Abstract
A critical challenge for all organisms is to carefully control the amount of lipids they store. An important node for this regulation is the protein coat present at the surface of lipid droplets (LDs), the intracellular organelles dedicated to lipid storage. Only limited aspects of this regulation are understood so far. For the probably best characterized case, the regulation of lipolysis in mammals, some of the major protein players have been identified, and it has been established that this process crucially depends on an orchestrated set of protein-protein interactions. Proteomic analysis has revealed that LDs are associated with dozens, if not hundreds, of different proteins, most of them poorly characterized, with even fewer data regarding which of them might physically interact. To comprehensively understand the mechanism of lipid storage regulation, it will likely be essential to define the interactome of LD-associated proteins.
Previous studies of such interactions were hampered by technical limitations. Therefore, we have developed a split-luciferase based protein-protein interaction assay and test for interactions among 47 proteins from Drosophila and from mouse. We confirmed previously described interactions and identified many new ones. In 1561 complementation tests, we assayed for interactions among 487 protein pairs of which 92 (19%) resulted in a successful luciferase complementation. These results suggest that a prominent fraction of the LD-associated proteome participates in protein-protein interactions.
In targeted experiments, we analyzed the two proteins Jabba and CG9186 in greater detail. Jabba mediates the sequestration of histones to LDs. We successfully applied our split luciferase complementation assay to learn more about this function as we were e.g. able to map the interaction between Jabba and histones. For CG9186, expression levels affect the positioning of LDs. Here, we reveal the ubiquitination of CG9186, and link this posttranslational modification to LD cluster induction.
Lipid droplets (LDs)1 are the principal lipid storage organelles from bacteria to humans (1, 2). A hydrophobic core harboring the storage lipids is surrounded by a phospholipid hemimembrane associated with proteins. During the last decade, numerous proteomics studies have characterized the LDs isolated from a wide variety of sources, including cultured cells, various tissues, organs, and organisms (examples include: (3–13)). These proteomes typically share a core-set of about 50 metabolic and structural proteins, and in addition encompass several hundred proteins that vary depending on the source of LDs and the mass spectrometry technique used. The best characterized LD proteins are the mammalian members of the Perilipin family (14) and the proteins involved in stimulated lipolysis in white adipocytes (15). But the vast majority of LD-associated proteins still awaits a detailed functional characterization.
Our understanding of interactions among LD proteins is even less advanced. The mammalian lipolysis machinery can be considered as a multienzyme complex that depends on highly regulated protein-protein interactions (15). But apart from these interactions only few interactions among LD-associated proteins have been identified so far. In part, this dearth of knowledge may be because of the fact that classical yeast-2-hybrid interaction assays do not work well for membrane-associated proteins. In addition, abundant lipid stores are typically absent in many experimental systems, unless fatty acids are supplied exogenously.
In order to circumvent such obstacles, we adapted a split Gaussia princeps luciferase complementation assay (16) to Drosophila melanogaster tissue culture cells. This assay system allows the detection of protein-protein interactions (PPI) within intact cells or cell lysates. The highly sensitive luciferase amplification reaction yields high signal-to-background ratios. This assay was able to correctly reproduce known interactions between mammalian LD proteins, such as for the Perilipin5 - CGI-58 - ATGL ternary complex (17–20). We then generated a library of LD-associated proteins from Drosophila based on a previous proteomics screen which had characterized LDs isolated from the lipid storage organs of Drosophila third instar larvae (6). This library includes both characterized and uncharacterized Drosophila and mammalian LD proteome constituents. In total, we performed more than 1500 PPI assays testing for interactions among 487 luciferase fragment fusion protein pairs (covered by 830 different construct combinations). Of those, 85 (17.45%) resulted in a significant luciferase complementation. In proof-of-principle experiments, we confirmed selected interactions by independent, secondary assays and employed the assay to identify structural features necessary for the protein-protein interactions.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
The main body of the study consists of the split luciferase protein-protein interaction assay. During assay development and screening, we used untransfected cells, or cells transfected with only one construct (thus unable to show complementation), as baseline negative controls. Additionally, we used the previously described homotypic yeast GCN4 leucine zipper dimerization (16) as positive control and for thresholding purposes. In every assay, all samples were tested in triplicate. Triplicates showed very little variability. Based on the low assay-to-assay variance (e.g. Fig. 3), as well as the general reproducibility and dynamic range of the assay (see below), the complementation experiments result in high-confidence data. The confocal microscopy data show representative images of single cells. Comparable localizations and results were obtained in at least three biologically independent experiments.
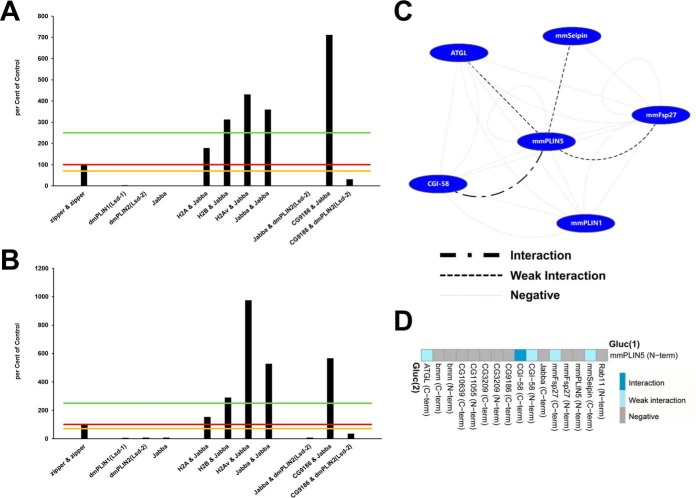
Fig. 3.
The split luciferase complementation assay shows excellent reproducibility and dynamic range. In order to test the performance and reproducibility of the Gaussia princeps split luciferase complementation PPI assay, we transfected cells with the same set of constructs in two biologically independent experiments (A, B): the known leucine zipper interaction pair, single expression plasmids—which thus could not result in a complementation and serve as a negative control—as well as several cotransfections with interacting as well as noninteracting protein pairs. Luciferase reads were normalized to total cellular protein and expressed as percent of the leucine zipper reference protein-protein interaction results. Different significance levels for interaction scoring were introduced: 0 to 70% of zipper-zipper—negative interaction; 70–100% of zipper-zipper—weak interaction; 100–250% of zipper-zipperinteraction; above 250% of zipper-zipper—strong interaction. Significance threshold levels are marked in (A, B) by orange, red, and green lines, respectively. The replicate experiment resulted in concordant scoring of the respective interaction tests. Absolute values, however, can differ between independent experiments (e.g. H2Av&Jabba, or CG9186&Jabba). C, Interactions among mammalian LD-associated proteins. Nodes represent proteins, edges symbolize luciferase complementation test results. A legend for the different edge types is provided. D, Luciferase complementation assay results for the murine PLIN5 protein tagged at the N terminus with the Gluc(1) fragment and different putative interactors. A color-code of the complementation assay results is provided. For protein combinations tested multiple times, the highest luciferase complementation result is visualized.
Drosophila Tissue Culture
Drosophila Kc167 cells obtained from the Harvard RNAi screening center were cultured in 25 cm2 T-flasks (Sarstedt, Nümbrecht, Germany) at 25 °C with no CO2 in Schneider's Drosophila medium (PAN Biotech, Aidenbach, Germany), containing 10% heat-inactivated FCS (PAN Biotech) and Penicillin/Streptomycin (Gibco BRL by Thermo Fisher Scientific Inc., Waltham, MA).
Protein Localization Studies
Drosophila Kc167 cells were seeded in eight-well chamber slides (Sarstedt, Nümbrecht, Germany) and transfected with the respective expression construct using the Qiagen Effectene (Qiagen, Hilden, Germany) transfection reagent according to the manufacturer's instructions. Cells were incubated for two to 3 days either in the absence or the presence of 400 μm oleic acid in order to induce lipid droplets. Cells were subsequently fixed with 4% paraformaldehyde and stained with 0.5 μg/ml BODIPY498/503 and with Hoechst33258 (both Molecular Probes/Invitrogen by Thermo Fisher Scientific Inc., Waltham, MA), to detect lipid droplets or nuclei/DNA, respectively. Cells were imaged using a Zeiss LSM780 confocal microscope and a 40× water immersion objective.
Molecular Biology
Protein coding sequences were cloned by polymerase chain reaction using oligonucleotides with AscI and NotI restriction sites at the 5′ or 3′ end of the genes respectively. As template, we employed cDNA preparations prepared from RNA samples originating from different Drosophila life stages or Drosophila tissue culture cell samples. The amplified sequences having or lacking Stop codons (for N- or C-terminal fusions, respectively) were directionally cloned into the pENTR Gateway plasmid system (Invitrogen, Carlsbad, CA). These Entry plasmids were subsequently recombined using the Gateway recombination cloning system (Invitrogen) in custom-made destination plasmids suitable for fusion protein expression. These plasmids were generated on the basis of a construct containing the Drosophila ubiquitin promotor (kind gift from A. Herzig). We also incorporated Gaussia princeps luciferase fragments (which we amplified by PCR or generated by gene synthesis (Genscript, Piscataway, NJ)), as published (16). The final constructs promote expression of N- or C-terminal fusion proteins, respectively (Fig. 2A). For quality control, all Entry and destination plasmids were sequenced on both strands. The recombined expression clones were confirmed by restriction digests and analytical gel electrophoresis.
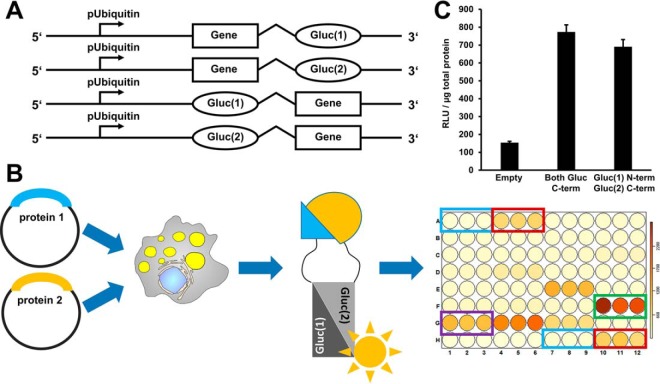
Fig. 2.
A protein-protein interaction assay based on luciferase fragment complementation. A, On the basis of a published luciferase fragment complementation assay, we constructed Gateway recombination cloning vectors suitable for the expression of luciferase fragment fusion proteins in Drosophila cells. These fusion proteins carry at the N- or C terminus either the N-terminal half of the Gaussia princeps luciferase protein (Gluc(1)) or the C-terminal half of the luciferase protein (Gluc(2)). B, In order to test for protein interactions, two plasmids encoding the potential interaction partners fused to Gluc(1) or Gluc(2), respectively, are coexpressed in cells. If the proteins interact, the luciferase fragments can assemble into an enzymatically active enzyme. Luciferase activity is measured in a 96-well plate assay format using an injector equipped luminometer plate-reader. Our standard assay plate design covered every condition/plasmid combination in triplicate. We included untransfected cells as negative controls (blue rectangle), coexpression of a leucine zipper as a positive control and for thresholding purposes (red rectangle), and the interaction between the Jabba and CG9186 proteins as a LD specific positive control (green rectangle). Protein combinations resulting in luciferase activity reads exceeding a certain threshold were scored as interaction (e.g. purple rectangle). For the different thresholding levels cf. material and methods. C, We used the dimerization of the GCN4 leucine zipper protein as a positive control. Luciferase complementation readings normalized to total protein content (RLU/μg total protein) were about ten times higher than background levels of untransfected cells. In this case, luciferase fragment positioning did not have a prominent influence on the results.
The CG9186 variant marked by the replacement of all lysine by arginine residues was generated by gene synthesis (Genscript), re-cloned into pENTR and incorporated into the different destination vectors as described above. The truncated protein variants and internal deletions of CG9186 and Jabba were generated by PCR mutagenesis. All oligonucleotides are listed in supplemental Table S1.
Gaussia-princeps Split Luciferase Protein-Protein Interaction (PPI) Assay, Data Analysis and Data Visualization
For the Gaussia princeps split luciferase PPI assay we transfected Drosophila Kc167 cells in tissue culture treated 96-well plates (Sarstedt, Nümbrecht, Germany) using the Qiagen Effectene (Qiagen, Hilden, Germany) transfection reagent. Each plasmid combination was tested in triplicate in each assay plate, which always included controls in the form of (1) untransfected cells, (2) the published Gcn4 leucine zipper interaction (16) for thresholding purposes, and (3) the robust interaction between the LD-associated proteins Jabba (21) and CG9186 (22). For the standard assay, we added 400 μm OA during the transfection procedure to the cells to induce LD deposition. However, the assay also works in the presence of higher OA amounts as well as in the absence of OA (Fig. 4C, supplemental Table S2).
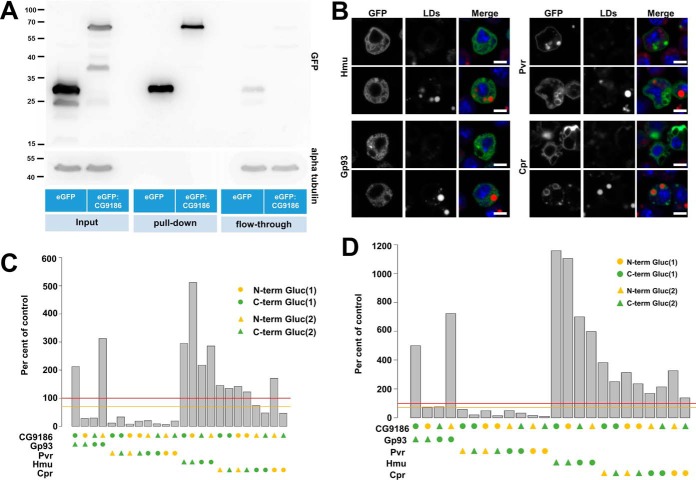
Fig. 4.
Identification and confirmation of CG9186 interaction partners by mass spectrometry and the luciferase complementation PPI assay. A, Extracts of Drosophila Kc167 cells stably expressing either GFP alone or GFP fused to CG9186 were subjected to immunoprecipitation using GFP-specific antibodies. Western blot analysis of SDS-PAGE separated samples using a GFP specific antiserum demonstrated efficient purification of GFP alone and GFP-tagged CG9186 (compare enrichment between pull-down and flow-through). Probing the membrane with an alpha tubulin-specific serum (shown below) demonstrates absence of tubulin in the pull-down fractions and comparable amounts in the input and flow-through fractions. B, Localization of candidate CG9186 interactors (supplemental Fig. S2) in Drosophila Kc167 tissue culture cells. GFP fusions of Hemomucin (Hmu), Cytochrome P450 reductase (Cpr), PDGF- and VEGF-receptor related (Pvr) or the Glycoprotein 93 (Gp93) were expressed in the presence (top rows) or absence of OA (bottom rows). In the absence of OA Hmu, Cpr, and Gp93 localize in a reticular pattern. Pvr is enriched at the plasma membrane, as reported previously (72, 73), and on intracellular membranes. The localization of Gp93 and Pvr does not change with OA addition, whereas Cpr and Hmu are now found in the vicinity of LDs. (C, D) In the split luciferase complementation assay tested against CG9186, Pvr did not show complementation of the luciferase activity, whereas Hmu, Cpr or Gp93, displayed significant luciferase activity, both in the absence (C) and presence (D) of LDs. Threshold levels are indicated as in Fig. 3. Scale bars represent 5 μm.
After a total of 4 days incubation, the cells were lysed with the lysis buffer included with the Gaussia princeps Coelenterazine substrate (Perbio by Thermo Fisher Scientific Inc.). Flash luciferase activity was subsequently measured in white opaque 96-well assay plates (Brand, Wertheim, Germany) using an injector equipped BioTek Synergy Mx multimode plate-reader (BioTek, Winooski, VT). Luciferase activity could also be measured in intact cells, and we tested formats up to 384-well plates successfully (data not shown). To be able to normalize luciferase reads to total protein, we used the lysis dependent protocol in this study. For this purpose, we measured total protein content per well using a second aliquot of the cell extracts by standard BCA protein assays (Perbio by Thermo Fisher Scientific Inc). Data analysis was performed with Microsoft Excel and the statistical R programming language with custom scripts for visualization (available upon request).
The dynamic range of the assay separated negative from positive interactions very well. For example, the leucine zipper dimerization scored with an average of 1113.30 relative light units per μg total protein (RLU/μg protein; standard error of the mean (S.E.) = 84.60) which was on average about ten times above the background signal of untransfected cells (134.42 RLU/μg protein; S.E. = 5.45). In order to catch the complete dynamic range of the assay also in terms of the interaction classification, we selected the following (arbitrary) thresholds after normalization to the leucine zipper complementation (set to 100%): (1) 0% < 70%: negative; (2) 70%≤100%: weak interaction; (3) 100%<250%: interaction; (4) ≥250%: strong interaction. Interaction networks were assembled using the Cytoscape software package (version 3.3) (23).
Affinity Purification of CG9186 Interaction Partners
We generated polyclonal Drosophila Kc167 cell lines expressing either GFP alone or a GFP-tagged version of the LD-associated protein CG9186 (22) by cotransfection of the respective expression plasmids and the Blasticidin-resistance mediating pCoBlast plasmid (Invitrogen) according to standard procedures. Both proteins were expressed constitutively under control of an ubiquitin promotor, and cells were grown in 75 cm2 flasks in the presence or absence of 400 μm OA for 24 h before we homogenized the cells in Pierce IP Lysis Buffer (Thermo Fisher Scientific Inc., Waltham, MA, USA). For the immunopurification, we used GFP-Trap_A beads (ChromoTek, Planegg-Martinsried, Germany) according to the manufacturer's instructions. Beads were incubated over night at 4 °C to allow protein binding. Samples of cells grown in the absence of OA were washed three times with 150 mm NaCl buffer, whereas LD samples were washed once with 150 mm and twice with 200 mm NaCl buffer to increase stringency. In the end, we eluted bound proteins by 200 mm glycine pH 2.5 and analyzed them by mass spectrometry.
Liquid Chromatography and Mass Spectrometry
Samples from affinity enrichment (see previous paragraph) were loaded on a 4–12% polyacrylamide gel and separated for 5 min. Each condition (GFP or GFP-tagged CG9186 as a bait in the presence or absence of oleic acid) was tested once. After silver staining, the protein containing bands were excised and processed as described (24). Briefly, bands were destained, washed, reduced with dithiothreitol, alkylated with iodoacetamide and digested with 0.05 μg trypsin (Serva, Heidelberg, Germany) in 50 mm NH4HCO3 overnight at 37 °C. Resulting peptides were extracted with 1:1 (v/v) 0.1% Trifluoroacetic acid (TFA)/acetonitrile, and after acetonitrile removal by vacuum concentration, the peptides were resuspended in 0.1% TFA.
Subsequently, the peptide mixture was analyzed with a reverse phase liquid-chromatography system coupled to a Q Exactive plus hybrid quadrupole-orbitrap mass spectrometer (Thermo Scientific, Bremen, Germany). For peptide separation, an UltiMate 3000 RSLCnano system (Thermo Scientific, Dreieich, Germany) was used. Peptides were first loaded on an Acclaim PepMap100 trap column (3 μm C18 particle size, 100 Å pore size, 75 μm inner diameter, 2 cm length, Thermo Scientific, Dreieich, Germany) for 10 min at a flow rate of 6 μl/min using 0.1% TFA as mobile phase and then separated at 60 °C on an analytical column (Acclaim PepMapRSLC, 2 μm C18 particle size, 100 Å pore size, 75 μm inner diameter, 25 cm length, Thermo Scientific, Dreieich, Germany) at a flow rate of 300 nl/min using a 2 h gradient.
The liquid chromatography system was coupled to the mass spectrometer via a nano electrospray ionization source equipped with distal coated SilicaTip emitters (New Objective, Woburn, MA). A spray voltage of 1400 V was applied and the capillary temperature set to 250 °C. The Q Exactive was operated in positive mode and full scans recorded in the orbitrap analyzer in profile mode with a resolution of 70,000 over a scan rage from 350 to 2000 m/z with a maximum ion time of 80 ms and the target value for the automatic gain control set to 3,000,000. Subsequently, up to ten precursors (charge states 2 and 3) were isolated within a 2 m/z isolation window, fragmented via higher-energy collisional dissociation and MS/MS spectra recorded in centroid mode with a maximal ion time of 60 ms and the target value for the automatic gain control set to 100,000. Here, the resolution was 17,500 at an available scan range of 200 to 2000 m/z. Already fragmented precursors were excluded from further isolation for the next 100 s.
Acquired spectra were searched within the MaxQuant version 1.5.2.8 framework against the UniProtKB Drosophila melanogaster proteome dataset (release 2015_03, 21361 sequences), including an additional entry for the green fluorescent protein (P42212). Carbamidomethyl at cysteines was set as fixed modification and acetylation at protein N termini, oxidation at methionine and lysine ubiquitinylation (glygly, mass 114.0429 Da) were considered as variable modifications as well as tryptic cleavage specificity (cleavage behind K and R) with a maximum of two missed cleavage sites. Predefined values were used for other parameters including a false discovery rate of 1% on peptide and protein level, a main search precursor mass tolerance of 4.5 ppm and mass tolerance of 20 ppm for fragment spectra. Proteins with at least two peptides were reported to be identified.
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (25) partner repository with the data set identifier PXD003944.
RESULTS
A Library of LD-associated Proteins
In order to obtain a diverse collection of proteins to assay for interactions, we chose 36 LD-associated proteins, based on a previous proteomics screen (6) and literature reports. The collection covers proteins lacking a detailed functional characterization or annotation as well as previously characterized LD-associated proteins of Drosophila such as the Perilipins dmPLIN1 and dmPLIN2 (also known as Lsd-1 and Lsd-2) (26–28), CG9186 (22), the Drosophila adipose triglyceride lipase (ATGL; (29)) homolog Brummer (30), the esterase CG1112 (31), the proteins Jabba (21) and dmSeipin (32) (see supplemental Table S3 for an overview of all tested proteins). Additionally, we cloned known mammalian LD-associated proteins, such as the Perilipins mmPLIN1 and mmPLIN5 (14), ATGL (29), its colipase CGI-58 (33), and the mmSeipin (34) and mmFsp27 (35) proteins.
We first examined the localization of the Drosophila LD-associated proteins in Drosophila Kc167 cells, by monitoring the expression of the GFP-tagged fusion proteins by confocal microscopy. LDs were induced by the addition of 400 μm oleic acid (OA). Under these conditions, we observed LD association for the bona-fide Drosophila LD-associated proteins CG9186, dmPLIN1 and dmPLIN2, CG2254, CG1112, CG11055 (the presumptive homolog of the hormone sensitive lipase) (36) and dmSeipin (32) (Fig. 1A). The previously uncharacterized proteins CG5958 and CG10691 also targeted LDs (Fig. 1B). The ATGL homologous Brummer lipase, however, could not be found at LDs under the standard 400 μm OA feeding regimen. However, Bmm:GFP positive cells showed no or only very little LD deposits, suggesting that overexpression of the active lipase depleted the lipid stores. Consistent with this hypothesis, we found a clear-cut localization of Brummer to LDs when we treated the cells with a short pulse of high OA amounts prior to fixation of the cells (Fig. 1C).
Fig. 1.
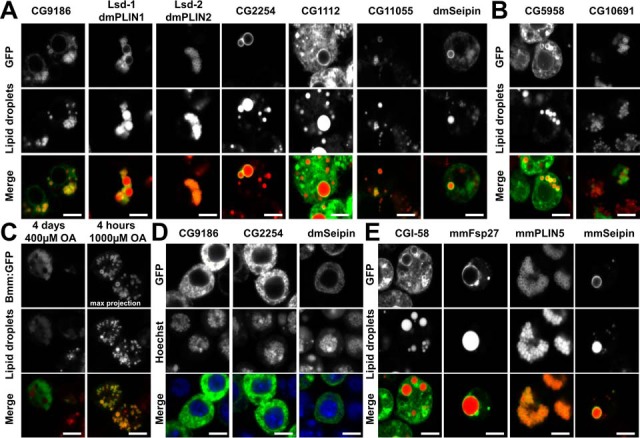
Expression of LD-associated proteins in Drosophila tissue culture cells. Expression of GFP-tagged candidate LD-associated proteins in Drosophila Kc167 tissue culture cells. Unless otherwise noted, cells were grown in the presence of 400 μm OA to induce LDs. A, Bona-fide Drosophila LD-associated proteins show the expected ring-shaped localization around LDs, characteristic of most LD proteins. Some proteins (e.g. CG1112) additionally localize in a broader pattern and appear to target membranes in addition to the LDs. B, The previously uncharacterized proteins CG5958 or CG10691 also target LDs in Drosophila tissue culture cells. C, Expression of the Brummer lipase (Bmm) results in an almost complete depletion of lipid stores, and thus no LD-localization, when the standard experimental regimen (4 days long incubation in the presence of 400 μm OA) is used. However, LD localization of Bmm is apparent when the cells are exposed to a short pulse of 1000 μm OA. D, In the absence of LDs several otherwise LD-associated proteins target the endoplasmic reticulum (ER). Expression of the CG9186, CG2254, or Seipin in the absence of OA results in a prominent reticular localization pattern. For CG9186, we had previously demonstrated that this pattern reflects ER localization (22). E, GFP-tagged mammalian LD-associated proteins heterologously expressed in Drosophila Kc167 tissue culture cells. The proteins localize in the same way as described for homologous expression in mammalian cell systems. Scale bars represent 5 μm.
In the absence of LDs, some LD-associated proteins, as for example CG9186 (22), are known to localize to the endoplasmic reticulum (ER). Indeed, CG9186, CG2254, and dmSeipin displayed a reticular pattern in cells lacking LDs consistent with ER Localization (Fig. 1D).
We also expressed known mammalian LD-associated proteins in the Drosophila Kc167 cells. The murine proteins CGI-58, mmFsp27, mmPLIN5 and mmSeipin localized to LDs as expected (Fig. 1E). As for the Brummer lipase, overexpression of ATGL resulted in highly reduced lipid storage levels (data not shown), but with a short pulse of OA feeding, ATGL localized clearly to LDs (supplemental Fig. S1).
Detecting Protein-Protein Interactions by a Split Gaussia Princeps Luciferase Assay
Our assay is based on the luciferase enzyme of the copepod Gaussia princeps which is a monomeric protein of 185 amino acids. This luciferase does not require cofactors for activity and emits blue light with a peak at 480 nm (16). Its substrate coelenterazine is cell permeable and diffuses into all cellular compartments, enabling quantitative measurements in live cells (16). In order to test for interactions among the LD-associated proteins we adapted a published split Gaussia princeps luciferase protein protein interaction (PPI) assay (16), where the luciferase enzyme is split into two similarly sized parts, to the Drosophila tissue culture system. Both luciferase fragments were cloned in Gateway recombination cloning vectors suitable to drive expression from a constitutive Drosophila ubiquitin promotor (see experimental procedures section; Fig. 2A). Luciferase fragments were positioned either at the N- or C terminus. This allowed us to test interactions with a cis- (i.e. tags on both proteins at N- or C terminus) and trans- (i.e. one protein carries tag at the N terminus and the other one at the C terminus) configuration. Thus, we could investigate whether the relative positioning of the luciferase fragments influences the signal strength, or even discriminates between a successful or lack of complementation. In our constructs, a flexible glycine-rich linker connects the proteins of interest and the luciferase fragments, to allow for a bigger steric flexibility (16) (Fig. 2A, 2B). Plasmids encoding the fusion proteins were cotransfected in Drosophila cells in a 96-well plate format to allow medium- to high-throughput screening (Fig. 2B). All transfections were tested in triplicate on each plate. A homotypic GCN4 leucine zipper interaction served as a positive control and to normalize readings on each plate (16). For the leucine zipper, there was only a mild effect of placing the luciferase fragments in cis- or trans-configuration (Fig. 2C), presumably because it is relatively small (5.8 kDa).
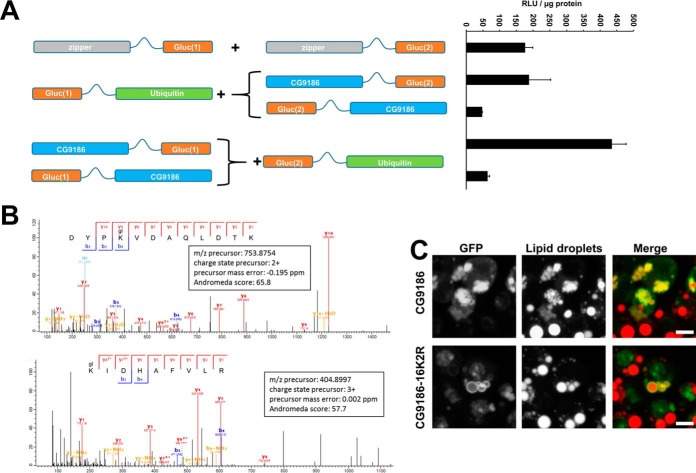
Early on, we detected a strong interaction among the Drosophila proteins Jabba (21) and CG9186 (22) (Fig. 3A, 3B), which resulted in luciferase readings clearly surpassing the ones received by the leucine zipper interaction. To capture the whole range of signal strengths observed, we normalized all readings to the values of the homotypic leucine zipper dimerization (set as 100%) and sorted interactions into three classes (whose limits were arbitrarily chosen): levels between 70 and 100% of the homotypic zipper interactions were classified as “weak interactions”, levels between 100 and 250% as “interactions,” and above 250% as “strong interactions.”
To determine the reproducibility of the assay, we repeated some interaction tests multiple times. During the course of the screen, the interaction between Jabba and CG9186 was scored positive 54 out of 57 times. Individual luciferase fragment fusions as well as several negative interactions consistently showed negligible complementation (an example of a replicate assay is shown in Fig. 3A, 3B). These results suggest that the amount of false negative and false positive interactions because of assay fluctuations is low.
The Assay Correctly Identifies Known Protein Interactions Among Mammalian LD-associated Proteins
As a first test, we assayed for interactions among selected mammalian LD-associated proteins. These proteins provide an important test case because many interactions between them had already been identified by other means (15). We tested, for example, for interactions among murine mmPLIN1 (37), mmPLIN5 (20), ATGL (33), and CGI-58 (33), which represent key components of mammalian lipolysis regulation (15). Further, we included the mmSeipin (38) and mmFsp27 (39) proteins, which are involved in LD biogenesis and LD size control. We recapitulated the published interactions between mmPLIN5 and CGI-58 (20) as well as between mmPLIN5 and ATGL (18–20) (Fig. 3C), demonstrating that interactions among mammalian proteins can be detected in our heterologous cell system. Although interaction tests between mmPLIN5 and eight additional proteins were negative (Fig. 3D), we also observed interactions between mmPLIN5 and mmFsp27 as well as between mmPLIN5 and mmSeipin (Fig. 3C, 3D), suggesting that mmPLIN5 is involved in more interactions than previously known.
Mass-spectrometry Based Identification of CG9186 Interaction Partners and Verification of Identified Candidates Via the Split-luciferase Based Assay
We next tested whether the split luciferase interaction assay is suitable to validate mass-spectrometry based identifications of de novo identified candidate interaction partners. To identify such candidates for CG9186, we expressed a GFP-fusion of the LD-associated protein CG9186 (22) in Drosophila Kc167 cells and performed anti-GFP pull-down experiments (Fig. 4A). CG9186 traffics between the ER and LDs depending on LD availability (22) (Fig. 1). We therefore performed the assay both in presence and absence of OA to probe CG9186 interactions in the two different compartments. Cells expressing GFP only served as control. Out of 412 copurifying proteins (Pride Accession # PXD003944; supplemental Table S4), we focused on four (supplemental Fig. S2): Hemomucin (Hmu), glycoprotein of 93 kDa (Gp93), Cytochrome P450 reductase (Cpr), and PDGF- and VEGF-receptor related (Pvr). Hmu, Gp93, and Cpr were previously identified in the LD proteome of third instar larval fat bodies (6). RNA-mediated knock-down of Pvr in Kc167 cells induces LD-clustering (40), similar to the phenotype upon CG9186 overexpression (22).
We first expressed the four proteins as GFP fusion proteins in Drosophila Kc167 cells. In the absence of OA, Hmu and Gp93 showed a reticular pattern throughout the cell, reminiscent of ER localization (cf. Fig. 1D). Pvr was mostly enriched at the plasma membrane, but also found in intracellular vesicles and membranes. Cpr showed a perinuclear enriched membrane localization. The presence of OA did not alter the distribution of Gp93 or Pvr, but Cpr and Hmu now displayed localization around LDs (Fig. 4B).
Next, we tested the proteins with CG9186 for luciferase complementation in the absence (Fig. 4C) or presence (Fig. 4D) of OA. For Pvr no interaction was detected, but the other three proteins showed robust interaction, irrespective of the cells' nutritional status. For Gp93, the configuration of the luciferase fragments made a dramatic difference: a significant interaction was apparent in only two of the four tested configurations. Thus, the luciferase-fragment configuration indeed can affect the outcome of the interaction tests. Also in the interactions discovered in the later experiments we often saw an impact of the luciferase fragment positioning. Thus, it is advisable to test different variants, if possible. Additionally, the results revealed that our assay is likely able to detect interactions at different membrane-bound compartments, namely the ER and LD, and therefore should be able to detect interactions in many different experimental settings.
A Screen to Detect LD-associated Protein Interactions
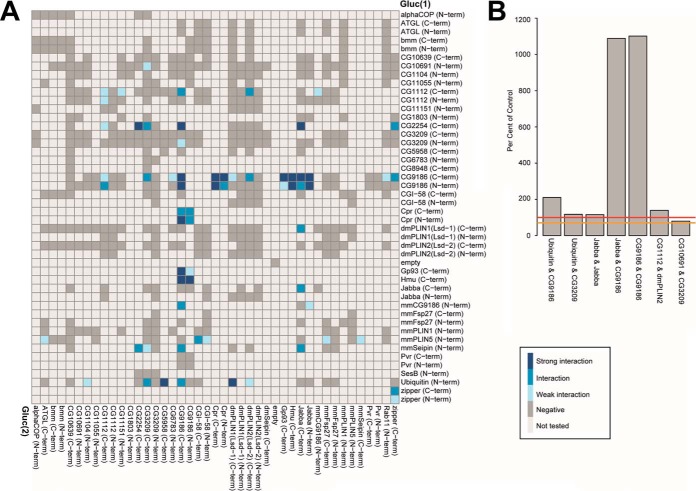
We next tested for interactions among our panel of LD-associated proteins. To focus on those interactions that might occur at the LD surface, we performed the majority of complementation assays with cells grown in the presence of 400 μm OA. Most of the tested protein combinations did not result in a relevant luciferase complementation signal and were thus classified as negative interactions (Fig. 5A). In total, we performed 862 interaction tests with this experimental regimen, covering 299 protein combinations of which 11.8% scored as “interactions” or “strong interactions” (or 17.4% if we include “weak interactions”). Examples of significant Drosophila LD-associated protein-protein interactions are shown in Fig. 5B.
Fig. 5.
Split luciferase complementation PPI results. A, Split luciferase complementation assay results for coexpression of the indicated proteins in the presence of 400 μm OA for 4 days in Drosophila Kc167 cells. A legend explaining the color coding is included in the panel. Interactions of PLIN5 were partially shown in Fig. 3D. For protein combinations tested multiple times, the highest luciferase complementation result is visualized. B, Representative interactions between Drosophila LD-associated proteins. Threshold levels are indicated as in Fig. 3.
Characterization of Selected Protein-Protein Interactions
In the following, we present follow-up experiments for a few selected examples, either to characterize the interaction further or to utilize the split luciferase complementation assay to answer biological questions. We focus on interactions involving CG9186 (22) and Jabba (21). Both proteins showed prominent homo- and heterotypic interactions in our assay (Fig. 5A) and thus were amenable to test for a possible mapping of the interactions to distinct amino acid stretches of the respective protein. These proteins also function in biological processes that are not yet well understood: Jabba sequesters nuclear proteins to LDs (21), and CG9186 protein expression levels somehow modulate the intracellular positioning of LDs (22). Deeper analysis of these two proteins therefore has the potential to illuminate the mechanistic basis for these processes.
(1) Mapping of Interaction Domains Among the CG9186 and Jabba Proteins
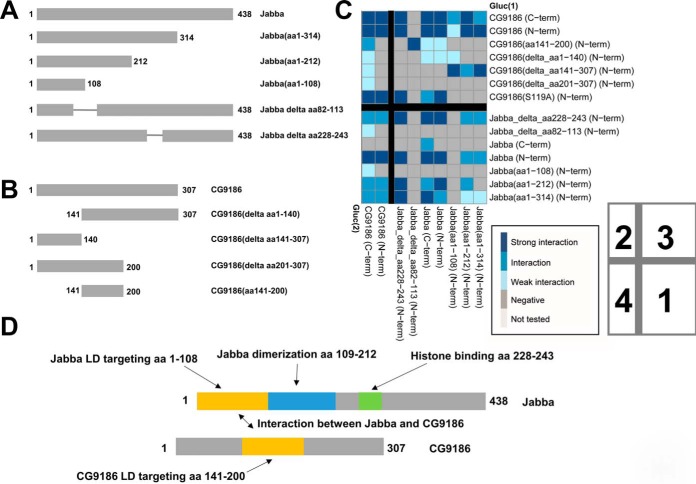
The Jabba (21) and CG9186 (22) proteins showed prominent homo- and heterotypic interactions (Fig. 5A, 5B). We thus employed the split luciferase assay to map protein regions necessary for self or mutual binding. We employed the mutant Jabba protein variants shown in Fig. 6A as well as previously generated (22) deletion constructs of CG9186 (Fig. 6B). We first asked which regions were sufficient to target GFP to the surface of LDs. For CG9186, we had previously identified amino acids 141–200 to be necessary and sufficient for LD targeting (22). For Jabba, amino acids 1–108 are sufficient (supplemental Fig. S3), though the localization to LDs was imperfect. Hydropathy analysis reveals that Jabba(1–108) breaks within an extremely hydrophobic region of Jabba (supplemental Fig. S4). Targeting to LDs often involves such highly hydrophobic motifs (41). We therefore generated a version of Jabba lacking amino acids 82 to 113, and thus deleting the entire hydrophobic region. Indeed, this variant localizes mostly in the cytoplasm and only occasionally reaches LDs (supplemental Fig. S3).
Fig. 6.
Mapping of interacting sequences using protein deletion variants and the split luciferase PPI assay. A, Truncated Jabba protein variants were generated and tested in the PPI assay as well as for their subcellular localization (supplemental Fig. S3 and supplemental Fig. S4). B, For CG9186 we utilized previously described protein variants (22). C, Matrix of interaction tests using full length and truncated variants of the CG9186 and Jabba proteins. For protein combinations tested multiple times, the highest luciferase complementation result is visualized. Schemes with the color code for the interaction scorings, and the numbering of the four quadrants as described in the main text, are included in the lower right. D, Schematic representation of the Jabba and CG9186 proteins and functional annotation of the amino acid stretches identified in our study.
For the dimerization of Jabba, complementation values are higher when the Gluc(1) fragment is placed at the N terminus (Fig. 6C; quadrant 1). Dimerization apparently depends on amino acids beyond residue 108 because a minimal LD targeting construct covering only amino acids 1 to 108 is not sufficient to allow the dimerization, yet amino acids 1 to 212 of Jabba dimerize. Dimerization additionally seems to depend on LD localization of the Jabba protein, because the mutated protein lacking amino acids 82 to 113 fails to show complementation with any other Jabba protein tested. Removal of the charged amino acid residues 228 to 243 (supplemental Fig. S4) of Jabba, in contrast, does not affect the complementation with the truncated Jabba protein versions.
CG9186 dimerization was unaffected by the placement of the luciferase fragments (Fig. 6C; quadrant 2). Each of the deletions tested was still able to dimerize, suggesting that dimerization does not depend on any single sequence stretch. However, every truncation of the protein resulted in lower luciferase activity, and complementation was only above the 70% threshold when the Gluc(2) fragment was placed at the C terminus of CG9186. Thus, dimerization may be driven by multiple additive interactions or each truncation may partially destabilize CG9186's conformation.
For the interaction between Jabba and CG9186 (Fig. 6C, quadrants 3 and 4) the placement of the luciferase fragments did not make much of a difference in the case of CG9186, whereas the Gluc(1) fragment had to be placed at the N terminus of Jabba for the interaction to be detectable. Amino acids 141 to 200 of CG9186 are sufficient for Jabba binding. This region is also necessary and sufficient for LD targeting (22). For Jabba, the smallest truncated variant spanning amino acids 1 to 108 is sufficient to interact with CG9186. Intriguingly, the CG9186 deletion construct missing amino acids 141 to 307 (22) fails to complement with the full length Jabba proteins, but interacts with all tested Jabba truncated protein versions. Because this CG9186 deletion is cytosol-localized, these results can be explained if the truncated Jabba proteins are incompletely localized to the LD surface, thus allowing interactions between two protein truncations in the cytosol. A scheme summarizing our findings is provided as Fig. 6D.
(2) Interaction Between Jabba and Histone Proteins
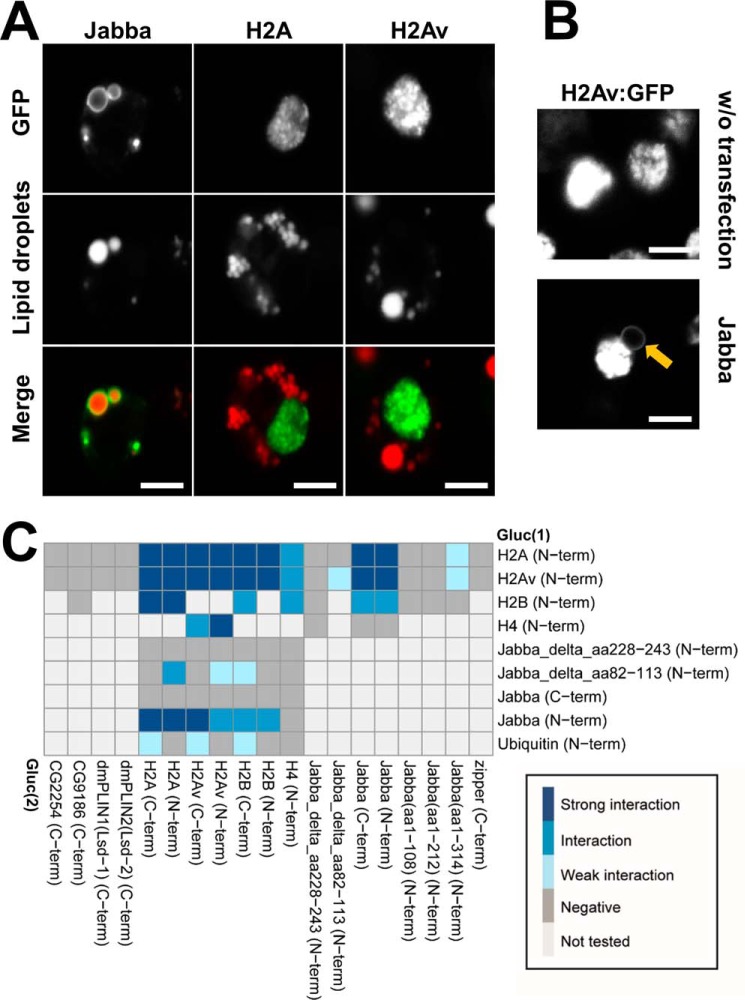
In early embryos, the LD-associated Jabba protein anchors the Histones H2A, H2B, and H2Av to LDs, allowing droplets to transiently sequester excess amounts of histones important for early embryogenesis (21). This interaction apparently does not require any embryo-specific factors as we were able to recreate it in tissue culture cells. When we expressed Jabba, H2A, or H2Av individually in Drosophila Kc167 cells in the presence of 400 μm OA, Jabba targeted the LDs, whereas the Histone proteins were found in the nucleus, as expected (Fig. 7A). However, when we coexpressed an untagged Jabba variant and the H2Av protein, the histone was not only found in the nucleus but also in the ring pattern indicative of LD localization (Fig. 7B).
Fig. 7.
Jabba recruits Histones to LDs. Characterizing Jabba's Histone recruiting ability (21) in cultured cells. A, Expression of GFP tagged Jabba, H2A, and H2Av proteins in Drosophila Kc167 cells in the presence of 400 μm OA. B, Coexpression of untagged Jabba protein and the GFP-tagged H2Av protein results in recruitment of H2Av to a ring-shaped subcellular localization reminiscent of LDs (highlighted by the orange arrow). C, Matrix summarizing the interaction tests between Jabba and the Histone proteins. A legend for the color coding of interaction strengths is provided. For protein combinations tested multiple times, the highest luciferase complementation result is visualized. Scale bars represent 5 μm.
In embryos, Jabba and H2Av coimmunoprecipitate, suggesting recruitment of histones to LDs is mediated by physical interaction with Jabba (21). This intimate interaction between Jabba and H2Av was also evident in the luciferase complementation assay (Fig. 7C). In addition, Jabba displayed interactions with histones H2A and H2B, but not H4 (Fig. 7C). These results reflect Jabba's specificity in vivo, where H2A, H2B, and H2Av, but not H4 (21), are found on LDs. The H4 constructs are indeed competent to interact, as we detected complementation of H4 with the histone proteins H2A, H2B, and H2Av. Overall, we detected extensive interactions among all the histone proteins (Fig. 7C), as expected from their proximity to each other in nucleosomes. H2A and H2Av did not display detectable interactions with other LD-associated proteins tested, such as the fly Perilipins dmPLIN1 and dmPLIN2, CG2254, or CG9186 (Fig. 5A). These observations support specificity of the interaction between Jabba and histones.
Histones are highly positively charged proteins, and histone binding to LDs—and thus presumably to Jabba—depends on electrostatic interactions (4). We therefore speculated that negative charges in Jabba play an important role in Jabba-histone interactions. Charge distribution in Jabba is highly asymmetric (supplemental Fig. S4C): the region between residues 225 and 268 is devoid of positively charged amino acids, and the density of negative charges is high. In particular, there is a highly acidic stretch between residues 228 and 243: 11 out of 16 amino acids are negatively charged (supplemental Fig. S4). A Jabba protein lacking amino acids 228–243 is able to bind to LDs (supplemental Fig. S3) and displays robust interactions with CG9186 (Fig. 6C, quadrants 3 and 4). However, it showed no interactions with the histone proteins, suggesting that the acidic stretch is indeed necessary for Histone binding (Figs. 7C and supplemental Fig. S4).
The Jabba locus encodes multiple protein isoforms that all share a common N-terminal region and differ in their C termini (21) (supplemental Fig. S4A). In the studies reported, we employed isoform I. However, all the structural elements we mapped (for dimerization, LD targeting, histone interaction) are present in the shared region and thus likely function similarly in all the isoforms.
(3) Ubiquitination of CG9186 is Important for its LD-cluster Inducing Phenotype
Numerous experiments suggest important roles for ubiquitin in LD biology. A number of LD-associated proteins are ubiquitinated (42–45) and components of the ubiquitination machinery localize to the LD surface (46–50). For example, ubiquitination triggers the degradation of certain LD-associated proteins in the absence of LDs (42, 45, 51) and can alter the spatial distribution of LDs within cells (44). How ubiquitin promotes these changes is unknown, but possibilities include the homodimerization of ubiquitinated proteins or interactions between the ubiquitinated proteins and ubiquitin binding proteins.
In Kc167 cells, a GFP-tagged Ubiquitin protein localizes in proximity of LDs (supplemental Fig. S5), suggesting that some LD proteins are ubiquitinated also in these cells. To identify candidates for ubiquitination, we tested for complementation between various proteins in our library and a Drosophila Ubiquitin protein (Fig. 5). We tagged ubiquitin at the N terminus with either Gluc(1) or Gluc(2). The C terminus of ubiquitin needs to be free, because it is usually linked to the epsilon amino group of a substrate lysine residue through an isopeptide bond (52). Once this modified ubiquitin gets attached to a target protein, complementation with the respective luciferase fragment should result in a positive read-out.
Histones served as a positive control (Fig. 7C), because ubiquitination is known to remove excess histone proteins which otherwise can become toxic within cells (53). We indeed found ubiquitination of the histone H2A, H2B, and H2Av proteins but only when the Gluc(2) fragment was fused to the C terminus of the histone proteins.
We identified a potential ubiquitination of several LD-associated proteins including CG9186 (Figs. 5A and 8A). As an independent test, we analyzed immunopurified GFP-tagged CG9186 by mass spectrometry and found that the lysine residues at amino acid positions 271 and 280 are indeed ubiquitinated (Fig. 8B and supplemental Fig. S6).
Fig. 8.
CG9186 is ubiquitinated and ubiquitination is important for the induction of LD clusters. A, Schematic representation of the constructs utilized in the split luciferase complementation assay testing for an interaction between CG9186 and Ubiquitin. Exemplary assay results are provided on the right hand side. B, Mass spectra matching to two different peptides from CG9186 (Uniprot accession Q9W0H3). Both matched peptides include a ubiquitination (glygly, mass shift of 114.04 Da, marked as gl) at a lysine residue at amino acid positions 271 and 280 of the CG9186 primary sequence, respectively. C, Overexpression of the GFP-tagged wildtype CG9186 protein results in smaller and clustered LDs. Conversely, cells with overexpression of the GFP-tagged mutated CG9186 variant carrying a total 16 lysine to arginine exchanges (CG9186–16K2R) are indistinguishable from untransfected cells. Expression of both proteins in Kc167 cells in the presence of 800 μm OA. Scale bars represent 5 μm.
Overexpression of the wildtype CG9186 protein results in a clustering of LDs (22) (Fig. 8C). To test the functional significance of CG9186 ubiquitination, we overexpressed an GFP-tagged variant of CG9186 where all lysine residues were changed to arginines to prevent ubiquitination. LD clustering was completely abolished, suggesting that the ubiquitination is indeed important for the LD clustering phenotype (Fig. 8C).
DISCUSSION
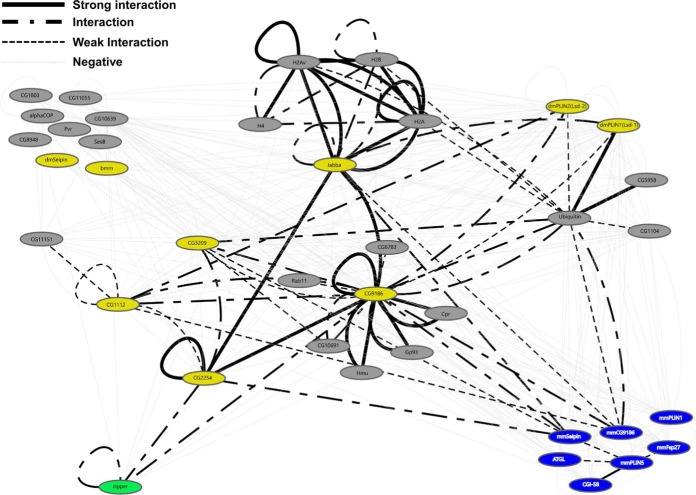
Here we report the utilization of a Gaussia princeps split luciferase complementation based protein-protein interaction (PPI) assay to study interactions of LD-associated proteins. We are able to detect interactions among a collection of mammalian and Drosophila proteins (Fig. 3C and Fig. 5), suggesting that this assay has broad applicability. Based on our findings, the assay is well suited to detect protein interactions in different membrane-bound subcellular compartments, such as the ER or LDs; this property represents an advantage over assays depending on a transcription-based reporter read-out as well as assays that necessitate removing the proteins from the membrane compartment. Our analysis (Fig. 9) greatly expands the number of identified interactions between LD-associated proteins and suggests that the assay is suitable to map interactions among the whole LD-associated proteome with high confidence.
Fig. 9.
The Interaction Network identified by the split luciferase protein interaction assay. Interaction tests based on the split-luciferase interaction assay were visualized using the Cytoscape software package. Groups of interacting proteins are highlighted. Each node represents an individual protein and edges represent results of the interaction tests. Blue nodes represent mammalian LD-associated proteins. Yellow nodes represent previously described LD-associated proteins of Drosophila. Gray nodes represent previously uncharacterized Drosophila proteins. Green node represents the GCN4 leucine zipper protein. Solid black edges: strong interactions. Point-Dash-Point edges represent interactions. Black dotted edges represent weak interactions. Light gray dotted edges represent negative interaction tests. Nodes can be connected by multiple edges based on the different luciferase fragment combinations/configurations tested.
In this study, we focus on proof-of-principle experiments to test the utility of the luciferase fragment complementation assay to identify and map interactions between LD proteins. In the future, this approach can be further refined to address more detailed mechanistic questions. For example, we induced LDs with OA, the standard experimental approach in the field. However, under native conditions, the broad range of FAs available likely results in a complex lipid composition of the LD core. Because its lipid composition might modulate LD protein binding (54), it will be interesting to investigate whether LD targeting and protein-protein interactions are altered when other lipids are used (such as palmitate, cholesterol, or complex lipid mixtures). In addition, overexpression of some proteins can prominently affect lipid storage levels (e.g. Brummer/ATGL, Fig. 1C, supplemental Fig. S1) and subcellular LD distribution (e.g. CG9186 or dmPLIN2; Fig. 1A); these factors might in turn indirectly modulate how likely the proteins interact. As a first step to untangle this, it will be important to determine how triglyceride levels and LD morphology change when certain protein combinations are expressed.
We employed the complementation assay to gain further insight into the LD protein CG9186. CG9186 is broadly conserved (from yeast to mammals) (22), binds LDs with exceptional strength (55), modulates the positioning of LDs in fly and mammalian cells (22), and can promote triglyceride storage as RNAi-mediated CG9186 depletion in whole flies results in reduced triglyceride levels (22). However, the molecular function of CG9186 remains unresolved. Although originally annotated as serine hydrolase or lipase (22) overexpression experiments and directed enzyme activity tests detected no evidence for an activity as neutral lipid lipase (22). For the murine CG9186 homolog (MGI:1916082; LDAH) a weak cholesterylester hydrolase activity in macrophages was reported (56). Our characterization of candidate CG9186 interaction partners might provide inroads into CG9186 function by linking it to other proteins with annotated or characterized functions.
One putative CG9186 interactor is Hemomucin (Hmu). The Hmu protein sequence contains a strictosidine synthase and a mucin domain and has relatives in plants and animals. Under OA loading conditions, Hmu accumulated partially around LDs (Fig. 4B). Intriguingly, the mouse Hmu homolog adipose plasma membrane-associated protein (APMAP) was identified in a LD proteomics screen (13), and strictosidine synthase was detected on the LD-related oil bodies in maize embryos (59). Because strictosidine synthases activity promotes Alkaloid synthesis (57), CG9186 might be involved with the production of secondary metabolites or their storage in lipophilic environments. In addition, Hmu is linked to immune response induction in the adult gut and in hemocytes, the Drosophila blood cells that mediate cellular immunity (58). Together with the observation that LDAH, the murine homolog of CG9186, is expressed in macrophages (56), this might indicate an immune function for CG9186.
A possible role of CG9186 in immune regulation is also supported by the identification of Pvr as potential interactor because this protein is important for macrophage lineage development (60–62). However, in the complementation experiments geared to validate the putative binding partners of GFP-tagged CG9186, the interaction between Pvr and CG9186 could not be confirmed. Whether this lack of interaction represents a false-negative outcome of the complementation assay or is due to a false-positive identification in the coimmunoprecipitation experiment needs to be investigated in the future. Discrepancies between different protein-protein interaction assay methodologies could, for example, result from an altered stoichiometry based on the overexpression of only one (coimmunoprecipitation) or both (luciferase complementation) interactors. Based on the relatively few well characterized LD-associated protein-protein interactions known, it is not yet possible to estimate false-negative and false-positive rates.
Two other proteins interacted with CG9186 both in the coimmunoprecipitation and luciferase complementation assay tests: glycoprotein of 93 kDa (Gp93) and Cytochrome P450 reductase (Cpr). Both proteins—as well as Hmu—were previously identified in a LD proteomics screen performed with fat body LDs of Drosophila 3rd instar larvae (6). Gp93 and other ER-derived chaperones are often found in mass spectrometry analyses of LD preparations (e.g. (63–65)). It is still not clear whether ER chaperones represent contaminants from the ER or whether they might be involved in quality control functions during LD biogenesis safeguarding e.g. the localization of only appropriately folded proteins to nascent LDs.
The putative CG9186 interaction partner Cpr is of particular interest. Although the Drosophila Cpr gene has not yet been analyzed in depth, the murine cytochrome P450 reductase (POR) localizes to the ER and functions in drug metabolism and sterol and bile acid synthesis (66). Cytochrome P450 reductases act in concert with squalene epoxidase and lanosterol synthase; in particular Cytochrome P450 reductases provide necessary reduction equivalents for squalene epoxidase. Squalene epoxidase and lanosterol synthase catalyze the first step of sterol biosynthesis. Both enzymes localize in the ER and on LDs (67). It was previously proposed that LD targeting of squalene epoxidase negatively regulates its activity, as it was thought that cytochrome P450 reductase proteins are not present on LDs (67). Our results, however, suggest that Cpr can indeed translocate to LDs (Fig. 4B). This finding is supported by the identification of Cytochrome P450 reductases in the LD proteomes of different mammalian cells (3, 12, 68).
The interaction between a presumptive cholesterol releasing enzyme such as LDAH and a potential cholesterol biogenesis cofactor such as cytochrome P450 reductase thus could represent a mechanism to shut down cholesterol synthesis by sequestration of an important cofactor in case cholesteryl esters are sufficient to fuel the cholesterol demand. However, because insects lack the sterol biosynthesis pathway (69–71), the interaction between Cpr and CG9186 might have a more ancestral regulatory function.
Because CG9186 interacts with multiple proteins, it might represent an interaction hub coordinating LD protein association. CG9186's interaction partner Jabba probably also has multiple partners: LDs isolated from Jabba mutant embryos lack not only histones, but also other proteins (21). In addition, our complementation assay revealed multiple putative Jabba interaction partners (Fig. 5). Further interaction tests will reveal, whether interaction hubs among the LD proteome constituents exist and how such hub proteins coordinate and regulate different LD functions.
Our results suggest that complementation efficacy depends on the positioning of the luciferase fragments (cis- or trans), especially for bigger interactors. Examples where the luciferase fragment configuration affects complementation efficacy include the interaction between CG9186 and CG1112 (Fig. 5A), the ubiquitination of Histone proteins (Fig. 7C) and the ubiquitination of CG9186 (Fig. 8A). For interactions among small proteins—such as the leucine zipper or histones—the position of the luciferase fragments appears to be less important (Fig. 7C).
In proof-of-principle experiments, we characterized several interactions revealed by the PPI screen in greater detail. The dimerization of Jabba depends on amino acids 109–212 because a minimal LD-associated Jabba version spanning amino acids 1–108 was no longer able to interact with the full-length protein (Fig. 6C), while it still targeted LDs (supplemental Fig. S3). We were unable to identify a single amino acid stretch mediating the dimerization of CG9186. All the constructs examined displayed reduced interactions. We speculate that all these deletions alter CG9186's conformation sufficiently to impair dimerization or that the interaction sequences act in an additive manner. Amino acids 141–200 of CG9186 and amino acids 1–108 of Jabba are sufficient to mediate both LD localization ((22) and supplemental Fig. S3) and the interaction between the two proteins (Fig. 6C, quadrants 3 and 4; and Fig. 6D). However, all truncated Jabba proteins only displayed imperfect LD targeting. Consistent with this, they—but not the full length Jabba—interacted with the cytosolic CG9186 protein lacking amino acids 141 to 307 (“CG9186(delta_aa141–307)”) (Fig. 6C, quadrant 3).
The sequestration of histone proteins to the LDs of early Drosophila embryos is a mechanism to protect the organism from histones in excess (21). Because the sequestration is reversible, the LD-localized histone proteins can act as a reservoir to supply histones when demand for chromatin assembly is high (21). Here, we show that Jabba expression in Drosophila cells is sufficient for the recruitment of histone proteins to LDs (Fig. 7B). An internal deletion of Jabba lacking amino acids 228–243 was no longer able to interact with H2A, H2B, or H2Av (Fig. 7C). Thus, the split luciferase PPI assay reveals that histone recruitment depends crucially on this stretch of 16 amino acids (supplemental Fig. S4).
Finally, we analyzed the ubiquitination of CG9186. Overexpression of CG9186 results in a strong LD clustering phenotype (22). Clustering depends on the C-terminal third of the protein, as the overexpression of a protein lacking this region does no longer result in the clustering phenotype (22). The predicted catalytically active serine at position 119 appears to counteract clustering because a serine to alanine exchange enhances LD clustering (22).
In this study, we employed the split luciferase PPI assay to identify that CG9186 interacts with ubiquitin (Fig. 5A). This interaction likely represents an ubiquitination because mass spectrometry revealed two ubiquitinated lysine residues in CG9186 (Fig. 8B). Overexpressing a CG9186 protein variant lacking all lysine residues (CG9186–16K2R) failed to induce LD clusters (Fig. 8C). These results suggest that the C-terminal ubiquitination sites—and perhaps additional ubiquitination sites not yet mapped—are important for the LD cluster induction. Ubiquitination was previously shown to be important for LD cluster induction by AUP1, a protein unrelated to CG9186 (44). We speculate that ubiquitin-dependent LD clustering might be mediated by ubiquitination induced homo-dimerization or by recruitment of an ubiquitin binding protein. Because CG9186–16K2R, which is presumably unable to be ubiquitinated, not only fails to induce LD clusters but also shows impaired homo-dimerization (supplemental Fig. S5B), we currently favor a role for homo-dimerization in LD cluster induction.
Although full-length CG9186 interacted with all CG9186 variants tested, CG9186–16K2R only interacted with a small subset, namely full length CG9186, the mutated CG9186 S119A protein, and the CG9186 protein variant missing amino acids 1 to 140 (Fig. 6). Our results therefore suggest that the dimerization of CG9186 is based on the interaction of the ubiquitinated C-terminal residues and/or the C-terminal amino acids 201–307. A construct lacking these amino acids failed to interact with CG9186–16K2R, and a LD-associated construct encompassing amino acids 141–200 of CG9186 was not sufficient for CG9186–16K2R binding. Intriguingly, the CG9186–16K2R variant also completely lost the ability to interact with any Jabba variant protein (supplemental Fig. S5B).
An important goal for the future will be to unravel the mechanism by which ubiquitination affects both LD clustering and CG9186 protein binding. Perhaps ubiquitination not only affects CG9186's ability to interact but also directly affects its affinity for the LD membrane, which might explain the loss of interaction with Jabba.
The PPI assay presented herein is therefore a promising tool to identify additional protein-protein interactions between LD proteins and additionally serve as a rapid method for functional studies.
Supplementary Material
Acknowledgments
We thank Erika Zernickel who helped in the early phase of the project. Additionally, we thank Alf Herzig, Remy Michnick, and Thomas Klein for sharing reagents and materials.
Footnotes
Author contributions: P.K., M. Werthebach, G.P., M. Welte, K.S., and M.B. designed research; P.K., M. Werthebach, A.v., G.P., L.C., and M.B. performed research; M. Werthebach, A.v., G.P., and M.B. analyzed data; G.P., M. Welte, and M.B. wrote the paper.
* This project was financed by funds from the Strategic Research Funds of the University of Duesseldorf (SFF; F2012/279-6; M. Beller), the Deutsche Forschungsgemeinschaft (DFG; BE4597/2-1; M. Beller), the German Federal Ministry of Education and Research (BMBF; 031A306; M. Beller) and the National Institutes of Health (NIH; 1R01 GM102155; M.A. Welte). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- LD
- lipid droplets
- PPI
- protein-protein interactions.
REFERENCES
- 1. Farese R. V., and Walther T. C. (2009) Lipid droplets finally get a little R-E-S-P-E-C-T. Cell 139, 855–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beller M., Thiel K., Thul P., and Jäckle H. (2010) Lipid droplets: A dynamic organelle moves into focus. FEBS Letters 584, 2176–2182 [DOI] [PubMed] [Google Scholar]
- 3. Larsson S., Resjö S., Gomez M. F., James P., and Holm C. (2012) Characterization of the lipid droplet proteome of a clonal insulin-producing β-cell line (INS-1 832/13). J. Proteome Res. 11, 1264–1273 [DOI] [PubMed] [Google Scholar]
- 4. Cermelli S., Guo Y., Gross S., and Welte M. (2006) The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 16, 1783–1795 [DOI] [PubMed] [Google Scholar]
- 5. Schmidt C., Ploier B., Koch B., and Daum G. (2013) Analysis of yeast lipid droplet proteome and lipidome. Methods Cell Biol. 116, 15–37 [DOI] [PubMed] [Google Scholar]
- 6. Beller M., Riedel D., Jänsch L., Dieterich G., Wehland J., Jäckle H., and Kühnlein R. P. (2006) Characterization of the Drosophila lipid droplet subproteome. Mol. Cell. Proteomics 5, 1082–1094 [DOI] [PubMed] [Google Scholar]
- 7. Zhang P., Na H., Liu Z., Zhang S., Xue P., Chen Y., Pu J., Peng G., Huang X., Yang F., Xie Z., Xu T., Xu P., Ou G., Zhang S. O., and Liu P. (2012) Proteomic study and marker protein identification of Caenorhabditis elegans lipid droplets. Mol. Cell. Proteomics 11, 317–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu P., Ying Y., Zhao Y., Mundy D., Zhu M., and Anderson R. (2004) Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J. Biol. Chem. 279, 3787–3792 [DOI] [PubMed] [Google Scholar]
- 9. Zehmer J., Huang Y., Peng G., Pu J., Anderson R., and Liu P. (2009) A role for lipid droplets in inter-membrane lipid traffic. Proteomics 9, 914–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krahmer N., Hilger M., Kory N., Wilfling F., Stoehr G., Mann M., Farese R. V., and Walther T. C. (2013) Protein correlation profiles identify lipid droplet proteins with high confidence. Mol. Cell. Proteomics 12, 1115–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Du X., Barisch C., Paschke P., Herrfurth C., Bertinetti O., Pawolleck N., Otto H., Rühling H., Feussner I., Herberg F., and Maniak M. (2013) Dictyostelium lipid droplets host novel proteins. Eukaryotic cell 12, 1517–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bouchoux J., Beilstein F., Pauquai T., Guerrera I. C., Chateau D., Ly N., Alqub M., Klein C., Chambaz J., Rousset M., Lacorte J.-M. M., Morel E., and Demignot S. (2011) The proteome of cytosolic lipid droplets isolated from differentiated Caco-2/TC7 enterocytes reveals cell-specific characteristics. Biol. Cell 103, 499–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ding Y., Wu Y., Zeng R., and Liao K. (2012) Proteomic profiling of lipid droplet-associated proteins in primary adipocytes of normal and obese mouse. Acta Biochim. Biophys. Sin. 44, 394–406 [DOI] [PubMed] [Google Scholar]
- 14. Sztalryd C., and Kimmel A. R. (2014) Perilipins: lipid droplet coat proteins adapted for tissue-specific energy storage and utilization, and lipid cytoprotection. Biochimie 96, 96–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lass A., Zimmermann R., Oberer M., and Zechner R. (2011) Lipolysis – A highly regulated multi-enzyme complex mediates the catabolism of cellular fat stores. Progress Lipid Res. 50, 14–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Remy I., and Michnick S. (2006) A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat. Methods 3, 977–979 [DOI] [PubMed] [Google Scholar]
- 17. Granneman J., Moore H.-P., Krishnamoorthy R., and Rathod M. (2009) Perilipin controls lipolysis by regulating the interactions of AB-hydrolase containing 5 (Abhd5) and adipose triglyceride lipase (Atgl). J. Biol. Chem. 284, 34538–34544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Granneman J., Moore H.-P., Mottillo E., and Zhu Z. (2009) Functional interactions between Mldp (LSDP5) and Abhd5 in the control of intracellular lipid accumulation. J. Biol. Chem. 284, 3049–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Granneman J., Moore H.-P., Mottillo E., Zhu Z., and Zhou L. (2011) Interactions of perilipin-5 (Plin5) with adipose triglyceride lipase. J. Biol. Chem. 286, 5126–5135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang H., Bell M., Sreenivasan U., Sreenevasan U., Hu H., Liu J., Dalen K., Londos C., Yamaguchi T., Rizzo M. A., Coleman R., Gong D., Brasaemle D., and Sztalryd C. (2011) Unique regulation of adipose triglyceride lipase (ATGL) by perilipin 5, a lipid droplet-associated protein. J. Biol. Chem. 286, 15707–15715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li Z., Thiel K., Thul P. J., Beller M., Kühnlein R. P., and Welte M. A. (2012) Lipid droplets control the maternal histone supply of Drosophila embryos. Curr. Biol. 22, 2104–2113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thiel K., Heier C., Haberl V., Thul P. J., Oberer M., Lass A., Jäckle H., and Beller M. (2013) The evolutionarily conserved protein CG9186 is associated with lipid droplets, required for their positioning and for fat storage. J. Cell Sci. 126, 2198–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., and Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Poschmann G., Seyfarth K., Besong Agbo D., Klafki H.-W. W., Rozman J., Wurst W., Wiltfang J., Meyer H. E., Klingenspor M., and Stühler K. (2014) High-fat diet induced isoform changes of the Parkinson's disease protein DJ-1. J. Proteome Res. 13, 2339–2351 [DOI] [PubMed] [Google Scholar]
- 25. Vizcaíno J. A., Csordas A., Del-Toro N., Dianes J. A. A., Griss J., Lavidas I., Mayer G., Perez-Riverol Y., Reisinger F., Ternent T., Xu Q.-W. W., Wang R., and Hermjakob H. (2016) 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 44, D447–D456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Beller M., Bulankina A. V., Hsiao H.-H. H., Urlaub H., Jäckle H., and Kühnlein R. P. (2010) PERILIPIN-dependent control of lipid droplet structure and fat storage in Drosophila. Cell Metab. 12, 521–532 [DOI] [PubMed] [Google Scholar]
- 27. Grönke S., Beller M., Fellert S., Ramakrishnan H., Jäckle H., and Kühnlein R. P. (2003) Control of fat storage by a Drosophila PAT domain protein. Curr. Biol. 13, 603–606 [DOI] [PubMed] [Google Scholar]
- 28. Teixeira L., Rabouille C., Rørth P., Ephrussi A., and Vanzo N. F. (2003) Drosophila Perilipin/ADRP homologue Lsd2 regulates lipid metabolism. Mech. Dev. 120, 1071–1081 [DOI] [PubMed] [Google Scholar]
- 29. Zimmermann R., Strauss J., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., Lass A., Neuberger G., Eisenhaber F., Hermetter A., and Zechner R. (2004) Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 306, 1383–1386 [DOI] [PubMed] [Google Scholar]
- 30. Grönke S., Mildner A., Fellert S., Tennagels N., Petry S., Müller G., Jäckle H., and Kühnlein R. P. (2005) Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 1, 323–330 [DOI] [PubMed] [Google Scholar]
- 31. Birner-Gruenberger R., Bickmeyer I., Lange J., Hehlert P., Hermetter A., Kollroser M., Rechberger G. N., and Kühnlein R. P. (2012) Functional fat body proteomics and gene targeting reveal in vivo functions of Drosophila melanogaster α-Esterase-7. Insect Biochem. Mol. Biol. 42, 220–229 [DOI] [PubMed] [Google Scholar]
- 32. Tian Y., Bi J., Shui G., Liu Z., Xiang Y., Liu Y., Wenk M., Yang H., and Huang X. (2011) Tissue-autonomous function of Drosophila Seipin in preventing ectopic lipid droplet formation. PLoS Genet. 7, e1001364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lass A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J., Gorkiewicz G., and Zechner R. (2006) Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metabolism 3, 309–319 [DOI] [PubMed] [Google Scholar]
- 34. Fei W., Li H., Shui G., Kapterian T. S., Bielby C., Du X., Brown A. J., Li P., Wenk M. R., Liu P., and Yang H. (2011) Molecular characterization of seipin and its mutants: implications for seipin in triacylglycerol synthesis. J. Lipid Res. 52, 2136–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gong J., Sun Z., Wu L., Xu W., Schieber N., Xu D., Shui G., Yang H., Parton R., and Li P. (2011) Fsp27 promotes lipid droplet growth by lipid exchange and transfer at lipid droplet contact sites. J. Cell Biol. 195, 953–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bi J., Xiang Y., Chen H., Liu Z., Grönke S., Kühnlein R. P., and Huang X. (2012) Opposite and redundant roles of the two Drosophila perilipins in lipid mobilization. J. Cell Sci. 125, 3568–3577 [DOI] [PubMed] [Google Scholar]
- 37. Greenberg A. S., Egan J. J., Wek S. A., Garty N. B., Blanchette-Mackie E. J., and Londos C. (1991) Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J. Biol. Chem. 266, 11341–11346 [PubMed] [Google Scholar]
- 38. Bi J., Wang W., Liu Z., Huang X., Jiang Q., Liu G., Wang Y., and Huang X. (2014) Seipin promotes adipose tissue fat storage through the ER Ca2+-ATPase SERCA. Cell Metab. 19, 861–871 [DOI] [PubMed] [Google Scholar]
- 39. Jambunathan S., Yin J., Khan W., Tamori Y., and Puri V. (2011) FSP27 Promotes lipid droplet clustering and then fusion to regulate triglyceride accumulation. PLoS ONE 6, e28614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Werthebach M., Mailliet J., and Beller M. (2016) Automatisierte Mikroskopie und Bildanalyse der zellulären Lipidspeicherung. Biospektrum 22, 392–394 [Google Scholar]
- 41. Kory N., Farese R. V., and Walther T. C. (2016) Targeting fat: Mechanisms of protein localization to lipid droplets. Trends Cell Biol. 26, 535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu G., Sztalryd C., and Londos C. (2006) Degradation of perilipin is mediated through ubiquitination-proteasome pathway. Biochim. Biophys. Acta 1761, 83–90 [DOI] [PubMed] [Google Scholar]
- 43. Jo Y., Hartman I. Z., and DeBose-Boyd R. A. (2013) Ancient ubiquitous protein-1 mediates sterol-induced ubiquitination of 3-hydroxy-3-methylglutaryl CoA reductase in lipid droplet-associated endoplasmic reticulum membranes. Mol. Biol. Cell 24, 169–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lohmann D., Spandl J., Stevanovic A., Schoene M., Philippou-Massier J., and Thiele C. (2013) Monoubiquitination of ancient ubiquitous protein 1 promotes lipid droplet clustering. PLoS ONE 8, e72453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu G., Sztalryd C., Lu X., Tansey J. T., Gan J., Dorward H., Kimmel A. R., and Londos C. (2005) Post-translational regulation of adipose differentiation-related protein by the ubiquitin/proteasome pathway. J. Biol. Chem. 280, 42841–42847 [DOI] [PubMed] [Google Scholar]
- 46. Alberts P., and Rotin D. (2010) Regulation of lipid droplet turnover by ubiquitin ligases. BMC Biol. 8, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hooper C., Puttamadappa S. S., Loring Z., Shekhtman A., and Bakowska J. C. (2010) Spartin activates atrophin-1-interacting protein 4 (AIP4) E3 ubiquitin ligase and promotes ubiquitination of adipophilin on lipid droplets. BMC Biol. 8, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nian Z., Sun Z., Yu L., Toh S. Y., Sang J., and Li P. (2010) Fat-specific protein 27 undergoes ubiquitin-dependent degradation regulated by triacylglycerol synthesis and lipid droplet formation. J. Biol. Chem. 285, 9604–9615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Eastman S. W., Yassaee M., and Bieniasz P. D. (2009) A role for ubiquitin ligases and Spartin/SPG20 in lipid droplet turnover. J. Cell Biol. 184, 881–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Olzmann J. A., Richter C. M., and Kopito R. R. (2013) Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proc. Natl. Acad. Sci. U.S.A. 110, 1345–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Masuda Y., Itabe H., Odaki M., Hama K., Fujimoto Y., Mori M., Sasabe N., Aoki J., Arai H., and Takano T. (2006) ADRP/adipophilin is degraded through the proteasome-dependent pathway during regression of lipid-storing cells. J. Lipid Res. 47, 87–98 [DOI] [PubMed] [Google Scholar]
- 52. Finley D., Ulrich H. D., Sommer T., and Kaiser P. (2012) The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics 192, 319–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Singh R. K., Kabbaj M-HMH, Paik J., and Gunjan A. (2009) Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat. Cell Biol. 11, 925–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hsieh K., Lee Y. K., Londos C., Raaka B. M., Dalen K. T., and Kimmel A. R. (2012) Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. J. Cell Sci. 125, 4067–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kory N., Thiam A.-R. R., Farese R. V., and Walther T. C. (2015) Protein crowding Is a determinant of lipid droplet protein composition. Dev. Cell 34, 351–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Goo Y.-H. H., Son S.-H. H., Kreienberg P. B., and Paul A. (2014) Novel lipid droplet-associated serine hydrolase regulates macrophage cholesterol mobilization. Arterioscler. Thromb. Vasc. Biol. 34, 386–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fabbri M., Delp G., Schmidt O., and Theopold U. (2000) Animal and plant members of a gene family with similarity to alkaloid-synthesizing enzymes. Biochem. Biophys. Res. Commun. 271, 191–196 [DOI] [PubMed] [Google Scholar]
- 58. Theopold U., Samakovlis C., Erdjument-Bromage H., Dillon N., Axelsson B., Schmidt O., Tempst P., and Hultmark D. (1996) Helix pomatia lectin, an inducer of Drosophila immune response, binds to hemomucin, a novel surface mucin. J. Biol. Chem. 271, 12708–12715 [DOI] [PubMed] [Google Scholar]
- 59. Tnani H., López I., Jouenne T., and Vicient C. M. (2011) Protein composition analysis of oil bodies from maize embryos during germination. J. Plant Physiol. 168, 510–513 [DOI] [PubMed] [Google Scholar]
- 60. Brückner K., Kockel L., Duchek P., Luque C. M., Rørth P., and Perrimon N. (2004) The PDGF/VEGF receptor controls blood cell survival in Drosophila. Dev. Cell 7, 73–84 [DOI] [PubMed] [Google Scholar]
- 61. Wang C.-W. W., Purkayastha A., Jones K. T., Thaker S. K., and Banerjee U. (2016) In vivo genetic dissection of tumor growth and the warburg effect. Elife 5, e18126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sansone C. L., Cohen J., Yasunaga A., Xu J., Osborn G., Subramanian H., Gold B., Buchon N., and Cherry S. (2015) Microbiota-Dependent Priming of Antiviral Intestinal Immunity in Drosophila. Cell Host Microbe 18, 571–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Khan S. A., Wollaston-Hayden E. E., Markowski T. W., Higgins L., and Mashek D. G. (2015) Quantitative analysis of the murine lipid droplet-associated proteome during diet-induced hepatic steatosis. J. Lipid Res. 56, 2260–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang W., Wei S., Li L., Su X., Du C., Li F., Geng B., Liu P., and Xu G. (2015) Proteomic analysis of murine testes lipid droplets. Sci Rep 5, 12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. D'Aquila T., Sirohi D., Grabowski J. M., Hedrick V. E., Paul L. N., Greenberg A. S., Kuhn R. J., and Buhman K. K. (2015) Characterization of the proteome of cytoplasmic lipid droplets in mouse enterocytes after a dietary fat challenge. PLoS ONE 10, e0126823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Porter T. D., Banerjee S., Stolarczyk E. I., and Zou L. (2011) Suppression of cytochrome P450 reductase (POR) expression in hepatoma cells replicates the hepatic lipidosis observed in hepatic POR-null mice. Drug Metab. Dispos. 39, 966–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Goodman J. M. (2009) Demonstrated and inferred metabolism associated with cytosolic lipid droplets. J. Lipid Res. 50, 2148–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang H., Wang Y., Li J., Yu J., Pu J., Li L., Zhang H., Zhang S., Peng G., Yang F., and Liu P. (2011) Proteome of skeletal muscle lipid droplet reveals association with mitochondria and apolipoprotein a-I. J. Proteome Res. 10, 4757–4768 [DOI] [PubMed] [Google Scholar]
- 69. Carvalho M., Schwudke D., Sampaio J. L., Palm W., Riezman I., Dey G., Gupta G. D., Mayor S., Riezman H., Shevchenko A., Kurzchalia T. V., and Eaton S. (2010) Survival strategies of a sterol auxotroph. Development 137, 3675–3685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rawson R. B. (2003) The SREBP pathway–insights from Insigs and insects. Nat. Rev. Mol. Cell Biol. 4, 631–640 [DOI] [PubMed] [Google Scholar]
- 71. Clark A. J., and Block K. (1959) The absence of sterol synthesis in insects. J. Biol. Chem. 234, 2578–2582 [PubMed] [Google Scholar]
- 72. Garlena R. A., Lennox A. L., Baker L. R., Parsons T. E., Weinberg S. M., and Stronach B. E. (2015) The receptor tyrosine kinase Pvr promotes tissue closure by coordinating corpse removal and epidermal zippering. Development 142, 3403–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Harris K. E., Schnittke N., and Beckendorf S. K. (2007) Two ligands signal through the Drosophila PDGF/VEGF receptor to ensure proper salivary gland positioning. Mech. Dev. 124, 441–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.