Abstract
Extracellular vesicles (EVs) are membrane-coated objects such as exosomes and microvesicles, released by many cell-types. Their presence in body fluids and the variable surface composition and content render them attractive potential biomarkers. The ability to determine their cellular origin could greatly move the field forward. We used multiplex proximity extension assays (PEA) to identify with high specificity and sensitivity the protein profiles of exosomes of different origins, including seven cell lines and two different body fluids. By comparing cells and exosomes, we successfully identified the cells originating the exosomes. Furthermore, by principal component analysis of protein patterns human milk EVs and prostasomes released from prostate acinar cells clustered with cell lines from breast and prostate tissues, respectively. Milk exosomes uniquely expressed CXCL5, MIA, and KLK6, whereas prostasomes carried NKX31, GSTP1, and SRC, highlighting that EVs originating from different origins express distinct proteins. In conclusion, PEA provides a powerful protein screening tool in exosome research, for purposes of identifying the cell source of exosomes, or new biomarkers in diseases such as cancer and inflammation.
It is broadly accepted that cells continuously secrete molecules such as amino acids, RNA, and proteins, protein complexes and lipids packaged into extracellular vesicles (EVs)1 with potential roles in intercellular communication (1). EVs, encompassing subcategories such as exosomes, microvesicles and apoptotic bodies, constitute structures secreted from cells and are surrounded by a phospholipid bilayer membrane whose constituents may reflect their cells of origin (2, 3). It has been demonstrated that the content of exosomes may be selectively incorporated (4, 5), with examples of oncogenic proteins enriched in exosomes compared with their cells of origin (6). The largest of the EVs are apoptotic bodies and microvesicles, both originating from the plasma membrane (7), whereas smaller EVs, so called exosomes, are formed intracellularly by multiple invaginations of the late endocytic membrane, leading to formation of vesicle-containing endosomes called multivesicular bodies (8, 9).
Several studies have suggested that EVs are suitable as biomarkers because of their biological relevance and because of their convenient accessibility from a broad range of body fluids (10). For instance, tumor cells secrete exosomes that contain and transport tumor antigens (11) and integrins (12), and therefore represent promising markers for predicting tumor progression and metastasis. Furthermore, it has been shown that cancer cells release more exosomes and microvesicles than healthy cells (13–16) and EVs have also been associated with a wide range of diseases including Alzheimers disease (17), prion disease (18, 19), sarcoidosis (20), and cardiac disease (21). Despite their potential as biomarkers, it has been challenging to demonstrate the cellular origin of EVs in a multicellular environment.
To validate large sets of EV associated proteins, highly specific and sensitive multiplex detection techniques with low sample consumption are required. The affinity-based proximity ligation and extension technologies exhibit key advantages in high-throughput analyses with minimal sample requirements (22–24). In the proximity assays the target proteins are recognized by pairs or trios of affinity reagents such as antibodies, conjugated to DNA oligonucleotides. Upon target recognition the DNA oligonucleotides are brought in proximity to either be ligated to each other in the presence of a connector DNA oligonucleotide such as in proximity ligation assay (PLA), or to anneal and be extended as in proximity extension assay (PEA), forming a amplifiable reporter DNA template. These assays are also suitable for multiplexing because only cognate reagent pair give rise to detectable signals (25). Here, we characterize proteins of EVs using PEA, where antibody-mediated protein detection is combined with integrated fluidic circuit real-time PCR to measure multiple proteins simultaneously using minimal amounts of sample (26). The PEA technology has primarily been applied to screen protein biomarkers in blood, but recent demonstrations highlight its utility also to detect cellular proteins, even in single cells (27). Using this technology, we characterize proteins associated with exosomes from different sources, allowing us to identify the cellular origin of the exosomes.
EXPERIMENTAL PROCEDURES
Cell Cultures
Prostatic cell lines PC3 (CRL-1435) and DU145 (HTB-81), breast cancer cell line MCF7 (HBT-22), colon cancer cell line HCT116 (CCL-247), lymphoma cell line U937 (CRL-1593.2), lymphoblast cells K562 (CCL-243) and epidermoid carcinoma cell line A431 (CRL-1555), all from ATCC (Manassas, VA), were cultured according to manufacturer's instructions in culture medium supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 100 U/ml penicillin-streptomycin (all from Sigma-Aldrich; St. Louis, MO) and maintained at 37 °C in 5% CO2. All cells were tested for mycoplasma using the Mycoplasma Detection Kit-Quick Test (Biotool; Houston,TX). Prior to isolation of EVs, the cell lines were grown to 75% confluence, washed with phosphate buffered saline (PBS; pH 7.6) and FBS-free medium was added to the cells for 4 h. The cell medium was removed and fresh FBS-free medium was added. After 24 h the conditioned medium was collected and subjected to EV isolation. Cell lysates were prepared with lysis buffer containing 50 mm Tris (pH 7.4), 150 mm NaCl, 1 mm EDTA (pH 8), 1% Triton X-100, and 0.1% sodium deoxycholate. Protease inhibitor (Complete Mini, Roche; Basel, Switzerland) was added, the samples were vortexed and protein concentration was measured using a bicinchoninic acid (BCA) assay (Thermofisher; Waltham, MA).
Exosome Isolation from Cultured Cells
The conditioned medium was centrifuged for 10 min at 3,000 × g, and the supernatant was centrifuged for 10 min at 10,000 × g, followed by filtering of the resulting supernatant through a 0.45 μm PES filter (VWR). The filtrate was centrifuged for 2 h at 100,000 × g, and the pellet was washed in sterile filtered PBS. Pelleted exosomes were resuspended in a small amount of PBS supplemented with a protease inhibitor (Complete Mini), and total protein concentration was determined by the BCA assay (Thermofisher) to 10–30 μg/ml. Samples were stored at −70 °C until further analysis.
Prostasome Isolation from Seminal Plasma
Seminal plasma (100 ml) obtained from healthy volunteers at Uppsala University Hospital, was collected as previously described (3) and kept at −20 °C. The plasma was centrifuged for 10 min at 3,000 × g, followed by centrifugation of the supernatant for 30 min at 10,000 × g. The resulting supernatant was subjected to ultracentrifugation for 2 h at 100,000 × g, using Rotor 90Ti (Beckman Coulter; Brea, CA) (28). The pellet was resuspended in PBS and loaded onto an XK16/70 Superdex 200 gel column (GE Healthcare) (29). Fractions were collected at a flow rate of 5 ml/h where the absorbance at 260 nm (for nucleic acid) and at 280 nm (for proteins) indicated the presence of prostasomes. Fractions with elevated absorbance were pooled and ultracentrifuged again for 2 h at 100,000 × g. The resulting pellet was resuspended in PBS and floated on a step gradient of 1, 1.5, and 2 molar sucrose and ultracentrifuged for 20 h at 85,000 × g using an SW28.1 rotor (Beckman Coulter). This was followed by additional ultracentrifugation for 2 h at 100,000 × g where the main fraction at 1.5 m (density range 1.13–1.19 g/ml) was pelleted. The pellet was resuspended in PBS and the protein concentration was determined to 8–10 mg/ml using the BCA assay (Merck-Millipore), and adjusted to a concentration of 2 mg/ml. Isolated prostasomes were kept at −70 °C until further use.
Exosome Isolation from Human Breast Milk
Human breast milk (50–150 ml) was collected from healthy mothers in sterile tubes using a manual breast pump and prepared directly or stored at 4 °C less than 12 h. The milk was diluted 2× in PBS and centrifuged for 10 min at 300 × g, followed by filtration of the supernatant through a 100 μm filter before centrifugation for 30 min at 3,000 × g. The supernatants were stored at −80 °C until further use. The supernatants were then centrifuged for 30 min at 10,000 × g using an Ti45 rotor (Beckman Coulter), and the resulting supernatants were filtered through a 0.22 μm vacuum filter system (TPP Rapid-filtermax) before exosomes were pelleted for 70 min at 100,000 × g, washed with PBS and resuspended in a small volume of PBS. Exosome protein concentrations were determined using the BCA protein assay (Bio-Rad; Hercules, CA, USA) to be between 40–50 mg/ml and adjusted to a concentration of 2 mg/ml. The resuspended material was stored at −80 °C until further use.
Ethical Approval
The study was approved by the local ethics committees and informed consent was obtained from donors of both seminal fluid and breast milk.
Lysis of Exosomes
Exosomes were lysed in lysis buffer (50 mm Tris pH 7.4, 150 mm NaCl, 1 mm EDTA (pH 8), 1% Triton X-100, and 0.1% sodium deoxycholate), Complete Mini protease inhibitor was added before use and samples were vortexed.
Multiplex PEA
Proteins were measured using multiplex PEA, which allows detection of 92 proteins simultaneously in one μl samples. Six different panels were used, from which three are commercially available at Olink Proteomics (Uppsala, Sweden). The Proseek Multiplex Oncology I v2 panel comprised cancer-related human proteins, the Proseek Multiplex Inflammation I panel represents analytes involved in inflammation, and the Proseek Multiplex CVD I panel contains markers of relevance in cardiovascular disease. Individual immunoassays in the commercial panels have been assessed by the manufacturer for specificity, sensitivity, dynamic range, and for matrix effects (www.olink.com). In addition to the commercially available panels, three other noncommercial, experimental PEA panels were used. The assays targeted proteins active in cellular processes (e.g. cell cycle), and cancer and neuro-oncology related pathways (27). The custom panels were validated using cell lysates diluted to contain the equivalent of 1, 10, 100, and 1,000 cells, and a pool of recombinant antigens representing a subset of the targets for the assays. All assays were able to detect proteins in cell lysates from 1,000 cells or less. The full list of analyzed proteins are summarized in supplemental Tables S1–S3. Each panel includes 92 assays and four spike-in controls consisting of two recombinant non-human proteins as incubation controls, an extension control and a PCR/detection control (26). To compensate for variation within and between runs the Cq values, obtained by realtime PCR, were normalized against the background and the extension control to generate normalized protein expression (NPX, log2-scale) values according to the manufacturer's instructions. In brief, 1 μl of sample or negative control was incubated with a panel of oligonucleotide-conjugated antibodies (final concentration 100 pm) at 4 °C overnight. Lysis buffer was used as a negative control. When a pair of antibodies bind their target, oligonucleotides coupled to the respective antibodies anneal and can be extended by polymerization to create a DNA reporter molecule that can be amplified. Because of the requirement for dual recognition the risk for antibody cross-reactivity and thereby unspecific signal is substantially reduced. Moreover, because only cognate pairs of antibodies can give rise to detectable reporter molecules, the multiplex format does not result in increasing risks of cross reactive detection (25).
Western Blot
Exosome proteins were extracted using 1x RIPA buffer, followed by sonication and vortexing to isolate total proteins. 20 μg of EV proteins (milk exosomes or prostasomes) and mouse bone marrow derived dendritic (BMDC) cell lysate were run on Mini Protean TGX precast gels (Any kDa, Bio-Rad Laboratories) and blotted to Trans-Blot Mini PVDF membranes using the Trans-Blot Turbo™ Transfer system (Bio-Rad). Incubation with blocking buffer (5% non-fat milk/PBST) was performed overnight at 4 °C. Anti-calnexin antibody (Endoplasmic Reticulum marker, 1:1000; sc-11397, Santa Cruz) was incubated, followed by donkey-anti-rabbit secondary antibody (1:10,000; NA9340, GE Healthcare) and visualized by enhanced chemiluminescence (GE Healthcare), the ChemiDoc™ MP Imaging System and Image Lab™ software version 4.1 (both from Bio-Rad).
PEA Data Analysis
The limit of detection (LOD) and NPX values for each protein in each sample (both in log2 scale) were calculated as previously described (26), and were used as input for a data analysis pipeline. From this point all further analyses were done using R programming language (RStudio). LOD values were comparable across plate replicates. All data sets were individually corrected for background noise by the following steps (supplemental Fig. S1): (1) The background LOD level was subtracted from the NPX values for each protein, respectively; (2) Next, a cut-off of 2-fold the LOD standard deviation (comprising a 95% confidence interval of background noise) was subtracted from the NPX values; (3) Negative subtracted NPX values were set to 0. Proteins were further selected for analysis only if present in more than 50% of samples from at least 1 sample group.
The resulting NPX values were used for preparing heatmaps and performing principal component analysis (PCA) with and without ComBat batch removal. Statistical uncertainty of hierarchical clustering was estimated using a bootstrapping algorithm from pvclust package (30). Uncentered Pearson correlation was used as metrics in combination with the complete-agglomerative hierarchical clustering algorithm. For tracing cell origin of exosomes (see “Results”), we removed protein patterns conserved among cell lysates or among exosomes, using the ComBat function from the SVA package (31). Cells and exosomes were input as “batches” for the ComBat function to balance the differences in expression values between these groups. The obtained NPX values after the ComBat step were used strictly for comparing sample distances by PCA and hierarchical clustering.
In data set 3, after removing background noise, data distribution was corrected using quantile normalization to allow comparison between milk exosomes and prostasomes. Sample comparison was performed using empirical Bayes statistics for differential expression (eBayes) function from the limma package (32). Proteins were considered differentially expressed when fold change > 4 (log2FC > 2) and false discovery rate (FDR) < 0.01.
RESULTS
Exosomes Can Be Analyzed by PEA and This Can Reveal Cellular Origin of Exosomes
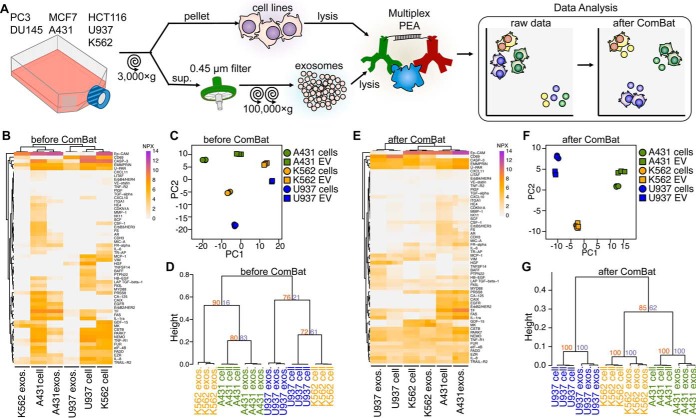
To compare cell lines and their exosomes, we applied the epidermoid carcinoma cell line A431, the lymphoma cell line U937 and the lymphoblast cell line K562 to the commercially available PEA panel (Proseek Oncology 1 v2) containing markers for cancer (supplemental Table S1). Cells were cultured, exosomes and cell lysate were obtained as schematically presented (Fig. 1A). First, we needed to confirm if exosomes could be analyzed using PEA, and whether differences could be detected between their protein profiles (Data set 1, supplemental Table S1). Among 88 proteins analyzed, we were able to detect 58 proteins from the cell lysates and 37 from their exosomes, corresponding to a coverage of 64.4% and 41.1%, respectively (Table I). We observed that the analyzed cell lysates and their respective exosomes displayed shared patterns of detected proteins (Figs. 1B and supplemental Fig. S1). Sample comparison using PCA and hierarchical clustering preferentially clustered by sample type, i.e. cell lysates versus exosomes, rather than associating exosomes with their cell lines of origin (Figs. 1C and 1D). We considered that this clustering could result from some proteins only being detectable in either cell lysates or exosomes (Fig. 1B). To overcome this, we removed protein patterns that were present in cell lines but not exosomes and vice versa, using the SVA ComBat method commonly applied in transcriptome data analysis for removing batch effects (31). This resulted in a distinct clustering of cell lysates with their respective exosomes, regardless of cells of origin (Fig. 1E). PCA and hierarchical clustering further confirmed that after ComBat, sample distances largely depend on the cell line of origin, rather than sample type (Figs. 1F and 1G). These results demonstrate that measurement of a set of proteins combined with ComBat method allows tracing the cell lines of origin for the analyzed exosomes.
Fig. 1.
Cell line derived exosome protein patterns cluster with their originating cell lysates. A, Schematic illustration of the experimental setup. Exosomes were isolated from seven cell lines, and, together with the corresponding cell lysates, subjected to multiplex PEA. B, Heatmap of protein patterns of lysates from the A431, K562 and U937 cell lines and from their respective exosomes. C, PCA of cell lysates and exosomes. D, Hierarchical clustering of cell lysate and their corresponding exosomes. E, Heatmap (F) PCA plot and (G) hierarchical clustering of proteins in lysates from cells and their corresponding exosomes after removal of cell- and exosome-specific proteins using the ComBat analyses, resulting in analyses of only proteins shared by both sample types. For dendrograms, heights represent dissimilarity among clusters and numbers on the plots indicate approximate unbiased p values (orange) and bootstrap probability (purple). Three to five biological replicates were analyzed for each sample type.
Table I. Summary of the number of proteins analyzed for each dataset.
| Data set1 | Data set2 | Data set3 | |
|---|---|---|---|
| Number of samples used | 18 | 25 | 8 |
| Total proteins tested | 90 | 402 | 238 |
| Proseek Multiplex Oncology I V2 panel | 90 | 67 | 90 |
| Proseek Multiplex Inflammation I panel | - | 80 | - |
| Proseek Multiplex CVD I panel | - | 49 | - |
| Cancer panel | - | 71 | 72 |
| Cellular Pathways panel | - | 77 | 76 |
| Neurology panel | - | 58 | - |
| Total proteins detecteda | 58 | 264 | 127 |
| Cell lines | |||
| Number of cell line groups tested | 3 | 7 | - |
| Proteins detecteda | 58 | 257 | - |
| EVs | |||
| Number of exosome groups tested | 3 | 7 | 2 |
| Proteins detecteda | 37 | 134 | 127 |
a Number of proteins detected in more than 50% of the sample from at least one group.
Extended Protein Panels for Tracing Cell Origin of Exosomes
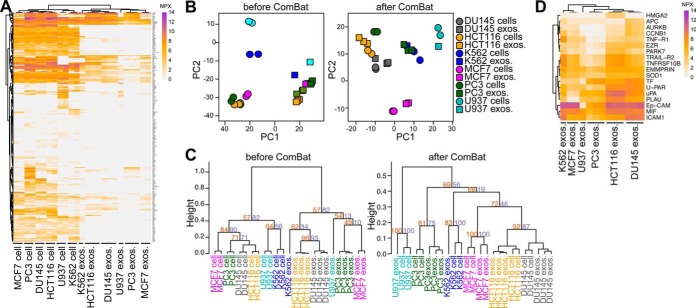
To further explore the potential of PEA for tracing the cell line origin of exosomes, the same analysis was applied on extended data for lysates and exosomes (Data set 2, supplemental Table S2), from the lymphoma cell line U937 and the lymphoblast cell line K562 as above and also from four additional cell lines; two human prostate cancer cell lines PC3 (bone metastasis) and DU145 (brain metastasis), the human epithelial colorectal carcinoma HCT116; and the mammary gland epithelial breast cancer cell line MCF7 (Fig. 2a). Overall, we detected 257 proteins from the cell lysates and 134 from their EVs (Table I), corresponding to a coverage of 63.9% and 33.3%, respectively, out of 402 proteins tested (supplemental Table S2), thus demonstrating a high detection sensitivity. Again, all samples presented a clear clustering according to sample type: cell lysate versus exosomes (Fig. 2A), supporting the data presented in Fig. 1B. As previously shown, ComBat analysis allowed exosomes and their originating cells to cluster together, suggesting the possibility to trace exosomes to their cells of origin because of their shared proteins (Figs. 2B and 2C). Some proteins were detected among all exosomes analyzed regardless of cell line of origin, such as; EpCAM, ICAM1, MIF, ITGB1, and U-PAR, suggesting basal similarities of the these exosomes (Figs. 2D and supplemental Fig. S2). CD69 was present both in cell lysates and exosomes from U937 and K562 (supplemental Fig. S2). IL-18R1 was present both in cell lysates and their exosomes from HCT116 and U937 (supplemental Fig. S2). NTRK1 was also detected in cell lysates as well as exosomes from PC3 and MCF7 cell lines. The U937 cell line was unique in showing detectable levels of CD244 and MPO (supplemental Fig. S2). Thus, by using multiplex PEA we were able to detect proteins characteristic for certain cell lines and their exosomes as well as those common to all cell lines and exosomes. Moreover, a majority of proteins were detected in cell lysates but absent in the respective exosomes, indicating selective loading of certain proteins into exosomes.
Fig. 2.
PEA identifies shared proteins among exosomes from six different cell lines. U937, K562, HCT116, DU145, MCF7, and PC3 cell lines were cultivated and used for purification of exosomes, which were further submitted for multiplex PEA. A, Heatmap for proteins detected in cell lysates and their respective exosomes. (B) PCA and (C) hierarchical clustering comparing cell line lysates and their respective exosomes before and after removal of cell- or EV-specific proteins using ComBat analyses. D, Heatmap for proteins detected (NPX > 0) in all exosome samples, regardless of cell line of origin. Protein levels are shown as NPX values. In dendrograms, heights represent dissimilarity among clusters and numbers on the plots indicate approximate unbiased p value (orange) and bootstrap probability (purple). Three to five biological replicates were analyzed for each sample type.
Protein Detection of Body Fluid-derived Exosomes Allowed Predicting Their Tissues of Origin
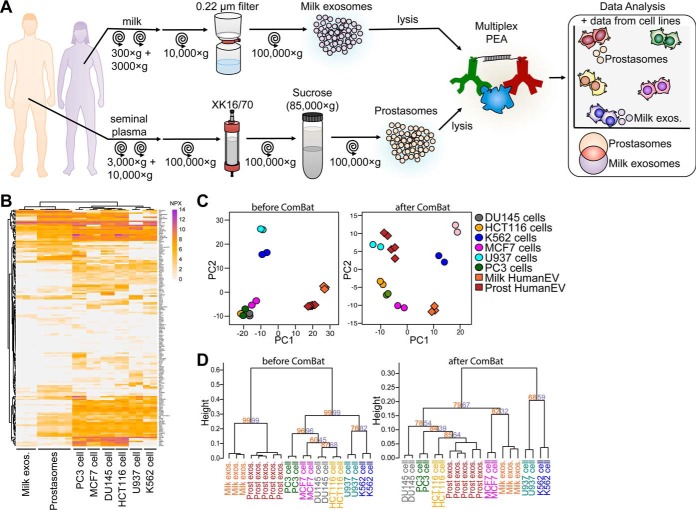
To investigate the feasibility of using PEA for analysis of body fluid exosomes, and to identify the tissue origin of exosomes, we compared the protein composition of exosomes from body fluids to those of the cell lines. We isolated body fluid exosomes from healthy individuals, from breast milk (n = 3) and seminal plasma (prostasomes) (n = 5) (Data set 3, Fig. 3A and Table I). As expected, samples representing the same biological fluid shared similar protein profiles regardless of donors (Figs. 3B and 4B). We then sought to trace what tissue or cell type these human-derived exosomes could be associated with. We analyzed the NPX values from the cell lysate samples (Data set 2, Fig. 2A) together with the milk- and prostate-derived exosome samples (Data set 3, Fig. 3B). As above, proteins shared between cell lysates and exosomes were removed using ComBat. Strikingly, we observed by PCA and hierarchical clustering that milk exosomes grouped closest to the breast cancer MCF7, whereas prostasomes clustered with the prostate cancer cell lines PC3 and DU145, respectively (Fig. 3C and 3D). Our results indicate that applying ComBat analysis to PEA data enables tracing of milk exosomes and prostasomes to the cellular origin.
Fig. 3.
Comparison of protein profiles in lysates from cell lines and in body fluid-derived exosomes reveals tissue origin of exosomes in milk and seminal fluid. A, Illustration of experimental layout. Briefly, breast milk-derived exosomes and prostasomes isolated from healthy donors were submitted to multiplex PEA, and further compared with six cell line lysates (see Fig. 2). B, Heatmap for proteins detected in U937, K562, HCT116, DU145, MCF7 and PC3 cell line lysates together with milk exosomes and prostasomes. C, PCA comparing samples before and after removal of cell- or exosome-specific proteins using ComBat analyses. D, Hierarchical clustering for cell lysates and body fluid-derived exosomes before and after removal of cell- or exosome-specific proteins with ComBat. Protein levels are shown as NPX values. For dendrogram, heights represent dissimilarity among clusters and numbers on the plot indicate approximate unbiased p value (orange) and bootstrap probability (purple). Two to five biological replicates were analyzed for each sample type.
Fig. 4.
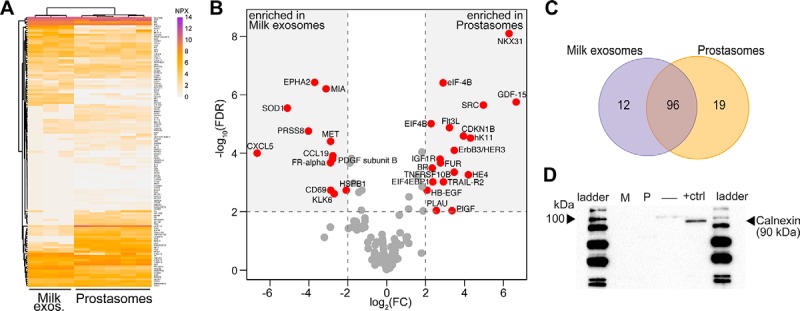
Comparison between milk exosomes and prostasomes protein profiles. A, Heatmap for prostasomes and milk exosomes. Protein levels are shown as normalized NPX values. B, Volcano plot comparing normalized NPX values for all proteins detected in milk-derived exosomes and prostasomes. Gray dashed lines represent the 2 log2 (fold change) and FDR < 0.01 to distinguish proteins significantly enriched in each group (red dots), or proteins shared between the two groups not differentially expressed (gray dots). C, Venn diagram summarizing the number of proteins significantly differentially expressed in B. D, Western-blot for calnexin (90 kDa) in milk exosomes (M) prostasomes (P), an empty well (–) and the cell lysate (+ctrl). Two to five biological replicates were analyzed for each sample type.
Milk- and Prostate-derived Exosomes Present Distinct Proteomic Profiles
To investigate whether PEA could reveal unique proteins in milk exosomes and prostasomes (Data set 3, supplemental Table S3) we compared results from their analyses (Fig. 4). Differences in protein profiles by PEA were observed between exosomes from breast milk and prostate, indicating unique protein compositions (Fig. 4A). Out of a total of 238 screened proteins, 127 (53.3%) gave signals above the LOD in both milk exosomes and prostasomes (Table I and supplemental Table S4). Overall, we observed that milk exosomes and prostasomes shared several detectable proteins (Fig. 4B and Table II). Some unique proteins were also identified. Twelve proteins were enriched in milk exosomes and 19 proteins were enriched in prostasomes (Fig. 4C and Table II; full list is presented in supplemental Table S4). The proteins CXCL5, MIA and KLK6 were only detected in milk EVs, whereas NKX31, GSTP1, and SRC were detected specifically in prostasomes (supplemental Fig. S3 and Table II). Vesicle purity was validated by Western blot using the lack of calnexin, an endoplasmic reticulum marker, as evidence for purity. A positive signal was seen in the cell lysate control, but not in the pools of breast milk exosomes or prostasomes (Fig. 4D), indicating pure exosomes, preferentially of endosomal origin. The parallel protein analysis via PEA thus served to identify proteins that could possibly be used to distinguish exosomes from distinct biological sources.
Table II. List of proteins with significantly different NPX values between Prostasomes and Milk exosomes.
| Protein symbol | Average NXP milk exosomes | Average NPX prostasomes | log2FCa | FDR |
|---|---|---|---|---|
| Enriched in milk exosomes | ||||
| CXCL5 | 0.94 | 7.61 | −6.67 | <0.001 |
| SOD1 | 1.09 | 6.20 | −5.11 | <0.001 |
| PRSS8 | 4.02 | 8.04 | −4.03 | <0.001 |
| EPHA2 | 0.24 | 3.95 | −3.71 | <0.001 |
| MIA | 0.17 | 3.29 | −3.12 | <0.001 |
| FR-alpha | 4.59 | 7.49 | −2.89 | <0.001 |
| MET | 0.67 | 3.56 | −2.89 | <0.001 |
| CD69 | 1.09 | 3.97 | −2.88 | 0.002 |
| PDGF subunit B | 2.67 | 5.46 | −2.79 | <0.001 |
| CCL19 | 0.43 | 3.20 | −2.77 | <0.001 |
| KLK6 | 0.17 | 2.87 | −2.70 | 0.002 |
| HS1 | 2.20 | 4.28 | −2.08 | 0.002 |
| Enriched in prostatasomes | ||||
| GDF-15 | 9.39 | 2.73 | 6.67 | <0.001 |
| NKX31 | 6.72 | 0.42 | 6.30 | <0.001 |
| SRC | 5.40 | 0.42 | 4.98 | <0.001 |
| hK11 | 6.38 | 2.07 | 4.31 | <0.001 |
| HE4 | 5.87 | 1.67 | 4.21 | 0.001 |
| CDKN1B | 4.38 | 0.42 | 3.95 | <0.001 |
| ErbB3/HER3 | 6.34 | 2.86 | 3.48 | <0.001 |
| TNFRSF10B | 5.53 | 2.06 | 3.48 | <0.001 |
| PlGF | 6.59 | 3.22 | 3.36 | 0.009 |
| Flt3L | 3.66 | 0.42 | 3.23 | <0.001 |
| TRAIL-R2 | 4.82 | 1.90 | 2.92 | 0.001 |
| eIF-4B | 3.33 | 0.42 | 2.90 | <0.001 |
| FUR | 4.84 | 2.07 | 2.77 | <0.001 |
| IGF1R | 4.63 | 1.89 | 2.75 | <0.001 |
| PLAU | 6.51 | 3.95 | 2.55 | 0.009 |
| EIF4EBP1 | 3.26 | 0.89 | 2.37 | 0.001 |
| BR | 2.77 | 0.42 | 2.34 | <0.001 |
| EIF4B | 2.71 | 0.42 | 2.29 | <0.001 |
| HB-EGF | 2.50 | 0.42 | 2.07 | 0.002 |
a log2 fold changes for the comparison between prostasomes versus milk exosomes. Proteins were considered significantly enriched when log2FC > 2 and FDR < 0.01.
DISCUSSION
In this study, we applied multiplex PEA for proteomic profiling of seven different cell lines and their associated exosomes, as well as of exosomes from two different body fluids. Applying antibody-based panels for immunology, cardiovascular diseases, and oncology, our goal was to evaluate the potential of multiplex PEA to be used as a screening method in EV research, and also to compare individual protein profiles from different cellular sources or body fluids.
We demonstrate for the first time the possibility to use multiplex protein detection to trace EVs to their originating parent cells based on their protein profile. Other protein screening methods available such as 2D gel electrophoresis and mass spectrometry (33), are more time consuming, are poorly suited for high-throughput screening of many samples, and their sensitivity and reproducibility may prove limiting. However, milk EVs previously subjected to MS/MS for unbiased proteome profiling (34, 35) have revealed several markers in common with our study, such as MIF, EpCAM, ICAM, and ITGB2. We also observed distinct protein profiles in cell lines and their corresponding exosomes, in line with previous findings using an antibody microarray called DotScan (36). The presence of the membrane glycoprotein Thy-1 in milk EVs and prostasomes has been confirmed in a recent study where a Thy-1 antibody was included in a set of probes to selectively detect prostasomes using the flow cytometry-based ExoPLA technique. Similarly, the granulocyte colony-stimulating factor receptor, CD114, was detected in EVs isolated from the cell line U937, confirming our observation in this study (37). This illustrates that PEA results are in accordance with results from previous proteomic analyses.
The tissue-specific protein composition of EVs provides opportunities to identify cell type-specific signatures to be used as diagnostic markers. We demonstrate that multiplex PEA is suitable for identification, analysis, and validation of potential EV-associated markers. We were able to both verify the presence of proteins previously identified in EVs, such as EpCAM (36), TRAIL (38), uPAR (39) and also to detect proteins that to our knowledge have not formerly been described as associated with EVs, such as human kallikrein 11 (hK11) and placental growth factor (PlGF), among others. Noteworthy, we were also able to detect BIRC5, mainly described as a nuclear protein. However, BIRC5 has previously also been shown to be actively secreted via exosomes and could potentially be responsible for driving tumor progression (14). It is well established that certain proteins (40) and/or RNA molecules (41, 42) are selectively sorted into vesicles. Comparison of EVs from healthy individuals to those from patients may enable identification of disease specific molecular patterns that may be possible to trace in specific tissues involved in disease. Importantly, analysis of such patterns are promising for both diagnostic, prognostic and therapeutic purposes.
Moreover, general similarities across all cell lines suggest that factors such as in vitro immortalization and culture conditions may influence EV protein composition. For example, the colon carcinoma cell line HCT116 cluster with the prostate cancer cell lines PC3 and DU145 and hence appear to be similar, suggesting either that (1) the HCT116 cell line might be closely related to the PC3 and DU145 cell lines and/or that (2) the panels used in this study were not sufficient to highlight the differences between these cell lines.
The comparison between prostasomes and milk exosomes revealed some biologically interesting differences. For instance, CXCL5, an attractor of neutrophils that can also be protective of inflammation (43) was detectable in milk exosomes but not in prostasomes. CXCL5 on exosomes in milk could possibly have a role in regulating the immune system including influencing inflammation in the newborn gut. Further, CCL19, present in milk exosomes, is a ligand to CCR7, present on e.g. B-cells, dendritic cells and CCR7+ central memory T cells, and could help attracting these cells to the gut. This could lead to CCR7-mediated immune cell regulation in the infant (44). In addition, KLK6, detected in milk but not in prostasomes, is an enzyme important for immune cell differentiation and survival (45). All these factors may have a role in the development of the infant immune system. In contrast, prostasomes contain the immune stimulatory factor Flt3l that has been described to promote dendritic cell development (46) and NF-κB activation (47), and PDGFB known to promote embryonic development and vascularization (48) These findings support the notion that milk exosomes may help in growth and development of the infant and its immune system, whereas prostasomes may stimulate implantation by supporting inflammation and vascularization.
We further detected integrins such as ITGA1 and ITGB1 present in the exosomes of both cell line and body fluid origin. The presence of integrins on exosomes has previously been reported (6, 49). Exosome-associated integrins are important in cancer metastasis and preferential exosome uptake by specific cells (12). ITGB1 is abundant in urine exosomes of patients with metastatic disease (50), indicating that ITGB1 or other integrins might serve as biomarkers and/or prognostic markers for progressive disease. Integrins were similarly detected in both milk exosomes and prostasomes. Although the present study examined exosomes from cell lines of distinct organs, follow-up studies are required for a detailed organ-specific mapping of the EV proteome from a broader set of cells and organs, which could result in identification of cell/tissue specific EV markers. Importantly, our ability to trace cellular origin could be extended to plasma-derived EVs, facilitating efficient, non-invasive diagnostic strategies, such as for early disease diagnosis.
A limitation of the current study is the fact that the PEA panels used were not designed to target proteins of relevance for exosomes, which complicates both technical validation and overall interpretation of results. Nonetheless, the sensitivity of PEA allows for detection of less abundant EV proteins than established methods, complicating validation by other antibody-based methods such as Western blot. The present study should therefore be considered as a basis for further investigation and optimization of PEA in larger-scale screening of larger patient cohorts and additional body fluids. The assays served to trace the cellular origin of the investigated exosome samples. Using this information, dedicated assays may be developed to monitor tissue specific EVs in fluids such as plasma, as possible markers for cancer in specific tissues.
In conclusion, we demonstrate for the first time the ability to trace cellular origin of EVs and that PEA can be used for EV screening for biomarker-identification purposes. This may prove valuable in a clinical setting by serving to identify diagnostic and prognostic markers for disease diagnosis and evaluation of progression and outcome.
Supplementary Material
Footnotes
Author contributions: P.L., L.W., M.K.-M., S.G. conceived and designed the study; P.L., L.W., L.L. performed the experiments and parts of the analyses; P.C. performed bioinformatics data analysis; J.O., E.F. contributed with initial data analysis; K.G.R., L.D., C.G., A.L., G.R., U.L. provided reagents; P.L., L.W., P.C., M.E., E.V., G.R., U.L., S.G., M.K.-M. contributed to the scientific discussions and development of the project; P.L., L.W., P.C., K.G.R., U.L., S.G., M.K.-M. wrote the manuscript with the help of all other authors.
* This work was supported by the Swedish Research Council, the European Commission's Seventh Framework Programme (FP7/2007–2013) under the grant agreements DIATOOLS (project n° 259796), GastricGlycoExplorer (316929), European Research Council under the European Union's Seventh Framework Programme (FP/2007–2013) ProteinSeq (294409), IngaBritt och Arne Lundbergs Forskningsstiftelse. The Swedish Cancer Foundation 2013/867, The Cancer Research Foundations of Radiumhemmet 131082, The Stockholm County Council 20140405, The Swedish Heart-Lung Foundation 20140497, The Centre for Allergy Research and the ChAMP consortium at the Karolinska Institute, the Hesselman Foundation, the KID grant of the Karolinska Institute, The Milk drop foundation and the Cancer and Allergy foundation.
 This article contains supplemental material.
This article contains supplemental material.
Competing financial interests: UL is founder and stockholder of Olink Proteomics, having rights to the PEA technology used in this paper.
1 The abbreviations used are:
- EV
- extracellular vesicle
- PEA
- proximity extension assay
- PLA
- proximity ligation assay
- PCA
- principal component analysis
- hK11
- human kallikrein 11
- PlGF
- placental growth factor
- MIF
- migration inhibitory factor.
REFERENCES
- 1. Stoorvogel W., Kleijmeer M. J., Geuze H. J., and Raposo G. (2002) The biogenesis and functions of exosomes. Traffic 3, 321–330 [DOI] [PubMed] [Google Scholar]
- 2. Gyorgy B., Szabo T. G., Pasztoi M., Pal Z., Misjak P., Aradi B., Laszlo V., Pallinger E., Pap E., Kittel A., Nagy G., Falus A., and Buzas E. I. (2011) Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell. Mol. Life Sci. 68, 2667–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ronquist G. K., Larsson A., Ronquist G., Isaksson A., Hreinsson J., Carlsson L., and Stavreus-Evers A. (2011) Prostasomal DNA characterization and transfer into human sperm. Mol. Reprod. Dev. 78, 467–476 [DOI] [PubMed] [Google Scholar]
- 4. Simons M., and Raposo G. (2009) Exosomes–vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 21, 575–581 [DOI] [PubMed] [Google Scholar]
- 5. Eldh M., Ekstrom K., Valadi H., Sjostrand M., Olsson B., Jernas M., and Lotvall J. (2010) Exosomes communicate protective messages during oxidative stress; possible role of exosomal shuttle Rna. PLoS One 5, e15353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Keerthikumar S., Gangoda L., Liem M., Fonseka P., Atukorala I., Ozcitti C., Mechler A., Adda C. G., Ang C. S., and Mathivanan S. (2015) Proteogenomic analysis reveals exosomes are more oncogenic than ectosomes. Oncotarget 6, 15375–15396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cocucci E., and Meldolesi J. (2015) Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol., 25, 364–372 [DOI] [PubMed] [Google Scholar]
- 8. Thery C., Ostrowski M., and Segura E. (2009) Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 9, 581–593 [DOI] [PubMed] [Google Scholar]
- 9. Pan B. T., and Johnstone R. M. (1983) Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell 33, 967–978 [DOI] [PubMed] [Google Scholar]
- 10. Tavoosidana G., Ronquist G., Darmanis S., Yan J., Carlsson L., Wu D., Conze T., Ek P., Semjonow A., Eltze E., Larsson A., Landegren U. D., and Kamali-Moghaddam M. (2011) Multiple recognition assay reveals prostasomes as promising plasma biomarkers for prostate cancer. Proc. Natl. Acad. Sci. U. S. A. 108, 8809–8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wolfers J., Lozier A., Raposo G., Regnault A., Thery C., Masurier C., Flament C., Pouzieux S., Faure F., Tursz T., Angevin E., Amigorena S., and Zitvogel L. (2001) Tumor-derived exosomes are a source of shared tumor rejection antigens for Ctl cross-priming. Nat. Med. 7, 297–303 [DOI] [PubMed] [Google Scholar]
- 12. Hoshino A., Costa-Silva B., Shen T. L., Rodrigues G., Hashimoto A., Tesic Mark M., Molina H., Kohsaka S., Di Giannatale A., Ceder S., Singh S., Williams C., Soplop N., Uryu K., Pharmer L., King T., Bojmar L., Davies A. E., Ararso Y., Zhang T., Zhang H., Hernandez J., Weiss J. M., Dumont-Cole V. D., Kramer K., Wexler A., Narendran L. H., Schwartz G., KHealey J. H., Sandstrom P., Labori K. J., Kure E. H., Grandgenett P. M., Hollingsworth M. A., de Sousa M., Kaur S., Jain M., Mallya K. S., Batra K., Jarnagin W. R., Brady M. S., Fodstad O., Muller V., Pantel K., Minn A. J., Bissell M. J., Garcia B. A., Kang Y., Rajasekhar V. K., Ghajar C. M., Matei I., Peinado H., Bromberg J., and Lyden D. (2015) Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nilsson J., Skog J., Nordstrand A., Baranov V., Mincheva-Nilsson L., Breakefield X. O., and Widmark A. (2009) Prostate cancer-derived urine exosomes: A novel approach to biomarkers for prostate cancer. Br. J. Cancer 100, 1603–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khan S. J., Jutzy M., Valenzuela M. M., Turay D., Aspe J. R., Ashok A., Mirshahidi S., Mercola D., Lilly M. B., and Wall N. R. (2012) Plasma-derived exosomal survivin, a plausible biomarker for early detection of prostate cancer', PLoS One 7, e46737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Doormaal F. F., Kleinjan A., Di Nisio M., Buller H. R., and Nieuwland R. (2009) Cell-derived microvesicles and cancer. Neth. J. Med. 67, 266–273 [PubMed] [Google Scholar]
- 16. Graves L. E., Ariztia E. V., Navari J. R., Matzel H. J., Stack M. S., and Fishman D. A. (2004) Proinvasive properties of ovarian cancer ascites-derived membrane vesicles. Cancer Res. 64 7045–9 [DOI] [PubMed] [Google Scholar]
- 17. Lugli G., Cohen A. M., Bennett D. A., Shah R. C., Fields C. J., Hernandez A. G., and Smalheiser N. R. (2015) Plasma exosomal mirnas in persons with and without Alzheimer Disease: altered expression and prospects for biomarkers', PLoS One 10, e0139233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fevrier B., Vilette D., Archer F., Loew D., Faigle W., Vidal M., Laude H., and Raposo G. (2004) Cells release prions in association with exosomes. Proc. Natl. Acad. Sci. U. S. A. 101, 9683–9688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wik L., Klingeborn M., Willander H., and Linne T. (2012) Separate mechanisms act concurrently to shed and release the prion protein from the cell. Prion 6, 498–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Admyre C., Grunewald J., Thyberg J., Gripenback S., Tornling G., Eklund A., Scheynius A., and Gabrielsson S. (2003) Exosomes with major histocompatibility complex class ii and co-stimulatory molecules are present in human bal fluid. Eur. Respir. J. 22, 578–583 [DOI] [PubMed] [Google Scholar]
- 21. Amabile N., Rautou P. E., Tedgui A., and Boulanger C. M. (2010) Microparticles: key protagonists in cardiovascular disorders. Semin. Thromb. Hemost. 36, 907–916 [DOI] [PubMed] [Google Scholar]
- 22. Fredriksson S., Dixon W., Ji H., Koong A. C., Mindrinos M., and Davis R. W. (2007) Multiplexed protein detection by proximity ligation for cancer biomarker validation. Nat. Methods 4, 327–329 [DOI] [PubMed] [Google Scholar]
- 23. Darmanis S., Nong R. Y., Hammond M., Gu J., Alderborn A., Vanelid J., Siegbahn A., Gustafsdottir S., Ericsson O., Landegren U., and Kamali-Moghaddam M. (2010) Sensitive plasma protein analysis by microparticle-based proximity ligation assays. Mol. Cell. Proteomics 9, 327–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lind A. L., Wu D., Freyhult E., Bodolea C., Ekegren T., Larsson A., Gustafsson M. G., Katila L., Bergquist J., Gordh T., Landegren U., and Kamali-Moghaddam M. (2016) A multiplex protein panel applied to cerebrospinal fluid reveals three new biomarker candidates in Als but none in neuropathic pain patients. PLoS One 11, e0149821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Darmanis S., Nong R. Y., Vanelid J., Siegbahn A., Ericsson O., Fredriksson S., Backlin C., Gut M., Heath S., Gut I. G., Wallentin L., Gustafsson M. G., Kamali-Moghaddam M., and Landegren U. (2011) Proteinseq: high-performance proteomic analyses by proximity ligation and next generation sequencing', PLoS One 6, e25583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Assarsson E., Lundberg M., Holmquist G., Bjorkesten J., Thorsen S. B., Ekman D., Eriksson A., Rennel Dickens E., Ohlsson S., Edfeldt G., Andersson A. C., Lindstedt P., Stenvang J., Gullberg M., and Fredriksson S. (2014) Homogenous 96-plex pea immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One, 9, e95192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Darmanis S., Gallant C. J., Marinescu V. D., Niklasson M., Segerman A., Flamourakis G., Fredriksson S. Assarsson E., Lundberg M., Nelander S., Westermark B., and Landegren U. (2016) Simultaneous multiplexed measurement of rna and proteins in single cells. Cell Rep. 14, 380–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ronquist G., and Brody I. (1985) The prostasome: its secretion and function in man. Biochim. Biophys. Acta 822, 203–218 [DOI] [PubMed] [Google Scholar]
- 29. Stegmayr B., and Ronquist G. (1982) Promotive effect on human sperm progressive motility by prostasomes. Urol. Res. 10, 253–257 [DOI] [PubMed] [Google Scholar]
- 30. Suzuki R., and Shimodaira H, (2006) Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542 [DOI] [PubMed] [Google Scholar]
- 31. Leek J. T. Johnson W. E. Parker H. S., Jaffe A. E. and Storey J. D. (2012) The Sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ritchie M. E., Phipson B., Wu D., Hu Y., Law C. W., Shi W., and Smyth G. K. (2015) Limma powers differential expression analyses for rna-sequencing and microarray studies. Nucleic Acids Res. 43, e47–e47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zichi D., Eaton B., Singer B., and Gold L. (2008) Proteomics and diagnostics: Let's get specific, again. Curr. Opin. Chem. Biol. 12, 78–85 [DOI] [PubMed] [Google Scholar]
- 34. Admyre C., Johansson C. M., Qazi K. R., Filen J. J., Lahesmaa R., Norman M., Neve E. P., Scheynius A., and Gabrielsson S. (2007) Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 179, 1969–1978 [DOI] [PubMed] [Google Scholar]
- 35. van Herwijnen M. J., Zonneveld M. I., Goerdayal S., Nolte-'t Hoen E. N., Garssen J., Stahl B., Maarten Altelaar A. F., Redegeld F. A., and Wauben M. H. (2016) Comprehensive proteomic analysis of human milk-derived extracellular vesicles unveils a novel functional proteome distinct from other milk components. Mol. Cell. Proteomics 15, 3412–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Belov L., Matic K. J., Hallal S., Best O. G., Mulligan S. P., and Christopherson R. I. (2016) Extensive surface protein profiles of extracellular vesicles from cancer cells may provide diagnostic signatures from blood samples. J. Extracell. Vesicles, 5, 25355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lof L., Ebai T., Dubois L., Wik L., Ronquist K. G., Nolander O., Lundin E., Soderberg O., Landegren U., and Kamali-Moghaddam M. (2016) Detecting individual extracellular vesicles using a multicolor in situ proximity ligation assay with flow cytometric readout. Sci. Rep. 6, 34358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stenqvist A. C., Nagaeva O., Baranov V., and Mincheva-Nilsson L. (2013) Exosomes secreted by human placenta carry functional fas ligand and trail molecules and convey apoptosis in activated immune cells, suggesting exosome-mediated immune privilege of the fetus. J. Immunol. 191, 5515–5523 [DOI] [PubMed] [Google Scholar]
- 39. Thuma F., Heiler S., Schnolzer M., and Zoller M. (2016) Palmitoylated claudin7 captured in glycolipid-enriched membrane microdomains promotes metastasis via associated transmembrane and cytosolic molecules. Oncotarge 7, 30659–30677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thery C., Regnault A., Garin J., Wolfers J., Zitvogel L., Ricciardi-Castagnoli P., Raposo G., and Amigorena S. (1999) Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein Hsc73. J. Cell Biol. 147, 599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van Balkom B. W., Eisele A. S., Pegtel D. M., Bervoets S., and Verhaar M. C. (2015) Quantitative and qualitative analysis of small Rnas in human endothelial cells and exosomes provides insights into localized rna processing, degradation and sorting. J. Extracell. Vesicles 4, 26760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Skog J., Wurdinger T., van Rijn S., Meijer D. H., Gainche L., Sena-Esteves M., Curry W. T. Jr., Carter B. S., Krichevsky A. M., and Breakefield X. O. (2008) Glioblastoma microvesicles transport rna and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 10, 1470–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rousselle A., Qadri F., Leukel L., Yilmaz R., Fontaine J. F., Sihn G., Bader M., Ahluwalia A., and Duchene J. (2013) Cxcl5 limits macrophage foam cell formation in atherosclerosis. J. Clin. Invest. 123, 1343–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Forster R., Davalos-Misslitz A. C., and Rot A. (2008) Ccr7 and its ligands: balancing immunity and tolerance. Nat. Rev. Immunol. 8, 362–371 [DOI] [PubMed] [Google Scholar]
- 45. Scarisbrick I. A., Epstein B., Cloud B. A., Yoon H., Wu J., Renner D. N., Blaber S. I., Blaber M., Vandell A. G., and Bryson A. L. (2011) Functional role of kallikrein 6 in regulating immune cell survival. PLoS One 6, e18376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maraskovsky E., Brasel K., Teepe M., Roux E. R., Lyman S. D., Shortman K., and McKenna H. J. (1996) Dramatic increase in the numbers of functionally mature dendritic cells in Flt3 ligand-treated mice: multiple dendritic cell subpopulations identified. J. Exp. Med. 184, 1953–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Karin M., and Ben-Neriah Y. (2000) Phosphorylation meets ubiquitination: The control of Nf-[Kappa]B activity. Annu. Rev. Immunol. 18, 621–663 [DOI] [PubMed] [Google Scholar]
- 48. Guo P., Hu B., Gu W., Xu L., Wang D., Huang H. S., Cavenee W. K., and Cheng S. Y. (2003) Platelet-derived growth factor-b enhances glioma angiogenesis by stimulating vascular endothelial growth factor expression in tumor endothelia and by promoting pericyte recruitment. Am. J. Pathol. 162, 1083–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tauro B. J., Mathias R. A., Greening D. W., Gopal S. K., Ji H., Kapp E. A., Coleman B. M., Hill A. F., Kusebauch U., Hallows J. L., Shteynberg D., Moritz R. L., Zhu H. J., and Simpson R. J. (2013) Oncogenic H-ras reprograms Madin-Darby canine kidney (Mdck) cell-derived exosomal proteins following epithelial-mesenchymal transition. Mol. Cell. Proteomics 12, 2148–2159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bijnsdorp I. V., Geldof A. A., Lavaei M., Piersma S. R., van Moorselaar R. J., and Jimenez C. R. (2013) Exosomal Itga3 interferes with non-cancerous prostate cell functions and is increased in urine exosomes of metastatic prostate cancer patients. J. Extracell. Vesicles, 2, doi: 10.3402/jev.v2i0.22097 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.