Abstract
Male sex predisposes to many kidney diseases. Considering that androgens exert deleterious effects in a variety of cell types within the kidney, we hypothesized that dihydrotestosterone (DHT) would alter the biology of the renal tubular cell by inducing changes in the proteome. We employed stable isotope labeling with amino acids (SILAC) in an indirect spike-in fashion to accurately quantify the proteome in DHT- and 17β-estradiol (EST)-treated human proximal tubular epithelial cells (PTEC). Of the 5043 quantified proteins, 76 were differentially regulated. Biological processes related to energy metabolism were significantly enriched among DHT-regulated proteins. SILAC ratios of 3 candidates representing glycolysis, N-acetylglucosamine metabolism and fatty acid β-oxidation, namely glucose-6-phosphate isomerase (GPI), glucosamine-6-phosphate-N-acetyltransferase 1 (GNPNAT1), and mitochondrial trifunctional protein subunit alpha (HADHA), were verified in vitro. In vivo, renal GPI and HADHA protein expression was significantly increased in males. Furthermore, male sex was associated with significantly higher GPI, GNPNAT1, and HADHA kidney protein expression in two different murine models of diabetes. Enrichment analysis revealed a link between our DHT-regulated proteins and oxidative stress within the diabetic kidney. This finding was validated in vivo, as we observed increased oxidative stress levels in control and diabetic male kidneys, compared with females. This in depth quantitative proteomics study of human primary PTEC response to sex hormone administration suggests that male sex hormone stimulation results in perturbed energy metabolism in kidney cells, and that this perturbation results in increased oxidative stress in the renal cortex. The proteome-level changes associated with androgens may play a crucial role in the development of structural and functional changes in the diseased kidney. With our findings, we propose a possible link between diabetic and non-diabetic kidney disease progression and male sex hormone levels. Data are available via ProteomeXchange (https://www.ebi.ac.uk/pride/archive/) with identifier PXD003811.
Chronic Kidney Disease (CKD)1 often results in irreversible deterioration of renal function that can progress to renal failure (1). Sex plays a relevant role in the progression and severity of many kidney diseases (2). At a clinical level, it is generally accepted that male sex is a risk factor for CKD (3). In addition, sex differences impact the progression of renal disease in animal models of non-diabetic (4) and diabetic nephropathy (5–7).
Experimental evidence suggests that androgens play a deleterious role in kidney disease. Testosterone promoted apoptosis in immortalized and primary human proximal tubule cells (8). In addition, preincubation with the antiandrogen flutamide prevented the apoptotic effects of testosterone, indicating that deleterious effects of androgens are mediated by testosterone conversion to DHT and binding to androgen receptor (AR). In turn, EST significantly attenuated the fibrotic effect of TGFβ-1 in mesangial cells (9). The counter-regulatory effects of androgens and estrogens have also been observed in podocytes, where EST prevented the testosterone-induced increase in the percentage of TUNEL-positive cells. In this sense, both estrogen receptor alpha (ERα) deficiency and testosterone administration were associated with podocyte loss and augmented apoptosis in vivo (10). In type 1 diabetic castrated male rats, low dose of DHT attenuated, whereas high dose accentuated, the severity of several hallmarks of kidney disease such as glomerulosclerosis and tubulointerstitial fibrosis (11).
Androgens and estrogens have shown to trigger genomic and non-genomic events within the renal cortex (12) and in renal tubular cells in culture (13, 14). Genomic actions of sex hormones are mediated by primary interactions with their specific nuclear receptors (AR for androgens and ERα/ERβ for estrogens) (15). In addition, estrogens promote rapid extranuclear signaling upon binding to ERα, ERβ or G protein coupled estrogen receptor 1 (GPER) localized on the cell membrane (16), promoting several downstream events such as changes in cAMP (17, 18), recruitment and activation of the MAPK/ERK signaling cascade (19), and induction of PI3K/AKT signaling to activate eNOS (20). Androgens can also stimulate MAPK/ERK (21) and AKT pathways (22) after interacting with the AR or the G-coupled receptor GPRC6A on the cell membrane (23, 24).
Taken together, in vitro and in vivo data suggest that androgen signaling pathways in the renal cortex play a harmful effect on the progression of CKD. However, the specific mechanisms by which androgens detrimentally regulate kidney cells over estrogens remain unclear. To enhance our understanding of sex effects on CKD progression, we aimed to perform an in-depth analysis of the sex hormone-regulated proteome in human proximal tubular epithelial cells (PTEC) after stimulation with DHT or EST. We hypothesized that male sex hormones induce changes in the proteome of the renal tubular cell in a more detrimental manner than estrogens.
Quantitative proteomics is widely considered as an invaluable approach to explore biological systems in vitro and in vivo (25–27). To achieve our aim, we carried out a quantitative strategy by using stable isotope labeling of amino acids in cell culture (SILAC) in combination with a high-resolution mass analyzer. To quantify the proteome of a single cell line, the SILAC experiment can be performed in a classical or in a spike-in format (28). One of the advantages of spike-in SILAC is that the experimental cells are maintained in their normal state and are not affected by the special media required for SILAC or by the use of dialyzed serum. Furthermore, the same standard can be used for multiple samples and similar cell lines, and there is therefore no need to label all of them (29). Single cell line spike-in standard has been previously and efficiently used when multiple samples with high similarity were studied, or in cases where growing the cells in SILAC media was challenging (30).
Because SILAC-labeling of primary PTEC is not trivial and the several passages needed for the full incorporation of the labeled amino acids may lead to cell culture-induced loss of differentiation, we have employed spike-in SILAC to accurately quantify their proteome after sex hormone incubation. In particular, we SILAC-labeled immortalized human kidney HK-2 cells, and used their “heavy” proteome as internal standard to quantify the treated proteome from PTEC. Both PTEC and HK-2 are epithelial cells and originate from human renal proximal tubule. Thus, we reasoned that HK-2 proteome would allow us to accurately quantify a large proportion of PTEC proteome. In addition, HK-2 cells have been reported to show excellent labeling efficiency (31). However, HK-2 cells are immortalized cells and may differ from tubular cells in vivo in several important aspects, such as the changes in their proteome in response to certain stimuli (32, 33). We thus selected the primary PTEC cells for stimulation experiments, which were previously shown to respond to sex hormones (8, 14). Among all different cell types in the kidney, we performed our study in proximal tubular cells because tubules constitute most of the renal parenchymal mass, and tubular atrophy with interstitial fibrosis carries prognostic significance, and represents the common final pathway of most causes of CKD (36, 37).
To our knowledge, the effect of sex hormones on the proteome of human kidney cells has not been previously explored. In addition, we are the first to use a spike-in SILAC quantitative proteomic approach between two different renal cell lines. In this discovery-based study, we provide a detailed portrait of the key biological processes impaired by DHT and not EST. By performing bioinformatics analyses and the corresponding verification and validation experiments, we demonstrate: (1) a sex-specific regulation of three candidate proteins related to energy metabolism; and (2) an association between male sex and increased oxidative stress levels within the kidney. In this article we suggest for the first time that male sex hormones induce perturbations in the metabolism of the tubular cell that may ultimately lead to impaired oxidative stress and hypertrophy in the kidney and consequently increase the susceptibility to renal disease progression in males, especially in the context of diabetes. With our findings, we provide a novel link between male sex hormones and impaired energy metabolism in the kidney that may shed new light on how to explore the mechanisms involved in the more rapid progression of CKD ascribed to male sex.
EXPERIMENTAL PROCEDURES
Experimental Design and Statistical Rationale
To cover the intrinsic variability associated to cell culture across passages, sex hormone stimulation experiments in PTEC were conducted in two different passages (passage 4 and 5). In each passage, incubation of PTEC with sex hormones or ethanol was performed in two separate experiments, aiming to obtain biological replicates that allowed us to achieve adequate reliability and consistency in further quantitative proteomics profiling.
Cell Culture
PTEC were purchased from Lonza Walkersville Inc (Walkersville, MD). They were cultured in custom-made Dulbecco's modified Eagle's medium (DMEM), and supplemented with 10% v/v dialyzed fetal bovine serum (FBS), 10 ng/ml EGF, 5 μg/ml transferrin, 5 μg/ml insulin, 0.05 μm hydrocortisone, 50 units/ml penicillin, and 50 μg/ml streptomycin, as previously described (38). Cells were serum starved for 18 h and treated with 100 nm DHT (n = 4) or EST (n = 3) for 10min (aiming to detect AKT and ERK phosphorylation as control experiments) or 8h (aiming to activate both nongenomic and genomic signaling for proteomic analysis). Ethanol treated cells were used as controls (CONT, n = 4). After stimulation, cells were washed three times with PBS, harvested with trypsin, and snap-frozen at −80 °C until further analysis.
HK-2 cells used for SILAC labeling were provided by Dr. López Novoa (Instituto de Investigación Biomédica de Salamanca, Salamanca, Spain). They were cultured in DMEM/F12 (1:1) free of arginine, lysine, methionine and leucine (AthenaES, Baltimore, MD), and supplemented with 10% v/v dialyzed FBS, 50units/ml penicillin, 50 μg/ml streptomycin, 2 mm glutamine, 5 μg/μl transferrin, 5 μg/μl insulin, 0.05 μm hydrocortisone, 1 nm T3 hormone, 10 ng/ml EGF, 147.5 mg/L heavy arginine (13C6), 91.25 mg/L heavy lysine (13C615N2), 17.24 mg/L light methionine, and 59.05 mg/L light leucine. After 5, 6, 8 and 10 cell population doublings of SILAC labeling, HK-2 cells were serum starved for 18h and cell pellets were collected as mentioned above for further spike-in experiments. At each point, 1×105-2×105 cells were separated and collected as a different pellet for determination of labeling efficiency. Percent label incorporation for individual peptides was calculated manually using the equation (Intensity H/Total Intensity) * 100. 100% labeling efficiency was considered for peptides with intensity equal to 0 in the light form. Results from labeling efficiency experiments are summarized in supplemental Table S1. Both PTEC and HK-2 cells were cultured in a humidified incubator at 37 °C and 5% CO2. All media were freshly made and filtered using a 0.22 μm syringe filter.
Sample Preparation for Proteome Analysis
Cell pellets from treated PTEC (light) and labeled HK-2 cells (heavy) were thawed on ice, resuspended in 200 μl of 0.1% w/v acid-labile detergent RapiGest SF (Waters, Milford, MA) in 25 mm ammonium bicarbonate, vortexed, and sonicated three times for 30 s. All lysates were centrifuged for 20 min at 15,000 × g at 4 °C. Total protein concentration was measured using a Coomassie (Bradford) protein assay reagent (Pierce, Waltham, MA). One hundred and fifty micrograms of protein from heavy HK-2 cells were spiked to 150 μg of each sample from light treated PTEC (1:1 mixing ratio) (Fig. 1). Proteins in detergent solution were denatured at 60 °C, and the disulfide bonds were reduced with 10 mm dithiothreitol. Following reduction, the samples were alkylated with 20 mm iodoacetamide. Proteins were then digested overnight at 37 °C with sequencing grade modified trypsin (Promega, Madison, WI). A trypsin/total protein ratio of 1:50 (w/w) was used. After digestion, RapiGest SF detergent was cleaved with trifluoroacetic acid, 1% (v/v) final concentration, and samples were centrifuged for 15 min at 15,000 rpm at 4 °C.
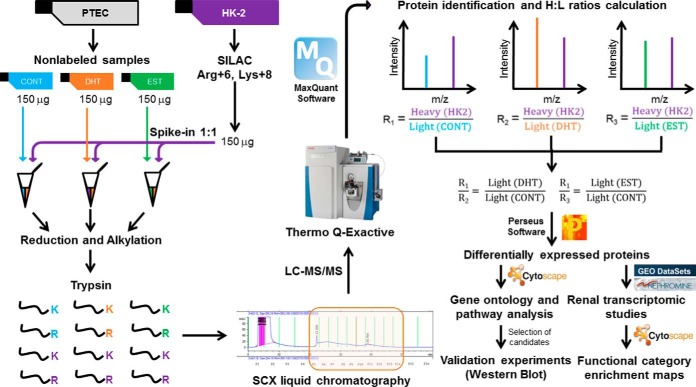
Fig. 1.
Experimental scheme. The figure shows a simplified workflow, including sex hormone treatment to PTEC, SILAC labeling of HK-2 cells, 1:1 mixing of labeled and nonlabeled proteins, protein digestion, SCX fractionation followed by LC-MS/MS, data analysis by MaxQuant, assignment of heavy/light protein ratios, calculation of DHT/CONT and EST/CONT ratios, selection of differentially regulated proteins, validation studies and bioinformatics analyses.
Proteome Analysis of DHT- and EST-stimulated PTEC Using Two-dimensional LC-MS/MS
Upon removal of Rapigest, tryptic peptides were diluted to 500 μl with strong cationic exchange (SCX) mobile phase A (0.26 m formic acid in 5% v/v acetonitrile; pH2) and loaded directly onto a 500 μl loop connected to a PolySULFOETHYL A column (2.1-mm inner diameter × 200 mm, 5 μm, 200 Å, The Nest Group Inc. (Southborough, MA)). The SCX chromatography and fractionation were performed on an HPLC system (Agilent 1100) using a 60 min two-step gradient. An elution buffer that contained all components of mobile phase A with the addition of 1 m ammonium formate was introduced at 10min and increased to 20% at 30 min and then to 100% at 45 min. Fractions were collected every 3 min from the 20 min time point onward. The resulting 10 fractions (600 μl each) corresponding to chromatographic peaks of eluting peptides were collected. Peptides in each fraction were identified by LC-MS/MS as described previously (39). Briefly, peptides were extracted with OMIX C18 MB tips (Agilent Technologies, Lake Forest, CA), eluted in 3 μl of 65% v/v acetonitrile, diluted to 41 μl with 0.1% v/v formic acid in pure water, and loaded onto a 3.3 cm C18 pre-analytical column (IntegraFrit capillary, New Objective; inner diameter of 150 μm; 5 μm bead size; Agilent Pursuit C18, Agilent Technologies, Santa Clara, CA). Eluted peptides from the pre-analytical column were loaded onto a resolving analytical column with dimensions 15 cm × 75 μm ID (PicoTip emitter, 8 μm tip, New Objective Agilent Pursuit C18, 3 μm bead size). The trap and analytical columns were operated on the EASY-nLC1000 system (Thermo Fisher Scientific, San Jose, CA), and this liquid chromatography setup was coupled on line to a Q Exactive Plus hybrid quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific, San Jose, CA) using a nano-ESI source (Proxeon Biosystems, Odense, Denmark). Each fraction was run using a 60 min gradient, and analyzed in a data-dependent mode in which a full MS1 scan acquisition from 400–1500 m/z with resolution 70,000 was acquired. This was followed by MS2 scan acquisition of the top 12 parent ions with resolution 17,500. The following parameters were enabled: monoisotopic precursor selection, charge state screening, and dynamic exclusion (enabled for 45 s), MS1 target value of 3e6 and maximum injection time (IT) of 100 ms, MS2 target value of 5e4 with maximum IT of 50 ms, isolation window of 1.6 Da, normalized collision energy (NCE) of 27, peptide match set to preferred, underfill ratio set to 2% (underfill ratio of 2e4), exclude isotopes set to ON. In addition, charge states of +1, >4, and unassigned charge states were not subjected to MS2 fragmentation. For protein identification and data analysis, Xcalibur software (version 3.0.63; Thermo Fisher) was utilized to generate RAW files of each MS run. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (1) partner repository (https://www.ebi.ac.uk/pride/archive/) with the data set identifier PXD003811.
MS Data Analysis
The MS files were processed with the MaxQuant software version 1.5.2.8 (40) and searched with Andromeda search engine (41) against the human UniProt database (42) (release 01_07_2015, 89,649 entries). The raw files from all biological replicates were analyzed simultaneously with MaxQuant (Fig. 1). To assess the false-positive rate, a reverse hit database was created by MaxQuant. The false discovery rates at the protein and peptide level were set to 1%. First search peptide tolerance was set to 20 ppm against a small “human-first-search” database for the purpose of mass recalibration and for main search precursor ion mass deviation of 4.5 ppm was allowed, while fragment mass deviation of 20 ppm was used. The minimum peptide length was set to 7 amino acids and strict specificity for trypsin cleavage was required, allowing up to two missed cleavage sites. Searches were performed with fixed carbamidomethylation of cysteines, variable oxidation of methionine and proline residues, and N-terminal acetylation. Multiplicity of 2 was used, selecting Arg (+6 Da) and Lys (+8 Da) as heavy labels. Re-quantification and matching between runs was selected. Protein was identified with a minimum of one unique peptide. Quantification was performed using unmodified unique and razor peptides and a minimum of one counted ratio.
SILAC Ratio Analysis
The reverse and common contaminant hits were removed from MaxQuant output. Only proteins identified with at least one peptide uniquely assigned to the respective sequence were considered for the analysis. The distributions of all Log2 transformed H/L and H/L normalized ratios were normal (supplemental Fig. S1), thus the H/L normalized ratios of biological replicates were used to calculate DHT/CONT and EST/CONT ratios as described in Fig. 1. These ratios express the fold change between two different conditions and were calculated in all four experiments. Significance A of DHT/CONT and EST/CONT ratios was calculated for all the quantified proteins. A protein was considered to be significantly differentially regulated if its ratio was significant by significance A with p < 0.01 in at least two experiments. Proteins were eliminated if their ratios in the other two experiments showed significant changes in the opposite direction, defined as ≥2-fold change in the opposite direction, or change in the opposite direction with p < 0.05. Only proteins with at least one unique peptide with two or more ratio counts were further pursued. In addition, if ratios of proteotypic (unique) peptides of the same protein were changing in opposite directions, these proteins were also discarded for the validation studies. A total of 76 proteins were found to be differentially regulated in at least one of the three comparisons (Table I).
Table I. Summarized median DHT/CONT and EST/CONT ratios for the 76 proteins that are significantly differentially expressed in primary proximal tubular cells.
| Gene names | DHT/CONT |
EST/CONT |
||||
|---|---|---|---|---|---|---|
| Median | S.E. | Significant | Median | S.E. | Significant | |
| ALDH1B1 | 7.4391 | 0.072 | YES (Upregulated) | 0.852 | 0.214 | NO |
| ALDH4A1 | 6.806 | 2.960 | YES (Upregulated) | 1.575 | 0.444 | NO |
| C14orf159 | 8.330 | 3.011 | YES (Upregulated) | 0.981 | 0.049 | NO |
| C19orf25 | 5.837 | 1.241 | YES (Upregulated) | 0.495 | NaN | NO |
| CAD | 3.704 | 3.848 | YES (Upregulated) | 1.100 | 14.303 | NO |
| CALR | 7.818 | 1.205 | YES (Upregulated) | 0.981 | 0.576 | NO |
| CASP6 | 5.212 | 1.577 | YES (Upregulated) | 1.232 | 0.428 | NO |
| CDC2;CDK1 | 20.344 | 10.175 | YES (Upregulated) | 1.386 | 1.118 | NO |
| CDK4 | 1.664 | 0.095 | NO | 4.016 | 2.809 | YES (Upregulated) |
| CDK5RAP3 | 8.448 | 0.470 | YES (Upregulated) | 0.947 | 0.021 | NO |
| CDKN2AIP | 6.325 | 0.988 | YES (Upregulated) | NaN | NaN | NO |
| CTSC | 8.085 | 2.518 | YES (Upregulated) | 0.943 | 0.410 | NO |
| CTSZ | 8.807 | 0.076 | YES (Upregulated) | 0.952 | 0.080 | NO |
| DDAH2 | 1.171 | 1.905 | NO | 7.923 | 2.302 | YES (Upregulated) |
| DSG2 | 3.466 | 1.034 | YES (Upregulated) | 2.618 | 0.891 | YES (Upregulated) |
| DUSP23 | 1.232 | 0.625 | NO | 2.664 | 0.351 | YES (Upregulated) |
| ECHS1 | 6.356 | 0.933 | YES (Upregulated) | 0.973 | 0.079 | NO |
| EDIL3 | 1.488 | 1.519 | NO | 2.216 | 0.611 | YES (Upregulated) |
| F11R | 1.955 | 0.596 | NO | 1.914 | 0.279 | YES (Upregulated) |
| FAM98B | 2.897 | 0.806 | NO | 10.421 | 8.163 | YES (Upregulated) |
| GATAD2B | 1.928 | 0.342 | NO | 2.380 | 0.192 | YES (Upregulated) |
| GGH | 6.630 | 0.150 | YES (Upregulated) | 0.875 | 0.182 | NO |
| GLDC | 1.483 | 0.505 | NO | 2.482 | 0.567 | YES (Upregulated) |
| GLS | 6.892 | 1.572 | YES (Upregulated) | 0.854 | 0.362 | NO |
| GLUD1;GLUD2 | 9.265 | 1.726 | YES (Upregulated) | 0.957 | 0.351 | NO |
| GNA13 | 17.180 | 4.978 | YES (Upregulated) | 9.777 | 9.754 | NO |
| GNPNAT1 | 5.626 | 1.631 | YES (Upregulated) | 2.171 | 0.001 | YES (Upregulated) |
| GPI | 12.068 | 9.117 | YES (Upregulated) | 1.242 | 3.165 | NO |
| HADHA | 8.027 | 1.105 | YES (Upregulated) | 0.882 | 0.683 | NO |
| HADHB | 7.514 | 1.955 | YES (Upregulated) | 0.990 | 0.981 | NO |
| HEXB | 7.279 | 2.790 | YES (Upregulated) | 0.475 | 0.225 | NO |
| HPCAL1;HPCA;NCALD | 6.180 | 0.548 | YES (Upregulated) | 0.848 | 0.831 | NO |
| HSPA14 | 2.183 | 2.053 | YES (Upregulated) | 2.030 | 0.477 | NO |
| HYOU1 | 11.009 | 0.051 | YES (Upregulated) | 0.790 | 0.057 | NO |
| KHSRP | 13.118 | 3.911 | YES (Upregulated) | 0.794 | 1.575 | NO |
| LETM1 | 6.306 | 0.888 | YES (Upregulated) | 0.974 | 0.618 | NO |
| LTN1 | 0.789 | 0.147 | NO | 2.683 | 0.257 | YES (Upregulated) |
| MAN2B1 | 4.416 | 2.410 | YES (Upregulated) | 1.669 | 0.359 | NO |
| MCM4 | 8.936 | 13.031 | YES (Upregulated) | 0.203 | 0.610 | NO |
| MDH2 | 9.249 | 2.752 | YES (Upregulated) | 0.792 | 0.351 | NO |
| MRPL40 | 2.273 | 0.496 | YES (Upregulated) | 3.368 | NaN | NO |
| MSH6 | 1.443 | 0.338 | NO | 2.255 | 3.419 | YES (Upregulated) |
| MYEF2 | 6.874 | 0.127 | YES (Upregulated) | 0.705 | NaN | NO |
| MYO5B | 2.852 | 0.321 | NO | 3.176 | 0.705 | YES (Upregulated) |
| NASP | 3.023 | 1.839 | YES (Upregulated) | 0.922 | 0.729 | NO |
| NUDT15 | 0.881 | NaN | NO | 2.576 | 0.008 | YES (Upregulated) |
| NUP188 | 2.734 | 1.835 | YES (Upregulated) | 0.767 | 0.062 | NO |
| PALLD | 5.965 | 3.219 | YES (Upregulated) | 2.502 | 5.275 | NO |
| PARK7 | 11.700 | 5.605 | YES (Upregulated) | 1.168 | 0.158 | NO |
| PCBD1 | 6.425 | 3.355 | YES (Upregulated) | 1.104 | 0.199 | NO |
| PDHB | 8.464 | 2.035 | YES (Upregulated) | 0.782 | 1.070 | NO |
| PLBD2 | 7.145 | 4.383 | YES (Upregulated) | 1.271 | 1.318 | NO |
| PPIL3 | 3.224 | 2.113 | YES (Upregulated) | 1.783 | 1.372 | NO |
| PPM1G | 15.859 | 1.541 | YES (Upregulated) | 3.151 | 1.825 | NO |
| PSAP | 9.220 | 3.803 | YES (Upregulated) | 1.091 | 0.865 | NO |
| PTBP1 | 10.770 | 4.759 | YES (Upregulated) | 0.947 | 2.358 | NO |
| QPRT | 3.164 | 2.123 | YES (Upregulated) | 1.187 | 0.490 | NO |
| RABGGTA | 2.152 | 1.583 | YES (Upregulated) | 1.495 | 0.121 | NO |
| RIPK1 | 6.101 | 4.253 | YES (Upregulated) | 1.347 | 0.465 | NO |
| SDF4 | 8.346 | 0.535 | YES (Upregulated) | 0.996 | 0.556 | NO |
| SFN | 2.435 | 0.263 | NO | 2.558 | 0.180 | YES (Upregulated) |
| SLC4A1AP | 2.952 | 0.878 | YES (Upregulated) | 1.078 | 0.047 | NO |
| SOD2 | 6.109 | 1.587 | YES (Upregulated) | 1.145 | 1.153 | NO |
| SRSF2;SRSF8 | 6.395 | 3.138 | YES (Upregulated) | 1.507 | 6.084 | NO |
| SUCLG2 | 7.160 | 0.323 | YES (Upregulated) | 0.926 | 0.626 | NO |
| TALDO1 | 7.875 | 3.891 | YES (Upregulated) | 1.025 | 0.292 | NO |
| TKT | 3.950 | 2.514 | YES (Upregulated) | 1.156 | 0.338 | NO |
| TMSB15B;TMSB15A | 0.585 | 2.257 | NO | 5.868 | 2.490 | YES (Upregulated) |
| TMSB4X | 15.253 | 9.388 | YES (Upregulated) | 1.263 | 0.124 | NO |
| TNRC11;MED12 | 7.550 | 1.477 | YES (Upregulated) | NaN | NaN | NO |
| TOP2A | 5.088 | 2.515 | YES (Upregulated) | 3.521 | 2.395 | NO |
| TUBB6 | 3.253 | 3.862 | YES (Upregulated) | 0.959 | 0.179 | NO |
| UACA | 2.560 | 5.024 | NO | 2.852 | 0.217 | YES (Upregulated) |
| UHRF1 | 1.279 | 0.522 | NO | 0.187 | 0.029 | YES (Downregulated) |
| VPS4A | 4.048 | 0.848 | YES (Upregulated) | 1.041 | 0.125 | NO |
| WDR5 | 2.186 | 2.707 | YES (Upregulated) | 1.485 | 0.757 | NO |
Experimental Animals
For the in vivo validation of the selected differentially expressed proteins found in SILAC experiments, we used 16-week-old C57BL/6 healthy female and male mice. To assess the effect of sex in the context of diabetes, we employed female and male streptozotocin (STZ)-induced mice. Diabetes was induced to 10-week-old mice following the High Dose STZ Induction Protocol from the Animal Models of Diabetic Complications Consortium with slight modifications. Briefly, 4-hour-fasted mice were given two intraperitoneal injections of 150 mg/Kg STZ (Sigma, Saint Louis, MO) in two consecutive weeks as previously reported (43, 44). We also studied 16-week-old female and male diabetic Akita (Ins2WT/C96Y) mice. Five to eight animals were included in each experimental group. Mice were anesthetized with isoflurane and sacrificed by terminal surgery. Kidneys were removed, weighted, snap frozen, and kept at −80 °C until further analysis. Half of the right kidney was maintained in 10% formalin solution and paraffin embedded for histological studies. Mice were housed in ventilated cages with full access to chow and water at the Division of Comparative Medicine at University of Toronto (control and Akita mice), or at the Animal Facility of Barcelona Biomedical Research Park (STZ-induced mice). All experiments were conducted under the guidelines of the University of Toronto Animal Care Committee and the Ethical Committee of Animal Experimentation of Barcelona Biomedical Research Park (CEEA-PRBB).
Western Blot
Proteins from sex hormone-treated cell pellets and mouse kidney cortex were solubilized in modified RIPA buffer (150 mm sodium chloride, 50 mm Tris-HCl (pH 7.4), 1 mm EDTA, 1% v/v Triton X-100, 1% w/v sodium deoxycholic acid, 0.1% v/v SDS) and extracted by sonication. Protein concentration was determined using a Coomassie (Bradford) protein assay reagent (Pierce, Waltham, MA). 15 μg of protein were loaded onto 12% acrylamide gels and separated by SDS-PAGE. Membranes were incubated with antibodies to pSer473 AKT (193H12, Cell Signaling, Danvers, MA), p44/42 ERK1/2 (9102, Cell Signaling), GPI (PA5–26787, Thermo Scientific, Waltham, MA), GNPNAT1 (HPA044647, Atlas Antibodies, Bromma, Sweden) and HADHA (ab54447, Abcam, Cambridge, UK). The secondary antibody was anti-rabbit antibody developed in goat (sc-2004, Santa Cruz Biotechnology, Delaware Ave). Control for protein loading was performed by reblotting membranes for β-actin using a mouse monoclonal antibody (A1978, Sigma) and an anti-mouse secondary antibody (sc-3697, Santa Cruz Biotechnology). Following detection, bands were quantified by densitometry with Image J software. For these in vitro verification and in vivo validation experiments, three to six PTEC replicates per condition and five to ten animals per group were studied, respectively.
Kidney Histology: Oxidative Stress and Immunolocalization Studies
For all kidney histology studies, paraffin blocks were cut into 3 μm sections, deparaffinized in xylene and rehydrated through graded alcohols.
Oxidative stress is generally identified by indirect markers such as peroxynitrite production. For this reason, nitrotyrosine staining was used to evaluate superoxide and peroxynitrite levels in renal tissues as previously described (45). Deparaffinized samples were boiled in 10 mm sodium citrate solution (pH 6.0) for antigen retrieval. Kidney sections were stained with rabbit antinitrotyrosine antibody (1:500, Millipore, Billerica, MA). Fifteen microphotographs at ×40 were taken for each sample, and brown-stained areas were quantified with ImageJ software. Data are expressed as percentage of positive area. All analyses were performed in a blinded fashion. For these experiments, six to eight animals per group were studied.
To localize top candidate proteins from our proteomic data set in the murine kidney, immunohistochemistry staining was also performed for HADHA, GPI and GNPNAT1. Samples were boiled in 10 mm sodium citrate solution (pH 6.0) for GNPNAT1 staining, whereas antigen retrieval for HADHA and GPI was performed in 10 mm Trizma® base, 0.5 m EDTA, 0.05% Tween buffer (pH 9.0). Sections were incubated with rabbit primary antibodies for HADHA (1:1000; ab54447, Abcam), GPI (1:1000; PA5–26787, Thermo Scientific) or GNPNAT1 (1:500; HPA044647, Atlas Antibodies). HRP-conjugated anti-rabbit (EnVisionTM, Dako, Santa Clara, CA) was used as secondary antibody. Slides were counterstained with hematoxylin. For these experiments, three to six animals per group were studied.
Statistical and Bioinformatics Analyses
For the in vitro proteomics analysis, Perseus software (version 1.5.1.6) was used for calculation of significance A of protein ratios. Benjamini-Hochberg FDR procedure was used to calculate p values and p < 0.01 was considered statistically significant. The corresponding gene IDs of the 76 sex hormone-regulated proteins were used to perform the gene ontology enrichment analyses using BiNGO plugin in Cytoscape software (version 3.2.1) (46, 47). For the statistical analyses in BiNGO, hypergeometric test with Benjamini-Hochberg correction was used and corrected p < 0.05 was considered significant. In addition, Enrichment Map plugin, was used to assess the significantly enriched functional categories among our DHT-regulated proteins and other previous data sets reflecting the effect of diabetes (48) on kidney transcriptome. These data sets were publically available in GEO and Nephromine online platforms. Hypergeometric test with FDR = 0.05 and Jaccard coefficient cutoff = 0.25 was set to build the enrichment map. Hypergeometric test cutoff of 0.05 was set for post-analysis involving comparison of our data set with the diabetic transcriptional data set above.
For the in vitro verification of SILAC ratios and the additional in vitro studies, densitometries of Western blot bands are reported as mean ± S.E. For the in vivo validation studies, values for physiological parameters, Western blot analysis and quantification of nitrotyrosine positive area are also reported as mean ± S.E. Statistical comparison between experimental groups was performed using a two-tailed Mann-Whitney U test. Spearman's correlation coefficient was calculated when appropriate. p values<0.05 were considered statistically significant.
RESULTS
Sex Hormone Stimulation of PTEC
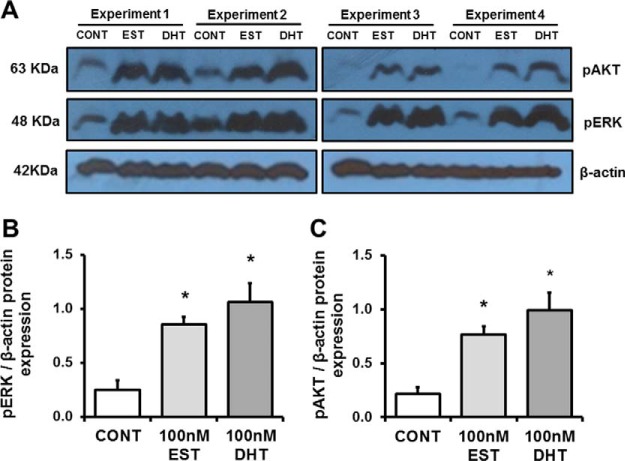
As shown in Fig. 1, PTEC at the 4th and 5th passage were split into T25 flasks, deprived of serum for 18 h and treated with ethanol (CONT), DHT, or EST for 8 h. Experiments 1 and 2 were performed at passage 4, whereas experiments 3 and 4 were performed at passage 5. Cell lysates were collected and lysed as described under Experimental Procedures. As a control experiment, we determined in each passage levels of phosphorylated AKT and ERK after 10min stimulation with DHT or EST. Western blot analysis showed that both sex hormones led to phosphorylation of ERK (Fig. 2A,B) and AKT (Fig. 2A, 2C) in comparison with the control cells. This control experiment demonstrated that DHT and EST engaged the androgen and estrogen receptor, respectively, leading to predictable signaling events in the cells. Although these phosphorylation events are two of many initiated by sex hormones, this was an important confirmation that PTEC exhibited a biological response to DHT and EST.
Fig. 2.
Control experiments. A, shows a representative Western blot demonstrating phosphorylation of AKT and ERK after stimulation with EST and DHT, but not in cells exposed to ethanol alone (CONT). Densitometry analysis of each band was performed using Image J software. Intensities for pERK (B) and pAKT (C) were calculated and normalized to β-actin. Data are expressed as means ± S.E. *p < 0.05 compared with control cells.
Spike-in SILAC Quantitative Analysis of Sex Hormone-treated PTEC Proteome
For accurate, in-depth characterization of the proteomes in these different conditions we employed the advantages of spike-in SILAC (29, 30). We thus labeled an immortalized proximal tubular cell line (HK-2) with heavy-SILAC labels, and used it in spike-in experiments for relative quantification. We first evaluated the efficiency of labeling of HK-2 cells. Although even five doublings demonstrated excellent labeling efficiency, we selected eight doublings, which showed the highest labeling efficiency (99.66%) (supplemental Table S1). Proteins from these cells were spiked into the lysate of each of the samples described above, acting as an internal heavy standard to quantify the light proteome of PTEC under each experimental condition. Mixed proteomes were then analyzed by online liquid chromatography mass spectrometry (LC MS/MS) (Fig. 1). EST-treated PTEC in experiment 1 ultimately showed poor protein recovery after peptide fractionation, and was thus eliminated from analysis.
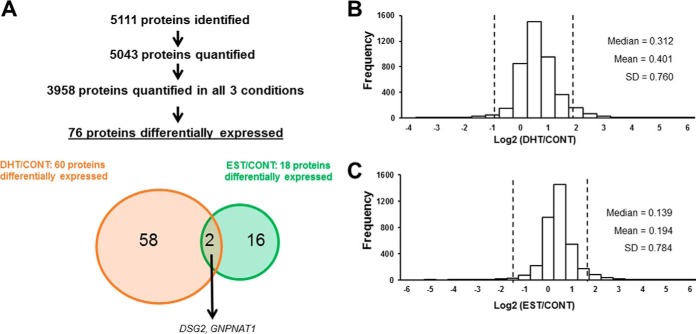
MS analysis identified 5111 proteins. Of those, 5043 were accurately quantified in at least one sample, and 3958 were accurately quantified in at least one replicate of each condition (Fig. 3A, supplemental Table S2). Log2 transformed ratios and their distributions were evaluated. Histograms showed normal distributions for the two ratios (Fig. 3B, 3C). However, histograms for DHT/CONT and EST/CONT were modestly shifted to the right (Fig. 3B, 3C), even after normalization. Spearman's rank correlations were calculated between H/L normalized ratios of two different samples, considering all possible combinations among the 11 samples. As shown in supplemental Table S3, correlation coefficients between biological replicates from the same passage were higher than the coefficients between biological replicates from different passages. This observation was true not only in treated but also in control cells, indicating an intrinsic variability of the PTEC proteome across passages. Similar findings had been reported previously (30). We decided to only consider those proteins that were differentially regulated in both passages, or that at least, did not change significantly in the opposite direction in different passages.
Fig. 3.
Identified, quantified and differentially expressed proteins in our spike-in SILAC approach. The scheme illustrates the number of identified and quantified proteins, followed by a Venn diagram representing the number and overlap of significantly regulated proteins according to significance A for DHT/CONT and EST/CONT ratio values (A). Histograms depicting the distributions of Log2 transformed DHT/CONT (B) and EST/CONT (C) ratios are shown. Vertical lines represent 1.96·S.D.
Based on significance A, a total of 76 proteins were differentially expressed between 2 conditions (Fig. 3A, Table I). The complete list of these proteins and their corresponding ratios in each experiment is in supplemental Table S4. Individual peptide ratios for all differentially regulated candidates were examined manually. Sixty proteins were found to be differentially expressed after treatment with DHT, compared with control cells. Treatment with EST resulted in 18 differentially expressed proteins compared with control cells. As shown in the Venn diagram, 2 proteins were found to be differentially regulated by both male and female sex hormones. Among these 76 proteins, cyclin-dependent kinases 1 (CDK1) and 4 (CDK4) were regulated by DHT and EST, respectively (supplemental Table S4A, S4B). Supporting our findings, CDK1 has been reported to phosphorylate AR preventing its degradation and thereby increasing AR stability and protein expression (49–51), whereas CDK4 was up-regulated after 17β-estradiol treatment in MCF-7 breast cancer cells (52) and rat vascular smooth muscle cells (53).
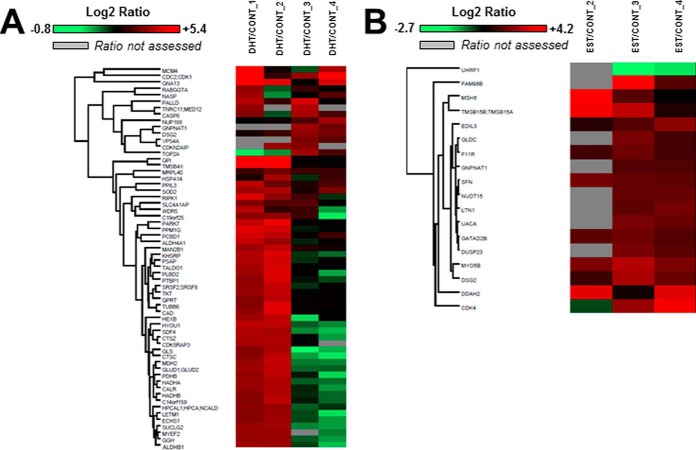
Log2 transformed normalized ratios are shown as heat maps for all DHT/CONT and EST/CONT differentially regulated proteins (Fig. 4). Interestingly, all DHT-regulated proteins were up-regulated relative to control (Fig. 4A, supplemental Table S4). Similarly, only RING-finger type E3 ubiquitin ligase (UHRF1), an important nuclear protein that that plays critical roles in regulating various processes such as DNA methylation maintenance and G1/S transition (54), was found to be downregulated after EST treatment (Fig. 4B, supplemental Table S4). The intrinsic variability across passages is also reflected in the values of the ratios between conditions, as replicates 1 and 2 represent passage 4, and replicates 3 and 4 represent passage 5.
Fig. 4.
Heat map representation of sex-regulated proteins in PTEC. Heat maps were constructed using Perseus software and reflect the value of Log2 transformed normalized DHT/CONT (A) and EST/CONT (B) ratios for each individual protein (rows) in each experiment (columns). The color intensity of red and green boxes correlates with the degree of protein over- and under expression, respectively. A gray box indicates that the ratio was not assessed for that particular protein in that particular experiment.
Gene Ontology and Molecular Pathway Analysis
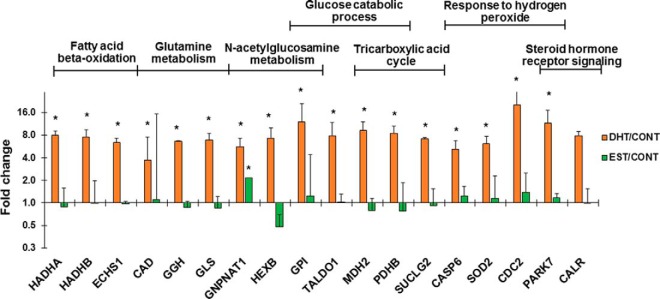
We next evaluated Gene Ontologies of our proteins differentially regulated by sex hormones. Some of the most relevant and significant biological processes enriched among all DHT-regulated proteins were fatty acid beta-oxidation (FAO), glutamine metabolic process, N-acetylglucosamine metabolic process, glucose catabolic process, tricarboxylic acid (TCA) cycle, and response to hydrogen peroxide, indicating DHT-induced alterations in energy metabolism (Fig. 5, supplemental Table S5). In this sense, gene ontology revealed mitochondrion as the most enriched cellular organelle (supplemental Table S5). Steroid hormone receptor signaling biological process was also significantly enriched by DHT treatment (Fig. 5). In addition, positive regulators of androgen signaling pathway such as protein deglycase DJ-1 (PARK7) (55, 56) and calreticulin (CALR) (57) were up-regulated, providing further confirmation of the PTEC biological response to DHT treatment. Biological processes linked to apoptosis were also significantly enriched among EST-regulated proteins (supplemental Table S5).
Fig. 5.
Enriched biological processes among the 60 proteins differentially regulated by DHT. Bar graphs illustrating the Log2 transformed DHT/CONT and EST/CONT ratios of several DHT-regulated proteins involved in the most representative, informative and significantly enriched biological processes according to gene ontology analysis using BinGO plugin in Cytoscape. An asterisk indicate that these protein was significant by significance A with p < 0.01 in at least 2 experiments.
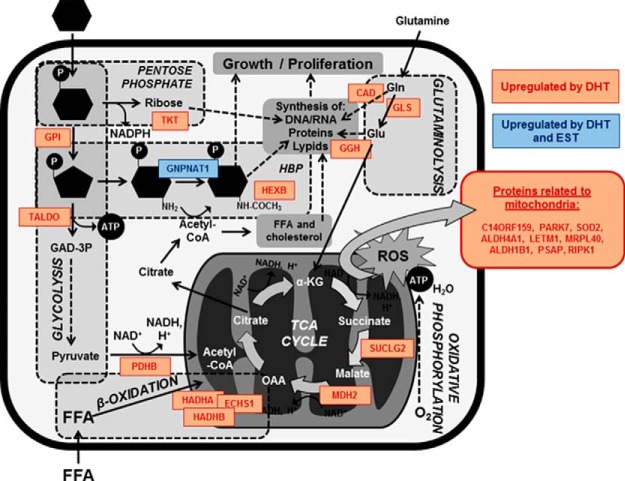
Dysregulated metabolism is a key factor in many kidney diseases (58). In DKD, increased intracellular glucose metabolism leads to excessive activation of hexosamine and protein kinase C pathways, as well as accentuated non-enzymatic glucose oxidation to advanced glycation end products (59). As a consequence, mitochondrial oxidative phosphorylation and reactive oxygen species (ROS) release are promoted. Increased FAO and acetyl-CoA production in TCA cycle accentuate these processes in the mitochondria (60). DHT alone was able to dysregulate processes that are also altered by diabetes such as glucose metabolism, N-acetylglucosamine metabolism (a relevant step in hexosamine biosynthetic pathway, HBP), and FAO. Thus, we selected GPI, GNPNAT1 and HADHA as candidates for validation that represented each one of these three processes. It is remarkable that GNPNAT1 protein expression was increased in both DHT- and EST-treated PTEC, compared with control cells. Fig. 6 illustrates the role of the most representative up-regulated proteins in cellular energy metabolism pathways.
Fig. 6.
Role of DHT- and EST-regulated proteins in energy metabolism. The figure illustrates a simplification of several processes related to energy metabolism and mitochondrial compartment. Proteins that appeared to be up-regulated by DHT (orange), or both DHT and EST (blue) are also represented. GAD-3P, glyceraldehyde-3-phosphate; FFA, free fatty acids; HBP, hexosamine biosynthetic pathway; Gln, glutamine; Glu, glutamate; ROS, reactive oxygen species; α-KG, alpha-ketoglutarate; OAA, oxaloacetate.
In Vitro and In Vivo Validation
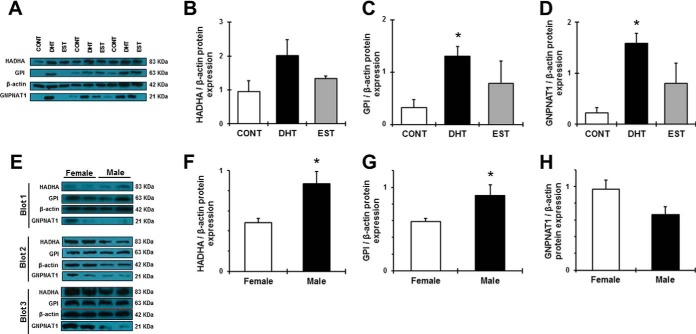
We next aimed to confirm SILAC ratios by Western blot analysis in the same protein extracts used for the proteomic study. In consistence with our findings from SILAC, Western blot analysis in treated PTEC confirmed a significant up-regulation of HADHA, GPI and GNPNAT1 by DHT. To a lesser extent, these proteins were also up-regulated by EST (Fig. 7A–7D). We next examined whether these observations were consistent in new PTEC stimulation experiments, at both passages studied in SILAC experiments. Cells at passage 4 and 5 were thus serum-starved for 18 h and then treated with control, DHT or EST for 8h (n = 5–6 in each passage). Expression of proteins HADHA, GPI and GNPNAT1 was numerically increased in DHT-treated cells at passage 4 (supplemental Table S6). At passage 5, both DHT- and EST-treated PTEC exhibited a significant increase in GNPNAT1 protein expression, a result concordant with SILAC protein expression data. However, HADHA and GPI at passage 5 were not up-regulated by DHT, simulating our SILAC experiments (supplemental Table S4).
Fig. 7.
In vitro and in vivo validation studies for HADHA, GPI, and GNPNAT1. A, shows the immunoblots representing HADHA, GPI, and GNPNAT1 protein expression in the same treated PTEC used for the spike in SILAC study. Intensities for HADHA (B), GPI (C), and GNPNAT1 (D) were calculated and normalized to β-actin (B, C, D). E, shows the immunoblots representing HADHA, GPI, and GNPNAT1 protein expression in the kidney of healthy female and male mice. Densitometry analysis of each band was performed using Image J software. Intensities for HADHA (F), GPI (G) and GNPNAT1 (H) were calculated and normalized to β-actin (F, G, H). Data are expressed as means ± S.E. *p < 0.05 compared with control cells.
We next aimed to examine whether our in vitro observations in PTEC have relevance in vivo. With this purpose, we determined expression of HADHA, GPI and GNPNAT1 proteins in the kidney of healthy female and male mice by immunohistochemistry. As shown in supplemental Fig. S2, both sexes showed strong positive staining for all three candidates in cells from the proximal tubule and other tubular segments. These findings suggest that changes in renal HADHA, GPI and GNPNAT1 across sexes will be mostly ascribed to sex-specific regulation of these proteins in the tubular compartment, strengthening the significance of our in vitro observations. In concordance with our results from SILAC proteomics, Western blot analyses revealed that, under physiological conditions, renal HADHA and GPI were significantly increased in males, compared with females. In contrast, GNPNAT1 was not significantly different between female and male mice (Fig. 7E–7H). Expression of HADHA and GPI renal proteins strongly and significantly correlated with the animal kidney weights (r = 0.675, p = 0.023; r = 0.772, p = 0.005; respectively) which were significantly increased in males, compared with females (supplemental Table S7A).
Biological Significance: Validation of Top Candidates in the Diabetic Kidney
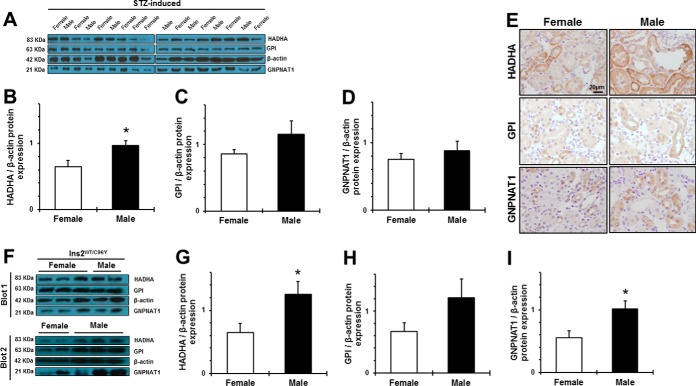
Because our top candidate proteins belong to biological processes critical for DKD (59), namely glycolysis, HBP and FAO, we evaluated the effect of sex on renal HADHA, GPI, and GNPNAT1 protein expression in female and male mice of two different models of diabetes: the STZ-induced and the Akita (Ins2WT/C96Y) models of type 1 diabetes. We specifically chose diabetes as an experimental model because of the significant role of metabolism and the poorly understood impact of sex on the natural history of diabetic nephropathy. Of importance, both STZ-induced and Ins2WT/C96Y diabetic males showed accentuated hyperglycemia compared with females, as well as a more severe renal hypertrophy in terms of kidney weight and kidney to body weight ratio (supplemental Table S7B–S7C). In the setting of type 1 diabetes, male sex was associated to increased HADHA, GPI and GNPNAT1 protein expression, compared with females (Fig. 8). Specifically, renal HADHA was significantly increased, whereas GPI and GNPNAT1 were modestly higher, in STZ males than in females (Fig. 8A–8D). We demonstrated that the expression of these three proteins was predominantly in renal tubules, similar to the control animals (Fig. 8E). These findings were further validated in diabetic Akita mice, a genetic model of type 1 diabetes. Akita males showed significantly higher protein levels of HADHA and GNPNAT1, as well as a modest increase in GPI protein expression (Fig. 8F–8I). Correlation analyses in all the studied diabetic mice revealed a moderate but significant association between HADHA protein expression and kidney weight (r = 0.471, p = 0.013).
Fig. 8.
Protein expression and localization of HADHA, GPI, and GNPNAT1 in the mouse diabetic kidney. The expression of the 3 candidate proteins was evaluated in the renal cortex of female and male diabetic STZ-induced (A–E) and Ins2WT/C96Y (F–I) mice. A, and F, show the immunoblots representing sex differences in HADHA, GPI, and GNPNAT1 protein expression in kidneys of STZ-induced and Ins2WT/C96Y mice, respectively. Densitometry analysis of each band was performed using Image J software. Intensities for HADHA (B, G), GPI (C, H) and GNPNAT1(D, I) were calculated and normalized to β-actin. Renal immunolocalization of the three top candidate proteins was also assessed in STZ-diabetic females and males. Scale Bar = 20 μm. Original magnification ×40. Data are expressed as means ± S.E. *p < 0.05 compared with females.
Enriched Functional Category Analysis
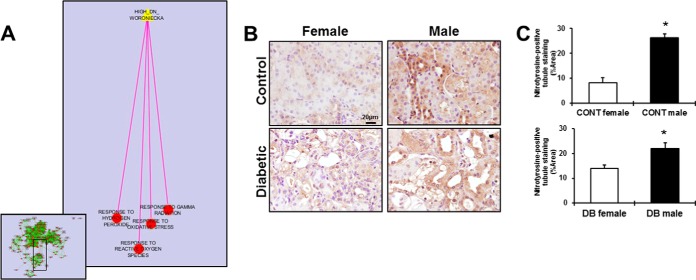
We explored enriched biological processes in our SILAC data that were reproducible in prior studies of diabetes to assess the association between the modulations in renal energy metabolism with DHT, male sex and its deleterious effects on the kidney. For this purpose, we employed Enrichment Map plugin in Cytoscape and evaluated functionally enriched genes between our sex hormone proteomic dataset and previous transcriptomic datasets in kidney tissue (48, 61). Specifically, we superimposed a network of enriched GO terms among our 60 proteins up-regulated by DHT with the gene set up-regulated in human diabetic versus control kidneys (48), and we found that several processes associated with oxidative stress were significantly enriched in both data sets (Fig. 9A). Specifically, response to gamma radiation (p = 0.0056), response to oxidative stress (p = 0.032), hydrogen peroxide (p = 0.032), and response to reactive oxygen species (p = 0.032), were significantly enriched in both proteomic and transcriptomic data sets. Given the recognized importance of oxidative stress in diabetic nephropathy, and its link to our dataset of DHT regulated proteins in kidney cells, we then examined whether oxidative stress was increased in male compared with female mouse kidneys.
Fig. 9.
Functional enrichment of oxidative stress among our DHT-regulated proteins. The enrichment map (EM) was constructed imposing our list of DHT-regulated proteins to a renal transcriptomic data set from human diabetic kidneys in Cytoscape (A). EM is depicted in the left bottom, in the panel. Each circle represents an enriched GO term, with red color indicating significance. Green edges indicate that up-regulated proteins are shared by the two GO terms they connect. The analysis revealed that functional groups associated with oxidative stress (A) were shared with our data set. The pink edge represents overlap between the target gene set and the enriched biological processes from our experiment. To validate the link between male sex and renal oxidative stress, nitrotyrosine immunostaining was used to evaluate superoxide and peroxynitrite levels in kidney sections from female and male, control and STZ-diabetic mice (B). Scale Bar = 20 μm. Original magnification ×40. Brown-stained areas were quantified with ImageJ software (C). Data are expressed as means ± S.E. of positive area fraction (%). *p < 0.05 compared with females.
Examination of kidney tissues of male and female control mice demonstrated significantly increased nitrotyrosine staining in male kidneys (Fig 9B, 9C). Similarly, nitrotyrosine staining was significantly increased in kidneys of diabetic males compared with females (Fig 9B, 9C).
DISCUSSION
This study was designed to shed new light on the biological processes and cellular compartments involved in the deleterious effects of androgens in CKD by studying proteins regulated by male and female sex hormones in the kidney. Our specific goal was to capture a DTH and EST “protein signature” in vitro that reflected an early and consistent response of the PTEC to sex hormones.
To define the proteome of sex hormone-stimulated PTEC we utilized SILAC methodology, which is the current standard for relative quantification of cellular proteomes by MS (25). We identified and quantified over 5000 proteins, thus providing a unique depth of insight into PTEC responses to sex hormones. The strengths of our approach include good proteome coverage, use of instruments with high sensitivity and accuracy, consideration of the effect of cell passage on protein expression, and control experiments to minimize false-positive hits. To the best of our knowledge, this is the first effort to date to characterize proteomic responses of human kidney cells to DHT and EST stimulation.
We first noticed that among our DHT-differentially regulated proteins, many were enzymes that have been shown to play a role in CDK and DKD. As an example, in cultured fibroblasts from type 1 diabetic patients with and without nephropathy, mRNA levels of GPI and other enzymes of the glycolytic pathway were found to be increased in the context of DKD (62). In turn, HADHA and HADHB, as subunits of the mitochondrial trifunctional protein, are involved not only in FAO but also in molecular events relevant to kidney disease, such as lysine acetylation (63), and modulation of renin expression (64), respectively. In addition, GNPNAT1 (which catalyzes a reaction critical for HBP activation) participates in proliferative and hypertrophic processes associated with diabetes (65).
Our next aim was to use systems biology to demonstrate biological processes and molecular pathways fundamental in PTEC response to DHT and EST. Three of the dominant biological processes identified among our 60 DHT/CONT differentially regulated proteins were glucose catabolic process, N-acetylglucosamine metabolic process, and FAO, and one of the most enriched cellular compartments was the mitochondrion. In concordance with our results from SILAC proteomics and MS, validation experiments by Western blot confirmed that GPI and HADHA protein levels were augmented in DHT-treated renal cells and in kidneys from C57BL/6 males, supporting the idea that androgens impair carbohydrate and fatty acid metabolism in the kidney through the action of DHT.
It is known that alterations in the processing of carbohydrates and fatty acids may ultimately compromise the mitochondrial function and the oxidative stress status within the cell (59). Thus, the increase in glycolytic enzymes and proteins responsible for FAO and glutamine metabolism after DHT treatment may explain the up-regulation of several TCA cycle enzymes such as malate dehydrogenase, as well as the increased expression of many proteins related to the mitochondrial compartment (Fig. 6). Furthermore, in this work, we demonstrate that perturbed energy metabolism and up-regulation of mitochondrial proteins by male sex hormones are associated with increased oxidative stress levels in male kidneys under non-pathological conditions. Therefore, our findings suggest that these processes may play a critical role in mediating the deleterious effects of androgens in the physiology of the renal cells. In this sense, DHT has been shown to alter glycolytic metabolism and impair mitochondrial function in other cell types (66, 67). As recently reported by Wang et al., pancreatic islets from DHT-treated rats showed significant changes in the levels of several key genes involved in mitochondrial biogenesis, mitochondrial oxygen consumption rate, and ATP production. The authors also demonstrated that androgens can directly impair beta cell function by inducing mitochondrial dysfunction in vitro in an AR-dependent manner (66). In a different context, DHT enhanced glucose consumption and lactate export in prostate cancer cells (67).
We speculate that increased metabolism of sugar, lipid and amino acids in DHT-treated PTEC induced an imbalance on the levels of several metabolites such as pyruvate, α-ketoglutarate and acetyl-CoA. Thus, the increase in the protein expression of pyruvate dehydrogenase and other enzymes involved in N-acetylglucosamine metabolism and TCA cycle may be explained, at least in part, as an attempt of the tubular cell to counterbalance the abnormal accumulation of these metabolites. For example, GNPNAT1, one of the key enzymes of the HBP that was found to be significantly increased by DHT in both SILAC and Western blot experiments, uses acetyl-CoA as the acetyl group donor for the conversion of glucosamine-6-phosphate to N-acetylglucosamine-6-phosphate.
The enrichment in energy metabolism processes after DHT stimulation, together with the association between male sex and renal oxidative stress levels, pointed out a possible activation of the classic and well defined mitochondrial-dependent apoptotic pathways (68–70). It has been reported that long exposure to testosterone (24 h to 48 h) promoted apoptosis in HK-2 cells and PTEC (8, 13). We decided to treat PTEC for only 8 h, a time that we considered long enough to observe both genomic and nongenomic actions of sex hormones, but still prevented us from the interference of androgen-induced apoptosis and cell death in our cell culture. Thus, the lack of a more significant number of apoptotic players among our DHT proteomic signature despite the increase in proteins related to energy metabolism may be indicative of an early, adaptive response of the cell to DHT signaling, preceding the gradual development of pathological consequences because of a continuous impaired metabolism and subsequent accumulation of ROS. In this sense, protein expression of the antioxidant enzyme SOD2 was also found to be up-regulated by DHT, together with CASP6 and PARK7 as part of response to hydrogen peroxide. SOD2 is responsible for the conversion of superoxide anion to hydrogen peroxide (71). Decreases in SOD2 expression and activity have been associated with exacerbated levels of oxidative stress and mitochondrial dependent apoptosis in HK-2 cells and murine kidneys after long exposures to high glucose, angiotensin II (72), or nephrotoxic reagents such as cisplatin (73). From this point of view, evidence for not decreased but increased SOD2 after 8 h of DHT treatment in PTEC suggests that oxidative stress and apoptosis are partially blunted by a physiological early response of the tubular cell. Furthermore, several proteins related to mitochondrial p53-mediated apoptosis, namely BAX, BCLAF1, BCL2L13, TP53BP1, and TP53BP2, were detected and quantified in all four experiments but were not affected by DHT treatment (data available in Proteome Xchange).
Cumulative evidence has linked altered energy metabolism and mitochondrial modification to the pathogenesis of diabetes and its complications (74). In this sense, proteomic efforts in different tissues from mouse models of type 1 diabetes (T1DM) detected increased expression of several mitochondrial proteins related to FAO, TCA cycle and oxidative stress (75, 76). Within the diabetic kidney, label-free quantification of the mitochondrial proteome revealed increased FAO protein content and induction of TCA cycle enzymes (76). In this sense, renal mitochondrial complex III of the respiratory chain, one of the major sites for ROS generation (71), was found to be altered in the early stage of STZ-induced T1DM in rats (77). We provide strong evidence supporting that these metabolic processes relevant to diabetes are impaired by male sex hormones and increased in diabetic males, compared with females. In addition, the increase in renal oxidative stress levels in diabetic males indicates that these alterations may ultimately lead to a more rapid kidney disease progression in males. In accordance with our results, male sex has been associated with higher urinary and kidney levels of oxidative stress in animal models of hypertension (4) and renal ischemia (78), respectively. To our knowledge, we are the first to report a sexual dimorphism in renal nitrotyrosine levels in the diabetic kidney.
As defended by Forbes and coauthors, it is important for the cells to maintain glucose homeostasis when exposed to hyperglycemic conditions by reducing the transport of glucose inside the cells. However, certain cell groups are unable to decrease glucose concentration, and are thus susceptible to damage (79). From this perspective, it is conceivable that several molecular pathways related to energy metabolism are activated in the tubular cell as a mechanism to compensate the tremendous increase in glucose influx under diabetic conditions. Among these pathways, the role of HBP in the pathophysiology of diabetic cardiorenal disease has been extensively studied (80–82). UDP-GlcNAc, the donor sugar for O-GlcNAcylation of proteins, is synthesized from glucose, glutamine, and UTP via the HBP. In turn, GNPNAT1 requires acetyl-CoA, which can be generated through FAO or transformation of citrate from TCA cycle by ATP-citrate lyase activity. Thus, it is generally accepted that HBP sits at the nexus of glucose, nitrogen, fatty acid and nucleic acid metabolism, which are altered in the context of diabetes (83). According to our SILAC data, these metabolic pathways were enhanced in DHT- but not EST-treated PTEC. This may explain the fact that renal GNPNAT1 appeared to be higher in our diabetic but not control male mice as compared with females.
It is of importance that diabetic male mice developed renal hypertrophy and accentuated hyperglycemia in comparison to diabetic females. In addition, significant correlations between renal HADHA and GPI protein expression and kidney weight were found in the present study, even under non-diabetic conditions. In this sense, metabolic remodeling has been extensively associated to maladaptative hypertrophic processes in other organs such as the heart (84), the pancreas (85) and the adipose tissue (86). In particular, remodeling of glucose (87) and fatty acid (88) metabolism promoted ventricular hypertrophy in experimental models of heart dysfunction. When accompanied by increased oxidative stress levels, these hypertrophic changes are more likely to become maladaptive and lead to organ failure (89). In the endocrine pancreas of mice exposed to a high-fat diet, metabolic changes associated to hypertrophy were found to be more accentuated in males (85). Alterations in processes related to mitochondrial biogenesis also play a role in hypertrophy (90). Interestingly, these processes can be differentially regulated by female and male sex hormones (91). In the kidney, increased amino acid delivery to the proximal tubule cells has been associated with renal growth and hypertrophy; regulation of molecular mechanisms such as AKT-pathways, that are also regulated by sex hormones, play a role in these renal alterations (92). It has been proposed that HBP activation leading to O-GlcNAcylation is also involved in hypertrophy, especially under hyperglycemic conditions (65). Therefore, we presume that glucose influx into HBP was augmented in the kidneys of diabetic males, probably contributing to the increase in protein expression of renal GNPNAT1 and the accentuated renal hypertrophy. Overall, strong experimental evidence demonstrating a link between these hypertrophic processes and perturbations in bioenergetics in the kidney is still lacking. With our data, we reinforce the idea that the maladaptive hypertrophy as a consequence of metabolic remodeling observed in other organs under certain pathological conditions also occurs in the kidney, and in a sex-specific manner.
To date, little is known about the effect of sex on energy metabolism within the kidney. In this work, we are the first to provide strong evidence that, under physiologic conditions, DHT and male sex promote higher activation of energy metabolism in the tubular cell and in the renal cortex. We also demonstrate that renal expression of the three candidates representing glycolysis, N-acetylglucosamine metabolism and FAO was increased in males in the context of T1DM. Further bioinformatics analyses and validation experiments allowed us to link this effect of male sex hormones in renal energy metabolism with accentuated renal hypertrophy and oxidative stress levels. For the first time, we suggest that altered metabolic activity in male kidneys under physiologic conditions confer a major susceptibility to develop renal complications, especially in the context of diabetes.
The strengths of our work include (1) the use of a solid, robust and accurate quantitative proteomic approach that was not previously used in renal cells; (2) good proteome coverage by employing a mass spectrometer with high sensitivity and accuracy, (3) control experiments to minimize false-positive hits, (4) the use of two different models of DKD for validation; (5) the inclusion of publically available data on renal transcriptomics in our systems biology analyses, and (6) the discovery of a new, biologically relevant link between male sex and perturbed renal energy metabolism.
Despite the novelty of our findings, our study has several limitations. Most significantly, we studied PTEC responses to sex hormones at a single time point, and the expression levels of proteins are likely to be dynamic. Although we studied a single time point, the variety of identified processes has been implicated to CKD and DKD processes. In turn, the intrinsic variability of PTEC proteome across passages may have probably contributed to the relatively low number of differentially expressed proteins between conditions. The MS analysis of EST-treated PTEC in passage 4 was limited to a single experiment and thus could contribute only marginally to the overall findings.
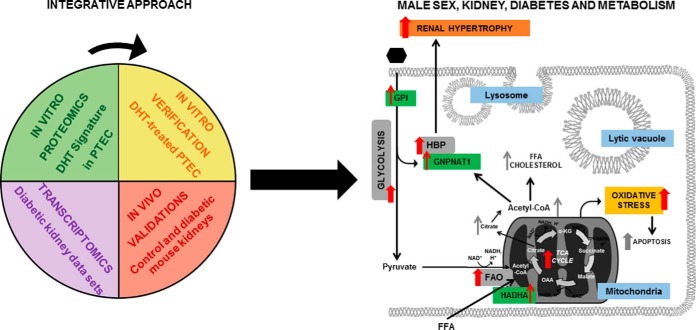
In conclusion, DHT alone led to dysregulated metabolic processes that are also altered in the diabetic kidney. These processes, including glucose metabolism, HBP and FAO, are associated with diabetes and may represent the link to understand the more rapid progression of CKD in males. Top candidate proteins representative of each of these processes were verified and validated in sex hormone-treated PTEC and in male and female control and diabetic mice. Specifically, sex-specific regulation of GPI, HADHA, and GNPNAT1 was demonstrated in vitro and in vivo. To the best of our knowledge, we are the first to demonstrate that GPI, HADHA, and GNPNAT1 are responsive to androgens. Our results suggest that detrimental effects of androgens in diabetic nephropathy and other kidney diseases are mediated, at least in part, by altered energy metabolism within the tubular cell (Fig. 10). Future studies focused on evaluating the influence of male sex hormones on the connection between these metabolic processes, together with other “omics” efforts in DKD approached from a “sex-specific perspective,” may potentially shed new light on the identification of new serum and urine biomarkers for sex-directed therapies in renal disease.
Fig. 10.
Proposed scheme for the role of male sex on energy metabolism in the diabetic kidney. By integrating our proteomic findings with previous transcriptomics data using bioinformatics tools, and supported by the corresponding in vitro and in vivo validation studies, we propose a model of interconnected enzymes, metabolites, biological processes, and pathophysiological events increased by male sex hormones and diabetes within the renal tubule. Red arrows indicate effects of DHT or male sex relevant to diabetes and demonstrated in the present study. Gray arrows depict hypothetical connections that have not been investigated in the present work. Green boxes indicate validated proteins in our study. Gray and blue boxes indicate significantly enriched biological processes and cellular components in our gene ontology analysis, respectively. Yellow boxes indicate processes that emerged from our functional enrichment analysis. Orange boxes indicate physiological events triggered by impaired energy metabolism observed in DKD and accentuated by androgens in the present study. FFA, free fatty acids; α-KG, alpha-ketoglutarate; OAA, oxaloacetate; HBP, hexosamine biosynthetic pathway: FAO, fatty acid beta-oxidation.
Supplementary Material
Acknowledgments
We thank Vanessa Williams, Dr. Dimitrios Korbakis, Julie Van, and Marta Rebull for technical assistance.
Footnotes
Authors' contributions to the study: Participated in research design: Sergi Clotet, Ana Konvalinka, James W. Scholey, Maria Jose Soler, Marta Riera. Conducted experiments: Sergi Clotet, Fei Fang, Joyce Zhou. Performed data analysis: Sergi Clotet, Ana Konvalinka, Ihor Batruch, Stella K. Vasiliou, Apostolos Dimitromanolakis. Wrote or contributed to the writing and editing of the manuscript: Sergi Clotet, Ana Konvalinka, James W. Scholey, Maria Jose Soler, Marta Riera, Stella K. Vasiliou, Clara Barrios, Julio Pascual, Eleftherios P. Diamandis.
* This work has been supported by operating grants from the Kidney Foundation of Canada (KFOC) Award (recipient: Dr. Ana Konvalinka), and from Heart & Stroke foundation, Canadian Institutes of Health Research (CIHR) and KFOC (recipient: Dr. James W. Scholey), ISCIII-FEDER FI11/00480) and ISCIII-FEDER PI14/00557 (recipient: Dr. María José Soler), Fundación SENEFRO 2010 (recipient: Dr. Marta Riera), and ISCIII-RETICS REDinREN RD12/0021/0024 (recipient: Dr. Julio Pascual). Dr. Konvalinka is further supported by the infrastructure and salary award from the Kidney Research Scientist Core Education and National Training (KRESCENT) program and CIHR.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- CKD
- chronic kidney disease
- AR
- androgen receptor
- ARE
- androgen response element
- DHT
- dihydrotestosterone
- DKD
- diabetic kidney disease
- ERα/ERβ
- estrogen receptor alpha/estrogen receptor beta
- ERE
- estrogen response element
- EST
- 17β-estradiol
- FAO
- fatty acid beta-oxidation
- GNPNAT1
- glucosamine-6-phosphate-N-acetyltransferase 1
- GPER
- G protein coupled estrogen receptor 1
- GPI
- glucose-6-phosphate isomerase
- HADHA
- mitochondrial trifunctional protein subunit alpha
- HBP
- hexosamine biosynthetic pathway
- HK-2
- immortalized human kidney cells
- LC-MS/MS
- liquid chromatography – tandem mass spectrometry
- PTEC
- proximal tubular epithelial cells
- ROS
- reactive oxygen species
- SILAC
- stable isotope labeling with amino acids in cell culture
- TCA
- tricarboxylic acid.
REFERENCES
- 1. Levey A. S., Eckardt K. U., Tsukamoto Y., Levin A., Coresh J., Rossert J., De Zeeuw D., Hostetter T. H., Lameire N., and Eknoyan G. (2005) Definition and classification of chronic kidney disease: a position statement from Kidney Disease: Improving Global Outcomes (KDIGO). Kidney Int. 67, 2089–2100 [DOI] [PubMed] [Google Scholar]
- 2. Neugarten J., and Golestaneh L. (2013) Gender and the prevalence and progression of renal disease. Adv. Chronic Kidney Dis. 20, 390–395 [DOI] [PubMed] [Google Scholar]
- 3. Neugarten J., Acharya A., and Silbiger SR. (2000) Effect of gender on the progression of nondiabetic renal disease: a meta-analysis. J. Am. Soc. Nephrol. 11, 319–329 [DOI] [PubMed] [Google Scholar]
- 4. Pendergrass K. D., Pirro N. T., Westwood B. M., Ferrario C. M., Brosnihan K. B., and Chappell M. C. (2008) Sex differences in circulating and renal angiotensins of hypertensive mRen(2). Lewis but not normotensive Lewis rats. Am. J. Physiol. Heart Circ. Physiol. 295, H10–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yamaleyeva L. M., Gilliam-Davis S., Almeida I., Brosnihan K. B., Lindsey S. H., and Chappell M. C. (2012) Differential regulation of circulating and renal ACE2 and ACE in hypertensive mRen2.Lewis rats with early-onset diabetes. Am. J. Physiol. Renal Physiol. 302, F1374–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Alencar Franco Costa D., Todiras M., Campos L. A., Cipolla-Neto J., Bader M., and Baltatu O. C. (2015) Sex-dependent differences in renal angiotensinogen as an early marker of diabetic nephropathy. Acta Physiol. 213, 740–746 [DOI] [PubMed] [Google Scholar]
- 7. Clotet S., Riera M., Pascual J., and Soler M. J. (2016) RAS and sex differences in diabetic nephropathy. Am. J. Physiol. Renal Physiol 00292 2015 [DOI] [PubMed] [Google Scholar]
- 8. Verzola D., Gandolfo M. T., Salvatore F., Villaggio B., Gianiorio F., Traverso P., Deferrari G., and Garibotto G. (2004) Testosterone promotes apoptotic damage in human renal tubular cells. Kidney Int. 65, 1252–1261 [DOI] [PubMed] [Google Scholar]
- 9. Silbiger S., Lei J., Ziyadeh F. N., and Neugarten J. (1998) Estradiol reverses TGF-beta1-stimulated type IV collagen gene transcription in murine mesangial cells. Am. J. Physiol. 274, F1113–F1118 [DOI] [PubMed] [Google Scholar]
- 10. Doublier S., Lupia E., Catanuto P., Periera-Simon S., Xia X., Korach K., Berho M., Elliot S. J., and Karl M. (2011) Testosterone and 17beta-estradiol have opposite effects on podocyte apoptosis that precedes glomerulosclerosis in female estrogen receptor knockout mice. Kidney Int. 79, 404–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xu Q., Prabhu A., Xu S., Manigrasso M. B., and Maric C. (2009) Dose-dependent effects of dihydrotestosterone in the streptozotocin-induced diabetic rat kidney. Am. J. Physiol. Renal Physiol. 297, F307–F315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maric C. (2009) Sex, diabetes and the kidney. Am. J. Physiol. Renal Physiol. 296, F680–F688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Verzola D., Villaggio B., Procopio V., Gandolfo M. T., Gianiorio F., Fama A., Tosetti F., Traverso P., Deferrari G., and Garibotto G. (2009) Androgen-mediated apoptosis of kidney tubule cells: role of c-Jun amino terminal kinase. Biochem. Biophys. Res. Commun. 387, 531–536 [DOI] [PubMed] [Google Scholar]
- 14. Reed D. K., and Arany I. (2014) Sex hormones differentially modulate STAT3-dependent antioxidant responses during oxidative stress in renal proximal tubule cells. In Vivo 28, 1097–1100 [PubMed] [Google Scholar]
- 15. Hammes S. R., and Davis P. J. (2015) Overlapping nongenomic and genomic actions of thyroid hormone and steroids. Best Pract. Res. Clin. Endocrinol. Metab. 29, 581–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cato A. C., Nestl A., and Mink S. (2002) Rapid actions of steroid receptors in cellular signaling pathways. Sci. STKE, re9. [DOI] [PubMed] [Google Scholar]
- 17. Hammes S. R., and Levin E. R. (2007) Extranuclear steroid receptors: nature and actions. Endocr. Rev. 28, 726–741 [DOI] [PubMed] [Google Scholar]
- 18. Hammes S. R., and Levin E. R. (2011) Minireview: Recent advances in extranuclear steroid receptor actions. Endocrinology 152, 4489–4495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Filardo E. J., Quinn J. A., Bland K. I., and Frackelton A. R. Jr. (2000) Estrogen-induced activation of Erk-1 and Erk-2 requires the G protein-coupled receptor homolog, GPR30, and occurs via trans-activation of the epidermal growth factor receptor through release of HB-EGF. Mol. Endocrinol. 14, 1649–1660 [DOI] [PubMed] [Google Scholar]
- 20. Ho K. J., and Liao J. K. (2002) Nonnuclear actions of estrogen. Arterioscler. Thromb. Vasc. Biol. 22, 1952–1961 [DOI] [PubMed] [Google Scholar]
- 21. Sen A., O'Malley K., Wang Z., Raj G. V., Defranco D. B., and Hammes S. R. (2010) Paxillin regulates androgen- and epidermal growth factor-induced MAPK signaling and cell proliferation in prostate cancer cells. J. Biol. Chem. 285, 28787–28795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Freeman M. R., Cinar B., Kim J., Mukhopadhyay N. K., Di Vizio D., Adam R. M., and Solomon K. R. (2007) Transit of hormonal and EGF receptor-dependent signals through cholesterol-rich membranes. Steroids 72, 210–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang C., Liu Y., Cao J. M. (2014) G protein-coupled receptors: extranuclear mediators for the non-genomic actions of steroids. Int. J. Mol. Sci. 15, 15412–15425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pi M., Parrill A. L., and Quarles L. D. (2010) GPRC6A mediates the non-genomic effects of steroids. J. Biol. Chem. 285, 39953–39964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., and Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 26. Mann M. (2006) Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell Biol. 7, 952–958 [DOI] [PubMed] [Google Scholar]
- 27. Lamond A. I., Uhlen M., Horning S., Makarov A., Robinson C. V., Serrano L., Hartl F. U., Baumeister W., Werenskiold A. K., Andersen J. S., Vorm O., Linial M., Aebersold R., and Mann M. (2012) Advancing cell biology through proteomics in space and time (PROSPECTS). Mol. Cell. Proteomics 11, O112.017731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Geiger T., Cox J., Ostasiewicz P., Wisniewski J. R., and Mann M. (2010) Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat. Methods 7, 383–385 [DOI] [PubMed] [Google Scholar]
- 29. Geiger T., Wisniewski J. R., Cox J., Zanivan S., Kruger M., Ishihama Y., and Mann M. (2011) Use of stable isotope labeling by amino acids in cell culture as a spike-in standard in quantitative proteomics. Nat. Protoc. 6, 147–157 [DOI] [PubMed] [Google Scholar]
- 30. Zanivan S., Maione F., Hein M. Y., Hernandez-Fernaud J. R., Ostasiewicz P., Giraudo E., and Mann M. (2013) SILAC-based proteomics of human primary endothelial cell morphogenesis unveils tumor angiogenic markers. Mol. Cell. Proteomics 12, 3599–3611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Quan H., Peng X., Liu S., Bo F., Yang L., Huang Z., Li H., Chen X., and Di W. (2011) Differentially expressed protein profile of renal tubule cell stimulated by elevated uric acid using SILAC coupled to LC-MS. Cell Physiol. Biochem. 27, 91–98 [DOI] [PubMed] [Google Scholar]
- 32. Pan C., Kumar C., Bohl S., Klingmueller U., and Mann M. (2009) Comparative proteomic phenotyping of cell lines and primary cells to assess preservation of cell type-specific functions. Mol. Cell. Proteomics 8, 443–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hornburg D., Drepper C., Butter F., Meissner F., Sendtner M., and Mann M. (2014) Deep proteomic evaluation of primary and cell line motoneuron disease models delineates major differences in neuronal characteristics. Mol. Cell. Proteomics 13, 3410–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Quinkler M., Bujalska I. J., Kaur K., Onyimba C. U., Buhner S., Allolio B., Hughes S. V., Hewison M., Stewart P. M. (2005) Androgen receptor-mediated regulation of the alpha-subunit of the epithelial sodium channel in human kidney. Hypertension 46, 787–798 [DOI] [PubMed] [Google Scholar]
- 35. Hsu S. C., Huang S. M., Lin S. H., Ka S. M., Chen A., Shih M. F., Hsu Y. J. (2014) Testosterone increases renal anti-aging klotho gene expression via the androgen receptor-mediated pathway. Biochem. J. 464, 221–229 [DOI] [PubMed] [Google Scholar]
- 36. Hodgkins K. S., and Schnaper H. W. (2012) Tubulointerstitial injury and the progression of chronic kidney disease. Pediatr. Nephrol. 27, 901–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zoja C., Abbate M., and Remuzzi G. (2015) Progression of renal injury toward interstitial inflammation and glomerular sclerosis is dependent on abnormal protein filtration. Nephrol. Dial. Transplant 30, 706–712 [DOI] [PubMed] [Google Scholar]
- 38. Konvalinka A., Zhou J., Dimitromanolakis A., Drabovich A. P., Fang F., Gurley S., Coffman T., John R., Zhang S. L., Diamandis E. P., and Scholey J. W. (2013) Determination of an angiotensin II-regulated proteome in primary human kidney cells by stable isotope labeling of amino acids in cell culture (SILAC). J. Biol. Chem. 288, 24834–24847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Makawita S., Smith C., Batruch I., Zheng Y., Ruckert F., Grutzmann R., Pilarsky C., Gallinger S., Diamandis E. P. (2011) Integrated proteomic profiling of cell line conditioned media and pancreatic juice for the identification of pancreatic cancer biomarkers. Mol. Cell. Proteomics 10, M111.008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cox J., and Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 41. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., and Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 42. UniProt Consortium (2010) The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 38, D142–D148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Soler M. J., Wysocki J., Ye M., Lloveras J., Kanwar Y., and Batlle D. (2007) ACE2 inhibition worsens glomerular injury in association with increased ACE expression in streptozotocin-induced diabetic mice. Kidney Int. 72, 614–623 [DOI] [PubMed] [Google Scholar]
- 44. Clotet S., Soler MS., Rebull M., Gimeno J., Gurley S., Pascual J., and Riera M. (2016) Gonadectomy prevents the increase in blood pressure and glomerular injury in angiotensin-converting enzyme 2 knockout diabetic male mice. Effects on renin-angiotensin system. J. Hypertens. 34(9), 1752–65 [DOI] [PubMed] [Google Scholar]
- 45. Riera M., Anguiano L., Clotet S., Roca-Ho H., Rebull M., Pascual J., and Soler M. J. (2016) Paricalcitol modulates ACE2 shedding and renal ADAM17 in NOD mice beyond proteinuria. Am. J. Physiol. Renal Physiol. 310, F534–F546 [DOI] [PubMed] [Google Scholar]
- 46. Maere S., Heymans K., and Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 47. Cline M. S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B., Hanspers K., Isserlin R., Kelley R., Killcoyne S., Lotia S., Maere S., Morris J., Ono K., Pavlovic V., Pico A. R., Vailaya A., Wang P. L., Adler A., Conklin B. R., Hood L., Kuiper M., Sander C., Schmulevich I., Schwikowski B., Warner G. J., Ideker T., and Bader G. D. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2, 2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Woroniecka K. I., Park A. S., Mohtat D., Thomas D. B., Pullman J. M., and Susztak K. (2011) Transcriptome analysis of human diabetic kidney disease. Diabetes 60, 2354–2369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chen S., Xu Y., Yuan X., Bubley G. J., and Balk S. P. (2006) Androgen receptor phosphorylation and stabilization in prostate cancer by cyclin-dependent kinase 1. Proc. Natl. Acad. Sci. U.S.A. 103, 15969–15974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Amin K. S., Jagadeesh S., Baishya G., Rao P. G., Barua N. C., Bhattacharya S., and Banerjee P. P. (2014) A naturally derived small molecule disrupts ligand-dependent and ligand-independent androgen receptor signaling in human prostate cancer cells. Mol. Cancer Ther. 13, 341–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Willder J. M., Heng S. J., McCall P., Adams C. E., Tannahill C., Fyffe G., Seywright M., Horgan P. G., Leung H. Y., Underwood M. A., and Edwards J. (2013) Androgen receptor phosphorylation at serine 515 by Cdk1 predicts biochemical relapse in prostate cancer patients. Br. J. Cancer 108, 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Prall O. W., Sarcevic B., Musgrove E. A., Watts C. K., and Sutherland R. L. (1997) Estrogen-induced activation of Cdk4 and Cdk2 during G1-S phase progression is accompanied by increased cyclin D1 expression and decreased cyclin-dependent kinase inhibitor association with cyclin E-Cdk2. J. Biol. Chem. 272, 10882–10894 [DOI] [PubMed] [Google Scholar]
- 53. Yang Z., Cheng B., Song J., Wan Y., Wang Q., Cheng B., and Chen X. (2007) Estrogen accelerates G1 to S phase transition and induces a G2/M phase-predominant apoptosis in synthetic vascular smooth muscle cells. Int. J. Cardiol. 118, 381–388 [DOI] [PubMed] [Google Scholar]
- 54. Zhang Z. M., Rothbart S. B., Allison D. F., Cai Q., Harrison J. S., Li L., Wang Y., Strahl B. D., Wang G. G., Song J. (2015) An Allosteric Interaction Links USP7 to Deubiquitination and Chromatin Targeting of UHRF1. Cell Rep. 12, 1400–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Takahashi K., Taira T., Niki T., Seino C., Iguchi-Ariga S. M., and Ariga H. (2001) DJ-1 positively regulates the androgen receptor by impairing the binding of PIASx alpha to the receptor. J. Biol. Chem. 276, 37556–37563 [DOI] [PubMed] [Google Scholar]
- 56. Tillman J. E., Yuan J., Gu G., Fazli L., Ghosh R., Flynt A. S., Gleave M., Rennie P. S., and Kasper S. (2007) DJ-1 binds androgen receptor directly and mediates its activity in hormonally treated prostate cancer cells. Cancer Res. 67, 4630–4637 [DOI] [PubMed] [Google Scholar]
- 57. Obakan P., Barrero C., Coker-Gurkan A., Arisan E. D., Merali S., Palavan-Unsal N. (2015) SILAC-based mass spectrometry analysis reveals that epibrassinolide induces apoptosis via activating endoplasmic reticulum stress in prostate cancer cells. PLoS One 10, e0135788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vaziri N. D. (2014) Role of dyslipidemia in impairment of energy metabolism, oxidative stress, inflammation and cardiovascular disease in chronic kidney disease. Clin. Exp. Nephrol. 18, 265–268 [DOI] [PubMed] [Google Scholar]
- 59. Reidy K., Kang H. M., Hostetter T., and Susztak K. (2014) Molecular mechanisms of diabetic kidney disease. J. Clin. Invest. 124, 2333–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shadel G. S., and Horvath T. L. (2015) Mitochondrial ROS signaling in organismal homeostasis. Cell 163, 560–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Si H., Banga R. S., Kapitsinou P., Ramaiah M., Lawrence J., Kambhampati G., Gruenwald A., Bottinger E., Glicklich D., Tellis V., Greenstein S., Thomas D. B., Pullman J., Fazzari M., and Susztak K. (2009) Human and murine kidneys show gender- and species-specific gene expression differences in response to injury. PLoS One 4, e4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Iori E., Millioni R., Puricelli L., Arrigoni G., Lenzini L., Trevisan R., James P., Rossi G. P., Pinna L. A., and Tessari P. (2008) Glycolytic enzyme expression and pyruvate kinase activity in cultured fibroblasts from type 1 diabetic patients with and without nephropathy. Biochim. Biophys. Acta 1782, 627–633 [DOI] [PubMed] [Google Scholar]
- 63. Vazquez E. J., Berthiaume J. M., Kamath V., Achike O., Buchanan E., Montano M. M., Chandler M. P., Miyagi M., and Rosca M. G. (2015) Mitochondrial complex I defect and increased fatty acid oxidation enhance protein lysine acetylation in the diabetic heart. Cardiovasc. Res. 107, 453–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Adams D. J., Beveridge D. J., van der Weyden L., Mangs H., Leedman P. J., and Morris B. J. (2003) HADHB, HuR, and CP1 bind to the distal 3′-untranslated region of human renin mRNA and differentially modulate renin expression. J. Biol. Chem. 278, 44894–44903 [DOI] [PubMed] [Google Scholar]
- 65. Masson E., Lagarde M., Wiernsperger N., and El Bawab S. (2006) Hyperglycemia and glucosamine-induced mesangial cell cycle arrest and hypertrophy: Common or independent mechanisms? IUBMB Life 58, 381–388 [DOI] [PubMed] [Google Scholar]
- 66. Wang H., Wang X., Zhu Y., Chen F., Sun Y., and Han X. (2015) Increased androgen levels in rats impair glucose-stimulated insulin secretion through disruption of pancreatic beta cell mitochondrial function. J. Steroid Biochem. Mol. Biol. 154, 254–266 [DOI] [PubMed] [Google Scholar]
- 67. Vaz C. V., Marques R., Alves M. G., Oliveira P. F., Cavaco J. E., Maia C. J., and Socorro S. (2015) Androgens enhance the glycolytic metabolism and lactate export in prostate cancer cells by modulating the expression of GLUT1, GLUT3, PFK, LDH and MCT4 genes. J. Cancer Res. Clin. Oncol. 142(1), 5–16 [DOI] [PubMed] [Google Scholar]
- 68. Sinha K., Das J., Pal P. B., and Sil P. C. (2013) Oxidative stress: the mitochondria-dependent and mitochondria-independent pathways of apoptosis. Arch. Toxicol. 87, 1157–1180 [DOI] [PubMed] [Google Scholar]
- 69. Jamil S., Lam I., Majd M., Tsai S. H., and Duronio V. (2015) Etoposide induces cell death via mitochondrial-dependent actions of p53. Cancer Cell Int. 15, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zou Z. Z., Nie P. P., Li Y. W., Hou B. X., Rui L., Shi X. P., Ma Z. K., Han B. W., and Luo X. Y. (2015) Synergistic induction of apoptosis by salinomycin and gefitinib through lysosomal and mitochondrial dependent pathway overcomes gefitinib resistance in colorectal cancer. Oncotarget. DOI: 10.18632/oncotarget.5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lejay A., Meyer A., Schlagowski A. I., Charles A. L., Singh F., Bouitbir J., Pottecher J., Chakfe N., Zoll J., and Geny B. (2014) Mitochondria: mitochondrial participation in ischemia-reperfusion injury in skeletal muscle. Int. J. Biochem. Cell Biol. 50, 101–105 [DOI] [PubMed] [Google Scholar]
- 72. Sun L., Xiao L., Nie J., Liu F. Y., Ling G. H., Zhu X. J., Tang W. B., Chen W. C., Xia Y. C., Zhan M., Ma M. M., Peng Y. M., Liu H., Liu Y. H., and Kanwar Y. S. (2010) p66Shc mediates high-glucose and angiotensin II-induced oxidative stress renal tubular injury via mitochondrial-dependent apoptotic pathway. Am. J. Physiol. Renal Physiol 299, F1014–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Yuan Y., Wang H., Wu Y., Zhang B., Wang N., Mao H., and Xing C. (2015) P53 Contributes to cisplatin induced renal oxidative damage via regulating P66shc and MnSOD. Cell Physiol. Biochem. 37, 1240–1256 [DOI] [PubMed] [Google Scholar]
- 74. Chen X., Wei S., and Yang F. (2012) Mitochondria in the pathogenesis of diabetes: a proteomic view. Protein Cell 3, 648–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shen X., Zheng S., Thongboonkerd V., Xu M., Pierce W. M. Jr, Klein J. B., and Epstein P. N. (2004) Cardiac mitochondrial damage and biogenesis in a chronic model of type 1 diabetes. Am. J. Physiol. Endocrinol Metab 287, E896–905 [DOI] [PubMed] [Google Scholar]
- 76. Bugger H., Chen D., Riehle C., Soto J., Theobald H. A., Hu X. X., Ganesan B., Weimer B. C., and Abel E. D. (2009) Tissue-specific remodeling of the mitochondrial proteome in type 1 diabetic akita mice. Diabetes 58, 1986–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Munusamy S., Saba H., Mitchell T., Megyesi J. K., Brock R. W., and Macmillan-Crow L. A. (2009) Alteration of renal respiratory Complex-III during experimental type-1 diabetes. BMC Endocr. Disord. 9, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Rodriguez F., Nieto-Ceron S., Fenoy F. J., Lopez B., Hernandez I., Martinez R. R., Soriano M. J., and Salom M. G. (2010) Sex differences in nitrosative stress during renal ischemia. Am. J. Physiol. Regul. Integr. Comp. Physiol. 299, R1387–95 [DOI] [PubMed] [Google Scholar]
- 79. Forbes J. M., Coughlan M. T., Cooper M. E. (2008) Oxidative stress as a major culprit in kidney disease in diabetes. Diabetes 57, 1446–1454 [DOI] [PubMed] [Google Scholar]
- 80. Karunakaran U., and Jeoung N. H. (2010) O-GlcNAc modification: friend or foe in and diabetic cardiovascular disease. Korean Diabetes J. 34, 211–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Goldberg H., Whiteside C., and Fantus I. G. (2011) O-linked beta-N-acetylglucosamine supports p38 MAPK activation by high glucose in glomerular mesangial cells. Am. J. Physiol. Endocrinol. Metab. 301, E713–E726 [DOI] [PubMed] [Google Scholar]
- 82. Banerjee P. S., Ma J., and Hart G. W. (2015) Diabetes-associated dysregulation of O-GlcNAcylation in rat cardiac mitochondria. Proc. Natl. Acad. Sci. U.S.A. 112, 6050–6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Slawson C., Copeland R. J., and Hart G. W. (2010) O-GlcNAc signaling: a metabolic link between diabetes and cancer? Trends Biochem. Sci. 35, 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chatham J. C., and Young M. E. (2012) Metabolic remodeling in the hypertrophic heart: fuel for thought. Circ. Res. 111, 666–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Oliveira R. B., Maschio D. A., Carvalho C. P., and Collares-Buzato C. B. (2015) Influence of gender and time diet exposure on endocrine pancreas remodeling in response to high fat diet-induced metabolic disturbances in mice. Ann. Anat. 200, 88–97 [DOI] [PubMed] [Google Scholar]
- 86. Cummins T. D., Holden C. R., Sansbury B. E., Gibb A. A., Shah J., Zafar N., Tang Y., Hellmann J., Rai S. N., Spite M., Bhatnagar A., and Hill B. G. (2014) Metabolic remodeling of white adipose tissue in obesity. Am. J. Physiol. Endocrinol. Metab. 307, E262–E277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kundu B. K., Zhong M., Sen S., Davogustto G., Keller S. R., and Taegtmeyer H. (2015) Remodeling of glucose metabolism precedes pressure overload-induced left ventricular hypertrophy: review of a hypothesis. Cardiology 130, 211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Talati M., and Hemnes A. (2015) Fatty acid metabolism in pulmonary arterial hypertension: role in right ventricular dysfunction and hypertrophy. Pulm. Circ. 5, 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Rawat D. K., Alzoubi A., Gupte R., Chettimada S., Watanabe M., Kahn A. G., Okada T., McMurtry I. F., and Gupte S. A. (2014) Increased reactive oxygen species, metabolic maladaptation, and autophagy contribute to pulmonary arterial hypertension-induced ventricular hypertrophy and diastolic heart failure. Hypertension 64, 1266–1274 [DOI] [PubMed] [Google Scholar]
- 90. Pennanen C., Parra V., Lopez-Crisosto C., Morales P. E., Del Campo A., Gutierrez T., Rivera-Mejias P., Kuzmicic J., Chiong M., Zorzano A., Rothermel B. A., and Lavandero S. (2014) Mitochondrial fission is required for cardiomyocyte hypertrophy mediated by a Ca2+-calcineurin signaling pathway. J. Cell Sci. 127, 2659–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Capllonch-Amer G., Llado I., Proenza A. M., Garcia-Palmer F. J., and Gianotti M. (2014) Opposite effects of 17-beta estradiol and testosterone on mitochondrial biogenesis and adiponectin synthesis in white adipocytes. J. Mol. Endocrinol. 52, 203–214 [DOI] [PubMed] [Google Scholar]
- 92. Al-Awqati Q. (2015) Kidney growth and hypertrophy: the role of mTOR and vesicle trafficking. J. Clin. Invest. 125, 3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.