Abstract
Recessive dystrophic epidermolysis bullosa (RDEB) is a complex inherited skin disorder caused by loss-of-function mutations in the COL7A1 gene. In order for an effective treatment of this disorder to be realized, both a thorough understanding of the regulation of COL7A1 and an understanding of the underlying nature of the complications of RDEB is needed. Currently, both post-transcriptional regulation of COL7A1 and the underlying causes of fibrosis in RDEB patients are poorly understood. Here, we describe a previously unknown mechanism of regulation by which miR-29 regulates COL7A1 in a complex network: both directly through targeting its 3’ UTR at two distinct seed regions and indirectly through targeting an essential transcription factor required for basal COL7A1 expression, SP1. In turn miR-29 itself is regulated by SP1 activity and TGF-β signaling. RDEB mice express high levels of TGF-β and significantly lower miR-29 when compared to wild-type cohorts. The sustained decrease in miR-29 in RDEB skin leads to an increase of miR-29 target genes expressed in the skin, including pro-fibrotic extracellular matrix collagens. Collectively, we identify miR-29 as an important factor in both regulating COL7A1 and inhibiting TGF-β mediated fibrosis.
INTRODUCTION
Recessive dystrophic epidermolysis bullosa (RDEB) is a severe genetic skin disorder characterized by chronic skin blistering and abnormal wound healing (Nyström et al. 2013). Many long-term complications of RDEB are a result of systemic inflammation and contractile fibrosis due, in part, to an increase in TGF-β signaling (Fritsch et al. 2008). Fibrosis leads to scarring, fusion of the digits and toes, and joint contractures. A number of cell-, protein-, and gene-based therapies are underway to correct the primary defect of RDEB, but even with their successes, the RDEB disease cascade, dominated by inflammation and fibrosis, will need additional therapies (Remington et al. 2009; Wagner et al. 2010). Further understanding of the biological mechanisms driving fibrosis in RDEB is required for any meaningful therapy.
The underlying defect of RDEB is loss-of-function mutations in the COL7A1 gene, which encodes for the structural protein, type VII collagen (C7) (Kern et al. 2009). C7 is the major component of anchoring fibrils, the function of which is to establish and maintain structural adhesion between the dermis and the epidermis. C7 is produced in the skin by both keratinocytes and fibroblasts, and then deposited at the dermal-epidermal junction where it plays a major role in skin integrity and wound repair. Regulation of the COL7A1 gene in keratinocytes and fibroblasts occurs at the transcriptional level through SP1, a transcriptional factor that is responsible for basal expression of many genes lacking a canonical TATA box (Vindevoghel et al. 1997). Further regulation of COL7A1 occurs during various forms of skin injury and wound healing. On the transcriptional level, COL7A1 is upregulated by TGF-β signaling via SMAD3/4 activity (Vindevoghel 1998). During inflammation, IL1β or TNF-α signaling leads to an increase in COL7A1 expression in fibroblasts but a decrease in COL7A1 expression in keratinocytes (Kon et al. 2005). However, post-transcriptional regulation of COL7A1 in the context of wound healing and fibrosis is poorly understood.
Fibrosis in other settings, such as pulmonary and renal fibrosis, is influenced by TGF-β activity (Qin et al. 2011). Pathological fibrosis in these settings is also heavily dependent on micro RNA (miR) activity, most importantly, miR-29(Parker et al. 2014). The miR-29 family has been shown to regulate extracellular matrix during fibrosis: reduction in miR-29 levels results in an increase in ECM proteins, which subsequently leads to fibrosis. In most cases, TGF-β signaling is responsible for the reduction in miR-29 levels during fibrosis, and either targeting the TGF-β pathway or artificially increasing miR-29 activity in the context of fibrosis has been shown to reduce disease severity (Zhou et al. 2012).
Here we demonstrate that miR-29 directly regulates COL7A1 (in part via targeting the 3’ UTR), and decreases SP1 expression (which leads to indirect regulation of COL7A1 transcription). We propose a mechanism whereby healthy wound healing in the skin leads to an increase in COL7A1 expression through TGF-β mediated repression of miR-29; whereas in the context of RDEB, pathological changes in TGF-β expression and long-term reduction in miR-29 levels promote fibrosis.
RESULTS
Identification of potential miR-COL7A1 interactions
To identify miRs most relevant to COL7A1 regulation, we utilized miR-mRNA target prediction software. Bioinformatics software tools are a well-established approach for predicting miR-mRNA interactions in-silico (Lewis et al., 2005). Said prediction tools rely on established interactions between miRs and their respective mRNA targets (Friedman et al., 2009). This interaction involves a 6–8 bp sequence in the mRNA target complementary to the 5’ end of the miR known as the seed region. Highly conserved seed regions in the 3’ UTR of an mRNA are hallmarks of genuine miR-mRNA interactions, representing evolutionary pressure to conserve miR regulation of a particular target mRNA.
We used prediction software to investigate whether the 3’ UTR of COL7A1 revealed any seed regions for known miRs, preferentially focusing on regions that were highly conserved among mammalian species (http://www.targetscan.org/). Targetscan software analysis predicted two seed regions in the 3’ UTR of COL7A1 that are broadly conserved among mammalian species for miR-29 family members. These two seed regions (Figure 1A) are located at 77–84 bp and 290–296 bp of the COL7A1 3’ UTR. Both sites are complementary to the 5’ end of miR-29 and are predicted to base-pair with miR-29. Furthermore, miR-29 has been shown to be expressed in normal dermal fibroblasts (Cheng et al., 2013), cells found near dermal-epidermal junction where fibroblasts secrete C7 and other ECM components. We focused on miR-29 above other potential candidates for further analysis due to the two predicted seed regions in the COL7A1 3’ UTR and prior studies demonstrating miR-29 regulation of other collagens (Qin et al., 2011).
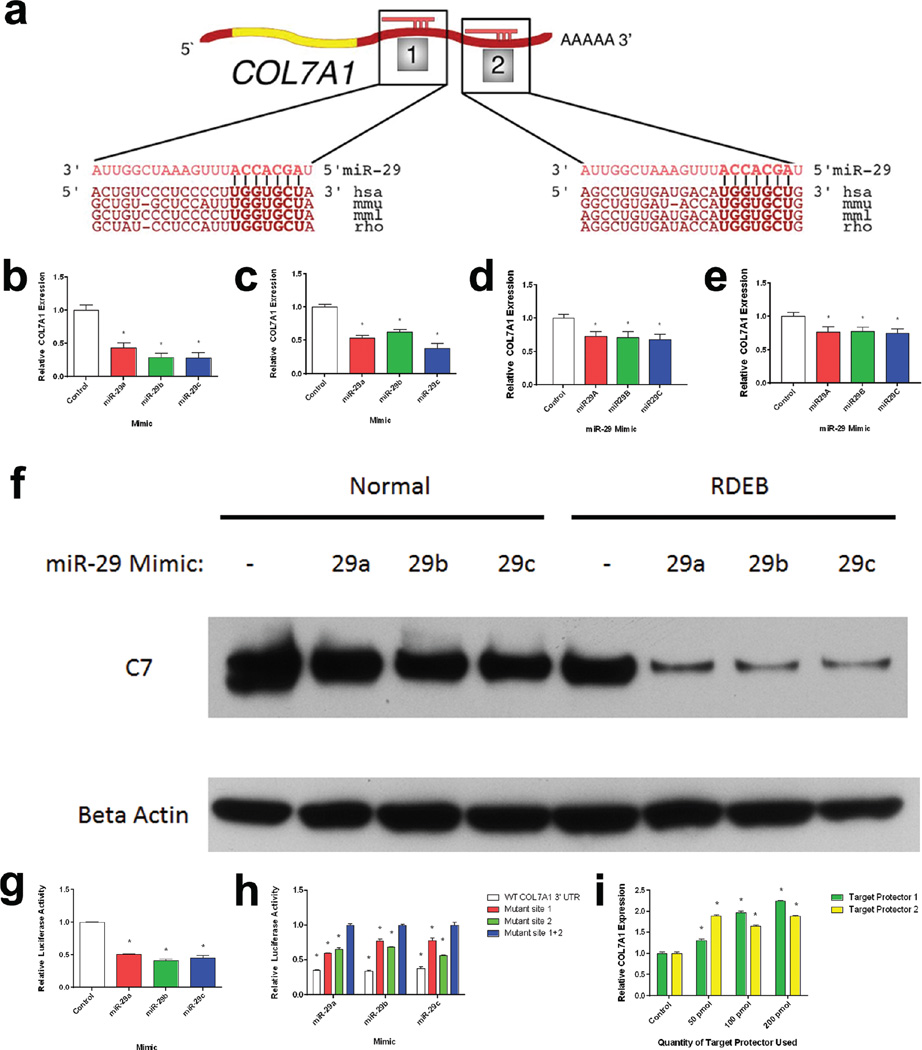
Figure 1. a–i: miR-29 Regulates COL7A1.
(a) A schematic alignment between miR-29c and two regions of the COL7A1 3’ UTR: 63–84 bp and 275–297 bp, respectively. Alignment based on Targetscan miR-mRNA predictions. Normal (b) and RDEB (c) dermal fibroblasts as well as normal (d) and RDEB (e) keratinocytes were transiently transfected with miR-29 mimics. Quantitative RT-PCR analysis of COL7A1 expression relative to GAPDH expression 24 h after transfection. N = 3. Values represent mean ± SE. *P < 0.05 (f) Normal and RDEB fibroblasts were transfected with miR-29 mimics and assayed for type VII collagen protein expression 72 h post transfection. Blot was stained for type VII collagen and Beta-actin as a loading control. (g) HEK-293 cells were co-transfected with a miR-29 mimic and a plasmid containing a firefly luciferase construct directly downstream of the COL7A1 3’ UTR and an independently regulated renilla luciferase construct. Luciferase levels were measured 24 h after transfection. Firefly luciferase levels were normalized to renilla luciferase levels. N = 3. Values represent mean ± SE. *P < 0.05. (h) 293T cells were co-transfected with both a plasmid containing a firefly luciferase construct containing the COL7A1 3’ UTR region and an independently regulated renilla luciferase construct in combination with a miR-29 mimic. Luciferase levels were measured 24 h post-transfection. COL7A1 3’UTR plasmids contained either wild-type sequences or mutations in either or both of the miR-29 seed regions. Firefly luciferase levels were normalized to renilla luciferase levels. Values represent mean ± SE. *P < 0.05, in comparison to mutant site 1+2. (i) HEK-293 cells were transiently transfected with miR-29 COL7A1 3’ UTR target protectors for either of the predicted miR-29 seed regions on the COL7A1 3’ UTR (Invitrogen). 24 h after transfection, qRT-PCR analysis was performed to determine COL7A1 expression. Quantitative RT-PCR analysis of COL7A1 expression relative to GAPDH expression 24 h after transfection. N = 3. Values represent mean ± SE. *P < 0.05. hsa, human; mmu, mouse; mml, rhesus; rho, rat; RDEB, recessive dystrophic epidermolysis bullosa; RT-PCR, reverse transcriptase PCR; SE, standard error.
miR-29 regulation of C7 collagen
The miR-29 family consists of three miRs (miR-29a/b/c) synthesized at two different genomic loci on chromosomes 1 and 7 (Jiang et al., 2014). The mature forms of each miR-29 (3’) species share the same 5’ sequence necessary for sequence-specific miR-mRNA interaction and consequently share overlapping function in regulating specific mRNA targets. It has been previously reported that miR-29 is responsible for regulating expression of multiple collagens, including COL1A1, COL1A2, COL3A1, COL4A1, COL5A1, COL5A2, COL15A1 (Luna et al., 2009, Liu et al., 2010, Cheng et al., 2013, Yang et al., 2013). Given this information and the results from target prediction software, we sought to determine whether miR-29 was capable of regulating COL7A1. To investigate this, we transfected synthetic mimics of miR-29 into dermal fibroblasts and assayed them for COL7A1 expression using qRT-PCR 24 h post-transfection. Transfection of dermal fibroblasts and keratinocytes with miR-29 mimics led to a decrease in COL7A1 expression levels in comparison to mimic controls (Figure 1B–1E). We observe similar results when measuring C7 protein following miR-29 mimic transfection as well (Figure 1F). This demonstrates that miR-29 activity inhibits COL7A1 expression.
To further define the relationship between COL7A1 and miR-29, we aimed to determine whether the regulation of miR-29 on COL7A1 was direct or indirect, as previous studies have demonstrated that miR-29 is capable of regulating other collagens either directly or indirectly. For example, in pulmonary fibrosis miR-29 has been shown to directly target COL1A1 via interaction with its 3’ UTR (Cushing et al., 2011). Conversely, in renal fibrosis miR-29 was shown to indirectly increase type I collagen production through the targeting of SP1, a transcription factor required for basal expression of COL1A1 (Cheng et al., 2013, Chen et al., 2014, Jiang et al., 2013). To determine whether miR-29 was capable of targeting the COL7A1 transcript, we generated luciferase reporter plasmids containing the 3’ UTR of COL7A1. Co-transfection of 293T cells with the COL7A1 3’ UTR reporter plasmid and synthetic miR-29 mimics led to a decrease in firefly luciferase activity in comparison to mimic controls (Figure 1G). This demonstrates that miR-29 is capable of regulating COL7A1, at least in part through direct interactions with its 3’ UTR.
As miR-29 is predicted to interact with the COL7A1 3’ UTR at two distinct seed regions (Figure 1A), we wished to determine how these seed regions mediate miR-29 regulation of C7. To that end we generated reporter constructs in which either or both of the seed regions were mutated to disrupt the predicted base-pairing between miR-29 and the 3’ UTR complementary sequence. Mutations in either of the seed sequences reduced the effect of miR-29 on the COL7A1 3’ UTR reporter construct, whereas mutations in both seed sequences abolished miR-29’s effect on the COL7A1 3’ UTR reporter altogether (Figure 1H). We found no significant differences in luciferase levels between experiments in which miR-29 seed sequence 1 was mutated versus the reporter construct in which seed sequence 2 was mutated. This demonstrates that both miR-29 seed sequences in the 3’ UTR of COL7A1 are required in regulating COL7A1 expression.
To further examine the functional specificity of each of the miR-29 seed sequences in COL7A1, we utilized 3’ UTR target protectors. These synthetic oligonucleotides consist of a single-stranded RNA molecule that is complementary to the 3’ UTR of interest. Specifically, we developed two distinct target protectors that effectively masked each seed region required for direct miR-29 regulation of the COL7A1 transcript. Upon transfection of these target protectors into dermal fibroblasts, we saw a dose-dependent increase in COL7A1 expression in comparison to non-transfected controls (Figure 1I). Collectively, these data demonstrate that miR-29 regulates C7 through directly targeting its 3’ UTR and confirm that both seed regions contribute to miR-29 regulation of C7.
miR-29 regulates COL7A1 indirectly via targeting its transcription factor SP1
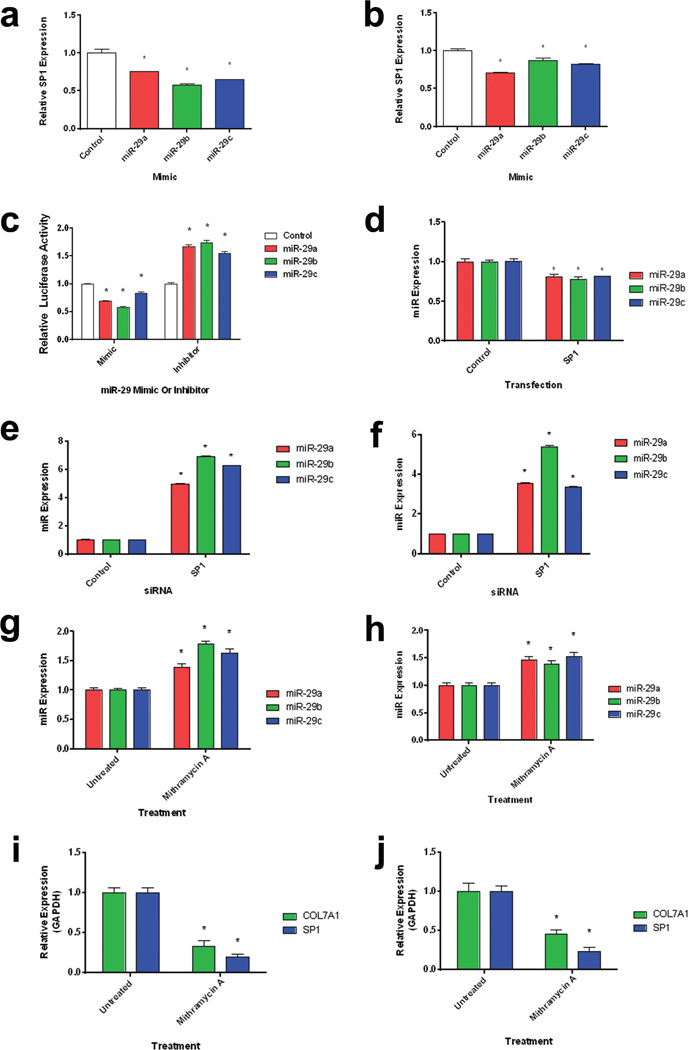
Previous studies have shown that miR-29 is capable of negatively regulating SP1 (Jiang et al., 2013). miR-29 has been shown to regulate COL1A1 directly in fibroblasts associated with fibrosis, as well as indirectly through SP1 in renal tubular epithelial cells (Cheng et al., 2013, Jiang et al., 2013). SP1 is a ubiquitously expressed transcription factor required for basal transcription of COL7A1; therefore factors that influence SP1 transcriptional activity are capable of indirectly influencing COL7A1 expression (Vindevoghel et al., 1997)(Vindevoghel et al. 1997). To investigate whether miR-29 was capable of regulating SP1, we transfected normal and RDEB dermal fibroblasts with synthetic mimics of miR-29 and assayed SP1 gene expression 24 h post-transfection. Transfection of dermal fibroblasts with miR-29 mimics led to a decrease in SP1 expression levels in comparison to controls (Figure 2A–B). To further refine the mechanism of SP1 regulation by miR-29, we utilized a reporter that contains five tandem repeats of the SP1 transcriptional response element and thus directly reflects the transcriptional activity of SP1. When we co-transfected this reporter construct with miR-29 mimics, we saw a decrease in relative luciferase activity (Figure 2C). Conversely, when we co-transfected this reporter with inhibitors of miR-29, we saw an increase in luciferase activity (Figure 2C). This confirms that miR-29 is capable of regulating genes indirectly through targeting the transcription factor SP1.
Figure 2. a–j: SP1 and miR-29 co-regulate each other and COL7A1.
Normal (a) and RDEB (b) dermal fibroblasts were transfected with miR-29 mimics. Quantitative RT-PCR analysis of SP1 expression relative to GAPDH expression 24 h after transfection. N = 3. Values represent mean ± SE. *P < 0.05. (c) 293T cells were transiently co-transfected with a luciferase reporter assay containing tandem repeats of the SP1 transcriptional response element in combination with either miR-29 mimics or inhibitors (Invitrogen). Luciferase levels were measured 24 h post-transfection. Firefly luciferase levels were normalized to renilla luciferase levels. N = 3. Values represent mean ± SE. *P < 0.05. (d) 293T cells were transfected with an SP1 overexpression vector for 24 h. Quantitative RT-PCR analysis of miR-29 levels relative to U6 levels. N = 3. Values represent mean ± SE. *P < 0.05. (e) Normal and (f) RDEB fibroblasts were transiently transfected with SP1 siRNA for 48 h. Quantitative RT-PCR analysis of miR-29 levels relative to U6 levels. N = 3. Values represent mean ± SE. *P < 0.05. Normal (g) and RDEB (h) fibroblasts were treated with mithramycin A (an inhibitor of SP1 binding) for 48 h. Quantitative RT-PCR analysis of miR-29 levels relative to U6 levels. N = 3. Values represent mean ± SE. *P < 0.05. Inhibition of SP1 binding decreases COL7A1 and SP1 Expression. Normal (i) and RDEB (j) fibroblasts were treated with mithramycin A (an inhibitor of SP1 binding) for 48 h. Quantitative RT-PCR analysis of COL7A1 and SP1 levels relative to GAPDH. N = 3. Values represent mean ± SE. *P < 0.05. RDEB, recessive dystrophic epidermolysis bullosa; RT-PCR, reverse transcriptase PCR; SE, standard error.
SP1 and miR-29 exhibit a co-inhibitory loop
Previous studies have shown that SP1 is capable of negatively regulating miR-29 expression (Liu et al., 2010, Li et al., 2012). To determine if SP1 regulates miR-29, we transfected 293-T cells with an SP1-expression cassette and measured miR-29 levels 24 h post transfection. Transfection with the SP1 cassette led to a decrease in miR-29 levels (Figure 2D). Conversely, transfection with a short interfering RNA targeting SP1 led to an increase in miR-29 levels (Figure 2E–F), demonstrating that SP1 regulates miR-29. To further delineate the roles of SP1 and miR-29 in regulation of COL7A1, we utilized mithramycin A, which binds to GC-rich sequences and subsequently inhibits the binding and activity of SP1 (Sleiman et al., 2011). Normal and RDEB fibroblasts treated with mithramycin A showed increased miR-29 expression (Figure 2G–H). Conversely, when normal and RDEB fibroblasts were treated with mithramycin A, we saw a decrease in both SP1 and COL7A1 expression (Figure 2I–J). These data demonstrate a negative regulatory loop by which miR-29 targets SP1 and leads to a decrease in SP1 expression and activity, whereas SP1 transcriptional activity leads to a decrease in miR-29 levels. Furthermore, since both SP1 and miR-29 are capable of regulating COL7A1 expression, changes in miR-29 or SP1 levels could amplify changes in COL7A1 expression due to changes in the miR-29/SP1 regulatory loop.
miR-29 levels are decreased in RDEB skin
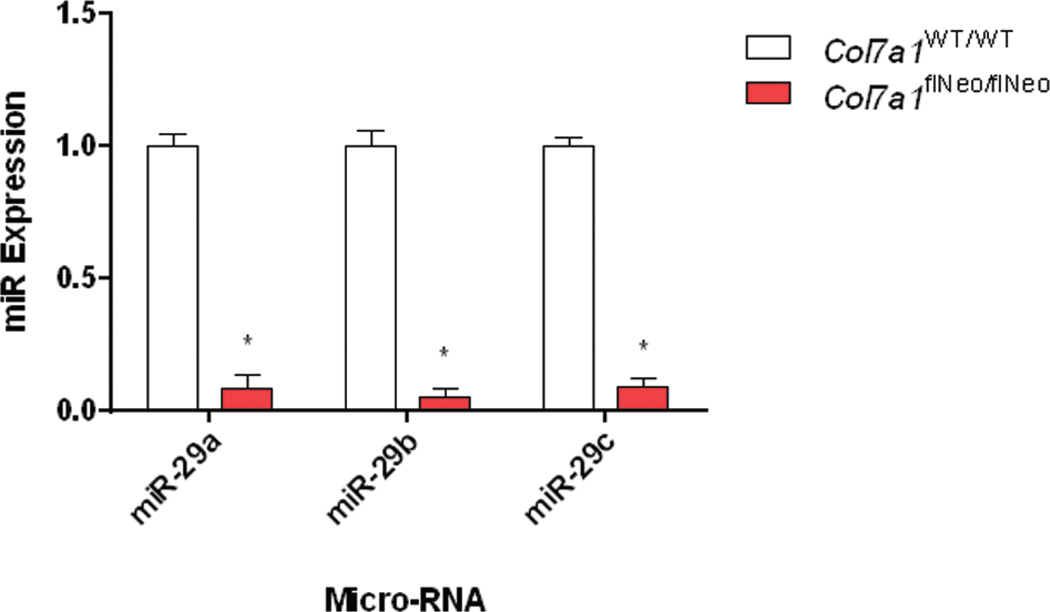
Hallmarks of the phenotype of RDEB in the skin include chronic skin blistering and scarring with non-productive wound healing. This is pronounced in the extremities, where the progression of chronic blistering in the digits leads to pseudosyndactyly. Characterization of these regions revealed TGF-β accumulation and aberrant contractile fibrosis in the dermis of Col7aflNeo/flNeo mice (Fritsch et al., 2008). Treatment of the RDEB mouse model using losartan led to a decrease in TGF-β signaling as well as a reduction in fibrosis and fusion of the digits (Nystrom et al., 2015). Based on work of others identifying miR-29 as a key player in TGF-β mediated fibrosis (van Rooij et al., 2008, Cushing et al., 2011, Qin et al., 2011)(van Rooij et al. 2008; Cushing et al. 2011; Qin et al. 2011), we set out to determine the levels of miR-29 in RDEB skin in comparison to healthy controls. We found that the miR-29 levels were significantly lower from skin isolated from COL7A1flNeo/flNeo mice in comparison to wildtype controls (Figure 3).
Figure 3. COL7A1flNeo/flNeo homozygous mice exhibit reduced miR-29 expression.
Small RNA was isolated from skin samples from both COL7A1flNeo/flNeo mice and wild-type controls. Quantitative RT-PCR analysis of miR-29 levels relative to U6 levels. N = 3. Values represent mean ± SE. *P < 0.05. RT-PCR, reverse transcriptase PCR; SE, standard error.
miR-29 is downregulated in normal dermal and RDEB fibroblasts and keratinocytes via TGF-β signaling
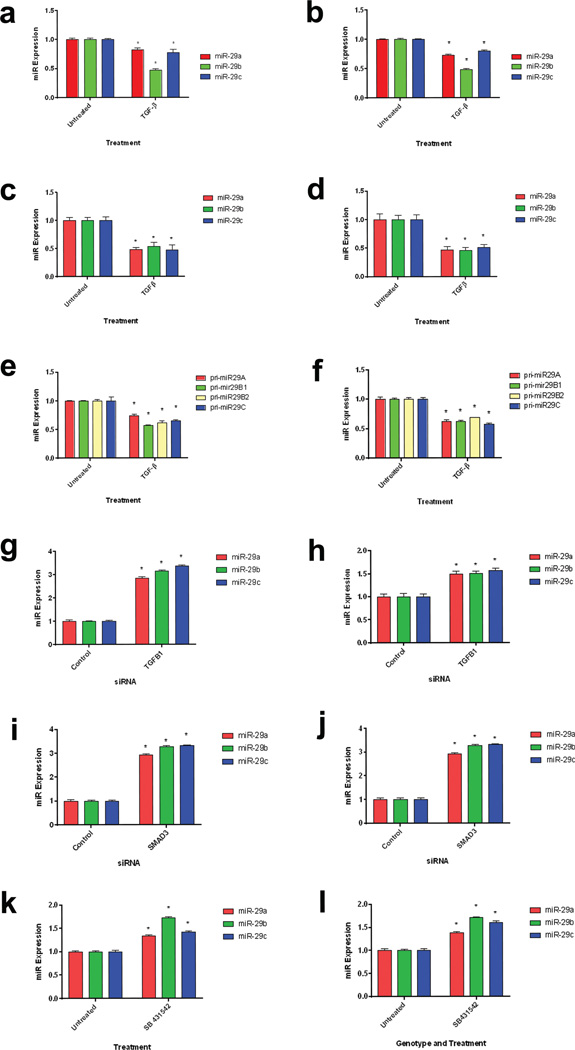
C7 expression is increased in response to injury to the skin (Vindevoghel et al., 1998, Nakano et al., 2001). In particular, C7 expression has been shown to increase following activation of TGF-β pathway (Vindevoghel et al., 1998). TGF-β signaling leads to phosphorylation of downstream effector SMAD proteins, of which, activated SMAD3/4 bind to COL7A1 promoter and promote COL7A1 transcription. Furthermore, TGF-β activation influences RDEB disease severity through increased type 1 collagen and decreased decorin expression (Odorisio et al., 2014). Previous studies have also demonstrated that TGF-β signaling results in a decrease in miR-29 expression through inhibition of transcription via binding of SMAD3 to the miR-29 promoter region (Qin et al., 2011, Zhou et al., 2012). SMAD activity on the miR-29 promoter region results in a decrease of the primary miR as well as downstream mature miR levels. Therefore, we sought to investigate if miR-29 levels changed in response to TGF-β in normal and RDEB fibroblasts and keratinocytes. Following exposure of normal and RDEB dermal fibroblasts and keratinocytes to TGF-β1, miR-29 levels decreased in comparison to untreated controls (Figure 4A–D). To determine if primary miR-29 levels were decreased in RDEB and normal fibroblasts following TGF-β exposure, we exposed RDEB fibroblasts for 24 h and subsequently assayed for primary miR-29 levels. Following exposure to TGF-β1, we saw a decrease in pri-miR-29A, pri-miR-29B1, pri-miR29-B2 and pri-miR-29C levels (Figure 4E–F). To further investigate whether endogenous levels of TGF-β signaling were sufficient for miR-29 regulation, we utilized siRNA knockdown of both TGFB1 as well as its downstream transcription factor, SMAD3. Upon transfection of both TGFB1 (Figure 4 G–H) and SMAD3 (Figure 4 I–J) knockdown we see an increase in miR-29 levels 48 h post transfection. Furthermore, inhibition of TGF-β receptor signaling by use of a TGF-β receptor inhibitor (SB 431542) results in an increase in miR-29 levels 24 h-post treatment (Figure 4 K–L). In vivo, however, TGF-β injection did not result in a significant decrease in miR-29 levels. This is likely due to multiple factors in RDEB skin contributing to miR-29 regulation as well as prolonged TGF-β signaling (Supplemental Figure 1). These data demonstrate that TGF-β signaling reduces miR-29 levels, which indirectly increases COL7A1 levels through derepression of miR-29 regulation. Furthermore, they demonstrate that TGF-β signaling alters COL7A1 expression at both the transcriptional and post-transcriptional level. This is further enhanced through the activity of SP1, both through COL7A1 transcription and miR-29 inhibition (Figure 5).
Figure 4. a–l: TGF-β downregulates miR-29.
Both normal (a) and RDEB (b) fibroblasts as well as normal (c) and RDEB (d) keratinocytes were treated with TGF-β1 for 24 h. Quantitative RT-PCR analysis of miR-29 levels relative to U6 levels. N = 3. Values represent mean ± SE. *P < 0.05. Normal (e) and RDEB (f) fibroblasts were treated with TGF-β1 for 24 h. Quantitative RT-PCR analysis of primary miR-29 levels relative to GAPDH levels. N = 3. Values represent mean ± SE. *P < 0.05. Normal (g) and RDEB (h) fibroblasts were transiently transfected with TGF-β1 siRNA for 48 h. Quantitative RT-PCR analysis of miR-29 levels relative to U6 levels. N = 3. Values represent mean ± SE. *P < 0.05. Normal (i) and RDEB (j) fibroblasts were transiently transfected with SMAD3 siRNA for 48 h. Quantitative RT-PCR analysis of miR-29 levels relative to U6 levels. N = 3. Values represent mean ± SE. *P < 0.05. Normal (k) and RDEB (l) fibroblasts were treated with a TGF-β1 receptor inhibitor, SB431542. Quantitative RT-PCR analysis of miR-29 levels relative to U6 levels. N = 3. Values represent mean ± SE. *P < 0.05. RDEB, recessive dystrophic epidermolysis bullosa; RT-PCR, reverse transcriptase PCR; SE, standard error.
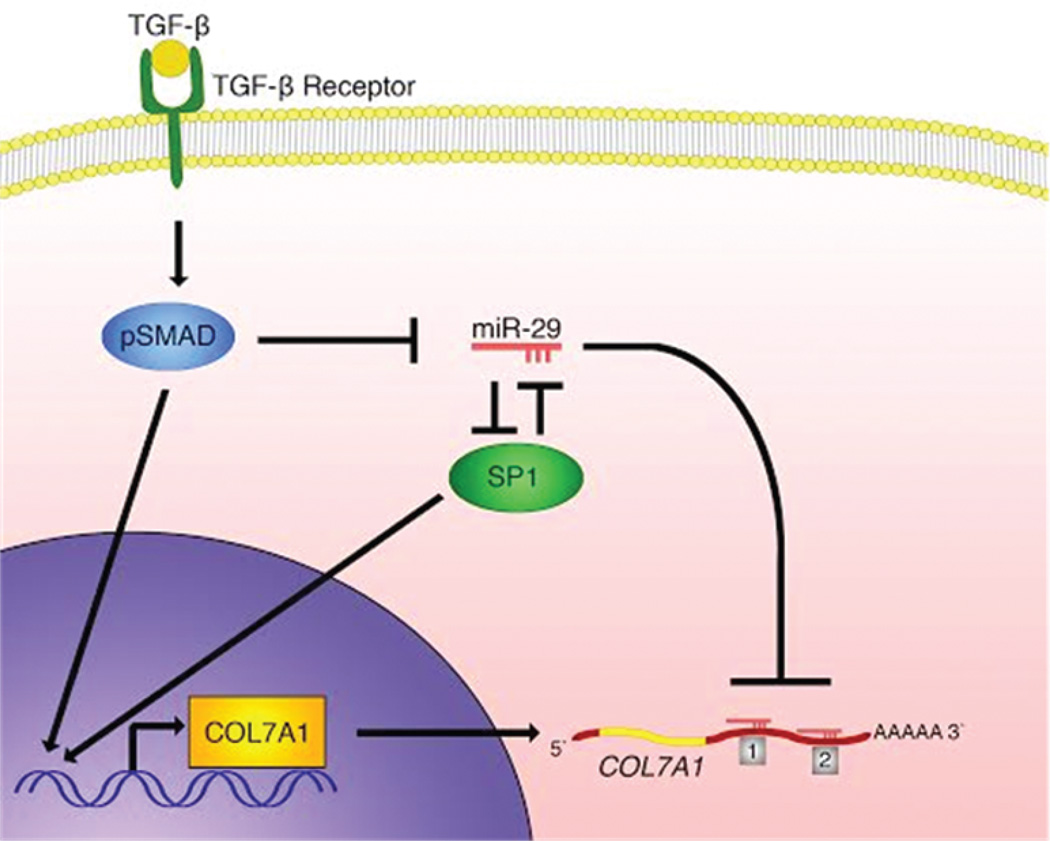
Figure 5. miR-29 Regulation of COL7A1.
A schematic diagram illustrating miR-29 regulation of COL7A1. miR-29 regulates COL7A1 directly through two seed sequences in the COL7A1 3’ UTR as well as indirectly through regulating its basal transcription factor, SP1. SP1 and miR-29 regulate each other in a negative fashion, establishing a co-inhibitory loop. During wound healing, TGF-β signaling leads to phosphorylation and activation of downstream SMAD transcription factors, which results in a subsequent decrease in miR-29 levels and an increase in SP1 and COL7A1 expression.
DISCUSSION
Previous reports have focused on the role of COL7A1 expression at the transcriptional level. Here, we demonstrate a mechanism by which COL7A1 is regulated at the transcriptional and post-transcriptional level by miR-29. Further, we confirm that TGF-β signaling results in a decrease in miR-29 levels, antagonizing regulatory control of miR-29 on COL7A1 and leading to de-repression of COL7A1. This regulation is coupled with an indirect regulation of miR-29 on COL7A1 through regulation of its basal transcription factor SP1. SP1 and miR-29 act in a negative co-regulatory loop that is perturbed by TGF-β signaling. Reduction of miR-29 in response to TGF-β signaling likely functions in skin homeostasis to increase ECM production during wound healing and re-epithelialization.
C7 plays a crucial role in both maintaining structural integrity of the skin as well as assisting in wound closure (Nystrom et al., 2013). Therefore, increasing C7 expression at both the transcriptional and post-transcriptional levels would be beneficial for tissue repair. However, prolonged reduction in miR-29 levels may also lead to complications such as the fibrosis that leads to joint contractures and pseudosyndactyly (Qin et al., 2011, Zhou et al., 2012). A mouse model for RDEB has demonstrated an increase in TGF-β1 in RDEB skin, resulting in an accumulation of myofibroblasts in the extremities which is responsible for driving contractile fibrosis in the digits (Fritsch et al., 2008). This is likely further exacerbated by a reduction in miR-29, which leads to an increase in multiple collagens, mainly COL1A1 and COL3A1. Furthermore, prolonged reduction in miR-29 levels may promote an increase in other miR-29 targets associated with the development of squamous cell carcinoma (SCC), such as MMP2 and SP1 (Jia et al., 2014, Lu et al., 2014).
In the context of wound healing, miR-29 is among a host of other miRs in the skin that regulate the wound healing process. During wound healing, miR-21 has been shown to regulate migration and re-epithelialization of keratinocytes within the wound bed (Yang et al., 2011). Also, miR-31 has also been shown to regulate keratinocyte proliferation and migration, although through regulating a different set of genes than miR-21 (Li et al., 2015). In this respect, miR-29 is among a network of other miRs that likely function in concert to regulate various aspects of wound healing, and how these miRs function together remains to be seen. Also, there are likely other miRs that are capable of regulating COL7A1, as well as other miRs that are dysregulated in the context of RDEB and may play a role in RDEB pathology as well.
While understanding COL7A1 regulation of miR-29 in the context of RDEB is important to understanding the disease pathology and nature or RDEB, understanding how other targets of miR-29 are expressed in the context of RDEB may prove to be useful as well. In the context of fibrosis, it is likely that dermal collagens such as COL1A1 and COL3A1 play a major role in fibrosis and are influenced by miR-29 regulation. Furthermore, miR-29 has been shown influence DNA methylation through targeting TET1 and TDG (Morita et al., 2013) as well as DNMT3A, DNMT3B and DNMT1 (Garzon et al., 2009). Whether there is any significant changes in DNA methylation in RDEB remains to be seen, but if other miR-29 targets are regulated in the skin in similar fashion to COL7A1 and SP1, there is likely to be dysregulation of DNA methylation in RDEB due to miR-29 activity.
Ideally, therapies for RDEB will result in an increase in functional, stable anchoring fibrils while reducing the amount of fibrosis and the incidence of SCC. Future clinical applications focused on increasing functional C7 in RDEB skin through manipulation of miR-29 will require a targeted approach by separating miR-29 regulation of COL7A1 versus reducing overall miR-29 levels. Specifically, COL7A1 target protectors, which disrupt miR-29 regulation of COL7A1 without interfering with miR-29 regulation of other targets genes, may be the preferred approach. Furthermore, miR-29 may be a suitable biomarker for overall skin integrity, and measuring miR-29 levels of the skin of RDEB patients may be an indicator for fibrosis.
MATERIALS AND METHODS
Cell culture
RDEB, normal human dermal fibroblasts, and HEK-293T cells were maintained in Dulbecco’s modified Eagle media supplemented with 10% fetal bovine serum, 100 U/ml nonessential amino acids, and 0.1 mg/ml each of penicillin and streptomycin (Invitrogen, Carlsbad, CA). Normal and RDEB keratinocytes were maintained in Epilife medium supplemented with 60 µM calcium, 1% Epilife Defined Growth Supplement, and 1% gentamicin/amphotericin solution (Invitrogen, Carlsbad, CA).
Inhibitors
Mithramycin A (150 ng/mL, GR305-0001, VWR Scientific, Radnor, PA) or SB 431542 (10 µM/mL, S-4317, Sigma-Aldrich, St. Louis, MO) were added directly to fibroblast media and cells were incubated for 24–48 hours prior to RNA or small RNA isolation.
Fibroblast transfection
RDEB and normal human dermal fibroblasts, as well as RDEB and normal keratinocytes, were transiently transfected utilizing the Neon Transfection System (Invitrogen). 2 × 105 fibroblasts were transfected with either 90 pmol of miRvana miRNA mimic (miR-29a, miR-29b, mir-29c, or mimic control; Invitrogen) or miRvana miRNA inhibitor (miR-29a, miR-29b, mir-29c, or mimic control; Invitrogen) or siRNA select (TGFβ1:s14056, SMAD3:s8402, and SP1:s13320) using the following settings: 1,500V, 20 ms pulse width, 1 pulse. 2 × 105 keratinocytes were transfected with 90 pmol of miRvana mimics using the following settings: 850V, 30 ms, 2 pulses.
293T transfection
2.5 × 105 HEK-293T cells were transiently transfected using Lipofectamine 2000 reagent (Invitrogen). For the luciferase assays, 1 µg of each luciferase reporter was used in combination with either 90 pmol of miR-29 miRvana mimic or inhibitor. For the target protector assays, either a target protector control or various concentrations (50 pmol, 100 pmol, and 200 pmol) of miScript Target Protector (Qiagen, Valencia, CA), which were designed to mask either the first or second mir-29 seed sequence in the COL7A1 3’ UTR, were used for transfection. Sequence for COL7A1 target protector 1: CCCACTGTCCCTCCCCTTGGTGCTAGAGGCTTGTGTGCAC and COL7A1 target protector 2: CCAAGCCTGTGATGACATGGTGCTGATTCTGGGGGCATT.
RDEB mouse strain miR analysis
All animal studies were approved by the University of Minnesota Institutional Animal Care and Use Committee. The RDEB hypomorphic mouse model, C57Bl/6-TgH(COL7A1flNeo)288LBT, was used for microRNA analysis. Full-thickness skin samples were isolated from RDEB hypomorphic pups and wild-type littermate controls. Samples were placed in RNAlater (Qiagen) prior to small RNA extraction. Skin samples were first processed using a Tissue-Tearor rotor/stator homogenizer for 1 minute in mirPremier miR lysis buffer prior to small RNA Isolation.
TGF-β injection
C57Bl/6 wild-type mice were injected with 800 ng carrier-free mouse TGF-β1 (Cell Signaling Technology, Danvers, MA) re-suspended in 20 µl PBS. Injections were done with a micro needle subcutaneously in the neck of neonatal mice. Following 3 daily injections, skin samples were taken from the injection site and processed as stated above for small RNA isolation.
RT-PCR
RNA was isolated using the RNeasy Plus Mini Kit (Qiagen). Subsequent first strand cDNA synthesis was performed using the SuperScript Vilo cDNA Synthesis Kit (Invitrogen). RT-PCR was performed using Taqman Gene Expression Assays and Taqman Universal Master Mix II, no UNG (Invitrogen). RT-PCR was performed on the StepOnePlus Real-Time PCR System (Applied Biosystems, Waltham, MA). The Taqman Gene Expression assays used were: COL7A1 (Hs00164310_m1), SP1 (Hs00916521_m1), miR-29a-pri (Hs03302672_pri), miR-29b1-pri (Hs03302748_pri), miR-29b2-pri (Hs03302750_pri), and miR-29c-pri (Hs04225365_pri). GAPDH (Hs02758991_g1) was used as an endogenous control. Small RNA from both mouse skin samples and cell culture was isolated using the mirPremier microRNA Isolation Kit (Sigma-Aldrich). Micro RNA was reverse transcribed using the TaqMan® MicroRNA Reverse Transcription Kit (Invitrogen). The Taqman Micro RNA assays used were: miR-29a (002112), miR-29b (000413) and miR-29c (00587). U6 (001973) was used as a positive control.
Immunoblot
Normal and RDEB fibroblasts transfected with miR-29 mimics or controls as stated above were incubated for 72 h prior to cell lysis. Cells were lysed in RIPA buffer containing a protease inhibitor cocktail (Sigma-Aldrich). Lysates were clarified and measured for protein concentration using a BCA protein assay prior to loading. Equal quantities of protein were loaded onto 3–8% tris-acetate gel. Following electrophoresis, protein was transferred onto a nitrocellulose membrane and incubated with anti-C7 antibody (a kind gift provided by Drs. David Woodley and Mei Chen) or anti-Beta Actin (A2228, Sigma-Aldrich) antibody overnight. The following day, membranes were incubated with goat anti-rabbit HRP-conjugated secondary antibody (sc-2005, Santa Cruz Biotechnology, Santa Cruz, CA). After incubation with secondary antibody, blots were developed using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fischer Scientific, Waltham, MA) and developed on X-ray film for imaging.
Luciferase reporter plasmids
Luciferase plasmid reporter constructs containing both the wild-type sequence of the COL7A1 3’ UTR or mutant derivatives containing changes in either or both of the miR-29 seed sequences were developed utilizing the pMIRglo Dual Luciferase Vector (Promega, Madison, WI). MiR-29 seed sequences (Figure 1) were mutated as follows: UGGUGCU to ACCACGA for both site 1 and site 2.
Statistics
All data are presented as mean ± SD of three independent biological replicates or more. Student’s T test was used to determine the significance between two groups or between an experimental variable and a control. P-values ≤ 0.05 were considered to be statistically significant.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the National Institutes of Health (R01 AR063070 and R01AR059947), and US Department of Defense (W81XWH-12-1-0609).
Abbreviations
- C7
type VII collagen
- miR
micro RNA
- RDEB
recessive dystrophic epidermolysis bullosa
- SCC
squamous cell carcinoma
Footnotes
CONFLICTS OF INTEREST
The authors state no conflict of interest.
REFERENCES
- Chen HY, Zhong X, Huang XR, Meng XM, You Y, Chung AC, et al. MicroRNA-29b inhibits diabetic nephropathy in db/db mice. Mol Ther. 2014;22:842–853. doi: 10.1038/mt.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Wang Y, Wang D, Wu Y. Identification of collagen 1 as a post-transcriptional target of miR-29b in skin fibroblasts: therapeutic implication for scar reduction. Am J Med Sci. 2013;346:98–103. doi: 10.1097/MAJ.0b013e318267680d. [DOI] [PubMed] [Google Scholar]
- Cushing L, Kuang PP, Qian J, Shao F, Wu J, Little F, et al. miR-29 is a major regulator of genes associated with pulmonary fibrosis. Am J Respir Cell Mol Biol. 2011;45:287–294. doi: 10.1165/rcmb.2010-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch A, Loeckermann S, Kern JS, Braun A, Bosl MR, Bley TA, et al. A hypomorphic mouse model of dystrophic epidermolysis bullosa reveals mechanisms of disease and response to fibroblast therapy. J Clin Invest. 2008;118:1669–1679. doi: 10.1172/JCI34292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia LF, Huang YP, Zheng YF, Lyu MY, Wei SB, Meng Z, et al. miR-29b suppresses proliferation, migration, and invasion of tongue squamous cell carcinoma through PTEN-AKT signaling pathway by targeting Sp1. Oral Oncol. 2014;50:1062–1071. doi: 10.1016/j.oraloncology.2014.07.010. [DOI] [PubMed] [Google Scholar]
- Jiang H, Zhang G, Wu JH, Jiang CP. Diverse roles of miR-29 in cancer (review) Oncol Rep. 2014;31:1509–1516. doi: 10.3892/or.2014.3036. [DOI] [PubMed] [Google Scholar]
- Jiang L, Zhou Y, Xiong M, Fang L, Wen P, Cao H, et al. Sp1 mediates microRNA-29c-regulated type I collagen production in renal tubular epithelial cells. Exp Cell Res. 2013;319:2254–2265. doi: 10.1016/j.yexcr.2013.06.007. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li D, Li X, Wang A, Meisgen F, Pivarcsi A, Sonkoly E, et al. MicroRNA-31 Promotes Skin Wound Healing by Enhancing Keratinocyte Proliferation and Migration. J Invest Dermatol. 2015;135:1676–1685. doi: 10.1038/jid.2015.48. [DOI] [PubMed] [Google Scholar]
- Li N, Cui J, Duan X, Chen H, Fan F. Suppression of type I collagen expression by miR-29b via PI3K, Akt, and Sp1 pathway in human Tenon's fibroblasts. Invest Ophthalmol Vis Sci. 2012;53:1670–1678. doi: 10.1167/iovs.11-8670. [DOI] [PubMed] [Google Scholar]
- Liu S, Wu LC, Pang J, Santhanam R, Schwind S, Wu YZ, et al. Sp1/NFkappaB/HDAC/miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer Cell. 2010;17:333–347. doi: 10.1016/j.ccr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Taylor NE, Lu L, Usa K, Cowley AW, Jr, Ferreri NR, et al. Renal medullary microRNAs in Dahl salt-sensitive rats: miR-29b regulates several collagens and related genes. Hypertension. 2010;55:974–982. doi: 10.1161/HYPERTENSIONAHA.109.144428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Xue X, Lan J, Gao Y, Xiong Z, Zhang H, et al. MicroRNA-29a upregulates MMP2 in oral squamous cell carcinoma to promote cancer invasion and anti-apoptosis. Biomed Pharmacother. 2014;68:13–19. doi: 10.1016/j.biopha.2013.10.005. [DOI] [PubMed] [Google Scholar]
- Luna C, Li G, Qiu J, Epstein DL, Gonzalez P. Role of miR-29b on the regulation of the extracellular matrix in human trabecular meshwork cells under chronic oxidative stress. Mol Vis. 2009;15:2488–2497. [PMC free article] [PubMed] [Google Scholar]
- Morita S, Horii T, Kimura M, Ochiya T, Tajima S, Hatada I. miR-29 represses the activities of DNA methyltransferases and DNA demethylases. Int J Mol Sci. 2013;14:14647–14658. doi: 10.3390/ijms140714647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano H, Gasparro FP, Uitto J. UVA-340 as energy source, mimicking natural sunlight, activates the transcription factor AP-1 in cultured fibroblasts: evidence for involvement of protein kinase-C. Photochem Photobiol. 2001;74:274–282. doi: 10.1562/0031-8655(2001)074<0274:uaesmn>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Nystrom A, Thriene K, Mittapalli V, Kern JS, Kiritsi D, Dengjel J, et al. Losartan ameliorates dystrophic epidermolysis bullosa and uncovers new disease mechanisms. EMBO Mol Med. 2015;7:1211–1228. doi: 10.15252/emmm.201505061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nystrom A, Velati D, Mittapalli VR, Fritsch A, Kern JS, Bruckner-Tuderman L. Collagen VII plays a dual role in wound healing. J Clin Invest. 2013;123:3498–3509. doi: 10.1172/JCI68127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorisio T, Di Salvio M, Orecchia A, Di Zenzo G, Piccinni E, Cianfarani F, et al. Monozygotic twins discordant for recessive dystrophic epidermolysis bullosa phenotype highlight the role of TGF-beta signalling in modifying disease severity. Hum Mol Genet. 2014;23:3907–3922. doi: 10.1093/hmg/ddu102. [DOI] [PubMed] [Google Scholar]
- Qin W, Chung AC, Huang XR, Meng XM, Hui DS, Yu CM, et al. TGF-beta/Smad3 signaling promotes renal fibrosis by inhibiting miR-29. J Am Soc Nephrol. 2011;22:1462–1474. doi: 10.1681/ASN.2010121308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleiman SF, Langley BC, Basso M, Berlin J, Xia L, Payappilly JB, et al. Mithramycin is a gene-selective Sp1 inhibitor that identifies a biological intersection between cancer and neurodegeneration. J Neurosci. 2011;31:6858–6870. doi: 10.1523/JNEUROSCI.0710-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vindevoghel L, Chung KY, Davis A, Kouba D, Kivirikko S, Alder H, et al. A GT-rich sequence binding the transcription factor Sp1 is crucial for high expression of the human type VII collagen gene (COL7A1) in fibroblasts and keratinocytes. J Biol Chem. 1997;272:10196–10204. doi: 10.1074/jbc.272.15.10196. [DOI] [PubMed] [Google Scholar]
- Vindevoghel L, Lechleider RJ, Kon A, de Caestecker MP, Uitto J, Roberts AB, et al. SMAD3/4-dependent transcriptional activation of the human type VII collagen gene (COL7A1) promoter by transforming growth factor beta. Proc Natl Acad Sci U S A. 1998;95:14769–14774. doi: 10.1073/pnas.95.25.14769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Liang Y, Lin Q, Liu J, Luo F, Li X, et al. miR-29 mediates TGFbeta1-induced extracellular matrix synthesis through activation of PI3K-AKT pathway in human lung fibroblasts. J Cell Biochem. 2013;114:1336–1342. doi: 10.1002/jcb.24474. [DOI] [PubMed] [Google Scholar]
- Yang X, Wang J, Guo SL, Fan KJ, Li J, Wang YL, et al. miR-21 promotes keratinocyte migration and re-epithelialization during wound healing. Int J Biol Sci. 2011;7:685–690. doi: 10.7150/ijbs.7.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Wang L, Lu L, Jiang P, Sun H, Wang H. Inhibition of miR-29 by TGF-beta-Smad3 signaling through dual mechanisms promotes transdifferentiation of mouse myoblasts into myofibroblasts. PLoS One. 2012;7:e33766. doi: 10.1371/journal.pone.0033766. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.