Abstract
Globally expected changes in environmental conditions, especially the increase of UV irradiation, necessitate extending our knowledge of the mechanisms mediating tree species adaptation to this stress. This is crucial for designing new strategies to maintain future forest productivity. Studies focused on environmentally realistic dosages of UV irradiation in forest species are scarce. Pinus spp. are commercially relevant trees and not much is known about their adaptation to UV. In this work, UV treatment and recovery of Pinus radiata plants with dosages mimicking future scenarios, based on current models of UV radiation, were performed in a time-dependent manner. The combined metabolome and proteome analysis were complemented with measurements of + physiological parameters and gene expression. Sparse PLS analysis revealed complex molecular interaction networks of molecular and physiological data. Early responses prevented phototoxicity by reducing photosystem activity and the electron transfer chain together with the accumulation of photoprotectors and photorespiration. Apart from the reduction in photosynthesis as consequence of the direct UV damage on the photosystems, the primary metabolism was rearranged to deal with the oxidative stress while minimizing ROS production. New protein kinases and proteases related to signaling, coordination, and regulation of UV stress responses were revealed. All these processes demonstrate a complex molecular interaction network extending the current knowledge on UV-stress adaptation in pine.
Forests are relevant players in ecosystems, providing food and raw material for processing industries. To satisfy the current and future demands of timber products, fast-growing species, such as Pinus radiata D. Don, are the current choice to increase forest productivity and long-term sustainability. Both natural and plantation forests are a significant part of the global carbon cycle and important to the well-being of our societies.
One of the future challenges facing forests is the increased UV radiation (280–320 nm), which has increased in recent years (1) and is modeled to continue increasing during the following decades (2). This situation is, and will be, especially acute in many countries, including South American and European (Chile has 1.5 million ha of this species (3); northwestern Spain has ∼300,000 ha (4)), with increments close to 12% in the incidence of radiation in some regions.
The effects of UV radiation in plants involve damage to macromolecules, including DNA and proteins, and the generation of reactive oxygen species (ROS)1 (5, 6), affecting growth and development (7), reproduction and survival (8), and, in consequence, significantly reducing tree productivity. The plant response is based on the activation of protective mechanisms, like altered morphology, photoprotection and photoinhibition, and antioxidant activities (9, 10). However, these processes are very complex. For instance, photoprotection leads not only to a metabolic rearrangement and physiological changes related to pigment content, but also to changes at the transcriptome (silencing of photosynthesis-related genes, activation of oxidative, wound, defense and stress genes) and proteome (degradation of PSII proteins, synthesis of light-protective proteins such as EARLY LIGHT INDUCIBLE PROTEINS (ELIPs), and synthesis of pigments) levels. Furthermore, UV mediates specific photomorphogenetic pathways for altering leaf morphology, and reducing its dry weight and surface (11).
Although many of these molecular responses to UV radiation have been reported mostly in Arabidopsis (12, 13), a small number of physiological and molecular responses to UV radiation have also been described in forest trees, including poplar (14, 15) and pine (16, 17). In Pinus radiata (16), these responses involve a decrease in photosynthetic parameters and photosynthesis-related gene expression, activation of stress-specific and photoprotective genes, and the accumulation of ELIP proteins. As previously shown in model organisms (13, 14), these observations suggest that UV-responsive mechanisms are the consequence of complex regulatory mechanisms, and that an active reprogramming of gene expression, proteome, and metabolome in trees is necessary to adapt to the environmental changes to ensure long-term survival. Despite the importance of these processes, most of the previous works focused on isolated pathways, providing very detailed information about specific responses, leaving aside its integration into a systemic response. The number of comprehensive studies covering system-level responses is scarce, focusing on targeting the transcriptome, proteome, or metabolome, but missing an effective integration of physiological and molecular multilevels (14, 18, 19). Furthermore, these studies were based on the application of high UV dosages, which will be very unlikely reached in the field, thus representing a limited translational application (20) and requiring more realistic designs with direct application to forest management and breeding programs.
To fill this gap, we have employed classical physiological measurements together with mass spectrometry-based, state-of-the-art analytical procedures for characterizing the changes in the metabolome and the proteome in a UV stress and recovery experiment. We employed the predicted daily radiation dosages for next decades (2), in terms of intensity (being modeled an increase of 8% over average current irradiation in European mid latitudes in summertime (21)), and applied for short-term for a maximum of 2 days, to study the changes involved in early responses to UV irradiation and to explore their role in the acquisition of UV tolerance (22). This knowledge may have potential application both for understanding tree physiology and for providing new tolerance markers for breeding programs. Needles were used for all the analyses as they are the photosynthetic organ of the tree and also the main radiation sensors, defining, in last term, both growth capacity and stress-responsiveness. The data were analyzed in a comprehensive manner following an integrative multivariate approach combining the proteomics and metabolomics data with a set of physiological measurements (maximal efficiency of PSII, quantum yield, pigment contents and enzyme activities). By mining the generated datasets, we depicted the responses of Pinus radiata to UV irradiation, showing the dynamic behavior of the metabolic, biochemical and signaling pathways, as well as connections between them. Altogether, this work provides new insights into the response and adaptation process to UV stress, identifying new targets for breeding and forest management programs.
EXPERIMENTAL PROCEDURES
Plant Material and Experimental Design
The assay was conducted on one-year-old seedlings. After cold stratification, the seeds were grown in peat/perlite/vermiculite (3:1:1) in 1.5 L pots. Plants were regularly watered at field capacity, properly fertilized (Osmocote, NPK 10:6:11) and grown under long photoperiod (16:8 h) at 24 ± 2 °C or 16 ± 2 °C (light/dark) in a controlled environment during 18 months prior to the assay. Same conditions were maintained during UV stress assay, in which plants were irradiated with 0.195 W m−2 UV light (250–350 nm; peaking at 300 nm). The UV irradiation started 3 h after the initiation of the light period. Samples, mature apical needles, were taken at different intervals (Fig. 1A): after 2 h (2h), 8 h (8h), and 8 h over two consecutive days (16h). A set of 16 h plants was cultivated under control conditions until they recovered (monitored by photosynthetic performance, see below) and then sampled (Rec). Control plants were maintained under the same greenhouse conditions without UV radiation (Control) (Fig. 1A). Mature needles that were closest to the apex were sampled and immediately frozen in liquid nitrogen. Samples were stored at −80 °C until use.
Fig. 1.
Experimental system and physiological measurements. A, Experimental system and aspect of plantlet tips after the treatment. B, Photosynthetic performance measured as phi PSII (Fv/Fm). Values were normalized regarding the Control. C, Photosynthetic performance measured as Quantum Yield (QY). Values were normalized regarding the Control. D, Needle content of Chlorophyll a, E, Chlorophyll b and F, Carotenoids in all the experimental situations. In all cases, the error bars show S.D., and the letters indicate significant differences (Kruskal-Wallis, p < 0.05).
Photosynthetic Performance Measurements and Needle Pigment Contents
Photosynthetic performance was measured immediately after the end of each treatment employing an imaging/pulse-amplitude modulation fluorimeter (OS1-FL, Opti-Sciences, Hudson) in ten replicates per experimental situation as described by Escandón et al. (23). Pigment contents were performed in four replicates of 100 mg of needles employing the protocol described by Sims et al. (24) with the modifications described in Escandón et al. (23).
Phenolics Autofluorescence Visualization
Needles from Control, 2h and + samples were harvested just after the treatment and immediately imaged by laser confocal microscopy (TCS-SP2-AOBS, Leica, Germany) using the settings described by Kaling et al. (14) for the visualization of phenolic compounds. Briefly, samples were introduced in a cryostat medium (Tissue-Tek, Killik; Sakura Finetek, Inc., Torrance, CA) and frozen at −23 °C. Sections of 50 μm were cut with a sliding cryotome CM1510S (2002 Leica Microsystems, Wetzlar, Germany) and imaged under a laser confocal microscope using a single-line excitation at 364 nm, and 505–530 nm and 385–470 nm as emission ranges of phenolic compound autofluorescence (blue and green channels). Images were processed using Fiji Software (25).
Enzymatic Activities
Enzymatic activities were measured in five replicates of each experimental situation following the protocol by Venisse et al. (26) with slight modifications. Briefly, crude extracts were obtained from 100 mg of fresh needles, ground in liquid nitrogen to a fine powder, and resuspended in extraction buffer (250 mm HEPES pH 7.4, 25 mm CHAPS, 1 mm PEG 8000, 1 mm PMSF and 8% (w/v) PVP 40,000). Proteins were quantified using the Bradford method (27).
Peroxidase (POX, EC 1.11.1.7) activity was measured by monitoring the 470 nm absorbance resulting from the formation of tetraguaiacol when 10 μl of crude extract were added to the reaction mixture containing 50 mm acetate buffer (pH 7.0), 25 mm guaiacol and 25 mm H2O2.
Glutathione-S-transferase (GST EC 2.5.1.18) activity was determined by measuring the absorbance at 340 nm when 10 μl of the crude extracts were added to the reaction mixture (0.1 m potassium phosphate pH 6.5, 3.6 mm reduced glutathione and 1 mm 1-chloro-2,4-dinitrobenzene).
Extraction of RNA, Proteins, and Metabolites for -Omics Analyses
RNA was extracted from three replicates of each situation according to Chang et al. (28) following the modifications described in Valledor et al. (29) and then quantified in a spectrophotometer. Its integrity was checked by agarose gel electrophoresis, and potential DNA contamination was assessed by PCR.
Proteins were precipitated from the phenolic phase obtained during RNA extraction by adding 3 volumes of 0.1 m ammonium acetate in methanol. The protein pellets were washed twice with methanol, rehydrated in 8 m Urea and 4% SDS and quantified using the BCA method (30).
Metabolites were extracted as described by Valledor et al. (31) starting from 40 mg of fresh weight. Polar and nonpolar fractions were recovered.
Quantitative Real-time PCR
The RNA (2 μg) was reversed transcribed using RevertAid (Thermo Scientific, Waltham, MA) and oligo (dT)18 as primers following the manufacturer's instructions.
RT-qPCR reactions were performed in an ABI Prism HT7900 Real Time PCR machine (Applied Biosystems, Foster City, United States) with Brilliant III Ultra-Fast SYBR Green QPCR Master Mix (Agilent, Santa Clara, United States); four biological and three analytical replicates were made. Normalized Relative Quantities (NRQ) and Standard Errors of RQ were determined according to Hellemans et al. (32). Expression levels of ACTIN (ACT) and UBIQUITINE (UBI) were used as endogenous controls. Detailed information about the primers used is available in supplemental Table S1.
Protein Identification and Quantitation (GeLC-Orbitrap/MS)
Proteins were processed and analyzed as described by Valledor et al. (33) with minor modifications. Three biological replicates of each treatment were analyzed. Protein samples (60 μg) were run 0.5 cm in 12% SDS-PAGE gels. Gels were fixed and stained with 0.1% CBB R-250 in methanol/acetic acid/water (40:10:50, v/v/v) for 30 min and destained in methanol/water (40:60, v/v). Protein bands were cleaned, digested with trypsin sequencing grade (Roche) and desalted as previously reported (33). The nLC-Orbitrap (Thermo) system was maintained as previously specified, with only a slight modification in the effective gradient, which was set to 90 min from 5 to 45% acetonitrile/0.1% formic acid (v/v) with a later column regeneration step of 27 min.
The MS/MS data were processed employing Proteome Discoverer 1.4 (Thermo) and identified using SEQUEST algorithm against a selection of NCBI taxa corresponding to Viridiplantae proteins (83269 entries). Furthermore, all available Pine and Picea protein, gene, nucleotide and EST sequences were downloaded from NCBI on early 2013 and then trimmed, contiged, 6-frame translated, and identified following the procedure described by Romero-Rodríguez et al. (34) to develop two specific databases belonging to Pinus and Picea genus (34063 and 67647 entries respectively) that were employed as an independent database. All sequences were downloaded the same date. In brief, identification confidence was set to a 5% FDR, Xcorr above the peptide-charge state to +0.25 and the variable modifications to acetylation of N terminus, oxidation of methionine, with a mass tolerance of 10 ppm for the parent ion and 0.8 Da for the fragment ion, and a maximum of one missed cleavage. Peptide list was generated considering specifically trypsin cleavage. No fixed modifications were considered in this analysis. Unspecific cleavages were not allowed. Peptides were matched against the mentioned databases plus decoys, considering a hit significant when its XCorr was greater than peptide-charge state +0.5 (supplemental Table S2). The results of the three previously described databases were searched in parallel nodes and integrated using the Percolator processing node available in Proteome Discoverer 1.4. Quantification was based on a label-free NSAF normalization strategy (35). Proteins were functionally classified according to Mapman (36) functional bins using a custom map generated for our databases employing the Mercator tool (37).
Metabolite Identification and Quantitation (GC-MS)
Sample derivatization or methyl-esterification and GC-MS measurements were carried out following the procedure previously developed by Furuhashi et al. (38). One μl of sample was injected on a GC-triple quad instrument (TSQ Quantum GC; Thermo). GC separation was performed splitless on a HP-5MS capillary column (30 m x 0.25 mm x 0.25 mm) (Agilent) over a 70 to 200 °C gradient at 3 °C min−1, and then to 250 °C at 10 °C min−1. The mass spectrometer was operated in electron-impact (EI) mode at 70 eV in a scan range of m/z 40–600.
Raw data were processed with Xcalibur Qual Browser 2.1 and LC-Quant 2.6 (both from Thermo Scientific), and metabolites were identified based on their mass spectra and GC retention time by comparison to reference databases (Fiehnlab (39), GMD (40), and inhouse) with NIST software. Typical chromatograms of polar and non-polar groups are shown in supplemental Fig. S1. Metabolite quantitation was performed using the peak-areas of one characteristic mass per metabolite, and peak integration was manually supervised to increase the reliability of the analysis.
Statistical Analyses and Data Integration
Proteome and metabolome datasets were preprocessed following the recommendations of Valledor et al. (41). In brief, missed values were imputed using a k-nearest neighbors approach, and variables were filtered out if they were not present in all replicates of one treatment or in, at least, 45% of the analyzed samples. Data were normalized following a NSAF approach in the case of proteins and a sample-centric approach followed by log transformation in the case of metabolites. Centered and scaled values (z-scores) were subjected to multivariate (Principal Component Analysis (PCA), k-means and Heatmap clustering) and univariate (one-way ANOVA; 5% FDR with an alpha = 0.05) analyses. sPLS was used to integrate physiological and molecular data with the aim to show the interactions between the different organization levels within the cell. Specifically, proteins data were used as predictors for metabolites and physiological data. Generated networks were visualized and filtered (only edges equal or higher than 0.8 were maintained) in Cytoscape v.2.8.3. All analyses were conducted employing R v3.2.3 (42) core functions, and the packages mixOmics v.4.0.2 (43), SeqKNN v.1.0.1 (44), ggplot2 (45), reshape2 (46), and FDRtool v.1.2.10 (47).
RESULTS
Increased UV Irradiation Induced System Level Changes in Pinus radiata Needles
We have studied the effect of a moderate UV irradiation in a five time-points experiment by mimicking the expected environmental dosage. The aim of this design, far from the classic configuration-based high-dosage experiments, was to deepen our understanding of the processes that mediate adaptation and acclimation to UV stress.
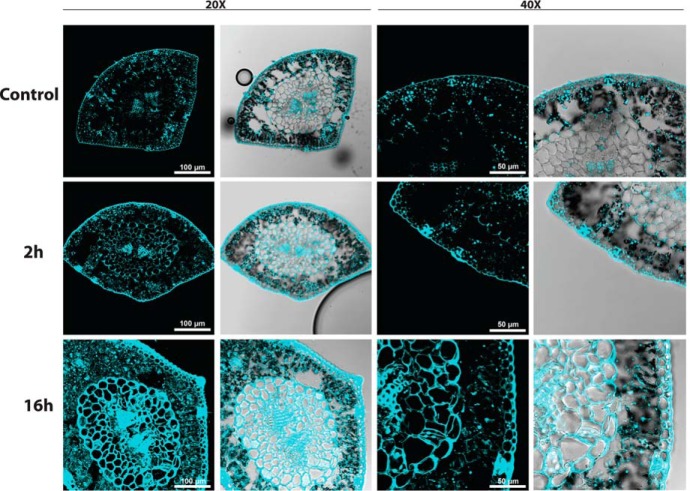
The applied UV dosage did not significantly alter the morphology of the plants, that showed none sign of external damage (Fig. 1A), although phenolic compounds progressively accumulated in the needles: first and quickly in epidermic cells and stomata, and throughout all the tissues in the long UV-stress exposure (Fig. 2). This protective response reflects severe changes at the physiological level, including reduced photosynthesis. Photosynthetic parameters Fv/Fm (Fig. 1B) and QY (Fig. 1C) were reduced significantly after 16 h of UV exposure, whereas the recovery period increased the Fv/Fm to control values. Chlorophylls (Fig. 1D, 1E) and carotenoids (Fig. 1F), which showed similar abundance than previously described in this species (16, 23), decreased in the longer exposure. Stress-recovered plants showed a higher chlorophyll content compared with controls. Furthermore, the applied irradiation also quickly trigger glutathione s-transferase (GST; Fig. 3A), whereas peroxidase (POX; Fig. 3B) showed a gradual decrease in 2h and 8h plants, recovering its activity after 16h of UV. The expression of the genes ASCORBATE PEROXIDASE (APX) and ELIP increased as a consequence of the UV radiation (Fig. 3C), as previously reported (16). However, the expression levels of SUPEROXIDE DISMUTASE (SOD) and GLUTATION-S-TRANSFERASE (GST) were not significantly altered by UV. On the other hand, photosynthesis- and carbohydrate-related genes (AOX, CA, RBCA, RBCS, and HXK1) showed a decrease of abundance after 8 h of irradiation. The latter four genes showed a correlated expression pattern of increased expression in recovered plants.
Fig. 2.
Tissue specific accumulation of phenolic compounds after UV irradiation. Phenolic compound autofluorescence (cyan signal) visualization by confocal laser scanning microscopy in transversal sections of needles from Control, 2h and 16h plants with 20X and 40X magnification. Merged fluorescence and differential interference contrast images are shown in every case.
Fig. 3.
Enzymatic assays and RT-qPCR. A, GST and B, POX-specific activities measured in the different experimental situations. Error bars show the standard error (S.E.). Different letters indicate significant differences (ANOVA followed by a TukeyHSD post-test, p < 0.05). C, Analysis of the relative quantity (RQ) in the different experimental situations of some genes belonging to the antioxidant system, photosynthesis machinery and UV-stress response measured by RT-qPCR. Expression levels are shown regarding the Control and were normalized using ACT and UBI as housekeeping genes. ASCORBATE PEROXIDASE (APX), GLUTATHION S-TRANSFERASE (GST), SUPEROXIDE DISMUTASE (SOD), EARLY LIGHT INDUCIBLE PROTEIN (ELIP), ALTERNATIVE OXIDASE (AOX), CARBONIC ANHYDRASE (CA), RUBISCO ACTIVASE (RBCA), RUBISCO SMALL (RBCS), HEXOKINASE 1 (HXK1), NAC DOMAIN PROTEIN 25 (NAC025), RAB GTPase 4 (RAB4). Error bars show the S.E. of normalized RQ for each gene and each sample scaled to the control.
Altogether, these results showed the triggering of changes affecting the physiology of the plant and involving the entire needle system in order to cope with mild UV irradiation, seeming that basal cell defense mechanisms cannot counterbalance the resultant damage. To further investigate these responses, we performed a comprehensive systemic analysis combining proteomics and metabolomics to understand the molecular mechanisms behind the adaptation to UV stress.
Integrated Proteomic, Metabolomic, and Phenotypic Analyses Revealed the Complexity of UV Stress Response in Pinus radiata
The results of high-throughput proteomic analyses rely on an adequate mass spectrometry and the availability of specific databases. Because Pinus radiata is a nonmodel species, we needed to combine standard with custom-built Pinus databases (34) to identify the proteins obtained in LTQ-Orbitrap instrument. This approach allowed the identification of 1646 protein species. Protein abundances were filtered to keep only those with efficient peptide counts to ensure accurate NSAF-based quantification, resulting in 1228 protein species for quantitative analyses. Proteins were annotated by sequence homology-based searches and identifications were manually curated, with a success annotation rate of 78.74%, with the remainder being unannotated or identified as unknown proteins (supplemental Table S3). After univariate statistical analyses, 747 proteins were considered as differentially accumulated during the time course (ANOVA, 5% FDR). The differentially accumulated proteins belonged to different pathways covering primary and secondary metabolism (Fig. 4), supporting the observations described in the previous section. Furthermore, biclustering heatmap analysis of proteins (Fig. 5; supplemental Fig. S2) showed a distinction of the irradiation time and UV adaptation process, differentiating sample groups and functional clusters, demonstrating the dramatic changes induced by the applied stress.
Fig. 4.
Metabolism overview. Overview of protein abundance changes in the different metabolic pathways accounting the differentially accumulated proteins (ANOVA, 5% FDR, Average of the three biological replicates). Protein abundances were normalized over a maximum value of 1. The number of proteins in each bin is indicated in brackets.
Fig. 5.
Heatmap representation of the proteomic dataset. Hierarchical clustering and heatmap of the analyzed proteins grouped by their function according to MapMan ontology. Manhattan distance and Ward's aggregation method were used for the hierarchical clustering. Numbers inside each box represent the average total abundance of the proteins belonging to each MapMan functional bin found in each experimental situation normalized by the total abundance of proteins in every bin.
GC-MS analyses resulted in the quantification of 118 metabolic compounds, identifying 86 of them after comparison to references. Half of the metabolites showed a differential accumulation at least in one sampling time (ANOVA, 5% FDR; supplemental Table S4), suggesting that UV irradiation-induced damage and defense requires a major reprogramming of metabolism.
The combined analysis of proteome and metabolome levels by PCA showed that the first two components of PCA accounted for almost 50% of the total variance, while grouping and distinguishing the irradiation times (Fig. 6A; supplemental Table S5). The physiological variance gathered by each component was explained by analyzing the variables exhibiting higher loadings to each component; PC1 potentially explained the adaptation to the stress: proteins related to protein folding, synthesis and degradation, like HEAT SHOCK PROTEINS (HSPs), chaperones and ribosomal proteins, positively correlate with this component, whereas photosynthesis, photorespiration, and glycolysis and respiration enzymes were negatively correlated. On the other hand, PC2 explained the adaptation to the stress related to active remodeling of the proteome, and it was positively correlated to MALATE DEHYDROGENASE, FRUCTOSE BISPHOSPATE (FBPase), TRIOSE PHOSPHATE ISOMERASE, and proteins involved in the active remodeling of the proteome. Specific proteases (CLP PROTEASE R SUBUNIT 4, PEPTIDASE FAMILY M48 FAMILY PROTEIN STE24, and CELL DIVISION CYCLE 48) and other proteome remodeling elements (EUKARYOTIC INITIATION FACTOR 3E and RIBOSOMAL PROTEIN S4), and several unannotated proteins were negatively correlated to this component. To address potential PCA bias caused by non-Gaussian effects related to the structure of the data, an Independent Component Analysis (ICA) was performed (Fig. 6B; supplemental Table S6). Independent component 1 (IPC1) gathers those variables related to metabolic adaption to stress that were also associated to PC2, showing the importance of this variables set. On the other hand, IPC2 positively correlated to central carbon metabolism (RuBIsCO, RBC ACTIVASE, ENOLASE, ATPases) and negatively to oxidative-stress related proteins (ACONITASE3, OXIDOREDUCTASES, SUPEROXIDE DISMUTASE, MALATE DEHIDROGENASE) and sucrose metabolism.
Fig. 6.
Multivariate analysis plots and network. Integration of proteomics and metabolomics data A, PCA plot of the first two principal components (PC), explaining almost 50% of the observed variance. B, ICA plot of the first two independent principal components (IPC). C, sPLS-base network constructed using quantified proteins as a predictive matrix for the changes observed in the metabolome and the physiological measures. Red edges represent positive correlations, whereas the green ones represent negative correlation values. Only those correlations equal or higher, in absolute value, than 0.8 are shown (Detailed plots of the different subnetworks are provided in supplemental Fig. S3). The network includes two main clusters, respectively center in EPOX, 1-naphthalenecarboxylic acid (1-NAA) and a couple of unknown metabolites, and in glycolic acid. The first is positively correlated to several ATPases and photosynthesis-related proteins, like RBCL or PSBB. On the contrary, glycolate is negatively correlated to those photosynthesis-related proteins, whereas positively correlates with photorespiration and photorespiration-related enzymes, like SHM4, CAT2 or COP8. Furthermore, there are several light-mediated signaling elements, consistent with the known light-dependent regulation of photorespiration. Other minor clusters represented in the network are centered in pyroglutamic acid, a subproduct of protein degradation, a main process in UV stress response, and in other metabolites related to stress, like phenol or lyxonic acid. It also highlights the presence all over the network of ribosomal proteins and several signaling proteins (kinases, phosphatases and GTPases), most of them revealed here to be relevant for stress response, although their signaling pathway/s are not known.
K-means clustering analysis of all the data (proteomic, metabolomic and physiological) was performed to test the complementarity of the data from the different levels. This analysis allowed the establishment of 30 clusters based on their quantitative trends during the experiment (Fig. 7, supplemental Table S7). The presence of different coexpression patterns showed those variables which are either coregulated or influenced (expressed or repressed by different mechanisms that acts coordinately) following the same dynamics, like light-signaling (COP8, PHOT2, EXL2, and NPH3) and antioxidant (APX) proteins related to photorespiration and the photorespiration enzyme SHM4 in cluster 2. Furthermore, this analysis showed a number of these clusters (i.e. 13, 16, 20, 25, and 27) that did not recover the abundance of controls in the recovered plants, suggesting a potential stress memory effect. Clusters 16 and 25, grouping proteins accumulated at levels higher than in the control after recovery, including central metabolism-related proteins, like ATP SYNTHASES and PS and Calvin cycle proteins; heat shock proteins, like CLPC1 or CPN60; redox-related proteins, such as CAT2, CAT3, and APX4, and the metabolites galactinol and tocopherol.
Fig. 7.
K-means clustering of the proteomic, metabolomics and physiological data. Graphic representation of the variables grouped in the 30 different coexpression clusters that were determined based on the different accumulation patterns showed during the UV experiment. To compare variables with different magnitudes, all variables were normalized to a maximum value of 1 to perform the clustering. The different clusters correspond to different quantitative trends showed by the different variables (physiological, proteomic and metabolomics) in the study. In summary, there are variables accumulated with the UV (cluster 1), peaking after 2 h of UV mainly (clusters 3 and 5), in 8h plants (clusters 2, 7, 12, and 30), in 16h plants (clusters 21 and 29) or showing an abundance peak after recovery (cluster 25). Likewise, there were clusters showing a decreasing with the stress (cluster 14), mainly in 2h plants (cluster 8), in 8h (cluster 9) or after 16 h of UV (cluster 19). Furthermore, there are more complex trends, like those with two abundance peaks at two different treatments (clusters 9 and 26).
Additionally, multivariate analyses based on sPLS was performed for integrating protein (supplemental Table S3), metabolite (supplemental Table S4) and physiological and biochemical data (supplemental Table S8). sPLS-based correlation networks (Fig. 6B) suggested the existence of two main UV-responsive clusters. The first cluster links primary metabolism activities (photosystems, Rubisco, FBAs, ATP synthases) and was centered in EPOX and 1NAA (supplemental Fig. S3A). The negative correlation of this cluster to the two minor nodes centered in lactate and pyroglutamic acid and connected to phenol (supplemental Fig. S3C, S3E) and linking protein targeting, folding and degradation proteins, respectively, highlights the same dynamics observed in both the heatmap and the multivariate analyses. The second main cluster, centered in glycolate, links proteins of the photorespiratory pathway and light-dependent signaling kinases (supplemental Fig. S3B) known to be involved in the transcriptional regulation of this pathway (48), the ascorbate-glutathione cycle, the proteolytic system and the amino acid, fatty acid and isoprenoid metabolism, processes taking place in or closely related to the peroxisome.
Statistical analyses showed that applied UV irradiation triggered abundance changes in a number of variables and pathways that can be, in consequence, considered as UV stress-responsive, for the applied dosage, in Pinus radiata. The changes in key pathways and novel discoveries are depicted below.
UV Induces an Adjustment of Photosystems and Electron Transfer Chain to Prevent Phototoxicity
Photosynthesis performance, as well as chlorophyll content, started decreasing after 16 h of UV radiation and returned to control levels in recovered plants (Fig. 1B–1E). This response was reported to be a consequence of phototoxicity of the radiation (49).
Photodamage under UV stress was compartmentalized into PSII, as indicated by the quick degradation of PSBO-1, PSBO-2, and CP43 after 2 h of UV exposure. This is aimed to protect PSI (50), which is described to remain relatively unaffected under UV radiation (51) (our data set showed that only PSAB and PSAA decreased after UV exposure). Furthermore, we found that other PSI protective elements such as PSBS, an important component for qE (50), was enhanced after long exposure times (1.78-fold) and in recovered plants (1.25-fold). UV also affected cyclic electron flow proteins: CYT B6 decreased under UV, whereas CYT F and PGR1 increased, with its peak observed in 8h plants. FNR2 decreased as consequence of the stress (-1.85-fold), but it increased again in 8h and 16h plants and after recovery (-1.25-fold), whereas FNR1 accumulated only after 8 h. This mechanism, besides protecting PSI (52), also has an important role for ATP production under stress situations (53).
The recovery of photosynthetic performance after stress depended on the activation of the repair systems. LHCB4 accumulation was reduced (-2-fold in 8h plants) by the radiation, whereas in the case of LHCA3 we observed a marked increase (1.96-fold) in 8h plants. The regulation of these mechanisms seems to be based on specific proteases (i.e. FtsH5), as it is discussed below.
Reduced Availability of Energy and Photorespiration Modulate Central Metabolism After UV Exposure
The reduction of the available energy and reducing equivalents as consequence of the photodamage induced to PSII, forced a remodeling of the central metabolic pathways to allow the survival of the cells with a minimum support to the growth.
UV affected Calvin cycle reactions, with a reduced RuBisCO activity based on a decrease of RCA observed in both protein (-1.52-fold) and transcript levels (Fig. 3C). Furthermore RBCS and RBCL, observed at the transcript level (Fig. 3C) and protein level (1.81-fold and −1.34-fold in 8h plants, respectively), showed an imbalanced RBCL/RBCS stoichiometry (supplemental Table S3), required for an adequate assembly of the holoenzyme (54). Oxidative damage seems also to increase RuBisCO oxygenase activity, because glycolate accumulated during the early stages of the stress (1.51-fold after 8 h of UV); the enzyme GLYCOLATE OXIDASE (GOX) followed a similar trend. CATALASE 2 (CAT2), the main photorespiration-related catalase (55), also accumulated during stress, possibly to cope with the increased H2O2 formed as a consequence of glycolate oxidation. This hypothesis is supported by sPLS-based interaction networks in which glycolate was positively correlated to SERINE HYDROXYMETHYLTRANSFERASE 4 (SHM4), which converts glycine into serine in the mitochondrial part of the photorespiration (56), and several light signaling proteins, such as CONSTITUTIVE PHOTOMORPHOGENIC 8 (COP8), PHOTOTROPINE 2 (PHOT2), PHOTOTROPIC-RESPONSIVE NPH3 FAMILY PROTEIN (NPH3), and EXORDIUM-LIKE 2 (EXL2), also related to this metabolite. The branches conformed by these enzymes are consistent with the proposed light-mediated regulation of this pathway (48, 57). Moreover, the positive correlation to enzymes of the ascorbate-glutathione cycle, like APX, transcriptionally induced with UV (Fig. 3C), highlights the involvement of photorespiration on ROS homeostasis. These light-signaling proteins, APX and SHM4 proteins, were grouped in the cluster 2 of K-means analysis (Fig. 7, supplemental Table S7).
As expected, the reductive phase of photosynthesis decreased under stress, since the key enzymes PHOSPHOGLYCERATE KINASE (PGK) and GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE (GAPDH), reduced their accumulation, although some of the GAPDH isoforms increased in 2h plants. Different plastidial isoforms of FRUCTOSE-BISPHOSPHATE ALDOLASE (FBA) were differentially accumulated: FBA6 increased after 2 h (1.88-fold) and FBA8 gradually decreased (-1.53-fold in 16h and recovered plants), showing the precise control of the isoforms of this family depending on the environmental and physiological circumstances, including the redox state of the cells, as has been previously reported for other abiotic stresses (58, 59).
Glycolysis was maintained during short-term exposure periods; long exposure times led to the accumulation of cytosolic FBA6, whereas plastidic FBA1 and FBA2 decreased. The pool of glucose-6P resulting from a block of glycolysis would enter in the pentose phosphate pathway (PPP) because 6-PHOSPHOGLUCONATE ISOMERASE (6PGI) accumulated after 8 h.
The longer-term decrease of RIBOSE 5P ISOMERASE (-1.2-fold) and PHOSPHOGLYCERATE KINASE (-1.53-fold) reduced the capacity of the oxidative pentose pathway (OPP), leading to the accumulation of Ribulose-5P. Because PHOSPHORIBULOKINASE accumulated, Ribulose-5P would enter into the chloroplast to be converted to 3-phosphoglycerate, allowing progression of the glycolytic pathway (60). This metabolic switch would provide NADPH and pyruvate, necessary for maintaining primary and secondary metabolism, thus compensating for the reduction of glycolysis and NADPH formation in PSI due to the increase of cyclic electron flow.
Additionally, enzymes belonging to the tricarboxylic acid (TCA) cycle showed an abundance peak after either 8 h or 16 h, despite short-term decreases after 2 h, consistent with their grouping in clusters such as 2, 21, 29, or 30 (Fig. 7, supplemental Table S7). Anaplerotic reactions showed a similar behavior: ASPARTATE AMINOTRANSFERASE 5 decreased after 2 h (-1.76-fold), although after 16 h its levels were higher than in the control plants. Both GLUTAMATE DEHYDROGENASES 1 and 2 (GDH1 and GDH2) decreased after 2 h and started to increase afterward. However, although GDH1 increased after 8 h (-1.19-fold), GDH2 (-1.16-fold) showed a partial recovery after 16 h (-1.14-fold). In both cases, the levels after recovery were lower than those in the control (-1.49-fold and −1.40-fold, respectively).
Secondary Metabolism is Rearranged to Increase the Accumulation of Photoprotective Pigments and Antioxidants
Secondary metabolism was rapidly altered after irradiation. Proteome analysis revealed a major rearrangement of secondary metabolism toward the production of pigments and antioxidants starting after UV exposure (Fig. 4). This was reflected at the metabolic level by the rapid accumulation of phenolic compounds (Fig. 2).
One of the functions of phenylpropanoids is acting as a natural sunscreen. Key enzymes leading to the biosynthesis of these compounds accumulated after irradiation: PHENYL AMMONIUM LYASE 4, caffeoyl-CoA 3 O-methyltransferase (1.64-fold) and Cinnamyl alcohol dehydrogenase 4CL1 and 4CL2. Metabolomic analyses showed that this activity is coincident with a peak of related primary metabolites, which may be priming the cell to support the later production of secondary metabolites. These phenomena occurred more rapidly than those previously described in Arabidopsis by Kusano et al. (2011) and may be the basis for the higher tolerance exhibited by pines.
Flavonoids, despite being the major UV filter, cannot be considered specific UV-B absorptive pigments because, except for their acetylated forms, do not maximally absorb in the 270–315 nm range (61). However, we observed an increase of FLAVONE 3-HYDROXYLASE, LEUCOANTHOCYANIDIN DIOXYGENASE, and ANTHOCYANIDIN REDUCTASE (2.08-fold, 1.26-fold, and 1.26-fold) after UV exposure, suggesting the accumulation of those flavonoids with higher photoprotective functions (i.e. flavan-3-ols, precursors of protoanthocyanidins which act as UV screens and antioxidants) (62, 63). This observation was supported at the metabolome level with an increased accumulation of catechins after UV exposure (3.72-fold). However, it is known that the main functions of flavonoids under UV stress are not exclusively related to sunscreen effects but also to antioxidant and developmental regulation functions (64). ISOFLAVONE REDUCTASE (-3.44-fold in 8h plants) catalyzes the last step in isoflavone biosynthesis and its levels decreased as consequence of the radiation; however, it did not reach control levels after recovery (-2.85-fold), suggesting that other flavonoids are required for further UV-stress adaptation processes.
The terpenoid biosynthetic machinery also responded to UV stress. DXP REDUCTOISOMERASE (DXR), one of the enzymes determining the rate-limiting steps in the non-mevalonate pathway (MEP) and affecting the levels of the derived isoprenoid compounds (carotenoids, tocopherols, phytols, giberellins, and ABA (65)), showed a peak in 2 h (1.72-fold), then decreased to control levels and accumulated again (1.41-fold) after recovery. This dynamic can explain the early signaling peak that should be mediated by ABA and other developmental regulators related to the first response to the stress, being in concordance with the pigment concentration measured during the experiment (Fig. 1D–1F).
Transcriptional, Ribosomal, and Proteasomal Rearrangements Mediate an UV-Responsive Active Remodeling of the Proteome
The metabolic and physiological changes conducted by pine trees to adapt and survive under an increased UV radiation (see above) can only occur after an active remodeling of the proteome, which is a consequence of a selective and tightly regulated synthesis/degradation of proteins. Both heatmap clustering (Fig. 5) and PCA (Fig. 6A; supplemental Table S5) revealed that a major proteome remodelling occurs after UV exposure, including key defense responses and secondary metabolism, as well as protein biosynthesis-degradation and transcriptional regulation.
Gene transcription was affected by UV exposure (supplemental Fig. S4), leading to a differential accumulation of related enzymes and transcription factors. NRPB5, a subunit of RNA Pol II and IV, that interacts with transcriptional activators (66) and has been proposed to be a mediator in SOS responses (67), accumulated after 8 h of UV. U2B″, an ABA-induced component of the stress response mediated by the U2 snRNP complex, playing a major role in post-transcriptional regulation of mRNA (68), was induced after 2 h of UV exposure.
Ribosomal and ribosome-translation associated protein levels were altered during stress response, maybe leading to more efficient forms under stressful environments (69). In chloroplasts or mitochondria, this response was characterized by the differential accumulation of proteins after short (RPL2.2, RPL22 and RPL4) or long (RPL11, RPS15, RPS20, and RPL5) irradiation times, suggesting that UV-responses are mediated by quick and retarded mechanisms.
Cytosolic ribosomes were more stable during our study; however, a set of ribosomal proteins was only detected after long exposure times (supplemental Table S3). Some of these proteins showed interesting connections in sPLS networks (Fig. 6B): RPS7E, found only in Control and in 16h plants (-1.63-fold), was closely related to phenol, whereas RPS11 and RPS18C were related to the EPOX cluster, and RPS19 to serine and lyxonic acid. However, the specific roles of these proteins are still unknown. Furthermore, subunits containing RPL14, reported to participate in UV responses by binding specific transcriptional activators and repressors (70, 71), and RPS19A (uncharacterized) were still maintained in recovered plants, suggesting their potential role not only in the UV stress response but also in adaptation and a likely memory of the stress.
Eukaryotic translation initiation factors, closely related to ribosomes, differentially accumulated during stress. EIF4A-III helicase, a protein that confers resistance to abiotic stress in tobacco (72, 73), accumulated after 2 h and 8 h, but was subsequently not detectable. In consequence, this factor may be regulating short responses, not being necessary in the longer term, which is consistent with its described down-regulation in long-term salt stress (74).
Protein Degradation Pathways are Required for Proteome Remodeling and UV Stress Adaptation
Specific protein degradation is also required for completing an active remodelling of the proteome. The importance of the proteasome pathway in the response to UV stress was demonstrated by the specific accumulation of three different ubiquitines (UBQ5, UBQ11 and UBQ13) and UBIQUITINE-SPECIFIC PROTEASE 6, an important modulator of the proteasome (75) that quickly increased after 2 h of irradiation (4.84-fold). Furthermore, SMALL UBIQUITIN-LIKE MODIFIER 2 (SUMO2) accumulated after long UV exposure times, preserving its conserved role as in other plant stress responses (76). The plasticity of this system is crucial for plant survival under stress (77, 78).
In chloroplasts, specific proteases constitute the main protein cleavage system. CHLOROPLAST PROTEASE C1 (CLPC1) increased after 2 h of exposure (1.72 fold), whereas P5 (CLPP5) and R4 (CLPR4) were detected after UV irradiation only. The FtsH family of chloroplastic metalloproteases was also responsive to stress, with FTSH2, FTSH5 and FTSH8 accumulating after 8 h of irradiation. FTSH8 is induced by UV-B through the UVR8 signaling pathway (6), whereas FTSH2 and FTSH5 have been related to D1 degradation (79). FtsH proteases are known for decoupling photoinactivated PSII through the removal of PSBA for decoupling LHCII (80). Our data are consistent with this function since the PSBA accumulation profile mirrored those observed for FtsH proteases. Short-term accumulation of PSBA would be in concordance both with the reduced abundance of FtsH proteases and an enhanced transcription of PSBA (81) and D1-synthesis-dependent repair of PSII occurring after UV-induced damage (82). The long-term accumulation of FTSH5 in the thylakoid (1–33-fold in the recovered plants) suggested the potential adaptive role of this protein.
DISCUSSION
The Use of a Realistic UV Dosage and an Integrative Systems Biology Approach Allowed the Identification of the Most Relevant Changes Involved in UV Stress
Unlike most of the studies on UV stress in tree species available in literature, focusing on the responses to high radiation dosages (6, 14, 83–85), in this study the experiments were set up for lower radiation dosages trying to mimic future likely scenarios in nature. This approach was aimed not only to deepen our knowledge on stress biology, but also to ease the translation of the obtained results to breeders and forest managers. Obtained data sets are unprecedented to a wide extent, being in concordance with the few studies published covering the effect of low UV rates, which described very limited changes in photosynthesis-related proteins, like those in LHCs, proteins involved in protein degradation, like UBQs, or secondary metabolism-related enzymes, like PAL (83, 86, 87).
The dosage used, despite not expected to cause any macromorphological damage to the plants, decreased the photosynthetic performance and triggered the accumulation of phenolic compounds all over the needle, indicating the trigger of stress sensing-responsing mechanisms. Stress (88), including UV stress (16), responses can be considered as systemic processes involving re-adjustments at almost all physiological and metabolic levels. In consequence, a systems biology approach was the most suitable methodology to cover the stress response stages, from the sensing and signaling phases to the triggering of the stress-responsive mechanisms (85, 89, 90). The functional categorization of both proteins and metabolites, along with their univariate analysis (ANOVA), allowed to determine which molecules and pathways were responsive to the stress. To increase the biological meaning of this datasets, multivariate analyses of the metabolites and proteins datasets allowed to get a holistic view helping in the determination of most relevant proteins and metabolites, as well as the functional bins and pathways playing a most relevant role in explaining the differences observed in the different experimental situations. K-means clustering analysis supported previous statistical approaches, increasing the consistency of the obtained results and showed graphically those molecules that were not re-established to control values in recovered plants. The construction of the sPLS-based network allowed, taking advantage of that data complementarity, to integrate the data from the different levels of complexity and perform new discoveries, as it was previously applied in other plant datasets (89, 90). Altogether provided an unprecedented holistic view of the UV stress response and adaptation process, having been unraveled the most important changes and inferred a set of key UV stress responses, described in the following sections.
UV Induced a Rearrangement of Central Metabolism to Reduce ROS Production While Supporting Biosynthetic Processes
Oxidative stress causes lipid peroxidation, oxidation of proteins, damage to nucleic acids, enzyme inhibition, activation of programmed cell death pathways and finally drives cells to death (91). At the metabolic level, the control of the production of ROS species is based on a precise rearrangement of key pathways in chloroplasts, mitochondria and peroxisomes. These changes were triggered by damage of the photosystems, as indicated by the decrease of PSII proteins. This damage would cause an imbalance in the electron transport chain in chloroplasts, leading to the formation of reactive oxygen species (ROS), one of the first consequences of any stress (92) and part of the role of the chloroplast as an environmental sensor (55).
Chloroplasts followed a double strategy for avoiding excessive ROS production. Short-term responses were mediated by an increase of cyclic electron flow (93), a strategy that would be valid for coping with normal irradiation changes that might occur during the day. However, when UV exposure persisted, cpFBPaseI, a negative regulator of the process (94), accumulated (1.4-fold after 8 h of exposure). Longer-term responses required a different strategy, based on energy dissipation by non-photochemical quenching. This was reflected by the accumulation of PSBS, which is an important component for qE (50), after long exposure times (1.56-fold after 8 h of UV). This may be a potential adaptive response as PSBS overaccumulated in recovered plants.
Under this situation, and even considering its high cost in terms of ROS production, photorespiration was enhanced for avoiding photoinhibition (95). The prominent role of glycolate in the statistical network (Fig. 6B), together with the accumulation of SHM4, GLUTAMATE:GLYOXYLATE AMINOTRANSFERASE 1 and the dynamics of glycine and serine supported this hypothesis. The proposed major role of this pathway in the adjustment of redox homeostasis (56, 95) and in the ROS-dependent activation of ROS scavenging mechanisms indicate the importance of this pathway in UV-stress responses. Interestingly, the maximum level of glycolate was reached after 8 h of exposure, coincident with the decrease in cyclic electron flow, which is consistent with the currently proposed scheme of events (95). Related enzymes also peaked after 16 h of UV, with its levels reduced to control levels in recovered plants, and showing that this is a longer-term response to stress, although it cannot be considered adaptive.
The adequate compartmenting and transport of reduced equivalents (NAD(P)H) is critical for energy supply and stress control. In this sense, mitochondrial malate valves (96) seemed to play an essential role. MITOCHONDRIAL MALATE DEHYDROGENASE showed a high loading in PCA, and specifically it showed a peak in 8h (1.34-fold) plants, which would lead to an absorbance of the excess of reduced equivalents formed in the chloroplast as a consequence of the reduced Calvin cycle, avoiding their possible photoinhibitory effect (97), and also have a role in maximizing photorespiration (98). Peroxisomal isoforms also accumulated after 8 h of UV, which supports the idea of releasing the oxidative pressure generated in the chloroplast. This is in concordance with the increases in malate found during stress, with a peak (1.44-fold) after 16 h of UV, and it is representative of the existing crosstalk between cell compartments towards an increased metabolome efficiency and adaptation.
Most of the proteins and metabolites involved in the above described processes were grouped in the same k-means cluster, the cluster 2, supporting the coordination of all of them, so the strength of these adjustments as a UV stress response for the reduction of the production of ROS. Glycolyc acid was in another cluster, 30, but showing also a peak after 8h of radiation.
Central metabolism also reflected a UV-induced reprogramming after 8h of exposure, involving glycolysis and pentose phosphate pathway, to provide NADPH and ensure that pyruvate pathways (99), necessary for lipid and secondary metabolism, more active in 8h plants. Furthermore, several transcription factors, longer-term accumulated and mainly of the NF-YBs family, are thought to play a role in the regulation of the secondary metabolism, as it was revealed in a study of the nuclear proteome of radiata pine under UV stress (100, 101).
UV Stress Responses are Coordinated by UV-, ROS-, and Light-dependent Signals
The remodelling of cell physiology was the result of an active adjustment of the proteome and metabolome. These adjustments can be considered either as general stress responses, as in the case of ROS-mediated, or stress-specific. In this sense, ULTRAVIOLET RECEPTOR (UVR8) is the only specific UV-signaling pathway that has been described so far (83). In this pathway, based on the activation of HY5 and HYH cascades by UVR8, we detected the accumulation and/or transcriptional activation of several proteins regulated by this pathway such as F3H (2.08-fold after 2 h of UV) or ELIP (2.82-fold in 16h plants) (6, 102). These signaling cascades are proposed to be controlled, among others, by the recruitment of RAN GTPases (103, 104). We identified one isoform of RAN GTPase activation protein only after UV exposure, whereas RAN3 was maintained during irradiation and was not identified in recovered plants. In addition, RAB4 transcript and protein showed accumulation trends induced by UV. This protein is known as a eukaryotic signaler of UV stress (105).
A similar pattern was observed for MIRO2, a mitochondrial calcium transducer GTPase previously described to be involved in ABA and salt stress signaling (106). However, given its location, it is presumably involved in the ROS signaling pathway more than in the direct signaling of UV-B light. A PHOTOTROPIN2 (PHOT2) isoform was only identified in longer-term irradiated plants, being positively correlated to glycolate in sPLS networks. This reveals a potential role of this kinase in ROS signaling cascades, which may be relevant for the crosstalk between the different organelles since this kinase was previously described for regulating chloroplast movements in response to light, including UV (107). There was also the accumulation of a glycolate correlated protein following the described trend, NONPHOTOTROPHIC HYPOCOTYL3 (NPH3), which is required for photo and gravitropic responses of roots mediated by PHOT1 (108). These correlations of PHOT2 and NPH3 with glycolate in the sPLS-network suggest a regulatory role of this transducer in needles, which integrate not only UV- but also both light- and ROS-dependent signaling.
Light is the other major regulator of photorespiration (109). We observed an accumulation of COP8 and EXORDIUM-LIKE 2 (EXL2) after 8 h of UV exposure, with these proteins related to light- (57) and low energy- (110) mediated photorespiration, respectively. Furthermore, EXL proteins are hypothesized to tune the balance between C supply and demand (110), suggesting a possible link between oxidative stress and the energetic state of the cell. This connection is supported by the shared correlations existing in our models between glycolate and inosose, an intermediate of the myo-inositol pathway (111), which has an emerging regulatory role under stress (112). On the other hand, HEXOKINASE1 (HXK1) was negatively correlated to both glycolate and inosose, which is in concordance with the proposed regulatory role of this kinase in maintaining ROS homeostasis and sugar sensing (113) and its relation to hormone signaling (114).
Our network also indicated the important role of CDPK-related kinase 3 (only observed in 8h plants), previously related to elevated ROS environments (115) and thought to be involved in nitrogen metabolism regulation (116). The accumulation of CALRETICULIN 1A and 1B after 8 and 16 h is consistent with previous reports of their increased expression under different biotic and abiotic stresses (117, 118).
Interestingly, and based on these networks, we can propose the signaling proteins PROTEIN PHOSPHATASE 2C FAMILY PROTEIN (294463732), PROTEIN KINASE SUPERFAMILY PROTEIN (294461783) and the transcription factor NAC025 (241926841) to be involved in UV stress response (119, 120) and presumably playing a relevant role, as their appearance in the high correlation networks supports. These proteins are the more interesting candidates for further studies, as they may be interesting novel target genes for the improvement of UV resistance in trees.
Altogether, these results showed that the response to UV involved a coordinate network of overlapping and interconnected light-, ROS- and UV-dependent signaling pathways. Such a coordinated action would allow the plant to sense the stress and trigger a different response and defense mechanism in an efficient way, which would ultimately lead to stress adaptation and survival. This would be dependent on the expression of the stress-responsive genes, mediated by specific transcription factors recruited as consequence of the triggered signaling cascades. In this respect, there are several transcription factors known to be involved in UV stress response in Pinus radiata (101), although the genes which expression regulate and the signaling pathway that enhances their recruitment needs further research to be determined.
Is It Possible to Cope with UV Stress Without Decreasing Productivity?
One of the major concerns about the effect of a slight-to-moderate increase in incident UV radiation is the expected negative impact on wood quality because of an increased accumulation of secondary metabolites (20). The productivity effects cannot be assessed because of the higher UV dosages applied in previous studies (20, 121). In this sense, our results showed that radiated pine plantlets exposed to realistic UV dosages (in the range of predicted levels for the near future) were able to survive, even recovering their normal physiology to some extent in terms of photosynthetic performance, which ultimately determines tree and forest productivity. In that sense, Pinus radiata can be considered a UV resistant species, which is in concordance with the known high resistance of conifers to this radiation (122). However, the adaptation had a cost in terms of productivity. Apart from the reduction in photosynthesis as consequence of the direct UV damage on the photosystems, the primary metabolism was rearranged to minimize ROS production. This adaptation was possible by favoring and enhancing photorespiration over carboxylation, and by shifting the sugar metabolism to produce the NADPH necessary to support the UV-enhanced secondary metabolism. These adaptations are the consequence of an active remodelling of the proteome, which was necessary to adapt and survive, requiring an effort in terms of resources. Furthermore, the recovered plants were not as the Control ones, as the separation of these samples from them in PCA, ICA and the Heatmap clustering, as well as the not reversibility of many proteins and metabolites to Control levels suggest the existence of a UV stress memory. The existence of that memory, although it has been little investigated and questioned, is supported by different lines of evidence, which specifically points to a chromatin-based epigenetic memory (123). In the particular case of Pinus radiata, this fact is supported by the importance of nucleosome remodelling and of histone-like NF-YBs transcription factors in the nuclear proteome under UV stress (100, 101). Furthermore, the proteins grouped in cluster 13 and 25, mainly HSPs and central metabolism and redox proteins, as well as the metabolites galactinol and tocopherol, all of them crucial in stress response and related to oxidative stress homeostasis, can be also related to stress memory, particularly to a higher resistance to oxidative damage. The determination of the epigenetic-based regulatory mechanisms responsible for that memory or of any other involved mechanisms would open the door to the use of short UV stress treatments for producing UV stress resistant plants.
In any case, the increased need for selecting and propagating tree genotypes adapted, or with a greater adaptation capacity, to UV stress is necessary and paramount to the forest practice in order to avoid a future decline in forest productivity. In this context, some of the molecules that proved to be determinant in stress response, such as PROTEIN PHOSPHATASE 2C FAMILY PROTEIN, CRK3, HXK1, EXL2, PSBS, the transcription factor NAC025, or the metabolites glycolic acid and malate seem to be ideal targets for breeders aiming to select UV tolerant genotypes. Likewise, for those proteins and metabolites (such as PROTEIN KINASE SUPERFAMILY PROTEIN (294461783) or several ribosomal proteins, like RPS19A or RPL14, or epicatechin and epigallocatechin, intermediates of the flavonoids biosynthesis pathway) that did not return to control levels after recovery, which possibly indicates they are part of a novel system for immediate response to further UV exposures or long term adaptive mechanisms. However, and despite further analyses are required to validate the suitability of these markers (i.e. comparing tolerant and susceptible varieties), these data provide a valuable step forward in our understanding of the UV-stress adaptive processes in Pinus radiata. The robustness of the combination of physiological, proteomics, and metabolomics data within a time course experiment allowed us to reveal the steps involved in UV stress-response and adaptation, promoting a better understanding of the process from a molecular standpoint and pinpointing on which aspects the selection and plant breeding programs should focus so as to produce UV stress resistant pine trees. Further studies including the comparison of the response to UV stress of provenances with different known susceptibility to UV stress will provide a deeper understanding of the revealed changes and markers.
Data Availability
RAW and MSF files corresponding to proteomic and metabolomics analyses are available at the Mass Spectrometry Interactive Virtual Environment (MassIVE; https://massive.ucsd.edu/; Datasets: MSV000080217, metabolites; MSV000080230, proteins).
Supplementary Material
Acknowledgments
We thank the anonymous reviewers for constructive and helpful comments and suggestions.
Footnotes
Author contributions: J.P., M.C., and L.V. designed research; J.P., M.E., M.M., and L.V. performed research; J.P., W.W., and L.V. contributed new reagents or analytic tools; J.P. and L.V. analyzed data; J.P., M.C., and L.V. wrote the paper.
* This work was supported by the Project financed by the Spanish Ministry of Economy and Competitiveness (MINECO) (AGL2011-27904, AGL2014-54995-P). J.P., M.E., M.M., and L.V. were respectively supported by FPU (AP2010-5857, Ministry of Education, Spain), Severo Ochoa (BP11117, Gobierno del Principado de Asturias, Spain), Ramón y Cajal (RYC-2014-14981; MINECO) and the Juan de la Cierva Programmes (JCI-2012-12444; MINECO).
 This article contains supplemental materials.
This article contains supplemental materials.
1 The abbreviations used are:
- ROS
- reactive oxygen species
- PCA
- principal component analysis
- ICA
- independent component analysis.
REFERENCES
- 1. McKenzie R. L., Aucamp P. J., Bais A. F., Bjorn L. O., Ilyas M., and Madronich S. (2011) Ozone depletion and climate change: impacts on UV radiation. Photochem. Photobiol. Sci. 10, 182–198 [DOI] [PubMed] [Google Scholar]
- 2. Montzka S., Reimann S., Engel A., Kruger K., Sturges W., Blake D., Dorf M., Fraser P., Froidevaux L., and Jucks K. (2011) Scientific assessment of ozone depletion: 2010. Global Ozone Research and Monitoring Project-Report No. 51 [Google Scholar]
- 3. Mead D. J. (2013) Sustainable management of Pinus radiate plantations. FAO Forestry Paper No. 170. Rome, FAO. 4–14. [Google Scholar]
- 4. MAGRAMA. (2014) Anuario de estadística forestal 2006. Madrid, Ministerio de Medio Ambiente y Medio Rural y Marino. [Google Scholar]
- 5. Brosche M., and Strid A. (2003) Molecular events following perception of ultraviolet-B radiation by plants. Physiol. Plant. 117, 1–10 [Google Scholar]
- 6. Brown B. A., Cloix C., Jiang G. H., Kaiserli E., Herzyk P., Kliebenstein D. J., and Jenkins G. I. (2005) A UV-B-specific signaling component orchestrates plant UV protection. Proc. Natl. Acad. Sci. U.S.A. 102, 18225–18230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krizek D. T. (1975) Influence of ultraviolet radiation on germination and early seedling growth. Physiol. Plant. 34, 182–186 [Google Scholar]
- 8. Day T., Ruhland C., Grobe C., and Xiong F. (1999) Growth and reproduction of Antarctic vascular plants in response to warming and UV radiation reductions in the field. Oecologia 119, 24–35 [DOI] [PubMed] [Google Scholar]
- 9. Cloix C., and Jenkins G. I. (2008) Interaction of the Arabidopsis UV-B-Specific Signaling Component UVR8 with Chromatin. Mol. Plant 1, 118–128 [DOI] [PubMed] [Google Scholar]
- 10. Kusano M., Tohge T., Fukushima A., Kobayashi M., Hayashi N., Otsuki H., Kondou Y., Goto H., Kawashima M., Matsuda F., Niida R., Matsui M., Saito K., and Fernie A. R. (2011) Metabolomics reveals comprehensive reprogramming involving two independent metabolic responses of Arabidopsis to ultraviolet-B light. Plant J. 67.2, 354–369. [DOI] [PubMed] [Google Scholar]
- 11. Teramura A. H. (1983) Effects of ultraviolet-B radiation on the growth and yield of crop plants. Physiol. Plant. 58, 415–427 [Google Scholar]
- 12. Hutin C., Nussaume L., Moise N., Moya I., Kloppstech K., and Havaux M. (2003) Early light-induced proteins protect Arabidopsis from photooxidative stress. Proc. Natl. Acad. Sci. U.S.A. 100, 4921–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ulm R., and Nagy F. (2005) Signalling and gene regulation in response to ultraviolet light. Curr. Opin. Plant. Biol. 8, 477–482 [DOI] [PubMed] [Google Scholar]
- 14. Kaling M., Kanawati B., Ghirardo A., Albert A., Winkler J. B., Heller W., Barta C., Loreto F., Schmitt-Kopplin P., and Schnitzler J.-P. (2014) UV-B mediated metabolic rearrangements in poplar revealed by non-targeted metabolomics. Plant, Cell Environ. 38, 892–904 [DOI] [PubMed] [Google Scholar]
- 15. Jia X., Ren L., Chen Q. J., Li R., and Tang G. (2009) UV-B-responsive microRNAs in Populus tremula. J. Plant Physiol. 166, 2046–2057 [DOI] [PubMed] [Google Scholar]
- 16. Valledor L., Canal M. J., Pascual J., Rodriguez R., and Meijon M. (2012) Early induced protein 1 (PrELIP1) and other photosynthetic, stress and epigenetic regulation genes are involved in Pinus radiata D. don UV-B radiation response. Physiol. Plant. 146, 308–320 [DOI] [PubMed] [Google Scholar]
- 17. Zu Y. G., Wei X. X., Yu J. H., Li D. W., Pang H. H., and Tong L. (2011) Responses in the physiology and biochemistry of Korean pine (Pinus koraiensis) under supplementary UV-B radiation. Photosynthetica 49, 448–458 [Google Scholar]
- 18. Casati P., Zhang X., Burlingame A. L., and Walbot V. (2005) Analysis of leaf proteome after UV-B irradiation in maize lines differing in sensitivity. Mol. Cell. Proteomics 4, 1673–1685 [DOI] [PubMed] [Google Scholar]
- 19. Lake J. A., Field K. J., Davey M. P., Beerling D. J., and Lomax B. H. (2009) Metabolomic and physiological responses reveal multi-phasic acclimation of Arabidopsis thaliana to chronic UV radiation. Plant, Cell Environ. 32, 1377–1389 [DOI] [PubMed] [Google Scholar]
- 20. Sullivan J. H. (2005) Possible impacts of changes in UV-B radiation on North American trees and forests. Environ. Pollution 137, 380–389 [DOI] [PubMed] [Google Scholar]
- 21. Organization W. H. (2003) INTERSUN: the Global UV Project: a guide and compendium. [Google Scholar]
- 22. Walley J. W., and Dehesh K. (2010) Molecular mechanisms regulating rapid stress signaling networks in Arabidopsis. J. Integrative Plant Biol. 52, 354–359 [DOI] [PubMed] [Google Scholar]
- 23. Escandón M., Cañal M. J., Pascual J., Pinto G., Correia B., Amaral J., and Meijón M. (2016) Integrated physiological and hormonal profile of heat-induced thermotolerance in Pinus radiata. Tree Physiol. 36, 63–77 [DOI] [PubMed] [Google Scholar]
- 24. Sims D. A., and Gamon J. A. (2002) Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sensing of Environment 81, 337–354 [Google Scholar]
- 25. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., and Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Venisse J.-S., Gullner G., and Brisset M.-N. (2001) Evidence for the Involvement of an Oxidative Stress in the Initiation of Infection of Pear by Erwinia amylovora. Plant Physiol. 125, 2164–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 28. Chang S., Puryear J., and Cairney J. (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Reporter 11, 113–116 [Google Scholar]
- 29. Valledor L., Jorriìn J. s. V., Rodriìguez J. L., Lenz C., Meijón M. n., Rodriìguez R., and Cañal M. J.s. (2010) Combined proteomic and transcriptomic analysis identifies differentially expressed pathways associated to Pinus radiata needle maturation. J. Proteome Res. 9, 3954–3979 [DOI] [PubMed] [Google Scholar]
- 30. Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., and Klenk D. C. (1985) Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 31. Valledor L., Escandón M., Meijón M., Nukarinen E., Cañal M. J., and Weckwerth W. (2014) A universal protocol for the combined isolation of metabolites, DNA, long RNAs, small RNAs, and proteins from plants and microorganisms. Plant J. 79, 173–180 [DOI] [PubMed] [Google Scholar]
- 32. Hellemans J., Mortier G., De Paepe A., Speleman F., and Vandesompele J. (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biology 8, R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Valledor L., and Weckwerth W. (2014) An Improved Detergent-Compatible Gel-Fractionation LC-LTQ-Orbitrap-MS Workflow for Plant and Microbial Proteomics. In: Jorrin-Novo, J. V., Komatsu, S., Weckwerth, W., and Wienkoop, S., eds. Plant Proteomics, pp. 347–358, Humana Press; [DOI] [PubMed] [Google Scholar]
- 34. Romero-Rodríguez M. C., Pascual J., Valledor L., and Jorrín-Novo J. (2014) Improving the quality of protein identification in non-model species. Characterization of Quercus ilex seed and Pinus radiata needle proteomes by using SEQUEST and custom databases. Journal of Proteomics 105, 85–91 [DOI] [PubMed] [Google Scholar]
- 35. Paoletti A. C., Parmely T. J., Tomomori-Sato C., Sato S., Zhu D., Conaway R. C., Conaway J. W., Florens L., and Washburn M. P. (2006) Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc. Natl. Acad. Sci. U. S. A. 103, 18928–18933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thimm O., Bläsing O., Gibon Y., Nagel A., Meyer S., Krüger P., Selbig J., Müller L. A., Rhee S. Y., and Stitt M. (2004) mapman: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939 [DOI] [PubMed] [Google Scholar]
- 37. Lohse M., Nagel A., Herter T., May P., Schroda M., Zrenner R., Tohge T., Fernie A. R., Stitt M., and Usadel B. (2013) Mercator: A fast and simple web server for genome scale functional annotation of plant sequence data. Plant, Cell Environ. 37, 1250–1258 [DOI] [PubMed] [Google Scholar]
- 38. Furuhashi T., Fragner L., Furuhashi K., Valledor L., Sun X., and Weckwerth W. (2011) Metabolite changes with induction of Cuscuta haustorium and translocation from host plants. J. Plant Interactions 7, 84–93 [Google Scholar]
- 39. Kind T., Wohlgemuth G., Lee D. Y., Lu Y., Palazoglu M., Shahbaz S., and Fiehn O. (2009) FiehnLib – mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-flight gas chromatography/mass spectrometry. Anal. Chem. 81, 10038–10048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hummel J., Selbig J., Walther D., and Kopka J. (2007) The Golm Metabolome Database: a database for GC-MS based metabolite profiling. In: Nielsen, J., and Jewett, M. C., eds. Metabolomics: A Powerful Tool in Systems Biology, pp. 75–95, Springer Berlin Heidelberg, Berlin, Heidelberg [Google Scholar]
- 41. Valledor L., and Jorrín J. (2011) Back to the basics: Maximizing the information obtained by quantitative two dimensional gel electrophoresis analyses by an appropriate experimental design and statistical analyses. J. Proteomics 74, 1–18 [DOI] [PubMed] [Google Scholar]
- 42. RDevelopment Core Team. (2012) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 43. González I., Lé Cao K.-A., and Déjean S. (2011) mixOmics: Omics Data Integration Project. [Google Scholar]
- 44. Ki-Yeol K., Gwan-Su Y., and CSBio lab I. a. C. U. (2008) Sequential KNN imputation method. [Google Scholar]
- 45. Wickham H. (2009) ggplot2: elegant graphics for data analysis, Springer Science & Business Media [Google Scholar]
- 46. Wickham H. (2012) reshape2: Flexibly reshape data: a reboot of the reshape package. R package version 1 [Google Scholar]
- 47. Klaus B., and Korbinian S. (2013) Signal identification for rare and weak features: higher criticism or false discovery rates? Biostatistics 14.1, 129–143 [DOI] [PubMed] [Google Scholar]
- 48. Foyer C. H., Bloom A. J., Queval G., and Noctor G. (2009) Photorespiratory metabolism: genes, mutants, energetics, and redox signaling. Ann. Rev. Plant Biol. 60, 455–484 [DOI] [PubMed] [Google Scholar]
- 49. Jansen M. A. K., Gaba V., and Greenberg B. M. (1998) Higher plants and UV-B radiation: balancing damage, repair, and acclimation. Trends Plant Sci. 3, 131–135 [Google Scholar]
- 50. Nishiyama Y., Allakhverdiev S. I., and Murata N. (2006) A new paradigm for the action of reactive oxygen species in the photoinhibition of photosystem II. Biochim. Biophys. Acta 1757, 742–749 [DOI] [PubMed] [Google Scholar]
- 51. Tikkanen M., Mekala N. R., and Aro E.-M. (2014) Photosystem II photoinhibition-repair cycle protects Photosystem I from irreversible damage. Biochim. Biophys. Acta 1837, 210–215 [DOI] [PubMed] [Google Scholar]
- 52. Joliot P., and Johnson G. N. (2011) Regulation of cyclic and linear electron flow in higher plants. Proc. Natl. Acad. Sci. U. S. A. 108, 13317–13322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Strand D. D., Livingston A. K., Satoh-Cruz M., Froehlich J. E., Maurino V. G., and Kramer D. M. (2015) Activation of cyclic electron flow by hydrogen peroxide in vivo. Proc. Natl. Acad. Sci. U. S. A. 112, 5539–5544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Recuenco-Muñoz L., Offre P., Valledor L., Lyon D., Weckwerth W., and Wienkoop S. (2015) Targeted quantitative analysis of a diurnal RuBisCO subunit expression and translation profile in Chlamydomonas reinhardtii introducing a novel Mass Western approach. J. Proteomics 113, 143–153 [DOI] [PubMed] [Google Scholar]
- 55. Kangasjärvi S., Neukermans J., Li S., Aro E.-M., and Noctor G. (2012) Photosynthesis, photorespiration, and light signalling in defence responses. J. Exp. Botany 63 (4): 1619–1636 [DOI] [PubMed] [Google Scholar]
- 56. Peterhansel C., Horst I., Niessen M., Blume C., Kebeish R., Kürkcüoglu S., and Kreuzaler F. (2010) Photorespiration. Arabidopsis Book, e0130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ma L., Zhao H., and Deng X. W. (2003) Analysis of the mutational effects of the COP/DET/FUS loci on genome expression profiles reveals their overlapping yet not identical roles in regulating Arabidopsis seedling development. Development 130, 969–981 [DOI] [PubMed] [Google Scholar]
- 58. Lu W., Tang X., Huo Y., Xu R., Qi S., Huang J., Zheng C., and Wu C-a. (2012) Identification and characterization of fructose 1,6-bisphosphate aldolase genes in Arabidopsis reveal a gene family with diverse responses to abiotic stresses. Gene 503, 65–74 [DOI] [PubMed] [Google Scholar]
- 59. van der Linde K., Gutsche N., Leffers H. M., Lindermayr C., Muller B., Holtgrefe S., and Scheibe R. (2011) Regulation of plant cytosolic aldolase functions by redox-modifications. Plant Physiol. Biochem. 49, 946–957 [DOI] [PubMed] [Google Scholar]
- 60. Eicks M., Maurino V., Knappe S., Flügge U.-I., and Fischer K. (2002) The plastidic pentose phosphate translocator represents a link between the cytosolic and the plastidic pentose phosphate pathways in plants. Plant Physiol. 128, 512–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Harborne J. B., and Williams C. A. (2000) Advances in flavonoid research since 1992. Phytochemistry 55, 481–504 [DOI] [PubMed] [Google Scholar]
- 62. Xie D.-Y., Sharma S. B., Paiva N. L., Ferreira D., and Dixon R. A. (2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299, 396–399 [DOI] [PubMed] [Google Scholar]
- 63. Pfeiffer J., Kühnel C., Brandt J., Duy D., Punyasiri P. A. N., Forkmann G., and Fischer T. C. (2006) Biosynthesis of flavan 3-ols by leucoanthocyanidin 4-reductases and anthocyanidin reductases in leaves of grape (Vitis vinifera L.), apple (Malus x domestica Borkh.) and other crops. Plant Physiol. Biochem. 44, 323–334 [DOI] [PubMed] [Google Scholar]
- 64. Agati G., and Tattini M. (2010) Multiple functional roles of flavonoids in photoprotection. New Phytologist 186, 786–793 [DOI] [PubMed] [Google Scholar]
- 65. Cordoba E., Salmi M., and León P. (2009) Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J. Exp. Botany 60, 2933–2943 [DOI] [PubMed] [Google Scholar]
- 66. Eun C., Lorkovic Z. J., Sasaki T., Naumann U., Matzke A. J. M., and Matzke M. (2012) Use of forward genetic screens to identify genes required for RNA-directed DNA methylation in Arabidopsis thaliana. Cold Spring Harbor Symposia on Quantitative Biol. 77, 195–204 [DOI] [PubMed] [Google Scholar]
- 67. Siaud N., Dubois E., Massot S., Richaud A., Dray E., Collier J., and Doutriaux M.-P. (2010) The SOS screen in Arabidopsis: A search for functions involved in DNA metabolism. DNA Repair 9, 567–578 [DOI] [PubMed] [Google Scholar]
- 68. Quaresma A. J. C., Sievert R., and Nickerson J. A. (2013) Regulation of mRNA export by the PI3 kinase/AKT signal transduction pathway. Mol. Biol. Cell 24, 1208–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Xue S., and Barna M. (2012) Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 13, 355–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ferreyra M. L., Biarc J., Burlingame A. L., and Casati P. (2010) Arabidopsis L10 ribosomal proteins in UV-B responses. Plant Signal. Behav. 5, 1222–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ferreyra M. L., Pezza A., Biarc J., Burlingame A. L., and Casati P. (2010) Plant L10 ribosomal proteins have different roles during development and translation under ultraviolet-B stress. Plant Physiol. 153, 1878–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Umate P., Tuteja R., and Tuteja N. (2010) Genome-wide analysis of helicase gene family from rice and Arabidopsis: a comparison with yeast and human. Plant Mol. Biol. 73, 449–465 [DOI] [PubMed] [Google Scholar]
- 73. Sanan-Mishra N., Pham X. H., Sopory S. K., and Tuteja N. (2005) Pea DNA helicase 45 overexpression in tobacco confers high salinity tolerance without affecting yield. Proc. Natl. Acad. Sci. U.S.A. 102, 509–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhao Q., Zhang H., Wang T., Chen S., and Dai S. (2013) Proteomics-based investigation of salt-responsive mechanisms in plant roots. J. Proteomics 82, 230–253 [DOI] [PubMed] [Google Scholar]
- 75. Hanna J., Meides A., Zhang D. P., and Finley D. (2007) A ubiquitin stress response induces altered proteasome composition. Cell 129, 747–759 [DOI] [PubMed] [Google Scholar]
- 76. Miura K., Sato A., Ohta M., and Furukawa J. (2011) Increased tolerance to salt stress in the phosphate-accumulating Arabidopsis mutants siz1 and pho2. Planta 234, 1191–1199 [DOI] [PubMed] [Google Scholar]
- 77. DeMartino G. N., and Gillette T. G. (2007) Proteasomes: machines for all reasons. Cell 129, 659–662 [DOI] [PubMed] [Google Scholar]
- 78. Hanna J., and Finley D. (2007) A proteasome for all occasions. FEBS Lett. 581, 2854–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Chi W., Sun X., and Zhang L. (2012) The roles of chloroplast proteases in the biogenesis and maintenance of photosystem II. Biochim. Biophys. Acta 1817, 239–246 [DOI] [PubMed] [Google Scholar]
- 80. Rodrigues R. A., Silva-Filho M. C., and Cline K. (2011) FtsH2 and FtsH5: two homologous subunits use different integration mechanisms leading to the same thylakoid multimeric complex. Plant J 65, 600–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Tyystjärvi T., Tuominen I., Herranen M., Aro E.-M., and Tyystjärvi E. (2002) Action spectrum of psbA gene transcription is similar to that of photoinhibition in Synechocystis sp. PCC 6803. FEBS Lett. 516, 167–171 [DOI] [PubMed] [Google Scholar]
- 82. Murata N., Takahashi S., Nishiyama Y., and Allakhverdiev S. I. (2007) Photoinhibition of photosystem II under environmental stress. Biochim. Biophys. Acta 1767, 414–421 [DOI] [PubMed] [Google Scholar]
- 83. Brown B. A., and Jenkins G. I. (2008) UV-B signaling pathways with different fluence-rate response profiles are distinguished in mature Arabidopsis leaf tissue by requirement for UVR8, HY5, and HYH. Plant Physiol. 146, 576–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Casati P., Campi M., Chu F., Suzuki N., Maltby D., Guan S., Burlingame A. L., and Walbot V. (2008) Histone acetylation and chromatin remodeling are required for UV-B-dependent transcriptional activation of regulated genes in Maize. Plant Cell 20, 827–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Casati P., Campi M., Morrow D., Fernandes J., and Walbot V. (2011) Transcriptomic, proteomic and metabolomic analysis of UV-B signaling in maize. BMC Genomics 12, 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Brosche M., Schuler M., Kalbina I., Connor L., and Strid A. (2002) Gene regulation by low level UV-B radiation: identification by DNA array analysis. Photochem. Photobiol. Sci. 1, 656–664 [DOI] [PubMed] [Google Scholar]
- 87. Hectors K., Prinsen E., De Coen W., Jansen M., and Guisez Y. (2007) Arabidopsis thaliana plants acclimated to low dose rates of ultraviolet B radiation show specific changes in morphology and gene expression in the absence of stress symptoms. New Phytol. 175, 255–270 [DOI] [PubMed] [Google Scholar]
- 88. Harfouche A., Meilan R., and Altman A. (2014) Molecular and physiological responses to abiotic stress in forest trees and their relevance to tree improvement. Tree Physiol. 34, 1181–1198 [DOI] [PubMed] [Google Scholar]
- 89. Valledor L., Furuhashi T., Hanak A.-M., and Weckwerth W. (2013) Systemic cold stress adaptation of Chlamydomonas reinhardtii. Mol. Cell. Proteomics 12, 2032–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Valledor L., Furuhashi T., Recuenco-Muñoz L., Wienkoop S., and Weckwerth W. (2014) System-level network analysis of nitrogen starvation and recovery in Chlamydomonas reinhardtii reveals potential new targets for increased lipid accumulation. Biotechnol. Biofuels 7, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Sharma P., Jha A. B., Dubey R. S., and Pessarakli M. (2012) Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J. Botany 2012, Article ID 217037, 26 pages [Google Scholar]
- 92. Suzuki N., Koussevitzky S., Mittler R. O. N., and Miller G. A. D. (2012) ROS and redox signalling in the response of plants to abiotic stress. Plant, Cell Environment 35, 259–270 [DOI] [PubMed] [Google Scholar]
- 93. Miyake C. (2010) Alternative electron flows (water–water cycle and cyclic electron flow around PSI) in photosynthesis: molecular mechanisms and physiological functions. Plant Cell Physiol. 51, 1951–1963 [DOI] [PubMed] [Google Scholar]
- 94. Livingston A. K., Cruz J. A., Kohzuma K., Dhingra A., and Kramer D. M. (2010) An Arabidopsis mutant with high cyclic electron flow around photosystem I (hcef) involving the NADPH dehydrogenase complex. Plant Cell Online 22, 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Voss I., Sunil B., Scheibe R., and Raghavendra A. S. (2013) Emerging concept for the role of photorespiration as an important part of abiotic stress response. Plant Biol. 15, 713–722 [DOI] [PubMed] [Google Scholar]
- 96. Scheibe R. (2004) Malate valves to balance cellular energy supply. Physiol. Plant. 120, 21–26 [DOI] [PubMed] [Google Scholar]
- 97. Raghavendra A. S., and Padmasree K. (2003) Beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Trends Plant Sci. 8, 546–553 [DOI] [PubMed] [Google Scholar]
- 98. Tomaz T., Bagard M., Pracharoenwattana I., Lindén P., Lee C. P., Carroll A. J., Ströher E., Smith S. M., Gardeström P., and Millar A. H. (2010) Mitochondrial malate dehydrogenase lowers leaf respiration and alters photorespiration and plant growth in Arabidopsis. Plant Physiol. 154, 1143–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Berg J., Tymoczko J., and Stryer L. (2002) The metabolism of glucose 6-phosphate by the pentose phosphate pathway is coordinated with glycolysis. Biochemistry, 5th Edition Ed., WH Freeman, New York [Google Scholar]
- 100. Alegre S., Pascual J., Naggler M., Escandón M., Annacondia M. L., Weckwerth W., Valledor L., and Cañal M. J. (2016) Dataset of UV induced changes in nuclear proteome obtained by GeLC-Orbitrap/MS in Pinus radiata needles Data in Brief. (Accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Pascual J., Alegre S., Nagler M., Escandón M., Annacondia M. L., Weckwerth W., Valledor L., and Cañal M. J. (2016) The variations in the nuclear proteome reveal new transcription factors and mechanisms involved in UV stress response in Pinus radiata. J. Proteomics 143, 390–400 [DOI] [PubMed] [Google Scholar]
- 102. Morales L. O., Brosché M., Vainonen J., Jenkins G. I., Wargent J. J., Sipari N., Strid Å Lindfors A. V., Tegelberg R., and Aphalo P. J. (2013) Multiple roles for UV RESISTANCE LOCUS8 in regulating gene expression and metabolite accumulation in Arabidopsis under solar ultraviolet radiation. Plant Physiol. 161, 744–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Favory J.-J., Stec A., Gruber H., Rizzini L., Oravecz A., Funk M., Albert A., Cloix C., Jenkins G. I., Oakeley E. J., Seidlitz H. K., Nagy F., and Ulm R. (2009) Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 28, 591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Jenkins G. (2009) Signal transduction in responses to UV-B radiation. Annu. Rev.. Plant Biol 60, 407–431 [DOI] [PubMed] [Google Scholar]
- 105. Vernoud V., Horton A. C., Yang Z., and Nielsen E. (2003) Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 131, 1191–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Sørmo C. G., Brembu T., Winge P., and Bones A. M. (2011) Arabidopsis thaliana MIRO1 and MIRO2 GTPases are unequally redundant in pollen tube growth and fusion of polar nuclei during female gametogenesis. PLoS ONE 6, e18530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Pontin M., Piccoli P., Francisco R., Bottini R., Martinez-Zapater J., and Lijavetzky D. (2010) Transcriptome changes in grapevine (Vitis vinifera L.) cv. Malbec leaves induced by ultraviolet-B radiation. BMC Plant Biol. 10, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Wan Y., Jasik J., Wang L., Hao H., Volkmann D., Menzel D., Mancuso S., Baluška F., and Lin J. (2012) The signal transducer NPH3 integrates the phototropin1 photosensor with PIN2-based polar auxin transport in Arabidopsis root phototropism. Plant Cell 24, 551–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Foyer C. H., and Noctor G. (2009) Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxidants Redox Signaling 11, 861–905 [DOI] [PubMed] [Google Scholar]
- 110. Schröder F., Lisso J., and Müssig C. (2012) Expression pattern and putative function of EXL1 and homologous genes in Arabidopsis. Plant Signaling Behavior 7, 22–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Loewus F. A., and Murthy P. P. (2000) myo-Inositol metabolism in plants. Plant Sci.e 150, 1–19 [Google Scholar]
- 112. Valluru R., and Van den Ende W. (2011) Myo-inositol and beyond – Emerging networks under stress. Plant Sci. 181, 387–400 [DOI] [PubMed] [Google Scholar]
- 113. Nagele T., and Weckwerth W. (2014) Mathematical modeling reveals that metabolic feedback regulation of SnRK1 and hexokinase is sufficient to control sugar homeostasis from energy depletion to full recovery. Frontiers Plant Sci. 5, 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Kushwah S., and Laxmi A. (2014) The interaction between glucose and cytokinin signal transduction pathway in Arabidopsis thaliana. Plant, Cell Environ. 37, 235–253 [DOI] [PubMed] [Google Scholar]
- 115. Munné-Bosch S., Queval G., and Foyer C. H. (2013) The impact of global change factors on redox signaling underpinning stress tolerance. Plant Physiol. 161, 5–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Li R.-J., Hua W., and Lu Y.-T. (2006) Arabidopsis cytosolic glutamine synthetase AtGLN1;1 is a potential substrate of AtCRK3 involved in leaf senescence. Biochem. Biophys. Res. Commun. 342, 119–126 [DOI] [PubMed] [Google Scholar]
- 117. Liu J.-X., and Howell S. H. (2010) Endoplasmic Reticulum Protein Quality Control and Its Relationship to Environmental Stress Responses in Plants. Plant Cell Online 22, 2930–2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Lim S. D., Lee C., and Jang C. S. (2013) The rice RING E3 ligase, OsCTR1, inhibits trafficking to the chloroplasts of OsCP12 and OsRP1, and its overexpression confers drought tolerance in Arabidopsis. Plant, Cell Environmen 37, 1097–1113 [DOI] [PubMed] [Google Scholar]
- 119. Olsen A. N., Ernst H. A., Leggio L. L., and Skriver K. (2005) NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87 [DOI] [PubMed] [Google Scholar]
- 120. Zhong R., Lee C., and Ye Z.-H. (2010) Functional characterization of poplar wood-associated NAC domain transcription factors. Plant Physiol. 152, 1044–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Wargent J. J., and Jordan B. R. (2013) From ozone depletion to agriculture: understanding the role of UV radiation in sustainable crop production. New Phytologist 197, 1058–1076 [DOI] [PubMed] [Google Scholar]
- 122. Karnosky D. F., Percy K. E., Mankovska B., Dickson R. E., Isebrands J. G., and Podila G. K. (2000) “Genetic implications for forest trees of increasing levels of greenhouse gases and UV-B radiation.” Forest Genetics and Sustainability, Springer; Netherlands: 111–124 [Google Scholar]
- 123. Müller-Xing R., Xing Q., and Goodrich J. (2014) Footprints of the sun: memory of UV and light stress in plants. Frontiers Plant Sci. 5, 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RAW and MSF files corresponding to proteomic and metabolomics analyses are available at the Mass Spectrometry Interactive Virtual Environment (MassIVE; https://massive.ucsd.edu/; Datasets: MSV000080217, metabolites; MSV000080230, proteins).