Summary
A global concern has emerged with the pandemic spread of Zika virus (ZIKV) infections that can cause severe neurological symptoms in adults and newborns. ZIKV is a positive-strand RNA virus replicating in virus-induced membranous replication factories (RFs). Here we used various imaging techniques to investigate the ultrastructural details of ZIKV RFs and their relationship with host cell organelles. Analyses of human hepatic cells and neural progenitor cells infected with ZIKV revealed endoplasmic reticulum (ER) membrane invaginations containing pore-like openings toward the cytosol, reminiscent to RFs in Dengue virus-infected cells. Both the MR766 African strain and the H/PF/2013 Asian strain, the latter linked to neurological diseases, induce RFs of similar architecture. Importantly, ZIKV infection causes a drastic reorganization of microtubules and intermediate filaments forming cage-like structures surrounding the viral RF. Consistently, ZIKV replication is suppressed by cytoskeleton-targeting drugs. Thus, ZIKV RFs are tightly linked to rearrangements of the host cell cytoskeleton.
Keywords: Zika virus, flavivirus, human neural progenitor cells, replication factories, replication organelles, microtubules, intermediate filaments, electron microscopy, electron tomography, live-cell imaging
Graphical Abstract
Highlights
-
•
ZIKV induces ER membrane invaginations similar to Dengue virus
-
•
ZIKV induces profound alterations of the cytoskeleton
-
•
Microtubules and intermediate filaments surround the ZIKV replication factory
-
•
ZIKV replication is sensitive to cytoskeleton-targeting drugs
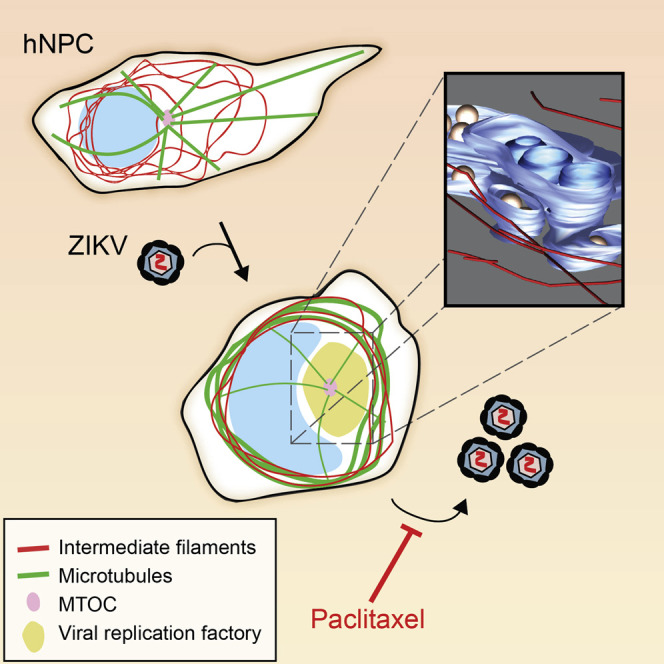
Cortese et al. show that ZIKV infection in both human hepatoma and neuronal progenitor cells induces drastic structural modification of the cellular architecture. Microtubules and intermediate filaments surround the viral replication factory composed of vesicles corresponding to ER membrane invagination toward the ER lumen. Importantly, alteration of microtubule flexibility impairs ZIKV replication.
Introduction
The pandemic outbreak of Zika virus (ZIKV), a member of the Flavivirus genus within the Flaviviridae family, and the association of ZIKV infections with severe disease raised intense concerns. Like other flaviviruses, ZIKV is primarily transmitted by mosquitoes, but more recently, unsuspected transmission modes and symptoms unique to ZIKV have been reported. Of note, infection of pregnant women with ZIKV can lead to congenital transmission and eventually to severe microcephaly in newborns (Pierson and Graham, 2016). Moreover, ZIKV is sexually transmissible through undefined mechanisms, and other neurological manifestations such as Guillain-Barré syndrome have been observed in infected individuals. Thus, intensive efforts have been undertaken to control this emerging disease, but neither antiviral therapies nor a prophylactic vaccine for ZIKV are currently available (Pierson and Graham, 2016). This unmet medical need is exacerbated by the spread of the Aedes mosquito vector beyond tropical countries.
ZIKV was identified almost 70 years ago in Uganda, but severe symptoms were never reported. However, the recent outbreaks of the contemporary Asian lineage in the Yap Islands and French Polynesia in 2007 and 2013, respectively, have raised major interest in ZIKV (Cao-Lormeau et al., 2014, Duffy et al., 2009). This Asian lineage is genetically closely related to the “Brazilian strain” currently spreading in the Americas and causing congenital microcephaly. Retrospective studies demonstrated that the French Polynesian strain also caused microcephaly (Cauchemez et al., 2016), while it is hypothesized that the African lineage does not. More recently, the use of developing brain organoid models showed that in contrast to the African strain MR766, the Brazilian strain (Asian lineage) caused apoptosis of neural progenitor cells (NPCs) (Cugola et al., 2016). However, the viral determinants governing ZIKV pathogenesis are still unknown.
Upon entry into the host cell and release of the positive-strand RNA genome into the cytoplasm, the genome is used for the synthesis of a polyprotein that is cleaved co- and post-translationally into 10 proteins (Acosta et al., 2014). These are the nonstructural proteins NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5 that are responsible for the replication of the viral RNA and the structural proteins Capsid, prM, and Envelope that assemble, together with the viral RNA genome, into infectious virions (Acosta et al., 2014). The genome of ZIKV is replicated via a negative-strand RNA intermediate.
Flavivirus-infected cells undergo a massive remodeling of the endoplasmic reticulum (ER) to form membranous replication factories (RFs) (Paul and Bartenschlager, 2013). Notably, flaviviruses, including ZIKV (Hamel et al., 2015), induce invaginations of the ER giving rise to clusters of vesicles designated vesicle packets (VPs), which are the presumed sites of viral RNA replication. Using electron tomography, several studies reported the 3D architecture of VPs formed during Dengue virus (DENV), West Nile virus (WNV), and tick-borne encephalitis virus (TBEV) infections (Paul and Bartenschlager, 2013). In most of these studies, the vesicles are linked to the cytoplasm via an ∼10-nm-wide opening. This pore might allow the exit of the positive-strand RNA genome into the cytoplasm to be used for polyprotein synthesis (RNA translation) or packaging into virus particles budding into the ER lumen at sites juxtaposed to the VPs (Welsch et al., 2009). In addition, in the case of WNV, vesicles were reported to be interconnected by a pore (Gillespie et al., 2010). Two additional ER-derived structures can be detected in flavivirus-infected cells: (1) convoluted membranes (CMs) accumulating in DENV-, TBEV-, and WNV-infected cells (Romero-Brey and Bartenschlager, 2014); and (2) DENV particles accumulating in regular arrays within enlarged ER cisternae. These “virus bags” might constitute a pool of assembled particles accumulating at this site prior to transport out of the cell. This transport is linked to proteolytic cleavage of the envelope glycoprotein by the cellular enzyme furin and it is required to render virions infectious (Acosta et al., 2014).
Although it has been reported that ZIKV induces VPs and CMs in infected cells (Hamel et al., 2015, Hanners et al., 2016, Offerdahl et al., 2017), the high-resolution 3D architecture of ZIKV RFs and their biogenesis are unknown. Importantly, these analyses were conducted in cancer-derived hepatic and neuronal cell systems, which may be considered physiologically distant to ZIKV target cells such as human NPCs (hNPCs).
In addition to the ER, other cellular organelles such as mitochondria, lipid droplets, and autophagosomes are morphologically remodeled and functionally perturbed by flaviviruses (Chatel-Chaix et al., 2016, Heaton and Randall, 2011, You et al., 2015). Furthermore, several studies identified a massive alteration of the cytoskeleton by some flaviviruses. For instance, intermediate filament and microtubule networks are reorganized in cells infected with DENV serotype 2 and their pharmacological disruption affects several steps of the viral replication cycle (Chen et al., 2008, Teo and Chu, 2014). Interestingly, the intermediate filament component vimentin associates with DENV NS4A and is important for the perinuclear localization of replication complexes (Teo and Chu, 2014). Moreover, electron tomography-based 3D reconstructions using TBEV-infected human neurons revealed a connection between microtubules and virion-containing vesicles (Bílý et al., 2015). Disrupting microtubules severely impeded virus production, highlighting the importance of this cytoskeleton network for the TBEV life cycle (Bílý et al., 2015). Whether ZIKV exploits similar or different strategies remains unknown.
Here we explored how host cell functions are altered to establish the membranous ZIKV RF. We compared intracellular remodeling induced by both the historical MR766 African strain and the contemporary Asian H/PF/2013 strain. Using Huh7 human hepatic cells and hNPCs, a physiologically relevant target cell of ZIKV, we demonstrate that independent of the lineage, ZIKV induces a drastic reorganization of intermediate filament and microtubule networks and reorganizes the ER to form VPs, zippered ER, and CMs. Cytoskeleton-targeting drugs suppress ZIKV replication, arguing that microtubules and intermediate filaments are critical for robust virus amplification.
Results
ZIKV Replication Depends on the Rearrangement of the Cytoskeletal Network
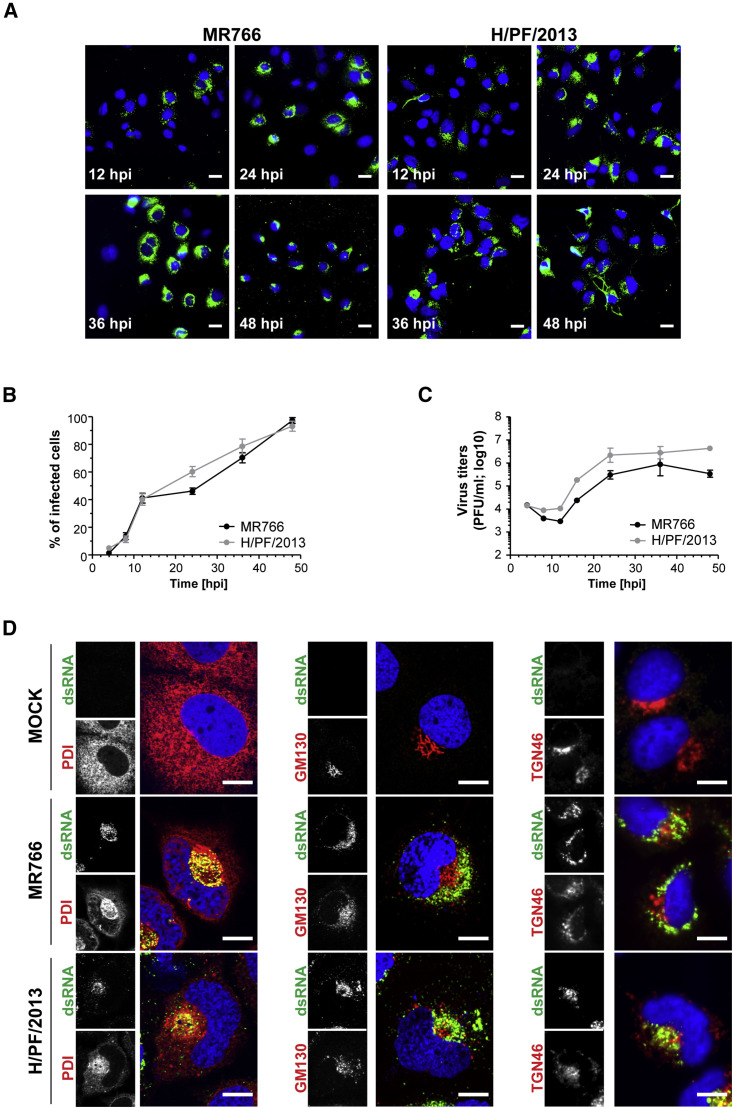
With the aim to study the reorganization of subcellular structures by ZIKV, we first determined the replication kinetics of two virus strains: the historical African strain MR766 and the contemporary Asian strain H/PF/2013. In addition to the brain and placenta, the liver is one of the organs targeted by ZIKV in vivo in both the dam and fetus (Adams Waldorf et al., 2016). Therefore, we used Huh7 human hepatoma cells, because they are highly susceptible to infection with several flaviviruses. Using immunofluorescence-based visualization of double-stranded (ds) RNA, a well-established replication intermediate, virus replication was detected as early as 4 hr post-infection in a low percentage of cells, with the percentage increasing up to 100% of cells 48 hr post-infection (Figures 1A and 1B). Interestingly, although the percentage of infected cells was similar for both strains, we retrieved up to 10-fold higher extracellular virus titers for the Asian H/PF/2013 strain (Figure 1C). This might be due to the stronger cytotoxic effects exerted by the African MR766 strain, as revealed by the higher number of dead cells at late time points after infection (data not shown).
Figure 1.
ZIKV Growth Kinetic in Huh7 Cells
(A) Time course of the spread of ZIKV infection (MOI = 5) in Huh7 cells as detected by immunofluorescence using a dsRNA-specific antibody (green). Nuclear DNA was stained with DAPI (blue). Scale bars, 20 μm. (B) Quantification of the percentage of infected cells shown in (A). Means ± SD from two independent experiments (four fields of view each) are shown. (C) Titers of ZIKV released from infected Huh7 cells. Supernatants from infected cells (MOI = 5) were collected at the indicated time points and titers were determined by plaque assay. Means ± SD from two independent experiments are shown. (D) Huh7 cells were infected with ZIKV (MOI = 5) for 36 hr. Cells were fixed and cellular proteins specified in the left of each panel (red) as well as dsRNA (green) were detected by immunofluorescence microscopy. Colored images are merges of the corresponding individual panels on the left. hpi, hours post-infection; scale bars, 10 μm.
To gain insight into the subcellular distribution of different organelles, we conducted immunofluorescence analyses using confocal microscopy of ZIKV-infected Huh7 cells (Figure 1D). Protein disulfide isomerase (PDI), GM130 protein, and TGN46 protein were used as markers of the ER, Golgi apparatus, and trans-Golgi network (TGN), respectively. Compared to mock cells, where the Golgi apparatus and the TGN showed the classic shape of stacked cisternae, these two organelles appeared fragmented in infected cells and localized toward the interior of the dsRNA-positive area. Additionally, the nuclear morphology was altered to form a kidney-shaped structure, with dense ER accumulating in the perinuclear region that was also enriched for dsRNA.
In addition to changes in the morphology of these organelles, we also observed alterations of the cytoskeleton network within infected cells. In mock-infected cells, both intermediate filaments and microtubules were tightly linked and were observed to completely enwrap the nucleus (Figures 2A and 2B). However, in infected cells, the cytoskeleton network was displaced by a perinuclear inclusion enriched for viral dsRNA and viral proteins (as revealed by staining for ZIKV NS3), both of which accumulated on the concave surface of the kidney-shaped nucleus (Figures 2A and 2B). Thicker cytoskeletal filaments surrounded the virus-induced structure (Figures 2A and 2B). Confocal microscopy and 3D reconstruction indicated the formation of a cage-like arrangement of cytoskeleton elements clustering in close proximity of viral RNA (Figure S1A). Of note, α-tubulin staining revealed a bright spot, corresponding to the microtubules organizing center (MTOC), frequently located in the center of the viral inclusion (Figure 2B).
Figure 2.
Rearrangement of the Cytoskeleton in ZIKV-Infected Huh7 Cells
(A and B) Huh7 cells were mock infected or infected for 36 hr with the MR766 or the H/PF/21013 ZIKV strain (MOI = 5) prior to methanol fixation. ZIKV infection was visualized by immunostaining for dsRNA and Cytokeratin 8 or NS3 and α-tubulin (red), respectively. Nuclear DNA was stained with DAPI (blue). Naïve cells (mock) served as the control. Representative view fields are shown. Yellow squares in the left panels indicate the magnified area shown in the corresponding right color panel. Scale bars, 10 μm. (C) Dynamics of tubulin in Huh7 GFP-tubulin-expressing cells infected with ZIKV MR766 (MOI = 10). Cropped sections of the GFP-tubulin channel recorded 3, 10, 20, 30, and 40 hr post-infection (see Movie S1). A representative infected cell (α) is highlighted to facilitate tracking. Images were taken every 30 min. Scale bars, 20 μm. (D) Titers of ZIKV particles released from Huh7 cells pre-treated 2 hr prior to infection or 3 hr post-infection with bafilomycin A1 (Baf; 2.5 nM). DMSO was used as the control. Supernatants were collected 24 hr post-infection and titers were determined using the plaque assay. Histogram bars represent the fold difference to DMSO-treated cells. Means ± SD from two replicates are shown. (E) Viability of Huh7 cells as determined by measurement of ATP level after a 24-hr treatment with 12.5 μM paclitaxel. Treatment with DMSO was used as the control. Means ± SD from three independent experiments are shown. (F) Huh7 cells were treated for 24 hr with 12.5 μM paclitaxel or with equal amounts of DMSO. α-tubulin was detected by immunofluorescence microscopy. Scale bars, 20 μm. (G) Titer of extracellular virus released from ZIKV-infected Huh7 cells treated with 12.5 μM paclitaxel for 20 hr. Cells were treated with the drug 3 hr after infection with ZIKV (MOI = 5). Histogram bars represent the fold difference to DMSO-treated cells. Means ± SD from three independent experiments are shown.
To monitor the dynamics of this cytoskeleton rearrangement, we performed live-cell imaging analysis of ZIKV-infected cells. A Huh7-derived cell line stably expressing tubulin-GFP was generated and infected with ZIKV. In mock cells, no apparent changes of the cytoskeletal network were observed within the 2-day observation period (Figure S1B). In contrast, in cells infected with ZIKV MR766, microtubule displacement and cage formation was first observed at ∼20 hr post-infection and became more prominent as infection proceeded (Figure 2C; Movie S1). At later time points after infection, vacuolization and microtubule disassembly were noticed, right before cell death as a result of virus-induced cytopathic effects (Movie S1).
To corroborate these findings in a non-carcinoma cell culture system that is physiologically relevant for ZIKV-associated neurological symptoms, we investigated ZIKV-induced subcellular alterations in hNPCs previously shown to be ZIKV target cells both in vitro and in vivo (Li et al., 2016a, Li et al., 2016b, Tang et al., 2016). Indeed, these cells were well permissive for both the MR766 and H/PF/2013 strains, with around 60% of cells becoming infected within 36 hr after infection (Figures S2A and S2B). Both strains replicated to comparable levels, as revealed by quantification of intracellular ZIKV RNA (Figure S2C), and virus titers were on the order of 106 PFU/mL (Figure S2D). Importantly, ZIKV also induced profound alterations of intermediate filaments and microtubules in this cell system, consistent with phenotypes detected in Huh7 cells (Figures S3A and S3B). Most striking was the effect on the intermediate filaments, as revealed by staining for nestin, an intermediate filament marker characteristic for neural stem and progenitor cells. As shown in Figure S3A, nestin filaments collapsed toward the perinuclear region, surrounding a dsRNA-positive structure presumably corresponding to the viral RFs (see below). Moreover, when visualized by stimulated emission depletion (STED) microscopy, which we used to discriminate between filamentous and non-filamentous structures, nestin filaments accumulated toward the periphery of the virus-induced structure, whereas the diffuse nestin signal present within the NS3-positive region suggested filament depolymerization within this particular area (Figure S3C).
Microtubule Stability Is Required for Efficient ZIKV Replication
Next we sought to define whether ZIKV-induced cytoskeleton remodeling was required for efficient replication. The observed microtubule displacement from the perinuclear region and the microtubule disassembly at the late time post-infection prompted us to evaluate the role of microtubules for ZIKV replication. To avoid any potential effect on viral entry, we first identified the endpoint of entry by using bafilomycin A1, an inhibitor of endosome acidification that prevents the fusion step between the viral envelope and the endosomal membrane (Yoshimori et al., 1991). Bafilomycin sensitivity was lost 3 hr post-infection, indicating that ZIKV entry and fusion were completed within this time span (Figure 2D). To examine the function of microtubules in ZIKV infection, we treated ZIKV-infected Huh7 cells with non-cytotoxic concentrations of paclitaxel (Figure 2E), a microtubule-stabilizing drug. Immunofluorescence microscopy staining of α-tubulin demonstrated that the drug performed as expected, causing the formation of thick bundles of microtubules (Figure 2F). The compound was then provided 3 hr after infection with ZIKV strains, and virus titers were analyzed 24 hr post-infection (Figure 2G). Strikingly, paclitaxel treatment strongly reduced viral titers, suggesting that microtubule dynamics is required for efficient ZIKV infection. Of note, the same phenotype was observed in hNPCs, supporting the conclusion that cytoskeleton flexibility is also required for the ZIKV replication cycle in human neural progenitor cells (Figures S3D and S3E).
Ultrastructural Analysis of Membrane Rearrangements Induced in ZIKV-Infected Cells
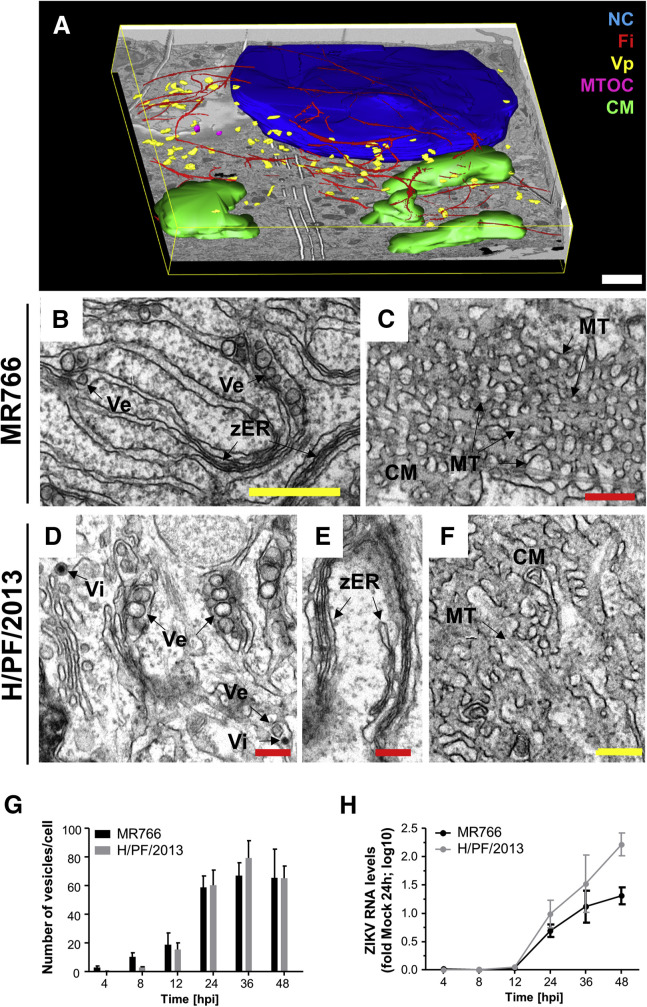
Considering the profound ZIKV-induced rearrangements of the cytoskeletal network, we characterized the ultrastructural details of these alterations in infected Huh7 cells. To this end, we applied focus ion beam (FIB)-scanning electron microscopy to obtain a 3D overview of the cytoplasmic organization of the infected cell. To identify infected cells for this analysis, we used a reporter cell line expressing the fluorescently labeled ER marker Sec61β (Chatel-Chaix et al., 2016). Given the close similarity of DENV and ZIKV, we assumed that this protein accumulates in CMs, analogous to what we reported earlier for DENV-infected cells (Chatel-Chaix et al., 2016). Huh7 cells were infected with the MR766 strain and 24 hr later, cells containing Sec61β clusters were selected by correlative light and electron microscopy (EM) and processed for subsequent FIB-scanning electron microscopy analysis (Figure 3 A). Following serial imaging and 3D reconstruction, the cytoplasm of the infected cells was found to display large areas covered by CMs with VPs located at the periphery of these structures and also accumulating in the perinuclear region (Figure 3A; Movie S2). Of note, the 3D reconstruction revealed cytoskeleton filaments surrounding most of the virus-induced VPs, while the large CM aggregates were located outside this region. This distribution nicely corroborated our immunofluorescence data described above and further implies that the perinuclear region containing both dsRNA and NS3 corresponds to the area where the majority of VPs accumulate.
Figure 3.
Ultrastructural Analysis of ZIKV-Infected Cells
(A) Huh7 YFP-Sec61β cells were infected with ZIKV MR766 (MOI = 5) for 24 hr and analyzed by FIB-scanning electron microscopy. Infected cells were identified by visual inspection for clusters of YFP-Sec61β corresponding to convoluted membranes (CMs; Chatel-Chaix et al., 2016) followed by correlative light and electron microscopy. Shown is a 3D-reconstruction of a representative infected cell: vesicle packets (Vp, yellow), cytoskeletal filaments (Fi, red), nucleus (NC, blue), convoluted membrane (green), and microtubule-organizing center (MTOC, pink). Scale bar, 2 μm. (B–F) Transmission electron microscopy (TEM) images of 70-nm-thin sections of resin-embedded Huh7 cells infected for 24 hr with ZIKV MR766 (B and C) or H/PF/2013 (D–F) (MOI = 5). (B and D) Virus-induced vesicles (Ve) clustering within the rough ER lumen. (B and E) Tightly juxtaposed and collapsed ER cisternae (zippered ER [zER]). (C and F) Microtubules (MTs) are often associated with convoluted membranes. Red scale bars, 200 nm; yellow scale bars, 500 nm. (G) Time course of accumulation of ZIKV-induced vesicles. Huh7 cells were infected with MR766 or H/PF/2013 prior to fixation at given time points and processed for TEM; 70-nm-thick sections were acquired. For quantification of vesicle numbers per cell, 10 cell profiles were randomly taken for each time point and the average number of vesicles per profile was plotted. Means ± SEM are shown. (H) Time course of accumulation of intracellular ZIKV RNA in infected Huh7 cells (MOI = 5) measured by two-step qRT-PCR. Values for each sample were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and are expressed relative to mock cells. Results from two independent experiments (three replicates each) are shown. Means ± SD are displayed.
Next, we focused our attention on the structure of ZIKV RFs and performed transmission electron microscopy (TEM) to achieve the resolution needed to analyze the ultrastructural details. Similar to our earlier observation with DENV, ZIKV-induced vesicles were found within the lumen of the rough ER. The average vesicle diameter was 80.82 ± 0.96 nm (n = 243) for the MR766 strain and 88.31 ± 1.25 nm (n = 219) for the H/PF/2013 strain (Figures 3B, 3D, and S4). Virus particles were found within the ER lumen or in isolated vesicles and had an outer diameter of around 40 nm (Figure 3D). Of note, CMs appeared to be crossed by microtubules (Figures 3C and 3F). This organization was not observed in DENV-infected cells (Welsch et al., 2009). In addition to these structures, we often observed tightly juxtaposed collapsed ER cisternae (Figures 3B and 3E) with a restricted luminal space that was reminiscent of the zippered ER found in cells infected with infectious bronchitis virus (IBV) (Maier et al., 2013).
Time-course analyses revealed that virus-induced vesicles formed as early as 4 hr and 8 hr post-infection for the MR766 and H/PF/2013 strains, respectively (Figure 3G). Vesicles accumulated through the course of infection, reaching a peak at 36 hr post-infection (Figure 3G). A sharp increase in vesicle numbers was observed between 12 hr and 24 hr post-infection, correlating with the accumulation of viral RNA levels in the infected cells (Figure 3H). This result confirms previous findings regarding the close relationship between viral replication and VP formation (Junjhon et al., 2014).
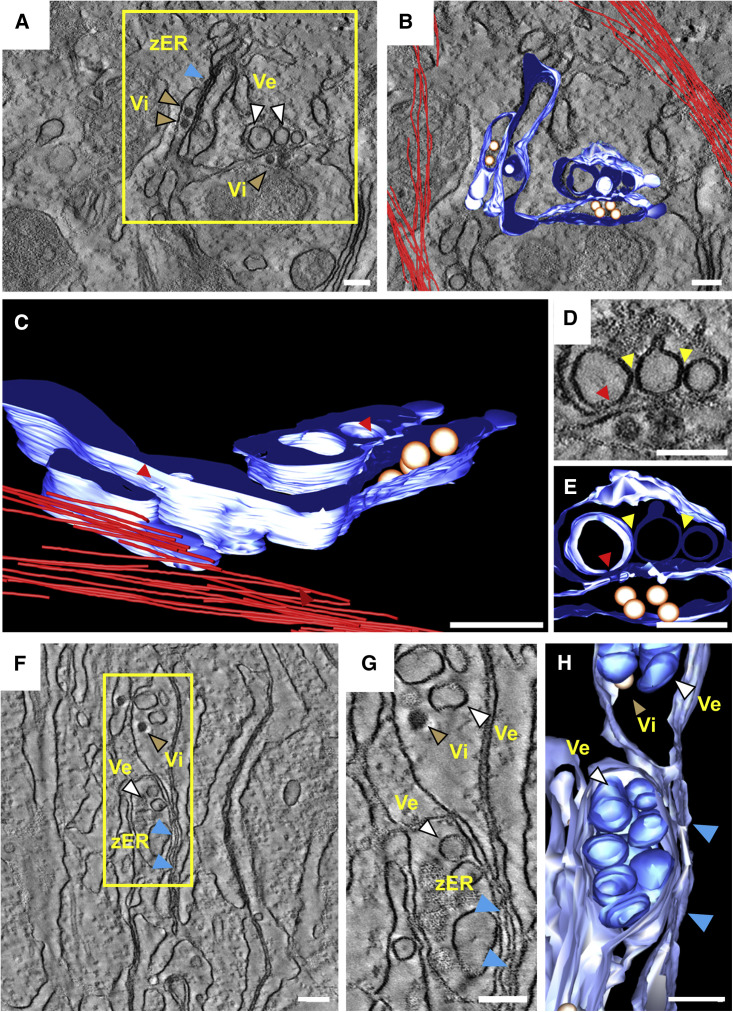
3D Architecture of ZIKV RFs
To determine the 3D organization of ZIKV RFs, we performed electron tomography on 250-nm-thick sections of Huh7 cells infected with the ZIKV H/PF/2013 strain (Figure 4 ). Consistent with the membranous alterations induced by the closely related DENV (Welsch et al., 2009) and other flaviviruses such as TBEV and WNV (Gillespie et al., 2010, Miorin et al., 2013), ZIKV induced an organelle-like compartment consisting of arrays of invaginated vesicles within dilated ER sheets; their decoration with ribosomes found in several instances (e.g., Figure 3B) argues for rough ER. Bundles of presumably intermediate filaments were observed in close proximity of the VPs (Figures 4A and 4B; Movie S3). As previously reported for other flavirivuses (Romero-Brey and Bartenschlager, 2014), the lumen of the invaginated vesicles was connected to the cytosol via a narrow pore-like structure. The calculated pore width of 10.9 ± 1.6 nm (n = 8) was comparable to the pore size observed in DENV-infected cells (11.2 ± 1.6 nm) (Welsch et al., 2009). Similar to DENV, ER sheets containing virus particles were often found adjacent to virus-induced invaginated vesicles on the opposite side of the pore (Figures 4D and 4E), suggesting that ZIKV particles bud in close proximity to the genome replication site. Although the pore-like structure was readily visible, a connection between adjacent vesicles was not observed (Figures 4D and 4E, yellow arrows). These results are similar to those previously described for DENV (Welsch et al., 2009) and TBEV (Miorin et al., 2013) but differ from what has been reported for WNV (Gillespie et al., 2010) or Langat virus (Offerdahl et al., 2012). 3D reconstruction of regions encompassing the zippered ER revealed that the collapsed ER was contiguous with regions containing invaginated vesicles (Figures 4F–4H; Movie S4), suggesting a transition from a swollen ER area filled with virus-induced vesicles to a luminally restricted area.
Figure 4.
Electron Tomography of ZIKV-Induced Vesicle Packets in Huh7 Cells
(A) Huh7 cells were fixed 24 hr post-infection, embedded in epoxy resin, and analyzed by electron tomography. A tomographic slice showing zippered ER, ZIKV virions (Vi), and virus-induced invaginated vesicles (Ve) within the ER is displayed. The yellow square represents the cropped section shown in (B) which is a 3D surface model of the ER lumen containing vesicles and virions. ER membranes are depicted in blue, virions in gold, and cytoskeletal filaments in red. (C) Side view of the 3D model showing the pore-like openings (red arrowheads) in the ER membrane. (D) Slice through the tomogram showing ER cisternae containing virions opposing ZIKV-induced vesicles. A close apposition of adjacent vesicles is visible (yellow arrowheads), but direct connections are not visible. Red arrowhead, vesicle pore. (E) Reconstruction of the area shown in (D). (F) Slice through a tomogram showing vesicle packets and details of the zippered ER (zER) (blue arrowheads). (G) Magnification of the yellow squared area shown in (F). (H) 3D reconstruction of the area in (F). Regions of zippered ER are indicated with blue arrowheads. Vesicles sharing the same ER lumen as a virion can be observed. Scale bars, 100 nm.
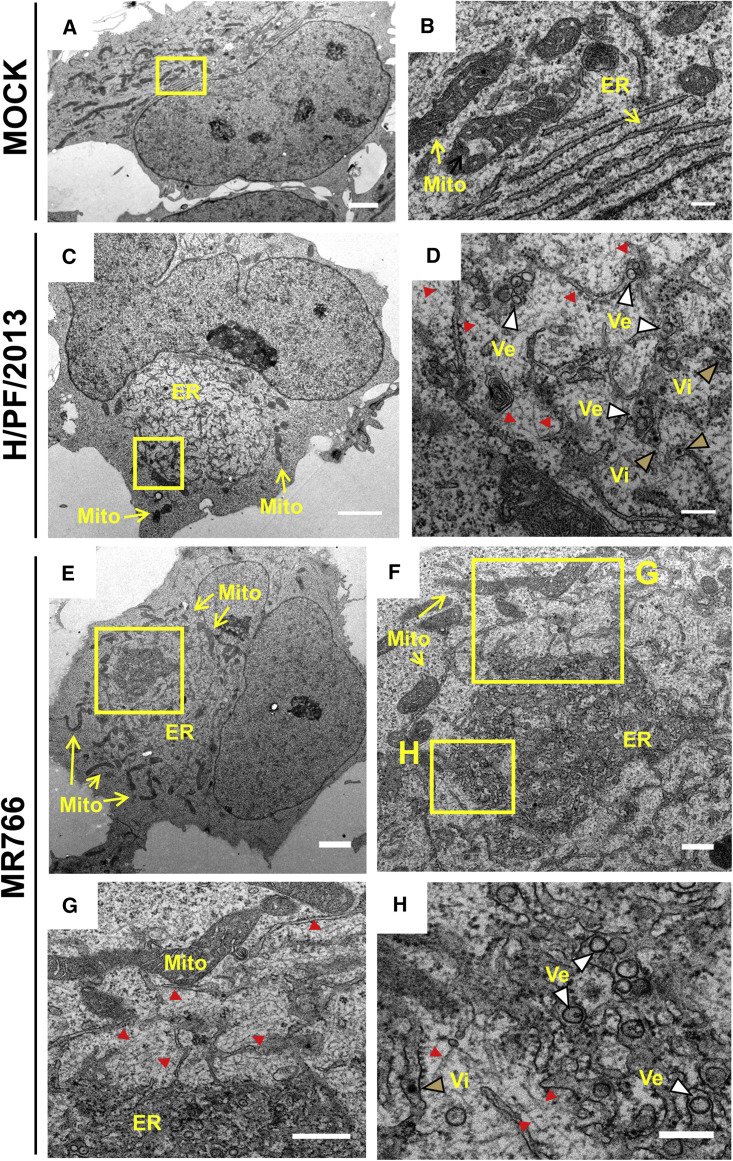
Next, we sought to analyze the ultrastructural alterations induced by ZIKV infection in hNPCs. To this end, hNPCs were infected with either ZIKV strains; 24 hr later, cells were fixed and analyzed by TEM. Inspection of mock cells revealed stacked rough ER cisternae forming an organized network surrounding the nucleus, with mitochondria dispersed throughout the cytoplasm and often in contact with the ER (Figures 5A and 5B). However, in infected cells, the rough ER appeared as a convoluted tangle of cisternae creating a large perinuclear inclusion residing on the concave side of the kidney-shaped nucleus (Figures 5C and 5E). Mitochondria accumulated on the periphery of the convoluted rough (r)ER region (Figures 5C, 5E, and 5F). Virus-induced vesicles were often observed to cluster within regions of more densely packed rER (Figures 5D, 5F, and 5G). Such areas were more frequent and prominent in MR766-infected cells (Figures 5E–5H). Cytoskeletal filaments were observed on the edge of the ER region (Figures 5D and 5G). The average diameters of ZIKV-induced vesicles was 62.80 ± 0.49 nm (n = 516) and 65.51 ± 0.56 nm (n = 457) for the African and the Asian strains, respectively. Compared to the vesicle diameters observed in Huh7 cells, ZIKV-induced vesicles in infected hNPCs appeared smaller (Figure S4), arguing that vesicle size might be cell-type dependent. Another difference among the two cell lines was the complete absence of convoluted smooth ER membranes and zippered rough ER in infected hNPCs.
Figure 5.
Ultrastructural Analysis of ZIKV-Infected hNPCs
(A–H) TEM images of 70-nm-thin sections of ZIKV-infected and resin-embedded hNPCs. Cells were mock infected (A and B) or infected (MOI = 5) with the H/PF/2013 (C and D) or MR766 (E–H) ZIKV strain for 24 hr. Yellow boxed areas are enlarged in the given panels. Overviews of ZIKV-infected cells (C and E) revealing incised nuclei and the ER inclusion. Mitochondria (mito) are located in the cell periphery adjacent to the outer rim of the ER (C and E). Virus-induced vesicles (Ve) and virions (Vi) are indicated in the magnified panels (D and H) and are marked with white and gold arrowheads, respectively. Red arrowheads, cytoskeletal filaments. Scale bars, 2 μm in (A), (C), and (E); 500 nm in (F) and (G); 200 nm in (B), (D), and (H).
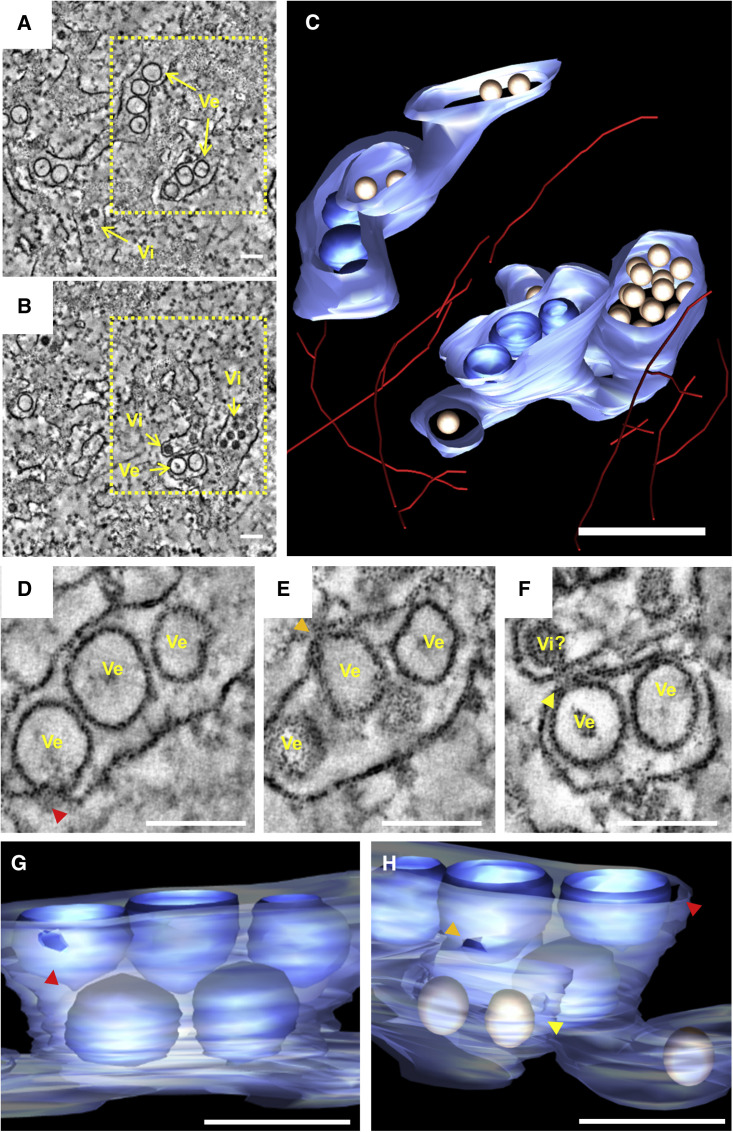
With the aim of revealing the 3D architecture of the viral RFs, we employed electron tomography analysis of hNPCs infected for 24 hr with the ZIKV H/PF/2013 strain (Figure 6 ; Movie S5). Virus-induced vesicles appeared as tightly stacked invaginations within the rough ER with narrow pores connecting the luminal side of the vesicles with the cytosol (Figures 6C–6H). Comparable to Huh7 cells, no connections were observed among adjacent vesicles in hNPCs. Quantification of pore sizes revealed an average width of 11.4 ± 1.2 nm and 10.7 ± 1.9 nm in H/PF/2013- and MR766-infected hNPCs, respectively. Interestingly, although vesicle diameters were different, pores sizes were comparable not only between hNPCs and Huh7 cells but also between the two virus strains, arguing that the vesicle pore is formed by viral proteins only or by a conserved cellular machinery. Consistent with previous observations, virions were found in close proximity to the invaginated vesicles, usually within tightly opposed ER sheets (Figures 6B and 6F). Diffuse electron density was observed on the cytosolic side of the vesicle pore, together with frequent virion budding events occurring within the opposed ER sheet (Figure 6F). 3D reconstruction of virion accumulations revealed that ZIKV particles stacked in regular arrays inside enlarged ER cisternae, which are contiguous and connected to VPs within the same ER compartment. In addition, an intricate network of intermediate filaments was present in close proximity of the ER membranes, although single filaments rather than bundles were observed.
Figure 6.
Electron Tomography of ZIKV-Induced Vesicle Packets in hNPCs
hNPCs were fixed 24 hr post-infection, embedded in epoxy resin, and analyzed by electron tomography. (A and B) A slice through a tomogram is shown depicting ZIKV-induced vesicles (Ve) within the rough ER as well as virions (Vi). (C) 3D surface model of the boxed area in (A), showing virus-induced vesicles, virions, and intermediate filaments. ER membranes are depicted in light blue, vesicles in dark blue, virus particles in gold, and filaments in red. ER tubules connected the virions with the invaginated vesicles. (D–F) Slice through the tomogram showing the pore-like openings (colored arrowheads) of ZIKV-induced vesicles toward the cytoplasm. A potential ZIKV budding event (Vi?) on the ER tubule opposing the vesicle pore can be observed in (F). (G and H) Reconstruction of the areas shown in (D) to (F). Colors of arrowheads refer to the vesicle pores marked in (D) and (E). Scale bars, 100 nm in (A), (B), and (D–H); 200 nm in (C).
In conclusion, these results reveal commonalities and differences of the 3D architecture of presumed ZIKV RFs in a human hepatic cell and hNPCs. While the overall architecture is well comparable to the one we reported earlier for DENV (Welsch et al., 2009), the utilization and perturbation of the cytoskeletal network by ZIKV sets it apart from DENV.
Discussion
In this study, we report the high-resolution 3D architecture of ZIKV RFs in Huh7 human hepatic cells (a standard cell system for flaviviruses) and, more importantly, in hNPCs, one of the physiological targets of ZIKV. While Offerdahl et al. (2017) recently reported low-resolution electron tomography of ZIKV-induced vesicles in infected neuroblastoma cells, our study unveils detailed features of ZIKV RFs in hNPCs. Similar to DENV, WNV, and TBEV (Bílý et al., 2015, Gillespie et al., 2010, Junjhon et al., 2014, Miorin et al., 2013, Welsch et al., 2009), ZIKV-induced VPs are also located within the ER lumen, implying that the vesicles originate from this organelle. Hence, all flaviviruses appear to share similar hallmarks of the architecture of their respective RFs. Since DENV VPs were shown to be associated with dsRNA (Junjhon et al., 2014, Welsch et al., 2009), these structures are believed to be the site of RNA replication. This spatial segregation would allow concentrating metabolites required for RNA replication while protecting viral RNA from cellular nucleases and innate immunity-triggering cytosolic RNA sensors. Furthermore, we observed a pore-like opening toward the cytosol allowing the exit of neo-synthesized positive-strand viral RNA into the cytoplasm, where it could be used for protein synthesis or virion formation. Very interestingly, electron-dense structures resembling virions could be detected in ER regions that were juxtaposed to the pore of VPs (Figures 6F and 6H). This suggests that particle assembly (including viral RNA encapsidation) and budding are spatially coordinated with RNA replication, reminiscent of what we reported for DENV (Welsch et al., 2009).
No obvious differences between the MR766 and H/PF/2013 strains were detected in terms of VP ultrastructural features, implying that the African and the Asian lineages use similar strategies to induce their RFs. The mean diameter of ZIKV-induced vesicles in Huh7 cells is 81 nm and 88 nm for MR766 and H/PF/2013, respectively, comparable with the reported size of DENV2 VPs (80–90 nm) (Welsch et al., 2009). Interestingly, the vesicle diameter was lower in NPCs, independent of the ZIKV lineage (63–66 nm), even though these cells were as permissive as Huh7 cells in terms of virus yield (Figures 1C and S2D). This result suggests that cell-specific determinants influence the morphogenesis of ZIKV RFs. Whether this is due to cell-specific differences in the stoichiometry of viral proteins, RNA, and host factors within ZIKV VPs remains to be determined.
In Huh7 cells, both MR766 and H/PF/2013 induced CMs comparable to DENV, WNV, and TBEV (Gillespie et al., 2010, Welsch et al., 2009). Interestingly, arrays of microtubules passing through CMs could be detected, a phenotype that was never observed in DENV-infected Huh7 cells (M.C. and R.B., unpublished data). Using live-cell imaging, we recently showed that DENV CMs are highly dynamic structures, which also undergo merging and fission events (Chatel-Chaix et al., 2016). Hence, in the case of ZIKV infection, CM-associated microtubules might influence mobility and morphology of this dynamic membranous structure.
The zippered ER observed in ZIKV-infected cells was reminiscent of structures observed during infection with IBV, an avian gamma-coronavirus generating ER-derived double-membrane vesicles and spherules (Maier et al., 2013). As for IBV, the precise role of zippered ER for ZIKV replication remains unknown, although they appear to be unique to ZIKV in the genus Flavivirus. Of note, CMs and zippered ER were not found in NPCs. This was probably not due to virus replication, since virus yields were comparable at the time point of TEM analysis (24 hr post-infection). This discrepancy might be due to cell-type-specific differences in the availability of host factors required for CM and zippered ER biogenesis. In any case, the physiological relevance of these structures for ZIKV-associated neurogenesis defects is questionable, although important roles for other aspects of viral pathogenesis cannot be excluded.
Taking advantage of various imaging techniques, we demonstrate that ZIKV induces major rearrangements of the cytoskeleton. Indeed, both intermediate filaments and microtubule networks are reorganized to form a cage-like structure in which dsRNA, NS3, and remodeled ER accumulate. Microtubule dynamics play a critical role in ZIKV replication, as deduced from the strong antiviral effect of the microtubule-stabilizing drug paclitaxel (Figures 2G and S3E). Interestingly, DENV, WNV, JEV, and TBEV also induce cytoskeleton remodeling (Chen et al., 2008, Chiou et al., 2003, Ng and Hong, 1989, Růzek et al., 2009, Teo and Chu, 2014). In fact, treatment with a microtubule-destabilizing drug enhances DENV production (Chen et al., 2008), suggesting that DENV and ZIKV have developed similar cytoskeleton remodeling strategies. Yeast two-hybrid-based interaction screenings identified the intermediate filament vimentin as an interactant of flaviviral NS5 (Le Breton et al., 2011). Moreover, vimentin was reported to interact with the N terminus of DENV NS4A and to colocalize with replication complexes (Teo and Chu, 2014). In contrast, we found that intermediate filaments surround ZIKV RFs in the perinuclear (MTOC-positive) area, arguing for differences between ZIKV and DENV cytoskeleton remodeling.
The cytoskeleton cage observed here for ZIKV might contribute to concentrate the different ZIKV-induced membranous structures and to spatially organize replication and assembly complexes in a closed environment to which RIG-like receptors have limited access. Interestingly, such a model was recently proposed for the related hepatitis C virus (HCV). Indeed, the HCV RF excludes retinoic acid-inducible gene I (RIG-I) by the recruitment of specific nuclear pore proteins (Neufeldt et al., 2016). Considering the accepted role of the cytoskeleton in innate immunity (Mostowy and Shenoy, 2015), it is tempting to speculate that ZIKV-mediated remodeling of the cytoskeleton might interfere with antiviral responses to favor viral replication. This might be either by direct contribution of the cytoskeleton or indirectly by promoting RF formation. In support to the latter assumption, evidence has been presented that both TBEV and the flavivirus Kunjin virus shield viral dsRNA in ER membrane invaginations from sensing by RIG-like receptors (Hoenen et al., 2007, Overby et al., 2010). Further work (e.g., by using pharmacological inhibitors) will be needed to unravel the possible role of the cytoskeleton in the biogenesis of the ZIKV RF and how ZIKV-induced cytoskeleton remodeling might interfere with the innate immune response.
Cytoskeleton dynamics is a crucial factor of neurogenesis. For instance, vimentin knockout mice have an altered morphology of Purkinje cells in the cerebellar cortex (Colucci-Guyon et al., 1999). In addition, neurofilaments, a sub-class of intermediate filaments, determine the diameter of myelinated axons (Elder et al., 1999) and a mutation in β3-tubulin is associated with defects of commissural axons and cranial nerves (Tischfield et al., 2010). Of note, intermediate filaments also possess cytoprotective functions such as apoptosis inhibition, organelle anchoring, and regulation of cell growth as well as motility (Snider and Omary, 2014, Toivola et al., 2010). It is therefore tempting to speculate that the remodeling of the cytoskeleton by ZIKV might contribute to the severe fetal neurogenesis defects following congenital viral transmission. Deciphering the molecular mechanisms regulating the biogenesis of ZIKV RFs might help to develop novel therapies suppressing virus replication and reducing the risk of congenital transmission and microcephaly.
Experimental Procedures
Further details of experimental procedures used are given in the Supplemental Experimental Procedures.
Cell Lines
Huh7, 293T, and VeroE6 cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich), 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM L-glutamine, and 1% non-essential amino acids (all from Gibco, Life Technologies). C6/36 mosquito cells were grown in Leibovitz L-15 medium (Gibco, Life Technologies) supplemented with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, 2 mM L-glutamine, and 10 mM HEPES (Gibco, Life Technologies). Huh7 yellow fluorescent protein (YFP)-Sec61β cells (Chatel-Chaix et al., 2016) were additionally supplemented with 1 μg/mL puromycin. Generation and maintenance of hNPCs are specified in the Supplemental Experimental Procedures.
Immunofluorescence Microscopy
For immunofluorescence microscopy, cells grown on glass coverslips were washed twice in cold PBS before fixation in 100% ice-cold methanol for 10 min at −20°C. Methanol was removed and samples were air-dried at room temperature. Fixed cells were incubated for 1 hr in blocking buffer (5% BSA in PBS). Samples were incubated with primary antibodies diluted in blocking buffer for 2 hr. After washing thrice with PBS, samples were incubated for 1 hr with Alexa Fluor 488-, 568-, or 647-conjugated secondary antibodies (Molecular Probes, Life Technologies) in the dark and subsequently washed three times with PBS for 10 min each. Nuclear DNA was stained with 4,6-diamidino-2-phenylindole, dihydrochloride (DAPI; Sigma-Aldrich) for 10 min, washed with PBS, and rinsed with ultrapure water. Coverslips were mounted on microscopy slides using Fluoromount G (SouthernBiotech). Fluorescence images were acquired with a Nikon Ti Eclipse microscope and analyzed with the NIS-Element AR software package. For 3D reconstruction, 130-nm optical sections were acquired with a Nikon TE2000-E confocal microscope equipped with an Ultraview ERS spinning disc (PerkinElmer). Images were deconvoluted with Autoquant X3 software (Media Cybernetics) and 3D reconstruction was performed with Imaris 8 (Bitplane).
TEM and Tomography
Huh7 cells were fixed with 2.5% glutaraldehyde in cacodylate buffer (50 mM cacodylate, 50 mM KCl, 2.6 mM CaCl2, 2.6 mM MgCl2, and 2% sucrose) for at least 1 hr at room temperature. Samples were incubated with 2% osmium tetroxide in 50 mM cacodylate buffer for 40 min on ice, washed three times with EM-grade water and incubated for 30 min in 0.5% uranyl acetate in water. Samples were rinsed three times with water, dehydrated in a graded ethanol series (from 40% to 100%) at room temperature before being embedded in epoxy resin, and polymerized for at least 48 hr at 60°C. Ultrathin sections of 70 nm were obtained by sectioning with a Leica EM UC6 ultramicrotome (Leica Microsystems) and a diamond knife. Sections were collected on EM grids, counterstained using lead citrate and uranyl acetate, and examined with a Zeiss 10C transmission electron microscope (80 kV; Carl Zeiss).
For electron tomography, 250-nm-thick sections were prepared. Counterstained and gold-labeled grids were placed in a high-tilt holder and digital images were recoded as single-axis tilt series over a −53° to 53° tilt range (increment, 1°) on an FEI Tecnai TF20 (Figure 4) or dual-axis tilt series over a −60° to 60° tilt range on an FEI Tecnai TF30 microscope (Figure 6). Tomograms were reconstructed using IMOD software (Kremer et al., 1996) (http://bio3d.colorado.edu/imod).
Author Contributions
M.C. and R.B. designed the study. M.C. and S.G. conducted most of the experiments. M.C. analyzed the data. E.G.A. performed drug treatments, C.J.N. performed part of the immunofluorescence microscopy, O.O. and M.L. performed STED microscopy, U.H. and C.F. provided expertise and technical assistance for EM sample preparation, C.F. acquired tomograms of infected Huh7 cells, N.S. conducted FIB-scanning electron microscopy analysis, M.S. and P.R. acquired tomograms of infected hNPCs, A.R. performed live-cell imaging, P.P. developed and provided hNPCs, and Y.S. provided equipment and expertise. M.C., A.R., L.C.-C., and R.B. wrote the manuscript. R.B. supervised the study.
Acknowledgments
We gratefully acknowledge the expert support provided by the Electron Microscopy Core Facility at Heidelberg University, the Flow Cytometry and FACS Core Facility of the Center for Molecular Biology of Heidelberg University (ZMBH), and the Electron Microscopy Core Facility at the European Molecular Biology Laboratory (EMBL) Heidelberg. We thank the Advanced Light Microscopy Facility (ALMF) at the European Molecular Biology Laboratory (EMBL) and Leica Microsystems for support. We are grateful to Dr. Vibor Laketa and the Infectious Diseases Imaging Platform (IDIP) at the Department of Infectious Diseases, University Hospital Heidelberg, for the excellent microscopy support. We are grateful to Hilmar Bading, in whose laboratory the hNPCs were generated, to Dr. Gualtiero Alvisi for constructing the YFP-Sec61b-encoding plasmid, and to the European Virus Archive (EVAg) for provision of ZIKV strains. We are very grateful to Philipp Klein for support in cell sorting and live-cell imaging. We thank Thais Moraes for virus stock production. This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 1129 to R.B and A.R). L.C.-C. holds a Discovery Grant from the Natural Sciences and Engineering Research Council of Canada (RGPIN-2016-05584). P.P. holds a fellowship by the Excellence Cluster Cell Networks at Heidelberg University. C.J.N is funded by a European Molecular Biology Organization (EMBO) Long-Term Fellowship (ALTF 466-2016).
Published: February 28, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and five movies and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2017.02.014.
Supplemental Information
Huh7 GFP-tubulin reporter cells were infected with ZIKV MR766 at a MOI of 10 pfu per cell and time lapse microscopy was started three hours later. Images were recorded in 30 min intervals until 48 h post infection. The 488 nm (GFP) channel is shown. Naïve Huh7 GFP-tubulin reporter cells were imaged under the same conditions and served as control. Scale bar, 20 μm.
Huh7 YFP-Sec61β cells infected with the MR766 strain using an MOI of 5 were fixed 24 hr after infection. Images stacks obtained by FIB-SEM with a pixel size of 5x5 nm and z step of 8 nm were acquired and used for 3D reconstruction. Cytoskeletal filaments are shown in red, the nucleus in blue, the microtubules organizing center in pink, convoluted membranes in green and packets of virus-induced vesicles in yellow. Note the distribution of cytoskeletal filaments surrounding the vesicle packets in the perinuclear region.
Three dimensional rendering of ZIKV-induced membranes. Intermediate filaments are shown in red, virus particles in gold and ER membranes and virus-induced invaginated vesicles in blue. Note the porelike opening connecting the vesicle lumen to the cytosol.
Colored overlay shows a 3D surface model of virus-induced membranes. Virus particles are depicted in gold, ER membranes in blue and virus-induced vesicles in dark blue. Note the continuity between swollen ER regions containing invaginated vesicles and areas of zippered ER in which the lumen volume is drastically reduced.
Three dimensional rendering of ZIKV-induced membranes in hNPCs. Intermediate filaments are shown in red, virus particles in gold, ER membranes in blue and virus-induced vesicles in dark blue. Note the membrane continuity between invaginated vesicles and virus containing ER cisternae. A putative budding event can be observed within the ER cisterna opposing the pore-like opening of an invaginated vesicle.
References
- Acosta E.G., Kumar A., Bartenschlager R. Revisiting dengue virus-host cell interaction: new insights into molecular and cellular virology. Adv. Virus Res. 2014;88:1–109. doi: 10.1016/B978-0-12-800098-4.00001-5. [DOI] [PubMed] [Google Scholar]
- Adams Waldorf K.M., Stencel-Baerenwald J.E., Kapur R.P., Studholme C., Boldenow E., Vornhagen J., Baldessari A., Dighe M.K., Thiel J., Merillat S. Fetal brain lesions after subcutaneous inoculation of Zika virus in a pregnant nonhuman primate. Nat. Med. 2016;22:1256–1259. doi: 10.1038/nm.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bílý T., Palus M., Eyer L., Elsterová J., Vancová M., Růžek D. Electron tomography analysis of tick-borne encephalitis virus infection in human neurons. Sci. Rep. 2015;5:10745. doi: 10.1038/srep10745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao-Lormeau V.M., Roche C., Teissier A., Robin E., Berry A.L., Mallet H.P., Sall A.A., Musso D. Zika virus, French Polynesia, South Pacific, 2013. Emerg. Infect. Dis. 2014;20:1085–1086. doi: 10.3201/eid2006.140138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cauchemez S., Besnard M., Bompard P., Dub T., Guillemette-Artur P., Eyrolle-Guignot D., Salje H., Van Kerkhove M.D., Abadie V., Garel C. Association between Zika virus and microcephaly in French Polynesia, 2013-15: a retrospective study. Lancet. 2016;387:2125–2132. doi: 10.1016/S0140-6736(16)00651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatel-Chaix L., Cortese M., Romero-Brey I., Bender S., Neufeldt C.J., Fischl W., Scaturro P., Schieber N., Schwab Y., Fischer B. Dengue virus perturbs mitochondrial morphodynamics to dampen innate immune responses. Cell Host Microbe. 2016;20:342–356. doi: 10.1016/j.chom.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Gao N., Wang J.L., Tian Y.P., Chen Z.T., An J. Vimentin is required for dengue virus serotype 2 infection but microtubules are not necessary for this process. Arch. Virol. 2008;153:1777–1781. doi: 10.1007/s00705-008-0183-x. [DOI] [PubMed] [Google Scholar]
- Chiou C.T., Hu C.C., Chen P.H., Liao C.L., Lin Y.L., Wang J.J. Association of Japanese encephalitis virus NS3 protein with microtubules and tumour susceptibility gene 101 (TSG101) protein. J. Gen. Virol. 2003;84:2795–2805. doi: 10.1099/vir.0.19201-0. [DOI] [PubMed] [Google Scholar]
- Colucci-Guyon E., Giménez Y Ribotta M., Maurice T., Babinet C., Privat A. Cerebellar defect and impaired motor coordination in mice lacking vimentin. Glia. 1999;25:33–43. [PubMed] [Google Scholar]
- Cugola F.R., Fernandes I.R., Russo F.B., Freitas B.C., Dias J.L., Guimarães K.P., Benazzato C., Almeida N., Pignatari G.C., Romero S. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016;534:267–271. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy M.R., Chen T.H., Hancock W.T., Powers A.M., Kool J.L., Lanciotti R.S., Pretrick M., Marfel M., Holzbauer S., Dubray C. Zika virus outbreak on Yap Island, Federated States of Micronesia. N. Engl. J. Med. 2009;360:2536–2543. doi: 10.1056/NEJMoa0805715. [DOI] [PubMed] [Google Scholar]
- Elder G.A., Friedrich V.L., Jr., Margita A., Lazzarini R.A. Age-related atrophy of motor axons in mice deficient in the mid-sized neurofilament subunit. J. Cell Biol. 1999;146:181–192. doi: 10.1083/jcb.146.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie L.K., Hoenen A., Morgan G., Mackenzie J.M. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J. Virol. 2010;84:10438–10447. doi: 10.1128/JVI.00986-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel R., Dejarnac O., Wichit S., Ekchariyawat P., Neyret A., Luplertlop N., Perera-Lecoin M., Surasombatpattana P., Talignani L., Thomas F. Biology of Zika virus infection in human skin cells. J. Virol. 2015;89:8880–8896. doi: 10.1128/JVI.00354-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanners N.W., Eitson J.L., Usui N., Richardson R.B., Wexler E.M., Konopka G., Schoggins J.W. Western Zika virus in human fetal neural progenitors persists long term with partial cytopathic and limited immunogenic effects. Cell Rep. 2016;15:2315–2322. doi: 10.1016/j.celrep.2016.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton N.S., Randall G. Dengue virus and autophagy. Viruses. 2011;3:1332–1341. doi: 10.3390/v3081332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenen A., Liu W., Kochs G., Khromykh A.A., Mackenzie J.M. West Nile virus-induced cytoplasmic membrane structures provide partial protection against the interferon-induced antiviral MxA protein. J. Gen. Virol. 2007;88:3013–3017. doi: 10.1099/vir.0.83125-0. [DOI] [PubMed] [Google Scholar]
- Junjhon J., Pennington J.G., Edwards T.J., Perera R., Lanman J., Kuhn R.J. Ultrastructural characterization and three-dimensional architecture of replication sites in dengue virus-infected mosquito cells. J. Virol. 2014;88:4687–4697. doi: 10.1128/JVI.00118-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., McIntosh J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- Le Breton M., Meyniel-Schicklin L., Deloire A., Coutard B., Canard B., de Lamballerie X., Andre P., Rabourdin-Combe C., Lotteau V., Davoust N. Flavivirus NS3 and NS5 proteins interaction network: a high-throughput yeast two-hybrid screen. BMC Microbiol. 2011;11:234. doi: 10.1186/1471-2180-11-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C., Xu D., Ye Q., Hong S., Jiang Y., Liu X., Zhang N., Shi L., Qin C.F., Xu Z. Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell. 2016;19:120–126. doi: 10.1016/j.stem.2016.04.017. [DOI] [PubMed] [Google Scholar]
- Li H., Saucedo-Cuevas L., Regla-Nava J.A., Chai G., Sheets N., Tang W., Terskikh A.V., Shresta S., Gleeson J.G. Zika virus infects neural progenitors in the adult mouse brain and alters proliferation. Cell Stem Cell. 2016;19:593–598. doi: 10.1016/j.stem.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier H.J., Hawes P.C., Cottam E.M., Mantell J., Verkade P., Monaghan P., Wileman T., Britton P. Infectious bronchitis virus generates spherules from zippered endoplasmic reticulum membranes. MBio. 2013;4:e00801–e00813. doi: 10.1128/mBio.00801-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miorin L., Romero-Brey I., Maiuri P., Hoppe S., Krijnse-Locker J., Bartenschlager R., Marcello A. Three-dimensional architecture of tick-borne encephalitis virus replication sites and trafficking of the replicated RNA. J. Virol. 2013;87:6469–6481. doi: 10.1128/JVI.03456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S., Shenoy A.R. The cytoskeleton in cell-autonomous immunity: structural determinants of host defence. Nat. Rev. Immunol. 2015;15:559–573. doi: 10.1038/nri3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeldt C.J., Joyce M.A., Van Buuren N., Levin A., Kirkegaard K., Gale M., Jr., Tyrrell D.L., Wozniak R.W. The hepatitis C virus-induced membranous web and associated nuclear transport machinery limit access of pattern recognition receptors to viral replication sites. PLoS Pathog. 2016;12:e1005428. doi: 10.1371/journal.ppat.1005428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M.L., Hong S.S. Flavivirus infection: essential ultrastructural changes and association of Kunjin virus NS3 protein with microtubules. Arch. Virol. 1989;106:103–120. doi: 10.1007/BF01311042. [DOI] [PubMed] [Google Scholar]
- Offerdahl D.K., Dorward D.W., Hansen B.T., Bloom M.E. A three-dimensional comparison of tick-borne flavivirus infection in mammalian and tick cell lines. PLoS ONE. 2012;7:e47912. doi: 10.1371/journal.pone.0047912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offerdahl D.K., Dorward D.W., Hansen B.T., Bloom M.E. Cytoarchitecture of Zika virus infection in human neuroblastoma and Aedes albopictus cell lines. Virology. 2017;501:54–62. doi: 10.1016/j.virol.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overby A.K., Popov V.L., Niedrig M., Weber F. Tick-borne encephalitis virus delays interferon induction and hides its double-stranded RNA in intracellular membrane vesicles. J. Virol. 2010;84:8470–8483. doi: 10.1128/JVI.00176-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D., Bartenschlager R. Architecture and biogenesis of plus-strand RNA virus replication factories. World J. Virol. 2013;2:32–48. doi: 10.5501/wjv.v2.i2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson T.C., Graham B.S. Zika virus: immunity and vaccine development. Cell. 2016;167:625–631. doi: 10.1016/j.cell.2016.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Brey I., Bartenschlager R. Membranous replication factories induced by plus-strand RNA viruses. Viruses. 2014;6:2826–2857. doi: 10.3390/v6072826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Růzek D., Vancová M., Tesarová M., Ahantarig A., Kopecký J., Grubhoffer L. Morphological changes in human neural cells following tick-borne encephalitis virus infection. J. Gen. Virol. 2009;90:1649–1658. doi: 10.1099/vir.0.010058-0. [DOI] [PubMed] [Google Scholar]
- Snider N.T., Omary M.B. Post-translational modifications of intermediate filament proteins: mechanisms and functions. Nat. Rev. Mol. Cell Biol. 2014;15:163–177. doi: 10.1038/nrm3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Hammack C., Ogden S.C., Wen Z., Qian X., Li Y., Yao B., Shin J., Zhang F., Lee E.M. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell. 2016;18:587–590. doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo C.S., Chu J.J. Cellular vimentin regulates construction of dengue virus replication complexes through interaction with NS4A protein. J. Virol. 2014;88:1897–1913. doi: 10.1128/JVI.01249-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischfield M.A., Baris H.N., Wu C., Rudolph G., Van Maldergem L., He W., Chan W.M., Andrews C., Demer J.L., Robertson R.L. Human TUBB3 mutations perturb microtubule dynamics, kinesin interactions, and axon guidance. Cell. 2010;140:74–87. doi: 10.1016/j.cell.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toivola D.M., Strnad P., Habtezion A., Omary M.B. Intermediate filaments take the heat as stress proteins. Trends Cell Biol. 2010;20:79–91. doi: 10.1016/j.tcb.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S., Miller S., Romero-Brey I., Merz A., Bleck C.K., Walther P., Fuller S.D., Antony C., Krijnse-Locker J., Bartenschlager R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimori T., Yamamoto A., Moriyama Y., Futai M., Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J. Biol. Chem. 1991;266:17707–17712. [PubMed] [Google Scholar]
- You J., Hou S., Malik-Soni N., Xu Z., Kumar A., Rachubinski R.A., Frappier L., Hobman T.C. Flavivirus infection impairs peroxisome biogenesis and early antiviral signaling. J. Virol. 2015;89:12349–12361. doi: 10.1128/JVI.01365-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Huh7 GFP-tubulin reporter cells were infected with ZIKV MR766 at a MOI of 10 pfu per cell and time lapse microscopy was started three hours later. Images were recorded in 30 min intervals until 48 h post infection. The 488 nm (GFP) channel is shown. Naïve Huh7 GFP-tubulin reporter cells were imaged under the same conditions and served as control. Scale bar, 20 μm.
Huh7 YFP-Sec61β cells infected with the MR766 strain using an MOI of 5 were fixed 24 hr after infection. Images stacks obtained by FIB-SEM with a pixel size of 5x5 nm and z step of 8 nm were acquired and used for 3D reconstruction. Cytoskeletal filaments are shown in red, the nucleus in blue, the microtubules organizing center in pink, convoluted membranes in green and packets of virus-induced vesicles in yellow. Note the distribution of cytoskeletal filaments surrounding the vesicle packets in the perinuclear region.
Three dimensional rendering of ZIKV-induced membranes. Intermediate filaments are shown in red, virus particles in gold and ER membranes and virus-induced invaginated vesicles in blue. Note the porelike opening connecting the vesicle lumen to the cytosol.
Colored overlay shows a 3D surface model of virus-induced membranes. Virus particles are depicted in gold, ER membranes in blue and virus-induced vesicles in dark blue. Note the continuity between swollen ER regions containing invaginated vesicles and areas of zippered ER in which the lumen volume is drastically reduced.
Three dimensional rendering of ZIKV-induced membranes in hNPCs. Intermediate filaments are shown in red, virus particles in gold, ER membranes in blue and virus-induced vesicles in dark blue. Note the membrane continuity between invaginated vesicles and virus containing ER cisternae. A putative budding event can be observed within the ER cisterna opposing the pore-like opening of an invaginated vesicle.