Abstract
Background
Cassava mosaic disease (CMD) is a major constraint to cassava production in sub-Saharan Africa. Under field conditions, evaluation for resistance to CMD takes 12–18 months, often conducted across multiple years and locations under pressure from whitefly-mediated transmission. Under greenhouse or laboratory settings, evaluation for resistance or susceptibility to CMD involves transmission of the causal viruses from an infected source to healthy plants through grafting, or by using Agrobacterium-mediated or biolistic delivery of infectious clones. Following inoculation, visual assessment for CMD symptom development and recovery requires 12–22 weeks. Here we report a rapid screening system for determining resistance and susceptibility to CMD based on virus-induced gene silencing (VIGS) of an endogenous cassava gene.
Results
A VIGS vector was developed based on an infectious clone of the virulent strain of East African cassava mosaic virus (EACMV-K201). A sequence from the cassava (Manihot esculenta) ortholog of Arabidopsis SPINDLY (SPY) was cloned into the CP position of the DNA-A genomic component and used to inoculate cassava plants by Helios® Gene Gun microparticle bombardment. Silencing of Manihot esculenta SPY (MeSPY) using MeSPY1-VIGS resulted in shoot-tip necrosis followed by death of the whole plant in CMD susceptible cassava plants within 2–4 weeks. CMD resistant cultivars were not affected and remained healthy after challenge with MeSPY1-VIGS. Significantly higher virus titers were detected in CMD-susceptible cassava lines compared to resistant controls and were correlated with a concomitant reduction in MeSPY expression in susceptible plants.
Conclusions
A rapid VIGS-based screening system was developed for assessing resistance and susceptibility to CMD. The method is space and resource efficient, reducing the time required to perform CMD screening to as little as 2–4 weeks. It can be employed as a high throughput rapid screening system to assess new cassava cultivars and for screening transgenic, gene edited and breeding lines under controlled growth conditions.
Electronic supplementary material
The online version of this article (doi:10.1186/s12985-017-0716-6) contains supplementary material, which is available to authorized users.
Keywords: Cassava mosaic disease, VIGS, SPINDLY, Resistance, Susceptible, Geminivirus
Background
The starchy storage root of cassava serves as a staple food for millions of people in Africa. In 2014, over 50% of the world’s cassava production took place in sub-Saharan Africa, where 146.8 million tons were harvested [1]. While cassava is resilient to abiotic stresses such as prolonged drought [2], its production is constrained by the two viral diseases, cassava mosaic disease (CMD) and cassava brown streak disease (CBSD) [3]. Cassava mosaic disease is caused by a cassava mosaic geminivirus (CMG) complex. CMGs are single-stranded bipartite DNA viruses in the family Geminiviridae, genus Begomovirus, comprised of 11 species, two of which are present in the Indian sub-continent, with the rest endemic to Africa [4, 5].
Improvement programs for development of CMD-resistant cassava germplasm include introgression of polygenic recessive resistance from the related species Manihot glaziovii (CMD1), identification of monogenic dominant resistance in West African cassava landraces (CMD2), and more recently, production of highly resistant cultivars carrying a quantitative trait loci (CMD3) [6–9]. Screening cassava germplasm for resistance to CMD traditionally involves cultivation under field conditions with exposure to transmission of CMGs mediated by the whitefly vector Bemisia tabaci for a growth cycle of 12–18 months [6, 10, 11]. Under contained conditions in the greenhouse or growth chamber, inoculation of cassava with CMGs can be achieved by a) graft inoculation from a CMG-infected host to healthy plants [12–14]; b) delivery of DNA genomes as infectious clones via microparticle bombardment [15–17]; c) Agrobacterium-mediated inoculation of plants with cloned infectious DNA genomes [18]; or d) mechanical transmission of cloned viral DNA genomes by abrasion [19]. Irrespective of the inoculation method employed, CMD symptom development and severity is scored over a period lasting 12–22 weeks from the time of inoculation through the potential disease recovery process [15, 18]. During this time the resistant/tolerant cultivars are identified based on displayed recovery phenotype on newly formed leaves, while the susceptible cultivars remain symptomatic throughout.
Methods currently available for evaluating resistance and susceptibility to CMD in new cultivars, breeding lines or transgenic and gene edited events are therefore lengthy, space inefficient and require frequent assessment of leaf symptoms by skilled personnel. We report here the development of a simple screening system for determining resistance or susceptibility to CMD that can be completed within 2–4 weeks from the time of inoculation. This rapid screening system is based on virus-induced gene silencing (VIGS) of an endogenous MeSPY gene. The method described saves time and space in the greenhouse and enhances capability to allow screening of a large number of plants in a short period of time.
Methods
Construction of infectious VIGS clones
VIGS clones were generated from the virulent infectious clone EACMV-K201 described previously by Patil and Fauquet [20]. EACMV-K201 was produced from East African cassava mosaic virus (EACMV-KE[KE:Msa:K201:02]), DNA-A GenBank: AJ717541; and DNA-B GenBank: AJ704953 [21]. The EACMV-K201 DNA-A infectious clone was digested with Hin dIII/Eco RI (2082 bp) and Eco RI/Bam HI (1157 bp), and cloned into pBlueScript vector (Stratagene) as Hin dIII/Bam HI in a three-way ligation. The resulting construct was named p8200. p8200 was modified to introduce restriction sites Nhe I and Avr II near the 5′-region, and Sbf I near the 3′-region within the coding sequence of the coat protein (CP) gene using QuickChange Multi Site-Directed Mutagenesis Kit (Agilent Technologies, Inc.) with the primers listed in Table 2. The mutagenized infectious clone was confirmed by sequencing and named p8202.
Table 2.
Primers used for in vitro mutagenesis, PCR, qPCR and RT-qPCR in this study
| Primer number | Sequence | Target Gene (construct) | Purpose | Reference |
|---|---|---|---|---|
| 823 | CTTCCCAACTCTATGGGTGATGGCTAGCCTAGGAGTAACATCACTGACACATCG | EACMV-K201 (DNA-A) | Mutagenesis primer (introduces Avr II and Nhe I) | This study |
| 824 | CGATGTGTCAGTGATGTTACTCCTAGGCTAGCCATCACCCATAGAGTTGGGAAG | |||
| 825 | ATAGATGCGTATTTTAAGCGTCGCACCTGCAGGATTTGAGGCATGTGTACATGC | EACMV-K201 (DNA-A) | Mutagenesis primer (introduces Sbf I) | This study |
| 826 | GCATGTACACATGCCTCAAATCCTGCAGGTGCGACGCTTAAAATACGCATCTAT | |||
| 857 | GCTAGCCCAACACTTGTCACTACTTTC | erGFP | GFP-VIGS | This study |
| 858 | CCTGCAGGGAAAGGGCAGATTGTGTGGA | |||
| 907 | GCTAGCGCTGAGTCATATCAGAAGGC | MeSPY1/Manes.09G052300.1 | MeSPY1-VIGS | This study |
| 908 | CCTGCAGGATACCTTGATTGATGTCTCC | |||
| 1260 | CCTGCAGGTAGACGTGAAACCAGATATTGTGC | MePSY2/Manes.01G124200.1 | MePSY2-VIGS | This study |
| 1261 | GCTAGCAACACTCATTAATCCAACCGTCCC | |||
| 1066 | CAAAGGTTAAATTGGAGGGAGACATC | MeSPY1/Manes.09G052300.1 | RT-qPCR | This study |
| 1067 | GCATTTCACCATATGCAACACCAAGA | |||
| 833 | TTGCAGAGGAAGATAGTGGGAATG | EACMV-K201 (DNA-A) | Probe for Southern blot | Beyene, et al. [15] |
| 834 | GAACGTGATGGGTTCCGCTG | |||
| 837 | TGCAAGGCTCACACTTTCATC | MePP2A/Manes.09G039900.1 | Reference gene for qPCR | Moreno, et al. [27] |
| 838 | CTGAGCGTAAAGCAGGGAAG | |||
| 1046 | GGTCTTCCCTGTACGACTATC | EACMV-K201 (DNA-A) | qPCR (virus load) | Beyene, et al. [15] |
| 1047 | GGAACTTGAAGTCTGGGTTTCC |
The Manihot esculenta homolog of A. thaliana SPINDLY (SPY) gene (MeSPY1 accession number Manes.09G052300.1) was cloned into p8202 at the CP site. A 452 bp fragment of MeSPY1 (406–857 counted from start codon ATG) was amplified from plasmid p8103 that harbors the 2781 bp coding sequence of MeSPY1 by introducing restriction sites Nhe I and Sbf I on the forward and reverse primer pairs, respectively (Table 2). The PCR product was cloned into Zero Blunt Topo (Invitrogen) first and positive clones digested and cloned into the Nhe I/Sbf I site of p8202 to generate the p8250 (hereafter named MeSPY1-VIGS). A non-target VIGS control was produced by amplification of 453 bp (235–687 bp counted from start codon ATG) from the erGFP [22] sequence of a binary vector erGFP-pCAMBIA2300 by introducing Nhe I and Sbf I sites on forward and reverse primer pairs, respectively, and cloned into p8202 to generate p8223 (GFP-VIGS). In order to generate a VIGS vector targeting cassava phytoene synthase (MePSY2 gene, Manes.01G124200.1), a 452 bp fragment (350–801 bp counted from start codon ATG) was amplified introducing a Sbf I and Nhe I restriction site and cloned into the Sbf I and Nhe I site of 8202 to produce p8375 (MePSY2-VIGS).
Cultivars and greenhouse growth conditions
Cassava cultivars used in this study are shown in Table 1 and included the CMD-susceptible cultivars 60444, Ebwanateraka and TME 7S; and CMD-resistant cultivars TME 3, TME 7 (Oko-iyawo), TME 204 and TMS 98/0505. Additional lines included CMD-susceptible plants [15] regenerated from friable embryogenic callus (FEC) of TME 204, TME 3 and TME 7 and CMD-resistant plants of cultivar TMS 98/0505 recovered from FEC following the method described by Chauhan, et al. [23]. All plants were micropropagated and established in the greenhouse as described previously [15, 24].
Table 1.
Cassava cultivars and lines used for MeSPY1-VIGS, GFP-VIGS and MePSY2-VIGS challenge
| Cultivar/line | Response to CMD | Remark |
|---|---|---|
| TME 204 | Resistant | Recovers from CMD |
| TME 3 | Resistant | Recovers from CMD |
| TME 7 | Resistant | Recovers from CMD |
| TMS 98/0505 | Resistant | Recovers from CMD |
| FEC- TMS 98/0505 | Resistant | Recovers from CMD |
| 60444 | Susceptible | Does not recover from CMD |
| TME 7S | Susceptible | Does not recover from CMD |
| Ebwanateraka | Susceptible | Does not recover from CMD |
| FEC-TME 204 | Susceptible | Does not recover from CMD |
| FEC-TME 3 | Susceptible | Does not recover from CMD |
| FEC-TME 7 | Susceptible | Does not recover from CMD |
Inoculation of VIGS clones and assessment of phenotype in the greenhouse
Four- to 6-week-old greenhouse-grown plants were inoculated with plasmid DNA of MeSPY1-VIGS, GFP-VIGS or MePSY2-VIGS vectors plus the DNA-B component of EACMV-K201 using a Helios® Gene Gun (BioRad, Hercules, California), following Beyene, et al. [15]. Approximately 75 ng each of the VIGS (DNA-A and DNA-B) components were used to inoculate each plant. Symptom and phenotype scoring after inoculation was performed in three manners. Plants challenged with GFP-VIGS were scored for development of CMD symptoms using an established visual scoring system with a scale of 0–5 [25]. Plants were scored starting 7–10 days post inoculation (DPI) and every 3–7 days thereafter for a total of 12 weeks [15]. For plants challenged with MePSY2-VIGS, visual assessment was made for presence or absence of chlorosis/bleaching. In the case of plants challenged with MeSPY1-VIGS, a new scoring system was employed by which plants were assessed for death of the shoot apical meristem (whole plant) starting 7–10 DPI and every 2–3 days thereafter for a maximum of 4 weeks. Data was expressed as incidence of plants showing shoot-tip necrosis/dead plants presented as the percent of the total number of plants inoculated at the end of the 4-week observation period.
Nucleic acid extraction, qPCR and RT-qPCR
Plants were sampled for nucleic acid extraction by collecting leaves and shoot material 9 DPI with VIGS constructs. Tissues from three plants were pooled in a 50 mL Falcon tube and immediately flash frozen in liquid nitrogen. A total of four pooled samples collected from 12 plants were analyzed per treatment. Frozen samples were ground in a mortar and pestle to fine powder and freeze-dried overnight. Approximately 20–30 mg of freeze-dried tissue were used for nucleic acid extraction. Genomic DNA was extracted using DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) and the resulting genomic DNA quantified and used for virus titer determination by qPCR and Southern blot analysis [15]. Total RNA was isolated using the Spectrum™ Plant Total RNA Kit, followed by on-column DNAse I treatment per manufacturer instructions (Sigma-Aldrich, St. Louis, MO). One microgram of total RNA was reverse transcribed using the PrimeScript™ RT reagent kit with gDNA Eraser (Takara Bio Inc., Japan). Expression levels of MeSPY in MeSPY1-VIGS-inoculated plants and control GFP-VIGS plants were quantified by RT-qPCR [26] using the primer pairs listed in Table 2, with approximately 10 ng reverse transcriptase template and SsoAdvanced™ Universal SYBER® Green Supermix (Bio-Rad Laboratories Inc.). The cassava PP2A gene was employed as an internal control for RT-qPCR [27].
Results
Production of EACMV-K201 VIGS clones and verification of infectivity
A VIGS system was developed by modifying the CP nucleotide sequence of the DNA-A component of the virulent infectious clone EACMV-K201 [20] to generate the VIGS construct p8202. Mutations were designed to introduce restriction sites Avr II, Nhe I and Sbf I (Fig. 1a) in order to allow introduction of target sequences. The introduced mutations were confirmed by sequencing both strands of the modified CP sequences as shown (Fig. 1a) and further confirmed by restriction analysis (data not shown). To check infectivity of the p8202 VIGS vector, a 452 bp fragment from the coding sequence of the cassava carotenoid biosynthetic gene phytoene synthase (MePSY2) was cloned at the Nhe I/Sbf I site of p8202 to generate MePSY2-VIGS. In addition, a GFP-VIGS DNA-A component was generated by cloning a 453 bp sequence from erGFP [22] into the restriction sites generated within the CP sequence.
Fig. 1.
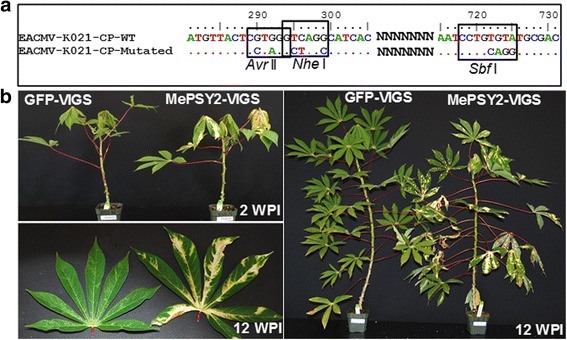
East African Cassava mosaic virus (EACMV-K201) infectious clone-based virus-induced gene silencing (VIGS) vector. a Coat protein (CP) nucleotide sequence of EACMV-K201 DNA-A showing position and mutated nucleotides that introduced Avr II and Nhe I near 5′-end and Sbf I near 3′-end. Nucleotide positions were counted from the start codon “ATG” of the CP. b Infectivity of the GFP-VIGS and MePSY2-VIGS targeting GFP and cassava phytoene synthase coding sequences, respectively, in wild-type cassava cultivar TME 7S. GFP-VIGS has no known target in the challenged cassava plant, and these plants display typical mosaic symptoms characteristic of CMD on leaves. MePSY2-VIGS-challenged plants show chlorosis on leaves. Pictures were collected at 2 and 12 weeks post inoculation (WPI)
Five-week-old plants of the CMD-susceptible cassava line TME 7S were inoculated by Helios® Gene Gun microparticle bombardment of the modified DNA-A component plus the infectious clone of DNA-B. TME 7S is a CMD-susceptible version of TME 7 previously described by Kuria, et al. [28]. Plants inoculated with MePSY2-VIGS developed visible chlorosis/bleaching on the challenged leaves and then subsequently on systemic leaves within 10–15 days after bombardment (Fig. 1b). Bleaching of leaves persisted throughout the experimental period of 12 weeks (Fig. 1b). Plants challenged with GFP-VIGS (which does not have a target gene sequence within the cassava genome) showed typical but mild CMD symptoms on systemic leaves that persisted throughout the study period (Fig. 1b). Response of inoculated plants to MePSY2-VIGS and GFP-VIGS confirms that the VIGS constructs made by modifying EACMV-K201 are both infectious and efficacious in silencing gene expression in cassava.
Silencing of MeSPY gene is lethal in CMD-susceptible cassava cultivars
Using the Arabidopsis SPY gene (AT3G11540.1) [29, 30] as a bait, two sequences, Manes.09G052300.1 (named MeSPY1) and Manes.08G028400.1 (named MeSPY2), were identified from the cassava v6.1 genome sequence [31]. The MeSPY1 and MeSPY2 sequences are 81% identical to Arabidopsis SPY at the amino acid level. Both MeSPY1 and MeSPY2 carry the conserved N-terminal tetratricopeptide repeat (TPR) domain, plus the novel serine and threonine O-linked N-acetylglucosamine transferase (OGT) [30] at the C-terminus of the protein (data not shown). The coding region of MeSPY1 and MeSPY2 genes are 91.80 and 90.78% identical to each other at the nucleotide and amino acid levels, respectively. The target sequence (452 bp) cloned into the VIGS vector from MeSPY1 was obtained closer to the 5′-end, within the conserved TPR region. As MeSPY1 and MeSPY2 share high sequence identity (94.69%) at this selected region, both can be targeted for simultaneous silencing by MeSPY1-VIGS.
The susceptible cassava cultivar TME 7S was challenged with MeSPY1-VIGS by Helios® microparticle bombardment. Inoculated plants did not show typical CMD symptoms but instead underwent wilting and withering of one or more leaves just above the inoculation site within 10–15 DPI. This was followed by shoot-tip necrosis within 12–21 DPI. Defoliation then progressed downwards to mature leaves below the inoculation site and along the main stem continuing until most of the plants had died within 2–4 weeks after inoculation with MeSPY1-VIGS (Fig. 2a). TME 7S plants challenged with the control GFP-VIGS showed typical CMD symptom development without shoot tip necrosis (Fig. 2a). In order to determine if this was a cultivar-specific response, the CMD2-type cultivar TME 204 (Table 1) was challenged with MeSPY1-VIGS and GFP-VIGS. Two types of TME 204 plants were used: wild-type TME 204, which shows development of CMD symptoms followed by recovery; and FEC-TME 204 lines. The latter TME 204 plants that were recovered through somatic embryogenesis lost their inherent resistance to CMD and showed no recovery from CMD after challenge with CMGs [15]. After challenge with MeSPY1-VIGS, all plants (10/10) of wild-type TME 204 displayed transient reduction in growth compared to the GFP-VIGS plants and then resumed normal growth without showing typical CMD symptoms. No shoot-tip necrosis was observed. In contrast, all FEC-derived TME 204 plants (8/8) displayed a similar phenotype to that described above for TME 7S, whereby elongation of the shoot was reduced, resulting in compact shoot-tips and wilting and withering of leaves just above the inoculation site. This was followed by death of the shoot-tip and wilting and defoliation of leaves that progressed downwards from the youngest to the oldest leaves. Time-lapse video of the process of shoot-tip necrosis and whole plant death in susceptible FEC-TME 204 and survival of the resistant wild-type TME 204 plant lines is presented (Additional file 1). All plants displayed shoot-tip necrosis with most plants dying within 2–4 weeks after challenge (Fig. 2b). Inoculation with GFP-VIGS resulted in plants of both the susceptible and resistant lines of TME 204 developing CMD symptoms. Wild-type TME 204 plants then underwent recovery from CMD to show no symptoms on newly formed leaves by 16 weeks after challenge. FEC-derived plants remained symptomatic throughout, in a manner consistent with previous observations [15]. This data indicates that silencing of MeSPY is lethal in the two susceptible cassava lines TME 7S and FEC-derived TME 204, but not in the resistant wild-type TME 204.
Fig. 2.

MeSPY1-VIGS-challenged susceptible and resistant cassava cultivars. a CMD-susceptible cassava cultivar TME 7S challenged with GFP-VIGS (left) and MeSPY1-VIGS (right) showing defoliation of leaves and death of shoot-tip at 31 days post-inoculation (DPI). b GFP-VIGS- (bottom) and MeSPY1-VIGS- (top) challenged wild-type TME 204 (resistant) and FEC-TME 204 (susceptible). Note symptoms of dead or dying plants in FEC-derived plants. Pictures captured at 24 DPI. All GFP-VIGS plants showed typical CMD symptoms on the leaves
Additional file 1: Time-lapse video showing response of wild-type TME 204 (resistant to cassava mosaic disease, CMD) and FEC-TME 204 plants (susceptible to CMD) to inoculation with MeSPY1-VIGS. Both plant types were challenged with the modified EACMV-K201 (MeSPY1-VIGS) along with infectious DNA B clone. Images of challenged plants were captured every hour beginning from the 5th day after inoculation for a total of 22 days using 5MP camera boards controlled by Raspberry Pi microcomputers. Images were then converted to a movie file using Apple iMovie. Only pictures collected every 2 hours and during daytime were presented. Note the susceptible FEC-TME 204 plants die while the wild-type TME 204 survive MeSPY1-VIGS challenge. (M4V 14035 kb)
MeSPY1-VIGS’s potential for use as a screening tool to determine resistance or susceptibility to CMD was investigated further by challenging additional cassava cultivars (Table 1). These included: a) CMD2-type resistant wild-type plants of cultivars TME 7 and TME 3, and CMD-susceptible plant lines derived from FEC of TME 7 and TME 3; b) CMD3-type resistant cultivar TMS 98/0505 (wild-type) and FEC-derived TMS 98/0505; and c) CMD-susceptible cassava cultivars 60444 and Ebwanateraka. Inoculation of these cassava accessions with MeSPY1-VIGS showed that CMD-susceptible germplasm (FEC-TME 3, FEC-TME 7, 60444 and Ebwanateraka) underwent wilting of leaves above the inoculation site and necrosis of shoot-tips within 10–21 DPI. This was followed by defoliation, culminating in death of the majority of plants within 4 weeks after inoculation (Figs. 3 and 4). The CMD-resistant wild-type cultivars TME 3, TME 7 and TMS 98/0505 survived challenge with MeSPY1-VIGS and displayed similar phenotype described above for wild-type TME 204 (Figs. 3 and 4). Plants of FEC-derived TMS 98/0505 behaved in a manner identical to wild-type TMS 98/0505 confirming their known resistance to CMD [15]. Likewise, wild-type and FEC-derived plants of CMD1-type cultivars NASE 3 (TMS 30572) and NASE 14 also survive MeSPY-VIGS (data not shown). All plant lines were also challenged with GFP-VIGS. Data collected for development of and recovery from CMD symptoms correlated with that from plants inoculated with MeSPY1-VIGS such that all plant lines that resisted challenge with MeSPY1-VIGS also showed recovery from CMD symptoms after inoculation with GFP-VIGS (Fig. 5).
Fig. 3.
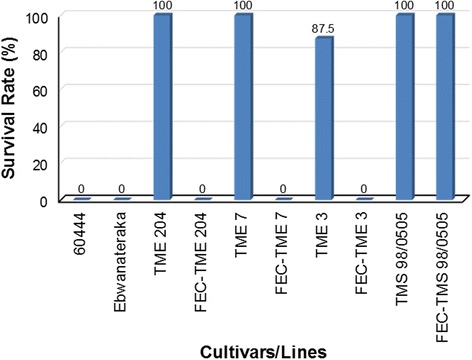
Survival rate of MeSPY1-VIGS-challenged cassava cultivars/lines. Four-week-old cassava seedlings were challenged with MeSPY1-VIGS and evaluated for survival at 4 weeks after challenge. Plants were considered dead if the shoot-tip had completely died and leaves had defoliated or were wilting and defoliating at evaluation period and are expressed as percent of challenged plants. Between 7 and 10 plants were challenged per cultivar/line and each experiment was repeated at least three times
Fig. 4.

MeSPY1-VIGS-challenged cassava cultivars/lines. a FEC-derived and wild-type TME 3, b FEC-derived and wild-type TME 7, c FEC-derived and wild-type TMS 98/0505, d CMD-susceptible cassava cultivars Ebwanateraka and 60444. The FEC-derived lines and wild-type susceptible cultivars exhibit wilting of leaves at 13 DPI and complete necrosis of shoot-tip by 22 DPI. Pictures were collected at 7, 13 and 22 DPI
Fig. 5.
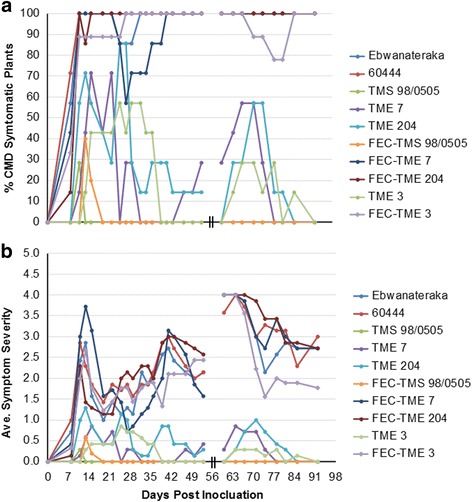
Response of cassava cultivars/lines to biolistic inoculation with GFP-VIGS under greenhouse conditions. a CMD incidence (b) average symptom severity scores (scale of 0–5). The resistant cassava cultivars showed full recovery from GFP-VIGS with 0 incidence and 0 average symptom severity at the end of the experiment (91 DPI), while the susceptible lines remained symptomatic with higher incidence (80–100%) and average severity score of 1.7–3.0 at the end of the experiment. Plant stems were cut back at 53 days after biolistic inoculation and CMD was assessed on new leaf growth. Breaks in the x-axis graph indicate lapse in shoot regrowth after this cut-back
Suppression of MeSPY and virus titer in challenged plants
Expression of MeSPY after inoculation with MeSPY1-VIGS was quantified by RT-qPCR in the CMD-resistant wild-type cultivar TME 7 and susceptible FEC-TME 7 plants at 9 DPI. At this time, plants challenged with the control GFP-VIGS showed systemic CMD symptoms on young leaves above the inoculation site, while the MeSPY1-VIGS leaves were smaller and shoot-tips were compact relative to the GFP-VIGS. Relative expression of the MeSPY gene was found to be significantly reduced (33%; p < 0.05) in FEC-derived TME 7 plants challenged with MeSPY1-VIGS when compared to TME 7 wild-type plants and to the corresponding GFP-VIGS controls (Fig. 5c). Virus DNA titer was determined by qPCR (Fig. 5b) and Southern blot analysis (Fig. 5a) from the same samples collected at 9 DPI. TME 7 wild-type plants challenged with MeSPY1-VIGS and GFP-VIGS had significantly lower relative virus loads (2.36-fold and 7.23-fold, respectively) compared to the FEC-derived CMD-susceptible TME 7 plants (16.08 and 29.39, respectively). Greater virus DNA was detected in GFP-VIGS inoculated lines than MeSPY-VIGS inoculated lines (Fig. 6a). qPCR virus titer data was seen to be consistent with Southern blot data showing relatively more virus titer in the susceptible FEC-derived TME 7 compared to the wild-type TME 7.
Fig. 6.
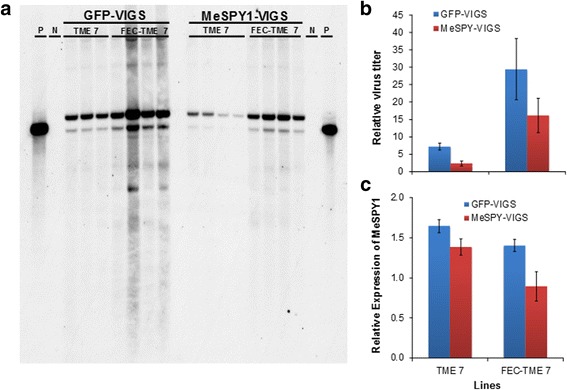
Virus titer in wild-type and FEC-derived TME 7 (Oko-iyawo) plants challenged with GFP-VIGS and MeSPY1-VIGS at 9 DPI. a Virus titer determination by Southern blot and b qPCR performed on total DNA extracted from leaves of wild-type (resistant to CMD) and FEC-derived (susceptible to CMD) plants. c RT-qPCR expression analysis of MeSPY. Higher virus titer is detected in both GFP-VIGS and MeSPY1-VIGS-challenged FEC-TME7 (susceptible plants) compared to the wild-type TME 7. Primers used for labelling probes for Southern blotting and for qPCR and RT-qPCR assays are shown in Table 1. Bars show SE (n = 4). P, positive control, N genomic DNA from unchallenged plants. Samples from three infected plants were pooled to make one sample, and a total of 4 samples (from 12 plants) were used per treatment combination
Discussion
Virus-Induced Gene Silencing (VIGS) has been used both in model and non-model plant systems to elucidate gene function [16, 32–35]. In cassava, a VIGS system was first reported based on an isolate of African cassava mosaic Cameroon virus (ACMV-CM) [16]. It was reported previously that this ACMV-CM infectious clone is less virulent than EACMV-K201 such that the CMD2-type cultivars TME 204, TME 3 and TME 7 (Oko-iyawo) are infected at low frequencies (0–30%) and develop only mild disease symptoms [15, 28]. This is less than ideal if robust suppression of gene expression is desired within an experimental system. The East African cassava mosaic virus isolate EACMV-K201 [21] is highly virulent and has been shown to infect all cassava genotypes [15, 28]. A new VIGS system was developed, therefore, based on EACMV-K201 by cloning target sequences into the coding region of the CP gene of the DNA-A component. Efficacy of this VIGS vector was confirmed by silencing the endogenous MePSY2 gene in the CMD-susceptible cultivar TME 7S, resulting in visually detectable bleaching and production of chlorotic tissues throughout a 12-week observation period (Fig. 1b).
Screening cassava germplasm for resistance to CMD under field conditions requires many months [10, 36]. Under laboratory or greenhouse conditions this evaluation period is shorter but still needs 12–22 weeks [15, 18] to allow for full disease establishment and expression of the recovery phenotype typical for most cassava cultivars. Data presented here shows that the MeSPY1-VIGS system can be used as a quick screening tool to determine resistance and susceptibility to CMD (Figs. 2, 3 and 4). This was achieved by targeting the cassava SPY gene using a newly developed EACMV-K201-based VIGS vector delivered by biolistic inoculation. The established CMD scoring system for cassava involves visual assessment of symptoms based on a scale of 0–5 [25]. Experienced personnel are required to capture accurate data due to subtle presentation of disease symptoms in some cultivars. The symptom scores are recorded for each individual plant in an experiment often at a frequency of 1–2 times a week for 12–22 weeks depending on the cultivar, virus species and isolate used. Using the MeSPY1-VIGS screening system, only shoot-tip necrosis/death or whole plant death needs be scored, and only once or twice up to 4 weeks post inoculation. On average, this saves 8–18 weeks per inoculation experiment allowing 3–5 times more plants to be tested in the same time period and the same available greenhouse space. This MeSPY-VIGS screening system has been developed using known CMD-resistant and CMD-susceptible cassava cultivars and plant lines that have been well characterized under field and greenhouse conditions [8, 11, 15, 28]. Data generated (Figs. 1, 2, 3 and 4) corroborates accurately with the known CMD response of these cultivars, as further validated with challenge experiment using GFP-VIGS (Fig. 5), showing the robustness of the screening system.
The cause of shoot-tip necrosis that eventually leads to whole plant death in CMD-susceptible cassava lines is not clear. Both virus DNA and MeSPY transcript quantification showed significant differences between the CMD-resistant and CMD-susceptible plant lines, with CMD-susceptible plant lines having greater virus load than resistant lines (Fig. 6a and b) in a manner consistent with our previous report [15]. This viral load corresponded with an expected and significant reduction in MeSPY transcript in CMD-susceptible plants (Fig. 6c). SPY is involved in diverse physiological and developmental roles, including suppression of GA signaling [30], promotion of cytokinin response [37, 38] and enhancement of sensitivity to drought and salinity stress [39]. A SPY mutation in Arabidopsis caused elongation of the stem that phenocopies wild-type plants treated with GA [29, 30]. In Arabidopsis, SPY and SECRET AGENT (SEC) [40, 41] are the only proteins known to have O-linked N-acetylglucosamine transferase activity involved in posttranslational modification of other proteins. We tested if silencing of cassava SEC (MeSEC) mimics what has been observed in plants challenged with MeSPY1-VIGS as shown in this study (Figs. 1, 2, 3 and 4). Silencing of the cassava ortholog of Arabidopsis SEC did not cause lethality or any different phenotype in susceptible or resistant TME 204 and TME 7 cassava cultivars as compared to GFP-VIGS (data not shown), suggesting that MeSPY and MeSEC may not be functionally redundant. The cause of death after challenge with MeSPY1-VIGSs might therefore be due to the crucial role MeSPY plays in plant function and/or due to an unknown interaction with the geminiviruses. Transgenic plants overexpressing MeSPY and RNAi cassava lines are being recovered to further elucidate a putative role of SPY in CMD resistance.
The ability to discriminate between CMD-resistant and-susceptible lines using a simple MeSPY1-VIGS challenge has significant practical application. We recently reported that the CMD2-type cultivars TME3, TME 7 and TME 204 lose inherent resistance to CMD after passage through the process of somatic embryogenesis, and that this phenomenon occurs at an early stage after culture of an explant on auxin-containing media [31]. Meristem tip culture for virus elimination in infected cassava plants to allow movement of plants between countries or within a country, in vitro germplasm preservation and production of transgenic and gene edited plants often employ tissue culture procedures [42, 43]. The rapid CMD screening system developed in this study can easily be applied for determining preservation of CMD resistance after such tissue culture manipulations. Furthermore, the technique can be applied for high throughput screening of large numbers of progenies in a breeding pipeline under controlled greenhouse conditions. Besides understanding the molecular mechanisms involved in the phenotype observed in susceptible cassava lines reported here, investigation into the utility of SPY-VIGS as a screening tool in other plant-virus interactions is needed.
Conclusion
A rapid and reliable screening method for determining resistance to CMD, one of the most important cassava viral diseases, has been developed. This screening system utilizes an EACMV-K201-based VIGS system to target the MeSPY gene and has proven effective across diverse cassava cultivars and lines. With this screening system in place, 3–5 times more plant lines can be screened with the same resources compared to previously available methods. It is therefore well suited for application for high throughput screening of breeding lines and transgenic and gene edited lines under controlled growth conditions.
Acknowledgments
We thank Amita Rai, Theodore Moll, Miriam Khalil, Maxwell Braud, Jackson Gehan, Collin Luebbert, Jennifer Winch, Jacquelyn Leise, Stephanie Lamb, Claire Albin and Mary Lyon at the Donald Danforth Plant Science Center for their technical assistance.
Funding
Funding for this research was provided by the Bill and Melinda Gates Foundation (OPPGD1485), the United States Agency for International Development from the American people (USAID Cooperative Agreement No. AID-EDH-A-00-09-00010), and the Monsanto Fund. The funders had no role in the design of the study; collection, analysis, or interpretation of data; nor in the writing of the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Authors’ contributions
GB and NT conceived the project. RC generated FEC-derived plants. GB, RC and NT designed the project, carried out the experiments, and generated and analyzed the data. GB, RC and NT wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Abbreviations
- ACMV-CM
African cassava mosaic Cameroon virus
- CBSD
Cassava brown streak disease
- CMD
Cassava mosaic disease
- CMG
Cassava mosaic geminivirus
- CP
Coat protein
- DPI
Days post inoculation
- EACMV
East African cassava mosaic virus
- FEC
Friable embryogenic callus
- MePSY2
Manihot esculenta phytoene synthase 2
- MeSEC
Manihot esculenta SECRET AGENT
- MeSPY
Manihot esculenta SPY
- SEC
SECRET AGENT
- SPY
SPINDLY
- TPR
Tetratricopeptide repeat
- VIGS
Virus-induced gene silencing
- WPI
Weeks post inoculation
References
- 1.FAOSTAT Database. FAO, Rome, Italy. 2015. http://faostat3.fao.org/home/E. Accessed 8 Sept 2016.
- 2.El-Sharkawy MA. Cassava biology and physiology. Plant Mol Biol. 2004;56:481–501. doi: 10.1007/s11103-005-2270-7. [DOI] [PubMed] [Google Scholar]
- 3.Legg J, Attiogbevi E, Barker S, Beach L, Ceballos H, Cuellar W, Elkhoury W, Gerling D, Helsen J, Hershey C, Jarvis A, Kulakow P, Kumar L, Lorenzen J, Lynam J, McMahon M, Maruthi G, Miano D, Mtunda K, Natwuruhunga P, Okogbenin E, Pezo P, Terry E, Thiele G, Thresh M, Wadsworth J, Walsh S, Winter S, Tohme J, Fauquet C. A global alliance declaring war on cassava viruses in Africa. Food Secur. 2014;6:231–248. doi: 10.1007/s12571-014-0340-x. [DOI] [Google Scholar]
- 4.Patil BL, Fauquet CM. Cassava mosaic geminiviruses: actual knowledge and perspectives. Mol Plant Pathol. 2009;10:685–701. doi: 10.1111/j.1364-3703.2009.00559.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patil BL, Fauquet CM. Studies on differential behavior of cassava mosaic geminivirus DNA components, symptom recovery patterns, and their siRNA profiles. Virus Genes. 2015;50:474–486. doi: 10.1007/s11262-015-1184-y. [DOI] [PubMed] [Google Scholar]
- 6.Akano O, Dixon O, Mba C, Barrera E, Fregene M. Genetic mapping of a dominant gene conferring resistance to cassava mosaic disease. Theor Appl Genet. 2002;105:521–525. doi: 10.1007/s00122-002-0891-7. [DOI] [PubMed] [Google Scholar]
- 7.Lokko Y, Danquah EY, Offei SK, Dixon AGO, Gedil MA. Molecular markers associated with a new source of resistance to the cassava mosaic disease. Afr J Biotechnol. 2005;4:873–881. [Google Scholar]
- 8.Okogbenin E, Egesi CN, Olasanmi B, Ogundapo O, Kahya S, Hurtado P, Marin J, Akinbo O, Mba C, Gomez H, de Vicente C, Baiyeri S, Uguru M, Ewa F, Fregene M. Molecular marker analysis and validation of resistance to cassava mosaic disease in elite cassava genotypes in Nigeria. Crop Sci. 2012;52:2576–2586. doi: 10.2135/cropsci2011.11.0586. [DOI] [Google Scholar]
- 9.Okogbenin E, Moreno I, Tomkins J, Fauquet CM, Mkamilo G, Fregene M. Marker-Assisted Breeding for Cassava Mosaic Disease Resistance. In: Varshney R, Tuberosa R, editors. Translational Genomics for Crop Breeding: Biotic Stress. Chichester: John Wiley & Sons Ltd; 2013. pp. 291–325. [Google Scholar]
- 10.Kawuki R, Pariyo A, Amuge T, Nuwamanya E, Ssemakula G, Tumwesigye S, Bua A, Baguma Y, Omongo C, Alicai T, Orone J. A breeding scheme for local adoption of cassava (Manihot esculenta Crantz) J Plant Breed Crop Sci. 2011;3:120–130. [Google Scholar]
- 11.Rabbi IY, Hamblin MT, Kumar PL, Gedil MA, Ikpan AS, Jannink J-L, Kulakow PA. High-resolution mapping of resistance to cassava mosaic geminiviruses in cassava using genotyping-by-sequencing and its implications for breeding. Virus Res. 2014;186:87–96. doi: 10.1016/j.virusres.2013.12.028. [DOI] [PubMed] [Google Scholar]
- 12.Vanderschuren H, Moreno I, Anjanappa RB, Zainuddin IM, Gruissem W. Exploiting the combination of natural and genetically engineered resistance to cassava mosaic and cassava brown streak viruses impacting cassava production in Africa. PLoS One. 2012;7:e45277. doi: 10.1371/journal.pone.0045277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagaba H, Beyene G, Trembley C, Alicai T, Fauquet C, Taylor N. Efficient transmission of Cassava brown streak disease viral pathogens by chip bud grafting. BMC Res Notes. 2013;6:516. [DOI] [PMC free article] [PubMed]
- 14.Monde G, Walangululu J, Bragard C. Screening cassava for resistance to cassava mosaic disease by grafting and whitefly inoculation. Arch Phytopathol Pfl. 2012;45:2189–2201. doi: 10.1080/03235408.2012.724969. [DOI] [Google Scholar]
- 15.Beyene G, Chauhan RD, Wagaba H, Moll T, Alicai T, Miano D, Carrington J, Taylor NJ. Loss of CMD2-mediated resistance to cassava mosaic disease in plants regenerated through somatic embryogenesis. Mol Plant Pathol. 2016;17:1095–1110. doi: 10.1111/mpp.12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fofana IB, Sangare A, Collier R, Taylor C, Fauquet CM. A geminivirus-induced gene silencing system for gene function validation in cassava. Plant Mol Biol. 2004;56:613–624. doi: 10.1007/s11103-004-0161-y. [DOI] [PubMed] [Google Scholar]
- 17.Pita JS, Fondong VN, Sangare A, Otim-Nape GW, Ogwal S, Fauquet CM. Recombination, pseudorecombination and synergism of geminiviruses are determinant keys to the epidemic of severe cassava mosaic disease in Uganda. J Gen Virol. 2001;82:655–665. doi: 10.1099/0022-1317-82-3-655. [DOI] [PubMed] [Google Scholar]
- 18.Vanderschuren H, Alder A, Zhang P, Gruissem W. Dose-dependent RNAi-mediated geminivirus resistance in the tropical root crop cassava. Plant Mol Biol. 2009;70:265–272. doi: 10.1007/s11103-009-9472-3. [DOI] [PubMed] [Google Scholar]
- 19.Ntui VO, Kong K, Khan RS, Igawa T, Janavi GJ, Rabindran R, Nakamura I, Mii M. Resistance to Sri Lankan cassava mosaic virus (SLCMV) in genetically engineered cassava cv. KU50 through RNA silencing. PLoS One. 2015;10:e0120551. doi: 10.1371/journal.pone.0120551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patil BL, Fauquet CM. Differential interaction between cassava mosaic geminiviruses and geminivirus satellites. J Gen Virol. 2010;91:1871–1882. doi: 10.1099/vir.0.019513-0. [DOI] [PubMed] [Google Scholar]
- 21.Bull SE, Briddon RW, Sserubombwe WS, Ngugi K, Markham PG, Stanley J. Genetic diversity and phylogeography of cassava mosaic viruses in Kenya. J Gen Virol. 2006;87:3053–3065. doi: 10.1099/vir.0.82013-0. [DOI] [PubMed] [Google Scholar]
- 22.Haseloff J, Siemering KR, Prasher DC, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci U S A. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chauhan RD, Beyene G, Kalyaeva M, Fauquet CM, Taylor N. Improvements in Agrobacterium-mediated transformation of cassava (Manihot esculenta Crantz) for large-scale production of transgenic plants. Plant Cell Tiss Org. 2015;121:591–603. doi: 10.1007/s11240-015-0729-z. [DOI] [Google Scholar]
- 24.Taylor N, Gaitan-Solis E, Moll T, Trauterman B, Jones T, Pranjal A, Trembley C, Abernathy V, Corbin D, Fauquet CM. A High-throughput platform for the production and analysis of transgenic cassava (Manihot esculenta) plants. Trop Plant Biol. 2012;5:127–139. doi: 10.1007/s12042-012-9099-4. [DOI] [Google Scholar]
- 25.Fauquet C, Fargette D. African cassava mosaic virus: etiology, epidemiology and control. Plant Dis. 1990;74:404–411. doi: 10.1094/PD-74-0404. [DOI] [Google Scholar]
- 26.Ogwok E, Alicai T, Rey MEC, Beyene G, Taylor NJ. Distribution and accumulation of cassava brown streak viruses within infected cassava (Manihot esculenta) plants. Plant Pathol. 2015;64:1235–1246. doi: 10.1111/ppa.12343. [DOI] [Google Scholar]
- 27.Moreno I, Gruissem W, Vanderschuren H. Reference genes for reliable potyvirus quantitation in cassava and analysis of Cassava brown streak virus load in host varieties. J Virol Methods. 2011;177:49–54. doi: 10.1016/j.jviromet.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 28.Kuria P, Ilyas M, Ateka E, Miano D, Onguso J, Carrington JC, Taylor NJ. Differential response of cassava genotypes to infection by cassava mosaic geminiviruses. Virus Res. 2016;227:69–81. doi: 10.1016/j.virusres.2016.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jacobsen SE, Olszewski NE. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell. 1993;5:887–896. doi: 10.1105/tpc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacobsen SE, Binkowski KA, Olszewski NE. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci U S A. 1996;93:9292–9296. doi: 10.1073/pnas.93.17.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bredeson JV, Lyons JB, Prochnik SE, Wu GA, Ha CM, Edsinger-Gonzales E, Grimwood J, Schmutz J, Rabbi IY, Egesi C, Nauluvula P, Lebot V, Ndunguru J, Mkamilo G, Bart RS, Setter TL, Gleadow RM, Kulakow P, Ferguson ME, Rounsley S, Rokhsar DS. Sequencing wild and cultivated cassava and related species reveals extensive interspecific hybridization and genetic diversity. Nat Biotechnol. 2016;34:562–570. doi: 10.1038/nbt.3535. [DOI] [PubMed] [Google Scholar]
- 32.Burch-Smith TM, Anderson JC, Martin GB, Dinesh-Kumar SP. Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 2004;39:734–746. doi: 10.1111/j.1365-313X.2004.02158.x. [DOI] [PubMed] [Google Scholar]
- 33.Kjemtrup S, Sampson KS, Peele CG, Nguyen LV, Conkling MA, Thompson WF, Robertson D. Gene silencing from plant DNA carried by a Geminivirus. Plant J. 1998;14:91–100. doi: 10.1046/j.1365-313X.1998.00101.x. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Schiff M, Dinesh-Kumar SP. Virus-induced gene silencing in tomato. Plant J. 2002;31:777–786. doi: 10.1046/j.1365-313X.2002.01394.x. [DOI] [PubMed] [Google Scholar]
- 35.Lu R, Malcuit I, Moffett P, Ruiz MT, Peart J, Wu AJ, Rathjen JP, Bendahmane A, Day L, Baulcombe DC. High throughput virus-induced gene silencing implicates heat shock protein 90 in plant disease resistance. EMBO J. 2003;22:5690–5699. doi: 10.1093/emboj/cdg546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawuki RS, Kaweesi T, Esuma W, Pariyo A, Kayondo IS, Ozimati A, Kyaligonza V, Abaca A, Orone J, Tumuhimbise R, Nuwamanya E, Abidrabo P, Amuge T, Ogwok E, Okao G, Wagaba H, Adiga G, Alicai T, Omongo C, Bua A, Ferguson M, Kanju E, Baguma Y. Eleven years of breeding efforts to combat cassava brown streak disease. Breed Sci. 2016;66:560–571. doi: 10.1270/jsbbs.16005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steiner E, Efroni I, Gopalraj M, Saathoff K, Tseng TS, Kieffer M, Eshed Y, Olszewski N, Weiss D. The Arabidopsis O-linked N-acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell. 2012;24:96–108. doi: 10.1105/tpc.111.093518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steiner E, Livne S, Kobinson-Katz T, Tal L, Pri-Tal O, Mosquna A, Tarkowska D, Mueller B, Tarkowski P, Weiss D. The putative O-linked N-acetylglucosamine transferase SPINDLY inhibits Class I TCP proteolysis to promote sensitivity to cytokinin. Plant Physiol. 2016;171:1485–1494. doi: 10.1104/pp.16.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qin F, Kodaira KS, Maruyama K, Mizoi J, Tran LS, Fujita Y, Morimoto K, Shinozaki K, Yamaguchi-Shinozaki K. SPINDLY, a negative regulator of gibberellic acid signaling, is involved in the plant abiotic stress response. Plant Physiol. 2011;157:1900–1913. doi: 10.1104/pp.111.187302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartweck LM, Genger RK, Grey WM, Olszewski NE. SECRET AGENT and SPINDLY have overlapping roles in the development of Arabidopsis thaliana L. Heyn. J Exp Bot. 2006;57:865–875. doi: 10.1093/jxb/erj071. [DOI] [PubMed] [Google Scholar]
- 41.Hartweck LM, Scott CL, Olszewski NE. Two O-linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics. 2002;161:1279–1291. doi: 10.1093/genetics/161.3.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mwangangi M, Ateka E, Nyende A, Kagundu A. Elimination of Cassava brown streak virus from infected cassava. J Biol Agric Healthc. 2014;4:34–40. [Google Scholar]
- 43.Damba Y, Quainoo A, Sowley E. Effectiveness of somatic embryogenesis in eliminating the Cassava Mosaic Virus from infected cassava (Manihot esculenta Crantz) plant materials. IJSTR. 2013;2:282–287. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.


