Summary
Microglia are tissue macrophages of the central nervous system (CNS). Their key tasks are immune surveillance as well as responding to infections or other pathological states such as neurological diseases or injury. In recent years it has been discovered that microglia are additionally crucial for the maintenance of brain homeostasis during development and adulthood by adjusting the neuronal network and phagocytosing neuronal debris. Microglia persist in the CNS throughout the life of the organism and self‐renew without engraftment of bone‐marrow‐derived cells. Until recently it remained unknown what controls their maturation and activation under homeostatic conditions. In this review we discuss new aspects of the interaction between host microbiota and brain function with special focus on the brain‐resident innate immune cells, the microglia.
Keywords: gut–brain axis, macrophage, microbiota, microglia, short‐chain fatty acids
Introduction
Man and mouse are occupied by trillions of commensal bacteria that co‐exist for potential mutual benefits. This host microbiota colonizes mainly the gut and the skin as well as several mucosal cavities (nasal, oral, vaginal and pulmonary). The microbiota is a necessary element for the synthesis of several vitamins (e.g. vitamins K and B), it provides energy for the host in terms of short‐chain fatty acids (SCFAs) by fermentation of otherwise indigestible carbohydrates and fibres, and is involved in the metabolism of bile acids, sterols and xenobiotics.1 In fact, there is some marked evidence that a complex composition and abundance of the gut microbiota is an essential element for the maintenance of the host's health. Altered microbial compositions have been linked to several human diseases,2 including cardiovascular disease,3 inflammatory bowel disease,4 and type 1 and type 2 diabetes.5, 6 Fairly unexpectedly, multifaceted interactions between the endogenous microbiota and the hosts' central nervous system (CNS) were discovered in the past decade. The influence of the host microbiota on CNS functions have been studied using several experimental approaches, including germ‐free (GF) animals7 that have never been exposed to bacteria and viruses from embryogenesis throughout their life, manipulation or eradication of the gut bacteria with antibiotics at an adult stage,8, 9, 10 transient colonization,11 defined limited colonization12 or faecal microbial transplantation.9 Over the last few years, several new publications have defined the impact of the gut microbiota on the innate and adaptive immune system.13
In this review we highlight and discuss recent findings concerning the environmental factor ‘host microbiota’ and its influences on the CNS with special focus on its resident tissue macrophages, the microglia.
Gut–brain axis: an important connection
First, it has been described that the microbiota affects several aspects of behaviour in mice and in humans in general,14 for example social interaction,15 stress responsiveness,16 depression‐like behaviour,17, 18 anxiety‐like behaviour19, 20, 21, 22, 23 and nociceptive responses.24 Interestingly, it was observed that the colonization of GF BALB/c mice with a strain‐specific microbiota from conventionally (specific pathogen‐free; SPF) raised NIH Swiss mice increased exploratory behaviour and hippocampal levels of brain‐derived neurotropic factor; however, GF NIH Swiss mice with a microbiota derived from conventionally housed BALB/c mice had the opposite effect and showed reduced exploratory behaviour,9 indicating that microbiota‐derived signals can shape host brain function in terms of behaviour. It was shown recently that the host microbiota has an effect on adult hippocampal neurogenesis leading to more immature neurons in young GF mice compared with conventionally colonized control mice, whereas the mechanisms and potential hippocampus‐related behavioural changes are only poorly understood.25, 26
In general, the mechanisms by which the microbiota influences the host are manifold and complex (Fig. 1).27 Commensal microbiota can either have a direct impact on the production of metabolic precursors like tryptophan and neurotransmitters (e.g. serotonin, noradrenaline or dopamine) or produce active mediators like SCFAs, which are commensal anaerobic bacteria‐derived fermentation products.14, 21, 28 SCFAs are also known to inhibit histone deacetylases and thereby result in epigenetic changes.29 Further, the gut is connected to the CNS through the vagus nerve, enabling a direct communication through a neurochemical pathway.30 A third pathway linking the microbiota and the CNS is the release of microbial‐associated molecular patterns (MAMPs) in the gut.14 MAMPs such as bacterial lipoproteins, double‐stranded RNA, lipopolysaccharide and many others are recognized by different receptors, mostly belonging to the Toll‐like receptors (TLRs) in association with the myeloid differentiation primary response gene, MyD88, signalling pathway, which is known to be involved in several aspects of the host immune response.31 A recently proposed mechanism as to how the host may control the microbiome is host epithelial derived miRNA, which is proposed to control bacterial gene expression.32
Figure 1.
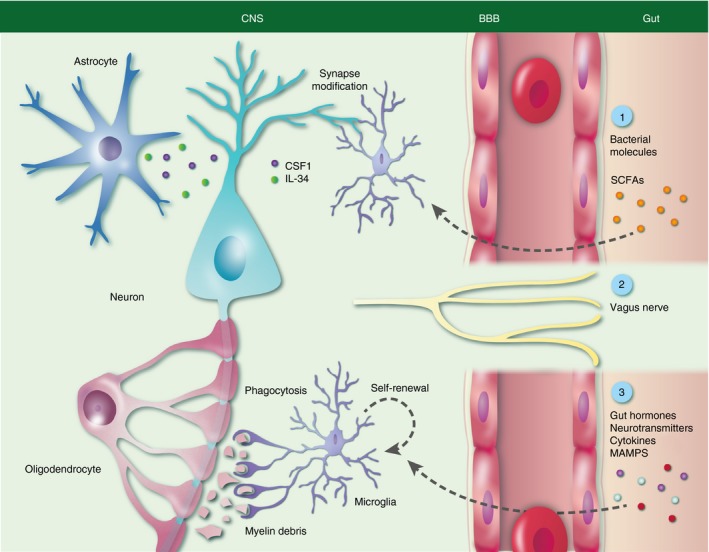
Microglia functions under homeostatic conditions and its modulation by host microbiota. During adulthood cortical microglia survey mature neurons and are involved in learning‐induced dendritic spine formation and nourish neurons. Furthermore, colony‐stimulating factor 1 (CSF‐1), interleukin‐34 (IL‐34) and short‐chain fatty acids (SCFAs) are important components for microglia function and maturation. Direct signalling via the vagus nerve and other molecules like gut hormones, neurotransmitters, cytokines or microbial‐associated molecular patterns (MAMPs) may also affect microglia.
It was suggested by Braniste and colleagues that blood–brain barrier (BBB) integrity in the frontal cortex, hippocampus and striatum is influenced by gut microbiota.33 They provided evidence that BBB permeability is increased in GF mice already during embryonal development – visible from E16.5 until adulthood – caused by reduced tight junction protein expression. Further, the impaired BBB integrity in GF mice could be rescued by recolonization with a complex intestinal microbiota or the application of butyrate. Pericyte numbers seem not to be altered but a more detailed analysis of all components of the BBB, namely endothelial cells, pericytes and astrocytes, including their function and mechanism by which SCFAs mediate BBB integrity remain to be investigated. Intriguingly, antibiotic‐induced alteration of the gut microbiota in adult mice also leads to decreased tight junction expression in the hippocampus, whereas in the amygdala an increased expression of tight junction proteins was observed,34 indicating potential region‐specific differences. Similar findings could be determined regarding the blood–testis barrier, as GF mice feature reduced expression of adherens and tight junctions proteins and increased blood–testis barrier permeability,35 suggesting the general involvement of the gut microbiota in the regulation of the blood–tissue barriers.36
Recent evidence demonstrated myelination being influenced by the microbiota under steady‐state conditions in some specific anatomical brain regions like the prefrontal cortex, but the (patho‐)physiological consequences are not yet examined.37
The impact of microbiota on neuropsychiatric disorders
The findings described above implicate that under homeostatic conditions brain function and architecture are highly influenced by gut microbiota. In addition, it has only recently been appreciated that commensals obviously influence CNS diseases as well.
A crucial role of the gut microbiota could be demonstrated in a relapsing–remitting (RR) mouse model (transgenic SJL/J anti‐MOG TCR transgenic RR mice) whereby mice spontaneously develop features similar to those of human multiple sclerosis.38 Mice that are kept under normal SPF conditions spontaneously develop disease hallmarks mediated by triggering peripheral immune processes driven by myelin‐specific CD4+ T cells, whereas mice housed in a sterile environment are nearly protected.38 It remains open, whether the microbiota influences directly the pathological process or the generally impaired immune system of GF mice is contributing to this observation.
Autism spectrum disorder (ASD) is a frequent neuropsychiatric disorder that is supposed to be caused by multifactorial aetiology.39 Affected people suffer from impaired sociability and communication, as well as frequent repetitive and stereotyped behaviours. Some potential risk factors for ASD are linked to the gut microbiota, such as perinatal infection, hospitalization or early antibiotic exposure.40 It was observed that SCFAs may trigger ASD in rodents41 and, supporting this finding, it has been described that children with ASD display increased faecal SCFA concentrations;42 however, the precise mechanism is not fully understood.40 A probiotic treatment with the human commensal Bacteroides fragilis was shown to ameliorate ASD‐linked behaviour in mice by normalizing 4‐ethylphenylsulphate levels.43
The presence of gut microbiota is also pathophysiologically relevant for Parkinson's disease as patients displayed an altered composition of the gut microbiota including reduced abundance of Prevotellaceae and increased abundance of Enterobacteriaceae compared with matched controls.44 Prevotellaceae are known to provide potential beneficial SCFAs, thiamine and folate, which are found to be decreased in patients with Parkinson's disease.44 The relative abundance of Enterobacteriaceae was associated with the severity of motor symptoms.44 Beyond these correlative data it is proposed that various α‐synuclein forms can spread from the gut to the brain by microtubule‐associated transport via the vagus nerve, suggesting that this pathway might be used for the transport of misfolded proteins as well. However, the detailed mechanism has yet to be determined.
Further, the gut microbiota may play a crucial role in the pathogenesis of other neurodegenerative diseases, such as Alzheimer's disease and amyotrophic lateral sclerosis45, 46 because it is known that neurons,21 astrocytes47 and microglia48 are shaped by the host microbiota and/or their metabolites, and microglial activation accompanied by the production of potential neurotoxic factors are common features of these diseases.49 Hence, it is likely that gut bacteria affect a wide range of neurological disorders, whereby it should be considered generally whether an observed altered composition of the microbiome is cause or consequence. A functional link between ascertained bacterial species and findings should be determined.
Factors modulating fate and function of tissue macrophages in the CNS
Different types of macrophages in the CNS are related to separate compartments. The non‐parenchymal macrophages comprise meningeal, perivascular and choroid plexus macrophages.49, 50 Under steady‐state conditions, microglia are the only myeloid cell type distributed throughout the CNS parenchyma with region‐specific varying numbers and with a wide range of morphologies.51, 52 These CNS immune cells belong to the large family of mononuclear phagocytes, which further includes peripheral tissue‐specific macrophages, different subsets of dendritic cells and circulating monocytes.53, 54 In mice all mentioned CNS macrophages – except choroid plexus macrophages – are derived from prenatal sources such as the yolk sac and self‐renew throughout life.55, 56, 57, 58, 59 Specifically, microglia originate in a c‐MYB‐independent but Runt‐related transcription factor 1 (RUNX1), interferon regulatory factor (IRF8) and PU.1‐dependent manner from CD45− Tie2+ c‐KIT+ F4/80−CX3CR1− erythromyeloid progenitors of the yolk sac during primitive haematopoiesis.55, 56, 60, 61 It is convincingly assumed that during embryonal development the forming CNS is uncoupled by the closing BBB starting from E14.5,62, 63 leading to a defined and restricted environment presumably excluding substantial immigration of peripheral myeloid cells from the definitive haematopoiesis.50, 55, 56, 60, 61 However, in a zebra fish model it was recently described by using high temporal–spatial resolution fate mapping that multiple sources may give rise to adult microglia.64
It is well known that under steady‐state conditions, microglia as innate immune cells actively survey their surrounding environment with their highly motile processes, as shown using in vivo time lapse two‐photon‐imaging of Cx3cr1 GFP mice.65, 66 In addition, several studies revealed that microglia are critical for maintaining tissue homeostasis during development and adulthood.49, 50, 67 In particular, microglia carry out distinct region‐specific tasks such as neuronal circuit development and modification of synapses in the cortex or phagocytosis of myelin debris in the white matter (Fig. 1).68, 69, 70, 71, 72, 73 Various pathological stimuli (such as bacterial or viral infection) or diseases (neuroinflammatory, neurodegenerative, neuro‐oncological or neuropsychiatric) can cause rapid recruitment of microglia to sites of injury, resulting in a resident innate immune response.50, 74, 75 In general, these microglial responses include characteristic macrophage functions such as phagocytosis, antigen presentation and the production and release of immunomodulatory factors.
Compared with their tissue relatives in peripheral organs, microglia exhibit a specific gene expression profile and a distinct chromatin state.76, 77, 78, 79, 80 In two large‐scale studies, Lavin et al.77 and Gosselin et al.78 demonstrated that gene expression patterns and epigenetic identities of macrophages are only partly shaped during development, but rather are shaped also by the local microenvironment, which is capable of reprogramming the genetic imprints, suggesting a remaining plasticity. In fact, Gosselin et al.78 could show that freshly isolated microglia and large peritoneal macrophages lose their specific gene expression patterns to a great extent in vitro. Furthermore, when large peritoneal macrophages were incubated with transforming growth factor‐β 1 (TGF‐β 1), which is important for microglial development and homeostasis,79 approximately 50% of genes usually expressed by microglia in vivo could be induced in these macrophages, indicating that environmental factors in the CNS have a determining influence to induce a microglia phenotype.78 Environmental signals that further shape microglia properties are certainly not limited to TGF‐β 1 and growth factors like colony‐stimulating factor 1 (CSF‐1) and interleukin‐34 (IL‐34) but rather include other factors that are essential for maintaining homeostasis of these unique CNS‐resident tissue macrophages.81
How host microbiota controls microglia maturation and immune function
It was recently shown that host microbiota is also an essential environmental factor shaping the brain innate immune system, in particular the maturation and function of microglia.48 Non‐colonized young adult GF mice exhibit stunted microglia under homeostatic conditions compared with microglia from mice bred under common SPF settings (Fig. 2).48 RNA‐sequencing analysis of FACS‐sorted CD11b+ CD45lo microglia from GF and SPF animals revealed marked differences regarding their cell differentiation, transcription factor binding and proliferation. Microglia expressed reduced mRNA levels for several activation markers under GF conditions (e.g. Il1α, Stat1, Jak3, B2m), whereas transcripts either for inhibitors of transcription, such as Nfkbiα (encodes IκBα), or the essential microglia transcription and survival factor Sfpi1, encoding PU.1, were up‐regulated. Further, the c‐fms gene, encoding for the integral tyrosine kinase transmembrane receptor CSF‐1 receptor (CSF‐1R), which is elementary for microglial proliferation, maturation, function and survival,82, 83, 84, 85 was found to be increased in GF microglia in addition to other genes controlling proliferation, cell cycle and apoptosis (e.g. Cdk9, Ccnd3, Bcl2). There is also evidence that microglia from CSF‐1Rop/op mice display reduced numbers of processes.86 Notably, it is known that DNA damage‐inducible transcript 4 (DDIT4, also known as REDD1) is induced by energy stress and essentially influencing cell growth, proliferation and survival via inhibition of the activity of the mammalian target of rapamycin complex (mTORC) 1 and activation of mTORC2.87, 88, 89 Germ‐free microglia express higher levels of Ddit4 compared with SPF mice. Accordingly, a salient hyper‐ramified morphology as well as an increased microglial density were observed in different brain regions in GF mice accompanied by an increased proliferation rate. In contrast, cell numbers of neuroectodermal cells, such as astrocytes, oligodendrocytes and neurons, were not altered. Although it was recently discovered that some brain regions show a leaky BBB,33 no lymphocytic infiltrates were detectable in GF brains.48
Figure 2.
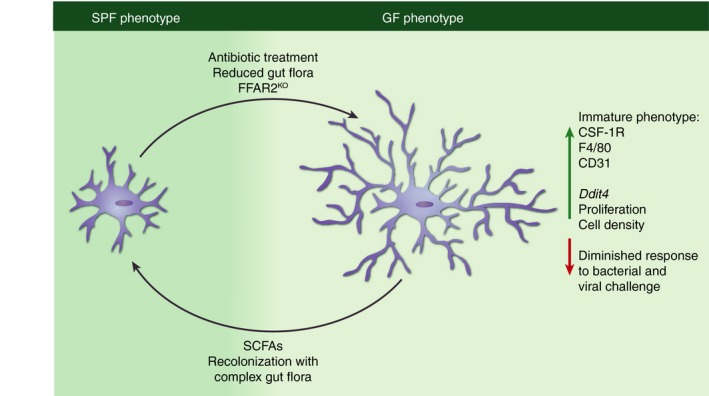
Immature microglia phenotype under germ‐free (GF) conditions. Mice raised under conventional (specific pathogen‐free; SPF) conditions exhibit mature microglia with ramified morphology that respect territorial boundaries (SPF; left). Microglia from GF mice (right) display a hyper‐ramified morphology, increased cell density accompanied by increased proliferation rate, up‐regulation of colony‐stimulating factor 1 receptor (CSF‐1R), F4/80 and CD31 and a changed gene expression profile more related to that of immature cells. Eradication of host microbiota with antibiotics, the presence of a gut flora with strongly limited diversity or FFAR2‐deficiency in the host are causing a microglial phenotype similar to that observed in GF mice, whereas recolonization with a highly diverse gut flora or the application of SCFA is able to re‐establish a mature microglial phenotype.
It is known that microglial numbers increase in the first 2 weeks after birth and then start to decline in the third postnatal week.68, 90 A higher microglia density therefore indicates an immature microglial attribute in GF mice. Remarkably, these findings contrast data from bone‐marrow‐derived haematopoietic cells, such as peripheral tissue macrophages, neutrophils and monocytes, which are most likely found to be diminished in GF mice (Table 1).10, 91, 92 Notably, reduced cell numbers for granulocytes, monocytes, granulocyte–monocyte progenitors and haematopoietic stem/progenitor cells (lineage − Sca1+ c‐kithi cells) corresponds to host microbiota complexity.93 F4/80hi and CD11b+ Gr1− F4/80lo splenic and bone marrow macrophages are described to be reduced in GF mice and in broad‐spectrum antibiotic (ABX)‐treated SPF mice.91 Moreover, it was shown that SCFAs and MAMPs were not sufficient to restore splenic macrophage numbers. Instead, only recolonization with a complex flora was able to rescue macrophage numbers and reduce bacterial burden upon Listeria monocytogenes infection in the spleen, indicating a restored proper innate immune response against infection.91 Further, Kupffer cells, the tissue macrophages of the liver, were reduced in GF and ABX‐treated Swiss Webster male mice94 and C57BL/6 mice, respectively.91 Interestingly, Kupffer cells exhibited a reduced MHC class II expression and an increased phagocytic capacity in GF animals.89 These ABX‐treated mice displayed mitigated liver damage in an ischaemia–reperfusion injury model.94 Contradictory data were recently published by Zhang et al. that did not find changed Kupffer cell numbers in the liver of ABX‐treated C57BL/6 mice.92 In addition, dermal macrophage populations were found to be decreased in GF mice95.
Table 1.
Effect of host microbiota on cell density in different innate immune cell populations
| Immune cell type | Cell density (GF) | Cell density (ABX) | Change of growth factors | References |
|---|---|---|---|---|
| Microglia | ↑ | No change |
CSF‐1: Slightly ↑ IL‐34: not changed |
Erny et al. (2015)48 |
| Neutrophils | ↓ | ↓ | CSF‐2↓ |
Zhang et al. (2015)92
Khosravi et al. (2014)91 Balmer et al. (2014)93 Deshmukh et al. (2014)10 |
| Monocytes | ↓ (spleen) | ↓ | Suggested CSF‐1 ↓ (data not shown) | Khosravi et al. (2014)91 |
| ↓ (bone marrow) ↓ (blood) | Not changed Not changed | Zhang et al. (2015)92 | ||
| Kupffer cells | ↓ | ↓ | Not changed |
Corbitt et al. (2013)94
Khosravi et al. (2014)91 |
| Not changed | Zhang et al. (2015)92 | |||
| Gut macrophages | ↓ | CSF‐1 ↓ | Muller et al. (2014)97 | |
| Splenic macrophages | ↓ | ↓ | Suggested CSF‐1 ↓ (data not shown) |
Khosravi et al. (2014)91
Zhang et al. (2015)92 |
| Bone marrow macrophages | ↓ | Suggested CSF‐1 ↓ (data not shown) | Khosravi et al. (2014)91 | |
| Skin macrophages | ↓ | Unknown | Tamoutounour et al. (2013)95 |
The often observed decrease in cell numbers might be in some cases caused by the fact that growth factors like CSF‐1 or CSF‐2 are reduced in the respective compartments or organs of GF animals (Table 1).10, 96, 97 However, brains of GF animals showed slightly increased Csf1 mRNA levels in the cortex and cerebellum whereas the second known ligand for the CSF‐1R Il34 was not affected by gut microbiota.98, 99
One common feature of microglia is that each cell covers its own defined territory.100 In contrast, GF microglia frequently crossed the neighbouring microglial territories and touched adjacent microglia, indicating a disturbed microglial network. In FACS analysis microglia from GF mice showed an up‐regulation of the surface markers CSF‐1R, F4/80 and CD31, which are known to be down‐regulated in mature adult microglia.55 In addition, the expression of the maturation marker Apoe was increased in GF microglia, usually declining during microglial maturation.79 Another crucial factor for the homeostasis of microglia is TGF‐β as Tgfβ1‐deficient mice show reduced microglia numbers in the CNS.79 Interestingly, we detected – in line with the already mentioned augmented microglial density – an increased Tgfβ1 expression in microglia from GF‐housed mice.79
To further investigate the bacterial factors mediating the maturation of microglia, mice harbouring a strongly reduced microbiota with a defined subset of known bacterial species, so‐called tri‐colonized altered Schaedler flora (ASF) mice, were analysed. These mice harboured only three bacterial strains, namely Bacteroides distasonis (strain ASF 519), Lactobacillus salivarius (strain ASF 361) and Clostridium cluster XIV (ASF 356),101 instead of the 400–1000 strains usually found in SPF mice.101 Although SPF controls and tri‐colonized ASF mice exhibited the same bacterial loads, microglia from the tri‐colonized ASF mice had still markedly altered microglia morphology, function and maturation, which indicates that bacterial complexity is essential for microglia prosperity.48 Importantly, recolonization with a diverse microbiota through SPF donors largely rescued the immature microglial phenotype, which could have major therapeutic implications as the full maturation of these cells can also be attained at later stages during adulthood. As GF mice have never been exposed to microbiota and therefore microglia development might also be influenced in those mice, the microbiota of adult SPF mice was eradicated by ABX. Microglia from these ABX‐treated mice displayed an immature phenotype of microglia reminiscent of the malformed microglia observed in GF animals, suggesting that continuous input signals from host microbiota are required for microglia properties postnatally, independent from developmental imprints. It was proposed that minocycline, a second‐generation tetracycline, inhibits microglial activation and therefore affects neurodegenerative, neuropsychiatric and inflammatory diseases.102, 103 However, it is not yet clarified how this ‘inhibition’ is generated. Orally administered minocycline can certainly affect the composition of the gut microbiota but is also well‐absorbed and is able to penetrate the brain parenchyma and could inhibit microglia independent of the abundance of the microbiota.104 There is some evidence that minocycline attenuates the nuclear factor‐κB pathway in microglia in vitro but the exact mechanism is not understood.105 The potential interaction of minocycline and microglia is further discussed elsewhere.106 Interestingly, in control experiments intraperitoneal administration of a mixture of non‐absorbable antibiotics (namely neomycin, bacitracin and pimaricin) to SPF mice or oral administration to GF mice did not influence behaviour.9 The antibiotic cocktail used in the study48 was composed of three antibiotics (cefoxitine, gentamicin and vancomycin) that are described not to reach the brain tissue,107 but the fourth substance metronidazole is considered to penetrate at least as poorly the CNS.107 Therefore, further experiments are necessary to clarify whether metronidazole is able to affect microglia directly.
The pathways mediating this gut–microglia connection are not yet known but in contrast to other myeloid cells92, 108 this is most likely independent of TLR‐signalling as microglia from mice deficient for TLR‐2, ‐3, ‐4, ‐7and ‐9109 have similar features to the respective wild‐type animals.48 Instead, SCFAs rather than TLR ligands were found to mediate signals to microglia in vivo.48 The oral application of a mixture of the three major SCFAs acetate, propionate and butyrate was sufficient to drive maturation of microglia. It is known that SCFAs are able to cross the BBB110, 111 and may therefore affect microglia directly. In general, SCFAs can also be recognized by specific receptors, such as free fatty acid receptor 2 (FFAR2) [also known as G protein‐coupled receptor (GPR) 43)] or FFAR3 (GPR41).112, 113 In general, GPRs are a large class of seven transmembrane spanning proteins that regulate a wide range of signalling events, whereas activation of FFAR2 by SCFAs is mediated through a dual coupling through Gi/o and Gq signalling.113 It is known that FFAR2 regulates immune system properties.114 Interestingly, FFAR2‐deficient mice displayed microglia reminiscent of those found in GF mice, although FFAR2 is not expressed in any adult brain cell including microglia and endothelial cells.76, 113 This indicates that an indirect signalling of SCFAs through peripheral myeloid or lymphocytic cells expressing FFAR2, such as splenic or enteric macrophages, to the adult brain is conceivable. Despite this possible connection, it has not been investigated whether FFAR2 is expressed at earlier stages during development. Other possible routes linking the gut and microglia such as direct signalling via the vagus nerve or other mediators like gut hormones, neurotransmitters, cytokines or MAMPs, which may also affect microglia, have not been investigated (Fig. 1).27
What are the functional consequences of microglial malformation and immaturity discovered in GF mice? Upon either bacterial (lipopolysaccharide) or viral (lymphocytic choriomeningitis virus) challenge, microglia from GF mice exhibited a restricted or disturbed innate immune response compared with microglia from SPF mice. Similar impaired immune responses are also described for other innate immune cells in ABX‐treated SPF or GF mice, such as peritoneal macrophages115 alveolar macrophages116 or natural killer cells.108
Concluding remarks
In the past few years, the link between the host microbiota and human health and disease has become apparent. An unbalanced composition of host microbiota is becoming recognized as a crucial environmental factor that strongly impacts the host immune system, metabolism and has an important role in many diseases, such as systemic diseases (obesity, diabetes), gut‐related irritable bowel syndrome and inflammatory bowel disease.117 A recent study by Tillisch and colleagues described how ingestion of probiotic bacteria alters brain function in humans.118 It can be assumed that numerous interactions between host microbiota and the CNS immune systems exist, which can essentially shape the outcome of a plethora of neuroinflammatory, neuro‐oncological and neurodegenerative CNS diseases. The future treatment of CNS disorders in man can certainly take advantage of the intimate and mutual interactions of the gut inhabitants with the brain that can be considered as an ‘axis of the good’.
Disclosures
The authors declare no competing financial interests.
Acknowledgements
MP was supported by the DFG (SFB 992, SFB 1160, Reinhart Koselleck Grant), the Fritz‐Thyssen Foundation, the European Union's Seventh Framework Program FP7 under Grant agreement 607962 (nEUROinflammation) and the Sobek Foundation.
References
- 1. Cummings JH, Macfarlane GT. Colonic microflora: nutrition and health. Nutrition 1997; 13:476–8. [DOI] [PubMed] [Google Scholar]
- 2. Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature 2012; 489:242–9. [DOI] [PubMed] [Google Scholar]
- 3. Howitt MR, Garrett WS. A complex microworld in the gut: gut microbiota and cardiovascular disease connectivity. Nat Med 2012; 18:1188–9. [DOI] [PubMed] [Google Scholar]
- 4. Sheehan D, Moran C, Shanahan F. The microbiota in inflammatory bowel disease. J Gastroenterol 2015; 50:495–507. [DOI] [PubMed] [Google Scholar]
- 5. Markle JG, Mortin‐Toth S, Wong AS, Geng L, Hayday A, Danska JS. γδ T cells are essential effectors of type 1 diabetes in the nonobese diabetic mouse model. J Immunol 2013; 190:5392–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, et al A metagenome‐wide association study of gut microbiota in type 2 diabetes. Nature 2012; 490:55–60. [DOI] [PubMed] [Google Scholar]
- 7. Smith K, McCoy KD, Macpherson AJ. Use of axenic animals in studying the adaptation of mammals to their commensal intestinal microbiota. Semin Immunol 2007; 19:59–69. [DOI] [PubMed] [Google Scholar]
- 8. Rakoff‐Nahoum S, Paglino J, Eslami‐Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll‐like receptors is required for intestinal homeostasis. Cell 2004; 118:229–41. [DOI] [PubMed] [Google Scholar]
- 9. Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, et al The intestinal microbiota affect central levels of brain‐derived neurotropic factor and behavior in mice. Gastroenterology 2011; 141:599–609. [DOI] [PubMed] [Google Scholar]
- 10. Deshmukh HS, Liu Y, Menkiti OR, Mei J, Dai N, O'Leary CE, et al The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med 2014; 20:524–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gomez de Aguero M, Ganal‐Vonarburg SC, Fuhrer T, Rupp S, Uchimura Y, Li H, et al The maternal microbiota drives early postnatal innate immune development. Science 2016; 351:1296–302. [DOI] [PubMed] [Google Scholar]
- 12. Schaedler RW, Dubos RJ. The fecal flora of various strains of mice. Its bearing on their susceptibility to endotoxin. J Exp Med 1962; 115:1149–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Geuking MB, Koller Y, Rupp S, McCoy KD. The interplay between the gut microbiota and the immune system. Gut Microbes 2014; 5:411–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sampson TR, Mazmanian SK. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe 2015; 17:565–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Desbonnet L, Clarke G, Shanahan F, Dinan TG, Cryan JF. Microbiota is essential for social development in the mouse. Mol Psychiatry 2014; 19:146–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Desbonnet L, Garrett L, Clarke G, Kiely B, Cryan JF, Dinan TG. Effects of the probiotic Bifidobacterium infantis in the maternal separation model of depression. Neuroscience 2010; 170:1179–88. [DOI] [PubMed] [Google Scholar]
- 17. Steenbergen L, Sellaro R, van Hemert S, Bosch JA, Colzato LS. A randomized controlled trial to test the effect of multispecies probiotics on cognitive reactivity to sad mood. Brain Behav Immun 2015; 48:258–64. [DOI] [PubMed] [Google Scholar]
- 18. Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, et al Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci USA 2011; 108:16050–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Selkrig J, Wong P, Zhang X, Pettersson S. Metabolic tinkering by the gut microbiome: implications for brain development and function. Gut Microbes 2014; 5:369–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Clarke G, Grenham S, Scully P, Fitzgerald P, Moloney RD, Shanahan F, et al The microbiome‐gut‐brain axis during early life regulates the hippocampal serotonergic system in a sex‐dependent manner. Mol Psychiatry 2013; 18:666–73. [DOI] [PubMed] [Google Scholar]
- 21. Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, et al Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci USA 2011; 108:3047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Neufeld KA, Kang N, Bienenstock J, Foster JA. Effects of intestinal microbiota on anxiety‐like behavior. Commun Integr Biol 2011; 4:492–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Messaoudi M, Lalonde R, Violle N, Javelot H, Desor D, Nejdi A, et al Assessment of psychotropic‐like properties of a probiotic formulation (Lactobacillus helveticus R0052 and Bifidobacterium longum R0175) in rats and human subjects. Br J Nutr 2011; 105:755–64. [DOI] [PubMed] [Google Scholar]
- 24. Amaral FA, Sachs D, Costa VV, Fagundes CT, Cisalpino D, Cunha TM, et al Commensal microbiota is fundamental for the development of inflammatory pain. Proc Natl Acad Sci USA 2008; 105:2193–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marin‐Burgin A, Schinder AF. Requirement of adult‐born neurons for hippocampus‐dependent learning. Behav Brain Res 2012; 227:391–9. [DOI] [PubMed] [Google Scholar]
- 26. Ogbonnaya ES, Clarke G, Shanahan F, Dinan TG, Cryan JF, O'Leary OF. Adult hippocampal neurogenesis is regulated by the microbiome. Biol Psychiatry 2015; 78:e7–9. [DOI] [PubMed] [Google Scholar]
- 27. Collins SM, Surette M, Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol 2012; 10:735–42. [DOI] [PubMed] [Google Scholar]
- 28. Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, et al Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015; 161:264–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu J, Zhou Z, Hu Y, Dong S. Butyrate‐induced GPR41 activation inhibits histone acetylation and cell growth. J Genet Genomics 2012; 39:375–84. [DOI] [PubMed] [Google Scholar]
- 30. Forsythe P, Bienenstock J, Kunze WA. Vagal pathways for microbiome‐brain–gut axis communication. Adv Exp Med Biol 2014; 817:115–33. [DOI] [PubMed] [Google Scholar]
- 31. Akira S, Hemmi H. Recognition of pathogen‐associated molecular patterns by TLR family. Immunol Lett 2003; 85:85–95. [DOI] [PubMed] [Google Scholar]
- 32. Liu S, da Cunha AP, Rezende RM, Cialic R, Wei Z, Bry L, et al The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe 2016; 19:32–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Braniste V, Al‐Asmakh M, Kowal C, Anuar F, Abbaspour A, Toth M, et al The gut microbiota influences blood–brain barrier permeability in mice. Sci Transl Med 2014; 6:263ra158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Frohlich EE, Farzi A, Mayerhofer R, Reichmann F, Jacan A, Wagner B, et al Cognitive impairment by antibiotic‐induced gut dysbiosis: analysis of gut microbiota‐brain communication. Brain Behav Immun 2016; 56: 140–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Al‐Asmakh M, Stukenborg JB, Reda A, Anuar F, Strand ML, Hedin L, et al The gut microbiota and developmental programming of the testis in mice. PLoS One 2014; 9:e103809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Al‐Asmakh M, Hedin L. Microbiota and the control of blood‐tissue barriers. Tissue Barriers 2015; 3:e1039691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hoban AE, Stilling RM, Ryan FJ, Shanahan F, Dinan TG, Claesson MJ, et al Regulation of prefrontal cortex myelination by the microbiota. Transl Psychiatry 2016; 6:e774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C, et al Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 2011; 479:538–41. [DOI] [PubMed] [Google Scholar]
- 39. Mulle JG, Sharp WG, Cubells JF. The gut microbiome: a new frontier in autism research. Curr Psychiatry Rep 2013; 15:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. MacFabe DF. Enteric short‐chain fatty acids: microbial messengers of metabolism, mitochondria, and mind: implications in autism spectrum disorders. Microb Ecol Health Dis 2015; 26:28177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. MacFabe DF, Cain DP, Rodriguez‐Capote K, Franklin AE, Hoffman JE, Boon F, et al Neurobiological effects of intraventricular propionic acid in rats: possible role of short chain fatty acids on the pathogenesis and characteristics of autism spectrum disorders. Behav Brain Res 2007; 176:149–69. [DOI] [PubMed] [Google Scholar]
- 42. Wang L, Christophersen CT, Sorich MJ, Gerber JP, Angley MT, Conlon MA. Elevated fecal short chain fatty acid and ammonia concentrations in children with autism spectrum disorder. Dig Dis Sci 2012; 57:2096–102. [DOI] [PubMed] [Google Scholar]
- 43. Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, et al Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013; 155:1451–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Scheperjans F, Aho V, Pereira PA, Koskinen K, Paulin L, Pekkonen E, et al Gut microbiota are related to Parkinson's disease and clinical phenotype. Mov Disord 2015; 30:350–8. [DOI] [PubMed] [Google Scholar]
- 45. Fang X. Potential role of gut microbiota and tissue barriers in Parkinson's disease and amyotrophic lateral sclerosis. Int J Neurosci 2015; 126: 771–6. [DOI] [PubMed] [Google Scholar]
- 46. Bhattacharjee S, Lukiw WJ. Alzheimer's disease and the microbiome. Front Cell Neurosci 2013; 7:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rothhammer V, Mascanfroni ID, Bunse L, Takenaka MC, Kenison JE, Mayo L, et al Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med 2016; 22: 586–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Erny D, Hrabe de Angelis AL, Jaitin D, Wieghofer P, Staszewski O, David E, et al Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci 2015; 18:965–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Prinz M, Priller J, Sisodia SS, Ransohoff RM. Heterogeneity of CNS myeloid cells and their roles in neurodegeneration. Nat Neurosci 2011; 14:1227–35. [DOI] [PubMed] [Google Scholar]
- 50. Prinz M, Priller J. Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat Rev Neurosci 2014; 15:300–12. [DOI] [PubMed] [Google Scholar]
- 51. Perry VH, Gordon S. Macrophages and microglia in the nervous system. Trends Neurosci 1988; 11:273–7. [DOI] [PubMed] [Google Scholar]
- 52. Lawson LJ, Perry VH, Dri P, Gordon S. Heterogeneity in the distribution and morphology of microglia in the normal adult mouse brain. Neuroscience 1990; 39:151–70. [DOI] [PubMed] [Google Scholar]
- 53. Ginhoux F, Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat Rev Immunol 2014; 14:392–404. [DOI] [PubMed] [Google Scholar]
- 54. Gomez Perdiguero E, Schulz C, Geissmann F. Development and homeostasis of “resident” myeloid cells: the case of the microglia. Glia 2013; 61:112–20. [DOI] [PubMed] [Google Scholar]
- 55. Kierdorf K, Erny D, Goldmann T, Sander V, Schulz C, Perdiguero EG, et al Microglia emerge from erythromyeloid precursors via Pu.1‐ and Irf8‐dependent pathways. Nat Neurosci 2013; 16:273–80. [DOI] [PubMed] [Google Scholar]
- 56. Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, et al Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 2010; 330:841–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Alliot F, Godin I, Pessac B. Microglia derive from progenitors, originating from the yolk sac, and which proliferate in the brain. Brain Res Dev Brain Res 1999; 117:145–52. [DOI] [PubMed] [Google Scholar]
- 58. Goldmann T, Wieghofer P, Muller PF, Wolf Y, Varol D, Yona S, et al A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat Neurosci 2013; 16:1618–26. [DOI] [PubMed] [Google Scholar]
- 59. Goldmann T, Wieghofer P, Jordao MJ, Prutek F, Hagemeyer N, Frenzel K, et al Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat Immunol 2016; 17: 797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schulz C, Gomez Perdiguero E, Chorro L, Szabo‐Rogers H, Cagnard N, Kierdorf K, et al A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 2012; 336:86–90. [DOI] [PubMed] [Google Scholar]
- 61. Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, et al Tissue‐resident macrophages originate from yolk‐sac‐derived erythro‐myeloid progenitors. Nature 2015; 518:547–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hagan N, Ben‐Zvi A. The molecular, cellular, and morphological components of blood–brain barrier development during embryogenesis. Semin Cell Dev Biol 2015; 38:7–15. [DOI] [PubMed] [Google Scholar]
- 63. Daneman R, Zhou L, Kebede AA, Barres BA. Pericytes are required for blood‐brain barrier integrity during embryogenesis. Nature 2010; 468:562–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xu J, Zhu L, He S, Wu Y, Jin W, Yu T, et al Temporal‐spatial resolution fate mapping reveals distinct origins for embryonic and adult microglia in Zebrafish. Dev Cell 2015; 34:632–41. [DOI] [PubMed] [Google Scholar]
- 65. Davalos D, Grutzendler J, Yang G, Kim JV, Zuo Y, Jung S, et al ATP mediates rapid microglial response to local brain injury in vivo . Nat Neurosci 2005; 8:752–8. [DOI] [PubMed] [Google Scholar]
- 66. Nimmerjahn A, Kirchhoff F, Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo . Science 2005; 308:1314–8. [DOI] [PubMed] [Google Scholar]
- 67. Goldmann T, Zeller N, Raasch J, Kierdorf K, Frenzel K, Ketscher L, et al USP18 lack in microglia causes destructive interferonopathy of the mouse brain. EMBO J 2015; 34:1612–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, et al Synaptic pruning by microglia is necessary for normal brain development. Science 2011; 333:1456–8. [DOI] [PubMed] [Google Scholar]
- 69. Wake H, Moorhouse AJ, Miyamoto A, Nabekura J. Microglia: actively surveying and shaping neuronal circuit structure and function. Trends Neurosci 2013; 36:209–17. [DOI] [PubMed] [Google Scholar]
- 70. Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR, 3rd Lafaille JJ, et al Microglia promote learning‐dependent synapse formation through brain‐derived neurotrophic factor. Cell 2013; 155:1596–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, et al Microglia sculpt postnatal neural circuits in an activity and complement‐dependent manner. Neuron 2012; 74:691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Fitzner D, Schnaars M, van Rossum D, Krishnamoorthy G, Dibaj P, Bakhti M, et al Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. J Cell Sci 2011; 124:447–58. [DOI] [PubMed] [Google Scholar]
- 73. Safaiyan S, Kannaiyan N, Snaidero N, Brioschi S, Biber K, Yona S, et al Age‐related myelin degradation burdens the clearance function of microglia during aging. Nat Neurosci 2016; 19: 995–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ransohoff RM, Perry VH. Microglial physiology: unique stimuli, specialized responses. Annu Rev Immunol 2009; 27:119–45. [DOI] [PubMed] [Google Scholar]
- 75. Hanisch UK, Kettenmann H. Microglia: active sensor and versatile effector cells in the normal and pathologic brain. Nat Neurosci 2007; 10:1387–94. [DOI] [PubMed] [Google Scholar]
- 76. Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, et al Gene‐expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol 2012; 13:1118–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lavin Y, Winter D, Blecher‐Gonen R, David E, Keren‐Shaul H, Merad M, et al Tissue‐resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 2014; 159:1312–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, et al Environment drives selection and function of enhancers controlling tissue‐specific macrophage identities. Cell 2014; 159:1327–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Butovsky O, Jedrychowski MP, Moore CS, Cialic R, Lanser AJ, Gabriely G, et al Identification of a unique TGF‐β‐dependent molecular and functional signature in microglia. Nat Neurosci 2014; 17:131–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hickman SE, Kingery ND, Ohsumi TK, Borowsky ML, Wang LC, Means TK, et al The microglial sensome revealed by direct RNA sequencing. Nat Neurosci 2013; 16:1896–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Amit I, Winter DR, Jung S. The role of the local environment and epigenetics in shaping macrophage identity and their effect on tissue homeostasis. Nat Immunol 2016; 17:18–25. [DOI] [PubMed] [Google Scholar]
- 82. Kierdorf K, Prinz M. Factors regulating microglia activation. Front Cell Neurosci 2013; 7:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Erblich B, Zhu L, Etgen AM, Dobrenis K, Pollard JW. Absence of colony stimulation factor‐1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLoS One 2011; 6:e26317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chitu V, Gokhan S, Nandi S, Mehler MF, Stanley ER. Emerging Roles for CSF‐1 receptor and its ligands in the nervous system. Trends Neurosci 2016; 39: 378–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Elmore MR, Najafi AR, Koike MA, Dagher NN, Spangenberg EE, Rice RA, et al Colony‐stimulating factor 1 receptor signaling is necessary for microglia viability, unmasking a microglia progenitor cell in the adult brain. Neuron 2014; 82:380–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sasaki A, Yokoo H, Naito M, Kaizu C, Shultz LD, Nakazato Y. Effects of macrophage‐colony‐stimulating factor deficiency on the maturation of microglia and brain macrophages and on their expression of scavenger receptor. Neuropathology 2000; 20:134–42. [DOI] [PubMed] [Google Scholar]
- 87. Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012; 149:274–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Sofer A, Lei K, Johannessen CM, Ellisen LW. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol Cell Biol 2005; 25:5834–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Weichhart T, Hengstschlager M, Linke M. Regulation of innate immune cell function by mTOR. Nat Rev Immunol 2015; 15:599–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Nikodemova M, Kimyon RS, De I, Small AL, Collier LS, Watters JJ. Microglial numbers attain adult levels after undergoing a rapid decrease in cell number in the third postnatal week. J Neuroimmunol 2015; 278:280–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Khosravi A, Yanez A, Price JG, Chow A, Merad M, Goodridge HS, et al Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe 2014; 15:374–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zhang D, Chen G, Manwani D, Mortha A, Xu C, Faith JJ, et al Neutrophil ageing is regulated by the microbiome. Nature 2015; 525:528–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Balmer ML, Schurch CM, Saito Y, Geuking MB, Li H, Cuenca M, et al Microbiota‐derived compounds drive steady‐state granulopoiesis via MyD88/TICAM signaling. J Immunol 2014; 193:5273–83. [DOI] [PubMed] [Google Scholar]
- 94. Corbitt N, Kimura S, Isse K, Specht S, Chedwick L, Rosborough BR, et al Gut bacteria drive Kupffer cell expansion via MAMP‐mediated ICAM‐1 induction on sinusoidal endothelium and influence preservation‐reperfusion injury after orthotopic liver transplantation. Am J Pathol 2013; 182:180–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Tamoutounour S, Guilliams M, Montanana Sanchis F, Liu H, Terhorst D, Malosse C, et al Origins and functional specialization of macrophages and of conventional and monocyte‐derived dendritic cells in mouse skin. Immunity 2013; 39:925–38. [DOI] [PubMed] [Google Scholar]
- 96. Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, et al Microbiota‐dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science 2014; 343:1249288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Muller PA, Koscso B, Rajani GM, Stevanovic K, Berres ML, Hashimoto D, et al Crosstalk between muscularis macrophages and enteric neurons regulates gastrointestinal motility. Cell 2014; 158:300–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wang Y, Szretter KJ, Vermi W, Gilfillan S, Rossini C, Cella M, et al IL‐34 is a tissue‐restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat Immunol 2012; 13:753–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Wei S, Nandi S, Chitu V, Yeung YG, Yu W, Huang M, et al Functional overlap but differential expression of CSF‐1 and IL‐34 in their CSF‐1 receptor‐mediated regulation of myeloid cells. J Leukoc Biol 2010; 88:495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Kettenmann H, Hanisch UK, Noda M, Verkhratsky A. Physiology of microglia. Physiol Rev 2011; 91:461–553. [DOI] [PubMed] [Google Scholar]
- 101. Stecher B, Chaffron S, Kappeli R, Hapfelmeier S, Freedrich S, Weber TC, et al Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog 2010; 6:e1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Yrjanheikki J, Tikka T, Keinanen R, Goldsteins G, Chan PH, Koistinaho J. A tetracycline derivative, minocycline, reduces inflammation and protects against focal cerebral ischemia with a wide therapeutic window. Proc Natl Acad Sci USA 1999; 96:13496–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Zhang L, Zhao J. Profile of minocycline and its potential in the treatment of schizophrenia. Neuropsychiatr Dis Treat 2014; 10:1103–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Yong VW, Wells J, Giuliani F, Casha S, Power C, Metz LM. The promise of minocycline in neurology. Lancet Neurol 2004; 3:744–51. [DOI] [PubMed] [Google Scholar]
- 105. Kobayashi K, Imagama S, Ohgomori T, Hirano K, Uchimura K, Sakamoto K, et al Minocycline selectively inhibits M1 polarization of microglia. Cell Death Dis 2013; 4:e525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Moller T, Bard F, Bhattacharya A, Biber K, Campbell B, Dale E, et al Critical data‐based re‐evaluation of minocycline as a putative specific microglia inhibitor. Glia 2016; doi: 10.1002/glia.23007. [DOI] [PubMed] [Google Scholar]
- 107. Nau R, Sorgel F, Eiffert H. Penetration of drugs through the blood–cerebrospinal fluid/blood–brain barrier for treatment of central nervous system infections. Clin Microbiol Rev 2010; 23:858–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ganal SC, Sanos SL, Kallfass C, Oberle K, Johner C, Kirschning C, et al Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity 2012; 37:171–86. [DOI] [PubMed] [Google Scholar]
- 109. Conrad ML, Ferstl R, Teich R, Brand S, Blumer N, Yildirim AO, et al Maternal TLR signaling is required for prenatal asthma protection by the nonpathogenic microbe Acinetobacter woffii F78. J Exp Med 2009; 206:2869–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Frost G, Sleeth ML, Sahuri‐Arisoylu M, Lizarbe B, Cerdan S, Brody L, et al The short‐chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat Commun 2014; 5:3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Huuskonen J, Suuronen T, Nuutinen T, Kyrylenko S, Salminen A. Regulation of microglial inflammatory response by sodium butyrate and short‐chain fatty acids. Br J Pharmacol 2004; 141:874–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Layden BT, Angueira AR, Brodsky M, Durai V, Lowe WL Jr. Short chain fatty acids and their receptors: new metabolic targets. Transl Res 2013; 161:131–40. [DOI] [PubMed] [Google Scholar]
- 113. Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D, et al The Orphan G protein‐coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem 2003; 278:11312–9. [DOI] [PubMed] [Google Scholar]
- 114. Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, et al Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009; 461:1282–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, et al Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 2012; 37:158–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Schuijt TJ, Lankelma JM, Scicluna BP, de Sousa EMF, Roelofs JJ, de Boer JD, et al The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut 2016; 65:575–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Bull MJ, Plummer NT. Part 1: the human gut microbiome in health and disease. Integr Med (Encinitas) 2014; 13:17–22. [PMC free article] [PubMed] [Google Scholar]
- 118. Tillisch K, Labus J, Kilpatrick L, Jiang Z, Stains J, Ebrat B, et al Consumption of fermented milk product with probiotic modulates brain activity. Gastroenterology 2013; 144:1394–401, 1401 e1391‐1394. [DOI] [PMC free article] [PubMed] [Google Scholar]


