Abstract
Background
Proteases represent one of the most abundant classes of enzymes in eukaryotes and are known to play key roles in many biological processes in plants. However, little is known about their functions in fruit ripening and disease resistance, which are unique to flowering plants and required for seed maturation and dispersal. Elucidating the genetic mechanisms of fruit ripening and disease resistance is an important goal given the biological and dietary significance of fruit.
Results
Through expression profile analyses of genes encoding tomato (Solanum lycopersicum) cysteine proteases, we identify a number of genes whose expression increases during fruit ripening. RNA interference (RNAi)-mediated repression of SlVPE3, a vacuolar protease gene, results in alterations in fruit pigmentation, lycopene biosynthesis, and ethylene production, suggesting that SlVPE3 is necessary for normal fruit ripening. Surprisingly, the SlVPE3 RNAi fruit are more susceptible to the necrotrophic pathogen Botrytis cinerea. Quantitative proteomic analysis identified 314 proteins that differentially accumulate upon SlVPE3 silencing, including proteins associated with fruit ripening and disease resistance. To identify the direct SlVPE3 targets and mechanisms contributing to fungal pathogen resistance, we perform a screening of SlVPE3-interacting proteins using co-immunoprecipitation coupled with mass spectrometry. We show that SlVPE3 is required for the cleavage of the serine protease inhibitor KTI4, which contributes to resistance against the fungal pathogen B. cinerea.
Conclusions
Our findings contribute to elucidating gene regulatory networks and mechanisms that control fruit ripening and disease resistance responses.
Electronic supplementary material
The online version of this article (doi:10.1186/s13059-017-1178-2) contains supplementary material, which is available to authorized users.
Keywords: Fruit ripening, Disease resistance, Protease, Vacuolar processing enzyme, RNA interference, Quantitative proteome, Protease inhibitor, Tomato
Background
Fruit are highly specialized plant organs that play a central role in seed maturation and dispersal in angiosperms. They are also valuable components of human diets, providing essential nutrients and a wide range of “bioactive” compounds that are important for human health [1]. The ripening of fleshy fruit is a genetically programmed process that is associated with changes in color, texture, flavor, and susceptibility to pathogen infection [2, 3]. Substantial insights have been made into the mechanistic basis of ripening, including its regulation by transcription factors and the gaseous hormone ethylene, which plays a central role in the ripening of climacteric fruit [4, 5]. The ethylene biosynthetic and signal transduction pathway have been well characterized [6–8]. Recently, significant progress has been made in understanding the transcriptional control of fruit ripening using tomato (Solanum lycopersicum) as a model system. The transcription factors RIPENING-INHIBITOR (RIN) [9], NONRIPENING (NOR) [10], and COLORLESS NONRIPENING (CNR) [11] function as global regulators of ripening and act upstream of ethylene, while additional transcription factors that are required for normal ripening include TOMATO AGAMOUS-LIKE1 (TAGL1) [12, 13], HD-ZIP HOMEOBOX PROTEIN-1 (HB-1) [14], APETALA2a (AP2a) [15, 16], ETHYLENE RESPONSE FACTOR6 (ERF6) [17], ARABIDOPSIS PSEUDO RESPONSE REGULATOR2-LIKE (APRR2-Like) [18], and FRUITFULL (TDR4/FUL1 and MBP7/FUL2) [19]. In addition to transcriptional regulation, gene expression in fruit ripening can also be modulated by epigenetic or translational control [20–22]. However, unlike the relatively well studied and defined changes in the fruit transcriptome during ripening, little is known about its post-transcriptional regulation.
One mode of post-translational control involves proteases, enzymes that are widely distributed in all living organisms and that catalyze the hydrolysis of peptide bonds during or after translation [23]. Proteases specifically cleave protein substrates either from the N- or C- terminus (aminopeptidases and carboxypeptidases, respectively) or within the molecule (endopeptidases) [24]. Based on the active site residues or ions that carry out catalysis, proteases are generally grouped into five classes: aspartic, cysteine, metallo-, serine, and threonine proteases [25]. Although initially considered to be purely degradative enzymes involved in intracellular protein turnover, it is now clear that they participate in the regulation of many critical physiological and cellular processes [23, 25]. The information housed in the MEROPS peptidase database [26] suggests that a plant genome sequence encodes hundreds of proteases belonging to dozens of unrelated families. These enzymes have been associated with a wide variety of biological processes, including programmed cell death (PCD), meiosis, seed coat formation, cuticle deposition, stomata development, flowering, pollen or embryo development, and chloroplast biogenesis [27]. However, the molecular mechanisms of plant proteases in these processes, such as their substrates, remain elusive. Moreover, the functional roles of proteases in fruit ripening have not been well defined. Our previous study has shown that specific E2 ubiquitin-conjugating enzymes, the components of an ubiquitin-proteasome system, contribute to the ripening of tomato fruit [28]. From this we infer that proteolysis is involved in the regulation of fruit ripening, but how proteases participate in this process and the nature of the underlying mechanisms are not known.
In the study presented here, we identified a number of cysteine protease genes whose expression increases during tomato fruit ripening. We then assessed the physiological importance for fruit ripening of one of these genes, SlVPE3, which encodes a vacuolar processing enzyme (VPE). VPEs are canonical cysteine proteases, harboring active cysteines in the catalytic site, and are responsible for the maturation or activation of specific vacuolar proteins in plants [29]. Here, we show that suppression of SlVPE3 by RNA interference (RNAi) in tomato delays ripening-related traits, including lycopene accumulation and ethylene synthesis. Interestingly, the tomato fruit with reduced SlVPE3 expression exhibited increased susceptibility to the necrotrophic pathogen Botrytis cinerea, despite showing a reduced ripening phenotype. This was unexpected since unripe green fruit are typically less susceptible than ripe fruit to pathogen infections [3, 30]. To investigate how SlVPE3 affects these processes, we performed a comparative proteomic analysis using isobaric tags for relative and absolute quantification (iTRAQ). This suggested a large number of proteins as potential SlVPE3 targets, including some involved in fruit ripening and disease responses. We also carried out a screen for SlVPE3-interacting proteins to identify the direct targets of SlVPE3. We provide evidence that a Kunitz trypsin inhibitor 4, a serine protease inhibitor, functions downstream of SlVPE3 to regulate disease resistance in tomato fruit.
Results
Expression of the protease SlVPE3 increases steadily during fruit ripening
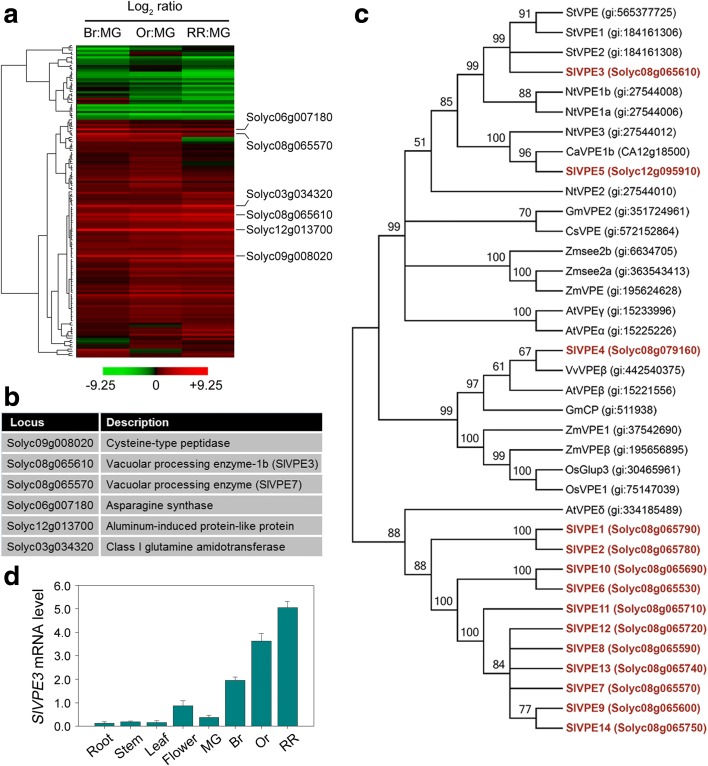
The tomato genome encodes more than 900 predicted proteases of diverse catalytic classes, based on the MEROPS protease database [26], but in this study we focused on cysteine proteases, a class that has been shown to take part in a variety of biological processes [27]. A total of 167 non-redundant cysteine proteases, belonging to 19 families, were identified from the tomato genome using the MEROPS database (Additional file 1: Table S1). Expression analysis of the corresponding genes using quantitative reverse transcriptase PCR (RT-PCR) (Additional file 2: Table S2) and hierarchical clustering analysis [31] revealed several genes whose transcript levels increased during fruit ripening (Fig. 1a). Those whose expression increased more than tenfold are shown in Fig. 1a, b. Of these, two encoded VPEs, a class of proteins that were originally identified as cysteine proteases responsible for the maturation of seed storage proteins [32]. They were later reported to be the plant functional orthologs of animal caspases, which are essential for the initiation and execution of PCD [29, 33, 34]. In addition, the transcript levels of a VPE gene from Citrus sinensis have been observed to increase during fruit ripening [35], which when taken together with our results suggests that VPE proteins might contribute to ripening in a range of species.
Fig. 1.
Expression analyses of tomato cysteine proteases reveal the involvement of SlVPE3 in fruit ripening. a Expression profiles of tomato cysteine protease genes during fruit ripening, as determined by quantitative RT-PCR. The ACTIN gene was used as the internal control. The stages of fruit development analyzed were mature green (MG), breaker (Br), orange (Or), and red ripe (RR). Expression ratios were calculated using the earliest stage (MG) as the denominator and plotted in a heat map on a log2 scale. Each row in the color heat map represents a single cysteine protease gene. The green and red colors indicate down- and up-regulation, respectively, at an indicated ripening stage relative to the MG stage. Black indicates no significant expression change. Data from biologically repeated samples are averaged and the detailed information is listed in Additional file 2: Table S2. The genes whose mRNA levels increased more than tenfold are shown. b Gene identifiers (Solyc numbers) and functional annotations of the cysteine protease genes whose mRNA levels increased more than tenfold during tomato fruit ripening as revealed by quantitative RT-PCR. c Phylogenetic analysis of plant vacuolar proteases. The phylogenetic tree was produced using MEGA version 5.2. Bootstrap values from 1000 replications for each branch are shown. Tomato proteins are indicated in red. Species names are abbreviated as follows: St, Solanum tuberosum; Sl, S. lycopersicum; Nt, Nicotiana tabacum; Ca, Capsicum annuum; Gm, Glycine max; Cs, Citrus sinensis; Zm, Zea mays; At, Arabidopsis thaliana; Vv, Vitis vinifera; Os, Oryza sativa. The accession numbers are indicated in parentheses. d Gene expression of SlVPE3 in vegetative and reproductive tomato organs as determined by quantitative RT-PCR. The ACTIN gene was used as an internal control. Values are means ± standard deviation of three independent experiments
According to the MEROPS protease database, the tomato genome has 14 VPE genes, five of which have previously been identified and named SlVPE1 to SlVPE5 [36]. We named the other nine VPE genes SlVPE6 to SlVPE14 on the basis of their chromosomal location (Additional file 3: Table S3). All these VPE proteins are predicted to contain two conserved cysteine residues in the active sites (Additional file 4: Figure S1). Phylogenetic analysis revealed that tomato VPE proteins can be divided into several subgroups, with >50% bootstrap support (Fig. 1c), and high sequence similarity among the proteins was observed (Additional file 5: Table S4), suggesting gene duplications. We selected SlVPE3 for functional analysis because its expression was not only higher in fruit than in other organs, such as root, stem, and leaf, but also increased gradually during fruit ripening (Fig. 1d). SlVPE3 has been shown to be involved in controlling sugar accumulation [36], but its function in fruit ripening and the underlying molecular mechanisms are unclear.
SlVPE3 is required for tomato fruit ripening
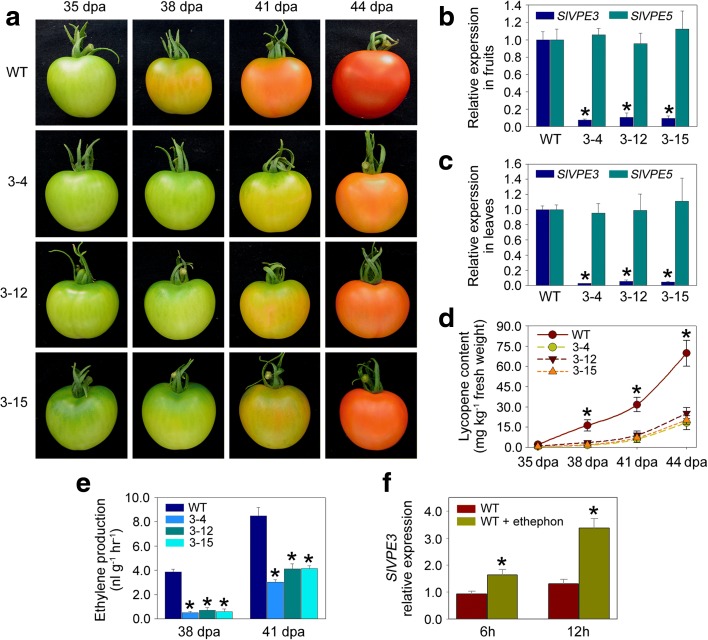
To gain insight into the function of SlVPE3, we generated a SlVPE3 RNAi construct under the control of a 35S cauliflower mosaic virus promoter and transformed it into the wild-type tomato cultivar Ailsa Craig. Three independent transgenic lines (3-4, 3-12, and 3-15) with confirmed transgene integration showed distinct and similar ripening-related phenotypes (Fig. 2a). The differences in fruit ripening between the SlVPE3 RNAi lines and wild-type became apparent at 38 days post-anthesis (dpa). A visible color change could be observed at this stage in the wild-type fruit, whereas SlVPE3 RNAi tomatoes were still green. At 41 dpa, the wild-type fruit had a homogenous orange color, while fruit from the SlVPE3 RNAi lines were only just starting to change color. To verify the specific repression of SlVPE3 in the RNAi lines, total RNA was extracted from fruit and leaves of wild-type and transgenic lines and submitted to quantitative RT-PCR analysis. The transcript levels of SlVPE3 were shown to be strongly reduced in both organs of transgenic lines compared with the wild-type (Fig. 2b, c). Sixteen potential off-targets for the RNAi construct were identified (Additional file 4: Figure S2) using the computational tool pssRNAit [37]. However, quantitative RT-PCR analysis indicated that none of these showed reduced expression in leaves of the SlVPE3 RNAi lines (Additional file 4: Figure S2). This result indicated that the RNAi construct was specific for the target gene. We also measured the expression of SlVPE5 because it is the most closely related tomato gene to SlVPE3 (Fig. 1c). The levels of SlVPE5 mRNA in all three RNAi lines (3-4, 3-12, and 3-15) were not significantly different from wild type (Fig. 2b, c), demonstrating the specificity of the SlVPE3 RNAi construct for the target gene. The three lines (3-4, 3-12, and 3-15) were selected for further analysis.
Fig. 2.
SlVPE3 is necessary for normal tomato fruit ripening. a Ripening phenotype of SlVPE3 RNAi lines. Fruit at 35, 38, 41, and 44 days post-anthesis (dpa) from wild-type (WT) and SlVPE3 RNAi lines (3-4, 3-12, and 3-15) are shown. b Expression of SlVPE3 and SlVPE5 in fruit of WT and SlVPE3 RNAi lines as determined by quantitative RT-PCR. c Expression of SlVPE3 and SlVPE5 in leaves of WT and SlVPE3 RNAi lines. In b and c, the gene transcript levels were normalized against the ACTIN gene, followed by normalization against WT expression. Values are shown as the means ± standard deviation (SD). Asterisks indicate P value <0.05 (t-test) when comparing values for each measurement between the SlVPE3 RNAi lines and WT plants. d Lycopene accumulation in WT and SlVPE3 RNAi fruit during ripening. e Ethylene generation in WT and SlVPE3 RNAi fruit at 38 and 41 dpa. In d and e, values are shown as the means ± SD. Asterisks indicate significant differences (P < 0.05; t-test) between WT and SlVPE3 RNAi fruit at an indicated ripening stage. f Expression of SlVPE3 in WT fruit (35 dpa) 6 or 12 h after untreated or treated with ethephon, as determined by quantitative RT-PCR. Gene transcript levels were normalized against expression of the ACTIN gene, followed by normalization against expression in the WT without ethephon treatment. Values are shown as the means ± SD. Asterisks indicate significant differences (P < 0.05; t-test) at an indicated time point after treatment
The color change of ripening tomatoes from green to red is largely due to the degradation of chlorophyll and the accumulation of lycopene, which accounts for 70–90% of the total carotenoids in most tomato varieties [38, 39]. To establish the underlying causes of the color differences observed between wild-type and SlVPE3 RNAi ripe fruit, we measured the levels of lycopene. As shown in Fig. 2d, levels of lycopene in fruit from the SlVPE3 RNAi lines were <10% of wild-type levels at 38 dpa, suggesting that SlVPE3 expression affects lycopene accumulation during fruit ripening.
As with all climacteric fruit, those of tomato require an increase in ethylene biosynthesis to ripen [4]. We investigated whether the delay in fruit ripening in the SlVPE3 RNAi lines correlated with ethylene production. It was observed that fruit of the repressed lines (3-4, 3-12, and 3-15) produced less ethylene than wild-type fruit at the same ripening stages (Fig. 2e). Ethylene regulates the expression of many genes associated with fruit ripening [40] and we observed that the expression of SlVPE3 increased during tomato fruit ripening (Fig. 1d). To determine whether the increased expression of SlVPE3 was ethylene-dependent, wild-type fruit at 35 dpa were treated with the ethylene precursor ethephon, which resulted in a substantial increase in expression (Fig. 2f). Taken together, these results suggest that ethylene induces the expression of SlVPE3, which in turn regulates ethylene synthesis in tomato fruit by a positive feedback loop.
Disease resistance is impaired in SlVPE3 silenced fruit
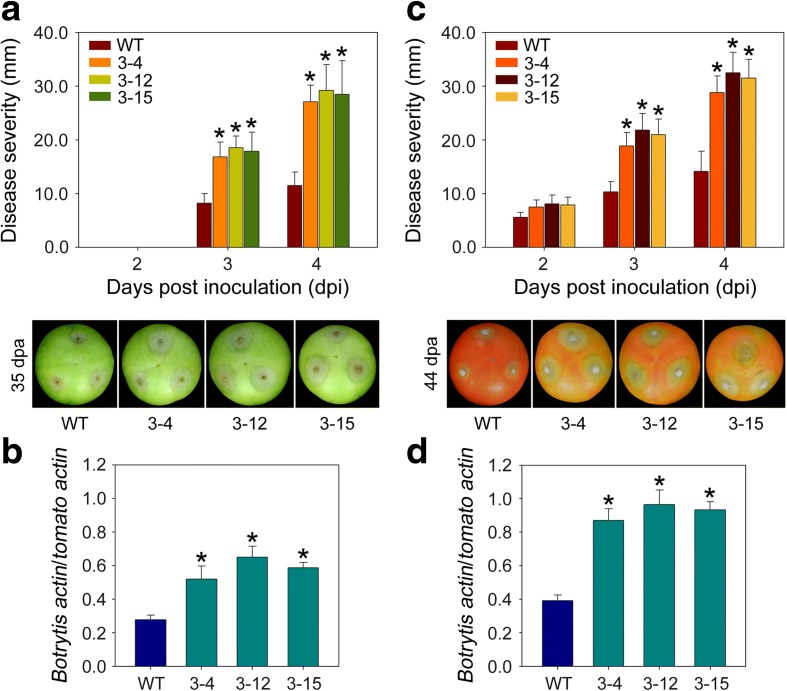
Fruit ripening involves the regulation of numerous biochemical pathways associated with pigmentation, cell wall metabolism, and the production of aromatic and nutritionally important compounds [13]. Additionally, ripening is characterized by a major increase in susceptibility to necrotrophic pathogens [3, 30]. Since repression of SlVPE3 resulted in a delay in fruit ripening, we investigated whether fruit from the SlVPE3 RNAi lines were also less susceptible to pathogen infection. Fruit at 35 and 44 dpa were inoculated with B. cinerea, a commercially important tomato postharvest pathogen that causes gray mold disease [30]. As shown in Fig. 3a, soft rot symptoms occurred at the third day post-inoculation in the fruit of both wild-type and the SlVPE3 RNAi lines (3-4, 3-12, and 3-15) at 35 dpa. An approximately twofold increase in lesion development was observed in SlVPE3 RNAi fruit compared with the wild type. Fungal growth, assessed based on the quantitative PCR amplification of B. cinearea ACTIN 2 (BC1G_08198), was significantly greater in the SlVPE3 RNAi fruit (Fig. 3b). In contrast, tissue rotting was evident on the second day post-inoculation in wild-type and SlVPE3 RNAi fruit at 44 dpa. Similarly, a significant increase in disease severity and fungal growth was observed in the SlVPE3 RNAi fruit (Fig. 3c, d). These data indicated that silencing SlVPE3 expression makes the fruit more susceptible to pathogen infection, even though fruit ripening was delayed. Since this finding was counter-intuitive and contradictory to previous observations that unripe fruit are usually less susceptible than ripe fruit to infection [3, 30], we hypothesized that SlVPE3 may target downstream proteins associated with pathogen resistance in tomato fruit.
Fig. 3.
Suppressing SlVPE3 enhances susceptibility of tomato fruit to B. cinerea. a Disease severity for fruit inoculated with B. cinerea at 35 days post-anthesis (dpa). Representative fruit at 3 days post-inoculation (dpi) for wild-type (WT) and SlVPE3 RNAi lines (3-4, 3-12, and 3-15) are shown. b Fungal growth during infection of tomato fruit at 35 dpa. Growth of B. cinerea at 3 dpi in infected WT and SlVPE3 RNAi fruit was measured based on expression levels of B. cinerea ACTIN 2 relative to tomato ACTIN gene. c Disease severity in fruit inoculated with B. cinerea at 44 dpa. Representative fruit at 3 dpi for WT and SlVPE3 RNAi lines (3-4, 3-12, and 3-15) are shown. d Fungal growth during infection of tomato fruit at 44 dpa. Growth of B. cinerea at 3 dpi in infected WT and SlVPE3 RNAi fruit was measured based on quantitative PCR amplification of B. cinerea ACTIN 2 relative to tomato ACTIN gene. Values are shown as the means ± standard deviation. Asterisks indicate significant differences (P < 0.05; t-test) between WT and SlVPE3 RNAi fruit at a given time point after inoculation
Quantitative proteome analysis identifies proteins that differentially accumulate upon SlVPE3 silencing
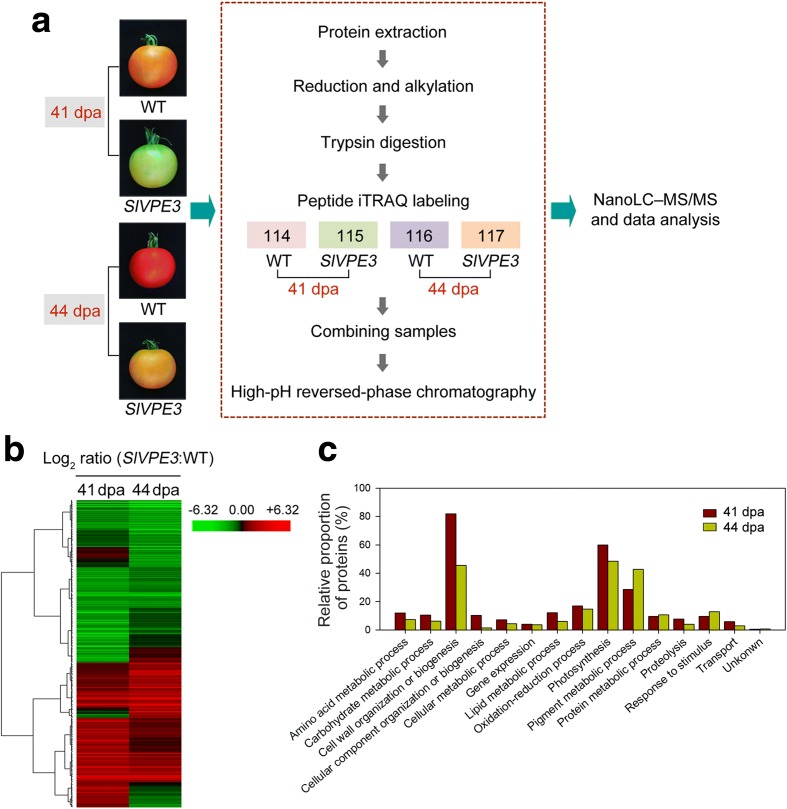
Protease target identification is essential to understand the molecular mechanism by which it regulates biological processes. An iTRAQ-based quantitative proteomic approach was employed to identify the proteins that differentially accumulate upon SlVPE3 silencing. The general design of the experiment and experimental workflow is shown in Fig. 4a. Proteins were isolated from SlVPE3 RNAi fruit (T1 generation) and wild-type fruit at 41 and 44 dpa and labeled with 4-plex iTRAQ reagents. The iTRAQ experiment was repeated with an independent biological replicate. For each replicate, pericarp tissue of fruit from five plants was pooled to account for variation among individuals. Using the S. lycopersicum protein database ITAG2.4_proteins_full_desc.fasta, a total of 3802 and 3840 proteins were identified in biological replicates 1 and 2, respectively, with a global false discovery rate (FDR) of <1% in both. A twofold cut-off was then used as a determinant for whether the changes in protein abundance were significant. The levels of 314 proteins were significantly different in the SlVPE3 RNAi fruit compared with those of the wild type at one ripening stage or both. Additional file 6: Table S5 lists these proteins, along with the associated identification information and the ratio of iTRAQ reporter ion intensities. To identify the proteins with similar expression profiles, hierarchical clustering [31] was applied and the 314 proteins were divided into two main clusters, representing those that had higher or lower abundance in the SlVPE3 RNAi fruit than in wild type (Fig. 4b). The Blast2go [41] web-based bioinformatic tool, which categorizes proteins according to their Gene Ontology annotations, classified the proteins into 15 functional categories (Fig. 4c). The functional classes “cell wall organization or biogenesis”, “photosynthesis”, and “pigment metabolic process” contained the largest number of proteins at both ripening stages (41 and 44 dpa).
Fig. 4.
Quantitative proteome analysis reveals the potential targets of SlVPE3. a Workflow of the quantitative proteome analysis. Proteins were isolated from wild-type (WT) and SlVPE3 RNAi fruit at 41 or 44 days post-anthesis (dpa), and subjected to isobaric tags for relative and absolute quantification (iTRAQ) labeling coupled with nanoLC–MS/MS. b A total of 314 proteins showing differential expression in the SlVPE3 RNAi fruit compared to the WT were identified. The expression patterns of the proteins were hierarchically clustered based on the expression ratio as a log2 scale. Each row in the color heat map represents a single protein. The green and red colors indicate down- and up-regulation, respectively, in the SlVPE3 RNAi fruit relative to the WT. Black represents no significant expression change. Values from biological replicates were averaged and the detailed information associated with the identified proteins is listed in Additional file 6: Table S5. c Functional categories of the proteins that changed abundance in the SlVPE3 RNAi fruit at 41 or 44 dpa compared with WT
SlVPE3 modulates the accumulation of proteins associated with fruit ripening and disease resistance
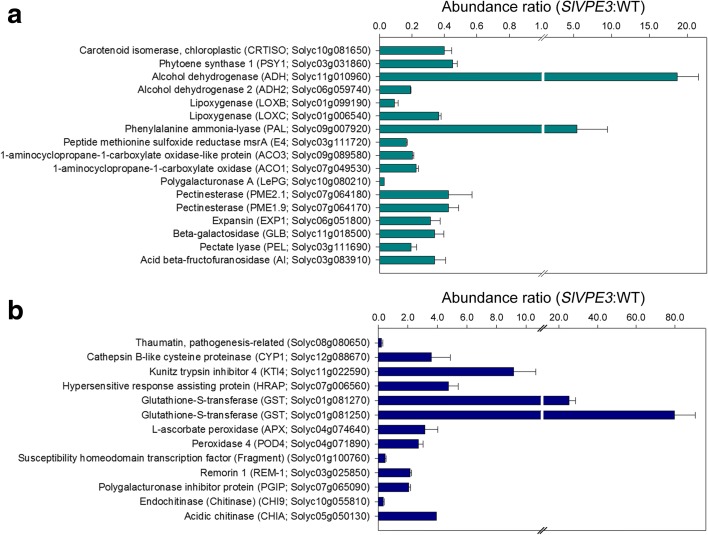
The proteins associated with fruit ripening that showed significant differences in abundance in the SlVPE3 RNAi fruit compared with wild type included some involved in cell wall degradation, such as polygalacturonase A (LePG, Solyc10g080210); ethylene synthesis, such as 1-aminocyclopropane-1-carboxylate oxidase (ACO1, Solyc07g049530); aroma formation, such as lipoxygenase (LoxC, Solyc01g006540); and carotenoid biosynthesis, such as phytoene synthase 1 (PSY1, Solyc03g031860) (Fig. 5a). The levels of most of these proteins were lower in the SlVPE3 RNAi fruit, consistent with the delayed ripening phenotype. We also identified a set of proteins associated with disease responses, such as endochitinase (CHI9, Solyc10g055810), polygalacturonase inhibitor protein (PGIP, Solyc07g065090), and thaumatin (Solyc08g080650) (Fig. 5b), although among proteins were examples with either decreased or increased abundance in the SlVPE3 RNAi fruit. Strikingly, the list of differentially abundant proteins included acid beta-fructofuranosidase (AI, Solyc03g083910), the tomato ortholog of AtFruct4, which is a downstream target of Arabidopsis thaliana VPEγ [29]. This highlighted the value of quantitative proteome analysis for identifying putative protease targets. It should be noted that, besides SlVPE3 targets, we also identified other proteins that differentially accumulated due to the delayed ripening.
Fig. 5.
SlVPE3 RNAi fruit showed altered levels of proteins associated with fruit ripening and disease resistance. Proteins isolated from wild-type (WT) and SlVPE3 RNAi fruit were subjected to isobaric tags for relative and absolute quantification (iTRAQ) labeling coupled with nanoLC–MS/MS. The abundance ratio of proteins in the SlVPE3 RNAi fruit versus WT at 41 days post-anthesis (dpa) is shown. Values represent means of two biological replicates, and error bars represent standard deviation. a Identification of proteins associated with fruit ripening in the iTRAQ analysis. b Identification of proteins associated with disease resistance in the iTRAQ analysis. The gene identifiers (Solyc numbers) and the functional annotations are indicated. Detailed protein information is listed in Additional file 6: Table S5
SlVPE3 affects gene transcript levels indirectly
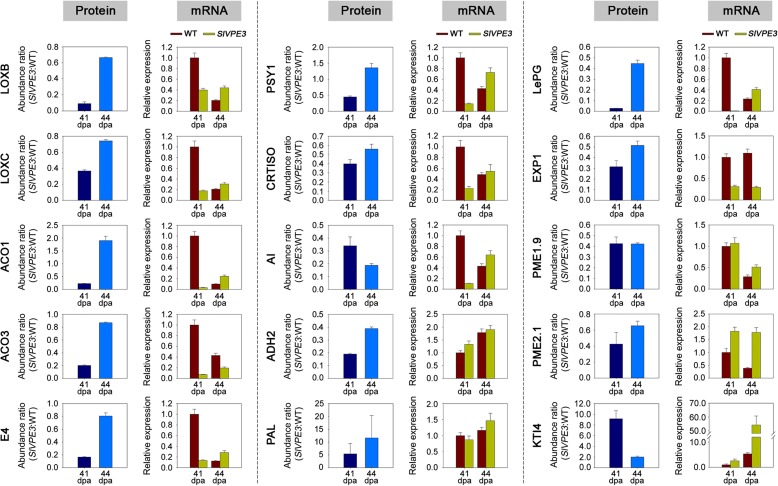
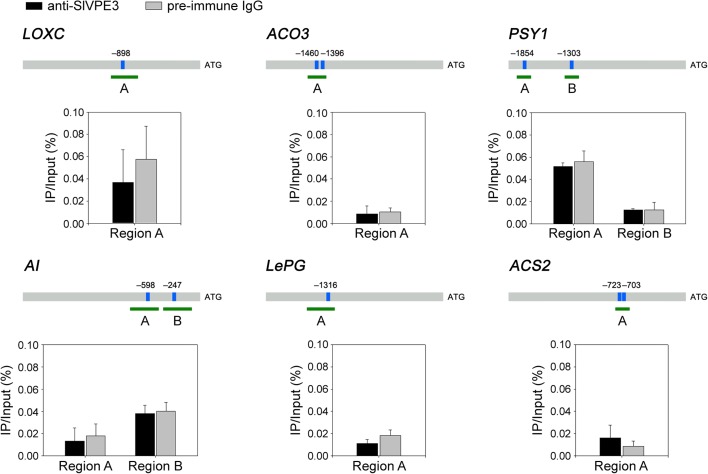
A previous study demonstrated that SlVPE5 (formerly named LeCp), a paralog of SlVPE3, can act as a transcription factor [42]. LeCp was reported to specifically bind to the promoter of ACC SYNTHASE 2 (ACS2), a key gene in ethylene biosynthesis, and this binding correlated induction of ACS2 in tomato leaves by the fungal elicitor Ethylene-inducing xylanase (EIX) [42]. To determine whether SlVPE3 acts as a transcription factor, we first investigated whether the protein expression patterns observed in the quantitative proteomic analysis correlated with cognate transcript levels. Of the 15 genes selected for quantitative RT-PCR analysis, the expression of 11, including LoxC, ACO1, E4, PSY1, CRTISO, and LePG, was consistent with the variations in protein abundance (Fig. 6). Next, we performed a chromatin immunoprecipitation (ChIP) assay to investigate whether SlVPE3 could bind directly to the promoters of these genes. A search for the putative DNA binding site in the 2000-bp upstream regions starting from the translational start sites revealed that only five of the genes (LoxC, ACO3, PSY1, AI, and LePG) contained the putative TAAAATAT binding motif [42] (Additional file 7: Table S6). For the ChIP assay, the cross-linked DNA–protein complexes were immunoprecipitated using an affinity-purified polyclonal antibody raised against SlVPE3. Specific primers (Additional file 8: Table S7) were designed for the five genes (LoxC, ACO3, PSY1, AI, and LePG) to amplify promoter sequences surrounding the putative binding sites from the immunoprecipitated DNA. We were not able to detect any specific enrichment for the promoters of these genes (Fig. 7). We also did not observe an enrichment of the ACS2 promoter (Fig. 7), although it has been shown to be bound by LeCp [42]. These data suggest that SlVPE3 affects gene transcript levels indirectly and so SlVPE3 may function at the post-transcriptional level, rather than directly regulating gene expression as a transcription factor.
Fig. 6.
Comparison of expression profiles at the protein and mRNA levels. The protein abundance in wild-type (WT) and SlVPE3 RNAi fruit at 41 and 44 days post-anthesis (dpa) was assessed by isobaric tags for relative and absolute quantification (iTRAQ)-based quantitative proteome analysis. Transcript abundance was evaluated by quantitative RT-PCR. The gene transcript levels are normalized against the ACTIN gene, followed by normalization against the expression in WT. Values are shown as the means ± standard deviation
Fig. 7.
SlVPE3 does not bind to the promoters of genes with abnormal expression in SlVPE3 silenced fruit. For the ChIP assay, genomic DNA and proteins from pericarp of tomato fruit at 41 dpa were cross-linked, and the chromatin complexes were co-immunoprecipitated with anti-SlVPE3 antibodies. The promoter regions of the analyzed genes are indicated. Blue boxes represent the binding motifs and numbers indicate the position of these motifs relative to the translational start site. The green fragments with upper-case letters represent the regions used for ChIP-quantitative PCR. Values represent the percentage of DNA fragments that were co-immunoprecipitated with anti-SlVPE3 antibodies or preimmune serum (rabbit IgG) relative to the input DNA. Error bars represent the standard deviation of three independent experiments
Screening for SlVPE3-interacting proteins
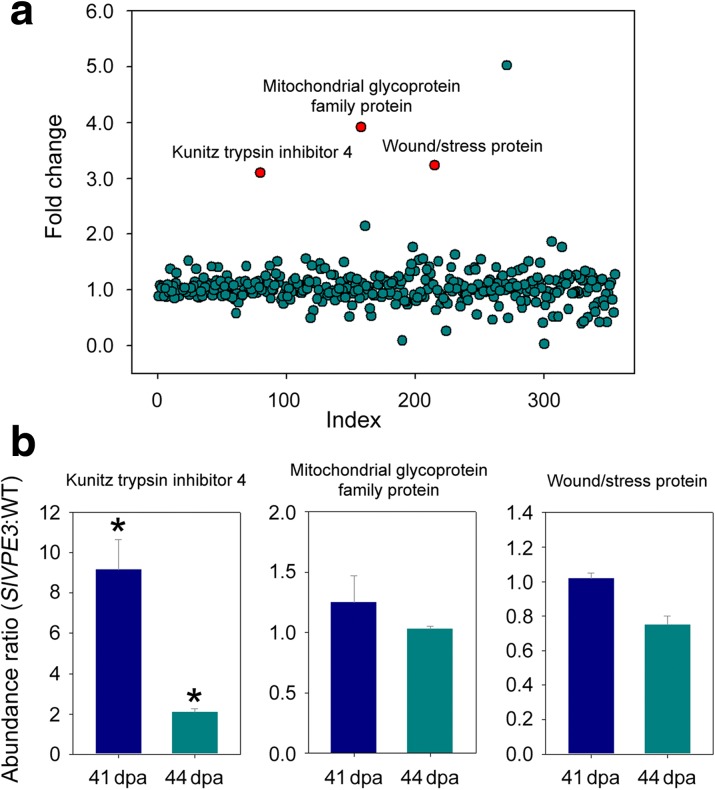
To identify the direct targets of SlVPE3, we screened for SlVPE3-interacting proteins by analyzing proteins that were immunoprecipitated with affinity-purified anti-SlVPE3 polyclonal antibody or pre-immune serum IgG (non-specific antibody; negative control) from 41-dpa wild-type fruit. The immunoprecipitated proteins were submitted to Sequential Window Acquisition of all Theoretical Mass Spectra (SWATH-MS) quantitative proteomic analysis [43], which measures quantitative changes in protein interactions [44]. This resulted in the identification of 355 proteins, of which 57 were also identified by the iTRAQ proteome analysis as showing significantly different abundance in SlVPE3 RNAi fruit compared with wild type (Additional file 9: Table S8). Most of the identified proteins had fold changes ~1 (Fig. 8a; Additional file 9: Table S8), indicating proteins that were immunoprecipitated regardless of whether anti-SlVPE3 or IgG was used, and so these proteins were considered to be nonspecific contaminants. Proteins with significantly increased abundance (P < 0.01) by using anti-SlVPE3 compared with pre-immune serum IgG included a mitochondrial glycoprotein family protein (Solyc03g079930), a wound/stress protein (Solyc03g093360), and a Kunitz trypsin inhibitor 4 (Solyc11g022590) (Fig. 8a), and these were designated as SlVPE3-interacting proteins. The Kunitz trypsin inhibitor 4 (KTI4) exhibited a significant difference in protein expression levels in SlVPE3 RNAi fruit compared with wild type in the iTRAQ analysis (Fig. 8b), further suggesting that KTI4 might serve as a substrate of SlVPE3. Notably, SlVPE3 itself was not identified in the SWATH-MS analysis, which we hypothesized was due to the experimental conditions used for the elution of the immunoprecipitated proteins. Indeed, when we changed the conditions, SlVPE3 was identified as an immunoprecipitated protein (Additional file 4: Figure S3).
Fig. 8.
Identification of endogenous SlVPE3-interacting proteins. a Proteins isolated from tomato fruit at 41 days post-anthesis (dpa) were immunoprecipitated with anti-SlVPE3 antibodies or pre-immune serum IgG (negative control). The enriched proteins were eluted and submitted to Sequential Window Acquisition of all Theoretical Mass Spectra (SWATH-MS) analysis. Each circle represents a single protein. Most identified proteins showed no difference in abundance, indicating nonspecifically bound proteins. Three proteins (red circle) were identified as the SlVPE3-interacting proteins with significant increases in abundance (P < 0.01; t-test) by using anti-SlVPE3 antibodies compared with pre-immune serum IgG. Additional information related to protein identification is listed in Additional file 9: Table S8. b The changes in protein abundance in the SlVPE3 RNAi fruit revealed by isobaric tags for relative and absolute quantification (iTRAQ) analysis. The abundance ratios of selected proteins in the SlVPE3 RNAi fruit versus wild-type (WT) at 41 and 44 dpa are shown. Values represent means of two biological replicates, and error bars represent standard deviation. Asterisks indicate significant differences at a twofold cut-off between WT and SlVPE3 RNAi fruit
We also performed a yeast two-hybrid (Y2H) screen to identify proteins that interact with SlVPE3 using the Matchmaker Library Construction and Screening Kits (Clontech). SlVPE3 was used as bait against a tomato cDNA library constructed from wild-type fruit at the breaker stage (38 dpa). A total of 179 positive colonies were analyzed, leading to the identification of 49 proteins (Additional file 10: Table S9) involved in various cellular processes. Notably, several proteins associated with proteolysis were identified, including KTI4, which was then targeted for further analysis.
SlVPE3 protein interacts directly with KTI4 in vacuoles
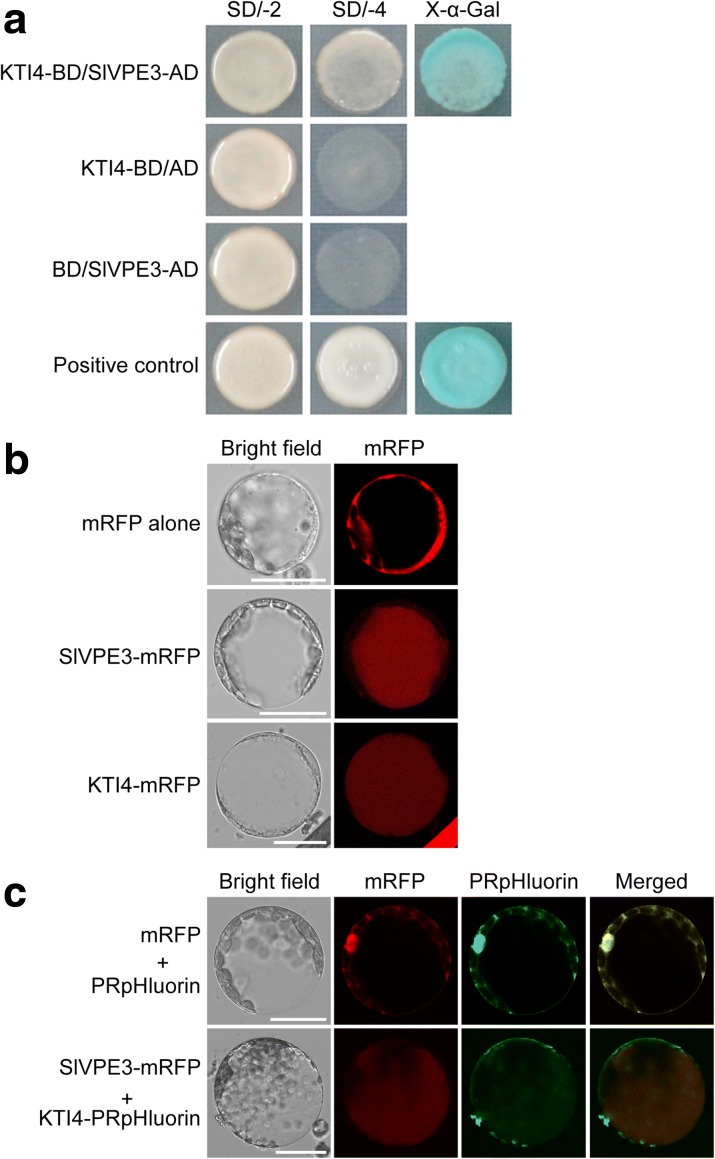
KTI4 belongs to the KTI family, members of which include serine protease inhibitors responsible for defense responses in leaves [45, 46]. However, little is known about the regulation of KTI genes and their functions in fruit disease resistance. To confirm the putative interaction between KTI4 and SlVPE3, we carried out a Y2H analysis. The open reading frame (ORF) of KTI4 and the cDNA fragment encoding the mature protein of SlVPE3 were cloned into the pGBKT7 (binding domain, BD) and pGADT7 (activation domain, AD) vectors, respectively. The resulting plasmids KTI4-BD and SlVPE3-AD were co-transformed into yeast, while yeast lines co-transformed with KTI4-BD and empty AD (KTI4-BD/AD) or SlVPE3-AD and empty BD (BD/SlVPE3-AD) were used as negative controls. KTI4-BD and SlVPE3-AD co-transformed yeast lines displayed normal growth and blue coloration when grown on the selective SD/-Leu/-Trp/-His/-Ade (SD/-4) medium containing X-α-Gal, whereas the control lines did not grow (Fig. 9a), further indicating that KTI4 interacts with SlVPE3.
Fig. 9.
SlVPE3 interacts with KTI4 in vacuoles. a Interactions between KTI4 and SlVPE3 in a Y2H analysis. The ORF of KTI4 and the cDNA fragment encoding the mature protein of SlVPE3 were cloned into the pGBKT7 (BD) and pGADT7 (AD) vectors, respectively, resulting in the KTI4-BD and SlVPE3-AD plasmids, which were co-transformed into yeast. As negative controls, KTI4-BD and AD or SlVPE3-AD and BD were co-transformed into yeast. The transformants were streaked on SD/-Leu/-Trp medium (SD/-2). Protein–protein interactions were assessed by examining growth on SD/-Leu/-Trp/-His/-Ade medium (SD/-4) and further confirmed by monitoring β-galactosidase activity (blue coloration). b Subcellular localization of SlVPE3 and KTI4 visualized by monomeric red fluorescent protein (mRFP) analysis. The constructs used for transformation are indicated (left): mRFP alone, control showing the signals throughout the cell except in the vacuolar lumen; SlVPE3-mRFP, signals from the SlVPE3-mRFP fusion protein; KTI4-mRFP, signals from the KTI4-mRFP fusion protein. Protoplasts of tobacco (Nicotiana benthamiana) leaves transiently expressing the mRFP-alone control, SlVPE3-mRFP, or KTI4-mRFP were isolated and observed under a Leica confocal microscope (Leica DMI600CS). c Subcellular colocalization of SlVPE3 and KTI4 determined using N. benthamiana leaf protoplasts co-expressing SlVPE3-mRFP and KTI4-PRpHluorin. The constructs used for transformation are indicated (left): mRFP + PRpHluorin, control showing the signals throughout the cell, except in the vacuolar lumen; SlVPE3-mRFP + KTI4-PRpHluorin, signals from SlVPE3-mRFP and KTI4-PRpHluorin fusion proteins. Colocalization is shown by merging mRFP and PRpHluorin images (Merged). Scale bars, 25 μm
To examine the intracellular localization of SlVPE3 and KTI4, their ORFs were separately introduced into a vector to generate a translational fusion with monomeric red fluorescent protein (mRFP) at the C-terminus. The constructs were separately introduced into Agrobacterium tumefaciens, which in turn was used to transform tobacco (Nicotiana benthamiana) leaves, from which mesophyll protoplasts were isolated. Protoplasts expressing mRFP alone served as a control. Confocal laser scanning microscopy showed that mRFP-tagged SlVPE3 (SlVPE3-mRFP) and KTI4 (KTI4-mRFP) both produced a strong signal in the vacuole, while the mRFP-only control displayed a fluorescent signal throughout the cell, but not from the vacuolar lumen (Fig. 9b). Notably, we were unable to detect the intracellular localization of SlVPE3 and KTI4 fused to green fluorescent protein (GFP) or mCherry, which might be due to degradation and/or protonation of GFP or mCherry under the acidic conditions that are found in vacuoles of higher plants [47].
To confirm the colocalization of SlVPE3 and KTI4, their ORFs were fused to mRFP and PRpHluorin (plant-solubility-modified ratiometric pH-sensitive mutants of green fluorescent protein), respectively, and the fusion proteins were co-expressed in tobacco leaves. PRpHluorin, derived from GFP, is a fluorescent pH sensor that has been successfully used to characterize the localization of vacuolar proteins [48]. We observed that the fluorescent signals of mRFP co-localized with those of PRpHluorin, suggesting the subcelluar colocalization of SlVPE3 and KTI4 (Fig. 9c).
SlVPE3 participates in the cleavage of KTI4
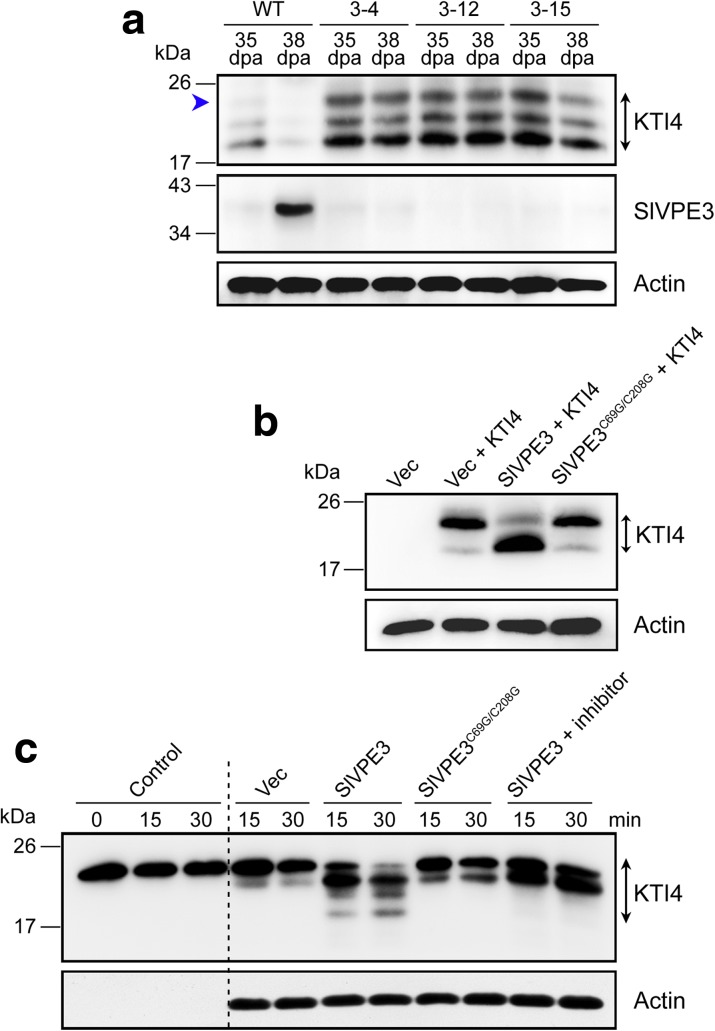
Having demonstrated that SlVPE3 protein interacts and colocalizes with KTI4, we then raised polyclonal antibodies against KTI4 to test whether SlVPE3 participates in the processing of KTI4. Polyclonal antibodies were affinity-purified and immunoblot analysis indicated no immunoreactive bands in extracts from wild-type tomato fruit at 35 dpa when pre-immune serum was used (Additional file 4: Figure S4), but a signal was detected by the affinity purified anti-KTI4 antibodies corresponding to the size (25 kDa) of the predicted full-length KTI4 polypeptide (Fig. 10a, blue arrowhead). The affinity purified anti-KTI4 antibodies recognized two additional bands with lower molecular mass (Fig. 10a), suggesting that KTI4 undergoes cleavage to form smaller peptides. We observed that KTI4 protein levels were higher in fruit of the SlVPE3-silenced lines (3-4, 3-12, and 3-15) and in wild type when SlVPE3 levels were low, at 35 dpa (Fig. 10a). Conversely, the accumulation of KTI4 was reduced when SlVPE3 levels were high (Fig. 10a). These results are consistent with SlVPE3 being involved in KTI4 processing. Notably, the total abundance of KTI4 in SlVPE3 RNAi fruit appeared to be higher than in the wild type, which is consistent with the iTRAQ analysis showing that KTI4 abundance increased significantly after SlVPE3 was repressed. The changes in protein levels of KTI4 might reflect direct proteolytic processing of KTI4 by SlVPE3, or by the increase in KTI4 transcript levels (Fig. 6), or both.
Fig. 10.
SlVPE3 is involved in KTI4 cleavage. a Immunoblot detection of KTI4 cleavage in tomato fruit. Total protein extracts from wild-type (WT) and SlVPE3 RNAi fruit (3-4, 3-12, and 3-15) at 35 and 38 days post-anthesis (dpa) were analyzed by immunoblot analysis using anti-KTI4 or anti-SlVPE3 antibodies. The predicted molecular mass of the full-length KTI4 is ~25 kDa, which is indicated by a blue arrowhead. b Determination of KTI4 cleavage in tobacco (N. benthamiana). Proteins were extracted from tobacco leaves expressing an empty vector (Vec), KTI4 alone (Vec + KTI4), SlVPE3 and KTI4 (SlVPE3 + KTI4), and mutated SlVPE3 and KTI4 (SlVPE3 C69G/C208G + KTI4) and subjected to immunoblot analysis with an anti-KTI4 antibody. The mutated form of SlVPE3 (SlVPE3C69G/C208G) was generated by site-directed mutagenesis. c Cell-free cleavage of His-tagged KTI4 proteins. His-KTI4 was expressed and purified from Escherichia coli and then incubated for 15 or 30 min at 20 °C with extracts of N. benthamiana expressing an empty vector (Vec), intact SlVPE3 (SlVPE3), or a mutated form of SlVPE3 (SlVPE3 C69G/C208G). An incubation of His-KTI4 with extraction buffer served as the control. The VPE inhibitor (biotin-YVAD-fmk) was applied to determine whether the His-KTI4 was directly cleaved by SlVPE3. In a–c, equal loading was confirmed with an anti-actin antibody. Similar results were obtained from three independent experiments and results from a representative experiment are shown
To verify that SlVPE3 cleaves KTI4, the SlVPE3 and KTI4 ORFs were transiently expressed in tobacco (N. benthamiana) leaves. As shown in Fig. 10b, a band of the predicted molecular mass of the full-length KTI4 (~25 kDa) was generated by the anti-KTI4 antibodies, together with an additional faint band with a lower molecular mass. When SlVPE3 was co-expressed with KTI4 in tobacco, the abundance of the 25-kDa band decreased, concomitant with an increase in the intensity of the lower molecular mass band (Fig. 10b), suggesting that the 25-kDa band constitutes a precursor of the lower molecular mass band. Importantly, when KTI4 was co-expressed with a mutated form of SlVPE3 (SlVPE3C69G/C208G) in which conserved cysteine (C) residues (C69 and C208) from the active site were replaced by glycine (G), the changes in abundance of the two bands were abolished (Fig. 10b), indicating that SlVPE3 must be catalytically active for KTI4 to be cleaved.
We subsequently examined the cleavage of KTI4 by SlVPE3 in vitro. The predicted mature SlVPE3 polypeptide and KTI4 without the signal peptide were independently expressed in Escherichia coli as fusion proteins with a His-tag. Substantial amounts of recombinant SlVPE3 protein were obtained but did not show VPE activity (data not shown). We also expressed SlVPE3 in Saccharomyces cerevisiae BY4741 and Pichia pastoris GS115 cells, but did not detect proteolytically active recombinant proteins (data not shown). We next investigated the cleavage of the His-tagged recombinant KTI4 proteins (His-KTI4) in cell-free extracts of tobacco expressing intact or mutated SlVPE3 (SlVPE3C69G/C208G). A 25-kDa band, in addition to a band with lower molecular mass, was detected by the anti-KTI4 antibodies when extracts of tobacco expressing the empty plasmid were incubated with His-KTI4 (Fig. 10c). Two additional bands were observed in extracts of tobacco expressing intact SlVPE3, suggesting the cleavage of KTI4 by SlVPE3 (Fig. 10c). Gradual processing of KTI4 was observed when the reaction time was prolonged. The processing could be blocked in extracts of tobacco expressing the mutated form of SlVPE3 (SlVPE3C69G/C208G), suggesting specific cleavage of KTI4 by SlVPE3. To determine whether KTI4 is directly cleaved by SlVPE3, biotin-YVAD-fmk [33], a VPE inhibitor that efficiently inhibits the activity of SlVPE3 (Additional file 4: Figure S5), was added to the reaction. Application of the inhibitor blocked KTI4 processing (Fig. 10c), further demonstrating that KTI4 is a direct target of SlVPE3.
KTI4 does not inhibit the activity of SlVPE3
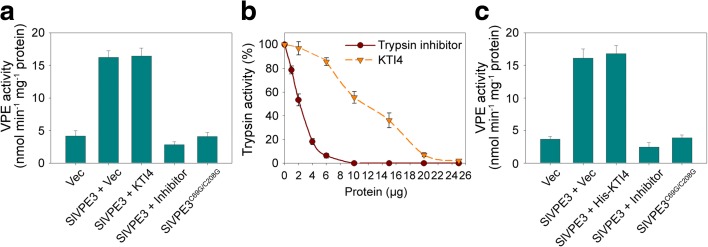
As a protease inhibitor, KTI4 has the potential to block protease activity. KTIs are specific for serine proteases, and have specific inhibitory activity solely against trypsin proteases that cleave polypeptides after Lys or Arg [45]. Accordingly, it is unlikely that KTI4 inhibits the activity of VPE proteins, as they are cysteine proteases. However, to eliminate the possibility that KTI4 inhibits SlVPE3 activity, we transformed KTI4 and SlVPE3 into tobacco and measured VPE activity with a fluorescent substrate. A fluorescent signal (3.34 nmol min−1 mg−1 protein) representing VPE activity was detected in tobacco leaves transformed with an empty plasmid control (Fig. 11a), indicating that tobacco itself contains VPE activity. However, when tobacco was transformed with SlVPE3, higher VPE activity (4.7 times; 15.6 nmol min−1 mg−1 protein) was detected, while tobacco expressing the mutated form of SlVPE3 (SlVPE3C69G/C208G) showed similar VPE activity to the empty vector control. The VPE activity was inhibited by the specific inhibitor biotin-YVAD-fmk. However, no significant differences in VPE activity were found in tobacco leaves co-expressing SlVPE3 and KTI4 compared with those expressing SlVPE3 alone (Fig. 11a), suggesting that KTI4 does not inhibit VPE activity.
Fig. 11.
KTI4 does not inhibit SlVPE3 proteolytic activity. a Determination of vacuolar processing enzyme (VPE) activity in tobacco (N. benthamiana). Proteins were extracted from N. benthamiana leaves expressing an empty vector (Vec), SlVPE3 alone (SlVPE3 + Vec), SlVPE3 and KTI4 (SlVPE3 + KTI4), and mutated SlVPE3 (SlVPE3 C69G/C208G) and subjected to a VPE activity assay with a VPE-specific fluorescent substrate. The mutated form of SlVPE3 (SlVPE3C69G/C208G) was generated by site-directed mutagenesis. The VPE inhibitor biotin-YVAD-fmk was added. Values are shown as the means ± standard deviation (SD). b The recombinant KTI4 protein from E. coli showed inhibitory activity against trypsin. Soybean trypsin inhibitor was used as a positive control. Trypsin activity was measured with the Nα-benzoyl-L-arginine ethyl ester substrate. Error bars represent the SD of three independent experiments. c VPE activity assay with recombinant KTI4 protein (His-KTI4). Extracts from N. benthamiana expressing an empty vector (Vec), intact SlVPE3, or a mutated form of SlVPE3 (SlVPE3 C69G/C208G) were incubated with recombinant KTI4 protein purified from E. coli, and then subjected to a VPE activity assay. Values are shown as the means ± SD
We also evaluated the effects of recombinant KTI4 proteins purified from E. coli on VPE activity. The recombinant KTI4 proteins exhibited activity against trypsin (Fig. 11b), indicating that they were active serine protease inhibitors. The incubation of the KTI4 proteins with extracts from tobacco leaves expressing intact or mutated SlVPE3 (SlVPE3C69G/C208G) did not significantly affect VPE activity, although this activity was inhibited by the specific inhibitor biotin-YVAD-fmk (Fig. 11c).
KTI4 contributes to resistance of fruit to pathogen infection
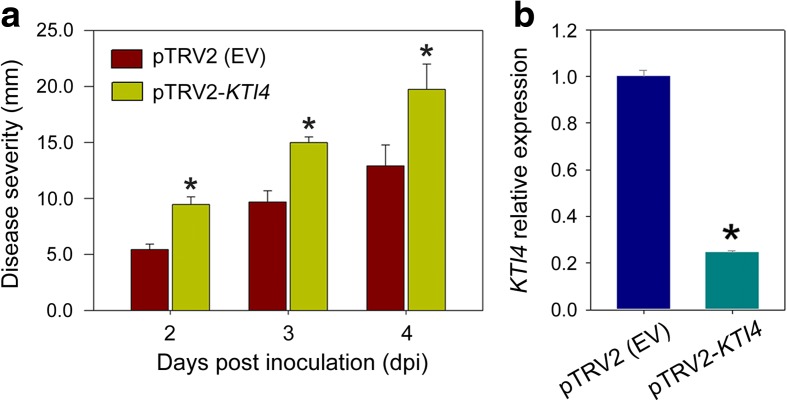
To examine whether KTI4 provides fruit with resistance to pathogen infection, we performed a virus-induced gene silencing (VIGS) assay [49, 50]. A KTI4 cDNA fragment was inserted into the pTRV2 vector, which was subsequently infiltrated into tomato fruit at the immature green stage. Fruit infiltrated with pTRV2 alone (empty vector) were used as a control. Fourteen days after infiltration, the fruit were inoculated with B. cinerea and the symptoms were visually inspected daily. As shown in Fig. 12a, fruit infiltrated with pTRV2-KTI4 showed increased disease severity compared with fruit infiltrated with pTRV2 alone (control). Gene expression analysis indicated that the KTI4 mRNA levels were reduced by ~75% compared with the control (Fig. 12b).
Fig. 12.
KTI4 influences disease resistance in tomato fruit. a Virus-induced gene silencing (VIGS) assay revealed that KTI4 affects disease resistance in tomato fruit. Immature green stage fruit were infiltrated with an empty vector control pTRV2 (EV) or pTRV2 containing a specific KTI4 sequence (pTRV2-KTI4). Fourteen days after infiltration, the fruit were inoculated with B. cinerea. Disease severity was observed daily after inoculation. b KTI4 gene expression after VIGS, as determined by quantitative RT-PCR. The gene transcript levels were normalized against expression of the ACTIN gene, followed by normalization against the control. Values are shown as the means ± standard deviation. Asterisks indicate significant differences (P < 0.05; t-test) between fruit infiltrated with pTRV2 (EV) and pTRV2-KTI4
SlVPE3 is regulated by the RIN transcription factor
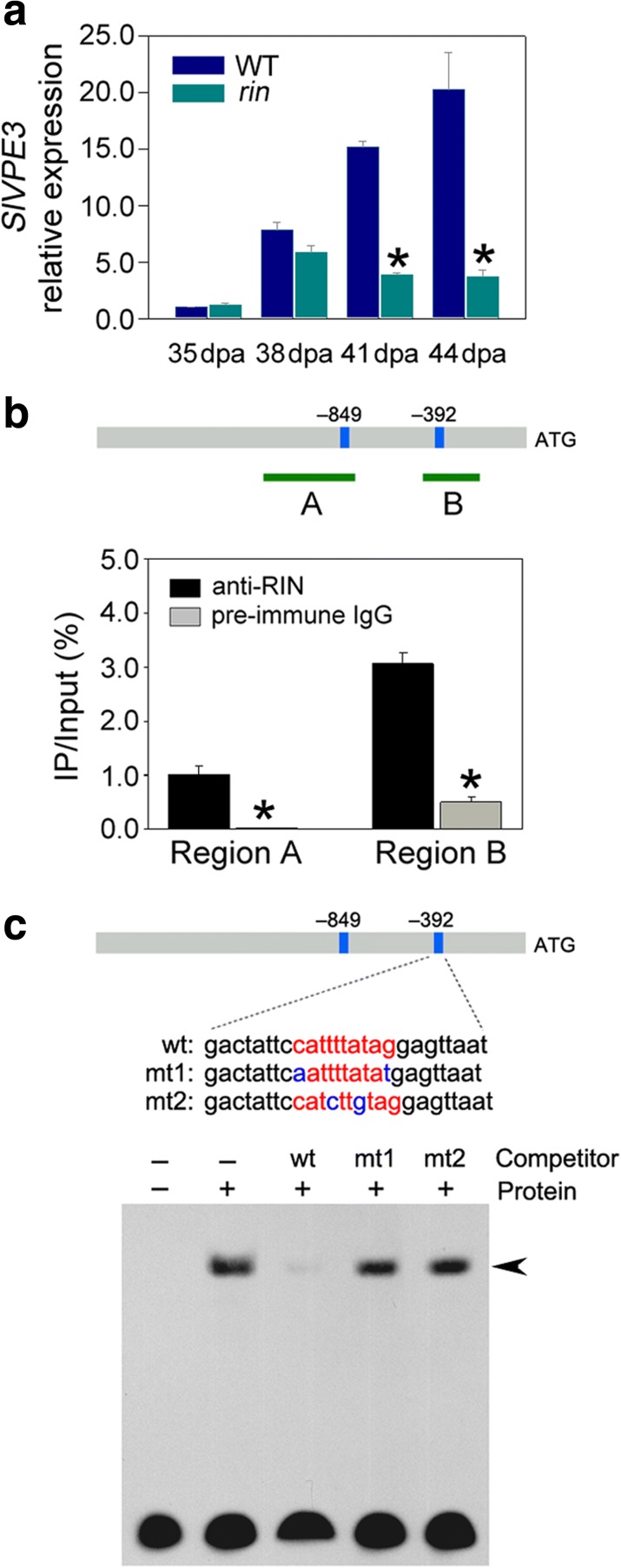
The activity of proteases needs to be tightly regulated due to their critical roles in diverse of biological processes. VPEs have been shown to be self-catalytically activated by sequential removal of C-terminal and N-terminal propeptides [51–53]. SlVPE3 shows a similar expression pattern to the transcription factor RIPENING INHIBITOR (RIN), a global regulator of tomato fruit ripening [1, 9]. Quantitative RT-PCR analysis indicated that the expression of SlVPE3 was decreased in fruit of the rin mutant tomato compared with wild type (Fig. 13a). To investigate whether RIN regulates the expression of SlVPE3 by directly binding to its promoter in vivo, a ChIP assay was performed. Using PlantCARE [54], two CArG-box elements, which are typical binding sites for RIN [55], were identified in the promoter region of SlVPE3 (Additional file 11: Table S10). For the ChIP assay, cross-linked DNA–protein complexes were immunoprecipitated with affinity-purified anti-RIN polyclonal antisera. The promoter sequences surrounding the CArG-box binding sites were amplified from the immunoprecipitated DNA using specific primers designed for SlVPE3, indicating that RIN binds to the promoter of SlVPE3 in vivo (Fig. 13b). Binding of RIN to the promoter of ACS2 was used as a positive control.
Fig. 13.
Regulation of SlVPE3 by the RIN transcription factor. a SlVPE3 expression in wild type (WT) and the rin mutant during fruit ripening, as determined by quantitative RT-PCR. The gene transcript levels were normalized against the ACTIN gene, followed by normalization against WT at 35 days post-anthesis (dpa). Values are shown as the means ± standard deviation (SD). Asterisks indicate significant differences (P < 0.05; t-test) between WT and the rin mutant. b ChIP-quantitative PCR assays indicated that RIN directly binds to the promoter of SlVPE3. The promoter structure of SlVPE3 is shown. Blue boxes represent CArG box elements and numbers indicate the position of these motifs relative to the translational start site. Green fragments with upper-case letters represent the regions used for ChIP-quantitative PCR. Values are shown as the means ± SD. Asterisks indicate significant differences (P < 0.05; t-test) between samples co-immunoprecipitated with anti-RIN antibodies and pre-immune serum. c Gel mobility shift assays revealed the direct binding of RIN to the CArG box element in the promoter region of SlVPE3. The probe sequences corresponding to the SlVPE3 promoter are shown, with red letters representing the CArG box. The mutated bases in the probes are represented by blue letters. wt, probe with intact CArG box element; mt, probe with mutated CArG box element. One thousand-fold excess amounts of unlabeled probes were added to the binding reaction as a competitor. The specific complexes formed are indicated by arrowheads
We also performed an electrophoretic mobility shift assay (EMSA) with the purified recombinant RIN protein. A band shift was observed when the purified RIN protein was mixed with the biotin-labeled probe (26-mer oligonucleotide) containing the CArG-box element. Binding of the RIN protein to the promoter fragment was out-competed by addition of an excessive amount of the corresponding unlabeled probe (competitor) containing an intact CArG box element, but not by the probe with a mutated CArG box element (Fig. 13c). This confirmed direct binding of RIN to the SlVPE3 promoter.
Discussion
In recent years, substantial progress has been made in understanding the transcriptional control of fruit ripening, but relatively little is known about the regulation of fruit ripening at the post-transcriptional level. In the present study, we show that SlVPE3, a cysteine protease, is involved in fruit ripening. Repression of SlVPE3 resulted in a delay in ripening and made the fruit more susceptible to pathogen infection. Via a quantitative proteome study we found that SlVPE3 affects the abundance of a set of proteins associated with fruit ripening and disease resistance. Further analysis indicated that SlVPE3 participates in the cleavage of KTI4, a protease inhibitor that contributes to fruit disease resistance.
SlVPE3 regulates fruit ripening at the post-transcriptional level
Proteases represent one of the most abundant classes of enzymes in eukaryotes and reside in various cellular compartments, such as the cytosol, mitochondria, vacuoles, lysosomes, endoplasmic reticulum, or the extracellular domain [23, 27, 56, 57]. In A. thaliana, more than 800 proteases have been identified, representing approximately 2% of the genome [27]. The distribution and the protease family size are well conserved within the plant kingdom [58]. However, even though they have been demonstrated to participate in a variety of biological processes [23, 27, 59–61], their roles in fruit ripening remain unclear. In this study, we showed that SlVPE3, a gene encoding a vacuolar processing enzyme, is required for normal tomato fruit ripening.
The iTRAQ-based quantitative proteome analysis represents a powerful technique for high-throughput nonbiased discovery of protease targets [62–64], and has provided increased coverage and sensitivity for quantitative proteomics [65]. Using this technique, more than 300 proteins that changed abundance upon SlVPE3 silencing were identified, of which a set was associated with fruit ripening. These results suggested that SlVPE3 regulates fruit ripening by affecting the accumulation of numerous ripening-related proteins. Notably, besides SlVPE3 targets, other proteins that changed abundance due to the delayed ripening were also identified. The proteins that differentially accumulated due to the activity of SlVPE3 itself might be identified by comparing the protein profiles between tomato leaves expressing an intact or mutagenized form of SlVPE3.
VPEs can degrade target proteins or activate substrate proteins from inactive precursor isoforms to active forms [23]. Interestingly, a homolog of SlVPE3, SlVPE5 (formerly named LeCp), was reported to function as a transcription factor and to regulate the expression of ACS2 in tomato and tobacco (N. tabacum) leaves upon induction by the fungal elicitor EIX [42]. LeCp was found to bind directly to the ACS2 promoter and it was proposed that LeCp functions as a protease in the cytoplasm and, upon elicitor signaling, enters the nucleus via binding by a small ubiquitin-related modifier (SUMO) protein, whereupon it acts as a transcription factor [42]. In our study, we found that SlVPE3 affects the transcript levels of several ripening-related genes (Fig. 6). However, ChIP-quantitative PCR analysis indicated that SlVPE3 does not bind to the promoters of these genes. In addition, we found no evidence for direct binding of SlVPE3 to the ACS2 promoter. These results indicated that although LeCp, the homolog of SlVPE3, exhibits transcription factor activity that induces ACS2 expression in tomato leaves, SlVPE3 does not act as a transcription factor in tomato fruit during ripening. Accordingly, SlVPE3 may regulate the ripening-related genes at the post-transcriptional levels rather than at the transcriptional levels. Alternatively, SlVPE3 may modulate the expression of ripening-related genes indirectly through processing of specific transcription factors, which in turn regulate the expression of these genes.
Protease inhibitor KTI4 functions downstream of SlVPE3 to regulate fruit disease resistance
The susceptibility of tomato fruit to necrotrophic pathogens increases during fruit ripening [3, 30], and we hypothesized that since suppressing the expression of SlVPE3 resulted in a delay in fruit ripening, it might also affect fruit susceptibility to pathogen infection. It has also been reported that simultaneously suppressing the expression of genes encoding the cell wall-modifying proteins polygalacturonase (LePG) and expansin (LeExp1) reduces the susceptibility of ripening tomato fruit to B. cinerea infection [3]. In this study, we found that a set of ripening-related proteins, including LePG and LeExp1, were less abundant in the SlVPE3 RNAi fruit than in wild type. Contrary to expectations, however, we observed that the SlVPE3 RNAi fruit were more susceptible to B. cinerea infection than the wild type (Fig. 3), suggesting that SlVPE3 may target proteins involved in defense responses, as well as those associated with ripening. Consistent with this idea, the proteomic data revealed that a number of proteins putatively associated with disease resistance have altered abundance in the SlVPE3 RNAi fruit. In A. thaliana, knocking out the expression of VPEγ was also demonstrated to increase the susceptibility of leaves to pathogens [34], but the underlying molecular mechanisms are not well understood.
To identify proteins directly responsible for the SlVPE3-regulated disease resistance, we screened for proteins that interact with SlVPE3 using quantitative affinity purification followed by mass spectrometry, which detects protein–protein interactions in the native cellular environment and is suitable for detecting weak and transient interactors. A protease inhibitor, KTI4, was identified as a SlVPE3-interacting protein and the interaction was confirmed by a Y2H assay (Figs. 8 and 9). Immunoblot analysis of fruit from wild-type tomato and SlVPE3 RNAi lines, and of tobacco leaves transiently expressing SlVPE3 and KTI4, using antibodies against KTI4 showed that SlVPE3 participates in the cleavage of KTI4 (Fig. 10a, b). Furthermore, KTI cleavage by SlVPE3 was confirmed by cell-free assays using recombinant KTI4 proteins (Fig. 10c). These data suggest that KTI4 is a direct target of SlVPE3.
KTIs are single-chain polypeptides of ~20 kDa belonging to the serine protease inhibitor family [45, 46]. A. thaliana KTI1 has been reported to be associated with leaf PCD in plant–pathogen interactions [45]; however, the function of KTI proteins in fruit disease resistance or their regulation has not been determined. In the present study, we found that KTI4 was processed by SlVPE3 (Fig. 10) and played a role in defense response of tomato fruit to B. cinerea (Fig. 12). Fruit in which KTI4 expression was suppressed by VIGS showed similar susceptibility to B. cinerea as those from the SlVPE3 RNAi lines (Figs. 3 and 12). Importantly, we observed that KTI4 does not inhibit SlVPE3 protease activity (Fig. 11). These data suggest that KTI4 functions as a downstream effector of SlVPE3 to regulate fruit disease resistance. Notably, suppression of KTI4 by VIGS did not affect fruit ripening (Additional file 4: Figure S6). The mechanism by which KTI4 affects pathogen infection remains unknown, but we speculate that it may target proteases that are secreted by fungal pathogens as virulence factors.
SlVPE3 is regulated at multiple levels
Due to the central roles of proteases in many biological processes and the irreversible nature of proteolysis, the action of proteases must be tightly controlled to prevent improper cleavage of substrate molecules [25]. Protease activities are regulated at multiple levels. The regulation can occur at the transcriptional level, and at the protein level by activation of inactive zymogens, or by the binding of inhibitors and cofactors [24]. Many proteases are synthesized as inactive zymogens, which contain inhibitory prodomains that must be removed for the protease to become active [25]. The activation can be either autocatalytic or performed by other proteases. VPEs have been reported to be self-catalytically activated from inactive proproteins into active VPEs [51–53]. In contrast to the understanding of post-transcriptional regulation of VPEs activity, little is known about the regulation of VPEs at the transcriptional level.
An analysis of the SlVPE3 promoter region revealed two CArG box elements, which are binding sites for the RIN transcription factor, suggesting that SlVPE3 may be transcriptionally regulated by RIN. Gene expression analysis, combined with ChIP and EMSA assays, supported this hypothesis, and indicated that RIN regulates the expression of SlVPE3 by directly binding to its promoter (Fig. 13). RIN has been reported to directly regulate genes involved in a wide variety of biological processes, such as lycopene accumulation, ethylene production, chlorophyll degradation, and aroma formation [66, 67], but little is known about the regulation of protein degradation by RIN. We previously demonstrated that genes encoding E2 ubiquitin-conjugating enzymes, the components of the ubiquitin-proteasome, serves as direct targets of RIN [28], suggesting that RIN might regulate ubiquitin-mediated proteolysis. In this study, we show that RIN directly regulates protein degradation by targeting a specific protease, SlVPE3, which represents a previously unreported RIN direct target in a study involving ChIP coupled with DNA microarray analysis [67]. Notably, the expression of SlVPE3 was only partially downregulated in the rin mutant fruit, suggesting that additional transcriptional regulators are necessary for SlVPE3 expression.
Conclusions
Our findings provide new insights into understanding the gene regulatory networks and proteolytic mechanisms that contribute to ripening and disease resistance in fruit. Future studies will investigate the target proteases of KTI4 and uncover the molecular mechanisms by which KTI4 regulates defense response in tomato fruit.
Methods
Plant material
Seeds of wild-type tomato (Solanum lycopersicum cv. Ailsa Craig) and the ripening mutant rin in the cv. Ailsa Craig background were kindly provided by Dr. James J. Giovannoni (Boyce Thompson Institute for Plant Research, Cornell University, Ithaca, NY, USA). Plants were grown in a greenhouse under standard culture conditions, with a regular supply of fertilizer and supplementary lighting when required. Flowers were tagged at anthesis to accurately follow fruit ages through development. Fruit ripening stages used in wild type were mature green (MG), breaker (Br), orange (Or), and red ripe (RR), which were defined on basis of the color, size, shape, seed development, and the development of locular jelly in the fruit [68]. These ripening stages (MG, Br, Or and RR) were on average 35, 38, 41, and 44 dpa, respectively. rin mutant fruit or transgenic lines were collected at the equivalent ripening stages, as determined by the number of dpa. Immediately after harvesting, pericarp tissue was collected, frozen in liquid nitrogen, and stored at −80 °C until use.
RNA isolation and quantitative RT-PCR analysis
RNA was isolated from the pericarp using the method of Moore et al. [69]. The extracted RNA was treated with DNase I (Promega) and reverse transcribed using an oligo (dT)18 primer with Moloney murine leukemia virus (M-MLV) reverse transcriptase (Promega) to synthesize cDNA. Quantitative RT-PCR was carried out with the SYBR Green PCR Master Mix (Applied Biosystems) using the StepOne Plus Real-Time PCR System (Applied Biosystems). Gene-specific primers (Additional file 12: Table S11) were designed with the help of the Primer Express software 3.0 (Applied Biosystems). The following program was applied for PCR amplification in a volume of 20 μL: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 30 s. Relative quantification of specific mRNA levels was performed using the cycle threshold (Ct) 2(−ΔCt) method [70]. Expression values were normalized using ACTIN (SGN-U580609). Each experiment had three biological repeats, each with three technical replicates.
Phylogenetic analysis
An alignment of the VPE protein sequences from tomato (S. lycopersicum) (Additional file 4: Supplementary text), rice (Oryza sativa), potato (Solanum tuberosum), tobacco (N. tabacum), pepper (Capsicum annuum), maize (Zea mays), soybean (Glycine max), citrus (C. sinensis), grape (Vitis vinifera) and A. thaliana was generated using ClustalX (version 2.1) software [71] with default multiple parameters and the PAM series protein weight matrix. The genedoc program was used to manually edit the alignment, which was then imported into MEGA (version 5.2) software [72] and the phylogenetic tree was constructed by the neighbor-joining statistical method using 1000 bootstrap replicates [72].
Construction of the RNAi vector and plant transformation
To construct the SlVPE3 RNAi plasmid, a 278-bp SlVPE3 fragment was amplified from cDNA of tomato fruit at 38 dpa with SlVPE3-specific primers (SlVPE3 RNAi/F: 5′-GTTCCCTCCACAGGGGTT-3′; SlVPE3 RNAi/R: 5′-AGATGAAAGAAAGTTTGTTCAGG-3′) and cloned into the pCR8/GW/TOPO Gateway entry vector (Invitrogen). The cloned fragment was subsequently transferred into the binary RNAi vector pK7GWIWG2D [73]. The resulting construct was sequence confirmed and transformed into A. tumefaciens strain GV3101 [74], which was subsequently transformed into tomato (S. lycopersicum cv. Ailsa Craig) according to the method of Fillatti et al. [75]. The presence of the transgene was verified by PCR in the T0 and T1 tomato generations.
Lycopene measurements
Pericarp lycopene content was measured as described by Sun et al. [76] and expressed as mg kg−1 fresh weight. Each sample contained three replicates with five fruit per replicate and the experiment was repeated twice.
Ethylene production assay and ethephon treatment
To measure ethylene biosynthesis, fruit from transgenic T1 lines and the wild-type tomato were harvested at 38 and 41 dpa and placed in open jars for 3 h to avoid measuring “wound ethylene”, transiently synthesized as a consequence of picking. The jars were then sealed, incubated at room temperate for 2 h and then 1 mL gas samples were taken and analyzed by a gas chromatograph (SQ-206, Beijing, China) equipped with an activated alumina column and a flame ionization detector. Ethylene concentrations were calculated by comparing the peak length from the gas chromatograph with reagent grade ethylene standards of known concentration and normalizing for fruit weight. Each sample contained three replicates with five fruit per replicate and the experiment was repeated twice.
For the ethephon treatment, tomato fruit at 35 dpa were immersed for 10 min in a 50 mM fresh aqueous solution of ethephon (Sigma) or in water for the control. Pericarp samples were collected at various time intervals.
Protein extraction, iTRAQ labeling, and NanoLC–MS/MS analysis
Proteins were extracted from tomato fruit pericarp at 41 and 44 dpa as previously described [77]. The isolated proteins were solubilized in protein buffer consisting of 500 mM triethylammonium bicarbonate (TEAB) and 1% SDS (w/v), pH 8.5, and the protein concentrations were determined by the Bradford method [78]. One-hundred micrograms of protein from each sample were reduced, alkylated, and digested using the filter-aided sample preparation (FASP) method [79]. The tryptic peptides were then labeled with the iTRAQ Reagents 4-plex Kit (Applied Biosystems) following the manufacturer’s protocol. Samples taken from wild-type and SlVPE3 RNAi fruit at 41 dpa were labeled with iTRAQ tags 114 and 115, respectively, while samples from wild-type and SlVPE3 RNAi fruit at 44 dpa were labeled with iTRAQ tags 116 and 117, respectively. The iTRAQ experiment was performed with two independent biological replicates. The iTRAQ-labeled samples were combined and subjected to high-pH reversed-phase chromatography. Briefly, the pooled iTRAQ-labeled peptides were reconstituted with buffer A (20 mM ammonium formate, pH 10, in water) and loaded onto a 4.6 × 250 mm, 150 Å size Durashell C18 (L) column containing 5 μm particles (Agela Technologies). The peptides were eluted at a flow rate of 0.8 mL min−1 using a gradient of 2% buffer B (20 mM ammonium formate in 80% acetonitrile, pH 10) for 5 min, 2–30% buffer B for 25 min, and 30–90% buffer B for 10 min. The system was then maintained in 90% buffer B for 10 min before equilibration with 2% buffer B for 10 min. The elution was monitored by measuring UV absorbance at 210 nm, and fractions were collected every 1 min. Forty-eight fractions were collected and pooled into a total of six fractions. After reconstitution in 0.1% formic acid, 8 μL of the combined iTRAQ-labeled peptides were submitted for NanoLC–MS/MS analysis.
The mass spectroscopy analysis was performed as previously described [80] using a NanoLC system (NanoLC–2D Ultra Plus, Eksigent) equipped with a Triple TOF 5600 Plus mass spectrometer (AB SCIEX). Protein identification and quantification for the iTRAQ experiments were performed using ProteinPilot™ 4.5 software (AB SCIEX). The mass spectra data were used to search the S. lycopersicum protein database ITAG2.4_proteins_full_desc.fasta, using the following parameters: (i) Sample type, iTRAQ 4-plex (Peptide Labeled); (ii) Cysteine alkylation, MMTS; (iii) Digestion, Trypsin; (iv) Instrument, TripleTOF 5600; (v) Species, None; (vi) Quantitate, Yes; (vii) Bias correction, Yes; (viii) Background correction, Yes; (ix) Search effort, Thorough; (x) FDR analysis, Yes. For iTRAQ quantification, the peptide for quantification was automatically selected using the Pro GroupTM algorithm (AB SCIEX) to calculate the reporter peak area. A reverse database search strategy [81] was used to estimate the global FDR for peptide identification. Only proteins identified below the 1% global FDR were ultimately exported for determining the meaningful cut-off value for the regulated proteins [82]. Hierarchical clustering (Pearson algorithm) was carried out with PermutMatrix software [31].
Preparation of polyclonal antibodies
For SlVPE3-specific antibody preparation, a truncated form of SlVPE3 lacking the conserved domain was amplified from tomato cDNA using primers F (5′-AGGATATCGAAAGACACAACCTGCG-3′) and R (5′-ACGTCGACCTAACCAGCAGGGCGGAC-3′) and inserted into the pET-30a vector (Merck KGaA). The resulting plasmid was transformed into E. coli BL 21 (DE3) competent cells. For recombinant protein expression, E. coli was cultured overnight and then diluted 1:100 in Luria Broth medium. The bacteria were incubated at 37 °C until A600 reached approximately 0.5, then isopropyl-1-thio-β-D-galactopyranoside (IPTG) was added to a final concentration of 1 mM to induce recombinant protein expression. The bacterial cells were incubated for an additional 3 h before the cells were collected by centrifugation for recombinant protein isolation. The recombinant protein was purified using Ni-NTA His Bind Resin according to the manufacturer’s manual (Merck KGaA), followed by further purification by 12% SDS-PAGE. The protein band corresponding to the predicted size of the recombinant SlVPE3 was excised from the gel and used to immunize rabbits at the Beijing Protein Institute Co., Ltd. Polyclonal antibodies that recognized SlVPE3 was affinity-purified from antisera using the AminoLink Plus Coupling Resin following the purification protocol (Thermo Scientific).
Polyclonal KTI4 antibodies were raised by injecting a rabbit with the synthetic peptide TYASVVDSDGNPVKAGAKYF followed by affinity-purification using the synthetic peptide.
Immunoprecipitation of SlVPE3-interacting proteins for SWATH-MS analysis
Proteins were isolated from 41 dpa tomato fruit using IP buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 50 μM MG132, 1 mM PMSF, and the protease inhibitor cocktail tablet (Roche). After centrifugation at 12,000 × g for 10 min, the supernatant containing the proteins was immunoprecipitated overnight at 4 °C with 50 μg of anti-SlVPE3 or pre-immune serum IgG (negative control) that was coupled to an agarose support, as described in the Pierce® Co-Immunoprecipitation (Co-IP) Kit (Pierce Biotechnology). The agarose beads were collected in spin columns and washed twice with IP buffer. The proteins were then eluted from the beads with 0.1 M glycine-HCl (pH 2.2), followed by reduction, alkylation, and digestion using the filter-aided sample preparation (FASP) method [79]. The resulting peptides were collected, dried under vacuum, and redissolved in 0.1% formic acid for NanoLC-MS/MS analysis. Quantitative analysis of SlVPE3-interacting proteins was performed using the SWATH-MS method [28]. Mass spectra were generated on a TripleTOF 5600 plus instrument (AB SCIEX) operating in the SWATH mode. Each sample contained three technical replicates and a t-test was used for statistical analysis. A P value <0.01 was considered to be significant.
Y2H analysis
Construction and two-hybrid screening of the tomato cDNA library was performed with the Matchmaker Library Construction and Screening Kit (Clontech). Total RNA was extracted from 38 dpa (Br stage) tomato fruit pericarp and mRNA was isolated and used for cDNA library construction. The tomato cDNA library constructed in the prey vector pGADT7 (AD) was screened with a SlVPE3 cDNA fragment encoding the mature protein cloned into the bait vector pGBKT7 (BD) in S. cerevisiae strain AH109 (Clontech), and positive clones were selected on SD/-Leu/-Trp/-His/-Ade medium supplemented with X-α-Gal.
To confirm the interactions between SlVPE3 and KTI4, the SlVPE3 cDNA fragment encoding the mature protein and the ORF of KTI4 were cloned into the AD and BD vectors, respectively, resulting in the SlVPE3-AD and KTI4-BD plasmids. Primers used for the vector construction are shown in Additional file 13: Table S12. The vectors were co-transformed into S. cerevisiae strain AH109 following the manufacturer’s manual (Clontech), and dripped on SD/-Leu/-Trp medium (SD/-2) and SD/-Leu/-Trp/-His/-Ade medium (SD/-4) containing X-α-Gal. As controls, AD and BD, KTI4-BD and AD, or SlVPE3-AD and BD were co-transformed. The experiments were repeated three times.
Subcellular colocalization
For subcellular localization analysis, the full-length SlVPE3 and KTI4 cDNAs were amplified by PCR (primer sequences in Additional file 13: Table S12) and individually cloned into the pCambia 2300-MCS-mRFP vector. The resulting plasmids were transformed into A. tumefaciens GV3101 [74], which was subsequently infiltrated into tobacco (N. benthamiana) leaves [83]. For colocalization analysis, N. benthamiana plants co-expressing mRFP-tagged SlVPE3 (SlVPE3-mRFP) and PRpHluorin-tagged KTI4 (KTI4-PRpHluorin) under the control of the CaMV 35S promoter were generated. The plasmid containing PRpHluorin was kindly provided by Dr. Liwen Jiang (School of Life Sciences, The Chinese University of Hong Kong, Shatin, New Territories, Hong Kong, China). The tobacco plants were kept in the greenhouse for two days, and then mesophyll protoplasts were isolated [84] and observed under a Leica confocal microscope (Leica DMI600CS).
Protein cleavage assay
Proteins from the pericarp of wild-type and SlVPE3 RNAi fruit at 35 and 38 dpa were extracted using a phenol extraction method [77]. The proteins were solubilized in lysis buffer consisting of 7 M urea, 2 M thiourea, 4% CHAPS, and 1% dithiothreitol (DTT). Protein concentrations were determined by the Bradford method [78]. The samples were separated by 12% SDS-PAGE and subjected to immunoblot analysis, as below.
For the KTI4 cleavage assay in tobacco (N. benthamiana), the full-length SlVPE3 and KTI4 were amplified using gene-specific primers (Additional file 13: Table S12) and cloned into the pCambia 2300 vector (Cambia) to generate the 35S:SlVPE3 and 35S:KTI4 constructs. A mutated form of SlVPE3 (SlVPE3C69G/C208G) was constructed by site-directed mutagenesis using the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies) according to the manufacturer’s instructions (primers are listed in Additional file 14: Table S13). The constructs were introduced into A. tumefaciens GV3101 [74] and then expressed transiently in N. benthamiana leaves [83] as described above. After 2 days, total proteins were extracted with an extraction buffer containing 25 mM Tris · HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 1% (v/v) TritonX-100, 1 mM PMSF, and a protease inhibitor cocktail tablet (Roche). The homogenates were centrifuged at 12,000 × g for 10 min at 4 °C, and then the supernatants were subjected to immunoblot analysis, as below.
Cell-free assays for protein cleavage
The cell-free cleavage assay was carried out as previously described [85]. The KTI4 coding sequence without the predicted signal peptide region was amplified from cDNA of tomato fruit using the primers KTI4-F (5′-GGGGTACCATGTCAACATTTTCTTCAGATCTT-3′) and KTI4-R (5′-GCGTCGACttaATCAGCCTTCTTGAAGTAA-3′) and cloned into the pCold I vector (Takara) to produce pCold I-KTI4. This construct allows an in-frame fusion of the coding region of KTI4 with an N-terminal His-tag. The plasmid was transformed into E. coli BL 21 (DE3) cells, and the recombinant protein expression and purification were performed as described above. For the cleavage assay, total proteins were extracted from N. benthamiana leaves that transiently expressed the intact or mutated form of SlVPE3 (SlVPE3C69G/C208G) using an extraction buffer containing 100 mM sodium acetate, pH 5.5, 1 mM PMSF, 100 mM DTT, and 100 μm E64-d. After two rounds of centrifugation at 12,000 × g for 10 min each at 4 °C, the supernatant was collected and the protein concentration was determined using the Bradford method [78]. The total protein extracts (250 μg) were incubated with 200 ng of purified recombinant His-KTI4 in 250 μL extraction buffer for 0, 15, and 30 min at 20 °C. Wherever indicated, the VPE inhibitor biotin-YVAD-fmk was used at 100 μM by pre-incubation with the total protein extracts for 30 min before incubation with His-KTI4. The mixtures were subjected to SDS-PAGE followed by immunoblot analysis, as below.
Immunoblot analysis
For immunoblot analysis, proteins were separated by 12% SDS-PAGE and electrotransferred to an Immobilon-P PVDF membrane (Millipore). The membranes were blocked for 2 h at room temperature with 5% BSA in a PBS-Tween buffer (137 mM NaCl, 2.7 mM KCl, 8.1 mM NaH2PO4, 1.5 mM KH2PO4 and 0.1% Tween-20). Immunoblotting was conducted at 4 °C overnight with anti-KTI4 (1:1000) or anti-SlVPE3 (1:1000) antibodies. The membranes were washed with PBS-Tween (3 × 10 min), and then the secondary antibodies conjugated to horseradish peroxidase (Abmart) were added (1:5000). Immunoreactive bands were visualized using a chemiluminescence detection kit (SuperSignal®, Pierce Biotechnology). Equal loading was confirmed with an anti-actin antibody (Abmart).
VPE activity assay
VPE activity was measured as previously described [86] using a synthesized fluorescent VPE-specific substrate, Ac-ESEN-MCA [Acetyl-Glutamyl-Seryl-Glutamyl-Asparagine α-(4-Metyl-Coumaryl-7-Amide)] (Shanghai Bootech BioScience & Technology Co., Ltd). Proteins were extracted with a buffer containing 100 mM sodium acetate, pH 5.5, 1 mM PMSF, 100 mM DTT, and 100 μm E64-d from leaves of N. benthamiana that transiently expressed intact SlVPE3 (alone or co-expressed with KTI4) or the mutated form of SlPVE3 (SlVPE3C69G/C208G) for 2 days. The samples were centrifuged at 12,000 × g for 15 min and the supernatants were used for VPE activity assays. The crude enzyme extract was incubated with 100 μM Ac-ESEN-MCA in an acidic buffer containing 100 mM sodium-acetate (pH 5.5) and 100 mM DTT for 2 h at 20 °C. The fluorescence intensity was measured using a Synergy™ H4 Multimode Microplate Reader (Bio-Tek Instruments, Inc.) with an excitation wavelength at 380 nm and an emission wavelength at 460 nm. VPE inhibitor biotin-YVAD-fmk was added to a final concentration of 100 μM by pre-incubating with the crude enzyme for 30 min before Ac-ESEN-MCA addition, wherever indicated.
The trypsin inhibitor activity of recombinant KTI4 was determined as previously described [45]. Trypsin (1 μg/μL, sigma) was pre-incubated with increasing concentrations of KTI4 in the assay buffer [45] for 30 min at 30 °C. The trypsin substrate N-benzoyl-L-arginine ethyl ester (Sigma) was then added to a final concentration of 0.25 mM and the changes in absorbance at 253 nm were monitored. Soybean trypsin inhibitor (Sigma) was used as a positive control. For the VPE activity assay with His-tagged recombinant KTI4 proteins, the purified KTI4 proteins (20 μg) were pre-incubated for 30 min with crude enzyme extracts from leaves of N. benthamiana that transiently expressed intact or the mutated form of SlPVE3 (SlVPE3C69G/C208G). Ac-ESEN-MCA was then added, as described above.
Virus-induced gene silencing
The VIGS assay was performed as previously described [87, 88]. The virus vectors pTRV1 and pTRV2 were provided by Dr. Daqi Fu (College of Food Science and Nutritional Engineering, China Agricultural University, Beijing, China). The specific KTI4 cDNA fragment was amplified using the primers F (5′-GGAATTCATGATGAAGAGCCTTGTTC-3′) and R (5′-CCGCTCGAGAGGACGTCCAGTGTTAAGTTCC-3′) and inserted into the pMD19-T vector (TaKaRa Bio). The resulting plasmid was transformed into E. coli and the sequence was verified. The cDNA fragment was subsequently cloned into the virus vector pTRV2 and then transferred to A. tumefaciens strain GV3101. For infiltration, equivalent aliquots of Agrobacterium strain GV3101 (with an optical density of 0.15 at 600 nm) containing pTRV1 or pTRV2 (empty or containing the insert) were mixed and injected into immature green stage tomato fruit. Agroinjected fruit were stored at room temperature (20 °C) for 14 days before infection with B. cinerea.
Microbial infection
Infection of tomato fruit by B. cinerea was carried out as previously described [89]. B. cinerea (1 × 105 conidia per ml) was applied to three sites that were equally spaced across the fruit surface. There were three replicates for each sample with at least ten fruit per replicate, and the experiment was repeated twice. Fungal growth was determined using quantitative PCR amplification of B. cinerea ACTIN 2 relative to the tomato ACTIN gene as described by Laluk and Mengiste [90]. The following primer pairs were used for quantitative PCR: B. cinerea ACTIN 2 forward (5′-ACTCATATGTTGGAGATGAAGCGCA-3′), reverse (5′-AATGTTACCATACAAATCCTTACGGA-3′); tomato ACTIN forward (5′-ACAACTTTCCAACAAGGGAAGAT-3′) and reverse (5′-TGTATGTTGCTATTCAGGCTGTG-3′).
ChIP and EMSA
For the ChIP assay, pericarps of fruit at 41 dpa were excised, fixed in 1% formaldehyde under a vacuum for 20 min, then powdered in liquid nitrogen, and chromatin complexes were isolated and sonicated as previously described [28]. The sonicated chromatin complexes were incubated with affinity purified polyclonal anti-SlVPE3/anti-RIN antibodies or pre-immune serum IgG (negative control) as previously described [28]. The cross-linking was then reversed, and the amount of each precipitated DNA fragment was determined by real-time PCR using specific primers (Additional file 8: Table S7). Values are expressed as the percentage of DNA fragments that co-immunoprecipitated with specific (anti-RIN) [66] or non-specific (IgG) antibodies relative to the input DNA.
For the EMSA, recombinant His-tagged RIN protein was prepared as previously described [28], and purified using Ni-NTA His Bind Resin according to the manufacturer’s instructions (Merck KGaA). The ability of RIN to bind to biotin-labeled oligonucleotide probes was determined with the Lightshift Chemiluminescent EMSA kit (Thermo Scientific), as previously described [28].
Data access
The mass spectrometry data have been deposited to the ProteomeXchange Consortium [91] via the PRIDE partner repository [92] with the dataset identifier PXD002980 (http://www.ebi.ac.uk/pride).
Acknowledgements
We would like to thank Dr. Zhuang Lu for analysis of MS/MS and Dr. Daqi Fu from the College of Food Science and Nutritional Engineering, China Agricultural University for assistance with VIGS. We also thank the PRIDE team for the deposition of our mass spectrometry proteomics data to the ProteomeXchange Consortium. We thank PlantScribe (http://www.plantscribe.com/) for editing this manuscript.
Funding
This work was supported by the National Basic Research Program of China (973 Program; grant number 2013CB127103), the National Natural Science Foundation of China (NSFC; grant numbers 31172004), and the Youth Innovation Promotion Association CAS.
Availability of data and materials
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository (http://www.ebi.ac.uk/pride) under accession number PXD002980.
Authors’ contributions
GQ conceived and designed the experiments. WW participated in quantitative RT-PCR analysis, RNAi vector construction, and plant transformation, ChIP assay, EMSA, iTRAQ analysis, and SlVPE3-interacting protein identification. JC carried out the Y2H assay, immunoblot analysis, enzyme activity assay, VIGS, and microbial inoculation. PW performed analyses related to lycopene production and ethylene biosynthesis. ST provided critical discussions. WW, JC, and GQ analyzed the data. GQ wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- AD
Activation domain
- BD
Binding domain
- Br
Breaker
- ChIP
Chromatin immunoprecipitation
- dpa
Days post-anthesis
- DTT
Dithiothreitol
- EMSA
Electrophoretic mobility shift assay
- FDR
False discovery rate
- GFP
Green fluorescent protein
- iTRAQ
Isobaric tags for relative and absolute quantification
- MG
Mature green
- mRFP
Monomeric red fluorescent protein
- MS
Mass spectroscopy
- Or
Orange
- ORF
Open reading frame
- PCD
Programmed cell death
- RNAi
RNA interference
- RR
Red ripe
- RT-PCR
reverse transcription polymerase chain reaction
- SWATH-MS
Sequential window acquisition of all theoretical mass spectra
- VIGS
virus-induced gene silencing
- VPE
Vacuolar processing enzyme
- Y2H
Yeast two-hybrid
Additional files
Tomato cysteine proteinase information. (XLS 51 kb)
Expression profiles of tomato genes encoding cysteine proteases during fruit ripening. (XLS 54 kb)
Tomato vacuolar processing enzymes (VPEs) information. (XLS 15 kb)
Supplementary figures S1–S6 and Supplementary text. Figure S1. Protein sequence alignment of tomato vacuolar processing enzymes (VPEs) using Clustal X. Figure S2. Prediction of the potential off-targets of the RNAi construct. Figure S3. Identification of SlVPE3 by mass spectrometry. Figure S4. Determination of KTI4 antibody specificity. Figure S5. Effects of biotin-YVAD-fmk on vacuolar processing enzyme (VPE) activity in tobacco (N. benthamiana) leaves transiently expressing SlVPE3. Figure S6. Virus-induced gene silencing (VIGS) of KTI4 in tomato has no effect on fruit ripening. Supplementary text Protein sequences of tomato vacuolar processing enzymes (VPEs). (PDF 1450 kb)
Similarities in the amino acid sequence of tomato vacuolar processing enzymes (VPEs). (XLS 16 kb)
Identification of differentially expressed proteins in the SlVPE3 RNAi tomato fruit using iTRAQ-based quantitative proteomic analysis. (XLS 73 kb)
Predicted vacuolar processing enzyme (VPE) binding motifs within the 2000-bp upstream region starting from ATG. (XLS 16 kb)
Primers used in the ChIP-quantitative PCR analysis. (XLS 16 kb)
Identification of proteins that interact with SlVPE3 using SWATH-MS. (XLS 168 kb)
Proteins interacting with SlVPE3 (bait) in a yeast two-hybrid screen against a tomato prey library. (XLS 19 kb)
Predicted RIN binding motifs within a 2000-bp upstream region starting from start ATG of the SlVPE3 gene coding sequence. (XLS 14 kb)
Primers used for quantitative RT-PCR analysis. (XLS 47 kb)
Primers used in the yeast two-hybrid (Y2H) analysis, subcellular colocalization of SlVPE3 and KTI4, and tobacco (N. benthamiana) expression of SlVPE3 and KTI4. (XLS 15 kb)
Primers used for SlVPE3 mutagenesis. (XLS 13 kb)
References
- 1.Seymour GB, Østergaard L, Chapman NH, Knapp S, Martin C. Fruit development and ripening. Annu Rev Plant Biol. 2013;64:219–41. doi: 10.1146/annurev-arplant-050312-120057. [DOI] [PubMed] [Google Scholar]
- 2.Giovannoni JJ. Genetic regulation of fruit development and ripening. Plant Cell. 2004;16(Suppl):S170–80. doi: 10.1105/tpc.019158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cantu D, Vicente AR, Greve LC, Dewey FM, Bennett AB, Labavitch JM, Powell AL. The intersection between cell wall disassembly, ripening, and fruit susceptibility to Botrytis cinerea. Proc Natl Acad Sci U S A. 2008;105:859–64. doi: 10.1073/pnas.0709813105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barry CS, Giovannoni JJ. Ripening in the tomato Green-ripe mutant is inhibited by ectopic expression of a protein that disrupts ethylene signaling. Proc Natl Acad Sci U S A. 2006;103:7923–8. doi: 10.1073/pnas.0602319103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kevany BM, Tieman DM, Taylor MG, Cin VD, Klee HJ. Ethylene receptor degradation controls the timing of ripening in tomato fruit. Plant J. 2007;51:458–67. doi: 10.1111/j.1365-313X.2007.03170.x. [DOI] [PubMed] [Google Scholar]
- 6.Guo H, Ecker JR. The ethylene signaling pathway: new insights. Curr Opin Plant Biol. 2004;7:40–9. doi: 10.1016/j.pbi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 7.Lin Z, Zhong S, Grierson D. Recent advances in ethylene research. J Exp Bot. 2009;60:3311–36. doi: 10.1093/jxb/erp204. [DOI] [PubMed] [Google Scholar]
- 8.Merchante C, Alonso JM, Stepanova AN. Ethylene signaling. simple ligand, complex regulation. Curr Opin Plant Biol. 2013;16:554–60. doi: 10.1016/j.pbi.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 9.Vrebalov J, Ruezinsky D, Padmanabhan V, White R, Medrano D, Drake R, Schuch W, Giovannoni J. A MADS-box gene necessary for fruit ripening at the tomato Ripening-inhibitor (Rin) locus. Science. 2002;296:343–6. doi: 10.1126/science.1068181. [DOI] [PubMed] [Google Scholar]
- 10.Giovannoni JJ. Fruit ripening mutants yield insights into ripening control. Curr Opin Plant Biol. 2007;10:283–9. doi: 10.1016/j.pbi.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 11.Manning K, Tor M, Poole M, Hong Y, Thompson AJ, King GJ, Giovannoni JJ, Seymour GB. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat Genet. 2006;38:948–52. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- 12.Itkin M, Seybold H, Breitel D, Rogachev I, Meir S, Aharoni A. TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 2009;60:1081–95. doi: 10.1111/j.1365-313X.2009.04064.x. [DOI] [PubMed] [Google Scholar]
- 13.Vrebalov J, Pan IL, Arroyo AJ, McQuinn R, Chung M, Poole M, Rose J, Seymour G, Grandillo S, Giovannoni J, Irish VF. Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell. 2009;21:3041–62. doi: 10.1105/tpc.109.066936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Z, Hong Y, Yin M, Li C, Zhang K, Grierson D. A tomato HD-Zip homeobox protein, LeHB-1, plays an important role in floral organogenesis and ripening. Plant J. 2008;55:301–10. doi: 10.1111/j.1365-313X.2008.03505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung MY, Vrebalov J, Alba R, Lee J, McQuinn R, Chung JD, Klein P, Giovannoni J. A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 2010;64:936–47. doi: 10.1111/j.1365-313X.2010.04384.x. [DOI] [PubMed] [Google Scholar]
- 16.Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, Fraser PD, Baxter C, Angenent GC, de Maagd RA. Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell. 2011;23:923–41. doi: 10.1105/tpc.110.081273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JM, Joung JG, McQuinn R, Chung MY, Fei Z, Tieman D, Klee H, Giovannoni J. Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J. 2012;70:191–204. doi: 10.1111/j.1365-313X.2011.04863.x. [DOI] [PubMed] [Google Scholar]
- 18.Pan Y, Bradley G, Pyke K, Ball G, Lu C, Fray R, Marshall A, Jayasuta S, Baxter C, van Wijk R, Boyden L, Cade R, Chapman NH, Fraser PD, Hodgman C, Seymour GB. Network inference analysis identifies an APRR2-like gene linked to pigment accumulation in tomato and pepper fruits. Plant Physiol. 2013;161:1476–85. doi: 10.1104/pp.112.212654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bemer M, Karlova R, Ballester AR, Tikunov YM, Bovy AG, Wolters-Arts M, Rossetto Pde B, Angenent GC, de Maagd RA. The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell. 2012;24:4437–51. doi: 10.1105/tpc.112.103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osorio S, Alba R, Damasceno CM, Lopez-Casado G, Lohse M, Zanor MI, Tohge T, Usadel B, Rose JK, Fei Z, Giovannoni JJ, Fernie AR. Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol. 2011;157:405–25. doi: 10.1104/pp.111.175463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhong S, Fei Z, Chen YR, Zheng Y, Huang M, Vrebalov J, McQuinn R, Gapper N, Liu B, Xiang J, Shao Y, Giovannoni JJ. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat Biotechnol. 2013;31:154–9. doi: 10.1038/nbt.2462. [DOI] [PubMed] [Google Scholar]
- 22.Liu R, How-Kit A, Stammitti L, Teyssier E, Rolin D, Mortain-Bertrand A, Halle S, Liu M, Kong J, Wu C, Degraeve-Guibault C, Chapman NH, Maucourt M, Hodgman TC, Tost J, Bouzayen M, Hong Y, Seymour GB, Giovannoni JJ, Gallusci P. A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc Natl Acad Sci U S A. 2015;112:10804–9. doi: 10.1073/pnas.1503362112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ehrmann M, Clausen T. Proteolysis as a regulatory mechanism. Annu Rev Genet. 2004;38:709–24. doi: 10.1146/annurev.genet.38.072902.093416. [DOI] [PubMed] [Google Scholar]
- 24.Turk B. Targeting proteases: successes, failures and future prospects. Nat Rev Drug Discov. 2006;5:785–99. doi: 10.1038/nrd2092. [DOI] [PubMed] [Google Scholar]
- 25.Sanman LE, Bogyo M. Activity-based profiling of proteases. Annu Rev Biochem. 2014;83:249–73. doi: 10.1146/annurev-biochem-060713-035352. [DOI] [PubMed] [Google Scholar]
- 26.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–33. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Hoorn RA. Plant proteases: from phenotypes to molecular mechanisms. Annu Rev Plant Biol. 2008;59:191–223. doi: 10.1146/annurev.arplant.59.032607.092835. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Wang W, Cai J, Zhang Y, Qin G, Tian S. Tomato nuclear proteome reveals the involvement of specific E2 ubiquitin-conjugating enzymes in fruit ripening. Genome Biol. 2014;15:548. doi: 10.1186/s13059-014-0548-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rojo E, Zouhar J, Carter C, Kovaleva V, Raikhel NV. A unique mechanism for protein processing and degradation in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2003;100:7389–94. doi: 10.1073/pnas.1230987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cantu D, Blanco-Ulate B, Yang L, Labavitch JM, Bennett AB, Powell AL. Ripening-regulated susceptibility of tomato fruit to Botrytis cinerea requires NOR but not RIN or ethylene. Plant Physiol. 2009;150:1434–49. doi: 10.1104/pp.109.138701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caraux G, Pinloche S. PermutMatrix: a graphical environment to arrange gene expression profiles in optimal linear order. Bioinformatics. 2005;21:1280–1. doi: 10.1093/bioinformatics/bti141. [DOI] [PubMed] [Google Scholar]
- 32.Hara-Nishimura I, Inoue K, Nishimura M. A unique vacuolar processing enzyme responsible for conversion of several proprotein precursors into the mature forms. FEBS Lett. 1991;294:89–93. doi: 10.1016/0014-5793(91)81349-D. [DOI] [PubMed] [Google Scholar]
- 33.Hatsugai N, Kuroyanagi M, Yamada K, Meshi T, Tsuda S, Kondo M, Nishimura M, Hara-Nishimura I. A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science. 2004;305:855–8. doi: 10.1126/science.1099859. [DOI] [PubMed] [Google Scholar]
- 34.Rojo E, Martín R, Carter C, Zouhar J, Pan S, Plotnikova J, Jin H, Paneque M, Sánchez-Serrano JJ, Baker B, Ausubel FM, Raikhel NV. VPEγ exhibits a caspase-like activity that contributes to defense against pathogens. Curr Biol. 2004;14:1897–906. doi: 10.1016/j.cub.2004.09.056. [DOI] [PubMed] [Google Scholar]
- 35.Alonso JM, Granell A. A putative vacuolar processing protease is regulated by ethylene and also during fruit ripening in Citrus fruit. Plant Physiol. 1995;109:541–7. doi: 10.1104/pp.109.2.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ariizumi T, Higuchi K, Arakaki S, Sano T, Asamizu E, Ezura H. Genetic suppression analysis in novel vacuolar processing enzymes reveals their roles in controlling sugar accumulation in tomato fruits. J Exp Bot. 2011;62:2773–86. doi: 10.1093/jxb/erq451. [DOI] [PubMed] [Google Scholar]
- 37.pssRNAit. Designing effective and specific plant RNAi siRNAs with genome-wide off-target gene assessment. http://plantgrn.noble.org/pssRNAit. [DOI] [PMC free article] [PubMed]
- 38.Ronen G, Carmel-Goren L, Zamir D, Hirschberg J. An alternative pathway to β-carotene formation in plant chromoplasts discovered by map-based cloning of Beta and old-gold color mutations in tomato. Proc Natl Acad Sci U S A. 2000;97:11102–7. doi: 10.1073/pnas.190177497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burns J, Fraser PD, Bramley PM. Identification and quantification of carotenoids, tocopherols and chlorophylls in commonly consumed fruits and vegetables. Phytochemistry. 2003;62:939–47. doi: 10.1016/S0031-9422(02)00710-0. [DOI] [PubMed] [Google Scholar]
- 40.Alexander L, Grierson D. Ethylene biosynthesis and action in tomato: a model for climacteric fruit ripening. J Exp Bot. 2002;53:2039–55. doi: 10.1093/jxb/erf072. [DOI] [PubMed] [Google Scholar]
- 41.Blast2go. A bioinformatics platform for high-quality functional annotation and analysis of genomic datasets. https://www.blast2go.com/.
- 42.Matarasso N, Schuster S, Avni A. A novel plant cysteine protease has a dual function as a regulator of 1-aminocyclopropane-1-carboxylic acid synthase gene expression. Plant Cell. 2005;17:1205–16. doi: 10.1105/tpc.105.030775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gillet LC, Navarro P, Tate S, Röst H, Selevsek N, Reiter L, Bonner R, Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11:O111.016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Findlay GM, Smith MJ, Lanner F, Hsiung MS, Gish GD, Petsalaki E, Cockburn K, Kaneko T, Huang H, Bagshaw RD, Ketela T, Tucholska M, Taylor L, Bowtell DD, Moffat J, Ikura M, Li SS, Sidhu SS, Rossant J, Pawson T. Interaction domains of Sos1/Grb2 are finely tuned for cooperative control of embryonic stem cell fate. Cell. 2013;152:1008–20. doi: 10.1016/j.cell.2013.01.056. [DOI] [PubMed] [Google Scholar]
- 45.Li J, Brader G, Palva ET. Kunitz trypsin inhibitor: an antagonist of cell death triggered by phytopathogens and fumonisin b1 in Arabidopsis. Mol Plant. 2008;1:482–95. doi: 10.1093/mp/ssn013. [DOI] [PubMed] [Google Scholar]
- 46.Huang H, Qi SD, Qi F, Wu CA, Yang GD, Zheng CC. NtKTI1, a Kunitz trypsin inhibitor with antifungal activity from Nicotiana tabacum, plays an important role in tobacco's defense response. FEBS J. 2010;277:4076–88. doi: 10.1111/j.1742-4658.2010.07803.x. [DOI] [PubMed] [Google Scholar]
- 47.Tamura K, Shimada T, Ono E, Tanaka Y, Nagatani A, Higashi SI, Watanabe M, Nishimura M, Hara-Nishimura I. Why green fluorescent fusion proteins have not been observed in the vacuoles of higher plants. Plant J. 2003;35:545–55. doi: 10.1046/j.1365-313X.2003.01822.x. [DOI] [PubMed] [Google Scholar]
- 48.Shen J, Zeng Y, Zhuang X, Sun L, Yao X, Pimpl P, Jiang L. Organelle pH in the Arabidopsis endomembrane system. Mol Plant. 2013;6:1419–37. doi: 10.1093/mp/sst079. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Butelli E, De Stefano R, Schoonbeek HJ, Magusin A, Pagliarani C, Wellner N, Hill L, Orzaez D, Granell A, Jones JD, Martin C. Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Curr Biol. 2013;23:1094–100. doi: 10.1016/j.cub.2013.04.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pombo MA, Zheng Y, Fernandez-Pozo N, Dunham DM, Fei Z, Martin GB. Transcriptomic analysis reveals tomato genes whose expression is induced specifically during effector-triggered immunity and identifies the Epk1 protein kinase which is required for the host response to three bacterial effector proteins. Genome Biol. 2014;15:492. doi: 10.1186/s13059-014-0492-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hiraiwa N, Nishimura M, Hara-Nishimura I. Vacuolar processing enzyme is self-catalytically activated by sequential removal of the C-terminal and N-terminal propeptides. FEBS Lett. 1999;447:213–6. doi: 10.1016/S0014-5793(99)00286-0. [DOI] [PubMed] [Google Scholar]
- 52.Kuroyanagi M, Nishimura M, Hara-Nishimura I. Activation of Arabidopsis vacuolar processing enzyme by self-catalytic removal of an auto-inhibitory domain of the C-terminal propeptide. Plant Cell Physiol. 2002;43:143–51. doi: 10.1093/pcp/pcf035. [DOI] [PubMed] [Google Scholar]
- 53.Hara-Nishimura I, Hatsugai N, Nakaune S, Kuroyanagi M, Nishimura M. Vacuolar processing enzyme: an executor of plant cell death. Curr Opin Plant Biol. 2005;8:404–8. doi: 10.1016/j.pbi.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 54.PlantCARE. A database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. http://bioinformatics.psb.ugent.be/webtools/plantcare/html/. [DOI] [PMC free article] [PubMed]
- 55.Ito Y, Kitagawa M, Ihashi N, Yabe K, Kimbara J, Yasuda J, Ito H, Inakuma T, Hiroi S, Kasumi T. DNA-binding specificity, transcriptional activation potential, and the rin mutation effect for the tomato fruit-ripening regulator RIN. Plant J. 2008;55:212–23. doi: 10.1111/j.1365-313X.2008.03491.x. [DOI] [PubMed] [Google Scholar]
- 56.Chapman HA, Riese RJ, Shi GP. Emerging roles for cysteine proteases in human biology. Annu Rev Physiol. 1997;59:63–88. doi: 10.1146/annurev.physiol.59.1.63. [DOI] [PubMed] [Google Scholar]
- 57.Turk B, Turk D, Turk V. Protease signalling: the cutting edge. EMBO J. 2012;31:1630–43. doi: 10.1038/emboj.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.García-Lorenzo M, Sjödin A, Jansson S, Funk C. Protease gene families in Populus and Arabidopsis. BMC Plant Biol. 2006;6:30. doi: 10.1186/1471-2229-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neurath H, Walsh KA. Role of proteolytic enzymes in biological regulation. Proc Natl Acad Sci U S A. 1976;73:3825–32. doi: 10.1073/pnas.73.11.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Urban S, Dickey SW. The rhomboid protease family: a decade of progress on function and mechanism. Genome Biol. 2011;12:231. doi: 10.1186/gb-2011-12-10-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Souza GA, Godoy LM, Mann M. Identification of 491 proteins in the tear fluid proteome reveals a large number of proteases and protease inhibitors. Genome Biol. 2006;7:R72. doi: 10.1186/gb-2006-7-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dean RA, Overall CM. Proteomics discovery of metalloproteinase substrates in the cellular context by iTRAQ labeling reveals a diverse MMP-2 substrate degradome. Mol Cell Proteomics. 2007;6:611–23. doi: 10.1074/mcp.M600341-MCP200. [DOI] [PubMed] [Google Scholar]
- 63.Morrison CJ, Butler GS, Rodríguez D, Overall CM. Matrix metalloproteinase proteomics: substrates, targets, and therapy. Curr Opin Cell Biol. 2009;21:645–53. doi: 10.1016/j.ceb.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 64.Prudova A, auf dem Keller U, Butler GS, Overall CM. Multiplex N-terminome analysis of MMP-2 and MMP-9 substrate degradomes by iTRAQ-TAILS quantitative proteomics. Mol Cell Proteomics. 2010;9:894–911. doi: 10.1074/mcp.M000050-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schilling O, Overall CM. Proteomic discovery of protease substrates. Curr Opin Chem Biol. 2007;11:36–45. doi: 10.1016/j.cbpa.2006.11.037. [DOI] [PubMed] [Google Scholar]
- 66.Qin G, Wang Y, Cao B, Wang W, Tian S. Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. Plant J. 2012;70:243–55. doi: 10.1111/j.1365-313X.2011.04861.x. [DOI] [PubMed] [Google Scholar]
- 67.Fujisawa M, Nakano T, Shima Y, Ito Y. A large-scale identification of direct targets of the tomato MADS box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell. 2013;25:371–86. doi: 10.1105/tpc.112.108118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ. Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell. 2005;17:2954–65. doi: 10.1105/tpc.105.036053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moore S, Payton P, Wright M, Tanksley S, Giovannoni J. Utilization of tomato microarrays for comparative gene expression analysis in the Solanaceae. J Exp Bot. 2005;56:2885–95. doi: 10.1093/jxb/eri283. [DOI] [PubMed] [Google Scholar]
- 70.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 71.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 72.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karimi M, Inzé D, Depicker A. GATEWAYTM vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–5. doi: 10.1016/S1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 74.Weigel D, Glazebrook J. Transformation of Agrobacterium using the freeze-thaw method. CSH Protoc. 2006;2006:pdb.prot4666. [DOI] [PubMed]
- 75.Fillatti JJ, Kiser J, Rose R, Comai L. Efficient transfer of a glyphosate tolerance gene into tomato using a binary Agrobacterium tumefaciens vector. Nat Biotechnol. 1987;5:726–30. doi: 10.1038/nbt0787-726. [DOI] [Google Scholar]
- 76.Sun Q, Zhang N, Wang J, Zhang H, Li D, Shi J, Li R, Weeda S, Zhao B, Ren S, Guo YD. Melatonin promotes ripening and improves quality of tomato fruit during postharvest life. J Exp Bot. 2015;66:657–68. doi: 10.1093/jxb/eru332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saravanan RS, Rose JK. A critical evaluation of sample extraction techniques for enhanced proteomic analysis of recalcitrant plant tissues. Proteomics. 2004;4:2522–32. doi: 10.1002/pmic.200300789. [DOI] [PubMed] [Google Scholar]
- 78.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 79.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–62. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 80.Qin G, Zhu Z, Wang W, Cai J, Chen Y, Li L, Tian S. A tomato vacuolar invertase inhibitor mediates sucrose metabolism and influences fruit ripening. Plant Physiol. 2016;172:1596–611. doi: 10.1104/pp.16.01269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–14. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 82.Gan CS, Chong PK, Pham TK, Wright PC. Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ) J Proteome Res. 2007;6:821–7. doi: 10.1021/pr060474i. [DOI] [PubMed] [Google Scholar]
- 83.Sparkers IA, Runions J, Kearns A, Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc. 2006;1:2019–25. doi: 10.1038/nprot.2006.286. [DOI] [PubMed] [Google Scholar]
- 84.Lei R, Qiao W, Hu F, Jiang H, Zhu S. A simple and effective method to encapsulated tobacco mesophyll protoplasts to maintain cell viability. MethodsX. 2014;2:24–32. doi: 10.1016/j.mex.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chi W, Li J, He B, Chai X, Xu X, Sun X, Jiang J, Feng P, Zuo J, Lin R, Rochaix JD, Zhang L. DEG9, a serine protease, modulates cytokinin and light signaling by regulating the level of ARABIDOPSIS RESPONSE REGULATOR 4. Proc Natl Acad Sci U S A. 2016;113:E3568–76. doi: 10.1073/pnas.1601724113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang Y, Zhu S, Liu S, Jiang L, Chen L, Ren Y, Han X, Liu F, Ji S, Liu X, Wan J. The vacuolar processing enzyme OsVPE1 is required for efficient glutelin processing in rice. Plant J. 2009;58:606–17. doi: 10.1111/j.1365-313X.2009.03801.x. [DOI] [PubMed] [Google Scholar]
- 87.Orzaez D, Medina A, Torre S, Fernández-Moreno JP, Rambla JL, Fernández-Del-Carmen A, Butelli E, Martin C, Granell A. A visual reporter system for virus-induced gene silencing in tomato fruit based on anthocyanin accumulation. Plant Physiol. 2009;150:1122–34. doi: 10.1104/pp.109.139006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fu DQ, Zhu BZ, Zhu HL, Jiang WB, Luo YB. Virus-induced gene silencing in tomato fruit. Plant J. 2005;43:299–308. doi: 10.1111/j.1365-313X.2005.02441.x. [DOI] [PubMed] [Google Scholar]
- 89.Zhang Z, Qin G, Li B, Tian S. Knocking out Bcsas1 in Botrytis cinerea impacts growth, development, and secretion of extracellular proteins, which decreases virulence. Mol Plant Microbe Interact. 2014;27:590–600. doi: 10.1094/MPMI-10-13-0314-R. [DOI] [PubMed] [Google Scholar]
- 90.Laluk K, Mengiste T. The Arabidopsis extracellular UNUSUAL SERINE PROTEASE INHIBITOR functions in resistance to necrotrophic fungi and insect herbivory. Plant J. 2011;68:480–94. doi: 10.1111/j.1365-313X.2011.04702.x. [DOI] [PubMed] [Google Scholar]
- 91.Vizcaíno JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Ríos D, Dianes JA, Sun Z, Farrah T, Bandeira N, Binz PA, Xenarios I, Eisenacher M, Mayer G, Gatto L, Campos A, Chalkley RJ, Kraus HJ, Albar JP, Martinez-Bartolomé S, Apweiler R, Omenn GS, Martens L, Jones AR, Hermjakob H. ProteomeXchange provides globally co-ordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;30:223–6. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vizcaino JA, Côté RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J, O’Kelly G, Schoenegger A, Ovelleiro D, Pérez-Riverol Y, Reisinger F, Ríos D, Wang R, Hermjakob H. The Proteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41:D1063–9. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository (http://www.ebi.ac.uk/pride) under accession number PXD002980.