Abstract
The highly conserved eukaryotic WD40 repeat protein, Receptor for Activated C Kinase 1 (RACK1), is involved in the abscisic acid (ABA) response in Arabidopsis. However, the regulation of RACK1 and the proteins with which it interacts are poorly understood. Here, we show that RACK1B is sumoylated at four residues, Lys50, Lys276, Lys281 and Lys291. Sumoylation increases RACK1B stability and its tolerance to ubiquitination-mediated degradation in ABA response. As a result, sumoylation leads to enhanced interaction between RACK1B and RAP2.6, an AP2/ERF family transcription factor. RACK1B binds directly to the AP2 domain of RAP2.6, which alters the affinity of RAP2.6 for CE1 and GCC cis-acting regulatory elements. Taken together, our findings illustrate that protein stability controlled by dynamic post-transcriptional modification is a critical regulatory mechanism for RACK1B, which functions as scaffold protein for RAP2.6 in ABA signaling.
Protein sumoylation, characterized by the covalent conjugation of a small ubiquitin-related modifier (SUMO) protein to a lysine residue in a target protein, is a key post-translational modification in plants. Eight SUMO isoforms, SUMO1–8, are found in Arabidopsis, of which SUMO1/2 is the main conjugate1. Similar to ubiquitination, sumoylation requires an E1-E2-E3 enzyme cascade to catalyze the activation, conjugation and ligation processes2,3. The E1, a heterodimer, activates the SUMO by thioester formation4. In Arabidopsis, two redundant genes, SAE1A and SAE1B, encode the smaller subunit of E1, whereas the larger subunit is encoded by a single gene, SAE24. The E2, encoded by AtSCE1, transfers the SUMO from E1 to the lysine site of the substrate4. In some cases, the E2 is able to directly recognize not only the lysine located in the conical consensus sequence (ΨKxE) but also other irregular lysine residues5. The E3 ligase functions as an additional platform that promotes the interaction between SUMO-charged E2 and targets6. SIZ1 and HPY2 were identified as functional E3 ligases for protein sumoylation in Arabidopsis7,8. Recently, two E4-type SUMO ligases, PIAL1 and PIAL2, were identified that catalyze SUMO chain formation9.
A wide range of environmental and developmental signals, such as abiotic stresses7,10,11, the innate immune response12, hormones13, flowering4, and meristem maintenance8, are coupled with massive and rapid changes of sumoylation in Arabidopsis. In the abscisic acid (ABA) response, sumoylation of ABI5 by SIZ1 increases its stability and negatively regulates seed germination and inhibits primary root growth14. Sumoylation of MYB30, which functions in parallel with the ABI5-mediated ABA signaling pathway during seed germination15, is also stabilized by SIZ1. These results indicate that sumoylation is a regulatory mechanism in ABA responses.
The modified stability of transcription factors is often concomitant with altered transcriptional activities. Several cis-regulatory elements are involved in ABA signaling, such as the ABA responsive element (ABRE)16, the coupling element (CE)17,18, MYC recognition sites (MYCRs) and MYB recognition sites (MYBRs)19,20 and NAC recognition sites (NACRs)21. Among them, the ABRE is the best characterized cis-acting regulatory element and is recognized by the ABRE-binding protein/ABRE binding factor (AREB/ABF) in ABA-dependent gene expression22,23,24,25. However, a single ABRE copy is not sufficient for ABA-induced gene transcription and coupling elements, such as CE1 and CE3, are required for an effective ABA-responsive complex17,18. Using gel shift assays and yeast one-hybrid screenings, a group of proteins which could interact with CE1 or a CE1-like element were identified and designated as CE1 binding proteins (CEBPs)26. These proteins belong to the APETALA2/Ethylene Response Factor (AP2/ERF) transcription factor family26. In addition, most CEBPs can interact with the GCC-box, which acts as a cis-acting regulatory element in ethylene and jasmonic acid responses27,28.
Receptor for Activated C Kinase 1 (RACK1) functions as a versatile scaffold protein in all well-studied eukaryotic organisms29. The structure of RACK1 is characterized as a seven-bladed β-propeller, which provides a platform for protein-protein interaction in both mammalian and plant cells30,31. RACK1 is a ribosomal protein but it possesses no known enzymatic activity32. In the Arabidopsis genome, there are three homologous RACK1 genes, RACK1A, RACK1B and RACK1C33. Accumulating evidence indicates that RACK1 plays pivotal roles in diverse physiological processes in plants. RACK1 regulates plant developmental processes, including rosette leaf production and root development34. RACK1 also mediates responsiveness to multiple phytohormones, such as brassinosteroid, gibberellins and abscisic acid, and functions in disease resistance and the innate immune pathway33. In addition, RACK1 is involved in several abiotic stresses, for example, water stress31.
It is well-known that Arabidopsis RACK1 participates in ABA signaling. Based on the observation that a rack1a single mutant, and rack1a/rack1b or rack1a/rack1c double mutants are hypersensitive to the inhibition effect of ABA in seed germination, cotyledon greening and root growth, it was proposed that AtRACK1 functioned as a negative regulator of the ABA response35. RACK1 affected the accumulation and processing of pri-miR159, which targets TF MYB33 and MYB65 in ABA responses36. On the other hand, ABA down-regulated the expression of RACK1 and eIF6, which is a regulator for 80S ribosome assembly in mammalian cells37. This supports the finding that ABA impaires biogenesis of 60S ribosome subunit and ribosome assembly, although ABA had no effect on the RACK1-eIF6 interaction37. These studies indicate that Arabidopsis RACK1 might be involved in an ABA-regulated feedback loop. Although RACK1s play important roles in the ABA response, little is known about their up- and downstream interacting partners or the roles they play in regulating the ABA response.
Here, we report that Arabidopsis RACK1B is sumoylated at four residues, K50, K276, K281 and K291. Sumoylation increases the stability of RACK1B by blocking Ub-conjugation. Increased levels of exogenous ABA interfere with the balance between sumoylation and ubiquitination of RACK1B and pushes the balance towards sumoylation, thereby enhancing RACK1B stability. In addition, we provided evidence that ABA-stimulated sumoylation increased the interaction between RACK1B and transcription factor RAP2.6, which is a member of AP2/ERF family. This interaction resulted in altered affinity of RAP2.6 for CE1 and the GCC-box, which are important responsive elements in ABA and ethylene/jasmonic acid signaling, respectively17,28,38.
Results
RACK1B is subjected to sumoylation
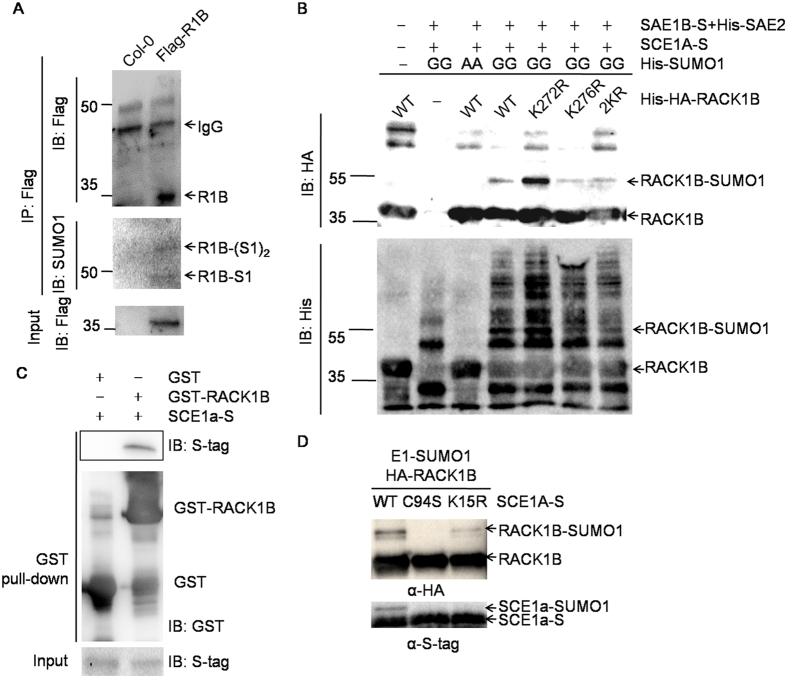
By analyzing protein structures and sequence homology of RACK1 from Arabidopsis and other species, two lysine residues, K272 and K276, were predicted to be potential sumoylation sites in AtRACK1B31. Further analysis with SUMOsp 2.0 software39 showed that both K276 and K272 have high probability as sumoylation sites and represent consensus and non-consensus sequences for sumoylation, respectively. To test whether RACK1B could be sumoylated, stable transgenic line expressing Flag-RACK1B in rack1b-2 background was used. The Flag tagged RACK1B was enriched by immunoprecipitation. Sumoylated RACK1B was detected by immunoblotting with an anti-Myc antibody (Fig. 1A). The identity of the protein corresponding to the immunoreactive signal at 50 kD was further confirmed to be SUMO1-conjugated RACK1B by tandem liquid chromatograph-mass spectrometry (LC-MS/MS) (Supplementary Table S1). These results suggested that RACK1B is indeed sumoylated by SUMO1 in vivo.
Figure 1. RACK1B is sumoylated by SUMO1.
(A) in vivo sumoylation assay. Seven-day-old Col-0 and 35S::Flag-RACK1B/rack1b-2 seedlings were used. Equal amounts of individual proteins were immunoprecipitated with anti-Myc antibodies and sumoylation was detected with the indicated antibodies. (B) In vitro sumoylation assay for RACK1B. Sumoylation of HA-RACK1B and K272R, K276R or K272R/K276R (2KR) mutants was detected by immunoblotting with anti-HA (upper panel) and anti-His antibodies (lower panel), respectively. GG and AA stand for WT forms of SUMO1 and SUMO1G92A-G93A, respectively. (C) GST pull-down analysis of interaction between SCE1A and RACK1B. GST-SCE1A or GST was used as bait to pull-down HA-RACK1B recombinant protein. Target proteins were eluted with 2 × sampling buffer and detected by immunoblot analysis with the antibodies indicated. (D) In vitro sumoylation assay with SCE1A mutants. S-tag fused SCE1AC94S or SCE1AK15R were used in the E. coli sumoylation system and compared with WT SCE1A. The sumoylation of HA-RACK1B was detected by immunoblot with anti-HA antibodies. SCE1A and SAE1B were detected with anti-S-tag antibodies.
Protein sumoylation can be accomplished either in an E3-dependent or an E3-independent manner40. To understand the RACK1B sumoylation mode and to verify the sumoylation site(s), an in vitro system consisting of E1, E2, SUMO and target protein was reconstituted in E. coli. The SUMO pathway-related proteins, SAE1B, SAE2, SCE1A and SUMO1(GG) were co-expressed with HA-RACK1B in E. coli. Note that SUMO1(GG) was used to mimic the mature form of SUMO1, which has C-terminal diglycine residues. A mutated version of SUMO1 containing two Ala residues [SUMO1(AA)], which cannot form a thioester bond, was used as a negative control6,41. In agreement with the in vivo experiment, both RACK1B and SUMO1-conjugated RACK1B could be detected (Fig. 1B). To our surprise, sumoylated RACK1B could still be detected in K272R and K276R single mutants and in the K272/K276R double mutant, suggesting that other undiscovered sumoylation sites may exist (Fig. 1B). Furthermore, RACK1B was not subjected to SUMO3 or SUMO5 conjugation in vitro, suggesting that RACK1B is specifically sumoylated by SUMO1 (Supplementary Fig. 1).
The fact that RACK1B can be sumoylated in vitro without the help of an E3 ligase indicates that SCE1A (E2) is able to directly recognize RACK1B. To test the hypothesis that SCE1A and RACK1B are components of one complex, we performed pull-down assays. The results showed that SCE1 directly interacted with RACK1B (Fig. 1C). SCE1A is self-sumoylated at Lys15 and at the catalytic Cys94 residue (within the active center)9; therefore, we investigated whether sumoylation of SCE1A is a prerequisite for sumoylation of RACK1B. Cys94 and Lys15 were mutated to Ser and Arg, respectively, via site-directed mutagenesis. As shown in Fig. 1D, RACK1B could be sumoylated by SCE1AK15R but not by the negative control SCE1AC94S, which suggested that SCE1A sumoylation at Lys15 was not necessary for RACK1B sumoylation. We also tested whether the two known E3 ligases contribute to the RACK1B sumoylation. The SIZ1 could interact with RACK1B and increased concentration of SIZ1 resulted in increased conjugation of SUMO1 on RACK1B in vitro (Supplementary Fig. 2A,B). However, sumoylated RACK1B remained detectable in the siz1–2 mutant (Supplementary Fig. 2C), suggesting that sumoylation of RACK1B is facilitated by but not rely on SIZ1. For the other E3 ligase, HPY2, due to severe dwarfism and defective meristems8, we failed to overexpress RACK1B in the hpy2–2 mutant. Therefore, a co-immunoprecipitation assay was performed to examine the interaction between RACK1B and HPY2. HPY2 was not precipitated with RACK1B and could not facilitate its sumoylation (Supplementary Fig. 2D–E), suggesting that HPY2 might not be involved in the RACK1B sumoylation complex. Taken together, these results suggested that RACK1B can be sumoylated via direct interaction between SCE1A and RACK1B.
RACK1B contains four lysine receptors of SUMO1
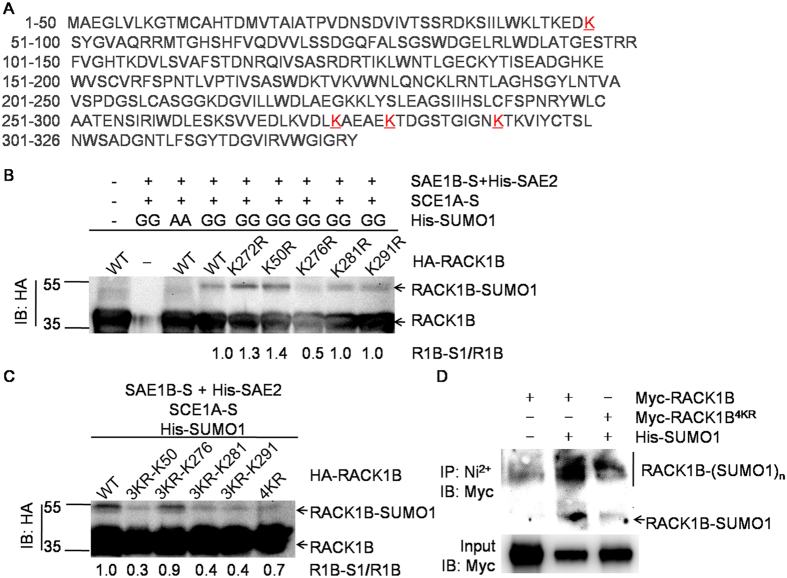
The sumoylation site(s) of RACK1B were identified using LC-MS/MS. In the E. coli sumoylation system, SUMO1(GG) was replaced by SUMO1 H89R(GG), which leaves a QTGG footprint on receptor Lys residues after digestion with trypsin42,43. The QTGG footprint facilitates the identification of sumoylated peptides via mapping the paralog-specific fragment ions. The sumoylated RACK1B was purified, separated and subjected to LC-MS/MS analysis (Supplementary Fig. 3). Four Lys residues, Lys50, Lys276, Lys281 and Lys291, conjugated to the QTGG footprint, were identified (Fig. 2A, Supplementary Fig. 4 and Supplementary Table S2). In addition to the consensus Lys276, Lys50 was located in a reverse consensus sequence (D/ExKΨ), and the other two Lys residues were considered as non-consensus sites. To our surprise, Lys272 was not sumoylated.
Figure 2. RACK1B has four major lysine residues.
(A) Amino acid sequence of RACK1B. Four lysine residues that could be sumoylated are underlined. (B,C) In vitro sumoylation assay. The sumoylation of RACK1B and its mutants was examined in the SUMO1-sumoylation system and was detected by immunoblotting with anti-HA antibodies. In (C), 3KR-K50, RACK1BK276,281,291R; 3KR-K276, RACK1BK50,281,291R; 3KR-K281, RACK1BK50,276,291R; 3KR-K291, RACK1BK50,276,281R; 4KR, RACK1BK50,276,281,291R. (D) In vivo sumoylation assay. 35S::Myc-RACK1B or 35S::Myc-RACK1B4KRwas co-transformed with 35S::His-SUMO1(GG) or empty vector in Col-0 protoplasts. The purified proteins were separated by SDS-PAGE and detected by immunoblotting with anti-Myc antibodies.
The identified Lys receptors were then verified by immunoblot analysis. Five single mutants HA-RACK1B (K272R, K50R, K276R, K281R or K291R), four triple mutants [K276,281,291R (3KR-K50), K50,281,291R (3KR-K276), K50,276,291R (3KR-K281), K50,276,281R (3KR-K291)], and a quadruple mutant [K50,276,281,291R (4KR)] were constructed and expressed in the E. coli sumoylation system. The sumoylation level of RACK1BK276R was severely decreased compared to the other four single mutants, while the sumoylation level of RACK1B3KR–K276 remained the highest among the four triple mutants (Fig. 2B,C), suggesting that SCE1A preferentially binds to the consensus Lys276 site in vitro. Mutation of the non-sumoylation site, K272R, increased the sumoylation level of RACK1B. The K50R mutation also slightly increased the level of sumoylation. Mutation of all four lysine residues severely decreased the sumoylation level both in vivo and in vitro although very low levels of sumoylation remained detectable (Fig. 2C,D). Taken together, these results indicated that RACK1B contains four sumoylation residues, Lys50, Lys276, Lys281 and Lys291.
Sumoylation increases the stability of RACK1B
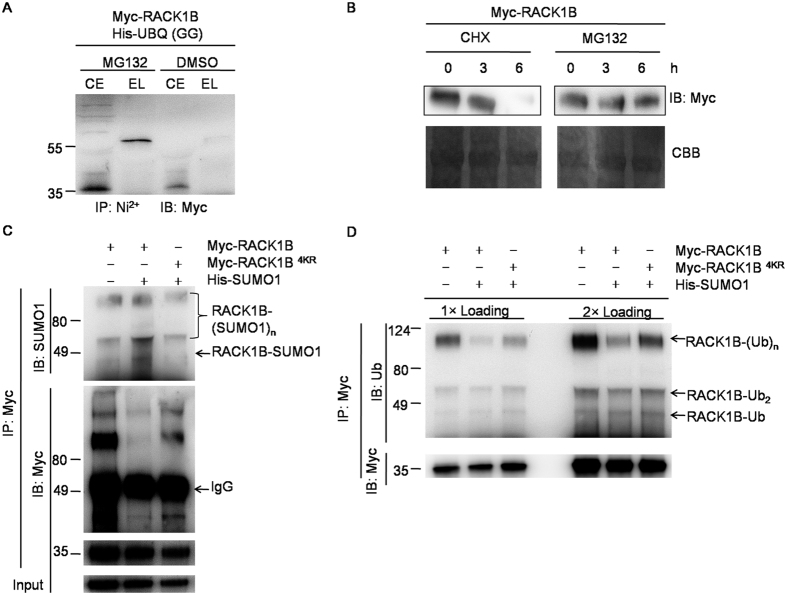
One consequence of sumoylation is a change in protein turnover rate5; therefore, we explored whether sumoylation influenced the stability of RACK1B. The degradation rates between transiently expressed RACK1B co-expressed with and without SUMO1(GG) were compared after treatment with cycloheximide (CHX), which inhibits new protein synthesis. As shown in Fig. 3, the level of RACK1B was significantly decreased at 3 h and was almost vanished at 5 h after CHX treatment in protoplasts expressing wild-type RACK1B alone. Co-transformation of SUMO1(GG) with RACK1B retarded the degradation of RACK1B and it remained detectable even 9 h after CHX treatment. Together, these results supported our postulation that sumoylation increases the stability of RACK1B. Note that we did not verify the degradation of RACK1B4KR as a negative control because mutation of these four lysine residues also reduced the level of ubiquitination (see below).
Figure 3. Sumoylation increases the stability of RACK1B.
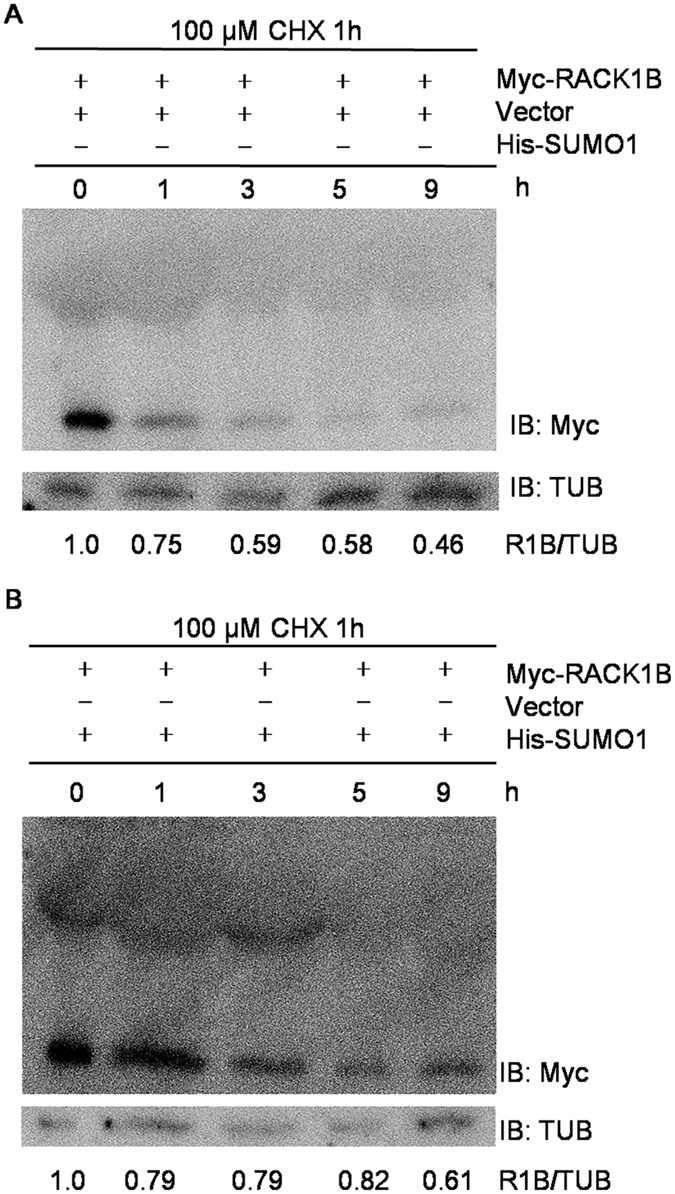
(A,B) Degradation assays. 35S::Myc-RACK1B co-transformed with 35S::His-SUMO1(GG) (A) or empty vector (B) in Col-0 protoplasts. After incubation for 20 h, the protoplasts were treated with 100 μM CHX and samples were harvested at 0, 1, 3, 5 and 9 h. The degradation of Myc-RACK1B was detected by immunoblotting with anti-Myc antibodies. Tubulin was used as a loading control. The signal intensities were analyzed and quantified using ImageJ. The Myc-RACK1B signal was normalized against that of tubulin and compared to signal at t = 0, which was set as 1.0. The numbers underneath represent the relative Myc-RACK1B abundance at each time point.
Sumoylation of RACK1B competes with its ubiquitination
Sumoylation of RACK1B increased its stability; therefore, we asked whether sumoylation prevents the degradation of RACK1B. First, we hypothesized that RACK1B is degraded by ubiquitination-mediated degradation. To confirm this, RACK1B and UBQ(GG) were co-expressed in Arabidopsis protoplasts. As shown in Fig. 4A, ubiquitinated RACK1B could only be detected after purification via Ni2+-chromatography with the addition of the proteasome inhibitor MG132 (Selleck, Shanghai, China). The unmodified form of RACK1B (35 kDa) was protected by MG132 (Fig. 4B). In contrast, over 95% of RACK1B was degraded after 6 h without MG132. These results indicated that RACK1B is degraded through the ubiquitination-mediated 26S proteosomal degradation pathway.
Figure 4. Sumoylation of RACK1B competes with ubiquitination at the same lysine residues.
(A) In vivo ubiquitination assay. 35S::Myc-RACK1B was co-transformed with 35S::His-UBQ(GG) in Col-0 protoplasts and treated with 50 μM MG132 or DMSO for 20 h. The ubiquitination of RACK1B was detected by immunoblotting with anti-Myc antibodies. CE, crude extract, EL, elutes from Ni2+-chromatography. (B) Degradation assay. 35S::Myc-RACK1B was transiently expressed in Col-0 protoplasts. After incubation for 20 h, the protoplasts were treated with 100 μM CHX or 50 μM MG132. The degradation rates of RACK1B were compared at the indicated time points. (C,D) In vivo immunoprecipitation assay. 35S::Myc-RACK1B or 35S::Myc-RACK1B4KRwas expressed with or without 35S::His-SUMO1(GG) in Col-0 protoplasts. RACK1B and RACK1B4KR were precipitated using anti-Myc antibodies and subjected to immunoblot analysis with anti-SUMO1 (C) or anti-ubiquitin (D) antibodies.
To confirm whether sumoylation prevents the degradation of RACK1B by competing with ubiquitination, RACK1B or RACK1B4KRwas transformed into protoplasts with SUMO1(GG). Overexpression of SUMO1(GG) increased the sumoylation level of RACK1B but not that of RACK1B4KR (Fig. 4C). Interestingly, the level of ubiquitinated RACK1B was significantly decreased in protoplasts overexpressing SUMO1(GG), whereas the level of ubiquitinated RACK1B4KR was slightly reduced and but still higher than that of RACK1B (Fig. 4D). This result suggested that other unidentified lysine receptors might also be potential ubiquitination sites, and the competition exists between sumoylation and ubiquitination of RACK1B.
ABA increases RACK1B stability by facilitating sumoylation
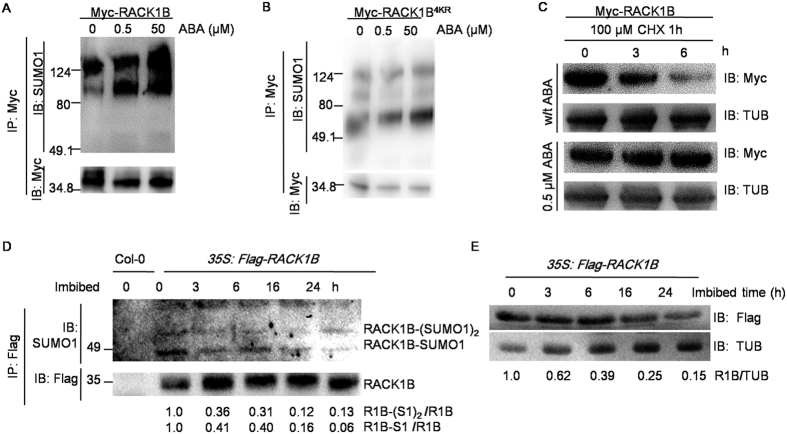
RACK1B is involved in the ABA signaling pathway35; therefore, we asked whether ABA regulates RACK1B at a post-translational level via sumoylation. Protoplasts expressing RACK1B were treated with different concentrations of ABA and, as expected, the sumoylation level of RACK1B significantly increased in a dose-dependent manner (Fig. 5A). In addition, the sumoylation level of Myc-RACK1B4KR was not affected by ABA, although a background of sumoylation still existed (Fig. 5B). Next, the degradation rates of RACK1B in protoplasts treated with exogenous ABA and in mock protoplasts were compared. The protoplasts were first treated with CHX to stop new protein synthesis and samples were harvested 3 and 6 h after incubation in the presence or absence of ABA. RACK1B was more resistant to degradation with the addition of ABA as shown by a higher abundance of RACK1B at 6 h (Fig. 5C). It is well-known that endogenous levels of ABA decrease during the first 24 h of seed imbibition44; therefore, we examined whether endogenous ABA regulates the sumoylation of RACK1B by using the transgenic line 35S::Flag-RACK1B/rack1b-2 (Supplementary Fig. 4). As expected, the levels of unmodified RACK1B and sumoylated RACK1B in germinating seeds gradually decreased with the time of imbibition (Fig. 5D). Furthermore, a co-immunoprecipitation assay was performed and optimized to achieve the same level of unmodified Flag-RACK1B in each well. Consistent with the unmodified RACK1B, the level of sumoylated RACK1B also decreased as endogenous ABA declined (Fig. 5E). Taken together, these results indicated that ABA enhances the stability of RACK1B by facilitating sumoylation. ABA increased the stability of RACK1B by up-regulating sumoylation and, at the same time, inhibiting ubiquitination-mediated degradation.
Figure 5. Sumoylation of RACK1B is regulated by ABA.
(A,B) In vivo sumoylation assay. 35S::Myc-RACK1B (A) and 35S::Myc-RACK1B4KR (B) were expressed in Col-0 protoplasts and treated with 0, 0.5 and 50 μM ABA for 20 h. Myc-RACK1B and Myc-RACK1B4KR were immunoprecipitated with anti-Myc antibodies. Protein sumoylation levels were detected by immunoblotting with anti-SUMO1 antibodies. (C) In vivo degradation assay. 35S::Myc-RACK1B or 35S::Myc-RACK1B4KR was expressed in Col-0 protoplasts and treated with or without 0.5 μM ABA for 20 h before 100 μM CHX treatment for 0, 3 and 6 h. Degradation of Myc-RACK1B was detected by immunoblotting with anti-Myc antibodies. Tubulin was used as a loading control. (D) The level of Flag-RACK1B in imbibed transgenic 35S::Flag-RACK1B seeds. The protein level of Flag-RACK1B was detected by immunoblotting with anti-Flag antibodies. (E) In vivo sumoylation assay. Individual proteins from (D) were subjected to immunoprecipitation using anti-Flag antibody-conjugated beads. The proteins were eluted and detected by immunoblotting with anti-SUMO1 and anti-Flag antibodies. In (D,E) the signal intensities were quantified using ImageJ. The numbers underneath represent the relative protein abundance at each time point, normalized against tubulin.
RACK1B interacts with RAP2.6 and alters its affinity to cis-elements
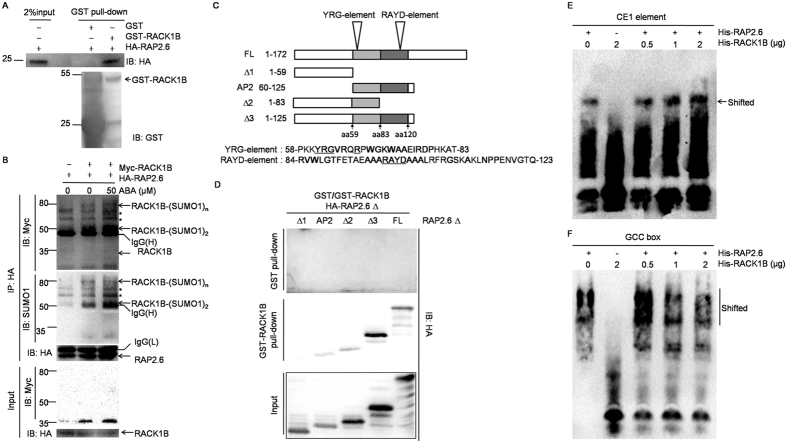
The transcription factor OsRAP2.6 interacts with OsRACK1 in the nucleus and cytoplasm45. Arabidopsis RAP2.6, which shares 94% amino acid identity with OsRAP2.6 in the AP2/ERF domain participates in ABA responses45. Because both RACK1 and RAP2.6 are nuclear localized36,45 and ABA enhanced the stability of RACK1B via sumoylation (Fig. 4), we assumed that AtRACK1B could interact with AtRAP2.6 and function as a scaffold protein in the ABA response. To test this hypothesis, a GST pull-down assay was performed and showed that RACK1B interacted with RAP2.6 in vitro (Fig. 6A). Furthermore, a co-immunoprecipitation assay showed that RACK1B and RAP2.6 could interact in planta (Fig. 6B). When RACK1B and RAP2.6 were co-expressed in mesophyll protoplasts, RACK1B could be precipitated with anti-HA antibodies that recognize the HA tag fused to RAP2.6. RAP2.6 co-precipitated both RACK1B and sumoylated RACK1B and the intensity of co-precipitation was enhanced by exogenous ABA treatment. In addition, a greater amount of sumoylated RACK1B could be precipitated compared to unmodified RACK1B, while only unmodified RACK1B could be detected in crude extracts.
Figure 6. Sumoylation of RACK1B enhanced its interaction with RAP2.6.
(A) In vitro GST pull-down assay. GST-RACK1B was used as bait to pull-down HA-RAP2.6 recombinant protein. The RACK1B-RAP2.6 interaction was detected by immunoblot analysis with anti-HA antibodies. (B) In vivo co-immunoprecipitation assay. 35S::Myc-RACK1B and 35S::HA-RAP2.6 were co-expressed in Col-0 protoplasts and thereafter incubated with or without 50 μM ABA for 20 h at normal growth conditions. A co-immunoprecipitation assay was performed with anti-HA antibodies and proteins detected by immunoblotting with indicated antibodies. (C) Schematic diagrams of truncated versions of RAP2.6. aa, amino acid. (D) In vitro GST pull-down assays. GST-RACK1B was used to pull-down truncated RAP2.6 as indicated in (C). FL, full-length. (E,F) Analysis of RAP2.6 binding to CE1 element and GCC-box by EMSA. Biotin-labeled CE1 (E) or GCC (F) was used as probe. Equal amounts of His-HA-RAP2.6 protein were used in all the binding assays. Recombinant protein His-HA-RACK1B was added at the amount indicated. The band shift of probes was detected by ECL. The experiment was repeated three times with same results.
To test which domain of RAP2.6 is responsible for the interaction with RACK1B, truncated versions of RAP2.6 were generated (Fig. 6C). Based on a previous report, two conserved motifs, YRG and RAYD elements, exist in the AP2 domain of twelve RAP2 proteins46. As shown in Fig. 6D, the interaction could be detected when the peptide contained an intact AP2 domain (both YRG and RAYD motifs) or the YRG element alone, but not the N-terminal region of RAP2.6. Collectively, these results suggest that RACK1B interacts with RAP2.6 by direct binding to its AP2 domain and that this interaction could be enhanced by sumoylation of RACK1B.
AtRAP2.6 can bind to the GCC-box and the CE1 element, which serve as cis-acting regulatory elements in various phytohormone signaling pathways47 The intact promoter recognized by RAP2.6 remains elusive; therefore, we performed Electrophoretic mobility shift assay (EMSA) to examine whether the RACK1B-RAP2.6 interaction affects the binding of RAP2.6 to the GCC-box and/or CE1 elements. RACK1B and RAP2.6 were fused to His-tags and purified by Ni2+-chromatography. The result showed that the binding affinity of RAP2.6 to the CE1 element gradually improved with increasing amounts of RACK1B (Fig. 6E). In contrast, RACK1B decreased the binding affinity of RAP2.6 to the GCC-box (Fig. 6F). Taken together, these results suggest that RACK1B functions as a scaffold protein for RAP2.6 in the ABA response and alters the affinity of RAP2.6 for downstream cis-elements.
Discussion
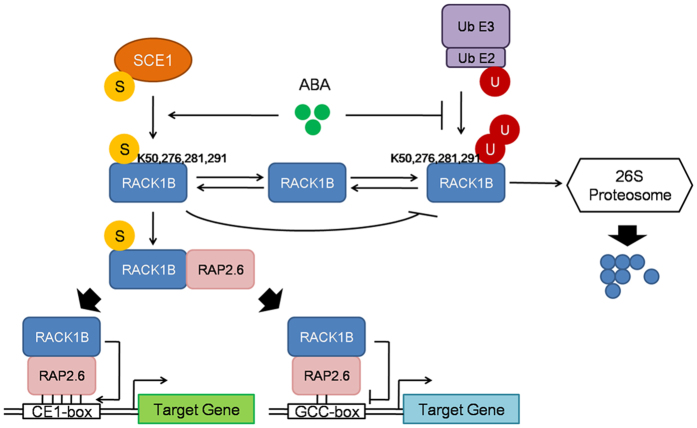
RACK1 proteins have been identified in all eukaryotic organisms and act as scaffold proteins that mediate protein-protein interactions or compete with other proteins for binding domains29. RACK1B participates in various hormone responses. For example, rack1a-1 and rack1a-2 mutants are insensitive to GA and BL, and produce reduced lateral roots when treated with exogenous 1-Naphthaleneacetic acid33. Here, we provide evidence for an expanded regulatory mechanism of ABA signaling that controls the stability of RACK1B, resulting in enhanced interaction with the transcription factor, RAP2.6. As summarized in Fig. 7, our study shows that RACK1B is sumoylated at four Lys residues via direct interaction with SCE1. The up-regulated sumoylation in response to ABA leads to increased stability and abundance of RACK1B, which results from competition with ubiquitination at the same residues. The interaction between RACK1B and RAP2.6 is also enhanced by sumoylation, which alters the affinity of RAP2.6 for its target cis-regulatory elements.
Figure 7. Working model for the ABA-induced increase in RACK1B stability via sumoylation.
Sumoylation of RACK1B is directly mediated by SCE1A at four residues, K50, K276, K281 and K291, resulting in increased stability of RACK1B. Covalent linkage of SUMO1 (S) on RACK1B blocks some of the UBQ (U) conjugation sites in RACK1B, which prevents protein degradation. ABA facilitates the sumoylation of RACK1B and simultaneously inhibits ubiquitination, and promotes the interaction between RACK1B and RAP2.6. The consequence of this interaction results in enhanced affinity of RAP2.6 with CE1 element (five solid lines) and reduced affinity with GCC-box (two solid lines).
Studies of Arabidopsis sumoylation revealed that a few substrates are sumoylated in a SIZ1-dependent manner10,14,48,49,50. The E2-dependent RACK1B sumoylation described here is different from that in these reports (Fig. 2). This is probably due to the high affinity between RACK1B and SCE1A, which is consistent with the finding that most E3 ligase-independent sumoylation occurs at non-consensus lysine residues. Indeed, two of four identified sumoylation sites in RACK1B are non-consensus lysine residues (Fig. 2A and Supplementary Fig. 4). Results of the in vitro sumoylation assay suggested that SIZ1 facilitated the sumoylation of RACK1B. However, we cannot rule out the possibility that SIZ1 contributes to the site selectivity since we failed to identified the sumoylated sites in vivo by MS analysis.
A consequence of sumoylation can be to alter substrate stability5. Many substrates are both sumoylated and ubiquitinated, and often at the same lysine residues. However, the inter-relationship between these two systems is substrate specific, and can act either synergistically or antagonistically51,52. We observed that sumoylation increased RACK1B stability by antagonizing ubiquitination. The possible explanation is either two modifications share several common sites or SUMO molecule spatially blocks the ubiquitination sites or both. Besides, a form of coordination may exist between the lysine residues to balance the levels of sumoylation and ubiquitination. However, the ubiquitin complex targeting RACK1B degradation is unknown and this hypothesis requires further investigation.
It is well-known that ABA triggers multiple post-transcriptional modifications of proteins, such as phosphorylation and ubiquitination53,54. Here, we showed that ABA is also able to regulate sumoylation of target proteins. Blocking ubiquitination sites via sumoylation might be one of the regulatory mechanisms by which ABA stabilizes RACK1B (Figs 4 and 5). However, we have no direct evidence that sumoylation and ubiquitination of RACK1B are mutually exclusive in the ABA response, although ABA can inhibit the ubiquitination of RACK1B (Supplementary Fig. 6). It is possible that down-regulated ubiquitination resulted from various regulatory events, such as inhibition of the ubiquitination complex, or unidentified forms of post-transcriptional modification, instead of from direct competition for the modification of lysine residues. The varied modification status of RACK1B suggests that RACK1B is subjected to the interplay between various post-transcriptional modifications in response to different signals. A similar example is that of RACK1A phosphorylation by WNK8, which tends to be degraded in glucose signaling55. However, neither the phosphorylation-mimic nor the dead form of RACK1B at Ser122 and Thr161 abolished RACK1B sumoylation, although both forms affect its degradation (Supplementary Fig. 7). Therefore, the crosstalk between distinct post-transcriptional modifications of RACK1B in ABA signaling needs to be further investigated.
The interaction between RACK1B and RAP2.6 sheds light on RACK1B function that binds to one of AP2/ERF family TFs in ABA response. RAP2.6 preferentially binds to sumoylated RACK1B rather than unmodified RACK1B in vivo, suggesting that sumoylation facilitates RACK1B interaction with its binding partners (Fig. 6B). Additionally, ABA stimulates RACK1B sumoylation and, therefore, plays a role in regulating the dynamics of the interaction between RACK1B and RAP2.6, which could influence the expression of downstream target genes in response to ABA. Three RACK1 proteins were regarded as negative regulators of the ABA response35,37. The expression of the ABA marker gene, RAB18, was induced by ABA in a rack1a mutant and a rack1a/rack1b double mutant as early as 3 h after treatment35. However, in RAP2.6 overexpressing plants, RAB18 was induced after 24 h ABA treatment47. These downstream events continue to obscure the function of RACK1 in the ABA response. One possible explanation is that RACK1B acts as a scaffold to a wide range of proteins for distinct TFs in ABA signaling and, therefore, induction of RAB18 in rack1 mutants is a cumulative effect of impaired activities of distinct TFs due to loss of the platform.
RACK1 is a highly conserved WD40 repeat family protein present in a wide range of eukaryotic species, from microorganisms to plants and animals29. RACK1 is a scaffold protein that acts as a hub for the interaction of signaling proteins, including kinases/phosphatases55,56, G proteins57, transcription factors58, cytoskeleton proteins29, and the ubiquitin complex59. Four main effects of RACK1 on interacting proteins have been described: diversifying intermolecular interaction, changing the activity of partners, regulating the stability of partners and shuttling partners from one location to another60. However, in contrast to the detailed understanding of RACK1 in animals, very few studies have investigated the molecular mechanisms of RACK1 in plants. In rice, OsRACK1 is a component of the Rac1 complex and plays a role in the production of reactive oxygen species, and is involved in resistance to rice blast infection61. Its homolog in Arabidopsis, AtRACK1, is involved in a novel immune pathway that consists of the G-protein-RACK1-MAPK cascade and which transmits the signal of pathogen-secreted proteases57. AtRACK1 also affects the accumulation and precision of pri-miRNAs by interacting with SERRATE and associating with the AGO1 complex36. Here, we postulate that a potential effect of RACK1B on RAP2.6 is to regulate its transcriptional activity because the in vitro EMSA result indicated that RAP2.6 has altered affinity to the GCC-box and CE1 elements once bound to RACK1B. This result also indicates that RACK1B and RAP2.6 are potential crosslink nodes between different abiotic and biotic signaling pathways. Identification of the target gene(s) of RAP2.6 will help explain the regulatory effect of RACK1B in signal transduction.
Sequence analysis revealed that RAP2 proteins are highly divergent except for the presence of at least one AP2 domain46. AP2/ERF proteins constitute a large family of plant specific transcription factors with more than 120 members in Arabidopsis62. Based on copies of the AP2 domain and sequence similarity, they can be categorized into three subfamilies, the AP2, ERF, and RAV subfamilies62,63. On the other hand, a number of cis-regulatory elements are mainly recognized by AP2/ERF transcription factors, including the drought responsive element/C-repeat responsive element (DRE/CRT)64, the ethylene responsive element (ERE or GCC-box)38, the JA and elicitor responsive element (JERE)65, the bipartite sequence66, and the ANT consensus site67. Because RACK1B binds directly to the AP2 domain within RAP2.6, we hypothesize that RACK1B is capable of binding different AP2/ERF members that contain an AP2 domain that shares high similarity with that of AtRAP2.6. High-throughput screening methods, such as yeast-two hybrid and co-immunoprecipitation, have identified potential binding partners29. Detailed analysis of these proteins will yield a better understanding of RACK1 function in plants.
Methods
Plant materials and growth conditions
Arabidopsis with a Col-0 ecotype was used in the study. The T-DNA mutants, siz1-2 (SALK_065397)7, hpy2-2 (SAIL_77_G06)8 and rack1b-2 (SALK_145920)34 used in this study were described previously.
Arabidopsis seeds were surface sterilized with 70% ethanol and subsequently soaked in 10% NaClO for 10 min and then washed with sterilized H2O five times. Seeds were germinated on MS medium with 0.7% agar. For seed propagation, one-week-old seedlings were transferred to soil and grown in a phytotron at 22 °C with light intensity of 120 μmolm−2s−1 and a 16/8 h photoperiod.
Generation of transgenic Arabidopsis plants
RACK1B coding sequences were amplified by PCR and cloned into pHB vector68 with Flag tag was fused to the N-terminus. The plasmids were transformed into Agrobaterium tumefaciens (EHA105) and plant transformation was performed by the floral dip method. Sequences of all the primers used in the study are listed in Supplementary Table S3.
Antibodies
Mouse anti-HA was purchased from Youke Biotechnology (Shanghai, China), mouse anti-Myc and anti-GST, and rabbit anti-S tag antibodies were from GenScript (Nanjing, China). Rabbit anti-SUMO1 and anti-UBQ antibodies were from Agrisera (Vännäs, Sweden). Rabbit anti-TUBB2A antibody was from Sigma-Aldrich (Saint Louis, USA). The secondary antibodies, goat anti-mouse IgG-HRP (sc-2005) and goat anti-rabbit IgG-HRP (sc-2004) were from Santa Cruz (Dallas, TX, USA).
Isolation of total RNA and first-strand synthesis of cDNA
Arabidopsis total RNA was isolated using TRIzol reagent (ThermoFisher, San Jose, CA, USA) according to the manufacturer’s instructions. One microgram of total RNA was used as template for first-strand cDNA synthesis using the PimeScript One Step RT-PCR Kit (Takara, Dalian, China). The cDNA was used as template for further PCR amplification of the coding sequences of indicated genes.
Molecular cloning of RACK1B and site directed mutagenesis
RACK1B was amplified from cDNA by PCR and cloned into pMD18 with HA tag fused at N-terminus. RACK1B sequences with the following mutations, K50R, K272R, K276R, K281R, K291R, 3KR-K50, 3KR-K276, 3KR-K281, 3KR-K291 and 4KR, were synthesized (GenScript) and cloned into pUC57 with HA tag fused at N-terminus. These plasmids were used as starting plasmids for other constructions.
The His89 of SUMO1 was mutated to Arg to generate SUMO1H89R(GG). At the same time, Gly92 and Gly93 were mutated to Ala to generate SUMO1H89R(AA). SCE1AK15R and SCE1AC94S were generated by site directed mutagenesis. PCR was used to perform the site directed mutagenesis using specific primers and cDNA as temperate.
Transient expression of proteins in protoplasts
Proteins were transiently expressed in Arabidopsis protoplasts. Four-week-old seedlings were used for mesophyll protoplast isolation and plasmid transfection according to a previously described method69. After incubation for 20 h in normal growth conditions, unless indicated otherwise, protoplasts were harvested for protein extraction.
Protein purification by Ni2+-chromatography
Total proteins were extracted from protoplasts using lysis buffer A containing 25 mM Tris-HCl, 200 mM NaCl pH 7.8, 2 mM DTT, 1% Triton X-100 and 1 mM PMSF. Protein concentration was measured with the Bradford method and 2 mg of total proteins were used for further purification.
For purification of recombinant proteins, 50 mL E. coli culture were collected for protein extraction. Proteins with the hexa-histidine tag were purified using Ni2+-NTA sepharose (Qiagen, Venlo, Limburg, Netherlands) according to the manufacturer’s instructions. The purified protein were separated by SDS-PAGE and analyzed by immunoblotting.
Immunoprecipitation of proteins and co-immunoprecipitation assay
Total proteins (1 mg) extracted from protoplasts were incubated with desired antibodies at 4 °C for 12 h with gentle shaking and then protein G agarose (Roche) slurry was added and incubation continued at 4 °C for another 4 h. The resin was collected and washed with 10-bed volumes of lysis buffer A three times. Proteins were then eluted with 2 × sampling buffer. For co-immunoprecipitation assays, the enriched proteins were separated by SDS-PAGE and detected by immunoblotting with another antibody. The chemiluminescence was visualized using a Tanon-5200 Chemiluminescent Imaging System (Tanon Science and Technology, Shanghai, China).
RACK1B sumoylation assay in Arabidopsis mesophyll protoplasts
His-SUMO1(GG), Myc-RACK1B, Myc-RACK1BK276R, Myc-RACK1B3KR-K276 and Myc-RACK1B4KR were amplified by PCR, and subsequently cloned into the expression vector, pGreenII 62-SK to generate 35S::His-SUMO1(GG), 35S::MYC-RACK1B, 35S::MYC-RACK1BK276R, 35S::MYC-RACK1B3KR-K276 and 35S::MYC-RACK1B4KR, respectively. His-SUMO1(GG) and different types of Myc-RACK1B proteins were transiently expressed in Arabidopsis protoplasts. The proteins sumoylated by His-SUMO1 were purified by Ni2+-chromatography. Sumoylation of RACK1B was detected by immunoblot analysis using anti-Myc antibodies.
RACK1B ubiquitination assays in Arabidopsis mesophyll protoplasts
His-UBQ(GG) was amplified by PCR and cloned into pGreenII 62-SK to generate 35S::His-UBQ(GG). His-UBQ(GG) was co-expressed with Myc-RACK1B in Col-0 protoplasts, which were then treated with 50 μM MG132 to block protein degradation or DMSO as control for 20 h. The ubiquitinated proteins were purified using Ni2+-chromatography and ubiquitination of RACK1B was detected by immuno blotting using anti-Myc antibodies.
Sumoylation of RACK1B in imbibed seeds
The WT Col-0 and Flag-RACK1B transgenic seeds (35S::Flag-RACK1B/rack1b-2) in the background of the T-DNA insertion mutant rack1b-2 were freshly harvested. For each individual sample, 50 mg seeds were used and imbibed in water for 0, 3, 6, 16 and 24 h. Proteins were extracted with lysis buffer A and filtrated through a 0.2 μM filter membrane (Millipore) to exclude the insoluble substances. Flag-RACK1B was immunoprecipitated with anti-DYKDDDDK beads (Clontech) and the sumoylation level of RACK1B was detected using anti-SUMO1 antibodies.
In vitro sumoylation assays
For sumoylation assay in E. coli system, pACYCDuet-1 and pCDFDuet-1 were used to reconstitute the Arabidopsis SUMO pathway in E. coli as described41. pACYCDuet-1 carries two subunits of E1 (SAE1B and SAE2). SAE1B and SAE2 were fused with an S-tag and His tag, respectively. pCDFDuet-1 carries a SUMO molecule [SUMO1H89R(GG), SUMO1H89R(AA), SUMO3(GG) or SUMO5(GG)] and E2 (SCE1A, SCE1AK15Ror SCE1AC94S). SUMO and SCE1A were fused with a His tag and S-tag, respectively. HA-RACK1B was cloned into pET28a such that the His tag was fused at the N-terminus. All three plasmids were co-transformed into E. coli BL21 (DE3) competent cells. The transformants were inoculated and cultured at 37 °C to an OD600 of approximately 0.6. IPTG was added to a final concentration of 0.2 mM. After induction for 12 h at 25 °C, the cells were harvested and re-suspended in PBS buffer. Cells were lysed by sonication, centrifuged; and the supernatant was collected. The proteins were detected by immunoblot analysis after separation by SDS-PAGE.
For in vitro sumoylation assay, the coding sequences of SUMO1, SAE1B, SAE2, SCE1, SIZ1 and HPY2 were cloned into pET28a. The expression, purification and in vitro sumoylation assay were performed as described7,70.
Identification of sumoylation sites using mass spectrometry
The crude extracts from in vitro sumoylation assays were purified by Ni2+-chromatography and separated by 10% SDS-PAGE. The gel was Coomassie Brilliant Blue-stained and the bands corresponding to sumoylated RACK1B were cut out (Supplemental Fig. 2). The gel slices were cleaned, desalted and concentrated via vacuum centrifugation and then digested with 5ng/μL trypsin at 37 °C for at least 20 h. Tryptic peptides were acidified with 100 μL extraction buffer (60% ACN, 0.1% TFA).
LC-MS/MS analyses were performed using QE mass spectrometry with a nano-electrospray ion source (ThermoFisher) coupled to a ThermoFisher Easy nLC1000 high performance liquid chromatography system equipped with a 0.15 mm × 150 mm Zorbax 300SB-C18 column. Peptides were separated using a linear gradient of 4–50% acetonitrile (0.1% formic acid) for 50 min, 50–100% acetonitrile (0.1% formic acid) from 50 min to 54 min, and 100% acetonitrile for 60 min. MS spectra were acquired in full-scan mode at a resolution of 100,000 and 10 MS/MS spectra were acquired per MS. MS/MS precursor selection met the criteria that the charge state be known and ≥ 2. Peptide sequences were searched using MASCOT 2.2 software against the Arabidopsis protein database and common contaminants. Search parameters included a precursor mass tolerance of 2.5 Da, fragment ion mass tolerance of 0.2 Da, up to two missed cleavages, oxidation of Met, variable modification of Lys residues by sumoylation (QTGG, +343.1492 m/z or pyroQTGG, +326.1226 m/z), and a filter by score ≥45.
In vitro GST pull-down assay
To verify the interaction of RACK1B with SCE1A and SIZ1, coding sequences of RACK1B, SCE1A and SIZ1 were cloned into pGS-21a (GenScript) to generate GST-RACK1B, GST-SCE1 and GST-SIZ1, respectively.
To verify the interaction between RACK1B and RAP2.6, HA tagged RAP2.6 was cloned into pET28a and pGreenII 62-SK to generate His-HA-RAP2.6 and 35S::HA-RAP2.6, respectively. To examine the domain of RAP2.6 that interacted with RACK1B, HA-tagged truncated peptides of RAP2.6 were generated by PCR and cloned into pET28a to generate expression vectors.
GST-RACK1B, GST-SIZ1, His-HA-RACK1B or SCE1A-S was transformed into E. coli BL21 (DE3). His-HA-RAP2.6 and truncated peptides were expressed in E. coli Rosetta (DE3). Transformed strains were first grown at 37 °C until OD600 reached 0.6~0.8. Cultures were then induced with 0.1 mM IPTG for 2 h at 25 °C. Cells were harvested in binding buffer (50 mM Tris-HCl pH 7.8, 200 mM NaCl, 1% Triton X-100, and 10% glycerol) at a ratio of 1:10. For the SIZ1-RACK1B interaction, 500 μL crude extract of GST-SIZ1 for each individual trial was incubated with 30 μL glutathione-Sepharose beads (GenScript) for 1 h on ice. After washing three times with 20 volumes of binding buffer, the beads were incubated with 200 μL crude extract of His-HA-RACK1B for another 1 h on ice. After another wash step, the bound proteins were eluted with 2 × SDS-PAGE sampling buffer. The same binding conditions were used for RACK1B-SCE1A and RACK1B-RAP2.6 (or truncated RAP2.6) interactions, except that 100 μL crude extract of GST-RACK1B was used as bait protein and equal volumes of crude extracts of SCE1A-S and His-HA-RAP2.6 (or truncated mutants) were used as prey proteins. The interactions were detected by immunoblot analysis using the antibodies indicated.
EMSA
Oligonucleotides 4 × CE1 and 4 × GCC element was synthesized and biotin labeled (Genewiz, Suzhou, China) with the sequences: 5′-GGAATTC TGCCACCGG TGCCACCGG TGCCACCGG TGCCACCGG TCTAGAGC-3′ and 5′-GGAATTC AGCCGCC AGCCGCC AGCCGCC AGCCGCC TCTAGAGC-3′, respectively47. The protein His-HA-RAP2.6 was expressed and induced as described above, and purified by Ni-NTA chromatography (Qiagen). In the binding assays, 1 μg His-RAP2.6 and 50 fmol biotin-labeled CE1 element, or 0.5 μg His-HA-RAP2.6 and 100 fmol biotin-labeled GCC-box was used. Purified His-RACK1B was added as indicated. The binding reaction (in a volume of 20 μl) was performed in binding buffer containing 10 mM Tris, 50 mM KCl, 1 mM DTT, 1 μg poly (dI·dC), pH 7.5. Binding reactions were incubated at room temperature for 20 min and analyzed by PAGE on 6% gels in 0.5 × TBE buffer. Gels were blotted on Hybond-N+ membranes (GE healthcare, Uppsala, Sweden) and signals were detected using a Light Shift Chemiluminescent EMSA Kit (ThermoFisher) according to the manufacturer’s instructions.
Data availability
The authors declare that the data supporting the findings of the study are available within the article and its Supplementary Information files or are available from the corresponding author upon request.
Additional Information
How to cite this article: Guo, R. and Sun, W. Sumoylation stabilizes RACK1B and enhance its interaction with RAP2.6 in the abscisic acid response. Sci. Rep. 7, 44090; doi: 10.1038/srep44090 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work is supported by the National Natural Science Foundation of China (31270873), Strategic Pioneer Project of CAS (XDA04020202-18), the China Manned Space Program (TG-2 mission).
Footnotes
The authors declare no competing financial interests.
Author Contributions R.G. and W.S. conceived the experiments, R.G. conducted the experiments, R.G. and and W.S. analyzed the data and wrote the manuscript. Both of the authors reviewed the manuscript.
References
- Kurepa J. et al. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem 278, 6862–6872 (2003). [DOI] [PubMed] [Google Scholar]
- Hay R. T. Protein modification by SUMO. Trends Biochem Sci 26, 332–333 (2001). [DOI] [PubMed] [Google Scholar]
- Melchior F. SUMO–nonclassical ubiquitin. Annu Rev Cell Dev Biol 16, 591–626 (2000). [DOI] [PubMed] [Google Scholar]
- Saracco S. A., Miller M. J., Kurepa J. & Vierstra R. D. Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol 145, 119–134 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R. & Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8, 947–956 (2007). [DOI] [PubMed] [Google Scholar]
- Flotho A. & Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem 82, 357–385 (2013). [DOI] [PubMed] [Google Scholar]
- Miura K. et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA 102, 7760–7765 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T. et al. SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell 21, 2284–2297 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomanov K. et al. Arabidopsis PIAL1 and 2 promote SUMO chain formation as E4-type SUMO ligases and are involved in stress responses and sulfur metabolism. Plant Cell 26, 4547–4560 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K. et al. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19, 1403–1414 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catala R. et al. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell 19, 2952–2966 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Burg H. A., Kini R. K., Schuurink R. C. & Takken F. L. Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell 22, 1998–2016 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Qi Y., Liu M. & Yang C. SUMO E3 ligase AtMMS21 regulates drought tolerance in Arabidopsis thaliana(F). J Integr Plant Biol 55, 83–95 (2013). [DOI] [PubMed] [Google Scholar]
- Miura K. et al. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci USA 106, 5418–5423 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Schumaker K. S. & Guo Y. Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc Natl Acad Sci USA 109, 12822–12827 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. K. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53, 247–273 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q. & Ho T. H. Functional dissection of an abscisic acid (ABA)-inducible gene reveals two independent ABA-responsive complexes each containing a G-box and a novel cis-acting element. Plant Cell 7, 295–307 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q., Zhang P. & Ho T. H. Modular nature of abscisic acid (ABA) response complexes: composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell 8, 1107–1119 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H. et al. Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9, 1859–1868 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe H. et al. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran L. S. et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H., Hong J., Ha J., Kang J. & Kim S. Y. ABFs, a family of ABA-responsive element binding factors. J Biol Chem 275, 1723–1730 (2000). [DOI] [PubMed] [Google Scholar]
- Uno Y. et al. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97, 11632–11637 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S. R., Rodriguez P. L., Finkelstein R. R. & Abrams S. R. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61, 651–679 (2010). [DOI] [PubMed] [Google Scholar]
- Yoshida T. et al. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61, 672–685 (2010). [DOI] [PubMed] [Google Scholar]
- Lee S. J., Park J. H., Lee M. H., Yu J. H. & Kim S. Y. Isolation and functional characterization of CE1 binding proteins. BMC Plant Biol 10, 277 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner M. & Singh K. B. Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC-box DNA-binding protein interacts with an ocs element binding protein. Proc Natl Acad Sci USA 94, 5961–5966 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R. L., Kazan K., McGrath K. C., Maclean D. J. & Manners J. M. A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol 132, 1020–1032 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islas-Flores T., Rahman A., Ullah H. & Villanueva M. A. The Receptor for Activated C Kinase in Plant Signaling: Tale of a Promiscuous Little Molecule. Front Plant Sci 6, 1090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta J. et al. Identification of the versatile scaffold protein RACK1 on the eukaryotic ribosome by cryo-EM. Nat Struct Mol Biol 11, 957–962 (2004). [DOI] [PubMed] [Google Scholar]
- Ullah H. et al. Structure of a signal transduction regulator, RACK1, from Arabidopsis thaliana. Protein Sci 17, 1771–1780 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams D. R., Ron D. & Kiely P. A. RACK1, A multifaceted scaffolding protein: Structure and function. Cell Commun Signal 9, 22 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. G. et al. RACK1 mediates multiple hormone responsiveness and developmental processes in Arabidopsis. J Exp Bot 57, 2697–2708 (2006). [DOI] [PubMed] [Google Scholar]
- Guo J. & Chen J. G. RACK1 genes regulate plant development with unequal genetic redundancy in Arabidopsis. BMC Plant Biol 8, 108 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. et al. RACK1 is a negative regulator of ABA responses in Arabidopsis. J Exp Bot 60, 3819–3833 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speth C., Willing E. M., Rausch S., Schneeberger K. & Laubinger S. RACK1 scaffold proteins influence miRNA abundance in Arabidopsis. Plant J 76, 433–445 (2013). [DOI] [PubMed] [Google Scholar]
- Guo J. et al. Involvement of Arabidopsis RACK1 in protein translation and its regulation by abscisic acid. Plant Physiol 155, 370–383 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi M. & Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Zhou F., Fu C., Xu Y. & Yao X. SUMOsp: a web server for sumoylation site prediction. Nucleic Acids Res 34, W254–257 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knipscheer P. et al. Ubc9 sumoylation regulates SUMO target discrimination. Mol Cell 31, 371–382 (2008). [DOI] [PubMed] [Google Scholar]
- Okada S. et al. Reconstitution of Arabidopsis thaliana SUMO pathways in E. coli: functional evaluation of SUMO machinery proteins and mapping of SUMOylation sites by mass spectrometry. Plant Cell Physiol 50, 1049–1061 (2009). [DOI] [PubMed] [Google Scholar]
- Miller M. J., Barrett-Wilt G. A., Hua Z. & Vierstra R. D. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci USA 107, 16512–16517 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galisson F. et al. A novel proteomics approach to identify SUMOylated proteins and their modification sites in human cells. Mol Cell Proteomics 10, M110 004796 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A. A. et al. Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8′-hydroxylase. Plant J 45, 942–954 (2006). [DOI] [PubMed] [Google Scholar]
- Wamaitha M. J. et al. OsRap2.6 transcription factor contributes to rice innate immunity through its interaction with Receptor for Activated Kinase-C 1 (RACK1). Rice (NY) 5, 35 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro J. K., Caster B., Villarroel R., Van Montagu M. & Jofuku K. D. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci USA 94, 7076–7081 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q. et al. The Arabidopsis AP2/ERF transcription factor RAP2.6 participates in ABA, salt and osmotic stress responses. Gene 457, 1–12 (2010). [DOI] [PubMed] [Google Scholar]
- Crozet P. et al. SUMOylation represses SnRK1 signaling in Arabidopsis. Plant J 85, 120–133 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X. L. et al. An Arabidopsis SUMO E3 Ligase, SIZ1, Negatively Regulates Photomorphogenesis by Promoting COP1 Activity. PLoS Genet 12, e1006016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B. S., Song J. T. & Seo H. S. Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nat Commun 2, 400 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy M. C. & Hay R. T. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat Rev Mol Cell Biol 10, 564–568 (2009). [DOI] [PubMed] [Google Scholar]
- Wilkinson K. A. & Henley J. M. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J 428, 133–145 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T. et al. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA 103, 1988–1993 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Wu Y. & Xie Q. Ubiquitin-Proteasome System in ABA Signaling: From Perception to Action. Mol Plant 9, 21–33 (2016). [DOI] [PubMed] [Google Scholar]
- Urano D., Czarnecki O., Wang X., Jones A. M. & Chen J. G. Arabidopsis receptor of activated C kinase1 phosphorylation by WITH NO LYSINE8 KINASE. Plant Physiol 167, 507–516 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long L. et al. Recruitment of phosphatase PP2A by RACK1 adaptor protein deactivates transcription factor IRF3 and limits type I interferon signaling. Immunity 40, 515–529 (2014). [DOI] [PubMed] [Google Scholar]
- Cheng Z. et al. Pathogen-secreted proteases activate a novel plant immune pathway. Nature 521, 213–216 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Chen C., Breslin M. B., Song K. & Lan M. S. Extra-nuclear activity of INSM1 transcription factor enhances insulin receptor signaling pathway and Nkx6.1 expression through RACK1 interaction. Cell Signal 26, 740–747 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. V. et al. RACK1 competes with HSP90 for binding to HIF-1alpha and is required for O(2)-independent and HSP90 inhibitor-induced degradation of HIF-1alpha. Mol Cell 25, 207–217 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J. J. & Xie D. RACK1, a versatile hub in cancer. Oncogene 34, 1890–1898 (2015). [DOI] [PubMed] [Google Scholar]
- Nakashima A. et al. RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell 20, 2265–2279 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Suzuki K., Fujimura T. & Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140, 411–432 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licausi F., Ohme-Takagi M. & Perata P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol 199, 639–649 (2013). [DOI] [PubMed] [Google Scholar]
- Mizoi J., Shinozaki K. & Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819, 86–96 (2012). [DOI] [PubMed] [Google Scholar]
- Turner J. G., Ellis C. & Devoto A. The jasmonate signal pathway. Plant Cell 14 Suppl, S153–164 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y., Ohmiya K. & Hattori T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res 27, 470–478 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek B. A. AINTEGUMENTA utilizes a mode of DNA recognition distinct from that used by proteins containing a single AP2 domain. Nucleic Acids Res 31, 1859–1868 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J., Zhang Y. C., Sang Y., Li Q. H. & Yang H. Q. From The Cover: A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc Natl Acad Sci USA 102, 12270–12275 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F. H. et al. Tape-Arabidopsis Sandwich - a simpler Arabidopsis protoplast isolation method. Plant Methods 5, 16 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Toh-e A. & Kikuchi Y. Comparative analysis of yeast PIAS-type SUMO ligases in vivo and in vitro. J. Biochem 133, 415–422 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of the study are available within the article and its Supplementary Information files or are available from the corresponding author upon request.