Abstract
Neurons affected in Alzheimer’s disease (AD) experience mitochondrial dysfunction and a bioenergetic deficit that occurs early and promotes the disease-defining amyloid β-peptide (Aβ) and Tau pathologies. Emerging findings suggest that the autophagy – lysosome pathway that removes damaged mitochondria (mitophagy) is also compromised in AD, resulting in the accumulation of dysfunctional mitochondria. Results in animal and cellular models of AD and in patients with sporadic late-onset AD suggest that impaired mitophagy contributes to synaptic dysfunction and cognitive deficits by triggering Aβ and Tau accumulation through increases in oxidative damage and cellular energy deficits; these, in turn, impair mitophagy. Interventions that bolster mitochondrial health and/or stimulate mitophagy may therefore forestall the neurodegenerative process in AD.
Keywords: neurodegeneration, Alzheimer’s disease, mitochondria, mitophagy, lysosomes, amyloid β peptide, Tau
Early Clues that Mitochondria are Central to AD
Alzheimer’s disease (AD) is the most common form of dementia and is characterized by a progression from episodic memory problems to severe cognitive decline and complete dependence of the patient on caregivers [1–3]. The disease-defining histopathological abnormalities, extracellular deposits of amyloid β-peptide (Aβ) and intraneuronal accumulation of hyperphosphorylated Tau (pTau) “spread” through the brain in a non-random manner with early pathology occurring in the entorhinal cortex and hippocampus [4, 5]. However, the alterations in cellular homeostasis that lead to the Aβ and pTau pathologies are unclear.
A widely documented abnormality in neuronal physiology that occurs prior to the onset of discernable cognitive deficits in those who develop AD is impaired glucose utilization as evaluated by 2-deoxy-D-glucose positron emission tomography (2DG-PET) brain imaging [6]. Disrupted mitochondrial health and neuronal metabolism as early features of AD were proposed as early as 1991 by Blass and Gibson [7]. More recently, however, there have been hundreds of studies documenting mitochondrial abnormalities in AD and elucidating the underlying molecular mechanisms and cellular consequences of mitochondrial deficits. Notably, in 2004 Swerdlow and Khan proposed a “mitochondrial cascade hypothesis”, which stated that each individual’s genetically determined and environmentally influenced mitochondrial function are primary factors influencing late-onset AD pathology [8]. Here we review emerging findings that suggest neurons affected in AD accumulate dysfunctional mitochondria in part due to impaired mitophagy, the process by which cells normally detect and remove mitochondria that have suffered molecular damage.
Mitochondria and neuroplasticity
Mitochondria are organelles referred to as the “powerhouses” of cells. Along with regulating calcium homeostasis and signaling to and from other organelles, mitochondria produce adenosine triphosphate (ATP) by oxidative phosphorylation, in which electrons are passed through the electron transport chain (ETC) from high energy substrates to oxygen. Impaired mitochondrial function may lead to a reduction in cellular energy levels; concomitant leakage of electrons promotes the formation of reactive oxygen species (ROS), which can damage proteins, membrane lipids and nucleic acids [9].
Mitochondria play key roles in developmental and adult neuroplasticity. For example, during early neuronal differentiation, mitochondria regulate the differentiation and growth of the axon by buffering cytosolic Ca2+ and thereby promoting polymerization of axonal microtubules [10]. Studies in which mitochondrial biogenesis is inhibited by RNA interference-mediated knockdown of peroxisome proliferator-activated receptor-y coactivator 1α (PGC-1α) suggest critical roles for mitochondria in the formation of synapses in developing neuronal circuits, and for the maintenance of synapses in the adult hippocampus [11]. The expression of PGC-1α and consequent mitochondrial biogenesis is stimulated by brain-derived neurotrophic factor (BDNF), a trophic factor known to play major roles in hippocampal synaptic plasticity, learning and memory and neuronal stress resistance [11]. Studies of affected patients and experimental models suggest that deficits in BDNF signaling contribute to synaptic dysfunction and neuronal degeneration in AD [12]. The mitochondrial protein deacetylase sirtuin 3 (SIRT3) is upregulated in response to exercise in a neuronal activity-dependent manner, which may modulate Ca2+ dynamics at glutamatergic synapses in ways that promote neuroplasticity and cellular stress resistance [11, 13]. Furthermore, mitochondria regulate Ca2+, and perturbed neuronal Ca2+ levels lead to neuronal death and are implicated in neurodegenerative diseases, including AD [14]. Importantly, damaged mitochondria can activate Caspase-9-dependent neuronal apoptosis by the release of cytochrome c. Thus, the maintenance of a healthy mitochondrial pool is essential for neuronal health. To this end, there exist many mitochondrial quality control pathways such as misfolded protein degradation, fission and fusion, and the engulfment and degradation of damaged mitochondria, termed mitophagy (Figure 1) [15, 16]. Mitophagy is important for neuronal survival and health; in addition, compromised mitophagy has been implicated in neurodegenerative diseases, including Parkinson’s disease and AD, as well as aging [17–19]. Collectively, mitochondria serve essential functions in neurons and mitochondrial maintenance directly affects neuronal development, function, and survival.
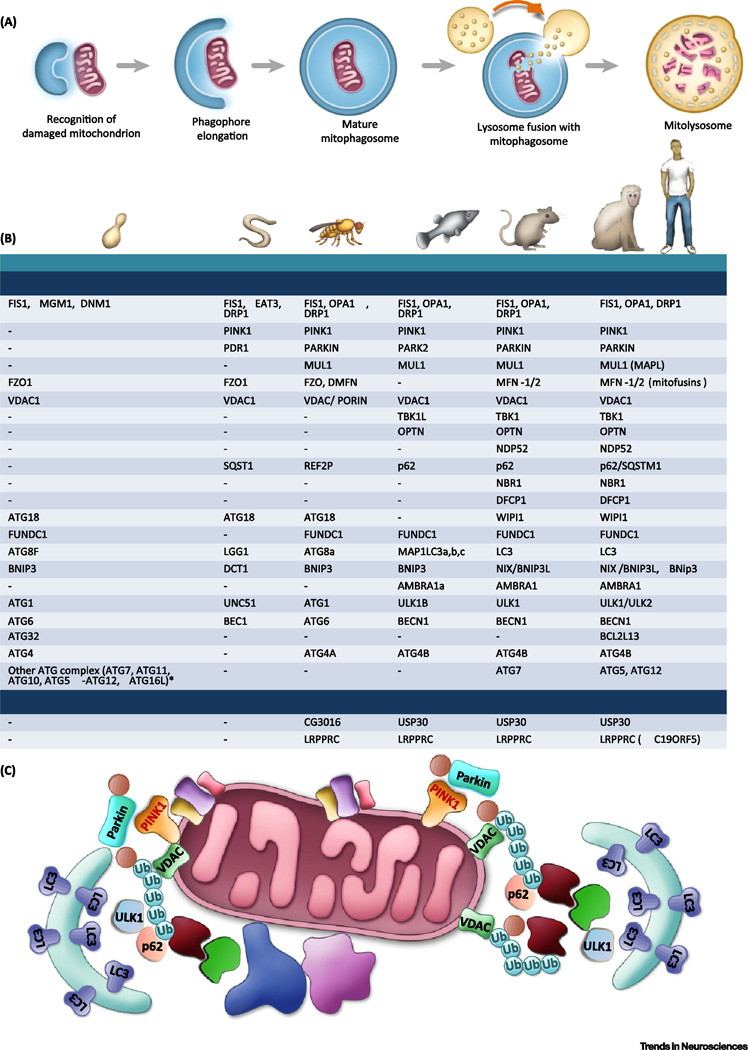
Figure 1. Mechanisms of mitophagy.
A. A simplified overview of the mitophagy pathway. A damaged mitochondrion is marked and recognized by the autophagic machinery, forming an autophagosome. This then fuses to a lysosome to be degraded.
B. Major autophagy/mitophagy proteins in the process of mitophagosome elongation and maturation across species. Major homology has been found in the higher eukaryotes; however, mammalian orthologues of many autophagy related proteins (ATGs) in yeast have not yet been identified. See Supplementary Table 1 for more details of the functions of each of the proteins. ‘-’, unknown.
*In yeast, ATG7 activates ATG12, passes to ATG10, then to ATG5-ATG12 complex. Yeast cells add ATG16L to form complex at membrane. This complex facilitates ATG8-phosphatidylethanolamine (PE) formation through stimulating ATG3 [122, 123].
C. Proteins involved in mitophagy in mammalian cells. PINK1 translocation into mitochondria is stopped upon loss of mitochondrial membrane potential, causing PINK1 to accumulate on mitochondria. Together with Parkin, these proteins form phospho-ubiquitin chains on mitochondrial outer membrane proteins such as VDAC1, which recruit autophagy receptors such as optineurin (OPTN) and NDP52, which bind to both ubiquitin and LC3 (an autophagosomal protein), inducing the formation of the autophagosome.
Abbreviations: FIS1- fission, mitochondrial 1; MGM1- mitochondrial dynamin-like GTPase; DNM1- dynamin 1; OPA1- Optic atrophy 1; DRP1- Dynamin-Related Protein; MARF- Mitochondrial assembly regulatory factor; PINK1- PTEN induced putative kinase 1; FZO1- mitofusin; VDAC1- voltage dependent anion channel 1; TBK1- TANK binding kinase 1; TAX1BP1- Tax1 binding protein 1; OPTN- Optineurin; NDP52(CALCOCO2)-calcium binding and coiled-coil domain 2; SQST1- SeQueSTosome related; REF2P–refractory to sigma P; NBR1- neighbor of Brca1 gene 1; DFCP1(ZFYVE1)- zinc finger FYVE-type containing 1; ATG- autophagy related gene; WIPI1- WD repeat domain, phosphoinositide interacting 1; FUNDC1- FUN14 domain containing 1; MAP1LC3-microtubule associated protein 1 light chain 3; BNIP3- BCL2 interacting protein 3; UNC51-Serine/threonine-protein kinase unc-51; ULKB- unc-51 like autophagy activating kinase 1; BEC1- Beclin homolog; BCL2L13- BCL2 like 13; PE- phosphatidylethanolamine; AMBRA1- autophagy and beclin 1 regulator 1; USP30- ubiquitin specific peptidase 30; LRPPRC- leucine rich pentatricopeptide repeat containing; P - Phosphate; Ub – Ubiquitin.
Mitophagy: Molecular machinery and regulatory mechanisms
Autophagy is an evolutionarily conserved process in which cytoplasmic substrates are engulfed in an autophagic vesicle, fused to lysosomes, and degraded. Autophagy is classified into various subgroups based on the mechanism of substrate delivery to lysosome; these groups are macroautophagy, chaperone-mediated autophagy, and microautophagy [18, 20]. We focus on macroautophagy (hereafter refer to as autophagy), in which engulfment by a double-membraned autophagosomal structure is followed by fusion into acidic lysosomes for bulk degradation. In mammalian cells, nascent autophagosomes can come from multifocal origins, encompassing mitochondria, endoplasmic reticulum (including omegasomes), Golgi complex, nucleus, and plasma membrane [18, 20]. Nutrient starvation leads to non-specific autophagy, while selective autophagy occurs in response to damaged cellular components, such as dysfunctional mitochondria (Figure 1A). Autophagy is necessary for cellular viability, and autophagic dysfunction has been linked to neurodegeneration, aging, and age-related diseases [18].
The molecular machinery that mediates the targeting of mitochondria to lysosomes has been actively elucidated in studies of yeast, worms (Caenorhabditis elegans), fruit flies (Drosophila melanogaster), zebrafish (Danio rerio), and mammals (Figure 1B) [21, 22]. When mitochondria become damaged, sustained depolarization of their inner membrane occurs and this stabilizes the protein PTEN-induced kinase 1 (PINK1) at the outer mitochondrial membrane (OMM). There, PINK1 phosphorylates mitofusin 2 (Mfn2) and ubiquitin which, in turn, recruit Parkin to the OMM. Parkin ubiquitylates several proteins which are then recognized by the ubiquitin-binding proteins optineurin (OPTN), p62, NDP52 and NBR1 that recruit the mitochondria to the autophagy pathway. Recent studies have suggested that p62 and NBR1 are dispensable for Parkin-mediated mitophagy [21]. In HeLa cells, PINK1 can also induce low-level mitophagy in a Parkin-independent manner [21]. While the importance of Parkin-mediated mitophagy has been demonstrated in vitro, a mitochondrial reporter mouse with mtDNA depletion failed to demonstrate Parkin recruitment to mitochondria [23]. Furthermore, studies with induced iPSC-derived neurons suggest physiological levels of Parkin may not be sufficient to induce mitophagy [24]. In addition to classical PINK1/Parkin-related mitophagy, other mitophagy receptors have been discovered, including AMBRA1, FUNDC1, and Nix/BNIP3L in mammals (Figure 1B). AMBRA1 can bind to LC3 and induce mitophagy in either a Parkin-dependent or Parkin-independent manner [25]. FUNDC1 is a mitochondrial outer membrane protein that can regulate mitochondrial fusion/fission (DRP1 and OPA1) to induce mitophagy in hypoxic conditions [26]. Nix/BNIP3L was originally reported as a mitophagy receptor to clear mitochondria in erythroid cells [27]; however, recent studies suggest that Nix can also induce mitophagy in other cell types, such as neurons, possibly as a downstream executor of the PINK1/Parkin pathway [28]. These mitophagy receptors bind to proteins associated with nascent autophagosomes via LC3-interacting region (LIR) motifs in LC3 and GABARAP family proteins that are covalently bound to the phagophore membrane lipid phosphatidylethanolamine. The formation of the protein bridges between the OMM and the phagophore membrane result in elongation (mediated by LC3 proteins) and closure (mediated by GABARAP proteins) of the phagophore membrane thereby completely engulfing the mitochondrion. The final stage of mitophagy is the fusion of the autophagosome with a lysosome, mediated by the phagophore LC3-binding proteins PLEKHM1 and HOPS, and the lysosome membrane-associated protein Rab7. Lysosomal hydrolases then degrade the mitochondrion (Figure 1A).
Accumulating evidence suggests that mitophagy functions throughout life, from fertilization and development through constitutive health maintenanceto prevention of age-related disease, including neurodegenerative diseases. Several interventions that are neuroprotective in experimental models of AD have also been reported to stimulate autophagy/mitophagy. These include the bioenergetic challenges of fasting, caloric restriction and exercise [29] and agents that inhibit the mTOR pathway including rapamycin [30] and the mitochondrial uncoupler 2,4-dinitrophenol [31]. On the other hand, conditions that impair autophagy/mitophagy include excessive dietary energy intake and diabetes [32, 33]. It is therefore important to understand the molecular mechanisms that may result in impaired mitophagy in AD, and whether stimulation of mitophagy might protect neurons against dysfunction and degeneration.
Mitochondrial dysfunction in AD
Studies in living AD patients and postmortem brain tissue have provided evidence that neurons in affected brain regions suffer from impaired mitochondrial function. PET brain scans reveal decreased radiolabeled glucose uptake into neurons and biochemical analyses demonstrate reduced activity of mitochondrial enzymes involved in oxidative phosphorylation and the TCA cycle [6]. Recent findings suggest that mitochondrial biogenesis is impaired in AD as indicated by reduced levels of the transcriptional regulator of mitochondrial biogenesis PGC1-α [34]. Consequently, dysfunctional mitochondria accumulate in neurons resulting in reduced cellular ATP levels and excessive ROS production which can exacerbate mitochondrial damage, leading to the aberrant amyloidogenic processing of APP and pTau and subsequent formation of AD-defining Aβ plaques and neurofibrillary tangles [10, 35].
Dysfunctional mitochondria upstream of Aβ and Tau pathologies
The β-amyloid precursor protein (APP) is a transmembrane protein that includes the 40–42 amino acid Aβ positioned partially within the membrane-spanning domain of APP. APP is cleaved by three enzymes, α-, β- and γ-secretases; α-secretase cleaves at the surface of the membrane to liberate a secreted form of APP, while intact A γ is generated by successive cleavages of APP at N- and C-termini of Aβ by β- and -secretases [35]. Genetic mutations in either APP or the γ-secretase enzyme presenilin 1 (PS1) can cause early-onset dominantly-inherited AD, in which the mutations result in accelerated age-dependent generation of Aβ1–42, which undergoes vigorous self-aggregation leading to neurotoxicity. Accumulating evidence suggests that mitochondrial dysfunction and associated oxidative stress, factors resulting from age, an inactive lifestyle, or excessive caloric intake, promote amyloidogenic APP processing and associated neuroinflammatory processes. When APP mutant transgenic mice are fed a diet with high levels of sucrose, they exhibit accelerated Aβ deposition and cognitive decline [36]. Increased caloric intake has been demonstrated to influence AD in humans as well. In 263 individuals homozygous or heterozygous for the E4 apolipoprotein allele (which increases AD susceptibility), those in the highest quartile for fat or calorie intake were over twice as likely to develop AD over 4 years than those in the lowest quartile [37]. This association was not seen in individuals with E2 or E3 alleles, however. Conversely, caloric restriction, which is known to stimulate mitophagy, ameliorates Aβ accumulation and cognitive deficits in several different mouse models of AD [38, 39]. Type 2 diabetes is a well-known consequence of obesity and sedentary lifestyles, and individuals with diabetes are more likely to develop AD [40]. Therefore, high-fat and high-calorie diets and the syndromes which result from them promote AD, while fasting, exercise, and other metabolism-inducing treatments such as insulin therapy may prevent AD [29, 41].
Evidence demonstrating that mitochondrial dysfunction promotes production of Aβ suggests a mito-centric chronological sequence of events in AD. Impaired mitochondrial function occurs prior to the accumulation of Aβ deposits in the brains of AD mouse models [42–44]. Experimental compromise of mitochondrial function by administration of toxins or genetic deletion of mitochondrial proteins that suppress ROS accelerates Aβ pathology [45, 46]. Studies of cultured neurons have shown that mitochondrial dysfunction promotes amyloid production through oxidative stress production. Oxidative stress increases γ-secretase activity by a mechanism involving covalent modification of the γ-secretase complex protein nicastrin by the membrane lipid peroxidation product 4-hydroxynonenal [47]. A major source of 4-hydroxynonenal is arachidonic acid, a lipid enriched in mitochondrial membranes. Altogether, accumulating evidence suggests amyloidogenic APP processing in late-onset AD occurs due to increased generation of ROS by dysfunctional mitochondria in neurons.
Microtubule associated protein Tau (MAPT) is another predominant player in AD. Microtubules are a component of the cellular cytoskeleton on which mitochondria, lysosomes, and other organelles are transported [48]. Tau binds and stabilizes microtubules, but when Tau is hyperphosphorylated it detaches from and thereby destabilizes microtubules resulting in their depolymerization. The accumulation of pTau aggregates and fibrils in the cell bodies and neurites of degenerating neurons is strongly correlated with cognitive deficits in AD (reviewed in [49]). Studies of experimental models have provided evidence that mitochondrial dysfunction can result in hyperphosphorylated Tau (pTau), microtubule depolymerization, and neurofibrillary tangle-like pathology. AD-like pTau occurs in hippocampal neurons with chronically elevated Ca2+ levels due to exposure to glutamate, a condition in which mitochondrial function is impaired. By initiating membrane lipid peroxidation, ROS generated by mitochondria may also promote pTau and its aggregation [50]. In rats, systemic brain infusion with rotenone, an inhibitor of complex 1 of the ETC, leads to pTau in neurons, astroglia and oligodendrocytes [51]. Similarly, the deletion of AFG3L2, a protease that processes misfolded proteins in the inner mitochondrial membrane, leads to mitochondrial network fragmentation, defective anterograde mitochondrial transport, and subsequent tau hyperphosphorylation in neurons [52]. Furthermore, genetic mitochondrial SOD2 deficiency causes AD-like pTau in brain neurons in mice, an effect counteracted by administration of an antioxidant [53]. The experimental induction of Tau pathology in neurons subjected to conditions associated with mitochondrial dysfunction, together with the fact that neurons accumulate Tau tangles in the absence of Aβ in frontotemporal dementia, suggests that mitochondria may be responsible for Tau pathology in AD.
Mitochondrial alterations downstream of Aβ and Tau
In support of a “vicious cycle” hypothesis, Aβ and pTau can adversely affect mitochondrial function and integrity in vitro and in vivo [10, 16]. Exposure of neurons to aggregating Aβ results in diminished mitochondrial ATP production, decreased activity of mitochondrial enzymes, and increased levels of mitochondrial ROS. Treatment of primary rat cortical neurons with Aβ1–42 also leads to opening of the mitochondrial permeability transition pore (mPTP); within one day of treatment transient mPTP opening occurs leading to decreased cell proliferation, while after 72 hours the mPTPs open irreversibly causing cell death [54]. In isolated rat mitochondria, Aβ decreases respiration rates,inhibits electron transport chain complex IV activity, and decreases the activities of the mitochondrial enzymes α-ketoglutarate dehydrogenase, cytochrome oxidase, and pyruvate dehydrogenase [55]. Cultured cells expressing mutant APP and producing high amounts of Aβ exhibit increased superoxide production and decreased ATP levels [56]. Synaptic mitochondria may be particularly susceptible to damage by aggregating Aβ because of their high energy demand and robust Ca2+ influx that occur during synaptic activation, particularly at glutamatergic synapses [57]. Indeed, direct exposure of synaptic terminals to aggregating Aβ results in membrane-associated mitochondrial dysfunction, oxidative stress, and impaired glutamate and glucose transport, thereby rendering the synapses vulnerable to excitotoxic degeneration [58].
Mice expressing P301L mutant human Tau that causes frontotemporal dementia exhibit reduced levels of mitochondrial complex V in affected brain regions, and a similar mitochondrial deficit was evident in brain tissue from patients harboring the same mutation [59]. Furthermore, decreased mitochondrial respiration and increased levels of ROS were observed in brain tissue of the Tau mutant mice [60]. The mechanism of this Tau-induced mitochondrial dysfunction remains to be established. Mitochondrial alterations may be secondary to microtubule depolymerization and a consequent inability of the cell to shuttle healthy mitochondria into axons and dendrites and remove dysfunctional mitochondria by mitophagy. Another possibility is that pTau inserts into the mitochondrial membrane and impairs Parkin-mediated mitophagy [61]. An N-terminal Tau fragment is enriched in mitochondria of human AD brains, and is related to impaired mitochondrial metabolism demonstrated by decreased expression of COXIV and cytochrome c oxidase [62]. Collectively, these data suggest that mitochondrial dysfunction can be an upstream inducer of Aβ aggregation and pTau, while Aβ aggregation and pTau can further exacerbate mitochondrial dysfunction, thus including a “vicious cycle” reaction in AD pathology.
Compromised autophagy and mitophagy in AD
Increasing evidence suggests that inhibition of the clearance of damaged mitochondria, along with concurrent increases in oxidative stress levels, results in the accumulation of dysfunctional neurons in AD. To be removed by mitophagy, the autophagosome containing the mitochondrion must fuse with a lysosome to form autolysosomes in which proteases degrade the mitochondrion (Figure 1A). Neurons exhibiting abnormal accumulation of autophagosomal vacuoles are a prominent feature in AD and their accumulation may result from lysosomal dysfunction (elevated pH), perhaps secondary to dysregulation of neuronal Ca2+ homeostasis [63]. The undegraded dysfunctional mitochondria accumulate in the soma which may result from the combination of local lysosome dysfunction and impaired mitochondrial transport [64]. It has been difficult to establish whether autophagic flux is reduced in neurons affected in AD, and to pinpoint the specific step(s) that is/are impaired. Recently, Bordi and colleagues interrogated the status of different steps in the autophagy/mitophagy pathway by performing microarray and immunochemical analyses of hippocampal CA1 neurons in postmortem tissue samples from AD subjects at different stages of the disease process [65]. Autophagy-related genes were upregulated beginning at early stages of AD, and increased lysosomal biogenesis was suggested by activation of the TFE3 transcription factor and several of its known target genes. Moreover, autophagic flux was apparently impeded by reduced substrate clearance (accumulation of LC3-II and p62 in autolysosomes). This gene expression analysis is supported by the finding that exosomes derived from AD patients, as well as from individuals ten years before AD diagnosis, had increased levels of cathepsin D, the primary protease in lysosomes; LAMP-1, a component of lysosomal membranes; and ubiquinylated proteins, all suggesting that lysosomal function is compromised and lysosomes containing undegraded cargo accumulate in neurons [66]. These findings suggest that autophagy/mitophagy is stimulated (perhaps secondary to mitochondrial dysfunction and fission) while lysosome function is impaired thereby contributing to the prominent accumulation of autophagosomes in neurons in AD.
Compromised mitophagy in AD may be caused by dysfunctional fusion between autophagosomes and lysosomes. For example, impairment of lysosomal function in healthy cells results in neuronal phenotypes like those in AD [63, 67]. Autophagosome accumulation in mouse cortical neurons occurs following oxidative stress, a condition associated with AD [68]. In 3xTgAD mouse brains as well as in blastocysts from PS1 knockout mice, LC3-II levels and LC3II-LC3I ratios were increased, suggesting that the autophagosomes that form in neurons may accumulate due to dysfunctional clearance via the lysosomal pathway [16, 69, 70]. Mutations in PS1 can impair autophagy/mitophagy [70]. Interestingly, wild type PS1 was reported to be required for lysosomal acidification, and familial AD PS1 mutations result in lysosomal alkalization and reduced lysosomal hydrolase activity [71]. Increased Parkin translocation to mitochondria, autophagosome accumulation and lysosomes containing undigested mitochondria occur in neurons in AD patient brains and in cultured cells overexpressing mutant APP, suggesting that autophagosome accumulation may represent deficient lysosomal efficiency [72]. Altogether, these findings suggest that defective mitophagy is involved in the pathogenesis of neuronal degeneration in AD (Figure 2).
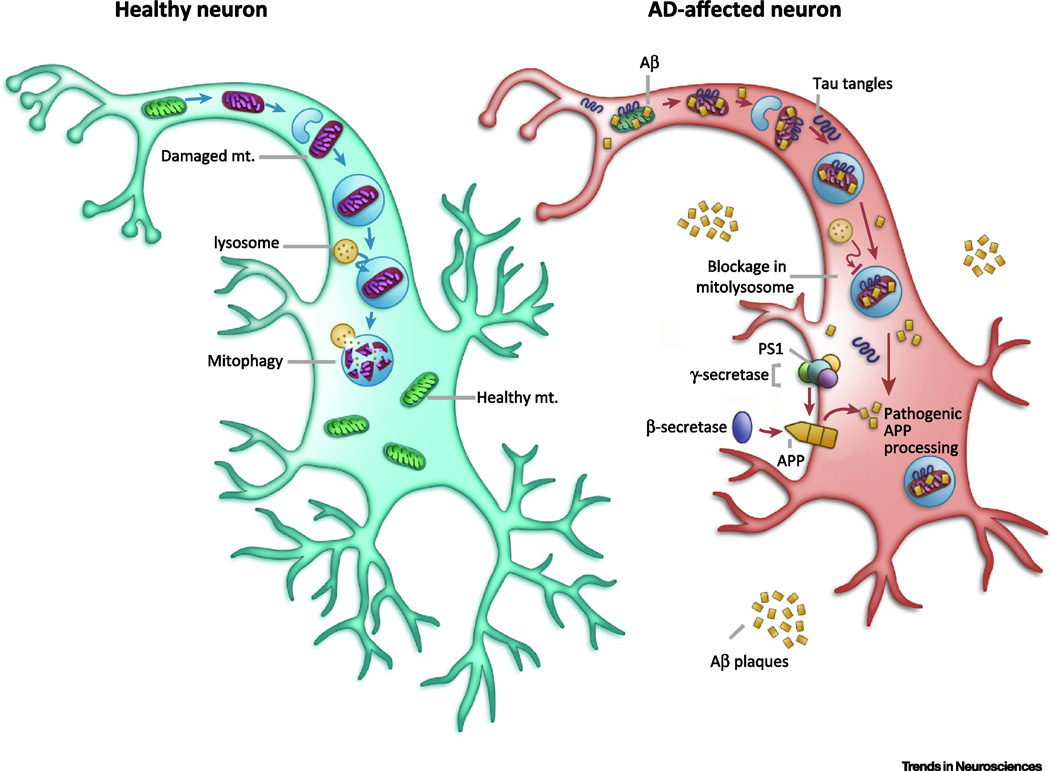
Figure 2. Mitochondrial dysfunction and compromised mitophagy in AD.
In unaffected neurons, healthy mitochondria are distributed through the neuron. When they become dysfunctional (shown here in purple), they are packaged into autophagosomes and trafficked to lysosomes to be degraded. In AD affected neurons, comprised mitophagy causes decreased energy production and increased oxidative stress. This leads to increased amyloidogenic processing of APP by β-secretase and γ-secretase/PS1 and, in parallel, the accumulation of pTau aggregates. Pathogenic Aβ and pTau can impair mitophagy, leading to a subsequent increase in damaged mitochondria, and initiation of a self-propagating vicious cycle. Mechanisms of compromised mitophagy in AD are still elusive.
Alterations of mitochondrial dynamics- and mitophagy-related proteins in AD
Mitophagy- and mitochondrial dynamics-related proteins affected in AD include those involved in mitochondrial fission and fusion (Drp1 and mitofusin), mitochondrial biogenesis (PGC-1α) and mitochondrial responses to bioenergetic and oxidative challenges (SIRT3 and SIRT1) [10, 11, 13]. Mitochondrial fission provides a mechanism to remove damaged and dysfunctional molecules from the mitochondrion. Mitochondrial fission and fusion are controlled by specialized proteins such as Fis1 and Drp1 [73, 74] and Opa1, respectively [75]; the levels of each of these proteins determine the amount of fission and fusion occurring. Prior to fission, damaged DNA and proteins are segregated to one side of the mitochondrion such that upon fission one of the ‘daughter’ mitochondrion contains damaged molecules and is targeted for mitophagy, while the other daughter mitochondrion is now pristine [10, 76]. Cell culture and animal models of AD have demonstrated excessive fission of mitochondria concomitant with accumulation of dysfunctional mitochondria, possibly as the result of lysosomal dysfunction [63, 77]. While some studies have demonstrated increased levels of Drp1 in the brains of AD patients and mice [78], others have shown conflicting results [79]. Analyses of postmortem brain tissue from AD patients and age-matched control subjects have revealed reduced expression of genes related to mitochondrial biogenesis including PGC-1α, TFAM and NRF2 [80]. Consistent with a role of impaired PGC-1α in AD pathology, upregulation of PGC-1α can inhibit AD progression. In APP23 transgenic mice, LV (lentiviral vector)-hPGC-1α injected in the hippocampus and cortex areas decreased Aβ plaques and improved spatial and recognition memory, possibly through downregulation of β-secretase [34].
The sirtuins (SIRTs) are a group of nicotinamide adenine dinucleotide (NAD)+-dependent enzymes that regulate multiple cellular pathways and can inhibit a series of metabolic and age-related diseases. Of the seven mammalian SIRTs, the nuclear SIRT1 and the mitochondrial SIRT3 have been linked to neuroprotection, and reduction of these proteins may contribute to neurodegenerative diseases, including AD [81]. Studies in AD patient brain tissues suggest reduced SIRT1 protein expression (−45%) in the parietal cortex, and this is closely associated with accumulation of Aβ and Tau tangles [82]. SIRT1 upregulates PGC-1α, and plays an important role in autophagy/mitophagy induction through deacetylation and activation of major autophagy proteins (ATG5, ATG7, ATG8/LC3), stabilization of PINK1, and upregulation of mitophagy receptor Nix/BNIP3L and LC3 [19, 83, 84]. SIRT3 activates FOXO3 to induce p62 (a major autophagy protein) clustering on ubiquitinated mitochondrial substrates and formation of autolysosomes [85]. SIRT3 levels are reduced in neurons of APP/PS1 double mutant AD mice [85–87]. SIRT3 may also protect mitochondria and neurons against excitotoxic and metabolic stress and apoptosis by a SOD2 and cyclophilin D deacetylation-dependent mechanism [13]. Thus, impaired SIRT1 activity can cause mitochondrial dysfunction and autophagy/mitophagy inhibition, leading to accumulation of damaged mitochondria as well as Aβ plaques and Tau tangles.
Neuronal NAD+ deficiency and impaired mitophagy in AD
As a basic cellular metabolite, NAD+ exists in all living cells including brain cells, where it plays fundamental roles in neuroplasticity and cellular stress resistance. In cell metabolism, NAD+, and its reduced form NADH, are necessary for glycolysis, the TCA cycle, oxidative phosphorylation and ATP production. Because neurons consume relatively large amounts of energy, they are extremely sensitive to the decreased NAD+ levels as well as the impairment of ATP production [84]. Furthermore, NAD+ affects neuronal health and survival through regulation of the balance between mitochondrial biogeneisis and mitophagy, which are controlled by the NAD+/SIRT1-PGC1α pathway and the DAF-16/FOXO3 pathway [83, 84]. Decreased NAD+ levels can compromise mitophagy and trigger the accumulation of misfolded proteins leading to neuronal death [81, 88].
Celluar NAD+ levels are affected by several NAD+-consuming enzymes which have links to AD. In addition to SIRTs, there are several other NAD+-dependent enzymes involved in neuronal stress responses including poly (ADP -ribose) polymerase 1 (PARP1), cyclic ADP ribose hydrolase (CD38), and CD157 [89]. NAD+ levels decline with age and in neurodegenerative conditions caused by DNA repair deficiency [84]. PARP1 is an enzyme that responds to DNA strand breaks by catalyzing polyADP-ribosylation (PARylation) of target proteins using NAD+ as a cofactor [17]. While PARP1 was first shown to be localized in the nucleus, it is now known also to localize to mitochondria in cells under stress where it can PARylate electron transport chain proteins. Levels of PARP1 activity are increased and PARylated proteins accumulated in brain tissue samples from vulnerable brain regions of AD patients [90]. Increased oxidative stress may underlie neuronal DNA damage in AD [91, 92]; therefore, oxidative stress is a likely trigger for PARP1 activation in AD which may occur both upstream and downstream of Aβ accumulation. CD38 is a multifunctional enzyme that catalyzes the synthesis and hydrolysis of cyclic ADP-ribose (cADPR) from NAD+ to ADP-ribose and mediates Ca2+ release from the endoplasmic reticulum. Importantly, PARP1 and CD38 activity may lead to decreased NAD+ levels and thus lower sirtuin activity. Treatment of neural cells with a PARP1 inhibitor can protect them against mitochondrial dysfunction and cell death caused by Aβ [93]. APP/PS1 double mutant transgenic mice that lack CD38 exhibit reduced levels of A in their brains and improved learning and memory [94], consistent with NAD+ depletion in promotion of amyloidogenesis in this mouse model of AD. The available data suggest that NAD+ deficiency in AD, possibly caused by PARP1 activation due to increased oxidative stress-mediated DNA damage, leads to decreased sirtuin activity, decreased mitophagy, and mitochondrial dysfunction.
DNA Damage and its possible relationships with mitophagy
Studies of AD patient brain tissue samples and animal models of AD suggest that the accumulation of unrepaired oxidatively damaged nuclear and mitochondrial DNA may occur early in the neurodegenerative process. There are two major DNA repair pathways in neurons, base excision repair (BER) and DNA double-strand break repair (DSBR), both of which are more comprised in AD brain cells than in normal aging [91, 95]. The DNA damage sensor PARP1 detects DNA damage in both BER and DSBR, suggesting that the chronic hyperactivation of PARP1 seen in AD reflects decreased operation of these DNA repair pathways [93, 96]. Decreased DNA repair function in AD has been shown to lead to increased neurodegeneration. Haplo-insufficiency of DNA polymerase β (Polβ), a key enzyme in neuronal BER, results in increased death of hippocampal neurons in 3xTgAD mice. Furthermore, the transcriptomic profiles of 3xTg/Polβ+/- mice are more similar to human AD patients than that of either 3xTg or Polβ+/- mice [92]. This Aβ and tau-independent increase in neurodegeneration caused by increased DNA damage may be mediated by impaired mitochondrial function caused by inhibition of the aforementioned NAD+/Sirtuin-PGC-1α pathway [81]. The connection between DNA repair and mitochondrial function is further suggested by DNA repair proteins such as ataxia telangiectasia mutated (ATM) with essential roles in autophagy and mitophagy [83]. The relationships, both direct and indirect, between DNA repair deficiency and mitophagy/ mitochondrial dysfunction in AD are a promising avenue of future research.
Novel experimental tools in AD
Using iPSCs to study mitochondrial function and mitophagy in AD
The mitochondrial etiology of AD discussed herein suggests animal or cellular models with AD-associated Aβ and tau pathologies, while of significant use, may not fully recapitulate important aspects of AD onset and progression. Studies using cells and tissue from human AD patients may better elucidate the nature and extent of mitophagy and mitochondrial dysfunction. Induced pluripotent stem cells (iPSCs) represent a useful tool for such studies. Since the discovery of iPSCs in 2006 by Yamanaka and colleagues [97], the field has evolved significantly and iPSCs have been used to study human cellular biology and the mechanisms of many human disorders. iPSCs are particularly useful in studying neurodegenerative disorders like AD, as neurons and glia from these patients are difficult or impossible to obtain. iPSCs from patients are increasingly available through open source platforms and large scale consortia such as HIPSCI and StemBANC [98], and the large scale expansion of iPSCs enables their use in high-throughput drug screens. The differences between animal models of AD and human cases are well-recognized [99], which supports the importance of iPSC-derived neurons in the investigation of mitophagic dysfunction and the mechanisms of compromised mitophagy in AD. However, modelling neurodegenerative disease in an iPSC-derived neurons does come with significant challenges and iPSC-derived neurons may revert to embryonic phenotypes, which is a caveat when modelling late-onset disease [100, 101]. The neuronal expression of progerin, a truncated form of lamin A which is defective in Hutchinson-Gilford progeria syndrome [102], and the direct reprogramming of fibroblasts are two recent approaches that have been used to impart a more ‘aged’ cellular phenotype. However, both approaches have significant limitations owing to discrepancies between progerias and normal aging and the limited number of expansions and less differentiation versatility in direct reprogramming. Further introduction of age-associated metabolic changes in iPSC-derived neuronal models may yield viable models to study AD and healthy aging in a human neurological system. Since DNA damage and mitochondrial dysfunction can be introduced experimentally and investigated on an AD relevant genetic background, this is an avenue that is worthy of further exploration. To the best of our knowledge, mitophagy has not yet been investigated in neurons derived from AD patients. However, advances made in studies of other diseases suggest that the iPSC system is useful for detecting mitophagic defects [103].
Animal models of Sporadic AD
Recent years have seen the utilization of several rat and mouse models which recapitulate AD symptoms such as neurodegeneration, pTau, oxidative stress, and Aβ aggregation without the need for expression familial AD-associated PS1 or APP mutations. Introduced in 1990 by Mayer [104], intracerebroventricular administration of low doses of streptozotocin leads to an insulin-resistant brain state, subsequent glucose hypometabolism and oxidative stress [105]. Treatment of rats with streptozotocin leads to Aβ and tau pathology within three months post-treatment, evidencing the primacy of dysregulated metabolism in AD development [106]. OXYS rats are an experimental model of accelerated aging which present learning and memory deficits and neurodegeneration [107]. These rats develop synaptic loss and mitochondrial abnormalities at 4 months of age, pTau at 3 months of age, but an increase in Aβ1-42 levels only at 15–25 months of age [108]. Other important models that may be relevant to sporadic AD are the senescence-accelerated mouse prone 8 strain (SAMP8) [109] and aldehyde-dehydrogenase knockout mice [110]. Such animal models will be useful for research into age-related abnormalities of mitophagy that may predispose the brain to late-onset AD.
Stimulating mitophagy as an approach for delaying and treating AD
Pharmacological agents and lifestyle interventions aimed at improving mitochondrial health and enhancing mitophagy have been evaluated in animal models and, in some cases, in mild cognitive impairment (MCI) or AD patients (Figure 3). Caloric restriction, intermitting fasting and vigorous exercise are bioenergetic challenges that can promote neuroplasticity (synapse formation, hippocampal neurogenesis, and learning and memory) and bolster neuronal stress resistance [111, 112]. Evidence from studies of rodents has shown that fasting and exercise affect signaling pathways in neurons in ways that reduce mitochondrial oxidative stress, stimulate mitochondrial biogenesis, and enhance autophagy [111]. For example, using GFP-LC3 transgenic mice, it was shown that fasting for 24 – 48 hours results in more autophagosomes in cerebral cortical neurons [113]. In addition, mice that run on wheels exhibit the neuroprotective effects of exercise mediated by increased expression of SIRT3 in hippocampal and cortical cells [13]. Exercise and fasting may stimulate mitochondrial biogenesis in neurons by a mechanism involving BDNF signaling and upregulation of PGC-1α, a pathway that plays important roles in synapse formation, maintenance and plasticity [11]. Thus, exercise and fasting may increase the numbers of well-functioning mitochondria in neurons by activating pathways that stimulate mitochondrial biogenesis and mitophagy.
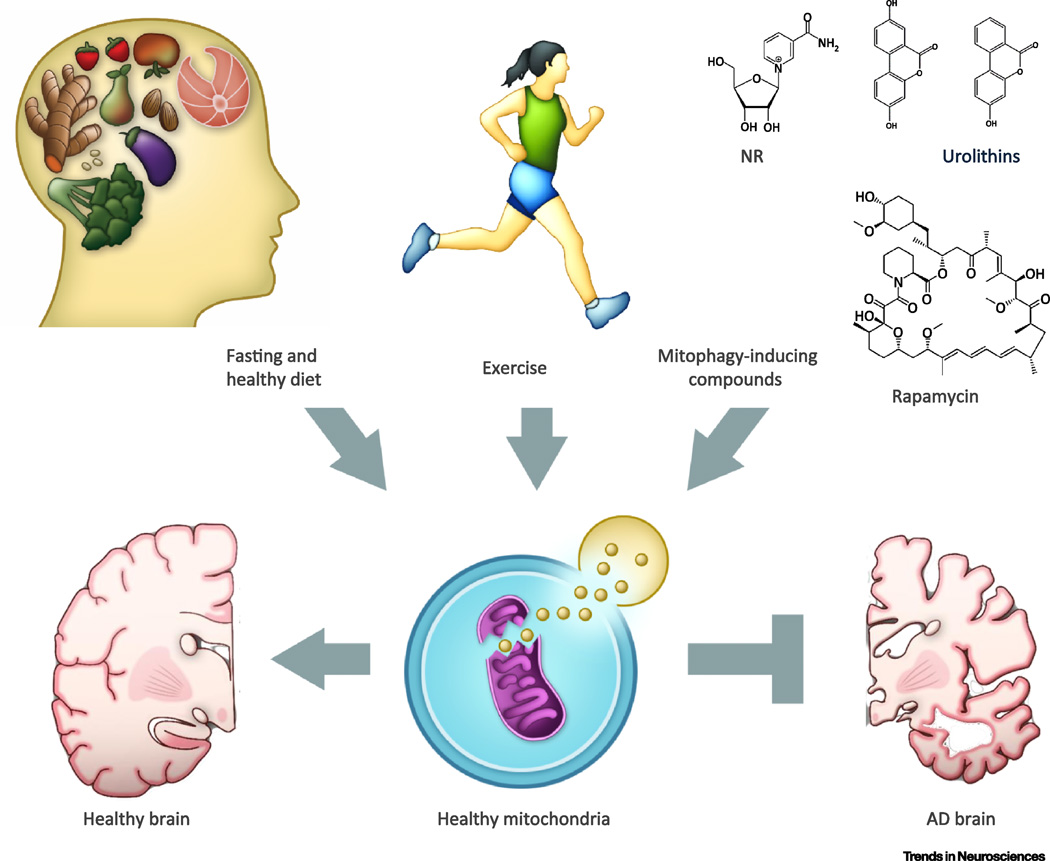
Figure 3. Physiological and pharmacological stimulation of mitophagy as potential therapeutic approaches for AD.
Maintenance of a healthy mitochondrial pool in neurons may be paramount in healthy aging and avoidance of AD development. We highlight several lifestyle factors that can contribute to mitochondrial health, such as decreased energy consumption and suitable exercise, as well as certain compounds that pharmacologically induce mitophagy and mitochondrial health, such as rapamycin, spermidine, and the urolithins, as possible methods of AD prevention. Research into candidate drugs that mimic these lifestyle changes and mitophagy-inducing compounds, holds promise in AD treatment.
Several compounds targeting mitochondrial health in AD have been examined in preclinical studies (see review [114]). One approach is the restoration of neuronal mitochondrial bioenergetics and SIRT3 and SIRT1 activities by elevation of cellular NAD+ levels. Treatment of 3xTgAD mice with nicotinamide, a precursor of NAD+ that enhances SIRT3 activity, improved Aβ and Tau pathologies and ameliorated learning and memory deficits [69]. This study also showed that nicotinamide increased mitochondrial resistance to oxidative stress, upregulated autophagy, and increased the activities of PI3K-Akt, MAPK/ERK1/2, SIRT1 and the transcription factor CREB. Nicotinamide treatment also reduced PARP-1 levels along with markers of oxidative stress and increased endogenous antioxidant enzyme activity in a rat model of Aβ neurotoxicity [115]. Moreover, treatment of APP mutant transgenic mice with nicotinamide riboside, an NAD+ precursor [84], elevated levels of NAD+ and PGC-1α in the cerebral cortex and ameliorated cognitive deficits [116]. The relative contributions of increased mitochondrial stress resistance and enhanced mitophagy to the beneficial effects of SIRT3 in AD models remain to be determined. Collectively, these data suggest that interventions that sustain neuronal NAD+ levels may benefit AD patients.
Additional pharmacological approaches that enhance mitophagy may prove beneficial in delaying or treating AD. Some such treatments include agents that induce mild bioenergetic stress or inhibit the mTOR pathway. Mitochondrial uncoupling agents such as 2,4-Dinitrophenol (DNP) can stimulate autophagy and are effective in preserving neuronal function in animal models of relevance to AD, including APP/PS1 transgenic mice [31]. Treatment of mice with 2-deoxyglucose, which induces mild bioenergetic stress and stimulates ketogenesis, protects neurons against dysfunction and degeneration in a mitochondrial toxin-based model of PD [117] and enhances mitochondrial function and stimulates autophagy and clearance of Aβ [118]. Another autophagy/mitophagy-inducing compound, the mTOR inhibitor rapamycin, ameliorated cognitive deficits and reduced Aβ pathology in an APP mutant mouse AD model [30]. Collectively, such findings provide a rationale for future testing of compounds that induce mitophagy, such as spermidine, urolithins and the antibiotic actinonin, in preclinical AD models [119–121].
Concluding Remarks
Significant progress has been made to ascertain the causes of the devastating neurodegenerative disorder AD. The accumulation of dysfunctional mitochondria has emerged as a common feature of neurons affected in patients and animal models that may occur prior to discernible cognitive deficits. Experimental manipulations that impair mitophagy can enhance Aβ and pTau pathologies, while interventions that stimulate mitophagy can preserve synaptic plasticity and cognitive function. In addition, aggregating Aβ can impair neuronal autophagy/mitophagy. These data suggest important roles for mitophagy deficits both upstream and downstream of Aβ and Tau. Thus, impaired mitophagy may be an important process in the vicious neurodegenerative cycle leading to synaptic dysfunction and neuronal death in AD. Further research into the molecular mechanisms of compromised mitophagy in AD laboratory models, AD iPSC-derived neurons, and AD patient samples are necessary and may provide novel therapeutic strategies for this widespread disease.
Supplementary Material
Trends.
Mitochondrial homeostasis is important for synaptic plasticity, learning, and memory.
Neurons affected in Alzheimer’s disease exhibit dysfunctional mitochondria.
Mitophagy plays important roles in mitochondrial homeostasis, neuroprotection, and resistance to neurodegeneration.
iPSC-derived neurons provide a powerful tool to investigate mitochondrial dysfunction and mitophagy with a human AD genetic background.
NAD+ depletion and impaired mitophagy may occur early in AD and contribute to synaptic dysfunction and neuronal degeneration.
Outstanding questions.
What are the molecular pathways of neuronal mitophagy in addition to the PINK1-Parkin pathway?
What are the molecular alterations that result in impaired mitophagy in AD?
How does DNA damage affect mitophagy?
Are mitochondrial dysfunction and impaired mitophagy upstream of Aβ and pTau pathologies?
How do neurons coordinate mitochondrial fusion-fission, transportation, and mitophagy in the maintenance of mitochondrial quality and homeostasis?
Will interventions that promote mitophagy benefit patients with, or at risk for AD?
Acknowledgments
We acknowledge the valuable work of the many investigators whose published articles we were unable to cite owing to space limitations. We thank Drs. Emmette Hutchison and Krisztina Marosi for critical reading of the manuscript, Dr. Nuo Sun for his comments and inputs, and Marc Raley for generation of the figures. This research was supported by the Intramural Research Program of the National Institute on Aging, including two NIA intra-laboratory grants (2015–2016, 2016–2017 to EFF/VAB).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Citron M. Alzheimer’s disease: strategies for disease modification. Nat Rev Drug Discov. 2010;9(5):387–398. doi: 10.1038/nrd2896. [DOI] [PubMed] [Google Scholar]
- 2.Hampel H, et al. The future of Alzheimer’s disease: the next 10 years. Prog Neurobiol. 2011;95(4):718–728. doi: 10.1016/j.pneurobio.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 3.Perry RJ, Watson P, Hodges JR. The nature and staging of attention dysfunction in early (minimal and mild) Alzheimer’s disease: relationship to episodic and semantic memory impairment. Neuropsychologia. 2000;38(3):252–271. doi: 10.1016/s0028-3932(99)00079-2. [DOI] [PubMed] [Google Scholar]
- 4.Braak H, Braak E. Staging of Alzheimer’s disease-related neurofibrillary changes. Neurobiol Aging. 1995;16(3):271–278. doi: 10.1016/0197-4580(95)00021-6. discussion 278–84. [DOI] [PubMed] [Google Scholar]
- 5.Price JL, et al. The distribution of tangles, plaques and related immunohistochemical markers in healthy aging and Alzheimer’s disease. Neurobiol Aging. 1991;12(4):295–312. doi: 10.1016/0197-4580(91)90006-6. [DOI] [PubMed] [Google Scholar]
- 6.Kapogiannis D, Mattson MP. Disrupted energy metabolism and neuronal circuit dysfunction in cognitive impairment and Alzheimer’s disease. Lancet Neurol. 2011;10(2):187–198. doi: 10.1016/S1474-4422(10)70277-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blass JP, Gibson GE. The role of oxidative abnormalities in the pathophysiology of Alzheimer’s disease. Rev Neurol (Paris) 1991;147(6–7):513–525. [PubMed] [Google Scholar]
- 8.Swerdlow RH, Khan SM. A “mitochondrial cascade hypothesis” for sporadic Alzheimer’s disease. Med Hypotheses. 2004;63(1):8–20. doi: 10.1016/j.mehy.2003.12.045. [DOI] [PubMed] [Google Scholar]
- 9.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283(5407):1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 10.Mattson MP, Gleichmann M, Cheng A. Mitochondria in neuroplasticity and neurological disorders. Neuron. 2008;60(5):748–766. doi: 10.1016/j.neuron.2008.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng A, et al. Involvement of PGC-1alpha in the formation and maintenance of neuronal dendritic spines. Nat Commun. 2012;3:1250. doi: 10.1038/ncomms2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marosi K, Mattson MP. BDNF mediates adaptive brain and body responses to energetic challenges. Trends Endocrinol Metab. 2014;25(2):89–98. doi: 10.1016/j.tem.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng A, et al. Mitochondrial SIRT3 Mediates Adaptive Responses of Neurons to Exercise and Metabolic and Excitatory Challenges. Cell Metab. 2016;23(1):128–142. doi: 10.1016/j.cmet.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bezprozvanny I, Mattson MP. Neuronal calcium mishandling and the pathogenesis of Alzheimer’s disease. Trends Neurosci. 2008;31(9):454–463. doi: 10.1016/j.tins.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menzies FM, Fleming A, Rubinsztein DC. Compromised autophagy and neurodegenerative diseases. Nat Rev Neurosci. 2015;16(6):345–357. doi: 10.1038/nrn3961. [DOI] [PubMed] [Google Scholar]
- 16.Cai Q, Tammineni P. Alterations in Mitochondrial Quality Control in Alzheimer’s Disease. Front Cell Neurosci. 2016;10:24. doi: 10.3389/fncel.2016.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang EF, et al. Nuclear DNA damage signalling to mitochondria in ageing. Nat Rev Mol Cell Biol. 2016;17(5):308–321. doi: 10.1038/nrm.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146(5):682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 19.Palikaras K, Lionaki E, Tavernarakis N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature. 2015;521(7553):525–528. doi: 10.1038/nature14300. [DOI] [PubMed] [Google Scholar]
- 20.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12(1):9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazarou M, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524(7565):309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12(2):119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 23.Sterky FH, et al. Impaired mitochondrial transport and Parkin-independent degeneration of respiratory chain-deficient dopamine neurons in vivo. Proc Natl Acad Sci U S A. 2011;108(31):12937–12942. doi: 10.1073/pnas.1103295108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rakovic A, et al. Phosphatase and tensin homolog (PTEN)-induced putative kinase 1 (PINK1)-dependent ubiquitination of endogenous Parkin attenuates mitophagy: study in human primary fibroblasts and induced pluripotent stem cell-derived neurons. J Biol Chem. 2013;288(4):2223–2237. doi: 10.1074/jbc.M112.391680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strappazzon F, et al. AMBRA1 is able to induce mitophagy via LC3 binding, regardless of PARKIN and p62/SQSTM1. Cell Death Differ. 2015;22(3):419–432. doi: 10.1038/cdd.2014.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen M, et al. Mitophagy receptor FUNDC1 regulates mitochondrial dynamics and mitophagy. Autophagy. 2016;12(4):689–702. doi: 10.1080/15548627.2016.1151580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandoval H, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454(7201):232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao F, et al. The mitochondrial protein BNIP3L is the substrate of PARK2 and mediates mitophagy in PINK1/PARK2 pathway. Hum Mol Genet. 2015;24(9):2528–2538. doi: 10.1093/hmg/ddv017. [DOI] [PubMed] [Google Scholar]
- 29.Mattson MP. Energy intake and exercise as determinants of brain health and vulnerability to injury and disease. Cell Metab. 2012;16(6):706–722. doi: 10.1016/j.cmet.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spilman P, et al. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer’s disease. PLoS One. 2010;5(4):e9979. doi: 10.1371/journal.pone.0009979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geisler JG, et al. DNP, mitochondrial uncoupling, and neuroprotection: A little dab’ll do ya. Alzheimers Dement. 2016 doi: 10.1016/j.jalz.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stranahan AM, et al. Diet-induced insulin resistance impairs hippocampal synaptic plasticity and cognition in middle-aged rats. Hippocampus. 2008;18(11):1085–1088. doi: 10.1002/hipo.20470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jung HS, Lee MS. Role of autophagy in diabetes and mitochondria. Ann N Y Acad Sci. 2010;1201:79–83. doi: 10.1111/j.1749-6632.2010.05614.x. [DOI] [PubMed] [Google Scholar]
- 34.Katsouri L, et al. PPARgamma-coactivator-1alpha gene transfer reduces neuronal loss and amyloid-beta generation by reducing beta-secretase in an Alzheimer’s disease model. Proc Natl Acad Sci U S A. 2016;113(43):12292–12297. doi: 10.1073/pnas.1606171113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mattson MP. Pathways towards and away from Alzheimer’s disease. Nature. 2004;430(7000):631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao D, et al. Intake of sucrose-sweetened water induces insulin resistance and exacerbates memory deficits and amyloidosis in a transgenic mouse model of Alzheimer disease. J Biol Chem. 2007;282(50):36275–36282. doi: 10.1074/jbc.M703561200. [DOI] [PubMed] [Google Scholar]
- 37.Luchsinger JA, et al. Caloric intake and the risk of Alzheimer disease. Arch Neurol. 2002;59(8):1258–1263. doi: 10.1001/archneur.59.8.1258. [DOI] [PubMed] [Google Scholar]
- 38.Halagappa VK, et al. Intermittent fasting and caloric restriction ameliorate age-related behavioral deficits in the triple-transgenic mouse model of Alzheimer’s disease. Neurobiol Dis. 2007;26(1):212–220. doi: 10.1016/j.nbd.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 39.Schafer MJ, et al. Reduction of beta-amyloid and gamma-secretase by calorie restriction in female Tg2576 mice. Neurobiol Aging. 2015;36(3):1293–1302. doi: 10.1016/j.neurobiolaging.2014.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sims-Robinson C, Kim B, Rosko A, Feldman EL. How does diabetes accelerate Alzheimer disease pathology. 2010;6(10):551–559. doi: 10.1038/nrneurol.2010.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker JM, Harrison FE. Shared neuropathological characteristics of obesity, type 2 diabetes and Alzheimer’s disease: impacts on cognitive decline. 2015;7(9):7332–7357. doi: 10.3390/nu7095341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao J, et al. Mitochondrial bioenergetic deficit precedes Alzheimer’s pathology in female mouse model of Alzheimer’s disease. Proc Natl Acad Sci U S A. 2009;106(34):14670–14675. doi: 10.1073/pnas.0903563106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leuner K, et al. Peripheral mitochondrial dysfunction in Alzheimer’s disease: focus on lymphocytes. Mol Neurobiol. 2012;46(1):194–204. doi: 10.1007/s12035-012-8300-y. [DOI] [PubMed] [Google Scholar]
- 44.Mao P, et al. Mitochondria-targeted catalase reduces abnormal APP processing, amyloid beta production and BACE1 in a mouse model of Alzheimer’s disease: implications for neuroprotec tion and lifespan extension. Hum Mol Genet. 2012;21(13):2973–2990. doi: 10.1093/hmg/dds128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esposito L, et al. Reduction in mitochondrial superoxide dismutase modulates Alzheimer’s diseaselike pathology and accelerates the onset of behavioral changes in human amyloid precursor protein transgenic mice. J Neurosci. 2006;26(19):5167–5179. doi: 10.1523/JNEUROSCI.0482-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen L, et al. NLRP3 inflammasome activation by mitochondrial reactive oxygen species plays a key role in long-term cognitive impairment induced by paraquat exposure. Neurobiol Aging. 2015;36(9):2533–2543. doi: 10.1016/j.neurobiolaging.2015.05.018. [DOI] [PubMed] [Google Scholar]
- 47.Gwon AR, et al. Oxidative lipid modification of nicastrin enhances amyloidogenic gamma-secretase activity in Alzheimer’s disease. Aging Cell. 2012;11(4):559–568. doi: 10.1111/j.1474-9726.2012.00817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buee L, et al. Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res Brain Res Rev. 2000;33(1):95–130. doi: 10.1016/s0165-0173(00)00019-9. [DOI] [PubMed] [Google Scholar]
- 49.Gong CX, Grundke-Iqbal I, Iqbal K. Targeting tau protein in Alzheimer’s disease. Drugs Aging. 2010;27(5):351–365. doi: 10.2165/11536110-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 50.Mattson MP, et al. 4-Hydroxynonenal, a product of lipid peroxidation, inhibits dephosphorylation of the microtubule-associated protein tau. Neuroreport. 1997;8(9–10):2275–2281. doi: 10.1097/00001756-199707070-00036. [DOI] [PubMed] [Google Scholar]
- 51.Hoglinger GU, et al. The mitochondrial complex I inhibitor rotenone triggers a cerebral tauopathy. J Neurochem. 2005;95(4):930–939. doi: 10.1111/j.1471-4159.2005.03493.x. [DOI] [PubMed] [Google Scholar]
- 52.Kondadi AK, et al. Loss of the m-AAA protease subunit AFG(3)L(2) causes mitochondrial transport defects and tau hyperphosphorylation. EMBO J. 2014;33(9):1011–1026. doi: 10.1002/embj.201387009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Melov S, et al. Mitochondrial oxidative stress causes hyperphosphorylation of tau. PLoS One. 2007;2(6):e536. doi: 10.1371/journal.pone.0000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hou Y, et al. Permeability transition pore-mediated mitochondrial superoxide flashes mediate an early inhibitory effect of amyloid beta1-42 on neural progenitor cell proliferation. Neurobiol Aging. 2014;35(5):975–989. doi: 10.1016/j.neurobiolaging.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Casley CS, et al. Beta-amyloid inhibits integrated mitochondrial respiration and key enzyme activities. J Neurochem. 2002;80(1):91–100. doi: 10.1046/j.0022-3042.2001.00681.x. [DOI] [PubMed] [Google Scholar]
- 56.Leuner K, et al. Mitochondrion-derived reactive oxygen species lead to enhanced amyloid beta formation. Antioxid Redox Signal. 2012;16(12):1421–1433. doi: 10.1089/ars.2011.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Du H, et al. Early deficits in synaptic mitochondria in an Alzheimer’s disease mouse model. Proc Natl Acad Sci U S A. 2010;107(43):18670–18675. doi: 10.1073/pnas.1006586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keller JN, et al. Impairment of glucose and glutamate transport and induction of mitochondrial oxidative stress and dysfunction in synaptosomes by amyloid beta-peptide: role of the lipid peroxidation product 4-hydroxynonenal. J Neurochem. 1997;69(1):273–284. doi: 10.1046/j.1471-4159.1997.69010273.x. [DOI] [PubMed] [Google Scholar]
- 59.Neurofibrillary tangles, amyotrophy and progressive motor disturbance in mice expressing mutant (P301L) tau protein. Nat Genet. 2000;26(1):127. doi: 10.1038/79109. [DOI] [PubMed] [Google Scholar]
- 60.David DC, et al. Proteomic and functional analyses reveal a mitochondrial dysfunction in P301L tau transgenic mice. J Biol Chem. 2005;280(25):23802–23814. doi: 10.1074/jbc.M500356200. [DOI] [PubMed] [Google Scholar]
- 61.Hu Y, et al. Tau accumulation impairs mitophagy via increasing mitochondrial membrane potential and reducing mitochondrial Parkin. Oncotarget. 2016;7(14):17356–17368. doi: 10.18632/oncotarget.7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Amadoro G, et al. A NH2 tau fragment targets neuronal mitochondria at AD synapses: possible implications for neurodegeneration. J Alzheimers Dis. 2010;21(2):445–470. doi: 10.3233/JAD-2010-100120. [DOI] [PubMed] [Google Scholar]
- 63.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med. 2013;19(8):983–997. doi: 10.1038/nm.3232. [DOI] [PubMed] [Google Scholar]
- 64.Ashrafi G, Schwarz TL. PINK1- and PARK2-mediated local mitophagy in distal neuronal axons. Autophagy. 2015;11(1):187–189. doi: 10.1080/15548627.2014.996021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bordi M, et al. Autophagy flux in CA1 neurons of Alzheimer hippocampus: Increased induction overburdens failing lysosomes to propel neuritic dystrophy. Autophagy. 2016:1–17. doi: 10.1080/15548627.2016.1239003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Goetzl EJ, et al. Altered lysosomal proteins in neural-derived plasma exosomes in preclinical Alzheimer disease. Neurology. 2015;85(1):40–47. doi: 10.1212/WNL.0000000000001702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nixon RA, Yang DS, Lee JH. Neurodegenerative lysosomal disorders: a continuum from development to late age. Autophagy. 2008;4(5):590–599. doi: 10.4161/auto.6259. [DOI] [PubMed] [Google Scholar]
- 68.Boland B, et al. Autophagy induction and autophagosome clearance in neurons: relationship to autophagic pathology in Alzheimer’s disease. J Neurosci. 2008;28(27):6926–6937. doi: 10.1523/JNEUROSCI.0800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu D, et al. Nicotinamide forestalls pathology and cognitive decline in Alzheimer mice: evidence for improved neuronal bioenergetics and autophagy procession. Neurobiol Aging. 2013;34(6):1564–1580. doi: 10.1016/j.neurobiolaging.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee JH, et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141(7):1146–1158. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Coffey EE, et al. Lysosomal alkalization and dysfunction in human fibroblasts with the Alzheimer’s disease-linked presenilin 1 A246E mutation can be reversed with cAMP. Neuroscience. 2014;263:111–124. doi: 10.1016/j.neuroscience.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ye X, et al. Parkin-mediated mitophagy in mutant hAPP neurons and Alzheimer’s disease patient brains. Hum Mol Genet. 2015;24(10):2938–2951. doi: 10.1093/hmg/ddv056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151(2):367–380. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Labrousse AM, et al. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell. 1999;4(5):815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- 75.Duvezin-Caubet S, et al. OPA1 processing reconstituted in yeast depends on the subunit composition of the m-AAA protease in mitochondria. Mol Biol Cell. 2007;18(9):3582–3590. doi: 10.1091/mbc.E07-02-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Burte F, et al. Disturbed mitochondrial dynamics and neurodegenerative disorders. Nat Rev Neurol. 2015;11(1):11–24. doi: 10.1038/nrneurol.2014.228. [DOI] [PubMed] [Google Scholar]
- 77.DuBoff B, Feany M, Gotz J. Why size matters - balancing mitochondrial dynamics in Alzheimer’s disease. Trends Neurosci. 2013;36(6):325–335. doi: 10.1016/j.tins.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 78.Cho DH, et al. S-nitrosylation of Drp1 mediates beta-amyloid-related mitochondrial fission and neuronal injury. Science. 2009;324(5923):102–105. doi: 10.1126/science.1171091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang X, et al. Dynamin-like protein 1 reduction underlies mitochondrial morphology and distribution abnormalities in fibroblasts from sporadic Alzheimer’s disease patients. Am J Pathol. 2008;173(2):470–482. doi: 10.2353/ajpath.2008.071208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rice AC, et al. Mitochondrial DNA copy numbers in pyramidal neurons are decreased and mitochondrial biogenesis transcriptome signaling is disrupted in Alzheimer’s disease hippocampi. J Alzheimers Dis. 2014;40(2):319–330. doi: 10.3233/JAD-131715. [DOI] [PubMed] [Google Scholar]
- 81.Fang EF, et al. Nuclear DNA damage signalling to mitochondria in ageing. Nat Rev Mol Cell Biol. 2016 doi: 10.1038/nrm.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Julien C, et al. Sirtuin 1 reduction parallels the accumulation of tau in Alzheimer disease. J Neuropathol Exp Neurol. 2009;68(1):48–58. doi: 10.1097/NEN.0b013e3181922348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fang EF, et al. NAD+ replenishment improves lifespan and healthspan in Ataxia telangiectasia models via mitophagy and DNA repair. Cell Metabolism. 2016;24(4):566–581. doi: 10.1016/j.cmet.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fang EF, et al. Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell. 2014;157(4):882–896. doi: 10.1016/j.cell.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tseng AH, Shieh SS, Wang DL. SIRT3 deacetylates FOXO3 to protect mitochondria against oxidative damage. Free Radic Biol Med. 2013;63:222–234. doi: 10.1016/j.freeradbiomed.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 86.Yang W, et al. Mitochondrial Sirt3 Expression is Decreased in APP/PS1 Double Transgenic Mouse Model of Alzheimer’s Disease. Neurochem Res. 2015;40(8):1576–1582. doi: 10.1007/s11064-015-1630-1. [DOI] [PubMed] [Google Scholar]
- 87.Young-Collier KJ, McArdle M, Bennett JP. The dying of the light: mitochondrial failure in Alzheimer’s disease. J Alzheimers Dis. 2012;28(4):771–781. doi: 10.3233/JAD-2011-111487. [DOI] [PubMed] [Google Scholar]
- 88.Zhou M, et al. Neuronal death induced by misfolded prion protein is due to NAD+ depletion and can be relieved in vitro and in vivo by NAD+ replenishment. Brain. 2015;138(Pt 4):992–1008. doi: 10.1093/brain/awv002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Verdin E. NAD(+) in aging, metabolism, and neurodegeneration. Science. 2015;350(6265):1208–1213. doi: 10.1126/science.aac4854. [DOI] [PubMed] [Google Scholar]
- 90.Strosznajder JB, et al. Poly(ADP-ribose) polymerase-1 in amyloid beta toxicity and Alzheimer’s disease. Mol Neurobiol. 2012;46(1):78–84. doi: 10.1007/s12035-012-8258-9. [DOI] [PubMed] [Google Scholar]
- 91.Chow HM, Herrup K. Genomic integrity and the ageing brain. Nat Rev Neurosci. 2015;16(11):672–684. doi: 10.1038/nrn4020. [DOI] [PubMed] [Google Scholar]
- 92.Sykora P, et al. DNA polymerase beta deficiency leads to neurodegeneration and exacerbates Alzheimer disease phenotypes. Nucleic Acids Res. 2015;43(2):943–959. doi: 10.1093/nar/gku1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Martire S, et al. Bioenergetic Impairment in Animal and Cellular Models of Alzheimer’s Disease: PARP-1 Inhibition Rescues Metabolic Dysfunctions. J Alzheimers Dis. 2016;54(1):307–324. doi: 10.3233/JAD-151040. [DOI] [PubMed] [Google Scholar]
- 94.Blacher E, et al. Alzheimer’s disease pathology is attenuated in a CD38-deficient mouse model. Ann Neurol. 2015;78(1):88–103. doi: 10.1002/ana.24425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hou Y, et al. Genome instability in Alzheimer disease. Mech Ageing Dev. 2016 doi: 10.1016/j.mad.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Martire S, Mosca L, d’Erme M. PARP-1 involvement in neurodegeneration: A focus on Alzheimer’s and Parkinson’s diseases. Mech Ageing Dev. 2015;146–148:53–64. doi: 10.1016/j.mad.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 97.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 98.Moran N. Banking iPS cells. Nat Biotech. 2013;31(1):11–11. [Google Scholar]
- 99.Jucker M. The benefits and limitations of animal models for translational research in neurodegenerative diseases. Nat Med. 2010;16(11):1210–1214. doi: 10.1038/nm.2224. [DOI] [PubMed] [Google Scholar]
- 100.Handel AE, et al. Assessing similarity to primary tissue and cortical layer identity in induced pluripotent stem cell-derived cortical neurons through single-cell transcriptomics. Hum Mol Genet. 2016;25(5):989–1000. doi: 10.1093/hmg/ddv637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang Y, et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 2013;78(5):785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Miller JD, et al. Human iPSC-based modeling of late-onset disease via progerin-induced aging. Cell stem cell. 2013;13(6):691–705. doi: 10.1016/j.stem.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sang-Ging Ong WHL, Kazuki Kodo Haodi Wu, Joseph CWu. Human Induced Pluripotent Stem Cells Reveal Mitophagy as an Essential Process Against Diabetic Cardiomyopathy. Circulation Research. 2015:117. (Abstracts From the American Heart Association’s Basic Cardiovascular Sciences 2015 Scientific Sessions: Pathways to Cardiovascular Therapeutics -Suppl 1) [Google Scholar]
- 104.Mayer G, Nitsch R, Hoyer S. Effects of changes in peripheral and cerebral glucose metabolism on locomotor activity, learning and memory in adult male rats. Brain Res. 1990;532(1–2):95–100. doi: 10.1016/0006-8993(90)91747-5. [DOI] [PubMed] [Google Scholar]
- 105.Salkovic-Petrisic M, et al. What have we learned from the streptozotocin-induced animal model of sporadic Alzheimer’s disease, about the therapeutic strategies in Alzheimer’s research. J Neural Transm (Vienna) 2013;120(1):233–252. doi: 10.1007/s00702-012-0877-9. [DOI] [PubMed] [Google Scholar]
- 106.Salkovic-Petrisic M, et al. Cerebral amyloid angiopathy in streptozotocin rat model of sporadic Alzheimer’s disease: a long-term follow up study. J Neural Transm (Vienna) 2011;118(5):765–772. doi: 10.1007/s00702-011-0651-4. [DOI] [PubMed] [Google Scholar]
- 107.Stefanova NA, et al. Senescence-accelerated OXYS rats: a model of age-related cognitive decline with relevance to abnormalities in A lzheimer disease. Cell Cycle. 2014;13(6):898–909. doi: 10.4161/cc.28255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stefanova NA, et al. Amyloid accumulation is a late event in sporadic Alzheimer’s disease-like pathology in nontransgenic rats. Oncotarget. 2015;6(3):1396–1413. doi: 10.18632/oncotarget.2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cheng XR, Zhou WX, Zhang YX. The behavioral, pathological and therapeutic features of the senescence-accelerated mouse prone 8 strain as an Alzheimer’s disease animal model. Ageing Res Rev. 2014;13:13–37. doi: 10.1016/j.arr.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 110.D’Souza Y, et al. Characterization of Aldh2 (−/−) mice as an age-related model of cognitive impairment and Alzheimer’s disease. Mol Brain. 2015;8:27. doi: 10.1186/s13041-015-0117-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Longo VD, Mattson MP. Fasting: molecular mechanisms and clinical applications. Cell Metab. 2014;19(2):181–192. doi: 10.1016/j.cmet.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mattson MP. Lifelong brain health is a lifelong challenge: from evolutionary principles to empirical evidence. Ageing Res Rev. 2015;20:37–45. doi: 10.1016/j.arr.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Alirezaei M, et al. Short-term fasting induces profound neuronal autophagy. Autophagy. 2010;6(6):702–710. doi: 10.4161/auto.6.6.12376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fernandez-Moriano C, Gonzalez-Burgos E, Gomez-Serranillos MP. Mitochondria-Targeted Protective Compounds in Parkinson’s and Alzheimer’s Diseases. Oxid Med Cell Longev. 2015;2015:408927. doi: 10.1155/2015/408927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Turunc Bayrakdar E, et al. Nicotinamide treatment reduces the levels of oxidative stress, apoptosis, and PARP-1 activity in Abeta(1–42)-induced rat model of Alzheimer’s disease. Free Radic Res. 2014;48(2):146–158. doi: 10.3109/10715762.2013.857018. [DOI] [PubMed] [Google Scholar]
- 116.Gong B, et al. Nicotinamide riboside restores cognition through an upregulation of proliferator-activated receptor-gamma coactivator 1alpha regulated beta-secretase 1 degradation and mitochondrial gene expression in Alzheimer’s mouse models. Neurobiol Aging. 2013;34(6):1581–1588. doi: 10.1016/j.neurobiolaging.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Duan W, Mattson MP. Dietary restriction and 2-deoxyglucose administration improve behavioral outcome and reduce degeneration of dopaminergic neurons in models of Parkinson’s disease. J Neurosci Res. 1999;57(2):195–206. doi: 10.1002/(SICI)1097-4547(19990715)57:2<195::AID-JNR5>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 118.Yao J, et al. 2-Deoxy-D-glucose treatment induces ketogenesis, sustains mitochondrial function, and reduces pathology in female mouse model of Alzheimer’s disease. PLoS One. 2011;6(7):e21788. doi: 10.1371/journal.pone.0021788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ryu D, et al. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat Med. 2016 doi: 10.1038/nm.4132. [DOI] [PubMed] [Google Scholar]
- 120.Sun N, et al. Measuring In Vivo Mitophagy. Mol Cell. 2015;60(4):685–696. doi: 10.1016/j.molcel.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Morselli E, et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol. 2011;192(4):615–629. doi: 10.1083/jcb.201008167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130(1):165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 123.Nakatogawa H, et al. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10(7):458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 124.Kanki T, Furukawa K, Yamashita S. Mitophagy in yeast: Molecular mechanisms and physiological role. Biochim Biophys Acta. 2015;1853(10 Pt B):2756–2765. doi: 10.1016/j.bbamcr.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 125.Wager K, Russell C. Mitophagy and neurodegeneration: the zebrafish model system. Autophagy. 2013;9(11):1693–1709. doi: 10.4161/auto.25082. [DOI] [PubMed] [Google Scholar]
- 126.Shen Q, et al. Mutations in Fis1 disrupt orderly disposal of defective mitochondria. Mol Biol Cell. 2014;25(1):145–159. doi: 10.1091/mbc.E13-09-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ziviani E, Tao RN, Whitworth AJ. Drosophila parkin requires PINK1 for mitochondrial translocation and ubiquitinates mitofusin. Proc Natl Acad Sci U S A. 2010;107(11):5018–5023. doi: 10.1073/pnas.0913485107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang M, et al. Remodeling of Mitochondrial Flashes in Muscular Development and Dystrophy in Zebrafish. PLoS One. 2015;10(7):e0132567. doi: 10.1371/journal.pone.0132567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sharp WW, et al. Dynamin-related protein 1 (Drp1)-mediated diastolic dysfunction in myocardial ischemia-reperfusion injury: therapeutic benefits of Drp1 inhibition to reduce mitochondrial fission. FASEB J. 2014;28(1):316–326. doi: 10.1096/fj.12-226225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Narendra D, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183(5):795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang L, et al. TBK1-like transcript negatively regulates the production of IFN and IFN-stimulated genes through RLRs-MAVS-TBK1 pathway. Fish Shellfish Immunol. 2016;54:135–143. doi: 10.1016/j.fsi.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 132.Matsumoto G, et al. TBK1 controls autophagosomal engulfment of polyubiquitinated mitochondria through p62/SQSTM1 phosphorylation. Hum Mol Genet. 2015;24(15):4429–4442. doi: 10.1093/hmg/ddv179. [DOI] [PubMed] [Google Scholar]
- 133.Heo JM, et al. The PINK1-PARKIN Mitochondrial Ubiquitylation Pathway Drives a Program of OPTN/NDP52 Recruitment and TBK1 Activation to Promote Mitophagy. Mol Cell. 2015;60(1):7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Paulus JD, Link BA. Loss of optineurin in vivo results in elevated cell death and alters axonal trafficking dynamics. PLoS One. 2014;9(10):e109922. doi: 10.1371/journal.pone.0109922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wong YC, Holzbaur EL. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A. 2014;111(42):E4439–E4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tsuyuki S, et al. Detection of WIPI1 mRNA as an indicator of autophagosome formation. Autophagy. 2014;10(3):497–513. doi: 10.4161/auto.27419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nagy P, et al. Different effects of Atg2 and Atg18 mutations on Atg8a and Atg9 trafficking during starvation in Drosophila. FEBS Lett. 2014;588(3):408–413. doi: 10.1016/j.febslet.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wei H, Liu L, Chen Q. Selective removal of mitochondria via mitophagy: distinct pathways for different mitochondrial stresses. Biochim Biophys Acta. 2015;1853(10 Pt B):2784–2790. doi: 10.1016/j.bbamcr.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 139.Yasuda M, et al. Adenovirus E1B-19K/BCL-2 interacting protein BNIP3 contains a BH3 domain and a m itochondrial targeting sequence. J Biol Chem. 1998;273(20):12415–12421. doi: 10.1074/jbc.273.20.12415. [DOI] [PubMed] [Google Scholar]
- 140.Kundu M, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112(4):1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wu W, et al. ULK1 translocates to mitochondria and phosphorylates FUNDC1 to regulate mitophagy. EMBO Rep. 2014;15(5):566–575. doi: 10.1002/embr.201438501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Shravage BV, et al. Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Development. 2013;140(6):1321–1329. doi: 10.1242/dev.089490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Otsu K, Murakawa T, Yamaguchi O. BCL2L13 is a mammalian homolog of the yeast mitophagy receptor Atg32. Autophagy. 2015;11(10):1932–1933. doi: 10.1080/15548627.2015.1084459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Low P, et al. Impaired proteasomal degradation enhances autophagy via hypoxia signaling in Drosophila. BMC Cell Biol. 2013;14:29. doi: 10.1186/1471-2121-14-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fujita N, et al. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell. 2008;19(11):4651–4659. doi: 10.1091/mbc.E08-03-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mai S, et al. Autophagy proteins LC3B, ATG5 and ATG12 participate in quality control after mitochondrial damage and influence lifespan. Autophagy. 2012;8(1):47–62. doi: 10.4161/auto.8.1.18174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hanada T, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282(52):37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 148.Benato F, et al. Ambra1 knockdown in zebrafish leads to incomplete development due to severe defects in organogenesis. Autophagy. 2013;9(4):476–495. doi: 10.4161/auto.23278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bingol B, et al. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014;510(7505):370–375. doi: 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- 150.Zou J, et al. Autophagy inhibitor LRPPRC suppresses mitophagy through interaction with mitophagy initiator Parkin. PLoS One. 2014;9(4):e94903. doi: 10.1371/journal.pone.0094903. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.