Abstract
PLIC, Protein Linking IAP (CD47) to Cytoskeleton, have long since been implicated in connecting the extracellular membrane to the intracellular cell cytoskeleton. This phenomenon is supposedly achieved by bridging a receptor protein CD47 to vimentin, an intermediate filament, which in turn regulates integrin dependent cell spreading. Since the discovery of these proteins, the molecular details of the above-mentioned interactions and the underlying complexes are yet to be characterized. Several independent studies have together emphasized PLIC/Ubiquilin’s role in the proteasomal degradation pathway. This seems to be in contrast to the purported initial discovery of PLIC as a cytoskeletal adaptor protein. In an effort to reconcile the different roles associated with the ubiquitous PLIC proteins, we tested the involvement of PLIC-2 both in the proteasomal degradation pathway and as a protein linking the cell cytoskeleton to the cytoplasmic tail of CD47. This was achieved thorough an in vitro investigation of their binding interface using a combination of biophysical techniques. Our results show that the two terminal domains of PLIC-2 interact weakly with each other, while the C-terminal UBA domain interacts strongly with ubiquitin. Interestingly, no perceptible interaction was observed for PLIC-2 with the cytoplasmic tail of CD47 questioning its role as a “PLIC” protein linking the cell membrane to the cytoskeleton.
Abbreviations: NMR, nuclear magnetic resonance spectroscopy; ITC, isothermal titration calorimetry; TM, transmembrane; CD47 or IAP, integrin associated protein; CT, cytoplasmic tail; ND, nanodiscs; PLIC, protein linking IAP and cytoskeleton; SPR, surface plasmon resonance; TMCT, transmembrane and cytoplasmic tail; UBL, ubiquitin-like domain; UBA, ubiquitin-binding domain; TEM, transmission electron microscopy; CSP, chemical shifts perturbation; PTM, post-translational modification
Keywords: CD47, PLIC, Ubiquilin, UBA, UBL, Nanodisc, Proteasomal degradation, Vimentin, Transmembrane, Cytoplasmic tail, Membrane proteins, NMR, ITC
Graphical abstract
Highlights
-
•
Pathway 1: Proteasomal Degradation Pathway – The two terminal domains of PLIC-2 UBA and UBL are involved in this pathway. The structure of UBL has been solved and its interaction with the proteasome has been established. In this paper we demonstrate the role of UBA in the degradation pathway: The two terminal domains UBL and UBA of PLIC-2, weakly interact with each other; the UBA domain interacts with mono-ubiquitin with an affinity that is several orders stronger than for the UBL domain. The binding interface of UBA to UBL or UBQ involves the conserved MGF loop and α3 helix consistent with the interactions seen among the family of its homologous proteins.
-
•
Pathway 2: As an adaptor protein involved in cytoskeletal regulation – PLICs/Ubiquilins were discovered as a Protein Linking IAP (CD47) in the cell membrane to cell Cytoskeleton (PLIC). Here we try to determine the molecular details of this interaction: We tried to analyze this interaction through the cytoplasmic tail of CD47 with the different domains of PLIC-2. No perceptible interactions were observed, both through NMR and ITC. To make the investigation thorough, we also utilized the trans-membrane and cytoplasmic form of CD47 in nanodiscs to identify interactions occurring at the membrane water interface. The results again failed to show any interaction.
-
•
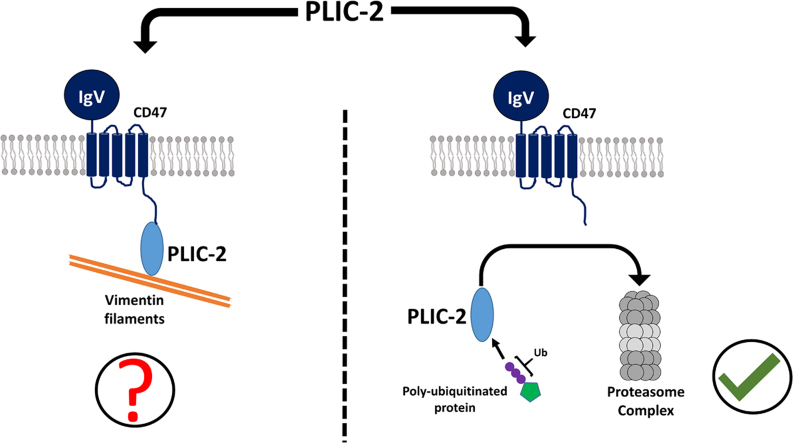
Overall, the results support the role of PLIC-2 in the proteasomal degradation pathway while suggesting that its role in the cytoskeletal regulation pathway as a “PLIC” protein is unlikely.
1. Introduction
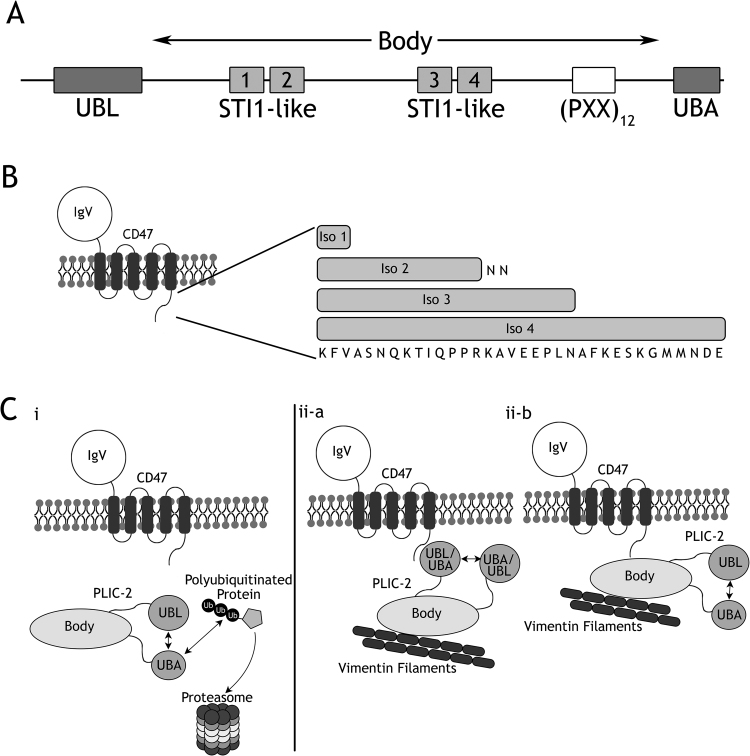
PLIC (or Protein Linking IAP and Cytoskeleton) proteins, also referred to as Ubiquilins, are human orthologs of the yeast Dsk2 family of proteins [1]. PLIC consists of a family of four homologous proteins, PLIC-1 through 4. All contain a ubiquitin-like (UBL) amino-terminal domain; a body region, which is predominantly predicted to be disordered by DISOPRED [2] and Jpred [3] containing STI1 (stress inducible) heat-shock protein binding motifs [4], [5]; and a carboxyl-terminal ubiquitin-associated (UBA) domain (Fig. 1A) [6]. PLIC-1 and 2 are localized to the cytoplasm [7], PLIC-3 [8] is found in the testis and PLIC-4 is localized to the cell nucleus [9]. PLICs were identified and subsequently named based on their ability to mediate an association between CD47 and Vimentin, an intermediate filament [10], forging a link between the outer membrane and cell cytoskeleton. CD47 (cluster of differentiation 47) is a ubiquitously present cell surface receptor. It is also known as Integrin associated protein (IAP) since it was initially identified through its association with integrin αvβ3 [11]. CD47 is primarily a cell surface receptor for thrombospondin-1 (THBS1) and signal regulatory protein-α (SIRPα). The engagement of CD47 with SIRPα leads to a “don’t eat me” signal that prevents cells from undergoing phagocytosis [12]. CD47 consists of an extracellular, glycosylated N-terminal IgV-like domain, five predicted transmembrane (TM) helices, and a short C-terminal cytoplasmic tail (CT) [13]. This tail is further alternatively spliced to produce four isoforms (Fig. 1B) with differential tissue expression [14]. Wu et al. had previously reported that PLIC-1 and 2 directly interact with the cytoplasmic tails of CD47. This interaction eventually enables Integrins to indirectly engage with the cell cytoskeleton and modulate cell spreading. Since then, these complexes remain uncharacterized and the exact details of their binary interfaces remains largely unknown.
Fig. 1.
(A) Schematic showing the different domains and motifs of PLIC-2. It contains a well characterized N-terminal UBL (ubiquitin like) domain and a C-terminal UBA (ubiquitin associated) domain. Interspersed between the two is the Body region which contains four Sti1 like domains and a repeat of twelve PXX motifs. (B) Schematic showing the overall structure of CD47. It consists of an N-terminal extracellular IgV domain, five trans-membrane helices and a C-terminal cytoplasmic tail. The tail is further differentially spliced from a single splice donor yielding the four isoforms of CD47. (C) A model representation showing the two pathways involving PLIC-2: (i) the proteasomal degradation pathway. This pathway is unrelated to the presence of CD47. Here the UBA domain of PLIC-2 binds to polyubiquitin chain bound to the substrate while the UBL domain interacts with the proteasome delivering the payload for degradation; (ii) the cytoskeletal regulation pathway. Central to this pathway is PLIC-2 which acts like an adaptor protein linking CD47 to Vimentin, an intermediate filament, hence by forging a link between the outer cell membrane and inner cell cytoskeleton. PLIC-2 has been proposed to interact with the cytoplasmic tail of CD47 [7]. We envision this interaction to occur in one of two ways: (ii-a) The UBA (or UBL) domain of PLIC-2 interacts with CD47, while the Body region interacts with vimentin filaments; (ii-b) the Body region of PLIC-2 interacts both with CD47 and Vimentin filaments. Meanwhile the UBL and UBA domains interact with each other.
The ubiquitin mediated proteasomal degradation pathway is a controlled mechanism for the proteolysis of cellular proteins. Key to this system is the comradery of three enzymes E1, E2 and E3; together they prime a protein for degradation through the addition of a polyubiquitin chain. The primary machinery of this mechanism resides in a 26S proteasomal complex, where target proteins meet their terminal fate. This complex is compartmentalized into two components: the 19S regulatory complex and the 20S core complex [15]. S5a/Rpn10, a subunit of the 19S component, has been shown to bind polyubiquitin chains. Additionally, and important to this study, are a class of proteins containing an N-terminus UBL (Ubiquitin Like) domain and a C-terminus UBA (Ubiquitin associated) domain. The UBL domain reversibly binds to the proteasome while the UBA domain binds to poly-ubiquitin chains. These UBL-UBA proteins shuttle their payloads to the 26S proteasome for degradation (Fig. 1C (i)). It has been shown that the S5a subunit of the 19S component binds to the UBL domain of hHR23a (mammalian homolog of Rad23) [16] and hPLIC-2 (mammalian homolog of Dsk-2) [17].
PLIC proteins derive its name from their purported role of being an adaptor protein linking the cell surface to the cell cytoskeleton. However, it has also been shown to be a proteasome shuttle factor for polyubiquitinated proteins, thereby regulating protein turnover [18]. It is puzzling how two diverse modalities are built into a single structural design of these proteins. It seems pertinent to ascertain whether the UBL/UBA system is directly involved in linking cell surface receptors to intermediate filaments or whether it acts as a shuttle protein delivering its payload to the proteasomal complex for degradation. In addition, PLIC-1, but not PLIC-2, has been shown to inhibit Jurkat T-cells migration by interacting with the βγ-subunit of the heterotrimeric G-protein [19]; and stabilize intracellular GABAA receptors to promote their accumulation within the plasma membrane [20]. Interestingly, PLIC-2, but not PLIC-1, down regulates endocytosis of the two unrelated GPCRs, V2 vasopressin receptor and beta-2 adrenergic receptor [21]. Though these studies highlight PLICs involvement in diverse cellular functions as a key regulatory molecule, they seem to further contradict its primary discovered role as a structural adaptor protein linking IAP/CD47 to cell cytoskeleton.
With the aim to reconcile these differences and understand which of the two pathways PLIC actually follows, we set to investigate the individual components involved in the interactions. First, we focused on the UBA domain of PLIC-2; since the structure of the UBL domain was already solved (PDB: 1J8C) and its interaction with the proteasome complex was clearly established [17]. Our data shows that there are inter-domain interactions within PLIC-2 where UBA binds weakly to the UBL domain which could be disrupted by its stronger binding affinity for Ubiquitin. Second, we sought to ascertain the primary binding interface between the cytoplasmic tail of CD47 and PLIC-2 as was initially observed [7]. We envisioned this interaction to occur via the three separate domains of PLIC-2 as demonstrated in Fig. 1C (ii-a and ii-b). We approached this problem in two ways, (i) we investigated the interaction of just the soluble cytoplasmic tail (CT) with the separate domains of PLIC-2 and (ii) we introduced the last transmembrane helix along with its cytoplasmic tail into nanodiscs to decipher any binding that might occur at the membrane/water interface in a near native membrane environment. Despite our rigorous multi-pronged approach, we were, quite disappointingly, unable to observe any specific interactions between the two proteins using both ITC and NMR. A full report of our findings is presented below.
2. Materials and methods
2.1. Cloning of PLIC-2 and CD47constructs
PLIC-2/UBLN-2 (Accession number: Q9UHD9) UBL domain UBLN21–107 present in pET23a vector, was kindly provided by Dr. Walters (NCI. USA). PLIC-2 UBA domain UBLN2578–624 was sub-cloned into pET15b vector using BamHI and HindIII restriction sites. PLIC-2 Body region UBLN2108–580 was sub cloned into pET15b using NdeI and BamHI restriction sites. To ensure accurate measurement of the recombinant protein concentration, a single Tryptophan residue was inserted into the corresponding vectors, for both UBL and UBA, following the six Histidine tags.
We used the longest isoform of CD47 (Isoform-4) as classified in the initial paper [7]. As per the Uniprot classification, this is annotated as Isoform-1. For this work, we will be following the classification from the original paper. Two constructs of CD47 isoform-4 (Accession no.: Q08722-1) were designed for this study, (i) CD47 290–323 containing the residues comprising its cytoplasmic tail (CD47-CT) and (ii) CD47 269–323 containing the residues comprising its last transmembrane helix along with its cytoplasmic tail (CD47-TMCT). The resulting amplicons were cloned into pET15b (for CT) (Novagen, USA) and pDB.His.MBP vector (for TMCT) (DNASU plasmid depository, USA) using the cutting sites NdeI and BamHI for the former and BamHI and HindIII for the latter. The pDB.His.MBP vector contains a Histidine-affinity tag and a MBP-fusion protein, separated from CD47′s sequence by a TEV protease cleavage site. Three additional glutamates were added before the N-terminal sequence to ensure proper accessibility of the cleavage site for proteolysis.
2.2. Expression and purification of the recombinant proteins
2.2.1. Expression
All constructs were expressed in E. coli BL21 (DE3) (NEB, USA). Cell cultures were grown in either LB or M9 minimal media supplemented with 15N-labeled ammonium chloride as the sole nitrogen source at 37°C. When the cells reached an optical density (OD600) of ~0.4 to 0.6, the cultures were transferred to a shaker at room temperature (for most proteins) or a 17°C shaker (for PLIC-2 Body region), and left overnight after induction with 0.5–1 mM IPTG.
2.2.2. Purification
UBA, UBL, Body, and CD47 constructs were purified using similar protocols with procedure variations outlined below. Harvested cells were resuspended and lysed using a French Press (Thermo Electron, USA). The supernatant was bound to Ni-NTA resin (Qiagen, USA) at 4°C for an hour, rinsed with ten column volumes of the wash buffer before elution. The eluate was further subjected to size exclusion chromatography (SEC) on a Superdex 75 (GE Healthcare, USA) for UBL and UBA domains, or Superdex 200 (GE Healthcare, USA) for CD47-TMCT. In addition to Ni-NTA purification the Body region was further purified using anion exchange chromatography on Resource Q (GE Healthcare, USA), with a 0–30% gradient of Buffer A (20 mM Tris, pH 8.0) and Buffer B (20 mM Tris, 1 M NaCl, pH 8.0). The Body containing fractions were further subjected to SEC on a Superdex 75 column. The exact buffer compositions are detailed in the Supplementary section.
CD47-CT expressed in inclusion bodies was purified under denaturing condition. Briefly, cells were resuspended in 6 M GuHCl in Tris buffer saline (TBS) buffer pH 8.0. Following lysis the supernatant was separated through centrifugation at 10,000 rpm and mixed with Ni-NTA resin at room temperature for 2 h. The resin was rinsed first with four column volumes of 8 M urea in TBS with 10 mM imidazole. Elution was carried out in the 8 M Urea buffer containing 400 mM imidazole. The eluate was loaded onto a Proto 300 C4 column (Higgins Analytical Inc., USA) and purified using a 10–40% gradient of (Buffer A: 90% H2O, 10% Acetonitrile, 0.1% TFA and Buffer B is 10% H2O, 90% Acetonitrile, 0.1% TFA). CD47-CT containing fractions were pooled and lyophilized (MillRock Tech., USA).
2.3. Nanodiscs preparation and Transmission Electron Microscopy (TEM)
Nanodiscs were prepared from a smaller truncated construct (D7) of MSP1D1 previously developed in our lab [22] as smaller discs are more conducive for NMR investigations. D7 was purified in the same manner as other MSP proteins forming nanodiscs [23]. D7 was mixed with purified 15N-CD47-TMCT in a molar ratio of two to one along with a twenty-fold molar excess of DMPC lipids (Avanti polar, USA) solubilized in a buffer containing sodium cholate. The reaction mixture was mixed for an hour at room temperature and later on mixed with wet Bio-beads SM2 (BIO-RAD, USA) for four hours. The resultant solution was loaded onto Superdex 200, to separate empty discs from protein incorporated ones. The MBP tag on the incorporated protein was cleaved using TEV protease and repurified on S-200 column to obtain discs containing the incorporated trans-membrane and cytoplasmic tail of CD47 Isoform-4. For TEM, 15N-CD47 TMCT nanodiscs were diluted several fold in water to obtain a well-dispersed population, applied to a glow discharged carbon-coated 400-mesh copper grid (Ted Pella Inc., USA), and negatively stained with freshly prepared 0.75% uranyl formate. Images were taken on a Tecnai G2 Spirit BioTWIN microscope (FEI, USA) at an accelerating voltage of 80 kV with a defocus of ∼−1.6.
2.4. Nuclear Magnetic Resonance Spectroscopy
15N-labeled proteins were overexpressed in M9 medium supplemented with 15NH4Cl as the sole nitrogen source. 15N NMR experiments were performed with a concentration of 75–150 µM on a Varian 600 MHz spectrometer equipped with an inverse triple-resonance cold probe at 25°C. All spectra were processed with NMR Pipe [24] and analyzed in CcpNmr software suite [25] made available through NMRBox.
For CD47-TMCT nanodiscs experiments, 15N-CD47-TMCT was titrated with unlabeled UBL at a ratio of 1:5 and Body region at 1:3. 15N-UBL was titrated with unlabeled CD47-TMCT at a ratio of 1:5, whereas 15N-UBA was titrated with unlabeled CD47-TMCT at a ratio of 2:5. For CD47-CT experiments, 15N-UBL was titrated with unlabeled CD47-CT at a ratio of 1:10 and 15N-UBA was titrated with unlabeled CD47-CT at a ratio of 1:10. For PLIC-2 self-association experiments, 15N-UBA was titrated with unlabeled UBL at ratios of 1:1, 1:3, 1:5, and 1:10. For UBA and ubiquitin experiments, 15N-UBA was titrated with unlabeled ubiquitin at ratios of 3:1, 2:1, 1:1, 1:2, 1:3, 1:5, 1:10. Kd values were determined form the chemical shift perturbations calculated using the following equation [26]:
| (1) |
where the scaling factor, α, was set to 0.15. The plot of the chemical shift perturbation vs. molar ratio was fitted to the equation below to define the Kd [27]:
| (2) |
where [P]0 is the concentration of 15N-UBA PLIC-2 and x is the molar ratio of UBL or ubiquitin to UBA.
2.5. Isothermal Titration Calorimetry
All ITC experiments were performed using a low volume Nano-ITC (TA instruments, USA). The titrations were done at 25°C with 200 RPM mixing, 300 s injection intervals, and 2.5 µL injections. The buffer used was 40 mM NaPO4, 20 mM NaCl, 0.5 mM EDTA, pH 7.0. The protein concentrations for the syringe and sample cell were as follows: 0.95 mM UBL PLIC-2 and 0.14 mM CD47-CT, 0.39 mM UBA PLIC-2 and 0.05 mM CD47-CT, 0.49 mM CD47-CT and 0.05 mM Body PLIC-2, and 0.49 mM CD47-CT and 0.04 mM Body PLIC-2. For nanodiscs, 0.35 mM UBL PLIC-2 and 0.07 mM CD47-TMCT; 0.32 mM UBA PLIC-2 and 0.07 mM CD47-TMCT. Data analysis was done with the NanoAnalyze Software (TA Instruments, USA) suite using “independent” model algorithm.
2.6. Molecular modeling
PLIC-2 UBA was modeled through Phyre 2 [28] web server using PLIC-1 UBA domain as a template which shares 98% identity. Models of the protein complexes (PLIC-2 UBA-UBL and PLIC-2 UBA-Ubiquitin) were calculated through the HADDOCK (High Ambiguity Driven protein-protein DOCKing) [29] webserver using the chemical shift perturbation data obtained from NMR HSQC titration experiments. The structures of the participating proteins were acquired from the RCSB database (PLIC-2 UBL: 1J8C, Ubiquitin: 2JY6, PLIC-2 UBA: 2JY5). Electrostatic surface potential showing charge distribution on the surface of the UBA domains of Dsk2 and PLIC-2 (Supplementary data) was calculated using Chimera [45].
3. Results
Solution NMR has been successfully applied to study protein interactions for numerous intrinsically disordered and membrane proteins [30], [31], [32], [33]. In our present study, we use NMR to investigate the interaction of PLIC-2 in two different pathways. PLIC-2 was chosen as a candidate over PLIC-1 since PLIC-1 exists in four isomeric forms (Accession No. Q9UMX0), potentially making the analysis cumbersome and inconclusive. PLIC-2 was compartmentalized into its individual domains: UBL, Body, and UBA. Each domain was used to assess their binding ability to its different partners.
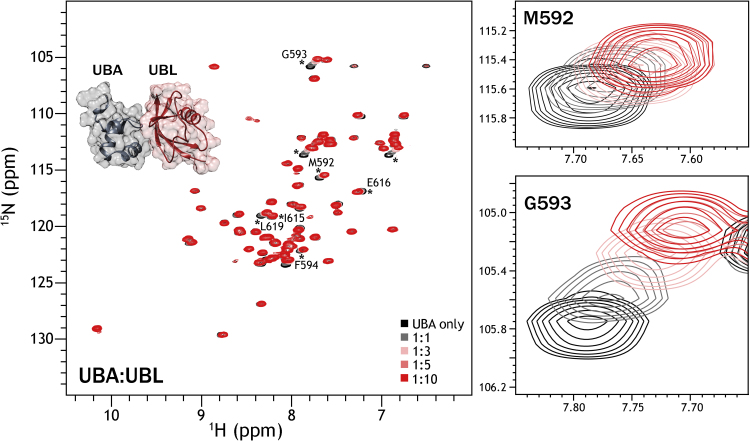
We first tried to ascertain the involvement of PLIC-2 in the proteasomal degradation pathway. We initiated this by determining whether the two terminal UBA and UBL domains of PLIC-2 interact with each other (Fig. 1C (i)). 15N-HSQC based chemical shifts perturbation (CSP) experiments were employed, where 15N labeled PLIC-2 UBA was titrated against unlabeled PLIC-2 UBL. Peaks representing maximum observed perturbations (marked with asterisk on Fig. 2), were used to calculate the Kd values using the equations summarized in the methods section. The average Kd value obtained was 175±25 µM, which is comparable to the previously reported Kd value of 80±15 µM measured for Dsk2, a yeast homolog of PLIC proteins, [34]. It is known that for UBL-UBA proteins involved in proteasomal degradation, UBL interacts with the proteasome while UBA binds to polyubiquitinated substrates [6]. Thus, we next examined the ability of PLIC-2 to bind ubiquitinated substrates through the interaction of PLIC-2 UBA domain with Ubiquitin (Ub). Our titration experiments demonstrate a stronger binding affinity of UBA for Ub, as exemplified by the pronounced CSPs in 15N-HSQC spectra (Fig. 3). The Kd value for this interaction was determined to be 8.1±1.5 µM by averaging fits from peaks with pronounced perturbations, which is similar to PLIC-1′s affinity for ubiquitin (Kd ~20 µM) [35]. Amide resonances were identified based upon assignments available for the almost identical UBA domain from PLIC-1 (PDB: 2JY5). CSP data was further utilized to generate a model of the binding interface between UBA-UBL and UBA-Ub using HADDOCK, which is presented as inset within the spectra in Fig. 2, Fig. 3. Intermolecular interaction between UBA and Ub is stronger than with UBL, and their binding interfaces involves the same core residues in addition to other exclusive to the UBA-Ub interactions reflective of stronger binding. Given that UBA and UBL domains associate with each other, a stronger affinity of the PLIC-2 UBA domain for Ubiquitin, implicates the disengagement of the UBA-UBL domains in the event of PLIC binding to the polyubiquitinated substrate en route to the proteasome.
Fig. 2.
15N-HSQC titration experiments of PLIC-2 15N-UBA domain in the presence of PLIC-2 UBL domain at a UBA to UBL molar ratio of 1:1, 1:3, 1:5, and 1:10. Partial amino acid assignments were achieved by aligning the 15N-HSQC to that of PLIC-1 UBA domain [35]. The inset shows a molecular model displaying the interaction of UBA (grey) and UBL (red) as determined by HADDOCK. Two residues M and G showing maximum perturbation are shown as slices on the right.
Fig. 3.
15N-HSQC titration experiments of PLIC-2 15N-UBA domain in the presence of ubiquitin (Ub) at a UBA to Ub molar ratios of 3:1, 2:1, 1:1, 1:2, and 1:3. Ratios 1:5 and 1:10 are not shown as the spectra reached a saturation at 1:3. A molecular model, determined by HADDOCK, of UBA PLIC-2 (grey) and ubiquitin (red) as inset in the spectra. On the right, specific peaks are shown separately to better capture the chemical shift perturbations. Two residues M and G displaying intermediate exchange are shown as slices on the right.
Next, we moved on to investigate the second half of our model, testing PLIC-2 as an adaptor protein linking the cell membrane to the cytoskeleton through the cytoplasmic tail of CD47 (Fig. 1Cii). This was achieved by determining interactions occurring between CD47 and (a) the two terminal UBA/UBL domains of PLIC-2 or (b) the Body region of PLIC-2. Two constructs of CD47 were utilized, one containing the cytoplasmic tail region (CT) and the other comprising of the last transmembrane helix in addition to the cytoplasmic tail (TMCT). This helped investigate interaction at both the aqueous and membrane/water interface. Since CD47 is a mammalian receptor protein, we assessed the possibility of posttranslational modifications (PTM) using several PTM databases from Uniprot. We found that although there are ten positions that potentially undergo PTM, the last is located at residue 206. As our longest CD47 construct (TMCT) starts from residue number 269, no PTMs affect the outcome of our study, validating the biological significance of our in-vitro analysis.
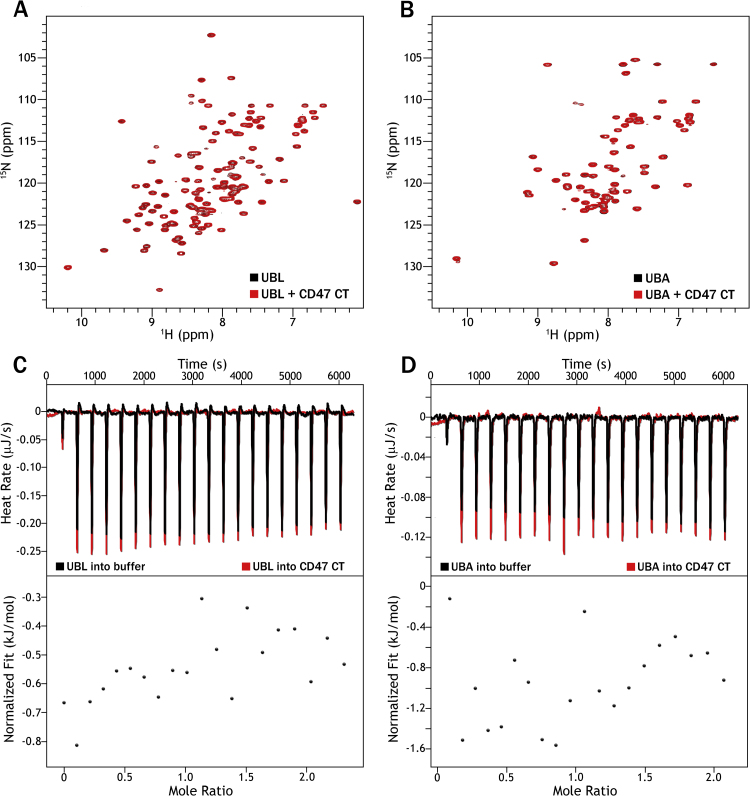
In order to ascertain whether CD47 interacts with the UBL and/or UBA domains of PLIC-2, we performed 15N-HSQC titration experiments with 15N-UBL or 15N-UBA. Figs. 4A and 4B present the spectra of the UBL and UBA domains, respectively. Both remained unperturbed by the addition of the cytoplasmic tail of CD47 (CD47-CT) with no CSPs or peak broadenings. To reinforce our results, we performed Isothermal Titration Calorimetric (ITC) experiments where CD47-CT was titrated against the UBL (Fig. 4C) or UBA domains (Fig. 4D). The acquired thermograms reflect the heats of dilution rather than specific enthalpy driven interactions consistent with the lack of binding.
Fig. 4.
(A)15N-HSQC of PLIC-2 15N-UBL alone (black) and in the presence of CD47-CT at a UBL to CD47 ratio of 1:10 in red. (B)15N-HSQC of PLIC-2 15N-UBA alone (black) and in the presence of CD47-CT at a UBA to CD47 ratio of 1:10 in red. (C) (Top) ITC thermogram of UBL titrated into buffer (black) and into CD47-CT (red). (Bottom) Integrated heats of the titration after correction for the heat of dilution. (D) (Top) ITC thermogram of UBA titrated into buffer (black) and into CD47-CT (red). (Bottom) Integrated heats of the titration after correction for the heat of dilution.
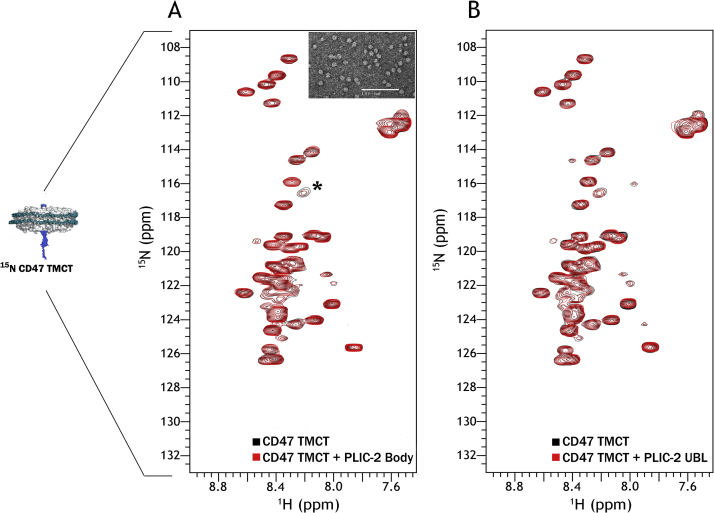
The other pertinent question to this interaction is the proximity of the tail region to the membrane surface, as in its native receptor state. To identify any interactions at this interface, we incorporated the CD47-TMCT, containing the last transmembrane helix, into small D7 nanodiscs [22]. Nanodiscs offer a soluble lipid bilayer system allowing the study of interactions at the membrane/water interface. We have previously successfully employed NMR for the study of the TMCT regions of Integrin β3 in nanodiscs [36]. The formation of discs was confirmed through TEM (Fig. 5A, inset). 15N-HSQC spectra were collected for labeled 15N-UBL (Supp. Fig. S1A) and 15N-UBA (Supp. Fig. S1B) in the presence of unlabeled CD47-TMCT followed by a further evaluation through ITC. No perceptible perturbations/peak broadenings were observed which was further corroborated by the ITC data where the heat differences were at the noise level for both UBL (Supp. Fig. S1C) and UBA (Supp. Fig. S1D). Once again, the results coincide with the results from previous section suggesting no discernable interactions occurring between CD47 and the UBL or UBA domains of PLIC-2.
Fig. 5.
15N-HSQC of labeled-CD47 TMCT. (A)15N-CD47 TMCT alone (black) and in the presence of PLIC-2 Body region at a CD47 to Body ratio of 1:3 in red. The amide peak with a slight intensity drop in the presence of Body is marked by the asterisk. Inset: Negatively stained TEM micrograph showing the formation of nanodiscs containing the incorporated 15N labeled protein (B)15N-CD47 TMCT alone (black) and in the presence of PLIC-2 UBL domain at a CD47 to UBL ratio of 1:5 in red.
Since no interactions were observed between the PLIC-2 UBA/UBL domains and CD47, we focused our attention on the PLIC-2 Body region. This is a 51.6 kDa intrinsically disordered protein which would provide a 15N-HSQC spectrum with a multitude of overlapping peaks corresponding to random coil frequencies making it difficult to observe any CSP or peak broadenings leading to inconclusive and ambiguous data. Therefore, we tried to analyze this interaction from the side of CD47 by titrating 15N-CD47-TMCT with unlabeled Body region. We also performed the same experiment with the unlabeled UBL domain serving as a control; and additionally confirming the above obtained results for the 15N-UBL and unlabeled CD47-TMCT experiment. Fig. 5 shows the spectra of 15N-CD47-TMCT alone (black) titrated against unlabeled proteins, Body (Fig. 5A) and UBL (Fig. 5B) shown in red. Both titration resulted in no pronounced changes in resonance frequencies or peak intensities of CD47. There was however, a slight intensity drop observed for the amide located at 8.20/116.56 ppm in the presence of the Body region (marked with the asterisk in Fig. 5A); but a minor intensity drop for a single peak does not substantiate a binding interaction between two proteins. The absence of binding was further confirmed through ITC measurements using the CD47-CT construct as described in the supplementary section, Fig. S2.
4. Discussion
Since the structure of the UBL domain from PLIC-2 was already available, we focused our attention on the UBA domain of PLIC-2. The human PLIC-2 UBA domain can be contrasted to its ortholog S. cerevisiae Dsk2 where similar UBA domain interactions have been observed. The UBA domains of these two proteins do not share much of a sequence homology, but they do have the same fold. Dsk2 UBA forms a dimer or higher order polymers [37], which is attributed to the presence of positive and negatively charged regions on the surface of the protein. PLIC-2 UBA, in contrast, lacks strong charges and is monomeric, as can be concluded from the SEC data (Supp. Fig. S3). From reliable partial assignments, we identified most of the PLIC-2 UBA residues which undergo perturbations in the presence of PLIC-2 UBL. These residues either belong to the loop region connecting helices α1 and α2 (M592-F594) or are located within helix α3 (I615-E616 and L619) of which I615 showed minimal shifts. These positions match well with the corresponding Dsk2 residues M342-F344 from the same loop region and L365-D366, L369 from the α3 helix. Defining the binding site are residues M342/592, which fits snugly in the hydrophobic cavity of UBL, and L365/I615 and L369/619, which reinforce the hydrophobic core. It is likely that the flexibility required for the two terminals domains to interact with each other is satisfied by having a Body region that is largely disordered.
For PLIC proteins to participate in the proteasomal degradation pathway, two events need to occur. First, the UBL domain needs to interact with the proteasome and, second, the UBA domain needs to interact with ubiquitin. The former has already been established for PLIC-2, where the UBL domain has been shown to complex with hRpn13 (Adrm1) [38], which is a subunit of the 19S regulatory particle of the 26S proteasome. Here we analyze the binding of the PLIC-2 UBA domain to ubiquitin. From our Kd estimation, the PLIC-2 UBA-Ub binds stronger than UBA-UBL. CSP data obtained from our titration experiments of 15N UBA with Ub highlights residues M592-F594 from the (helix α1/α2) loop region and A614-L619 belonging to helix α3 that are involved in this interaction. Majority of the perturbations belong to the fast exchange regime as only chemical shift perturbations were observed. However, residues M592-G593 displayed both chemical shift perturbations and peak broadening indicative of intermediate exchange. This is part of the canonical, M-G-F containing binding site conserved throughout UBA domains of PLIC/Dsk2 and hHR23/Rad23 proteins. In addition, I615-E616 and L619 from α3 showed significant perturbations, as has been observed for I580-E581 and L584 form PLIC-1 where they form close contacts with the hydrophobic patches of Ub [39]. Interestingly, T607, the last residue from the helix α2 showed shifts which can be attributed to its close proximity to A614 on α3 which exhibited strong CSP a feature also observed with PLIC-1. All together, we can conclude that the UBA-UBL and UBA-Ub interactions in PLIC-2 share the same binding interface, which is exemplified in the HADDOCK models presented as insets in Fig. 2, Fig. 3. This proves the role of PLIC-2 as a shuttle protein transferring its cargo to the proteasome for degradation.
It is important to note that the interactions reported here are for the isolated PLIC-2 UBA and UBL domains. The presence of an intervening Body region may affect the binding affinity of the two domains due to their localized proximity. There is evidence for PLIC-2 forming dimers/trimers [40] possibly through the Body region [41] promoting intermolecular UBA/UBL association. Moreover, a recent study has implicated the Body region to associate with heat shock protein 70 (HSP70) which is bound to an aggregated client protein all of which is transferred to the proteasomal complex for degradation. Specific mutations linked to amyotrophic lateral sclerosis were shown to abolish PLIC-2/HSP70 complex formation [40] highlighting the significance of recycling protein aggregates. The data suggests that the Body region is responsible for payload transfer and further consolidates PLIC-2′s role as a proteasomal shuttle factor.
The latter part of our study, which we hoped would lead to insightful results, was defining the role of PLIC-2 as an adaptor protein linking the outer cell membrane and cell cytoskeleton. The name PLIC is based on its function of being on a Protein that Links IAP (integrin associated protein/CD47) to Cell cytoskeleton, Vimentin, through its cytoplasmic tail. The only reported record identifying this association comes from a singular paper, where the name PLIC was conferred upon these adaptor proteins [10]. Almost two decades later, we tried to investigate this interaction with the aim of identifying the critical domains and residues involved in the process. Since the binding was shown to occur through the cytoplasmic tail of CD47, we employed two constructs of the tail region: (i) the tail by itself (CT), and (ii) along with the last transmembrane helix (TMCT) to capture interactions that might occur at the membrane water interface. The latter construct was inserted into small nanodiscs [22]. Even though we employed high sensitivity/ high-resolution biophysical technique like NMR, we did not observe direct interactions between the cytoplasmic tail of CD47 and any of the individual domains of PLIC-2. Both ITC and NMR results consistently refute the notion of a binary complex. The inference that the two may not be directly associated is not inconceivable when we factor in the following reasons.
First, several studies have shown the involvement of PLICs in alternative cellular processes. In particular, PLIC-2 has been linked to proteasomal degradation pathways [18], [42] and GPCR endocytosis [21], which are seemingly unrelated to cytoskeletal modulation. This potentially means that the purported role of PLIC linking CD47 to vimentin filaments is not its major cellular function. This, in part, is supported by another study where double labeled immune-fluorescence of PLIC-1 and vimentin filaments did not show any co-localization [43]. Additionally, the same group which published the original PLIC-2 paper later found that PLIC-1, but not PLIC-2, binds to Gβγ subunit of G proteins and interferes with its functionality. This was based on the observation that PLIC-2 was predominantly present in the cytoplasm and never co-localized to the plasma membrane [19]. They also reported that PLIC-2 binds to Eps15 and Epsin (1 and 2), all of which are UIM-containing adaptor proteins involved in clathrin mediated endocytosis and thereby in protein turnover [21]. Second, it’s likely that CD47 and PLIC-2 are part of a larger macromolecular assembly that may also contain, for example, integrin heterodimers [44]. The sole in vitro data available for PLIC’s interaction is from a pull down assay using recombinant GST-fused PLIC probed against CD47 from human placenta and analyzed through immunoblots [10]. This doesn’t rule out the presence of other proteins which could have been part of the pull down result and hence doesn’t exclude the possibility that other protein(s) might be involved in the complex formation. Silver staining of the pull down eluate followed by subsequent LC-MS might help identify these components. Since this experiment was not performed in the original study, the direct interaction of CD47 to PLIC remains a conjecture. Moreover, we were unable to find any interaction between the cytoplasmic tails of Integrin β3 and CD47 (data not shown) which are known to form a complex, most likely associating through their transmembrane region. This further rules out the possibility of CD47′s cytoplasmic tail acquiring a secondary structure through its interaction with the tail of Integrin β3 which would then bind to PLIC-2. Considering all the data, we are certain that PLIC-2 does not bind to the cytoplasmic tail of CD47.
In conclusion, we show that PLIC-2 belongs to the proteasomal degradation pathway and it does not interact with the cytoplasmic tail of CD47. This raises questions against its involvement as a protein linking the membrane to the cytoskeleton and leaves one to ponder whether the name Ubiquilins should be preferred over PLIC while referring to these adaptor proteins.
Acknowledgements
We would like to thank Drs. Kylie Walters, from the National Cancer Institute, and Andrew Wiemer, from the University of Connecticut, for kindly providing the PLIC-2 construct and pdb.His.MBP vector, respectively. This work has been supported by a large internal grant from the University of Connecticut. We would also like to thank Dr. Vitaliy Gorbatyuk for help with NMR experiments and NMRbox, the National Center for Biomolecular NMR Data Processing and Analysis, which is supported by NIH grant P41GM111135 (NIGMS)
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2017.01.013.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Biggins S., Ivanovska I., Rose M.D. Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J. Cell Biol. 1996;133(6):1331–1346. doi: 10.1083/jcb.133.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ward J.J. The DISOPRED server for the prediction of protein disorder. Bioinformatics. 2004;20(13):2138–2139. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- 3.Drozdetskiy A. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43(W1):W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaye F.J. A family of ubiquitin-like proteins binds the ATPase domain of Hsp70-like Stch. FEBS Lett. 2000;467(2–3):348–355. doi: 10.1016/s0014-5793(00)01135-2. [DOI] [PubMed] [Google Scholar]
- 5.Johnson B.D. Hop modulates Hsp70/Hsp90 interactions in protein folding. J. Biol. Chem. 1998;273(6):3679–3686. doi: 10.1074/jbc.273.6.3679. [DOI] [PubMed] [Google Scholar]
- 6.Elsasser S., Finley D. Delivery of ubiquitinated substrates to protein-unfolding machines. Nat. Cell Biol. 2005;7(8):742–749. doi: 10.1038/ncb0805-742. [DOI] [PubMed] [Google Scholar]
- 7.Wu A.L. Ubiquitin-related proteins regulate interaction of vimentin intermediate filaments with the plasma membrane. Mol. Cell. 1999;4(4):619–625. doi: 10.1016/s1097-2765(00)80212-9. [DOI] [PubMed] [Google Scholar]
- 8.Conklin D. Molecular cloning, chromosome mapping and characterization of UBQLN3 a testis-specific gene that contains an ubiquitin-like domain. Gene. 2000;249(1–2):91–98. doi: 10.1016/s0378-1119(00)00122-0. [DOI] [PubMed] [Google Scholar]
- 9.Davidson J.D. Identification and characterization of an ataxin-1-interacting protein: a1up, a ubiquitin-like nuclear protein. Hum. Mol. Genet. 2000;9(15):2305–2312. doi: 10.1093/oxfordjournals.hmg.a018922. [DOI] [PubMed] [Google Scholar]
- 10.Wu A.L. Ubiquitin-related proteins regulate interaction of vimentin intermediate filaments with the plasma membrane. Mol. Cell. 1999;4(4):619–625. doi: 10.1016/s1097-2765(00)80212-9. [DOI] [PubMed] [Google Scholar]
- 11.Brown E. Integrin-associated protein: a 50-kD plasma membrane antigen physically and functionally associated with integrins. J. Cell Biol. 1990;111(6 Pt 1):2785–2794. doi: 10.1083/jcb.111.6.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oldenborg P.A. Role of CD47 as a marker of self on red blood cells. Science. 2000;288(5473):2051–2054. doi: 10.1126/science.288.5473.2051. [DOI] [PubMed] [Google Scholar]
- 13.Lindberg F.P. Molecular cloning of integrin-associated protein: an immunoglobulin family member with multiple membrane-spanning domains implicated in alpha v beta 3-dependent ligand binding. J. Cell Biol. 1993;123(2):485–496. doi: 10.1083/jcb.123.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reinhold M.I. In vivo expression of alternatively spliced forms of integrin-associated protein (CD47) J. Cell Sci. 1995;108(Pt 11):3419–3425. doi: 10.1242/jcs.108.11.3419. [DOI] [PubMed] [Google Scholar]
- A. Ciechanover, A.L. Schwartz, The ubiquitin-proteasome pathway: the complexity and myriad functions of proteins death, in: Proceedings of the National Academy of Sciences of the United States of America, 1998. 95(6): p. 2727–30. [DOI] [PMC free article] [PubMed]
- 16.Mueller T.D., Feigon J. Structural determinants for the binding of ubiquitin-like domains to the proteasome. EMBO J. 2003;22(18):4634–4645. doi: 10.1093/emboj/cdg467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walters K.J. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry. 2002;41(6):1767–1777. doi: 10.1021/bi011892y. [DOI] [PubMed] [Google Scholar]
- 18.Grabbe C., Dikic I. Functional roles of ubiquitin-like domain (ULD) and ubiquitin-binding domain (UBD) containing proteins. Chem. Rev. 2009;109(4):1481–1494. doi: 10.1021/cr800413p. [DOI] [PubMed] [Google Scholar]
- 19.N'Diaye E.N., Brown E.J. The ubiquitin-related protein PLIC-1 regulates heterotrimeric G protein function through association with Gbetagamma. J. Cell Biol. 2003;163(5):1157–1165. doi: 10.1083/jcb.200307155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bedford F.K. GABA(A) receptor cell surface number and subunit stability are regulated by the ubiquitin-like protein Plic-1. Nat. Neurosci. 2001;4(9):908–916. doi: 10.1038/nn0901-908. [DOI] [PubMed] [Google Scholar]
- 21.N'Diaye E.N. The ubiquitin-like protein PLIC-2 is a negative regulator of G protein-coupled receptor endocytosis. Mol. Biol. Cell. 2008;19(3):1252–1260. doi: 10.1091/mbc.E07-08-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puthenveetil R., Vinogradova O. Optimization of the design and preparation of nanoscale phospholipid bilayers for its application to solution NMR. Proteins. 2013;81:1222–1231. doi: 10.1002/prot.24271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ritchie T.K. Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzym. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delaglio F. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 25.Vranken W., F. The CCPN data model for NMR spectroscopy. Proteins. 2005;59(59):687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 26.Williamson M.P. Using chemical shift perturbation to characterise ligand binding. Prog. Nucl. Magn. Reson. Spectrosc. 2013;73:1–16. doi: 10.1016/j.pnmrs.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Fielding L. NMR methods for the determination of protein-ligand dissociation constants. Curr. Top. Med. Chem. 2003;3(1):39–53. doi: 10.2174/1568026033392705. [DOI] [PubMed] [Google Scholar]
- 28.Kelley L.A. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10(6):845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Vries S.J., van Dijk M., Bonvin A.M. The HADDOCK web server for data-driven biomolecular docking. Nat. Protoc. 2010;5(5):883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 30.Tyukhtenko S. Characterization of the neuron-specific L1-CAM cytoplasmic tail: naturally disordered in solution it exercises different binding modes for different adaptor proteins. Biochemistry. 2008;47(13):4160–4168. doi: 10.1021/bi702433q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vinogradova O. A structural mechanism of integrin alpha(IIb)beta(3) "inside-out" activation as regulated by its cytoplasmic face. Cell. 2002;110(5):587–597. doi: 10.1016/s0092-8674(02)00906-6. [DOI] [PubMed] [Google Scholar]
- 32.J. Yang, et al., Structure of an integrin {alpha}IIb{beta}3 transmembrane-cytoplasmic heterocomplex provides insight into integrin activation, in: Proceedings of the National Academy of Sciences of the United States of America, 2009. 106(42): p. 17729–34. [DOI] [PMC free article] [PubMed]
- 33.Sahu D., Bastidas M., Showalter S.A. Generating NMR chemical shift assignments of intrinsically disordered proteins using carbon-detected NMR methods. Anal. Biochem. 2014;449:17–25. doi: 10.1016/j.ab.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowe E.D. Structures of the Dsk2 UBL and UBA domains and their complex. Acta Crystallogr. D. Biol. Crystallogr. 2006;62(Pt 2):177–188. doi: 10.1107/S0907444905037777. [DOI] [PubMed] [Google Scholar]
- 35.Zhang D., Raasi S., Fushman D. Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J. Mol. Biol. 2008;377(1):162–180. doi: 10.1016/j.jmb.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.R. Puthenveetil, K. Nguyen, O. Vinogradova, Nanodiscs and solution NMR: preparation, application and challenges, in Nanotechnology Reviews, 2016. [DOI] [PMC free article] [PubMed]
- 37.Lowe E.D. Structures of the Dsk2 UBL and UBA domains and their complex. Acta Crystallogr. Sect. D., Biol. Crystallogr. 2006;62(Pt 2):177–188. doi: 10.1107/S0907444905037777. [DOI] [PubMed] [Google Scholar]
- 38.Chen X. Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 reveal distinct binding mechanisms between substrate receptors and shuttle factors of the proteasome. Structure. 2016;24(8):1257–1270. doi: 10.1016/j.str.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang D., Raasi S., Fushman D. Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J. Mol. Biol. 2008;377(1):162–180. doi: 10.1016/j.jmb.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hjerpe R. UBQLN2 Mediates Autophagy-Independent Protein Aggregate Clearance by the Proteasome. Cell. 2016;166(4):935–949. doi: 10.1016/j.cell.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ford D.L., Monteiro M.J. Dimerization of ubiquilin is dependent upon the central region of the protein: evidence that the monomer, but not the dimer, is involved in binding presenilins. Biochem. J. 2006;399(3):397–404. doi: 10.1042/BJ20060441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su V., Lau A.F. Ubiquitin-like and ubiquitin-associated domain proteins: significance in proteasomal degradation. Cell Mol. Life Sci. 2009;66(17):2819–2833. doi: 10.1007/s00018-009-0048-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu S. Characterization of ubiquilin 1, an mTOR-interacting protein. Biochim. Biophys. Acta. 2002;1542(1–3):41–56. doi: 10.1016/s0167-4889(01)00164-1. [DOI] [PubMed] [Google Scholar]
- 44.Barazi H.O. Regulation of integrin function by CD47 ligands. Differential effects on alpha vbeta 3 and alpha 4beta1 integrin-mediated adhesion. J. Biol. Chem. 2002;277(45):42859–42866. doi: 10.1074/jbc.M206849200. [DOI] [PubMed] [Google Scholar]
- 45.Pettersen E.F. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material