Abstract
Background
Oleaginous organisms are a promising, renewable source of single cell oil. Lipid accumulation is mainly induced by limitation of nutrients such as nitrogen, phosphorus or sulfur. The oleaginous yeast Trichosporon oleaginosus accumulates up to 70% w/w lipid under nitrogen stress, while cultivation in non-limiting media only yields 9% w/w lipid. Uncoupling growth from lipid accumulation is key for the industrial process applicability of oleaginous yeasts. This study evaluates the effects of rapamycin on TOR specific signaling pathways associated with lipogenesis in Trichosporon oleaginosus for the first time.
Results
Supplementation of rapamycin to nutrient rich cultivation medium led to an increase in lipid yield of up to 38% g/L. This effect plateaued at 40 μM rapamycin. Interestingly, the fatty acid spectrum resembled that observed with cultivation under nitrogen limitation. Significant changes in growth characteristics included a 19% increase in maximum cell density and a 12% higher maximum growth rate. T. oleaginosus only has one Tor gene much like the oleaginous yeast Rhodosporidium toruloides. Consequently, we analyzed the effect of rapamycin on T. oleaginosus specific TORC signaling using bioinformatic methodologies.
Conclusions
We confirm, that target of rapamycin complex 1 (TORC1) is involved in control of lipid production and cell proliferation in T. oleaginosus and present a homology based signaling network. Signaling of lipid induction by TORC1 and response to carbon depletion to this complex appear to be conserved, whereas response to nitrogen limitation and autophagy are not. This work serves as a basis for further investigation regarding the control and induction of lipid accumulation in oil yeasts.
Electronic supplementary material
The online version of this article (doi:10.1186/s12896-017-0348-3) contains supplementary material, which is available to authorized users.
Keywords: Trichosporon oleaginosus, Rapamycin, TORC, Single cell oil, Oil yeast
Background
Production volumes of bio-based lipids have experienced a 65% increase over the last decade due to increasing demands by food, chemical- and pharmaceutical industries [1, 2]. Especially the application of natural oils for biofuels, oleochemicals and bioactive substances is expanding rapidly [3]. However, the application of plant oils for non-food use accelerates land use and climate change, which in turn negatively impacts on biodiversity. Further, the majority of pharmaceutically active high value lipids such as long chain polyunsaturated fatty acids are still sourced from fish and crustaceans, which negatively affects sensitive marine ecosystems.
Microbial oils have been designated as a sustainable alternative to plant and animal based lipids. In particular, oleaginous yeast, which can accumulate between 20 and 70% w/w lipids [4, 5] have gained increasing interest as providers for sustainable oleochemical building blocks in the biofuel, lubricant, food and cosmetics industry. However, induction of lipogenesis in microorganisms is linked to nutrient restriction (N or P), which results in significant reductions in cell growth. In order to design an economically relevant continuous production process that provides for high lipid and biomass formation the metabolic uncoupling of cell growth from intracellular lipid accumulation is a prerequisite. This aim however requires in depth knowledge of the cells regulatory mechanisms involved in lipogenesis.
The target of rapamycin complexes (TORCs) are central, highly conserved regulators in the control of cell proliferation, sexual development, cell skeleton organization, lipogenesis and other essential functions. TORCs draw their name from the TORC1 inhibiting heterocyclic makrolid rapamycin, derived from Streptomyces hygroscopicus. The complexes consist of different components depending on the organism. The key component is Tor1, a phosphatidylinositol 3 kinase-like serine/threonine kinase (mTOR in mammals).
Whereas some involvement of TORCs, such as TORC1 activation of translation and transcription are conserved from yeast over plants to mammals [6], other downstream processes are vastly different depending on the organism. It is reported that the signaling network plays a crucial role in the control of lipid homeostasis [7, 8]. Understanding the regulatory system of lipid accumulation would therefore contribute to the ability for targeted, genetic modification of single cell oil production strains.
Recently, the effects of rapamycin on the lipid accumulating microalgae Euglena gracilis in comparison to model algae Chlamydomonas reinhardtii and Cyanidioschyzon merolae were characterized [9]. Lipid content was significantly increased when the algae were exposed to minor concentrations of rapamycin. By contrast, higher rapamycin concentrations resulted in growth inhibition. While the effect of rapamycin has been well described in model yeasts, it has as of now not been evaluated in non-conventional oil forming yeasts strains. Therefore, this study evaluates the significance of TORC signaling pathways on lipogenesis in oleaginous yeast for the first time.
Methods
Strains and media
Wild type Trichosporon oleaginosus ATCC 20509 (DSM-11815), obtained from the “Deutsche Sammlung von Mikroorganismen und Zellkulturen” (DMSZ) (Braunschweig, Germany) was used for all experiments. Cultivation was done in YPD medium (glucose, 20 g/L; tryptone, 20 g/L; yeast extract, 10 g/L), nitrogen limitation medium [10] (glucose, 30 g/L; yeast extract, 0.5 g/L; (NH4)2SO4, 0.3 g/L; MgSO4•7H2O, 1.5 g/L; KH2PO4, 2.4 g/L; Na2HPO4 0.91 g/L; CaCl2•H2O, 0.22 g/L; ZnSO4•7H2O, 0.55 μg/L; MnCl2•4H2O, 22.4 μg/L; CuSO4•5H2O, 25 μg/L; FeSO4•7H2O, 25 μg/L, pH 6.1). Rapamycin (Tecoland, CA, USA) was solved in DMSO (10 mM stock) and added directly to the media in different concentrations after the inoculation.
Cultivation conditions
T. oleaginosus was cultivated as triplicate in yeast extract peptone dextrose medium (YPD) with different concentrations of rapamycin solution for 7 days at 28 °C in 500 mL baffled shake flasks. Cultivation was carried out in 100 mL YPD and nitrogen limitation medium (MNM) with glucose. Cells from an overnight culture grown in YPD medium under the same cultivation conditions were washed in ddH2O and used to inoculate all cultivations at OD600 = 0.5. Where applicable, rapamycin was added 8 h after inoculation and adjusted to varying concentrations. 6 mL samples were taken daily for analysis of cell-dry weight, lipid content and fatty acid distribution.
Biomass and lipid determination
Determination of cellular dry weight occurred by pelleting 2 mL samples (12,000 g for 10 min), washing cells with ddH2O and freeze drying at −80 °C for 24 h in pre-weighed microtubes. Cellular total lipid was obtained by extraction with chloroform and methanol by Folch et al. [11] (adapted). Cell pellets from 12 mL culture were washed with ddH2O twice and disrupted four times by french press (EmulsiFlex®-B15, Avestin) at 2400 bar. A triplicate of 4 mL cell lysate was transferred to glass vials with screw caps and mixed with 6 mL of Folch reagent (2:1 chloroform/methanol) each. After extraction by shaking at 900 rpm and room temperature for 1 h, 1 mL 0.9% NaCl was added to aid phase separation. Samples were vortexed, centrifuged at 1000 g and the chloroform phase was transferred to pre-weighed glass vials. After evaporation of the solvent und a constant stream of dried nitrogen, vials were weighed and lipid content was calculated per dry weight in % g/g.
Analysis of fatty acid composition
Triplicates of 2 mL culture were pelleted by centrifugation, washed with ddH2O and freeze dried at −80 °C. Between 10 and 20 mg were used for the fatty acid analysis. Fatty acid methyl esters (FAMES) were obtained by direct conversion of cell biomass by methanol transesterification [12].
FAMEs were analyzed on a GC-2025 gas chromatograph from Shimadzu (Nakagyo-ku, Kyōto, Japan) with flame ionisation detector and an AOC-20i auto injector (Shimadzu). 1 μl sample was applied onto a ZB-WAX column (30 m, 0.32 mm ID; 0.25 μm df; Phenomenex (Torrance, CA, USA)) with an initial column temperature of 150 °C (maintained for 1 min). A temperature gradient was applied from 150–240 °C (5 °C/min), followed by 6 min maintenance at 240 °C. Fatty acids were identified according to retention times of authentic standards.
Nile red assay
Semi-quantitative estimation of intracellular lipids was done by Nile Red staining according to Sitepu et al. [13]. 200 μL of culture sample was adjusted to OD600 = 1 and transferred to MaxiSorp F96 plates (Thermo Scientific Waltham, MA, USA) as 5 replicates. After addition of 25 μL DMSO, the blank measurement was taken. Subsequently, 25 μL of Nile Red staining solution (0.1 mg/mL in DMSO) were added and Nile Red fluorescence intensity was measured at 590 nm (excitation 530 nm) on an EnSpire 2 plate reader from Perkin Elmer (Waltham, MA, USA). Lipids were estimated by correcting FI (fluorescence intensity) by OD600.
Online OD measurements
Online OD measurements were conducted by measuring real-time backscatter at 525 nm with a Cell Growth Quantifier (Aquila biolabs - Baesweiler) using 100 mL YPD in 250 mL shake flask without baffles for 72 h. Rapamycin concentration was adjusted to 5 μM 8 h after inoculation. The backscatter signal was calibrated with a manual 2 point OD600 measurement by an HP 8453 photometer.
Statistical analysis
Statistical analysis and data visualization was done in R [14]. Data were fitted to a Richards’ growth curve using the grofit package [15]. All error bars show standard deviations. Stars show significance with p = 0.05 in comparison to untreated cultures.
Assembly of TORC-network
Previously published genomic and transcriptomic data of T. oleaginosus [16] were searched for TORC homologues using sequences from Schizosaccharomyces pombe and Saccharomyces cerevisiae, Candida curvata and Cryptococcus neoformans.
Results and discussion
Effect of rapamycin on nile red fluorescence
Cultivation of T. oleaginosus in full YPD medium does not lead to nutrient limitation and is associated with a low accumulation of intracellular lipids. Supplementation of this medium with rapamycin (20 μM, [9]) after 8 h cultivation time resulted in significantly higher nile red fluorescence (corrected for OD) signal indicating an increased lipid production (Fig. 1). The increase of 66% FI/OD at 72 h subsequently decreased to 44% (92 h) and 40% after 116 h respectively. During cultivation, no decrease in OD was observed, indicating that the cell growth was not affected by the applied rapamycin concentrations.
Fig. 1.
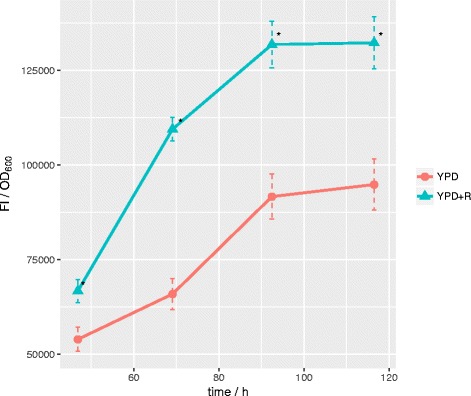
Effect of rapamycin on nile red fluorescence: T. oleaginosus grown in YPD without (YPD) and with 20 μM rapamycin (YPD + R) supplementation. At different time points, nile red fluorescence (FI) and OD600 were measured. The ratio between the latter is plotted on the y axis and is a semiquantitative indicator of lipid content. Stars show significance at p = 0.05 and error bars show standard deviation
It is reported that Nile Red is a semiquantitative lipid stain [17], as its specific fluorescent signal is dependent on the fatty acid profile, the type of lipid (phospho-, triacylglyceride or steran) and the protein content within intracellular lipid bodies.
Effect of rapamycin on lipid accumulation
To confirm an increase in lipid yield, a gravimetric biomass and lipid determination was conducted using cells grown in cultures containing between 0 and 40 μM rapamycin. The addition of 40 μM rapamycin resulted in a maximal lipid increase of 38% compared to controls. Moreover, at 5 μM rapamycin the total biomass production is significantly increased. This data contrasts reports on the effects of rapamycin on algae and bacterial growth, where supplementation of the compound resulted in decreased biomass formation in line with its established cell cycle inhibition effects [8, 18].
Due to the expected logarithmic dependency (Additional file 1), effects of rapamycin on lipid content (Fig. 2) saturate at low concentrations. To confirm the effect of rapamycin on intracellular lipogenesis, we applied a one tailed Welch’s t-test between samples in the absence and presence of rapamycin. The null hypothesis of both sample sets being of the same distribution is rejected with p = 0.003 and a confidence interval of 1.4–5% g/g increase in the absence and presence of rapamycin.
Fig. 2.
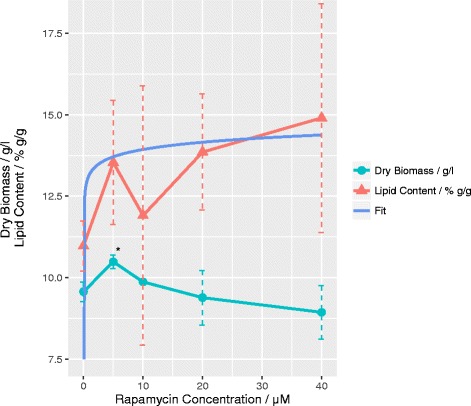
Effect of rapamycin on lipid content and biomass: Total lipid content (triangle) and dry biomass (circle) of T. oleaginosus after 72 h of cultivation in YPD are measured with different concentrations of rapamycin supplementation between 0 and 40 μM. Error bars show standard deviation and the star shows significance at p = 0.05 in comparison to culture without rapamycin supplementation. The blue line shows a robust logarithmic fit of the lipid content in dependence of rapamycin concentration as described in Additional file 1
Impact of rapamycin on T. oleaginosus growth kinetics
As our initial data suggested that at 5 μM rapamycin (at t: 8 h cultivation time) cell growth was enhanced, we applied a real-time backscatter measurement in order to compare cellular growth in the absence and presence of rapamycin (Fig. 3). Cell growth could be sufficiently described by a fit to a Richards’ curve [19], which allowed extraction of μmax (maximum growth rate), lambda (lag phase duration) and A (maximum OD600) values (Table 1).
Fig. 3.
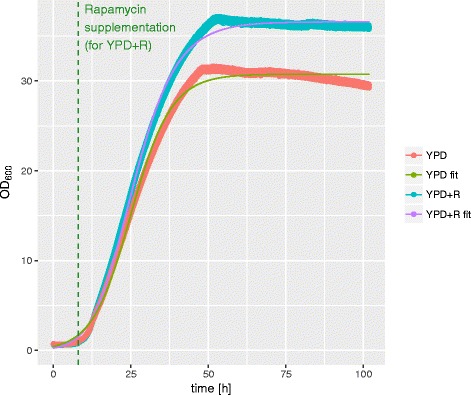
Online-OD measurement: Growth curves of T. oleaginosus without (YPD) and with 5 μM rapamycin (YPD + R) obtained by measuring backscatter with an online OD system. Thin lines (YPD fit, YPD + R fit) show fit of Richards’ growth curves
Table 1.
Comparison of growth curve parameters extracted from Richards’ fit: Maximum growth rate (μmax), lag phase (λ) and maximum cell density (A) of T. oleaginosus cultivated in YPD with and without rapamycin
| YPD | YPD + 5 μM rapamycin | Change / % | |
|---|---|---|---|
| μmax / OD600/h | 1.2068 ± 1.779*10−3 | 1.349 ± 1.362*10−3 | 11.79 ± 2.6*10−3 |
| λ / min | 12.433 ± 23.104*10−3 | 12.151 ± 5.776*10−3 | −2.27 ± 3.13*10−3 |
| A /OD600 | 30.754 ± 7.421*10−3 | 36.578 ± 6.73*10−3 | 18.93 ± 0.46*10−3 |
All values are given with standard deviations
Interestingly, we observed significant differences in the growth parameters of each culture. For the rapamycin treated culture, the maximum growth rate μmax (12% increase) and the maximum optical density (19% increase) were elevated compared to controls.
This translates to a change in the maximum growth rate from a nominal 1.2 to 1.35 OD600/h and an increase in the maximum cell density from OD 30.8 (61 g/L) to OD 36.6 (71 g/L) respectively. Hence, in the presence of 5 μM rapamycin the cell density increased by 19% concomitantly with a 25% lipid increase. These cumulative values translate to a 49% improved space time yield compared to controls in the absence of rapamycin.
The improved growth rate may be attributed to the upregulation of pathways relating to alternative nitrogen sources. A simultaneous assimilation of many different nitrogen sources could be advantageous in a high nutrient environment such as YPD. Furthermore, TORCs are known to affect cell cycle progression. Shortening of the G2 phase could lead to an increased growth rate, while sacrificing replication fidelity and long term offspring survival.
Fatty acid profile
Supplementation of rapamycin caused a non-concentration dependent shift in fatty acid spectrum (Fig. 4). Under these conditions, a major decrease of C18:0 in favor of C18:1 fatty acids was observed. Additionally, a minor decrease of C16:0 and a minor increase in C18:3 could be detected. These changes resemble the fatty acid profile obtained by cultivation in nitrogen limiting medium, thereby supporting the notion that rapamycin is at least partially simulating a low nutrient environment to the cells regulatory system. Fatty acid spectra show, that the effect of rapamycin saturates at comparatively low concentrations, confirming the fit in Fig. 2.
Fig. 4.
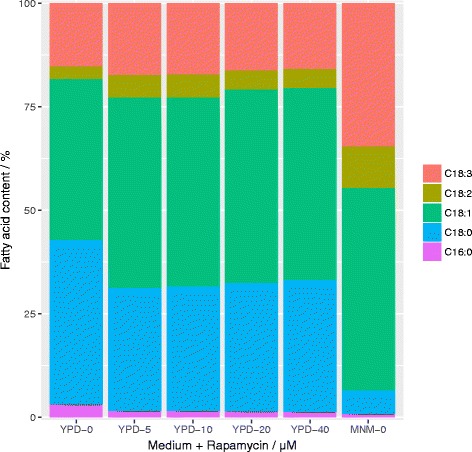
Effect of rapamycin on fatty acid content: Profile of the main fatty acids of T. oleaginosus after 72 h of cultivation in Minimal N Medium (MNM-0) or YPD with different rapamycin concentrations (YPD-0 – YPD-40)
In Euglena gracilis addition of rapamycin led to an increase in lipid amount but almost no change in fatty acid profile was reported [9]. The reason for this appears to be the different mechanism of rapamycin response between this algae and previously identified yeasts or animal cells [9]. This observation in this study motivated us to use a bioinformatics approach to investigate the effects of rapamycin on the Trichosporon cell signaling network and lipogenesis.
TORC-network
A homology-based TORC signaling network, including upstream and downstream elements (Fig. 5), was assembled. A table of all proposed pathway components can be found in Additional file 2.
Fig. 5.
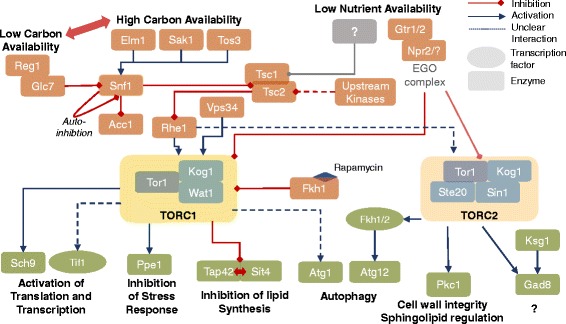
TORC signaling network in T. oleaginosus: Proposed signaling network surrounding TORC in T. oleaginosus inferred by homology and assembled using data obtained from S. pombe and S. cerevisiae
The SNF1/AMPK pathway is highly conserved from yeast to mammals, and homologues of its main components could also be detected in T. oleaginosus. These homologues termed Elm1, Sak1 and Tos3 are kinases which phosphorylate and activate Snf1 (orthologue to the mammalian AMPK), which in turn is a central regulator required for energy homeostasis. In S. cerevisiae Snf1 is mainly responsible for adaption to a glucose limiting environment [20]. Inactivation of Snf1 is caused by dephosphorylation initiated by the Reg1/Glc7 complex [20]. In a low carbon environment, phosphorylated Snf is translocated to the nucleus, where it phosphorylates the transcriptional activator Sip4. Subsequently, Sip4 activates the transcription of glucose-repressed genes [20]. This process is conserved in ascomycetes S. cerevisiae and S. pombe alike. Interestingly, activated Snf1 also inhibits Acc1p, which is responsible for catalysis of acetyl-CoA to malonyl-CoA, a main precursor for the production of fatty acids [21]. This indicates a direct link between metabolic signaling under nutrient stress conditions and the regulation of cellular lipid biosynthesis.
Furthermore, Snf1 is reported to inhibit the Tsc1/Tsc2 complex in model yeasts and in mammals. Moreover, the Tsc1/Tsc2 complex further integrates signals from other pathways, including the MAPK/ERK pathway [22], cytokines, hypoxia signals and Wnt signaling respectively. More recently, it has been suggested that in yeast, involvement of other factors, especially in reaction to low nutrient content is feasible [23]. However, a direct inhibition of TORC1 by Vps34, an essential gene which channels amino acid availability to the Tsc1/Tsc2 complex, has been reported in yeasts and mammals [22, 24].
More specifically, TORC1 itself receives inputs from RheB over the Tsc1/2 axis and directly from the EGO complex. In model yeasts this EGO is composed of the kinases Gtr1/2 and Npr2/3 respectively. However in T. oleaginosus, we could not detect homologous of the Npr3 or Ego1-Ego3 complexes, which indicates that the absence of amino acids in the medium are not sensed via the EGO involved signaling. Inhibition of the TORC1 complex by rapamycin occurs via initial formation of an Fkh1 protein-rapamycin complex (mammalian homologue Fkbp12), which then binds to TORC1. This mechanism is highly conserved throughout the microbial and animal kingdom. Expectantly, T. oleaginosus Tor1 contained the characteristic rapamycin binding motif [25]. In nitrogen limitied media, the addition of rapamycin has no effect on T. oleaginosus biomass formation, growth kinetics and intracellular lipid content. This indicates that TORC1 may already be blocked under nutrient limiting cultivation conditions. Consequently, rapamycin addition would have no effects on biomass or lipid formation in the non-conventional yeast T. oleaginosus.
In consensus with data from model yeasts, we could identify homologues components of the TORC1 complex, namely Tor1 (with strong similarity to Tor2 of S. pombe), Kog1 and Wat1 respectively. Wat1 is a scaffold protein facilitating the connection between Tor1 and downstream substrates, like Ppe1 and Sch9 [26]. Analogous to model yeasts and mammals the Wat1 protein in T. oleaginosus mainly contains 7 WD40 repeats. Specifically in mice, Wat1 was required for TORC2 but not TORC1 activity. Most interestingly, we could not detect any Tco89 homologue in T. oleaginosus, which indicates that a component of the TORC complexes found in model yeast S. cerevisiae is absent. Therefore, the TORC1 complex of T. oleaginosus more closely resembles the situation reported for S. pombe or the oleaginous yeast R. toruloides. Indeed, this resemblances is confirmed by the absence of a second Tor gene for TORC2. Furthermore, we could not identify any Avo2 or Bit61 homologues in T. oleaginosus, both of which are non-functional TORC2-binding structures [27]. Transcripts for the TORC2 component Sin4 could be identified in different splicing isoform, which is consistent with previous findings in S.cerevisiae [24]. Notably, in S. cerevisiae, mutation of Sin4 leads to rapamycin resistance [28]. In analogy, the presence of Sin4 isoforms detected in T. oleaginosus could render the TORC2 complex resistant to rapamycin.
Downstream of primary TORC effects, we could identify significant differences between signals in model yeasts and T. oleaginosus affecting autophagy. Particularly, genes essential to the autophagy signaling pathway, namely Atg13, Atg17, Atg31, Atg29 could not found in T. oleaginosus. In this respect, the T. oleaginosus system may resemble the regulatory system of Drosophila melanogaster, in which hyperphosphorylated Atg1 in conjunction with Atg13 are sufficient to inhibit autophagy. In the model yeast S. cerevisiae the autophagy signaling is by far more complex and therefore may not apply to T. oleaginosus [29]. Further, individual autophagy related homologues (Atg5, Atg6, Atg16) were found, but the actual signaling pathway appears to differ significantly from other yeast systems.
The other main signaling pathways for regulation of lipid biosynthesis, transcriptional and translational initiation appear to be conserved with high similarity.
In S. cerevisiae, TORC1 inhibition liberates Tap42 and Sit4 from being bound to each other. This in turn activates the downstream transcription factors, Gat1 and Gln3. After Gat1 and Gln3 transport to the nucleus, these transcription factors induce the accumulation of lipids. Our bioinformatics analysis indicates that a similar mechanism is likely for T. oleaginosus. The highly conserved Sch9 is homologous to the mammalian S6K, which is responsible for activation of ribosomal protein S6 and therefore directly controls translation. By contrast, no homologue to Gaf1, which in S. pombe is central for the response to nitrogen stress [30], could be identified. The absence of Gaf1 therefore may modulate the cell cycle in T. oleaginosus. Nonetheless, Ppe1 homologue a kinase acting within the S. pombe stress response which also affects the cell cycle [31] could be identified.
TORC2 activation, especially in yeast, remains elusive. In S. pombe, interaction with Rhe1 is confirmed [32]. Furthermore, it was reported in mammals, that activation can be achieved by growth factors (PI3K axis). Furthermore, ribosomal association of the complex suggests its activation by nutrients. Especially the latter is also likely for yeast [33], considering the effect on growth in S. pombe [34]. In T. oleaginosus however, little can be reported about the effects of downstream TORC2 elements due to the absence of detailed cell biology studies.
Active TORC2 activates Fkh1/2 in S. cerevisiae (FOXO genes in mammals), which affects autophagy related genes, life span and stress response [27, 35, 36]. TORC2 as well as the highly conserved kinase Ksg1 activate Gad8 by phosphorylation [37]. The subsequent cellular effects strongly depend on the organism. For S. cerevisiae, this impacts on actin organization and cell wall synthesis, whereas for C. elegans mainly lipid metabolism and growth are affected. For the related and well described yeast, S. pombe, changes in amino acid uptake and general changes in stress response are described [27]. Lack of Rho/Rac homologues indicates a strong difference in the regulation of actin organization from S. pombe.
A confirmed element of TORC2 is Pkc activation, as was described for the closely related and pathogenic Cryptococcus curvatus. Pkc itself is involved in regulation of spingholipid biosynthesis, which impacts the structural integrity of the cell wall [38]. Most recently, the GATA transcription factor Gaf1 was reported to be responsible for sexual development in yeast and upregulation of amino acid transporters [30]. It is activated as response to nitrogen stress about 10 to 120 min after the onset of nitrogen stress. Therefore it can be hypothesized that it is part of a first, reversible response to nutrient stress. Persisting lack of nitrogen would then trigger the second, delayed phase which includes elevated mating in S.pombe.
Two factors indicate, that the observed effects are not due to rapamycin involvement with TORC2: Rapamycin resistance of TORC2 could be structurally substantiated by Avo3 (Ste20), which wraps around the Fkpb-binding domain of Tor1/2 [39]. Prolonged exposure of certain mammalian cell types to rapamycin showed inhibited assembly of TORC2 [40], however this was not observed in unicellular organisms [27] and therefore appears unlikely for T. oleaginosus. Secondly, no obvious differences in cell morphology were observed using microscopy (Additional file 3) and FACS (data not shown), indicating that for T. oleaginosus, rapamycin does not impact on cytoskeleton and actin organization, which are commonly affected by TORC2.
Conclusions
For the first time, lipogenesis could be induced in an oleaginous yeast without compromising on growth, resulting in a 1.4 fold increase in total lipid yield. We observed an increase in growth as well as lipid content in the absence of nutrient limitation, using YPD as model substrate with high nutrient content. TORC1 in Trichosporon oleaginosus can be inhibited by rapamycin, impacting on growth characteristics and lipid accumulation. However, considering a lack of reduction in growth and comparatively minor increases in lipid accumulation, inactivation of TORC1 is not sufficient to induce a cell state resembling nitrogen starvation.
It is possible, that T. oleaginosus either relies on TORC2 inhibition, requires additive signals of both complexes or employs another unknown pathway for full activation of nitrogen stress response and associated lipid accumulation. However, Torc2 regulation of lipid synthesis and its strong involvement in the upregulation of aminoacid transporters [27], one of the defining features of lipid accumulation in T. oleaginosus, make TORC2 involvement in nutrient limitation response in this yeast likely.
Proteomic and transcriptomic approaches are excellent tools for elucidating how rapamycin impacts on T. oleaginosus physiology. Comparing these data with previously obtained information about transcriptomic changes [16] in the presence of nitrogen stress will allow for pinpointing more clearly the relevance of TORC1 for lipid accumulation. Using a metabolomic strategy is a promising approach for a more in-depth study of key intermediates, such as glutamine and glutamate as components of central nitrogen metabolism.
Acknowledgements
FB, FQ and TB would like to thank the German Federal Ministry of Education and Research for supporting the “Advanced Biomass –Value” research project (grant number: 03SF0446A). Information concerning the project is stated under: http://www.ibc.ch.tum.de/index.php?id=2042&L=1. Finally, the authors would like to express their gratitude to Dipl. Ing. (FH) Martina Haack for her kind help addressing analytical questions.
Funding
This work was supported by the BMBF (German Federal Ministry of Education and Research, 03SF0446A).
Availability of data and materials
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
Authors’ contributions
FB, VR and TB conceived the study and participated in its design and coordination. FB, VR, KB carried out the experimental work relating to the effects of rapamycin on Trichsporon oleaginousus growth. FQ supervised the experimental work and prepared scientific reports. FB carried out detailed bioinformatic studies to reconstruct the cell signaling pathways involved in rapamycin addition to the cultivation medium. The manuscript was drafted by FB and finalized by TB. The final manuscript version was approved by all authors.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable
Ethics approval and consent to participate
Not applicable
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- FACS
Fluorescence activated cell sorting
- FI
Fluorescence intensity
- MNM
Minimal nitrogen medium
- N
Nitrogen
- OD
Optical density
- P
Phosphorus
- S
Sulfur
- TORC
Target of rapamycin complex
- YPD
Yeast extract pepton dextrose medium
Additional files
Trend analysis: Analytical plots of robust regression using M-estimation of yeast lipid content in dependence of rapamycin concentration in cultivation medium (Fig. 2). Data points 9 and 7, both of which outliers causing deviation at [rapamycin] = 10 were excluded due to high Cook’s distance. The resulting fit was plotted in Fig. 2 and was based on the following formula: Lipid content = 13.1996 + 0.3197*log ([Rapamycin]). (DOCX 60 kb)
Table of T. oleaginosus homologues in TORC signaling network: Abbreviations: Trichosporon oleaginosus, TO; Schizosaccharomyces pombe, SP; Nitrogen Catabolite Repression, NCR; Saccharomyces cerevisiae, SC; Dictyostelium discodeum, DD. (DOCX 38 kb)
Fluorescence microscopy: 200 μL T. oleaginosus cells grown for 72 h in YPD with and without 5 μM rapamycin supplementation were pelleted, washed with ddH2O and resuspended in the same amount of water. 25 μL DMSO and 25 μL nile red (50 mg/ml) in DMSO were added and incubated in darkness for 10 min. Images were taken on a Zeiss Axio Lab A1 with an Axio Cam ICm1 (Oberkochen, Germany). Fluorescence was measured with a 525/25 filter with an exposure time of 500 ms. (DOCX 154 kb)
Contributor Information
Felix Bracharz, Email: Felix.Bracharz@tum.de.
Veronika Redai, Email: Veronika.Redai@tum.de.
Kathrin Bach, Email: kathrin.bach@online.de.
Thomas Brück, Email: brueck@tum.de.
References
- 1.Lu C, Napier JA, Clemente TE, Cahoon EB. New frontiers in oilseed biotechnology: meeting the global demand for vegetable oils for food, feed, biofuel, and industrial applications. Curr Opin Biotechnol. 2011;22:252–9. doi: 10.1016/j.copbio.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 2.Pfleger BF, Gossing M, Nielsen J. Metabolic engineering strategies for microbial synthesis of oleochemicals. Metab Eng. 2015;29:1–11. doi: 10.1016/j.ymben.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Fritsche UR, Sims REH, Monti A. Direct and indirect land-use competition issues for energy crops and their sustainable production - an overview. Biofuels Bioprod Biorefin. 2010;4:692–704. doi: 10.1002/bbb.258. [DOI] [Google Scholar]
- 4.Papanikolaou S, Aggelis G. Lipids of oleaginous yeasts. Part I: Biochemistry of single cell oil production. Eur J Lipid Sci Technol. 2011;113:1031–51. doi: 10.1002/ejlt.201100014. [DOI] [Google Scholar]
- 5.Ageitos JM, Vallejo JA, Veiga-Crespo P, Villa TG. Oily yeasts as oleaginous cell factories. Appl Microbiol Biotechnol. 2011;90:1219–27. doi: 10.1007/s00253-011-3200-z. [DOI] [PubMed] [Google Scholar]
- 6.de Virgilio C, Loewith R. The TOR signalling network from yeast to man. Int J Biochem Cell Biol. 2006;38:1476–81. doi: 10.1016/j.biocel.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 7.Aggarwal D, Fernandez ML, Soliman GA. Rapamycin, an mTOR inhibitor, disrupts triglyceride metabolism in guinea pigs. Metabolism. 2006;55:794–802. doi: 10.1016/j.metabol.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 8.Madeira JB, Masuda CA, Maya-Monteiro CM, Matos GS, Montero-Lomelí M, Bozaquel-Morais BL. TORC1 inhibition induces lipid droplet replenishment in yeast. Mol Cell Biol. 2015;35:737–46. doi: 10.1128/MCB.01314-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukaida S, Ogawa T, Ohishi K, Tanizawa Y, Ohta D, Arita M. The effect of rapamycin on biodiesel-producing protist Euglena gracilis. Biosci Biotechnol Biochem. 2016;80:1223–9. doi: 10.1080/09168451.2016.1141040. [DOI] [PubMed] [Google Scholar]
- 10.Görner C, Redai V, Bracharz F, Schrepfer P, Garbe D, Brück T. Genetic engineering and production of modified fatty acids by the non-conventional oleaginous yeast Trichosporon oleaginosus ATCC 20509. Green Chem. 2016;18:2037–46. doi: 10.1039/C5GC01767J. [DOI] [Google Scholar]
- 11.Folch J, Lees M, Sloane SGH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 12.Griffiths MJ, van Hille RP, Harrison STL. Selection of direct transesterification as the preferred method for assay of fatty acid content of microalgae. Lipids. 2010;45:1053–60. doi: 10.1007/s11745-010-3468-2. [DOI] [PubMed] [Google Scholar]
- 13.Sitepu IR, Ignatia L, Franz AK, Wong DM, Faulina SA, Tsui M, et al. An improved high-throughput Nile red fluorescence assay for estimating intracellular lipids in a variety of yeast species. J Microbiol Methods. 2012;91:321–8. doi: 10.1016/j.mimet.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.R Core Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing. 2013. http://www.R-project.org/. ISBN 3-900051-07-0.
- 15.Kahm M, Hasenbrink G, Lichtenberg-frate H, Ludwig J, Kschischo M. Grofit: fitting biological growth curves. J Stat Softw. 2010;33:1–21. doi: 10.18637/jss.v033.i07. [DOI] [Google Scholar]
- 16.Kourist R, Bracharz F, Lorenzen J, Kracht ON, Chovatia M, Daum C, et al. Genomics and transcriptomics analyses of the oil-accumulating basidiomycete yeast Trichosporon oleaginosus: Insights into substrate utilization and alternative evolutionary trajectories of fungal mating systems. MBio. 2015;6:1–13. doi: 10.1128/mBio.00918-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rumin J, Bonnefond H, Saint-Jean B, Rouxel C, Sciandra A, Bernard O, et al. The use of fluorescent Nile red and BODIPY for lipid measurement in microalgae. Biotechnol Biofuels. 2015;8:42. doi: 10.1186/s13068-015-0220-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown NF, Stefanovic-Racic M, Sipula IJ, Perdomo G. The mammalian target of rapamycin regulates lipid metabolism in primary cultures of rat hepatocytes. Metabolism. 2007;56:1500–7. doi: 10.1016/j.metabol.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 19.Richards FJ. A flexible growth function for empirical use. J Exp Bot. 1959;10:290–301. doi: 10.1093/jxb/10.2.290. [DOI] [Google Scholar]
- 20.Hedbacker K, Carlson M. SNF1/AMPK pathways in yeast. Front Biosci. 2008;13:2408–20. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M, Galdieri L, Vancura A. The yeast AMPK homolog SNF1 regulates acetyl coenzyme A homeostasis and histone acetylation. Mol Cell Biol. 2013;33:4701–17. doi: 10.1128/MCB.00198-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang R, Levine B. Autophagy in cellular growth control. FEBS Lett. 2010;584:1417–26. doi: 10.1016/j.febslet.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J, Brendan DM. The TSC1–TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2009;290:1717–21. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacinto E. What controls TOR? IUBMB Life. 2008;60:483–96. doi: 10.1002/iub.56. [DOI] [PubMed] [Google Scholar]
- 25.Choi J, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–42. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 26.Chantaravisoot N, Tamanoi F. mTOR signaling and human cancer. Struct. Funct. Regul. TOR complexes from Yeasts to Mamm. Part B. Cambridge: Academic Press; 2010. p. 301–16.
- 27.Cybulski N, Hall MN. TOR complex 2: a signaling pathway of its own. Trends Biochem Sci. 2009;34:620–7. doi: 10.1016/j.tibs.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 28.Zhang L, Liu N, Ma X, Jiang L. The transcriptional control machinery as well as the cell wall integrity and its regulation are involved in the detoxification of the organic solvent dimethyl sulfoxide in Saccharomyces cerevisiae. FEMS Yeast Res. 2013;13:200–18. doi: 10.1111/1567-1364.12022. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci. 2011;124:161–70. doi: 10.1242/jcs.064576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laor D, Cohen A, Kupiec M, Weisman R. TORC1 regulates developmental responses to nitrogen stress via regulation of the GATA transcription factor Gaf1. MBio Am Soc Microbiol. 2015;6:e00959–15. doi: 10.1128/mBio.00959-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yanagida M, Ikai N, Shimanuki M, Sajiki K. Nutrient limitations alter cell division control and chromosome segregation through growth-related kinases and phosphatases. Philos Trans R Soc Lond B Biol Sci. 2011;366:3508–20. doi: 10.1098/rstb.2011.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuo T, Otsubo Y, Urano J, Tamanoi F, Yamamoto M. Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast. Mol Cell Biol. 2007;27:3154–64. doi: 10.1128/MCB.01039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weisman R, Cohen A, Gasser SM. TORC2--a new player in genome stability. EMBO Mol Med. 2014;6:995–1002. doi: 10.15252/emmm.201403959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikai N, Nakazawa N, Hayashi T, Yanagida M. The reverse, but coordinated, roles of Tor2 (TORC1) and Tor1 (TORC2) kinases for growth, cell cycle and separase-mediated mitosis in Schizosaccharomyces pombe. Open Biol. 2011;1:110007. doi: 10.1098/rsob.110007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pic A, Lim FL, Ross SJ, Veal EA, Johnson AL, Sultan MR, et al. The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. EMBO J. 2000;19:3750–61. doi: 10.1093/emboj/19.14.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hollenhorst PC, Bose ME, Mielke MR, Müller U, Fox CA. Forkhead genes in transcriptional silencing, cell morphology and the cell cycle: overlapping and distinct functions for FKH1 and FKH2 in Saccharomyces cerevisiae. Genetics. 2000;154:1533–48. doi: 10.1093/genetics/154.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du W, Forte GM, Smith D, Petersen J. Phosphorylation of the amino-terminus of the AGC kinase Gad8 prevents its interaction with TORC2. Open Biol. 2016;6:150189. doi: 10.1098/rsob.150189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee H, Khanal Lamichhane A, Garraffo HM, Kwon-Chung KJ, Chang YC. Involvement of PDK1, PKC and TOR signalling pathways in basal fluconazole tolerance in Cryptococcus neoformans. Mol Microbiol. 2012;84:130–46. doi: 10.1111/j.1365-2958.2012.08016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaubitz C, Oliveira TM, Prouteau M, Leitner A, Karuppasamy M, Konstantinidou G, et al. Molecular basis of the rapamycin insensitivity of target of rapamycin complex 2. Mol Cell. 2015;58:977–88. doi: 10.1016/j.molcel.2015.04.031. [DOI] [PubMed] [Google Scholar]
- 40.Sarbassov DD, Ali SM, Sengupta S, Sheen J-H, Hsu PP, Bagley AF, et al. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–68. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.


