been linked to neuropathological disorders and brain tumors; moreover, disruption of the editing pathways in Caenorhabditis elegans and Drosophila results in behavioral and neural defects (Wang et al. 2000, 2004; Tonkin et al. 2002; Hartner et al. 2004, 2009; Jepson and Reenan 2009, 2010; Jin et al. 2009; Sebastiani et al. 2009). Despite having behavioral defects, C. elegans that lack ADARs are viable, making C. elegans a good model organism for studying the effects of RNA editing on gene expression and development. Currently, two ADAR genes are known in C. elegans, adr-1 and adr-2. Both enzymes are highly expressed in embryos, in the nervous system, and in the developing vulva (Tonkin et al. 2002). ADR-2 was found to be the only enzymatically active deaminase in C. elegans, whereas ADR-1 regulates RNA editing efficiency of specific adenosines by interacting with dsRNA and ADR-2 (Washburn et al. 2014). C. elegans mutated for either ADAR gene exhibit chemotaxis defects and reduced lifespan, as well as reduced expression of transgenes (Tonkin et al. 2002; Sebastiani et al. 2009). Because dsRNA is a substrate for both RNAi and RNA editing pathways, it is not surprising that several reports indicate that RNA editing can affect RNAi. However, the precise relationship between RNA editing and RNAi pathways is not well established. In a variety of organisms, microRNAs were shown to be edited with downstream effects on their targets (Alon et al. 2012; García-López et al. 2013). It was also shown that siRNAs can contain inosines (Zamore et al. 2000). Furthermore, studies in mouse showed that ADAR1 prevents interferon response and regulates the immune response to foreign dsRNA (Mannion et al. 2014; Liddicoat et al. 2015). Interestingly, all three phenotypes of C. elegans ADAR double mutants are rescued by introducing additional mutations in RNAi machinery factors (RDE-1 and RDE-4) (Tonkin and Bass 2003; Sebastiani et al. 2009). This suggests that RNAi and A-to-I RNA editing pathways have a competitive relationship and possibly regulate the same target transcripts. Indeed, studies in C. elegans identified a distinct set of loci in repetitive regions in the C. elegans genome in which siRNAs are markedly up-regulated in ADAR mutants. These loci are also highly edited, which suggests that RNA editing can prevent generation of siRNAs (Wu et al. 2011; Warf et al. 2012).
Although several studies found many A-to-I RNA editing sites in the C. elegans transcriptome (Wu et al. 2011; Washburn et al. 2014; Whipple et al. 2015; Zhao et al. 2015), the role of RNA editing remains largely unknown. Most of these editing sites reside in repetitive regions and do not explain the phenotypes of ADAR mutant worms. Additionally, no significant changes in gene expression were found in ADAR mutants.
To understand whether and in what way RNA editing affects gene expression, we performed a screen to identify A-to-I RNA editing sites in nonrepetitive genome regions and followed the expression pattern of edited genes and noncoding RNAs.
Results
Identifying A-to-I RNA editing sites in nonrepetitive regions
To study the effect of RNA editing on gene expression and function, we conducted a comprehensive search for RNA editing sites in the transcriptome of C. elegans. We concentrated mainly on nonrepetitive regions, assuming they are more likely to correlate with genes. Our approach was to perform a very restrictive search in contrast to the very promiscuous three-codon genome search (Wu et al. 2011). To this end, we eliminated single-nucleotide polymorphisms (SNPs) and changes found in ADAR mutant worms (Fig. 1).
Figure 1.
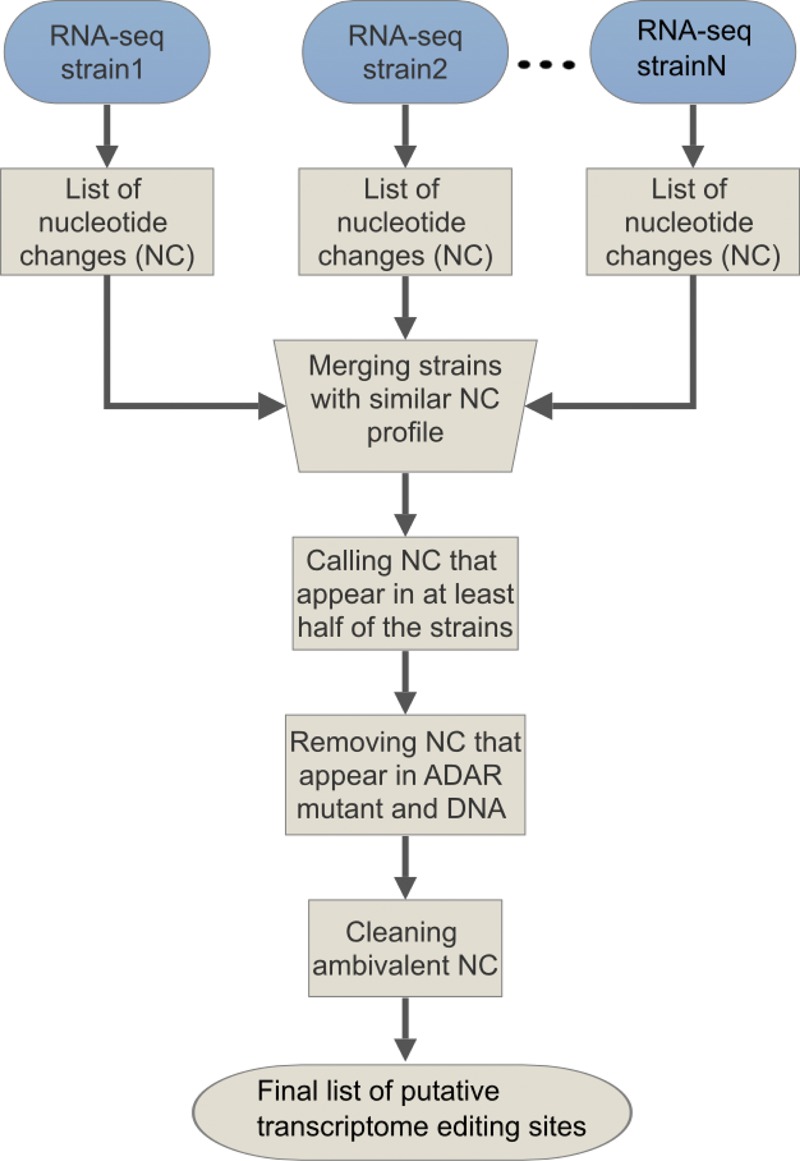
Schematic view of the computational pipeline for identification of A-to-I RNA editing sites.
To distinguish between SNPs and RNA editing sites, we chose to compare RNA-seq data from different C. elegans laboratories. The rationale was that although worms grown in different environmental conditions for many generations would probably present a very different SNP repertoire, a strong RNA editing site will appear in most or all strains. We obtained high-throughput RNA sequences from the Gene Expression Omnibus (GEO) from eight different laboratories (see Methods), and for each library we generated a list of nucleotide changes using the C. elegans WS220 genome as the reference genome. To prevent worm strains of related origin from causing bias, we compared the nucleotide change lists to each other to estimate the number of shared SNPs, and therefore the degree of relatedness between the strains (phylogenetic similarity). We merged similar strains and ended up with five RNA-seq libraries from worm strains that are far apart phylogenetically. We called a nucleotide change as a possible editing site if it appeared in at least half the libraries and in at least 5% of the reads covering this site, and if <1% of the reads at the site had non-A-to-I nucleotide changes. Next, we removed all nucleotide changes that were detected in DNA sequences from any of the strains, because they are probably SNPs and not RNA editing changes. We then removed nucleotide changes that were detected in RNA-seq data from adr-1−/−;adr-2−/− mutant strains, because they were probably not created by ADAR enzymes. In addition, we removed sites whose annotated location was ambivalent (see Methods).
We applied this pipeline (1) to detect RNA editing sites in repetitive regions, allowing 100 possible alignment sites in the genome for the RNA-seq reads; and (2) to detect RNA editing in nonrepetitive regions allowing not more than two possible alignment sites in the genome. In addition to running the pipeline on all obtained sequences, annotated as mix-stage, we separated the sequences into embryo-only and L4 larvae stage–only sets, at least four databases each. The results show clear enrichment in all databases for A-to-G and T-to-C changes over all other changes, which are expected to be RNA editing sites (Fig. 2A,B). The sequences were aligned to the genome and not to the cDNA to maximize information and eliminate biases; therefore, editing in transcripts that are transcribed in antisense to the reference genome were represented as T-to-C changes. Overall, we found 15,139 RNA editing sites in nonrepetitive regions and 69,698 sites in all regions. To ensure the correctness of the data analysis, we compared the results of the two pipelines. Only 682 sites (4.5% of the sites found by nonrepetitive analysis) were not found by the pipeline allowing repetitive regions (Fig. 2C). This is unsurprising considering we used the same data sets for both analyses. However, it provides a good indication for the consistency of the analysis. These undetected sites were probably excluded from the repetitive sites database because they did not reach the 5% changes cutoff.
Figure 2.
Nucleotide changes found by the computational pipeline. Bar graphs represent nucleotide changes found by using the computational pipeline at nonrepetitive regions (A) or repetitive regions (B). A-to-G and T-to-C changes (from antiparallel transcripts) indicate possible RNA editing sites. Sites found in mixed-stage worms (blue), embryos (green), and L4 larval stage (red). (C) A Venn diagram presenting the intersection between A-to-I editing sites found by repetitive (Rep align) and nonrepetitive alignments (Non-Rep align). Most of the nonrepetitive sites were also found by the repetitive analysis.
Unexpectedly, each of the stage-specific analyses yielded nearly twice as many editing sites as the large mixed-stage at the nonrepetitive analysis (Fig. 2A). One possible explanation for this result is the larger number of files required to achieve majority of editing site in mixed-stage than in the stage-specific analysis. Another possible explanation is that the mixed-stage sequences were not very heterogeneous and overrepresented developmental stages in which RNA editing is less frequent (Zhao et al. 2015).
Overall using this method, we found 57,117 novel editing sites by both repetitive and nonrepetitive pipelines, including all stages, of which 8685 sites reside in nonrepetitive regions (Supplemental Tables S1–S3).
Editing sites that reside in nonrepetitive regions in gene transcripts are mainly in 3′ UTR
We concentrated on editing sites in nonrepetitive regions to ensure a correct and unique position of each edited site. Gene annotation of the editing sites located in nonrepetitive regions revealed that most of the editing sites reside in introns and intergenic regions in the genome (Fig. 3A). It is possible that these intergenic regions are unannotated 3′ or 5′ untranslated regions (UTRs) of genes, because these regions are hard to annotate and can vary in size, in different gene isoforms, and in different tissues or developmental stages (Jan et al. 2011). Indeed, for some genes that have editing sites in the 3′ UTR, we found additional editing sites in close proximity in regions annotated as intergenic (see example in Supplemental Fig. S1).
Figure 3.

Most of the editing sites in nonrepetitive regions are located in intergenic and intronic regions or at 3′ UTR in gene transcripts. (A) Pie graph presenting the distribution of editing sites that were found by nonrepetitive alignment to the genome. Except for sites in intergenic regions that are not assigned to genes, all other sites were annotated as either sense or antisense (AS) to the assigned gene. (B) Distribution of editing sites located in gene transcripts, 5′ UTR, coding, or 3′ UTR regions. Sites were separated based on the region and orientation to the gene, sense, or antisense (AS). (C,D) Visualization of editing sites found at the 3′ UTR of the F48E8.4 gene. 3′ UTR of the gene is presented by arrows. (C) Sequences from wild-type worms. (D) Sequences from ADAR mutant worms. Red bars represent reads aligned to the 3′ UTR of F48E8.4 by nonrepetitive alignment from one library used in this study. Yellow lines are the predicted editing sites by this analysis. Blue dots on the sequences show A-to-G nucleotide changes, and orange dots show other nucleotide changes found in the sequences. Regions that do not have sequence coverage are repetitive regions. Therefore, sequences that aligned to them also aligned to other regions in the genome and were excluded from the analysis. The general abundance of A-to-G mismatches in the edited area can be observed in the wild type, in contrast to the complete lack of such changes in the ADAR mutant.
In gene transcripts, most of the RNA editing sites in nonrepetitive regions that are in the same orientation as the corresponding gene reside in the 3′ UTR regions (Fig. 3B). Overall, we found 258 nonrepetitive editing sites in the 3′ UTRs of 77 genes. In contrast to the 3′ UTR region, most of the editing sites in coding regions are dispersed. Only three protein-coding genes have clustered editing sites in exons: T05B4.13, C17C3.2, and nhr-231, whereas 30 genes have more than one editing site in their 3′ UTR (see example in Fig. 3C). RNA editing sites in all three genes change amino acid composition.
To validate our analysis, we chose five genes and performed Sanger sequencing to detect editing sites. All five genes were not found to undergo RNA editing by other studies (Supplemental Table S3). We were able to validate all five genes, although some of them undergo editing in a very low ratio (Supplemental Fig. S2).
Next, we searched for a common function or a similar expression pattern of the 3′ UTR-edited genes. Using defined gene mountains generated by grouping coexpressed genes in specific mutants (Kim et al. 2001), we found that genes that have editing sites in their 3′ UTRs are enriched for neuronal-related genes and germline oocyte genes (Table 1). The neuronal association of these genes fits in well with the association of RNA editing to neuronal behavior, both in mammals (Burns et al. 1997; Werry et al. 2008; Nicholas et al. 2010; Pullirsch and Jantsch 2010) and in C. elegans, where RNA editing is important for the sensory action of chemotaxis (Tonkin et al. 2002). Enrichment for germline genes was surprising; however, it might be associated with reduced life span phenotypes observed in ADAR mutant worms (Sebastiani et al. 2009).
Table 1.
3′ UTR-edited genes are enriched for neuronal genes

3′ UTR-edited genes and pseudogenes are differentially expressed in ADAR mutant worms due to RNAi
To study whether RNA editing of the 3′ UTR has a regulatory role, we evaluated changes in gene expression upon knockout of the A-to-I RNA editing genes. We generated RNA-seq libraries from wild-type and ADAR mutant worms grown in the same conditions. We collected the worms at the same developmental stage and compared the expression level of genes at embryo stage and L4 larval developmental stage separately. For each stage, we had at least four biological replicas. In ADAR mutant worms, we found a tendency for reduced expression of 3′ UTR-edited genes, in contrast to all genes, in comparison to their expression levels in wild-type worms. This tendency was noted in both L4 and embryo developmental stages (P-value <5 × 10−5) (Fig. 4A,B). We specifically tested the levels of 3′ UTR-edited genes with more than one editing site (clustered 3′ UTR-edited genes) and found them also to have reduced expression in ADAR mutant worms, both in L4 and in embryo developmental stages (P-value <6 × 10−7). Interestingly, in both L4 and embryo developmental stages, genes exhibiting the most reduced expression in ADAR mutant worms are pseudogenes and noncoding RNAs, for example, the long noncoding RNA rncs-1 (Supplemental Table S4). Therefore, to study if this is a global effect, we compared the expression levels of all annotated pseudogenes and lncRNAs in embryo and L4 stages between wild-type and ADAR mutant worms. Only pseudogenes and lncRNAs with significant expression levels were tested. This constituted more than 250 pseudogenes and lncRNAs in embryo stage and more than 350 in L4 stage. In embryos, pseudogenes had a significant overall reduction in expression in ADAR mutant worms compared to wild type (Fig. 4C). Surprisingly, in L4 larval stage, pseudogenes had a slight overall up-regulation in ADAR mutant worms when compared to all genes (P-value <0.001) (Fig. 4D).
Figure 4.
Genes edited at their 3′ UTR are down-regulated in ADAR mutant worms; pseudogenes are down-regulated only at embryo stage. Log scale plots presenting expression of genes in wild-type (N2) worms versus ADAR mutant worm at embryo stage (A,C) or L4 larval stage (B,D). Every dot in the graphs represents a gene. The red line is the regression line for all genes. 3′ UTR-edited genes with one editing site are in green, and the regression line is presented in green. 3′ UTR-edited genes with multiple editing sites are in blue, and the regression line is presented in blue. Purple dots indicate pseudogenes and lncRNAs, and the regression line is presented in purple.
These expression changes were validated by qPCR for both coding genes and pseudogenes at the embryo stage (Supplemental Fig. S3A; Supplemental Table S5) and at L4 stage (Supplemental Fig. S3B; Supplemental Table S6).
One explanation for the down-regulated expression of pseudogenes in the embryo of ADAR mutants could be the antagonistic relationship between ADARs and RNAi (Wu et al. 2011). Thus, it is possible that when A-to-I RNA editing is absent, the RNAi mechanism generates a large number of siRNAs that can target transcripts for silencing. Indeed, hundreds of loci in which siRNAs are up-regulated were identified in ADAR mutant worms (Wu et al. 2011). These loci have significant overlap with the sites that we identified in this screen (Supplemental Table S1). To test if RNAi is responsible for the observed down-regulation of pseudogenes in ADAR mutant worms, we generated RNA-seq libraries from strains having mutations in both RNA editing and RNAi genes (adr-1(−/−);adr-2(−/−);rde-1(−/−) and adr-1(−/−);adr-2(−/−);rde-4(−/−) strains) and evaluated the expression of pseudogenes at the embryo developmental stage. We found that when comparing the expression of pseudogenes in ADAR and RNAi mutant worms to wild-type worms, the down-regulation shows a substantial reversion in both strains (P-value was nonsignificant when comparing to all genes) (Fig. 5). In contrast to the embryo stage, in the L4 stage, pseudogenes in ADAR and RNAi mutants still show significant up-regulation when comparing to all genes (P-value <10−5 for both adr-1(−/−);adr-2(−/−);rde-1(−/−) and adr-1(−/−);adr-2(−/−);rde-4(−/−) strains) (Supplemental Fig. S4).
Figure 5.
Pseudogenes do not exhibit down-regulation in ADAR and RNAi mutants at the embryo stage. Log scale plots comparing gene expression of wild-type (N2) worms to adr-1(−/−);adr-2(−/−);rde-1(−/−) (A) or adr-1(−/−);adr-2(−/−);rde-4(−/−) (B) mutant worms at the embryo stage of development. (C) Log scale plot comparing primary siRNA expression of wild-type (N2) worms to adr-1(−/−);adr-2(−/−) mutant worms. Every dot represents a gene. The red line is the regression line for all genes. Purple dots indicate pseudogenes and the regression line is presented in purple.
To further test the idea that RNAi is responsible for the down-regulation of pseudogenes and lncRNAs in ADAR mutants, we compared siRNAs libraries generated from embryo stage worms by Wu et al. (2011) from wild-type worms and ADAR mutant worms. As expected, we found enrichment of siRNAs in ADAR mutant worms, when compared to wild type, in pseudogenes and lncRNAs (P-value = 6 × 10−7) (Fig. 5C).
To conclude, we found that A-to-I RNA editing promotes the expression of edited genes and pseudogenes in embryos, which is dependent on RNAi.
3′ UTR RNA editing is developmentally regulated
The screen to find editing sites in nonrepetitive regions was performed using transcriptome-wide sequencing of RNA isolated from two specific stages during worm development: embryo and larvae 4 (L4), and from a mixed stage, which contains sequences from all developmental stages. By comparing the editing of genes at different developmental stages, we could determine whether genes are edited only in a specific stage. Surprisingly, more than half of the 77 3′ UTR-edited genes were found to be stage-specifically edited (24 in L4, 21 in embryo) (Fig. 6A). Interestingly, most of the genes found to be edited at the 3′ UTR and were not detected by other studies are stage-specifically edited. This stage-specific editing could either be because the genes themselves are expressed in only one stage of development or because editing of the transcripts is regulated in a stage-specific manner. Expression analysis revealed that most of the stage-specifically edited genes are expressed in both embryo and L4 (Fig. 6B), indicating that editing is developmentally regulated.
Figure 6.
Some of the editing sites show developmental stage specificity, which is not caused by differential expression. (A) Overlap between 3′ UTR-edited genes found in each RNA-sequencing data set. The number of edited genes identified from RNA isolated from L4, embryo, and mixed stages is shown in purple, pink, and blue, respectively. The numbers inside the overlapping circles represent the intersection between the data sets. (B) Log scale plot presenting expression of genes at L4 and embryo stage in wild-type (N2) worms. Every dot represents a gene. Purple dots indicate L4-specific edited genes. Pink dots indicate embryo-specific edited genes. The green dot is egl-2 gene. The red line is the regression line for all genes.
One such example is the gene egl-2, which is expressed in both stages (Fig. 6B, green dot; Supplemental Fig. S5; Tonkin et al. 2002; LeBoeuf and Garcia 2012). Although egl-2 is frequently edited in its 3′ UTR in embryos, egl-2 transcripts completely lack editing at the L4 stage (Supplemental Fig. S6). In contrast, lem-2 is an example of a gene that undergoes RNA editing at its 3′ UTR similarly in embryo and L4 stages (Supplemental Fig. S6). lem-2 is ubiquitously expressed in C. elegans in all stages (Gruenbaum et al. 2002).
To eliminate bias from lowly expressed genes at a specific developmental stage, we further validated the editing results using the alternative approach of Sanger sequencing of clones generated by gene-specific PCR (Supplemental Table S7). Although, by sequencing many colonies, we could detect some editing in the absent developmental stage, the percentage of edited colonies was low.
We wondered if the tendency for down-regulated expression in ADAR mutant worms that we observed in 3′ UTR-edited genes occurs in all 3′ UTR-edited genes at both embryo and L4 stages or if the genes that are edited in a stage-specific manner also have stage specificity in the expression pattern. To answer this question, we separated the 3′ UTR-edited genes into three groups and analyzed the expression pattern of each group separately: (1) genes that are edited in both embryo and L4 stages; (2) genes that are edited only in the embryo stage; and (3) genes that are edited only in the L4 stage. Genes that are edited in both stages have significantly reduced expression in ADAR mutants (P-value = 9 × 10−8 and P-value = 4 × 10−8, respectively). In contrast, stage-specific edited genes show almost no significant reduction in the embryo stage (P-value <0.004, for embryo stage-specific edited genes and L4 stage-specific edited genes). In L4 stage, we did not observe expression changes for either stage-specific edited groups (nonsignificant P-value).
Thus, our results suggest that RNA editing is a highly regulated process, which results in a developmental stage–specific editome.
Discussion
In this study, we developed a screen to identify A-to-I RNA editing sites in nonrepetitive regions in the transcriptome. The screen was based on two major assumptions: (1) Using a variety of starting strains grown and sequenced in different laboratories will significantly reduce DNA polymorphisms and sequencing artifacts that are mistakenly identified as RNA editing sites by previous studies; and (2) using knockout strains for RNA editing enzymes will significantly improve detection of editing sites, because it is highly probable that any nucleotide change found by comparing RNA to the genome that is not found in the knockout strains is caused by the RNA editing. The screen we developed worked well for both nonrepetitive regions and repetitive regions in the transcriptome, whereas using a three-codon base genome screen, used in previous studies, only worked well on repetitive regions in the genome and is very effective at finding hyper-edited sites (Wu et al. 2011; Bazak et al. 2014). The screen we developed is not worm specific or A-to-I specific, although it heavily relies on the availability of knockout strains to reduce false positives. Using this screen, we found many novel RNA editing sites in nonrepetitive regions in the C. elegans transcriptome. As in human (Li et al. 2009b), most of the frequently clustered editing sites that we detected in gene regions were not in coding regions, but rather in introns and UTRs.
3′ UTR-edited genes are enriched for neuronal and germline genes
We found a group of genes that are edited in their 3′ UTR and are mainly expressed in neurons and germline. This observation can be linked to the phenotypes observed in worms lacking ADAR genes, such as changes in chemotaxis behavior and shortened lifespan. Added support to this conclusion is that knockout or down-regulation of several of these genes show similar phenotypes to ADAR mutant worms. For example, lem-2 mutant worms have reduced lifespan (Barkan et al. 2012), and rab-3 (Nonet et al. 1997) and olrn-1 (Bauer Huang et al. 2007) mutant worms exhibit chemotaxis defects. We also observed a tendency for down-regulated expression of these genes in ADAR mutant worms. It was previously shown that ADARs do not affect resistance of neurons to exogenously induced RNAi (Knight and Bass 2002); however, endogenous induced RNAi might still be affected by ADARs because triggering RNAi by dsRNA directly expressed in neurons can silence neuronal genes. The siRNAs databases generated by Wu et al. (2011) that we used in our analysis mainly consist of siRNAs generated by endogenous RNAi. The enhancement of siRNAs targeting pseudogenes and lncRNAs in ADAR mutants that we observed by using these databases further suggests that ADARs can interfere with the generation of siRNAs by endogenous RNAi.
RNA editing of mRNA is developmentally regulated
Concentrating on the genes edited in their 3′ UTR, we found that half of them are edited in a developmental stage–specific manner. It was observed previously that the overall frequency of RNA editing is higher in embryos than in other stages (Zhao et al. 2015), which correlates with the significant expression of ADARs in embryo compared to other developmental stages. However, we detected genes that undergo frequent RNA editing in their 3′ UTR in L4 developmental stage and almost none in embryo stage, although they are almost equally expressed in both stages. One possible explanation for the differential editing that we observed is that ADAR genes are not expressed uniformly in all tissues at similar levels (Tonkin et al. 2002) as are their targets, whereas our analysis was done on whole worms and did not distinguish between tissues. We do not favor this possibility because (1) some of the genes we found to be differentially edited were reported to have the same pattern of expression as ADAR genes, for example, egl-2 (Tonkin et al. 2002; LeBoeuf and Garcia 2012); and (2) we found L4-specific edited genes that are also expressed in embryo stage.
Another explanation is that the deamination activity is regulated. Thus, although ADR-2, the main active deamination enzyme is present, deamination does not occur. A good candidate for regulating deamination is ADR-1. ADR-1 was shown to affect editing efficiency, although it does not have deamination activity itself (Washburn et al. 2014). Other regulators of RNA editing were identified in C. elegans and other organisms and may regulate stage-specific editing. ADR-2 binding protein-1 (ADBP-1) was found to be essential for ADR-2 nuclear localization in C. elegans (Ohta et al. 2008). An important molecule that was found to be necessary for human ADAR2 folding is inositol hexakisphosphate (IP6). IP6 is located at the enzyme catalytic core and is required for its proper functionality (Macbeth et al. 2005). Another protein that seems to be involved in regulation of ADAR is the small ubiquitin-like modifier 1 (SUMO1). SUMO1 can modify human ADAR1 and lead to reduction in its activity, which suggests that sumoylation could regulate RNA editing (Desterro et al. 2005). The two possible explanations for the differential editing, regulation, and tissue-specific expression are not mutually exclusive. Interestingly, the down-regulated expression observed in genes edited in the 3′ UTR in ADAR mutant worms was mainly observed in genes that are edited in all developmental stages. This suggests that developmental-specific editing might be needed to regulate expression.
Down-regulation of pseudogene expression in worms lacking RNA editing is dependent on RNAi
Other than a tendency for down-regulated expression of 3′ UTR-edited genes, we observed extensive down-regulated expression of pseudogenes and lncRNAs in ADAR mutant worms. In contrast to 3′ UTR-edited genes, down-regulation of expression of pseudogenes and lncRNAs was observed only in the embryo developmental stage, which might be explained by low frequency of RNA editing of pseudogenes at L4 stage. Our method of detecting RNA editing sites was intended for nonrepetitive regions in the genome and was not as efficient for detecting RNA editing sites in pseudogenes and other repetitive elements. Therefore, although we cannot determine conclusively if these transcripts are less edited at L4 stage, this was observed by a different study (Zhao et al. 2015).
A high amount of siRNAs corresponding to specific highly edited repetitive regions in the genome were found in ADAR mutant worms (Wu et al. 2011). Therefore, it was suggested that RNA editing prevents RNAi from processing dsRNA. The study (Wu et al. 2011) also found that two transcripts that overlap enriched siRNAs regions are significantly down-regulated in ADAR mutant worms. Both of these transcripts, along with other pseudogenes and lncRNAs, were found to be edited by our screen and had decreased expression in ADAR mutant worms. Using siRNAs from Wu et al. (2011), we found a significant enrichment of siRNAs in ADAR mutant worms targeting the same pseudogenes and lncRNAs that we found to be down-regulated in ADAR mutant worms. This leads us to speculate that the extensive decrease in expression that we observed in pseudogenes and lncRNAs in ADAR mutant worms might be a result of siRNA generation. Corroboration of this idea comes from our results showing that when RNAi was knocked out in addition to RNA editing the decrease in expression was not observed.
3′ UTRs and pseudogenes have a tendency for secondary RNA structure and could be potential targets for endogenous RNAi. It is possible that one of the roles of RNA editing in C. elegans is to protect 3′ UTRs and genes from degradation by either destabilizing the dsRNA structure or by preventing RNAi components from processing the dsRNAs. We cannot determine from our analysis how RNA editing antagonizes RNAi, but it was shown in mammals that RNA editing events stabilize the double-stranded structure of Alu repeats (Athanasiadis et al. 2004) and that deamination of dsRNA inhibits RNAi in vitro (Scadden and Smith 2001). This favors the possibility that inosines prevent Dicer from processing the dsRNAs directly rather than by a change in the structure.
In summary, our findings suggest that RNA editing is a highly regulated process that has an important role in regulating RNAi to prevent degradation of “self” transcripts and thus plays a role in the surveillance of the transcriptome.
Methods
C. elegans strains
The following strains were used in this study: Bristol N2 (Brenner 1974), BB4 adr-1(gv6) I; adr-2(gv42) III (Tonkin et al. 2002), BB21 adr-1(tm668) I; adr-2(ok735) III (Hundley et al. 2008), BB23 adr-1(gv6); adr-2(gv42); rde-1(ne219) (Tonkin and Bass 2003), BB24 adr-1(gv6); adr-2(gv42); rde-4 (ne299) (Tonkin and Bass 2003).
mRNA-seq libraries preparation
All strains were raised at 20°C on enriched NGM with OP50 as food and cultured as described in Brenner (1974). Embryos were obtained by washing worms with M9 and sodium hypochlorite and were either resuspended in EN buffer and frozen into pellets with liquid nitrogen, or left overnight in M9 in a nutator at 20°C. The hatched synchronized L1 larva were placed on an enriched NGM plate until they reached the fourth larval stage (L4), at which point they were washed with EN buffer and frozen into pellets with liquid nitrogen. Frozen pellets were ground to powder with a liquid nitrogen chilled mortar and pestle. RNA in high and low molecular weight fractions was extracted by mirVana kit (Ambion). mRNA sequencing libraries were prepared from the high molecular weight fraction by means of Illumina TruSeq RNA Sample Preparation kit, automated by Agilent Bravo Automated Liquid Handling Platform and sequenced with an Illumina HiSeq 2500.
RNA editing sites identification
RNA sequences used to identify editing sites were generated for this study and obtained from Hillier et al. (2009), Mangone et al. (2010), Wu et al. (2011), Kim et al. (2013), Schmeisser et al. (2013a,b), Dallaire et al. (2014), Grün et al. (2014), Stoeckius et al. (2014), and Weimer et al. (2014). All reads were trimmed to 50 nt, and identical reads were merged. Sequences were aligned to the WS220 (WormBase; http://www.wormbase.org) genome with Bowtie (Langmead et al. 2009), allowing three mismatches and a maximum of two or of 100 alignments, for nonrepetitive or repetitive alignments, respectively. Sequences with mismatches at the first or last nucleotide were discarded to reduce splicing and poly(A) associated mismatches. The files were converted to the pileup format available in SAMtools 1.16 (Li et al. 2009a). Sequences that contained similar nucleotide changes profiles as compared to the reference genome were merged to remove noise. Data sets were separated based on developmental stage: (1) mixed stage that contains all data sets, (2) L4 stage, and (3) embryo stage. Nucleotide changes were identified in each of these data sets separately.
Nucleotide changes were selected if they met the following criteria: The change must be present in at least half of the files, with at least 5% of reads at that site having the change, and no more than 1% of reads at the site having different nucleotide changes. Reads from knockout mutants (BB21 and BB4) and DNA from N2 worms (a kind gift from Idan Gabdank and Andrew Fire, SRA:SRX1770065) were similarly processed to the point where pileup files were available. Any change that occurred in these two pileup files was removed from the list as not being edited.
Known variations that were published in WormBase and changes that are close to splice sites (potential splice artifacts) and right before start codons (potential SL1 artifacts) were removed. All remaining sequences were annotated based on WormBase assembly WS220. Sites with multiple annotations were removed. The final list is presented in Supplemental Table S2.
Detection of similarity between strains
To find and merge strains that are close in origin, phylogenetic profiling was performed. For this purpose, a list of nucleotide changes was prepared for each strain based on pileup files generated by RNA sequences alignment using Bowtie (Langmead et al. 2009) to the reference genome WS220 (WormBase, http://www.wormbase.org) and then SAMtools (Li et al. 2009a). Every change found so far was looked for in all N2 mRNA pileup files, and lists of changes each strain contains were compiled and sorted. Distance was measured by the Unix diff tool. Sequence files of the most similar samples were merged. Sequences generated by us and sequences from Wu et al. (2011) were known to be from the same strain, and therefore were the standard by which similarity was measured. Sequences were called to be from similar origin if the compared files had higher percentages of similarity than the standard that we set.
Gene expression analysis
At least four different biological RNA-seq samples were generated from N2, ADAR mutant worms (BB21 or BB4), and ADAR and RNAi mutant worms (BB23 or BB24), each at embryo and L4 stage. Technical replicas were also obtained to verify consistency; however, they were not used for gene expression evaluation. mRNA sequences were aligned to gene transcripts using Bowtie (Langmead et al. 2009), allowing multiple alignments in order to include multiple transcripts due to alternative splicing. DESeq (Anders and Huber 2010) package in R (R Core Team 2013) was used to identify differentially expressed genes. P-values were calculated by Welch two-sample t-test that was performed only on transcripts with significant padj value.
Random editing sites generation
Random controls for all the analyses were generated by a custom script that reads patterns from lists of edited files and generates similar patterns in 3′ UTRs of random genes. For each 3′ UTR-edited gene, 10 mock control editing sites were generated.
Data access
The sequence data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE83133. Sanger sequencing traces from this study have been submitted to the Trace Archive (https://trace.ncbi.nlm.nih.gov/Traces/trace.cgi) under TI numbers 2344111651–2344112080.
Supplementary Material
Acknowledgments
We thank Yomiran Nissan and Nabeel Ganem for critical reading of the manuscript; Idan Gabdank and Andrew Fire for sharing DNA sequences before publication; and the Caenorhabditis Genetics Center, National Bioresource Project for worm strains. This work was funded by The Israeli Centers of Research Excellence (I-CORE) program, (Center No. 1796/12 to A.T.L.), The Israel Science Foundation (grant no. 644/13 to A.T.L.), and the Binational Israel-USA Science Foundation (grant no. 2015091 to A.T.L.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.211169.116.
References
- Alon S, Mor E, Vigneault F, Church GM, Locatelli F, Galeano F, Gallo A, Shomron N, Eisenberg E. 2012. Systematic identification of edited microRNAs in the human brain. Genome Res 22: 1533–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiadis A, Rich A, Maas S. 2004. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol 2: e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak M, Levanon EY, Eisenberg E, Paz N, Rechavi G, Church GM, Mehr R. 2009. Evidence for large diversity in the human transcriptome created by Alu RNA editing. Nucleic Acids Res 37: 6905–6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan R, Zahand AJ, Sharabi K, Lamm AT, Feinstein N, Haithcock E, Wilson KL, Liu J, Gruenbaum Y. 2012. Ce-emerin and LEM-2: essential roles in Caenorhabditis elegans development, muscle function, and mitosis. Mol Biol Cell 23: 543–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL. 2006. How does RNA editing affect dsRNA-mediated gene silencing? Cold Spring Harb Symp Quant Biol 71: 285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer Huang SL, Saheki Y, VanHoven MK, Torayama I, Ishihara T, Katsura I, van der Linden A, Sengupta P, Bargmann CI. 2007. Left-right olfactory asymmetry results from antagonistic functions of voltage-activated calcium channels and the Raw repeat protein OLRN-1 in C. elegans. Neural Dev 2: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E, et al. 2014. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res 24: 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow M, Futreal PA, Wooster R, Stratton MR. 2004. A survey of RNA editing in human brain. Genome Res 14: 2379–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. 1997. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387: 303–308. [DOI] [PubMed] [Google Scholar]
- Dallaire A, Proulx S, Simard MJ, Lebel M. 2014. Expression profile of Caenorhabditis elegans mutant for the Werner syndrome gene ortholog reveals the impact of vitamin C on development to increase life span. BMC Genomics 15: 940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desterro JM, Keegan LP, Jaffray E, Hay RT, O'Connell MA, Carmo-Fonseca M. 2005. SUMO-1 modification alters ADAR1 editing activity. Mol Biol Cell 16: 5115–5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-López J, Hourcade Jde D, Del Mazo J. 2013. Reprogramming of microRNAs by adenosine-to-inosine editing and the selective elimination of edited microRNA precursors in mouse oocytes and preimplantation embryos. Nucleic Acids Res 41: 5483–5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Lee KK, Liu J, Cohen M, Wilson KL. 2002. The expression, lamin-dependent localization and RNAi depletion phenotype for emerin in C. elegans. J Cell Sci 115: 923–929. [DOI] [PubMed] [Google Scholar]
- Grün D, Kirchner M, Thierfelder N, Stoeckius M, Selbach M, Rajewsky N. 2014. Conservation of mRNA and protein expression during development of C. elegans. Cell Rep 6: 565–577. [DOI] [PubMed] [Google Scholar]
- Hartner JC, Schmittwolf C, Kispert A, Müller AM, Higuchi M, Seeburg PH. 2004. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem 279: 4894–4902. [DOI] [PubMed] [Google Scholar]
- Hartner JC, Walkley CR, Lu J, Orkin SH. 2009. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol 10: 109–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier LW, Reinke V, Green P, Hirst M, Marra MA, Waterston RH. 2009. Massively parallel sequencing of the polyadenylated transcriptome of C. elegans. Genome Res 19: 657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundley HA, Bass BL. 2010. ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem Sci 35: 377–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundley HA, Krauchuk AA, Bass BL. 2008. C. elegans and H. sapiens mRNAs with edited 3′ UTRs are present on polysomes. RNA 14: 2050–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Friedman RC, Ruby JG, Bartel DP. 2011. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature 469: 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepson JE, Reenan RA. 2009. Adenosine-to-inosine genetic recoding is required in the adult stage nervous system for coordinated behavior in Drosophila. J Biol Chem 284: 31391–31400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jepson JE, Reenan RA. 2010. Unraveling pleiotropic functions of A-to-I RNA editing in Drosophila. Fly (Austin) 4: 154–158. [DOI] [PubMed] [Google Scholar]
- Jin Y, Zhang W, Li Q. 2009. Origins and evolution of ADAR-mediated RNA editing. IUBMB Life 61: 572–578. [DOI] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. 2007. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 315: 1137–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Lund J, Kiraly M, Duke K, Jiang M, Stuart JM, Eizinger A, Wylie BN, Davidson GS. 2001. A gene expression map for Caenorhabditis elegans. Science 293: 2087–2092. [DOI] [PubMed] [Google Scholar]
- Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. 2004. Widespread RNA editing of embedded Alu elements in the human transcriptome. Genome Res 14: 1719–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Dh, Grün D, van Oudenaarden A. 2013. Dampening of expression oscillations by synchronous regulation of a microRNA and its target. Nat Genet 45: 1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinberger Y, Eisenberg E. 2010. Large-scale analysis of structural, sequence and thermodynamic characteristics of A-to-I RNA editing sites in human Alu repeats. BMC Genomics 11: 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Bass BL. 2002. The role of RNA editing by ADARs in RNAi. Mol Cell 10: 809–817. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBoeuf B, Garcia LR. 2012. Cell excitability necessary for male mating behavior in Caenorhabditis elegans is coordinated by interactions between big current and ether-a-go-go family K+ channels. Genetics 190: 1025–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. 2004. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol 22: 1001–1005. [DOI] [PubMed] [Google Scholar]
- Levanon K, Eisenberg E, Rechavi G, Levanon EY. 2005. Letter from the editor: adenosine-to-inosine RNA editing in Alu repeats in the human genome. EMBO Rep 6: 831–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009a. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Levanon EY, Yoon JK, Aach J, Xie B, LeProust E, Zhang K, Gao Y, Church GM. 2009b. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 324: 1210–1213. [DOI] [PubMed] [Google Scholar]
- Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH, Walkley CR. 2015. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349: 1115–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macbeth MR, Schubert HL, Vandemark AP, Lingam AT, Hill CP, Bass BL. 2005. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 309: 1534–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, et al. 2010. The landscape of C. elegans 3′UTRs. Science 329: 432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannion NM, Greenwood SM, Young R, Cox S, Brindle J, Read D, Nellåker C, Vesely C, Ponting CP, McLaughlin PJ, et al. 2014. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep 9: 1482–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas A, de Magalhaes JP, Kraytsberg Y, Richfield EK, Levanon EY, Khrapko K. 2010. Age-related gene-specific changes of A-to-I mRNA editing in the human brain. Mech Ageing Dev 131: 445–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet ML, Staunton JE, Kilgard MP, Fergestad T, Hartwieg E, Horvitz HR, Jorgensen EM, Meyer BJ. 1997. Caenorhabditis elegans rab-3 mutant synapses exhibit impaired function and are partially depleted of vesicles. J Neurosci 17: 8061–8073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta H, Fujiwara M, Ohshima Y, Ishihara T. 2008. ADBP-1 regulates an ADAR RNA-editing enzyme to antagonize RNA-interference-mediated gene silencing in Caenorhabditis elegans. Genetics 180: 785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osenberg S, Paz Yaacov N, Safran M, Moshkovitz S, Shtrichman R, Sherf O, Jacob-Hirsch J, Keshet G, Amariglio N, Itskovitz-Eldor J, et al. 2010. Alu sequences in undifferentiated human embryonic stem cells display high levels of A-to-I RNA editing. PLoS One 5: e11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Yaacov N, Levanon EY, Nevo E, Kinar Y, Harmelin A, Jacob-Hirsch J, Amariglio N, Eisenberg E, Rechavi G. 2010. Adenosine-to-inosine RNA editing shapes transcriptome diversity in primates. Proc Natl Acad Sci 107: 12174–12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullirsch D, Jantsch MF. 2010. Proteome diversification by adenosine to inosine RNA editing. RNA Biol 7: 205–212. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org/. [Google Scholar]
- Scadden AD, Smith CW. 2001. RNAi is antagonized by A→I hyper-editing. EMBO Rep 2: 1107–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser S, Priebe S, Groth M, Monajembashi S, Hemmerich P, Guthke R, Platzer M, Ristow M. 2013a. Neuronal ROS signaling rather than AMPK/sirtuin-mediated energy sensing links dietary restriction to lifespan extension. Mol Metab 2: 92–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeisser S, Schmeisser K, Weimer S, Groth M, Priebe S, Fazius E, Kuhlow D, Pick D, Einax JW, Guthke R, et al. 2013b. Mitochondrial hormesis links low-dose arsenite exposure to lifespan extension. Aging Cell 12: 508–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani P, Montano M, Puca A, Solovieff N, Kojima T, Wang MC, Melista E, Meltzer M, Fischer SEJ, Andersen S, et al. 2009. RNA editing genes associated with extreme old age in humans and with lifespan in C. elegans. PLoS One 4: e8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M, Grün D, Kirchner M, Ayoub S, Torti F, Piano F, Herzog M, Selbach M, Rajewsky N. 2014. Global characterization of the oocyte-to-embryo transition in Caenorhabditis elegans uncovers a novel mRNA clearance mechanism. EMBO J 33: 1751–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin LA, Bass BL. 2003. Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science 302: 1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin LA, Saccomanno L, Morse DP, Brodigan T, Krause M, Bass BL. 2002. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J 21: 6025–6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Khillan J, Gadue P, Nishikura K. 2000. Requirement of the RNA editing deaminase ADAR1 gene for embryonic erythropoiesis. Science 290: 1765–1768. [DOI] [PubMed] [Google Scholar]
- Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, Nishikura K. 2004. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem 279: 4952–4961. [DOI] [PubMed] [Google Scholar]
- Wang Q, Hui H, Guo Z, Zhang W, Hu Y, He T, Tai Y, Peng P, Wang L. 2013. ADAR1 regulates ARHGAP26 gene expression through RNA editing by disrupting miR-30b-3p and miR-573 binding. RNA 19: 1525–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf MB, Shepherd BA, Johnson WE, Bass BL. 2012. Effects of ADARs on small RNA processing pathways in C. elegans. Genome Res 22: 1488–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn MC, Kakaradov B, Sundararaman B, Wheeler E, Hoon S, Yeo GW, Hundley HA. 2014. The dsRBP and inactive editor, ADR-1, utilizes dsRNA binding to regulate A-to-I RNA editing across the C. elegans transcriptome. Cell Rep 6: 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer S, Priebs J, Kuhlow D, Groth M, Priebe S, Mansfeld J, Merry TL, Dubuis S, Laube B, Pfeiffer AF, et al. 2014. D-Glucosamine supplementation extends life span of nematodes and of ageing mice. Nat Commun 5: 3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werry TD, Loiacono R, Sexton PM, Christopoulos A. 2008. RNA editing of the serotonin 5HT2C receptor and its effects on cell signalling, pharmacology and brain function. Pharmacol Ther 119: 7–23. [DOI] [PubMed] [Google Scholar]
- Whipple JM, Youssef OA, Aruscavage PJ, Nix DA, Hong C, Johnson WE, Bass BL. 2015. Genome-wide profiling of the C. elegans dsRNAome. RNA 21: 786–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Lamm AT, Fire AZ. 2011. Competition between ADAR and RNAi pathways for an extensive class of RNA targets. Nat Struct Mol Biol 18: 1094–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. 2000. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33. [DOI] [PubMed] [Google Scholar]
- Zhao HQ, Zhang P, Gao H, He X, Dou Y, Huang AY, Liu XM, Ye AY, Dong MQ, Wei L. 2015. Profiling the RNA editomes of wild-type C. elegans and ADAR mutants. Genome Res 25: 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






