Abstract
Considering the biological roles of WNT4 and WNT5A involved in adipogenesis, we aimed to investigate whether SNPs in WNT4 and WNT5A contribute to obesity related traits in Han Chinese population. Targeted genomic sequence for WNT4 and WNT5A was determined in 100 Han Chinese subjects and tag SNPs were selected. Both single SNP and SNP × SNP interaction association analyses with body mass index (BMI) were evaluated in the 100 subjects and another independent sample of 1,627 Han Chinese subjects. Meta-analyses were performed and multiple testing corrections were carried out using the Bonferroni method. Consistent with the Genetic Investigation of ANthropometric Traits (GIANT) dataset results, we didn’t detect significant association signals in single SNP association analyses. However, the interaction between rs2072920 and rs11918967, was associated with BMI after multiple testing corrections (combined P = 2.20 × 10−4). The signal was also significant in each contributing data set. SNP rs2072920 is located in the 3′-UTR of WNT4 and SNP rs11918967 is located in the intron of WNT5A. Functional annotation results revealed that both SNPs might be involved in transcriptional regulation of gene expression. Our results suggest that a combined effect of SNPs via WNT4-WNT5A interaction may affect the variation of BMI in Han Chinese population.
Obesity is a complex medical condition that may lead to health problems, including heart disease, type 2 diabetes (T2D), and certain types of cancer1. Like many other complex diseases, obesity is the result of the combination of genetic susceptibility and environmental factors. Twin and family studies have shown that the heritability of body mass index (BMI) is 40–70%2,3, and other anthropometric measures of obesity have similar heritability2,3,4,5,6. Although genome-wide association studies (GWASs) have linked obesity with many genetic variants, known variants still account for only a small fraction of the heritability of obesity7. Therefore, more associated loci should be discovered.
The wingless-type MMTV integration site (WNT) signaling pathway plays important roles in regulating adipogenesis8. WNT molecules exert their effects through canonical WNT/β-catenin dependent or non-canonical WNT/β-catenin independent pathways. In vivo experiments have confirmed that both pathways are important in adipose tissue formation9. In rodents, inhibition of WNT10b of the canonical pathway could promote the differentiation of adipogenic precursor cells into mature adipocytes10. In humans, promotion of adipogenesis is related to the up-regulation of the Dickkopf-1, a known inhibitor of the canonical WNT signaling pathway11. WNT5A encodes a member of the WNT family. In mouse 3T3-L1 preadipocytes, Wnt5a is a positive regulator of adipogenesis at the beginning of adipocyte differentiation12. However, WNT5A signaling promotes human multipotent mesenchymal stem cells and human adipose tissue-derived mesenchymal stromal cells to undergo osteogenesis, while adipogenesis might be inhibited13,14. It is suggested that Wnt5a might inhibit adipogenesis through two mechanisms, suppressing the activity of Ppar-γ and enhancing the canonical WNT signaling through Lrp5/6 expression15. Therefore, WNT5A might be an important switch molecule in regulating the osteoblastogenesis and adipogenesis of multipotent stem cells. Genetic variants in Wnt5a have been associated with obesity in mice models16. However, the associations between WNT5A variations and obesity related traits in human subjects are still unclear. WNT4 encodes another WNT family member and it can also promote adipocyte differentiation in mouse 3T3-L1 preadipocytes at the initial stage of the differentiation12. In pancreatic islets of obese mice, WNT4 might inhibit the canonical WNT signaling17. A previous study suggested that a SNP in WNT4 is a susceptibility locus for fat distribution in European ancestry individuals when combined with endometriosis18. However, the associations of WNT4 polymorphisms with obesity related traits in subjects without special medical problems are still unknown.
It has been long known that genetic interactions can affect heritability calculations19. Generally, genetic interactions are not considered in SNP association analyses, leading to a substantial proportion of the missing heritability for complex diseases/traits20. Therefore, it is important and necessary to apply statistical methods to decipher genetic interactions and their relationships with disease susceptibility. Since WNT4 and WNT5A are both involved in the noncanonical WNT pathway, gene-gene interactions may contribute to their roles in adipogenesis. Currently, whether the interaction between WNT4 and WNT5A contributes to BMI variations is still unclear.
Since previous studies have implicated WNT5A and WNT4 in adipogenesis, we hypothesized that WNT5A and WNT4 might influence obesity related traits and might be candidate susceptibility genes for obesity. However, genetic variations contributing to their associations with obesity are still unclear. Therefore, in this study, we performed both single SNP and SNP × SNP interaction association analyses to investigate the effect of genetic variations of WNT5A and WNT4 on BMI in Han Chinese subjects.
Results
The basic characteristics of the subjects are listed in Table 1. The 100 unrelated subjects were sequenced successfully with the mean depth of at least 100.57 × and the coverage of target region for each sample was all over 92.5%. For WNT4, a total of 49 SNPs were identified (supplementary Table S1), with an average density of 1 SNP per 0.54 kb. 29 SNPs were identified for WNT5A, with the average density of 1 SNP per 0.76 kb (supplementary Table S1). The numbers of SNPs we identified were similar to the data from 1000 Genome phase III, which were 52 (WNT4) and 29 (WNT5A) in East Asian population, respectively. Using pairwise tagging with the r2 threshold of 0.8 in Haploview21, 10 and 8 tag SNPs were selected for WNT4 and WNT5A, respectively (supplementary Table S2 and Table 2). Therefore, 18 single SNP analyses and 80 (10 × 8) SNP × SNP analyses were performed. The significance threshold after multiple testing correction was set as combined P < 5.10 × 10−4 (0.05/98).
Table 1. Basic characteristics of all subjects.
| Sample 1 (100 Han Chinese) | Sample 2 (1,627 Han Chinese) | |||||
|---|---|---|---|---|---|---|
| Men (42) | Women (58) | Total (100) | Men (802) | Women (825) | Total (1,627) | |
| Age (years) | 49.67 ± 19.64 | 55.29 ± 11.82 | 52. 93 ± 15.75 | 31.43 ± 11.93 | 37.46 ± 13.77 | 34.49 ± 13.24 |
| Height (cm) | 171.52 ± 6.49 | 158.72 ± 6.10 | 164.10 ± 8.90 | 170.27 ± 5.96 | 158.38 ± 5.22 | 164.25 ± 8.16 |
| Weight (kg) | 74.36 ± 16.58 | 64.35 ± 11.58 | 68.56 ± 14.68 | 65.75 ± 9.64 | 54.63 ± 8.09 | 60.12 ± 10.48 |
| BMI (kg/m2) | 25.14 ± 4.63 | 25.55 ± 4.44 | 25.38 ± 4.50 | 22.66 ± 2.93 | 21.77 ± 3.05 | 22.21 ± 3.03 |
Data are shown as mean ± SD.
Table 2. Information of the tag SNPs.
| Chromosome | Position | Rs# | Region | Gene |
|---|---|---|---|---|
| 1 | 22444975 | rs10737462 | 3′-UTR | WNT4 |
| 1 | 22446265 | rs2072920 | 3′-UTR | WNT4 |
| 1 | 22449325 | rs59709264 | intron2 | WNT4 |
| 1 | 22450487 | rs2235529 | intron2 | WNT4 |
| 1 | 22451966 | rs10917155 | intron2 | WNT4 |
| 1 | 22455717 | rs77448785 | intron2 | WNT4 |
| 1 | 22455728 | rs56673898 | intron2 | WNT4 |
| 1 | 22462609 | rs12091003 | intron1 | WNT4 |
| 1 | 22463092 | rs2865175 | intron1 | WNT4 |
| 1 | 22469069 | rs60039305 | intron1 | WNT4 |
| 3 | 55499579 | rs589557 | downstream | WNT5A |
| 3 | 55501002 | rs669889 | 3′-UTR | WNT5A |
| 3 | 55502251 | rs3732750 | 3′-UTR | WNT5A |
| 3 | 55505390 | rs11918967 | intron4 | WNT5A |
| 3 | 55508102 | rs9818631 | intron4 | WNT5A |
| 3 | 55509750 | rs675575 | intron3 | WNT5A |
| 3 | 55510008 | rs12495121 | intron3 | WNT5A |
| 3 | 55519857 | rs648872 | intron1 | WNT5A |
Single SNP association analyses
For the single SNP association analyses, no significant association results were obtained in the meta-analysis (P > 0.05, Table S3). In addition, we checked the association results of these tag SNPs in the Genetic Investigation of ANthropometric Traits (GIANT) dataset for BMI in all ancestries published in 20157. Similarly, after multiple testing corrections, no significant association was detected.
SNP × SNP interaction analyses
We carried out SNP × SNP interaction analyses between the two genes to explore the underlying mechanism. Meta-analyses results showed that the SNP pair rs2072920-rs11918967 was associated with BMI after multiple testing corrections (combined P = 2.20 × 10−4, Table 3). These two SNPs are located in the 3′-UTR of WNT4 and intron4 of WNT5A, respectively. The interaction of rs2072920-rs11918967 was also significantly associated with BMI in each contributing data set, with the P values of 0.0122 in 100 Han Chinese and 0.0014 in 1,627 Han Chinese, respectively.
Table 3. Significantly associated SNP-SNP interactions in the two genes associated with BMI.
| SNP1-SNP2 | Combined P | 100 Han Chinese subjects | 1,627 Han Chinese subjects | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele1 | MAF1 | Allele2 | MAF2 | Beta | se | P | Allele1 | MAF1 | Allele2 | MAF2 | Beta | se | P | ||
| rs2072920-rs11918967 | 2.20 × 10−4 | G/A | 0.125 | C/G | 0.285 | 3.5 | 0.0271 | 0.0122 | G/A | 0.1172 | C/G | 0.2983 | 0.7157 | 0.0065 | 0.0014 |
Note: Only significantly associated SNP pairs after multiple testing corrections are shown. se: standard error; Allele1: Alleles of SNP1; Allele2: Alleles of SNP2; MAF1: minor allele frequency of SNP1; MAF2: minor allele frequency of SNP2.
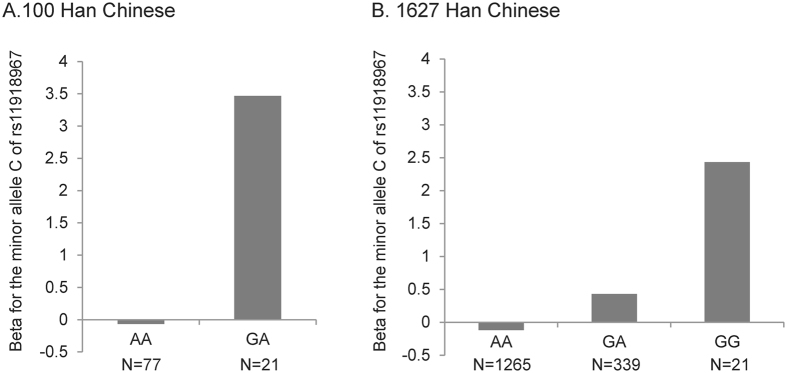
We further checked whether the effect of the minor allele “C” of rs11918967 on BMI was different between subjects carrying different genotypes of rs2072920 using the beta coefficient. As shown in Fig. 1A, in the 100 unrelated subjects, the minor allele “C” of rs11918967 was negatively associated with BMI in subjects carrying “AA” of rs2072920 (beta = −0.067, 95% CI = −1.653–1.518, standard error (se) = 0.809). However, it was positively associated with BMI in subjects carrying “GA” of rs2072920 (beta = 3.468, 95% CI = 1.454–5.481, se = 1.027). Similarly, in another sample of 1,627 Han Chinese subjects (Fig. 1B), the minor allele “C” of rs11918967 was also negatively associated with BMI in subjects carrying “AA” of rs2072920 (beta = −0.120, 95% CI = −0.350–0.110, se = 0.117). Consistently, it was positively associated with BMI in subjects carrying “GA” (beta = 0.430, 95% CI = −0.017–0.877, se = 0.228) and “GG” of rs2072920 (beta = 2.432, 95% CI = 0.797–4.068, se = 0.835). Therefore, the minor allele “C” of rs11918967 was positively associated with BMI in subjects with at least one copy “G” allele of rs2072920.
Figure 1.
(A) Association of the minor allele “C” of rs11918967 with BMI in subjects carrying different genotypes of rs2072920 in the 100 Han Chinese subjects. There were only two subjects with “GG” of rs2072920, so we didn’t analyze the association results in this subgroup. (B) Association of the minor allele “C” of rs11918967 with BMI in subjects carrying different genotypes of rs2072920 in the 1,627 Han Chinese subjects. The beta values of the association analyses results are shown in the y-axis.
Functional annotation
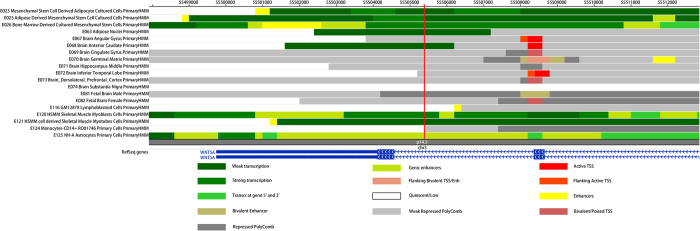
We used information from tissues/cell lines that might be relevant to obesity (supplementary Table S4) to annotate the selected SNPs. As shown in Fig. 2, rs2072920 was located in the region of strong transcription in adipose derived mesenchymal stem cell cultured cells (AMSC), bone marrow derived cultured mesenchymal stem cells (BMSC), adipose nuclei, brain germinal matrix, fetal brain female and Monocytes-CD14 + cells. It was also located in the genic enhancer region of GM12878. Of note, rs58543510, which was in complete linkage disequilibrium (LD) with rs2072920 (r2 = 1, D’ = 1), was located in the enhancer region of 10 tissues/cell lines. Ten other LD SNPs were also found in the enhancer region of at least one tissue/cell line. RNA binding protein (RBP) data analyses showed that rs2072920 and its LD SNPs were located in the poly(A) binding protein cytoplasmic 1 (PABPC1) binding region. We further checked whether the enhancer SNPs affect transcription factor binding to known motifs. As shown in Table S5, 9 of the 10 enhancer SNPs fell within at least one critical position in transcription factor binding motifs. Specifically, the effect of rs58543510 on the T3R motif has been validated in various cell lines, including some cells that might be related to obesity, such as GM12878, skeletal muscle myoblasts cells (HSMM), and HSMM cell derived skeletal muscle myotubes cells (HSMMtube).
Figure 2. Annotation of rs2072920 in WNT4 in tissues/cell lines that might be related to obesity.
The longest vertical red line refers to rs2072920. SNPs in LD with rs2072920 were shown with short vertical red lines. Primary HMM refers to the chromatin states predicted by hidden Markov model based on combinations of histone modification marks.
As shown in Fig. 3, rs11918967 was located in the region of strong transcription in AMSC, BMSC and HSMM. It was also located in the genic enhancer region of astrocytes. There were no SNPs in LD with rs11918967. RBP data analyses also showed that rs11918967 was located in the PABPC1 binding region. Motif analyses suggested that it might affect the binding motif of AP-2.
Figure 3. Annotation of rs11918967 in WNT5A in tissues or cell lines that might be related to obesity.
The vertical red line refers to rs11918967. Primary HMM refers to the chromatin states predicted by hidden Markov model based on combinations of histone modification marks.
Discussion
In this study, we aimed to investigate the genetic associations between WNT4 and WNT5A polymorphisms and BMI in Han Chinese subjects. We performed meta-analyses using two independent samples including 100 and 1,627 Han Chinese subjects and the results showed that the interaction between rs2072920 in WNT4 and rs11918967 in WNT5A was associated with BMI after multiple testing corrections. Our findings suggest that the interaction between WNT4 and WNT5A contributes to BMI variations in Han Chinese population.
Although WNT5A is a factor inhibiting adipogenesis in humans13,22, the associations between WNT5A genetic variations and BMI have not been reported before. Both WNT4 and WNT5A are known as noncanonical WNT genes23, and interaction between WNT4 and WNT5A protein has been proved by using high-throughput affinity-purification mass spectrometry24. Functional annotation analyses suggest that these two SNPs and their LD SNPs are located in strong transcription or enhancer region in at least one obesity related tissue/cell line. These regions could bind PABPC1, which is a poly(A) binding protein. Binding of PABPC1 to poly(A) tail of mRNA could promote translation initiation and it is also involved in the regulation of mRNA decay25. Since the SNPs we reported here are all located within or near the 3′-UTR regions, they may be involved in transcriptional regulation of gene expression through affecting the binding of PABPC1. Motif analyses results for SNPs in the enhancer region suggested that they may regulate gene expression through impacting the binding of transcription factors to known motifs. Further studies are needed to confirm the underlying mechanism of these SNPs in regulating gene expression.
We couldn’t detect any significant association results in single SNP association analyses in both WNT4 and WNT5A. This is different from previous studies since a SNP in WNT4 has been reported to be significantly associated with fat distribution in European ancestry individuals18. The inconsistence may be caused by the ethnic differences between European and Asian populations, since they have different LD structures and allele frequencies26. In addition, the association signal was detected in endometriosis patients18, which may lead to different results from our healthy subjects.
The detected interaction between rs2072920 and rs11918967 can explain 0.899% of the phenotypic variation. Given the sample size adopted, this study can achieve about 68.14% statistical power to detect the association signal that accounts for ~0.899% of the phenotypic variation. We acknowledge that this study is not powerful to detect association signals for variants with low effect size.
Limitations of the current study must be addressed. The two samples we used have notably different age and BMI distributions. We included age as covariate to adjust the BMI values, which could eliminate the effect of age to some extent. The SNP-SNP interaction association signals with P < 0.05 were also detected in each contributing data set, suggesting that the effect of rs2072920-rs11918967 interaction on BMI variations is independent from age and BMI distributions. We focused on the analyses in Han Chinese subjects and the results may not be applicable to other populations. Further studies are needed to investigate the association between WNT4-WNT5A interaction and BMI in other populations.
In summary, this study provides the evidence that the interactions between WNT4 and WNT5A could affect the variation of BMI in Han Chinese subjects. Further investigations are needed to clarify our findings in other populations.
Methods
Ethics, consent and permissions
This study was approved by the Institutional Review Boards of Xi’an Jiaotong University. Signed informed consent was obtained from all subjects. All experiments were performed in accordance with relevant guidelines and regulations.
Subjects
Detailed information of the subjects is described as follows:
Sample 1
100 unrelated healthy Han Chinese adults living in Xi’an and its neighboring areas were recruited. During physical examination of each individual, body weight and height were recorded. BMI was calculated as body weight (kg) divided by the square of height (m). Subjects with chronic diseases and conditions that affect fat metabolism were excluded. These disorders/conditions included diseases affecting vital organs (heart, lung, liver, kidney and brain) and severe endocrine, metabolic or nutritional diseases. The exclusion criteria were described in detail in previous studies27.
Sample 2
1,627 Han Chinese subjects were recruited from Xi’an and Changsha in Midwestern China. The exclusion criteria were the same as those used in the 100 unrelated subjects.
Targeted gene sequencing for the sample 1
Targeted gene sequencing was performed in the 100 unrelated subjects. WNT5A and WNT4 were provided to Roche NimbleGen, Inc. (Madison, WI, USA) for custom target region capture array design. Target region selection was accomplished by downloading the sequence and selecting the transcripts with the longest exons from the University of California Santa Cruz (UCSC) Genome Browser. These transcripts are NM_030761 for WNT4 and NM_003392 for WNT5A. The array was designed to target the whole transcripts of the two genes and ± 1,000 bp flanking regions. DNA was extracted from whole blood using a commercial isolation kit (Gentra systems, Minneapolis, MN, USA). Qualified genomic DNA was randomly fragmented into fragments with a base pair peak of 100 to 200 bp. A pair of adapters was ligated to both ends of the fragments, which were then amplified, purified, and hybridized to the custom array for enrichment. The resulting DNA library was subjected to paired-end sequencing with read length of 90 bp on the Illumina HiSeq 2000 platform.
Sequencing reads alignment and SNP detection in the sample 1
First, the adapter sequence in the raw data was removed, and nucleotides with a quality score lower than 20 were trimmed. The resulting filtered reads were mapped to the human reference genome (hg19) using the Burrows-Wheeler Aligner (version 0.7.10, command BWA-MEM)28. Sequence Alignment/Map (SAM) format alignment result files were imported to Samtools29 and the ‘rmdup’ command was used to remove potential PCR duplicates. SNPs were detected by SOAPsnp8 and annotated with ANNOVAR30. SNPs with minor allele frequencies (MAF) less than 0.05 and deviated from Hardy-Weinberg equilibrium (P < 0.001) were excluded. Haploview21 was used to select tag SNPs and only tag SNPs were used in the association analyses.
Genotyping in the sample 2
For the 1,627 Han Chinese subjects, SNP genotyping was performed using Genome Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA, USA), which has been detailed previously31. For SNPs which were not genotyped in the arrays, we imputed the genotypes with the IMPUTE32 to facilitate the comparison of association results. The 1000 Genome dataset was used as the reference data.
Statistical analyses
BMI was adjusted for age and sex in a linear regression model. The resulting residuals were tested for normality by Kolmogorov-Smirnov test and the residuals of BMI in both sample sets were normally distributed. The above analyses were performed with the software MINITAB (Minitab Inc., State College, PA, USA). The residuals were then used in subsequent association analyses. At the single-marker level, association analyses for all SNPs assuming additive models of inheritance were carried out using PLINK33. In this model, the beta coefficient represents the rate of changes of the response variable as a function of the changes in the independent variable. Pairwise SNPs interactions were then tested by a linear regression analysis which was also performed with PLINK33. Briefly, PLINK makes a model based on allele dosage for each SNP, which fits a linear regression model in the following equation:
Y ~ β + β1*SNP1 + β2*SNP2 + β3*SNP1 × SNP2 + e
For “two copies” of A allele (minor allele) of SNP2 (SNP2 = 2), the equation is:
Y ~ (β + 2β2) + (β1 + 2β3) *SNP1 + e
For “one copy” of A allele of SNP2 (SNP2 = 1), the equation is:
Y ~ (β + β2) + (β1 + β3) *SNP1 + e
For “zero copy” of A allele of SNP2 (SNP2 = 0), the equation is:
Y ~ β + β1*SNP1 + e
Summary statistics of association analyses from the two samples were subjected to meta-analysis using the METAL software (http://csg.sph.umich.edu/abecasis/Metal/) under the sample-size weighted model. Multiple comparison problems were adjusted using the Bonferroni method.
We estimated the statistical power of our study using the Quanto v1.2.4 software (http://biostats.usc.edu/Quanto.html). The conservative significance threshold was set at P < 5.10 × 10−4.
The Genetic Investigation of ANthropometric Traits (GIANT) dataset
The GIANT consortium is an international collaboration that aims to detect genetic loci associated with human anthropometric traits, including height and obesity related phenotypes. Summary statistics from large scale meta-analyses of genome wide single SNP association data are freely to access for all researchers. Here we downloaded the summary data for BMI from the article published in 20157, which incorporated results from 322,154 European and 17,072 non-European-descent individuals (total n = 339,224). We used the results from all ancestries to validate our single-SNP association results.
Functional annotation
In order to determine the potential regulatory function of SNPs associated with BMI, the SNPs were annotated with chromatin states predicted by hidden Markov model34 (HMM) based on combinations of histone modification marks, including H3K4me3, H3K4me1, H3K36me3, H3K27me3, and H3K9me3. The chromatin states data were obtained from the Roadmap project35. Detailed information of the states is shown in the Roadmap website (http://egg2.wustl.edu/roadmap/web_portal/chr_state_learning.html). Data from tissues or cell lines that might be relevant to obesity were obtained. Information for the cell lines or tissues we used is shown in supplementary Table S4. The annotation results were visualized in the WashU Epigenome Browser36.
We further used RNA binding protein (RBP) immunoprecipitation data from the ENCODE project37 to check whether the selected SNPs may affect gene expression through influencing protein binding. For the cell lines/tissues we selected for obesity (Table S4), only RBP data for GM12878 (B-lymphocyte, lymphoblastoid) are available now. The data were downloaded from the following URL: http://hgdownload.cse.ucsc.edu/goldenpath/hg19/encodeDCC/wgEncodeSunyAlbanyGeneSt/.
For SNPs in enhancer regions, we used HaploReg (v4.1)38 to check their effects on binding motifs. RegulomeDB39 was also used to check whether their effects on motifs binding were experimentally validated.
Additional Information
How to cite this article: Dong, S.-S. et al. SNP-SNP interactions between WNT4 and WNT5A were associated with obesity related traits in Han Chinese Population. Sci. Rep. 7, 43939; doi: 10.1038/srep43939 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This study is supported by the National Natural Science Foundation of China (31371278, 31471188, 81573241, 31511140285, 31301029); China Postdoctoral Science Foundation (2015M570819, 2016M602797); Natural Science Basic Research Program Shaanxi Province (2015JQ3089); and the Fundamental Research Funds for the Central Universities. The study was also funded by the grants from National Institutes of Health (P50AR055081, R01AG026564, R01AR050496, and R01AR057049).
Footnotes
The authors declare no competing financial interests.
Author Contributions Y.G. conceived the study. S.D. analyzed data and wrote the manuscript. T.Y. and Y.G. revised the manuscript. W.X.H., X.F.C., H.Y., X.D.C., L.T., Q.T. carried out experiments. H.D. designed the recruitment procedure used in sample 2. All authors had final approval of the submitted and published versions.
References
- Haslam D. W. & James W. P. Obesity. Lancet 366, 1197–1209, doi: 10.1016/S0140-6736(05)67483-1 (2005). [DOI] [PubMed] [Google Scholar]
- Stunkard A. J., Foch T. T. & Hrubec Z. A twin study of human obesity. JAMA 256, 51–54 (1986). [PubMed] [Google Scholar]
- Turula M., Kaprio J., Rissanen A. & Koskenvuo M. Body weight in the Finnish Twin Cohort. Diabetes Res. Clin. Pract. 10 Suppl 1, S33–36 (1990). [DOI] [PubMed] [Google Scholar]
- Selby J. V. et al. Evidence of genetic influence on central body fat in middle-aged twins. Hum. Biol. 61, 179–194 (1989). [PubMed] [Google Scholar]
- Rose K. M., Newman B., Mayer-Davis E. J. & Selby J. V. Genetic and behavioral determinants of waist-hip ratio and waist circumference in women twins. Obes. Res. 6, 383–392 (1998). [DOI] [PubMed] [Google Scholar]
- Malis C. et al. Total and regional fat distribution is strongly influenced by genetic factors in young and elderly twins. Obes. Res. 13, 2139–2145, doi: 10.1038/oby.2005.265 (2005). [DOI] [PubMed] [Google Scholar]
- Locke A. E. et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197–206, doi: 10.1038/nature14177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulides C., Lagathu C., Sethi J. K. & Vidal-Puig A. Adipogenesis and WNT signalling. Trends in endocrinology and metabolism: TEM 20, 16–24, doi: 10.1016/j.tem.2008.09.002 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudes M. Role of WNT signalling in the determination of human mesenchymal stem cells into preadipocytes. Journal of molecular endocrinology 46, R65–72, doi: 10.1530/JME-10-0169 (2011). [DOI] [PubMed] [Google Scholar]
- Ross S. E. et al. Inhibition of adipogenesis by Wnt signaling. Science 289, 950–953 (2000). [DOI] [PubMed] [Google Scholar]
- Christodoulides C. et al. The Wnt antagonist Dickkopf-1 and its receptors are coordinately regulated during early human adipogenesis. Journal of cell science 119, 2613–2620, doi: 10.1242/jcs.02975 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizuka M., Koyanagi A., Osada S. & Imagawa M. Wnt4 and Wnt5a promote adipocyte differentiation. FEBS letters 582, 3201–3205, doi: 10.1016/j.febslet.2008.08.011 (2008). [DOI] [PubMed] [Google Scholar]
- Bilkovski R. et al. Role of WNT-5a in the determination of human mesenchymal stem cells into preadipocytes. The Journal of biological chemistry 285, 6170–6178, doi: 10.1074/jbc.M109.054338 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos A., Bakker A. D., de Blieck-Hogervorst J. M. & Klein-Nulend J. WNT5A induces osteogenic differentiation of human adipose stem cells via rho-associated kinase ROCK. Cytotherapy 12, 924–932, doi: 10.3109/14653241003774011 (2010). [DOI] [PubMed] [Google Scholar]
- Okamoto M. et al. Noncanonical Wnt5a enhances Wnt/beta-catenin signaling during osteoblastogenesis. Scientific reports 4, 4493, doi: 10.1038/srep04493 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almind K. & Kahn C. R. Genetic determinants of energy expenditure and insulin resistance in diet-induced obesity in mice. Diabetes 53, 3274–3285 (2004). [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J. & Stoffel M. Regulation of wingless-type MMTV integration site family (WNT) signalling in pancreatic islets from wild-type and obese mice. Diabetologia 53, 123–127, doi: 10.1007/s00125-009-1578-2 (2010). [DOI] [PubMed] [Google Scholar]
- Rahmioglu N. et al. Genome-wide enrichment analysis between endometriosis and obesity-related traits reveals novel susceptibility loci. Human molecular genetics, doi: 10.1093/hmg/ddu516 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer D. & Mackay T. Introduction to Quantitative Genetics. (Longman, 1996). [Google Scholar]
- Zuk O., Hechter E., Sunyaev S. R. & Lander E. S. The mystery of missing heritability: Genetic interactions create phantom heritability. Proceedings of the National Academy of Sciences of the United States of America 109, 1193–1198, doi: 10.1073/pnas.1119675109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J. C., Fry B., Maller J. & Daly M. J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21, 263–265, doi: 10.1093/bioinformatics/bth457 (2005). [DOI] [PubMed] [Google Scholar]
- Bilkovski R. et al. Adipose tissue macrophages inhibit adipogenesis of mesenchymal precursor cells via wnt-5a in humans. International journal of obesity 35, 1450–1454, doi: 10.1038/ijo.2011.6 (2011). [DOI] [PubMed] [Google Scholar]
- Angers S. & Moon R. T. Proximal events in Wnt signal transduction. Nature reviews. Molecular cell biology 10, 468–477, doi: 10.1038/nrm2717 (2009). [DOI] [PubMed] [Google Scholar]
- Huttlin E. L. et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 162, 425–440, doi: 10.1016/j.cell.2015.06.043 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda N., Lejeune F. & Maquat L. E. Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. Molecular and cellular biology 26, 3085–3097, doi: 10.1128/MCB.26.8.3085-3097.2006 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Festen E. A. & Wijmenga C. Multi-ethnic studies in complex traits. Human molecular genetics 20, R206–213, doi: 10.1093/hmg/ddr386 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H. W. et al. A genomewide linkage scan for quantitative-trait loci for obesity phenotypes. American journal of human genetics 70, 1138–1151, doi: 10.1086/339934 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. & Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760, doi: 10.1093/bioinformatics/btp324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079, doi: 10.1093/bioinformatics/btp352 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Li M. & Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids research 38, e164, doi: 10.1093/nar/gkq603 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T. L. et al. Genetic variants in the SOX6 gene are associated with bone mineral density in both Caucasian and Chinese populations. Osteoporos Int 23, 781–787, doi: 10.1007/s00198-011-1626-x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J., Howie B., Myers S., McVean G. & Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913, doi: 10.1038/ng2088 (2007). [DOI] [PubMed] [Google Scholar]
- Purcell S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575, doi: 10.1086/519795 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J. & Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nature methods 9, 215–216, doi: 10.1038/nmeth.1906 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roadmap Epigenomics C. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330, doi: 10.1038/nature14248 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X. et al. The Human Epigenome Browser at Washington University. Nature methods 8, 989–990, doi: 10.1038/nmeth.1772 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium E. P. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306, 636–640, doi: 10.1126/science.1105136 (2004). [DOI] [PubMed] [Google Scholar]
- Ward L. D. & Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic acids research 40, D930–934, doi: 10.1093/nar/gkr917 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle A. P. et al. Annotation of functional variation in personal genomes using Regulome DB. Genome research 22, 1790–1797, doi: 10.1101/gr.137323.112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.