Abstract
Triple-negative breast cancer (TNBC), which lacks the expression of the estrogen, progesterone, and HER2 receptors, represents the breast cancer subtype with the poorest outcome1. No targeted therapy is available against this subtype due to lack of validated molecular targets. We previously reported that MYC signaling is disproportionally elevated in triple-negative (TN) tumors compared to receptor-positive (RP) tumors2. MYC is an essential, pleiotropic transcription factor that regulates the expression of hundreds of genes3. Direct inhibition of oncogenic MYC transcriptional activity has remained challenging4,5. The present study conducted an shRNA screen against all kinases to uncover novel MYC-dependent synthetic lethal combinations, and identified PIM1, a non-essential kinase. Here we demonstrate that PIM1 expression was elevated in TN tumors and was associated with poor prognosis in patients with hormone and HER2 receptor-negative tumors. Small molecule PIM kinase inhibitors halted the growth of human TN tumors with elevated MYC expression in patient-derived tumor xenograft (PDX) and MYC-driven transgenic breast cancer models by inhibiting oncogenic transcriptional activity of MYC while simultaneously restoring the function of the endogenous cell cycle inhibitor, p27. Our findings warrant clinical evaluation of PIM kinase inhibitors in patients with TN tumors that exhibit elevated MYC expression.
While deregulated MYC signaling has been identified in a variety of human malignancies, no targeted therapy has been clinically established to treat solid tumors with elevated MYC expression. To date, small molecules designed to directly inhibit MYC transcriptional activity have not been successful in preclinical animal studies4,5. An alternative approach is to indirectly inhibit oncogenic MYC activity by targeting druggable proteins that are essential for the viability of MYC-driven tumors but not for non-tumorigenic cells. This indirect treatment strategy has become known as a synthetic lethal approach6. To identify novel targets that are readily druggable for treating MYC-overexpressing TNBC, we conducted a kinome-wide MYC synthetic lethal shRNA screen in non-immortalized human mammary epithelial cells7 expressing a 4-hydroxytamoxifen (TAM)-activated MycER transgene8 (HMEC-MycER) (Fig. 1a). MYC-dependent synthetic lethal screens using mammary epithelial cell systems have been previously reported9,10, however, to our knowledge, our screen is the first that utilized early-passage non-immortalized HMEC cells, obtained through breast reduction mammoplasty, which show no detectable genomic alterations11–13. This approach allowed us to study the response of mammary epithelial cells to oncogenic MYC activation in a highly specific manner, independent of additional factors that could be due to in vitro cellular immortalization. Of 600 human kinases targeted by 2,000 individual shRNA clones, we identified 9 kinases that were selectively required for the survival of HMEC-MycER cells (Fig. 1a and Supplementary Table 1). Kinase components of NF-kappaB, mitogen ERK/JNK, PI3K/AKT and WNT signaling were identified, most of which had not been identified in prior synthetic-lethal screens. While any of these kinases could potentially serve as a druggable target for the treatment of MYC-overexpressing breast cancer, among these hits we decided to pursue further studies of PIM1. Knock-down of PIM1 had the greatest efficacy in causing cell death in the MYC-activated cells and had minimal inhibitory effects on the growth of the control cells (Fig. 1a and Supplementary Table 1). The dependency of the MYC-activated HMECs on PIM1 for survival was confirmed by treatment with four pooled PIM1-specific siRNAs (Fig. 1b–d), resulting in marked cell death in a MYC-dependent manner.
Figure 1. Loss of PIM1 induces synthetic lethality with MYC activation in a model human mammary epithelial cell system.
(a) Schematic representation of the human kinome MYC synthetic lethal shRNA screen conducted in this study. HMECs expressing a 4-Hydroxytamoxifen (TAM) activatable MycER transgene were first infected with individual shRNA viruses in a 96 well format (i.e., one shRNA clone per well) and then treated with −/+ TAM to induce MYC activation. Only genes targeted by at least two independent shRNA clones that selectively induced cell death in the MYC activated (i.e., +TAM) HMECs were identified as MYC synthetic lethal genes. Positive/total refers to the number of shRNA clones that induced MYC-dependent cell death/total number of shRNA clones available in the human kinome shRNA library that target a given gene. Cell death differential calculated as % growth - % death from averaged values in Supplementary Table 1.
(b) Representative pictures of HMEC-MycER cells treated with control or PIM1 specific siRNA and −/+ TAM.
(c) Western blots showing the effect of PIM1 specific siRNA on PIM1 protein expression in the MYC-activated HMECs. Actin expression serves as a loading control.
(d) The effect of PIM1 siRNA on the viability of HMEC-MycER cells −/+ TAM as determined by flow cytometry using Guava ViaCount cell viability assay. The assay was independently repeated three times in triplicate. Error bars represent means +/− S.E.M. ***** indicates P < 0.00001 as determined by two-tailed t-test.
PIM1 belongs to the PIM family of serine/threonine kinases consisting of three proto-oncoproteins (PIM1, 2, and 3)14. PIM triple knock-out mice are fully viable and exhibit only minor growth defects that result in smaller body size15. This has raised enthusiasm that PIM-inhibitors may perhaps induce less systemic toxicity than inhibitors of essential kinases. PIM kinases are thought to possess weak tumorigenic capability on their own16. However, PIM1 significantly enhances MYC-induced tumorigenesis in transgenic mouse models of lymphoma and prostate cancer16–19. Accordingly, PIM kinase inhibition has been shown to decrease growth of prostate cancer cells engineered to overexpress MYC and PIM120,21. However, the efficacy of PIM inhibition as a MYC synthetic-lethal therapeutic has not been stringently validated in preclinical animal models, particularly in breast cancer. Interestingly, one of the other hits in our screen was ETK/BMX (Fig. 1a and Supplementary Table 1), a non-receptor tyrosine kinase that physically interacts with PIM122, suggesting that multiple components of the PIM1 regulatory pathway may play a critical role in MYC-dependent breast tumor growth.
PIM kinase expression and function has been poorly described in human breast cancer. Thus, we first determined receptor status-specific expression of PIM1 in primary tumor samples across four distinct publically available clinical cohorts. We found that PIM1 mRNA expression was significantly elevated in TN tumors in all of the cohorts examined when compared to hormone receptor-positive/HER2-negative tumors, and in all but one when compared to HER2-positive tumors (Fig. 2a). Interestingly, a recent report showed that PIM1 expression could be induced by the activation of estrogen signaling in an estrogen receptor (ER)-responsive breast cancer cell line MCF7, but that depleting ERα in the absence of estrogen in cell culture media also resulted in up-regulation of PIM123. Thus, while the precise mechanism by which ER signaling may modulate PIM1 expression remains to be elucidated, it may be that the process of primary tumor development in which tumors lose hormone receptors, such as ERα, may contribute to up-regulation of PIM1 expression observed in primary human TN tumors. Consistent with a prior observation that PIM1 expression was associated with higher grade breast tumors23, we found that in two of the three clinical cohorts that had long-term patient outcome data, increased PIM1 expression was associated with poor prognosis, as defined by diminished recurrence-free and distant metastasis-free survival, respectively, in patients with hormone receptor-negative tumors (Fig. 2b).
Figure 2. PIM1 expression is disproportionally elevated in human primary TN tumor samples and is associated with poor clinical outcomes in patients with hormone receptor-negative breast cancer.
(a) PIM1 mRNA expression in primary breast tumor samples from TCGA, pooled node-negative chemotherapy naïve, pooled neoadjuvant chemotherapy (taxane-anthracycline) treated, and I-SPY1 cohorts, respectively, stratified by receptor status. Values are log2-transformed and median-centered. Bars representing patient groups are given a number that indicates sample size. Error bars represent means +/− S.E.M. ***** indicates P < 0.00001 as determined by pairwise two-tailed t-tests between the respective groups. N.S. = not significant.
(b) Kaplan-Meier graphs of the patients with hormone receptor-negative tumors, dichotomized by PIM1 mRNA expression at an optimal threshold, from node-negative chemotherapy-naïve (top), pooled neoadjuvant chemotherapy treated (middle), and I-SPY (bottom) cohorts. Samples with elevated PIM1 expression are represented with red lines.
(c) Multivariate analysis indicating the effect of PIM1 mRNA expression and MYC gene expression signature on survival in pooled node-negative chemotherapy-naïve, pooled neoadjuvant chemotherapy (taxane-anthracycline) treated, and I-SPY1 cohorts, specifically in hormone receptor-positive and receptor-negative subsets. PR status was not available for the pooled node-negative chemotherapy-naïve dataset, and as such ER−/ER+ subsets were considered instead. Hazard ratio is associated with a 1 standard deviation in PIM1 expression or change in the MYC centroid signature. P values are based on the likelihood ratio test.
We also examined receptor-status specific expression and prognostic significance of the other PIM kinase family members, PIM2 and PIM3. We found a tendency for PIM2 to be elevated in TN tumors, however, the extent of elevation was not as consistent as that found for PIM1 (Supplementary Fig. 1a), and neither was the prognostic significance of PIM2 (Supplementary Fig. 1b,c). We were unable to obtain consistent results for PIM3 primarily due to limited data availability, however, our analysis did not associate PIM3 expression with any specific receptor status and it did not reveal prognostic significance for PIM3 expression (Supplementary Fig. 2).
We next sought to determine if a MYC gene signature and PIM1 expression are independently associated with survival across several clinical cohorts using multivariate Cox proportional hazard modeling (Fig. 2c). In the hormone receptor-negative subset of the two largest clinical cohorts, we found increased PIM1 expression is associated with significantly higher risk of recurrence, independent of MYC signature. In the I-SPY1 dataset, the smallest cohort, the MYC signature was significantly associated with worse outcome2 independent of PIM1 expression (Fig. 2c). Thus, although both MYC and PIM are overexpressed in TNBCs (Fig 2a and Ref2), their association with patient outcome remains independent. This indicates that the poor outcome associated with PIM1 across several datasets is independent of MYC gene signature (Fig 2c).
To assess the feasibility of PIM1 inhibition as a therapy for TN tumors with elevated MYC expression, we treated a panel of breast cancer cell lines with PIM1 siRNA and studied its effect on cell proliferation and cell death (Fig. 3a–f). Our panel included a non-tumor cell line (HMEC) and 10 breast cancer cell lines with varying degrees of MYC and PIM1 expression (Fig. 3a). We found that PIM1 siRNA treatment significantly decreased the proliferation of a number of breast cancer cell lines (Fig. 3b,c), and it also induced appreciable amounts of cell death (Fig. 3e). We asked whether there is a correlation between the sensitivity of cell lines to PIM1 inhibition and protein expression levels of MYC and PIM1 in these lines. Our correlation studies based on the relative sensitivity of the cell lines (t ratio) revealed that the protein expression of MYC, and that of PIM1 to a lesser extent, were significantly correlated with the sensitivity of cell lines to PIM1 inhibition (Fig. 3d,f). We also observed correlation between MYC mRNA expression and sensitivity to PIM inhibitors in the panel of cell lines used in our study (Supplementary Fig. 3), though this was less significant than for MYC protein expression.
Figure 3. TNBC cells with elevated MYC expression are sensitive to PIM kinase inhibition in vitro and in vivo.
(a) Protein expression levels of PIM1 and MYC in a panel of breast cancer cell lines used in this study. Actin serves as a loading control.
(b) Protein expression levels of PIM in a panel of breast cancer cell lines treated with a pool of PIM1 scrambled siRNA or non-specific control siRNA for 60hrs. Actin serves as a loading control.
(c–f) The effects of knocking down PIM1 on the (c) proliferation as assessed by cell count, (e) induction of apoptosis as assessed by Annexin V/7-AAD staining, and the (d and f) correlation between protein expression levels of PIM1 and MYC, respectively, in a panel of breast cancer cell lines and their sensitivity to PIM1 inhibition as determined by Pearson correlation. The experiment in (c and e) was independently repeated three times in triplicate. Error bars represent means +/− S.E.M. P values were calculated by two-tailed t-test comparing the control siRNA treated group to each of the experimental groups. *P < 0.05, **P < 0.01, ***P < 0.001. N.S. = not significant.
(g) The effects of a small molecule pan-PIM kinase inhibitor SGI-1776 on the in vivo growth of xenografted MDAMB 231 and T47D tumors in nude mice. The tumor bearing mice were treated with SGI-1776 at 75mg/kg or its control diluent via oral gavage, daily, for two weeks. For MDAMB231, N=6 in each treatment group. For T47D, N=5 in each treatment group. Error bars represent means +/− S.E.M. P values were calculated by two-tailed t-test comparing the control treated group to the SGI-treated group.
(h) Protein expression levels of PIM1 and MYC in a panel of previously reported patient derived orthotopic breast tumor xenografts. Breast organoids originally collected through breast reduction mammoplasty were used to assess the expression levels of PIM1 and MYC in non-tumorigenic breast tissue. Numbers shown represent relative protein expression of MYC in these PDX tumors.
(i) PDX tumor HCI-002 ~ 2 mm chunks were orthotopically transplanted into the cleared mammary fat pads of female NOD/SCID mice and allowed to reach ~450mm3 in volume. The tumor bearing mice were then treated with either SGI-1776 at 75mg/kg or NVP-LGB321 at 100mg/kg, or with respective control diluent, via oral gavage, daily, for two weeks. Shown are the representative pictures of PDX tumors before (day 0) and at the end of the drug treatment experiment.
(j–k) The effects of PIM kinase inhibitors (j) SGI-1776 and (k) NVP-LGB321 on ethical endpoint (top) and in vivo growth (bottom). Fraction survival indicates the number of animals removed from the experiments when the tumor sizes reached the institutional limit of one dimension reaching or exceeding 2cm. Error bars represent means +/− S.E.M. P values were calculated by two-tailed t-test unless otherwise indicated.
(l) The effects of PIM kinase inhibitors on in vivo cell proliferation of PDX tumors as assessed by Ki67 staining. Shown are the representative pictures of PDX tumors collected 24 hours after the final drug treatment. Error bars represent means +/− S.E.M. P values were calculated by two-tailed t-test.
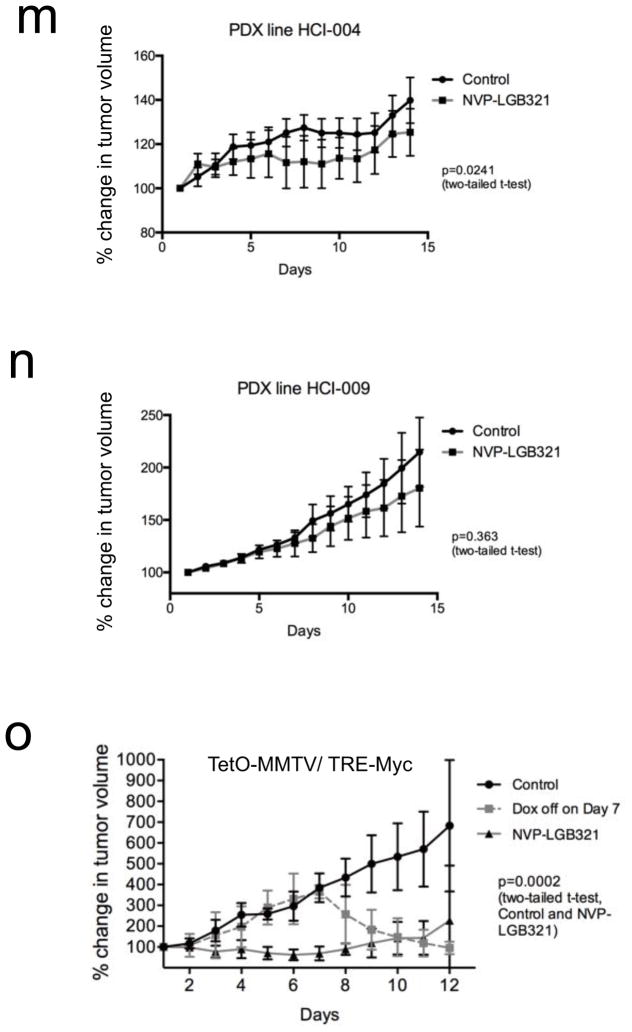
(m–n) Relative tumor volume of orthotopic (m) HCI-004 and (n) HCI-009 tumor xenografts in NOD/SCID mice that were treated with vehicle (N=5) or with NVP-LGB321 (N=3) at 100 mg/kg, daily for 2 weeks. Error bars represent means +/− S.E.M. P values were calculated by two-tailed t-test comparing the control treated group to the NVP-LGB321-treated group.
(o) Relative tumor volume of orthotopic TetO-MMTV/TRE-Myc allografts in FVB/N mice that were treated with vehicle (N=5) or with NVP-LGB321 (N=5) at 100 mg/kg, daily for 12 d, or were taken off doxycycline (N=5) for six days starting at day seven. Error bars represent means +/− S.E.M. P values were calculated by two-tailed t-test comparing the control treated group to the NVP-LGB321-treated group.
During these siRNA experiments, we noticed that the knock-down effect of PIM1 siRNA was rather short lived in some of the cell lines (Fig. 3b and Supplementary Fig. 4a). This is consistent with prior observations that the half-life of PIM1 mRNA and protein are short and that PIM kinases could be highly regulated at the levels of transcription and post-translational modifications to maintain elevated protein expression in tumors14. We also asked whether knocking-down PIM1 could influence expression of other PIM kinases, namely PIM2, whose expression was found elevated in TN tumors in some of the clinical cohorts (Supplementary Fig. 1a). We found that PIM1 siRNA treatment of MDAMB231 cells, which exhibited higher expression levels of both MYC and PIM1, resulted in acute and significant elevation in PIM2 protein expression, whereas PIM2 siRNA did not appear to activate a similar compensatory response with respect to PIM1 expression (Supplementary Fig. 4a). In MDAMB231 cells, we found that PIM2 siRNA was as effective as PIM1 siRNA in inhibiting cell proliferation and was more effective in inducing cell death (Supplementary Fig. 4b,c). These observations suggest that simultaneous inhibition of multiple PIM kinases may prove particularly efficacious in treating breast tumors with elevated MYC and PIM1 expression.
We next evaluated the effect of PIM kinase inhibition on the in vivo growth of breast cancer cell lines xenografted subcutaneously in immunocompromised mice, using a pan-PIM kinase inhibitor SGI-1776 24,25. The TN line MDAMB231 (high MYC/high PIM1) and the RP line T47D (low MYC/medium PIM1), which showed differential sensitivity to PIM1 siRNA in cell culture, were grown until tumors reached approximately 200 mm3 in size. Mice were subsequently treated with 75 mg/kg SGI-1776 or vehicle control daily via oral gavage for 14 days. We found that SGI-1776 significantly abrogated MDAMB231 xenograft tumor growth, but it did not have a significant effect on the growth of T47D xenograft tumors (Fig. 3g).
To conduct a more stringent preclinical assessment of the efficacy of PIM kinase inhibition in treating MYC-driven TN tumors, we took advantage of recently developed patient-derived xenograft (PDX) mouse models of breast cancer26, which exhibit varying expression levels of MYC and PIM1 (Fig. 3h). The HCI-002 PDX, which was derived from an original primary tumor collected prior to patient treatment with systemic chemotherapy, exhibits a poorly differentiated TN phenotype (Supplementary Table 2)26 and has the highest MYC expression of the three PDX models tested in this study (Fig. 3h). PDX tumors were orthotopically transplanted into the mammary gland of immunocompromised mice and grown to 400–450 mm3 before treatment was initiated with 75 mg/kg SGI-1776, 100 mg/kg of another structurally distinct, newer generation pan-PIM kinase inhibitor NVP-LGB32127,28 (Novartis) or vehicle control daily via oral gavage for 14 days. NVP-LGB321 is an experimental small molecule, and PIM44729 (Novartis), a clinical compound structurally similar to NVP-LGB321, is currently being evaluated in multiple Phase I-II trials against hematopoietic cancers. Vehicle-treated HCI-002 tumors rapidly reached the ethical end point of this study (Fig. 3i–k). In contrast, SGI-1776 markedly abrogated tumor growth, while NVP-LGB321 induced partial tumor regression (Fig. 3j,k). PIM inhibitor-treated tumors showed a significant decrease in the number of the Ki67-positive, proliferating cells (Fig. 3l), while TUNEL staining indicated that there was a modest increase in the number of apoptotic cells at the time of tumor sample collection (Supplementary Fig. 5). To confirm that PIM inhibitor sensitivity in vivo was dependent upon elevated MYC expression, we used two additional PDX tumor lines, HCI-004 and HCI-009. PDX tumors were orthotopically transplanted into the mammary gland of immunocompromised mice and grown to 400–450 mm3 before treatment was initiated with 100 mg/kg NVP-LGB321 or vehicle control daily via oral gavage for 14 days. NVP-LGB321 treatment of the HCI-004 tumors, which showed an intermediate level of elevated MYC expression (Fig. 3h), significantly attenuated tumor growth, although tumor growth was not completely inhibited (Fig. 3m). We found that the significant difference between the control- and drug-treated HCI-004 tumors observed during the course of treatment was maintained even after treatment was discontinued (Supplementary Fig. 6). In contrast, the HCI-009 line, which has low MYC expression similar to that found in non-tumor mammary tissue (Fig. 3h), did not show significant sensitivity to NVP-LGB321 during the course of treatment (Fig. 3n).
To further confirm our observations that TN tumors with elevated MYC expression are sensitive to PIM kinase inhibition, we examined the effects of PIM inhibition in a conditional transgenic mouse model of MYC-driven breast cancer (TetO-MMTV/TRE-Myc)30. In this model, tumor growth is dependent upon doxycycline-induced MYC expression and removal of doxycycline results in tumor regression (Fig. 3o). TetO-MMTV/TRE-Myc tumors were orthotopically transplanted into the mammary gland of isogenic mice and were grown to 400–500 mm3 before treatment was initiated with 100 mg/kg NVP-LGB321 or vehicle control daily via oral gavage for 12 days. NVP-LGB321 treatment nearly completely abolished the ability of MMTV-Myc allograft tumors to grow (Fig. 3o) as compared to control treated tumor-bearing mice. These data provide genetic evidence that MYC-driven breast tumors require PIM kinase activity to maintain tumorigenesis.
To understand the mechanisms by which PIM inhibition abrogated in vivo MYC-overexpressing tumor growth, we examined whether PIM kinase targets are affected in drug-treated HCI-002 tumors (Fig. 4a and Supplementary Fig. 7). PIM kinases are known for their role in phosphorylating (1) MYC on S62, which increases MYC transcriptional and oncogenic potential31–33, (2) the endogenous cyclin-dependent kinase (CDK) inhibitors p21 and p27 to negatively control their activities34,35, (3) the CDK phosphatases Cdc25A and Cdc25C, resulting in CDK activation36,37, (4) a pro-apoptotic BCL2 family member BAD to inhibit its function38,39, and (5) a regulator of cap-dependent protein synthesis 4EBP1 to increase protein synthesis40,41. Among these PIM substrates, we found that protein expression and/or phosphorylation status of MYC, p27, and BAD were most consistently and markedly altered in the drug-treated PDX tumors (Fig. 4a and Supplementary Fig. 7). Compared with the control-treated tumors, those treated with either PIM inhibitor showed marked reduction in total MYC expression, and significant reduction in the amount of S62 phosphorylated MYC was also observed for the tumors treated with NVP-LBG321 (Fig. 4b). Two weeks of drug treatment did not consistently alter the amount of T58 phosphorylation (red arrow, Fig. 4a), which negatively affects MYC transcriptional activity32,42. Intriguingly, some of the vehicle-treated tumors (i.e., 2 week control tumors) showed a marked decrease in T58 hyper-phosphorylation during primary tumor progression (Fig. 4a). Thus, these observations suggest that PIM inhibition resulted in an overall decrease in MYC activity while the control tumors exhibited increased MYC activity during continued growth. Consistent with this hypothesis, we found that the expression of a bona-fide MYC transcriptional miRNA target, miR-1843, was uniformly low in the drug-treated tumors compared to vehicle-treated tumors (Supplementary Fig. 8). A similar trend was observed for other MYC-activated miRNAs, miR-19b and miR-20a (Supplementary Fig. 8). Our results are consistent with the observation that the expression of a number of MYC signature genes was altered following PIM kinase inhibitor treatment in sensitive TNBC cell lines (A. Tutt and colleagues, personal communication, co-submitted manuscript).
Figure 4. The mechanisms of PIM inhibition mediated TN tumor growth abrogation involve simultaneous loss of MYC function and gain of p27 function.
(a) Protein expression levels and phospho-status of indicated PIM1 substrates in PDX tumors harvested before (Day 0) and after 2 weeks treatment with either SGI-1776 (75mg/kg, daily) or NVP-LGB321 (100mg/kg, daily), or with respective control diluents. Tumors were harvested 24 hours after the final drug treatment. Three tumor samples from three independent mice are represented for each time point and treatment condition. The Day 0 samples shown are identical across the two columns. Red arrow indicates the T58 hyper-phosphorylated form of MYC. Actin serves as a loading control.
(b) The effects of small molecule PIM inhibitors on the total protein expression of MYC (top) and on the amount of phosphorylated S62 (bottom). Total protein expression is based on the ratio between MYC signal and that of Actin. The bottom graphs show ratio of phospho-S62 to total MYC expression. Three tumor samples from three independent mice are represented in each treatment group. Error bars represent means +/− S.E.M. P values were calculated by two-tailed t-test.
(c) The effects of small molecule PIM inhibitors on the total protein expression of p27 (top) and on its regulatory phosphorylation sites (bottom). The bottom graphs show ratio of indicated p27 phospho-residue to total p27 band intensity. Three tumor samples from three independent mice are represented in each treatment group. Error bars represent means +/− S.E.M. P values were calculated by two-tailed t-test.
(d) The effects of small molecule PIM inhibitors on p27 nuclear accumulation in PDX tumors. Representative confocal images of control or PIM inhibitor treated PDX tumors stained for DAPI (blue) and p27 (green) (top), and quantification of the effects of SGI-1776 and NVP-LGB321 on p27 nuclear accumulation in PDX tumors (bottom). Scale bar represents 100μm. The percentage of Ki67 and p27 positive cells per high-power field (40x) is shown. Five high-power fields were taken from each of the three independent tumor samples per treatment group. Error bars represent means +/− S.E.M. P values were calculated by two-tailed t-test.
(e) p27 protein expression in a panel of breast cancer cell lines used in this study. Actin serves as a loading control.
(f) Time-dependent effects of SGI-1776 and NVP-LGB321 on protein expression of MYC and p27 in a panel of breast cancer cell lines. The cells were treated with the inhibitors at 10μM, unless otherwise noted, for the indicated amount of time. *HCC1143 cells were treated with SGI-1776 at 5μM for the indicated amount of time due to excessive amount of cell death observed with SGI-1776 at 10μM. Actin serves as a loading control.
(g) The effects of NVP-LGB321 at 10μM on p27 nuclear accumulation in a panel of TNBC cell lines. The cells were treated with the inhibitor for 48 hrs. Representative immunofluorescence images of DMSO or NVP-LGB321 treated cells stained for DAPI (blue) and p27 (green), and quantification of p27 nuclear accumulation (bottom) are shown. Scale bar represents 100μm. The number of nuclear p27-positive cells was counted in three high-power fields (20x) per sample/slide. N=6 for each experimental condition/cell line. Error bars represent means +/− S.E.M. P values were calculated by two-tailed t-test.
(h) The effects of combined MYC knock down and induced p27 up-regulation on the proliferation of a panel of TNBC cell lines. The TN lines containing a tet-regulatable p27 transgene were first transfected with either non-targeting control (CTL) or MYC siRNA. 24 hrs post siRNA transfection, the cells were treated with either doxycycline (Dox) or control diluent for 48 hrs. Error bars represent means +/− S.E.M. P values were calculated by Tukey’s multiple comparison test. Actin serves as a loading control.
(i) Schematic representation of the proposed mechanisms of PIM inhibitor action in MYC-driven TN tumors.
Consistent with a decrease in tumor cell proliferation, as measured by Ki67 staining (Fig. 3l), the drug treated PDX tumors showed significantly increased total p27 expression (Fig. 4a,c,d), whereas protein expression levels of CDKs, CDC25A/B and p21 were not altered (Supplementary Fig. 7). Regulation of p27 involves multiple steps of phosphorylation. Phosphorylation of p27 on S10 is required for its binding to the interphase CDKs bound to their respective cyclin partners, and results in inhibition of CDK activity. T187 phosphorylation, mediated by CDKs, is required for proteasomal degradation of p27. T198 is phosphorylated by AKT and also by PIM, and triggers p27 binding to 14-3-3, resulting in nuclear export of p27. Thus, while S10 phosphorylation of p27 is a positive regulatory modification, T187 and T198 phosphorylation are both negative regulatory modifications. In addition, S10 phosphorylation is a known prerequisite for subsequent phosphorylation on T187 and T19835,44–46. We found that levels of S10 phosphorylation mirrored that of total p27 (Fig 4a,c), indicating that the PIM inhibition did not interfere with S10 phosphorylation. In contrast, the patterns of T187 and T198 phosphorylation did not follow that of total p27 (Fig. 4a,c). The amount of T187- or T198-phosphorylated p27 found in the drug treated tumors was similar to that observed in the vehicle treated tumors and in tumors prior to drug treatment (i.e., Day 0 tumors) (Fig. 4a,c). These observations suggest that there could be a large pool of CDK-bound, T187/198 unphosphorylated p27 molecules trapped in the nucleus, interfering with the CDK-p27 recycling mechanism essential for cell proliferation. Indeed, co-immunostaining of the PDX tumors for DAPI and p27 revealed significant nuclear accumulation of p27 in the drug treated tumors (Fig. 4d).
The pro-apoptotic BCL2-family member BAD becomes activated upon dephosphorylation at conserved sites including S112, which in turn results in activation of BAX and BAK, leading to the induction of mitochondrial-dependent apoptosis38,39,47. We found that in vivo treatment of tumors with either PIM inhibitor resulted in decreased S112 phosphorylation, with NVP-LGB321 having a greater effect (Fig. 4a). This may, in part, account for the partial tumor regression and the increase in the number of apoptotic cells observed in the HCI-002 tumors treated with NVP-LGB231 (Fig. 3k and Supplementary Fig. 5).
We next asked whether anti-tumorigenic molecular effects, observed with PIM inhibitors in the PDX tumors, are also seen in treated tumor cell lines. To address this, we treated a panel of breast cancer cell lines, which exhibit differential sensitivity to PIM inhibitors (Supplementary Fig. 9), with different endogenous protein expression levels of PIM1 (Fig. 3a), MYC (Fig. 3a), and p27 (Fig. 4e) with the inhibitors and examined their effects on MYC and p27 expression. In these cell lines, we found that p27 protein expression was significantly lower in TN cell lines (Fig. 4e). Lower p27 mRNA was also observed in TNBC compared to hormone receptor positive tumors from clinical datasets (Supplementary Fig. 10). We found that PIM inhibitors reduced MYC protein expression in the panel of cell lines examined (Fig. 4f) and that this effect appeared to be independent of their receptor status or their dependence on MYC for cell proliferation (Supplementary Fig. 11a–c). PIM inhibition, however, did not significantly reduce MYC mRNA expression (Supplementary Fig. 12), suggesting that the reduction in MYC protein expression induced by PIM inhibition likely occurred via post-transcriptional mechanisms. In contrast, PIM inhibitors up-regulated p27 expression only in the TN cell lines which are dependent on MYC for cell proliferation (Supplementary Fig. 11a–c), but not in the RP cell lines tested (Fig. 4g). These molecular changes were reproducible with PIM1 specific siRNA (Supplementary Fig. 11a and Supplementary Fig. 13). Intriguingly, we found that the sensitivity to PIM1 inhibition of our panel of breast cancer cell lines was more strongly correlated with protein expression levels of MYC and PIM1 (positive correlation) than that of baseline p27 expression (negative correlation) (Fig. 3c–f and Supplementary Fig. 14). Finally, TN cell lines sensitive to PIM inhibitors induced nuclear p27 accumulation (Fig. 4g) as was also seen in vivo in the PIM inhibitor treated PDX tumors (Fig. 4d).
To understand whether elevated p27 expression alone is sufficient to inhibit TN cell proliferation, we studied the sensitivity of TN lines to increased p27 expression. We conditionally elevated p27 expression either in the presence or absence of MYC siRNA (Fig. 4h) using a doxycyclin-regulated approach. In doing so, efforts were made to achieve the extent of p27 up-regulation similar to what was observed in the PIM inhibitor treated PDX tumors in vivo (Fig. 4a). We found that increased p27 expression alone significantly diminished cell proliferation in all the lines tested (Fig. 4h). Inducing p27 overexpression together with MYC siRNA treatment resulted in further growth inhibition (Fig. 4h). This is consistent with diminished proliferation observed in PIM inhibitor treated PDX tumors (Fig. 3i–k). Thus, the near complete growth inhibition seen in the PIM inhibitor-treated PDX tumors is likely due to the cumulative anti-proliferative effects mediated by the inhibitor, which result in both diminished MYC activity, as well as elevated p27 expression (Fig. 4i).
Our observation that PIM inhibition resulted in diminished MYC activity in TN tumors, combined with the previous observation that PIM kinases are non-essential in mice15, makes PIM an attractive therapeutic target for breast cancer. Additionally, we found that PIM inhibition resulted in increased nuclear p27 of treated TN cells and PDX tumors. Previous observations in numerous breast cancer clinical cohorts have associated diminished p27 expression with poor prognosis48. The breast tumor cohort studies by the TCGA found that p27 was neither deleted nor mutated in the vast majority of TN tumor samples49, supporting the utility of PIM inhibition in restoring p27 expression and function in these aggressive tumors. In conclusion, our work identifies PIM1 kinase as a factor specifically upregulated in TNBC and also as a novel druggable target for patients with tumors that exhibit elevated MYC expression.
METHODS
Human mammary epithelial cells expressing MycER transgene
Primary human mammary epithelial cells (HMECs) were derived from histologically normal breast tissues and cultured as previously described7. The cells were infected with lentivirus encoding p16 shRNA53 and further infected with pBabe-MycER virus8. The resulting cells were named B1389-shp16-MycER (HMEC-MycER hereafter). While expressing p16 shRNA delays the onset of senescence in HMEC-MycER cells, the cells are not immortalized and undergo spontaneous senescence as the cells are continuously cultured13. The HMEC-MycER cells were used for the screen within 12 passages of derivation to minimize the accumulation of spontaneous genomic alterations unrelated to MYC activation. HMEC-MycER cells were treated with 4-Hydroxytamoxifen (TAM) at 500nM to induce MYC activation throughout this study.
Kinome MYC synthetic lethal shRNA screen
The screen in this study was performed using the Open Biosystems Human GIPZ Lentiviral Human Kinase Library v1 (RHS4808). In this library, approx. 600 kinases are targeted by 2,000 independent shRNA clones with each kinase being targeted by ~ 7 independent clones. The lentiviral supernatants were produced by the UCSF VIRACORE (viracore.ucsf.edu) in 96-well format by transfecting 293T cells with individual shRNA clones together with packaging plasmids VsVg and Delta 8.9. Approx. 90μl of unconcentrated viral supernatant was obtained form each well. HMEC-MycER cells were seeded onto 96-wells (2,000 cells/well), incubated with 40μl of viral supernatant in the presence of hexadimethrine bromide (Polybrene, Sigma) at 7μg/ml. The cells were incubated with the viruses for 20–21hrs, at which time the virus-containing medium was replaced by fresh medium −/+ TAM. After 48 hrs of incubation with −/+ TAM, the cells were subjected to CellTiter96 Aqueous One Solution Cell Proliferation assay (Promega) to determine −/+ cell proliferation from the time of MYC activation. % cell death was calculated by subtracting the values obtained prior to MYC activation from the final values, and % decrease in proliferation was determined by subtracting the final values in the +TAM plates from those in the –TAM plates. Those candidate synthetic lethal genes targeted by the shRNA clones that induced cell death in HMECs irrespective of the MYC activation status were not further pursued.
Bioinformatics analyses
Four different clinical datasets were used for bioinformatics analyses. The TCGA breast invasive carcinoma dataset was sourced from data generated by the TCGA Research Network: http://cancergenome.nih.edu/, made available on the University of California, Santa Cruz (UCSC) Cancer Browser. The chemotherapy-naïve dataset54 was obtained from the UCSC Cancer Browser. Series matrix files for I-SPY1 (Accession: GSE22226)55 and the pooled neoadjuvant chemotherapy-treated cohort (Accession: GSE25066)56 were downloaded from Gene Expression Omnibus (GEO) and extracted into a usable format with the GEOquery R package57. Multiple probes corresponding to the same gene were collapsed using the “MaxMean” method in the Weighted Gene Correlation Network Analysis (WGCNA) R package58,59.
To generate barplots, each of the datasets was stratified into three groups based on receptor status: samples that were positive for the HER2/ERBB2 receptor tyrosine kinase (HER2+), those that were positive for the estrogen and/or progesterone receptors (HR+), and those that were triple-negative (TN). Log-transformed and median-centered PIM1 expression values were derived for the respective groups and visualized using the ggplot2 R package60.
To generate Kaplan Meier plots, samples that were negative for both the estrogen and progesterone receptors (HR-) were isolated from each of the datasets, and dichotomized by PIM1 expression at an optimal threshold, yielding groups with the most significant difference in disease recurrence-free or distant metastasis-free survival based on the log-rank test. Kaplan Meier plots were then generated for the respective groups using the survival R package61.
Multivariate cox proportional hazards regression analysis was performed using the survival R package61 to assess the correlation of PIM1 expression and MYC gene signature expression2 as continuous variables to disease recurrence-free survival in the I-SPY1 and pooled neoadjuvant chemotherapy-treated cohorts, and to distance metastasis-free survival in the chemotherapy-naïve cohort respectively. To compute the MYC signature score, expression data was mapped onto the MYC gene signature score centroid62 by gene symbol; and the Pearson correlation of each tumor’s expression profile to the signature centroid was determined. This survival analysis was repeated specifically for hormone receptor-positive and receptor-negative subsets of these datasets as well. PR status was not available for the chemotherapy-naïve dataset, and as such ER−/ER+ subsets were considered instead.
Breast cancer cell lines
A panel of established human breast cancer cell lines and their culture conditions have been previously described63. A PCR-based method was routinely used to ensure that all the cells used in this study were mycoplasma free. The primers initially used are: forward-ACT CCT ACG GGA GGC AGC AGT A, reverse-TGC ACC ATC TGT CAC TCT GTT AAC CTC. Additionally, the Universal Mycoplasma Detection Kit (ATCC, 30-1012K) was used to ensure cells did not have mycoplasma infection.
Animal experiments
All protocols described in this and other sections regarding animal studies were approved by the UCSF Institutional Animal Care and Use Committee; ethical endpoint for mammary tumor transplantation experiments was reached when a tumor reached = 2.5 cm in any single dimension. To subcutaneously grow MDAMB231 and T47D tumors, respectively, the cells (3 × 106 for MDAMB231, and 1 × 107 for T47D cells) were subcutaneously injected into immuno-deficient female mice (BALB/c nude/nude) aged 6~8 weeks. The tumors were allowed to grow to 150~200mm3 in volume, at which time the drug treatment was initiated. The animals were treated with an experimental pan-PIM kinase small molecule inhibitor SGI-1776 (purchased from Selleckchem, S2198), dissolved in 5% dextrose/water solution, at 75mg/kg, or vehicle alone, daily, via oral gavage. SGI-1776 at 75mg/kg was previously shown to be effective in regressing the xenografted acute myeloid leukemia cells with higher PIM expression in mice64.
The PDX tumors were grown as previously described26. For the PIM inhibitor treatment, the PDX tumor chunks were transplanted into cleared mammary fad pads of the NOD-SCID immune-deficient female mice aged 4~4.5 weeks, and the grafted tumors were allowed to reach ~ 450mm3 in volume, at which time the drug treatment was initiated. The animals were treated with either SGI-1776 at 75mg/kg or a newer generation PIM kinase inhibitor NVP-LGB32127,28, which is under clinical development by and was provided by Novartis, or respective vehicle alone. The NVP-LGB321 solution was prepared in 50mM sodium acetate buffer, pH4.6, and was administered at 100mg/kg, daily, via oral gavage. For the HCI-004 study, some of the animals were monitored for additional 36 days after 2 weeks of drug treatment was completed.
TetO-MMTV/TRE-Myc mice were generated as previously described 30. Mice were bred and maintained off of doxycycline. At 12–15 weeks of age, female mice were put on doxycycline (200 mg/kg doxy chow, Bio-Serv) to induce MYC expression and tumorigenesis. Mice were sacrificed as per ethical guidelines (tumors reaching 2 cm in any single dimension) and harvested tumors were flash-frozen in liquid nitrogen. To perform orthotopic allograft studies, the flash-frozen MMTV-MYC tumors were thawed, cut into smaller chunks, and transplanted into cleared mammary fat pads of 4-week-old wild-type female FVB/N mice. The tumor-bearing FVB/N mice were treated with NVP-LGB321 as described for the PDX tumor-bearing NOD-SCID mice.
Western analyses
Tumor as well as cell lysates were prepared as previously described2. The primary antibodies used in this study are as follows: βActin (Clone AC-15, Sigma A1978), BAD (Clone Y208, abcam, ab32445), BADpS112 (Clone EPR1891(2), abcam, ab129192), Cdc25A (Sigma, HPA005855), Cdc25C (Clone 5H9, Cell Signaling, 4688), CDK1 (Clone 17, Santa Cruz Biotech., sc-54), CDK2 (Clone D-12, Santa Cruz Biotech., sc-6248), CDK4 (Clone C-22, Santa Cruz Biotech., sc-260), 4EBP1 (Clone 53H11, Cell Signaling, 9644), 4EBP1pT37/46 (Clone 236B4, Cell Signaling, 2855), 4EBP1pS65 (Cell Signaling, 9451), c-MYC (Clone Y69, Epitomics, 1472-1), c-MYCpT58 (Applied Biological Materials, Y011034), c-MYCpS62 (Clone 33A12E10, abcam, ab78318), p21 (BD Pharmingen, 556430), p27 (BD Transduction Lab., 610241), p27pS10 (Clone EP233(2)Y, abcam, ab62364), p27pT187 (abcam, ab75908), p27pT198 (R&D Systems, AF3994), PIM1 (Clone EP2645Y, abcam, ab75776, and GeneTex, GTX61985), PIM2 (clone EPR6987, abcam, ab129057, and clone D1D2, Cell Signaling, 4730). The secondary antibodies used are goat anti-mouse IgG-HRP (Santa Cruz Biotech., Sc-2055) and goat anti-rabbit IgG-HRP (Santa Cruz Biotech., Sc-2054). The ECL reaction was done using the Bio-Rad Clarity Western ECL Substrates and chemiluminescent signals were acquired with the Bio-Rad ChemiDoc XRS+ System or the Bio-Rad ChemiDoc Touch Imaging system equipped with a supersensitive CCD camera. Where indicated, unsaturated band intensities were quantified using BioRad Image Lab software. Actin signal was primarily used as loading control, however, the Bio-Rad TGX Stain-Free protein gels, which fluorescently label all the tryptophan residues of proteins in gels, were also used for normalization of western bands.
Indirect immunofluorescence microscopy
For immunostaining of the primary PDX tissues for Ki67 and p27 protein expression, paraffin embedded tissue samples were first sectioned, deparaffinized and rehydrated. Antigen retrieval was done using 10mM sodium citrate, pH6. Non-specific binding sites were blocked with 5% normal goat serum. The blocked samples were incubated with the primary antibodies against Ki67 (abcam, ab833) and p27 (BD Transduction Lab., 610241), respectively, over night at 4 degrees. After washing, the samples were incubated for 1 hour at room temperature with fluorescently labeled secondary antibodies goat anti-mouse IgG A488 (Invitrogen, A-11001) or goat anti-rabbit IgG A488 (Invitrogen, A-11008), respectively. The samples were then embedded in mounting medium containing DAPI (Dako) and the images were acquired using the Zeiss Axiovert 200M spinning disk confocal system equipped with the Micro Manager software. The acquired images were processed using ImageJ (Fiji). To quantify the percentage of Ki67 and p27 positive cells per high-power field (40x), at least five images per sample were taken from each of three drug-treated and three control treated samples. For each image, we determined the number of DAPI-positive nuclei using ImageJ and the number of Ki67 or p27 positive cells was counted manually.
For immunostaining of the established breast cancer cell lines for p27 protein expression, the cells were fixed with 4% paraformaldehyde in 6-wells, blocked with 3% normal goat serum, and treated with the p27 primary antibody (BD Transduction Lab., 610241) and with the secondary antibody (goat anti-mouse Alexa 488, Invitrogen, A-11029). Nuclear staining was done through the use of the mountant, ProLong Gold antifade reagent with DAPI (Life Technologies). The images were acquired using the Zeiss Axioplan 2 epi-fluorescence imaging system equipped with the Micro Manager software.
Real-time quantitative PCR
Total RNA from the PDX tumor samples was extracted using mirVanaTM mirna isolation kit (Ambion) according to the manufacturer’s instruction. Real-time PCR was carried out using TaqMan probes (Applied Biosystems) specific for miR-18a, miR-19b, and miR-20a, and were normalized against the RNU48 endogenous control gene.
siRNA experiments
Gene specific pools of scrambled siRNAs were purchased from GE Dharmacon (siGENOME SMARTpool siRNA) and siRNA transfection was performed using Lipofectamine RNAiMAX Transfection Reagent (Life Technologies) via reverse transfection method as previously described2. To sufficiently reduce PIM1 protein expression particularly in a panel of triple-negative breast cancer cell lines, it was necessary to use PIM1 siRNA at the highest suggested dose (120–150pmol per well in 6-well cell culture dishes). Even at these concentrations, we observed that some of the cell lines started recovering endogenous levels of PIM1 expression in less than 60 hours post siRNA reverse-transfection. PIM2 siRNA was used at 120pmol per well in 6-well cell culture dishes and the siRNA was validated by two independent PIM2 specific antibodies. The cellular phenotypes described in Supplementary Fig. 4 were more strongly correlated with the extent of PIM2 knock-down detected by clone EPR6987 (abcam, ab129057). To knock-down MYC expression in breast cancer cell lines, MYC siRNA was used at 60pmol per well in 6-well cell culture dishes. For control siRNAs, we used both Dharmacon siGENOME GAPD Control siRNA and siGENOME RISC-Free siRNA, both of which caused similar, acceptable levels of cytotoxicity.
In vitro cell proliferation and cell death assays
Unless otherwise noted, cell proliferation and cell death were assessed by performing a flow cytometry-based Guava ViaCount viability assay (Millipore). For the experiments shown in Fig. 3e, Guava Nexin reagent (Millipore) was used to specifically determine the number of apoptotic and dead cells positive for Annexin V and/or 7-AAD. For the experiments shown in Fig. 4i, cell number was determined by counting the cells using the Countess Automated Cell Counter (Life Technologies) according to the manufacture’s instruction.
Transgenic breast cancer cell lines
Doxycycline (tet)-regulated p27 expression construct was generated by subcloning a full-length human p27 cDNA with a Kozak sequence gccacc in front of the p27 start codon to the BamH1-EcoR1 sites located in the multiple cloning site of pLVX-Tight-Puro (Clontech) lentiviral expression plasmid. To produce recombinant lentiviral supernatants, 293FT cells were co-transfected with the ViraPower Lentiviral Packaging Mix (Life Technologies) and either pLVX-Tight-Puro-p27 or the trans-activator construct pLVX-Tet-ON Advanced using Lipofectamine 2000 Transfection Reagent. The breast cancer cell lines were co-incubated with the resulting Tight-p27 supernatant and the Tet-ON supernatant, mixed at the ratio of 3:1, in the presence of Polybrene at 6μg/ml for 24 hrs. The elevation in p27 protein expression induced by doxycycline at 100ng/ml was determined to resemble the extent of p27 up-regulation observed for the PIM inhibitor treated PDX tumor tissues. Thus, for the experiments shown in Fig. 4i, p27 up-regulation was achieved by treating the established transgenic cell lines with doxycycline at 100ng/ml.
TUNEL staining
TUNEL staining was carried out on the PDX tumor sections by using the ApopTag Peroxidase In Situ Apoptosis Detection Kit (Millipore) according to the manufacture’s instruction.
Statistics
All the results are shown as mean +/− S.E.M. unless otherwise indicated. Statistical analyses (two-tailed t-test, t ratio calculation, Pearson correlation, Log-rank test, and Tukey’s multiple test) were carried out using Prism 6 (Version 6.0f) from GraphPad Software, Inc. and R (Version 3.1.0), taking into consideration the assumptions required for the respective tests. P values <0.05 were considered to indicate statistical significance throughout the study. All the cell-based, in vitro experiments were independently repeated three times in triplicate. No statistical method was employed to predetermine sample size throughout this study. For animal experiments, efforts were made to achieve scientific goals of this study with the minimum number of animals. With respect to randomization, for animal experiments, the tumor-bearing mice of similar tumor burden were equally divided into control and experimental groups for subsequent drug treatment. For immuno-staining (i.e., p27) and TUNEL-staining of PDX tumor samples, the scoring process was carried out in a blinded fashion. No experimental samples were excluded throughout this study with the exception of the animals that experienced unexpected, acute illness and/or injury and thus were removed per veterinarian’s order.
Code availability
All custom computer code and datasets used for bioinformatics analyses, as outlined below, are freely available for download and/or use on Github at https://github.com/snjvb/pim_kinase. The raw data and processing routines are included for the following datasets (directory names for the respective datasets are indicated in parentheses): TCGA breast invasive carcinoma dataset (TCGA_BRCA_exp_HiSeqV2-2014-08-28), Chemotherapy-naïve dataset (YauGeneExp-2011-11-11), I-SPY1 dataset (ispy1_082814) and Pooled neoadjuvant chemotherapy-treated dataset (gse25066_091114). The core analysis pipeline, written in the R programming language, is included in the file pim_analysis.r. Additional code for multivariate analysis of MYC signature and PIM1 expression are available upon request.
Supplementary Material
Supplementary Table 1. List of the genes identified that induce MYC dependent synthetic lethality. Only genes targeted by at least two independent shRNA clones that selectively induced cell death in the MYC activated (i.e., +TAM) HMECs were identified as MYC synthetic lethal genes. Positive/total refers to the number of shRNA clones that induced MYC-dependent cell death/total number of shRNA clones available in the human kinome shRNA library that target a given gene. Decimals rounded to ones.
Supplementary Table 2. Clinical information on a panel of previously reported patient derived orthotopic breast tumor xenografts used in this study.
Supplementary Figure 1. PIM2 expression in human primary tumor samples and its prognostic significance.
(a) PIM2 mRNA expression in primary breast tumor samples from TCGA, pooled node-negative chemotherapy naïve, pooled neoadjuvant chemotherapy (taxane-anthracycline) treated, and I-SPY1 cohorts, respectively, stratified by receptor status. Values are log2-transformed and median-centered. Bars representing patient groups are given a number that indicates sample size. Error bars represent means +/− S.E.M. ***** indicates P < 0.00001 as determined by pairwise two-tailed t-tests between the respective groups. N.S. = not significant.
(b) Kaplan-Meier graphs of the patients with hormone receptor-negative tumors, dichotomized by PIM2 mRNA expression at an optimal threshold, from node-negative chemotherapy-naïve (top), pooled neoadjuvant chemotherapy treated (middle), and I-SPY (bottom) cohorts. Samples with elevated PIM2 expression are represented with red lines.
(c) Univariate analysis indicating the effect of PIM2 mRNA expression on survival in pooled node-negative chemotherapy-naïve, pooled neoadjuvant chemotherapy (taxane-anthracycline) treated, and I-SPY1 cohorts, specifically in hormone receptor-positive and receptor-negative subsets. PR status was not available for the pooled node-negative chemotherapy-naïve dataset, and as such ER−/ER+ subsets were considered instead. Hazard ratio is associated with a 1 standard deviation in PIM2 expression. P values are based on the likelihood ratio test.
Supplementary Figure 2. PIM3 expression in human primary tumor samples and its prognostic significance.
(a) PIM3 mRNA expression in primary breast tumor samples from TCGA and I-SPY1 cohorts, respectively, stratified by receptor status. Values are log2-transformed and median-centered. Bars representing patient groups are given a number that indicates sample size. Error bars represent means +/− S.E.M. * indicates P < 0.05 as determined by pairwise two-tailed t-tests between the respective groups. N.S. = not significant.
(b) Kaplan-Meier graphs of the patients with hormone receptor-negative tumors, dichotomized by PIM3 mRNA expression at an optimal threshold, from I-SPY cohort. Samples with elevated PIM3 expression are represented with red lines.
(c) Univariate analysis indicating the effect of PIM3 mRNA expression on survival in I-SPY1 cohort, specifically in hormone receptor-positive and receptor-negative subsets. PR status was not available for the pooled node-negative chemotherapy-naïve dataset, and as such ER−/ER+ subsets were considered instead. Hazard ratio is associated with a 1 standard deviation in PIM3 expression. P values are based on the likelihood ratio test.
Supplementary Figure 3. Correlation of MYC mRNA expression and sensitivity to PIM inhibition (t ratio) in triple-negative and receptor-positive cancer cell lines used in Figure 3a except HBL100 for which expression data was not publicly available. MYC mRNA expression data was extracted from the Cancer Cell Line Encyclopedia50 using cBioPortal (cbioportal.org)51,52. Pearson correlation and two-tailed t-test were used to generate the correlation coefficients and associated P values.
Supplementary Figure 4. siRNA mediated knock-down of PIM1 is accompanied by acute up-regulation of PIM2 in MDAMB 231 cells.
(a) The effects of knocking-down PIM1 and PIM2, respectively, on protein expression of one another, on (b) cell proliferation as assessed by cell count, and (c) induction of apoptosis as assessed by Annexin V/7-AAD staining in MDAMB 231 cells. The experiment in (b and c) was independently repeated three times in triplicate. Error bars represent means +/− S.E.M. P values were calculated by two-tailed t-test comparing the control siRNA treated group to each of the experimental groups. **P < 0.01, and N.S. = not significant.
Supplementary Figure 5. The effects of small molecule PIM kinase inhibitors on the induction of cell death in PDX tumors in vivo. HCI-002 tumors were treated with either SGI-1776 at 75mg/kg or NVP-LGB321 at 100mg/kg, or with respective control diluent, for 2 weeks and TUNEL staining was performed on the tumor samples harvested 24 hours after the final drug treatment. Representative images of control or NVP-LGB321 treated HCI-002 PDX tumors that were TUNEL stained are shown (top). Scale bar represents 100μm. The number of TUNEL-positive cells was counted in 10 high-power fields (40x) per tumor/animal (bottom). N=3 in each experimental group. Error bars represent means +/− S.E.M. P values were calculated by two-tailed t-test.
Supplementary Figure 6. Relative tumor volume of orthotopic HCI-004 PDX tumors in NOD/SCID mice that were treated with vehicle (N=5) or with NVP-LGB321 at 100 mg/kg (N=3), daily for 14 d. Vehicle-treated (N=4) and NVP-LGB321-treated (N=2) xenograft-bearing mice were followed for an additional 36 d. Error bars represent means +/− S.E.M. P values were calculated by two-tailed t-test comparing the two treatment groups.
Supplementary Figure 7. Protein expression levels and phosphorylation-status of the indicated PIM1 substrates as well as select CDKs in PDX tumors harvested before (Day 0) and after 2 weeks of treatment with either SGI-1776 (75mg/kg) or NVP-LGB321 (100mg/kg), or with respective control diluent, daily, via oral gavage. The tumors were harvested 24 hours after the final drug treatment. Three tumor samples from three independent mice are represented for each time point and treatment condition. The Day 0 samples shown are identical across the two columns. Actin serves as a loading control.
Supplementary Figure 8. Relative expression of miR-18a, miR-19b, and miR-20a in PDX tumors treated with SGI-1776, NVP-LGB321, or respective control diluents. Three tumor samples from three independent mice are represented in each treatment group. Error bars represent means +/− S.E.M. P values were calculated by two-tailed t-test.
Supplementary Figure 9. Sensitivity of non-tumorigenic and tumorigenic mammary cells to PIM kinase inhibitors.
(a) Sensitivity of HMEC-MycER cells to pan PIM kinase inhibitors, SGI-1776 and NVP-LGB321, in the presence or absence of exogenous MYC activation via the addition of 4-Hydroxytamoxifen. The cells were treated with the indicated drugs for 48 hours. Error bars represent means +/− S.E.M.
(b–c) Sensitivity of a panel of human breast cancer cell lines to PIM kinase inhibitors, SGI-1776 (a) and NVP-LGB321 (b). The cells were treated with the indicated drugs for 72 hours. Error bars represent means +/− S.E.M.
Supplementary Figure 10. P27 mRNA expression in primary breast tumor samples from TCGA, pooled node-negative chemotherapy naïve, pooled neoadjuvant chemotherapy (taxane-anthracycline) treated, and I-SPY1 cohorts, respectively, stratified by receptor status. Values are log2-transformed and median-centered. Bars representing patient groups are given a number that indicates sample size. Error bars represent means +/− S.E.M. *P < 0.05, **P < 0.01, ***P < 0.001, and N.S. = not significant as determined by pairwise two-tailed t-tests between the respective groups.
Supplementary Figure 11. A panel of breast cancer cell lines with elevated MYC expression that are sensitive to PIM1 inhibition are similarly sensitive to MYC inhibition.
(a) Protein expression levels of PIM and MYC in a panel of breast cancer cell lines treated with indicated siRNA for 60hrs. Actin serves as a loading control.
(b–c) The effects of knocking down MYC on the (b) proliferation as assessed by cell count, and the (c) induction of apoptosis as assessed by Annexin V/7-AAD staining, in a panel of breast cancer cell lines. The experiment was independently repeated three times in triplicate. Error bars represent means +/− S.E.M. P values were calculated by two-tailed t-test comparing the control siRNA treated group to each of the experimental groups. **P < 0.01, ***P < 0.001, and N.S. = not significant.
Supplementary Figure 12. Time dependent effects of PIM kinase inhibitor NVP-LGB321 on MYC mRNA expression in MDAMB231 and T47D cell lines.
A triple-negative cell line MDAMB231 and a receptor-positive cell line T47D were treated with NVP-LGB321 at 10μM for the indicated amount of time. The effect of PIM inhibition on MYC mRNA expression was determined using Real-Time PCR. The samples are normalized to time point 0 (hrs). The experiment was independently repeated at least three times. Error bars represent means +/− S.E.M. Statistical significance was evaluated by two-tailed t-test comparing the inhibitor treated samples collected at different time points as indicated. N.S. = not significant.
Supplementary Figure 13. The effects of siRNA mediated PIM1 knock down on p27 protein expression in a panel of TNBC cell lines. The indicated cell lines were treated with either control or PIM1 specific scrambled siRNA for 60 hours and were tested for protein expression of PIM1 and p27. Actin serves as a loading control.
Supplementary Figure 14. Correlation between protein expression levels of p27 in a panel of breast cancer cell lines, shown in Fig. 3a and Fig. 4e, and their sensitivity to PIM1 inhibition as determined by Pearson correlation.
Acknowledgments
This work was supported in part by grants from the US National Institute of Health (K99- and R00CA175700 to D.H., 5T32DK007418 to R.C., K99CA181490 to K.K., ES019458 to P.Y. and Z.W., U01CA168370 and P30DK63720 to M.T.M., R01CA180039 to Z.W., R01CA170447 to A.G.), Susan G. Komen Foundation (PDF15331114 to J.R.), the UCSF Program for Breakthrough Biomedical Research (M.T.M.), an Innovative, Developmental, and Exploratory Award from California Breast Cancer Research Program (17lB-0024 to A.G.), an Era of Hope Scholar Award from the CDMRP Breast Cancer Research Program (W81XWH-12-1-0272 to A.G.), an LLS Scholar Award and V-Foundation Award (to A.G.), the Breast Cancer Research Foundation (to H.S.R and A.G.), and the Northwestern Medicine Catalyst Funds (to D.H). The authors thank A. Welm for her guidance with the use of the patient derived orthotopic tumor xenograft models, J.W. Smyth for his assistance with the generation of the transgenic breast cancer cell lines, D.B. Udy, C.L. Hueschen, and A. Vasilopoulos for their assistance with microscopy. We thank Susan Samson, Carole Baas and Hannah Klein Connolly for consumer advocacy support and feedback related to this project, and J.M. Bishop for his insights into the project and for his mentorship to D.H.
Footnotes
AUTHOR CONTRIBUTIONS
D.H and A.G. conceived of the project. D.H. designed and executed the shRNA screen and the subsequent biological experiments, analyzed the data, and wrote the manuscript. A.V.B., M.T.M., and P.Y. provided materials for and contributed to designing and executing the screen. C.Y. and S.B. performed bioinformatics analyses. D.A.L., H.S.R. and Z.W. provided materials for and contributed to designing and executing the animal experiments involving the patient-derived orthotopic tumor xenografts. R.C., A.Y.Z., A.N.C., H.E., K.K., L.A.M., B.N.A., J.R. and R.K. contributed to executing the biological experiments. R.C., A.Y.Z., C.Y., S.B., L.A.M., B.N.A., P.Y., Z.W., and A.G. participated in writing the manuscript. A.G. supervised the project.
References
- 1.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 2.Horiuchi D, et al. MYC pathway activation in triple-negative breast cancer is synthetic lethal with CDK inhibition. J Exp Med. 2012;209:679–696. doi: 10.1084/jem.20111512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prochownik EV, Vogt PK. Therapeutic Targeting of Myc. Genes Cancer. 2010;1:650–659. doi: 10.1177/1947601910377494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKeown MR, Bradner JE. Therapeutic strategies to inhibit MYC. Cold Spring Harb Perspect Med. 2014;4:a014266. doi: 10.1101/cshperspect.a014266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaelin WG. The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 7.Mukhopadhyay R, et al. Promotion of variant human mammary epithelial cell outgrowth by ionizing radiation: an agent-based model supported by in vitro studies. Breast Cancer Res. 2010;12:R11. doi: 10.1186/bcr2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23:1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kessler JD, et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335:348–353. doi: 10.1126/science.1212728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sato M, et al. MYC is a critical target of FBXW7. Oncotarget. 2015;6:3292–3305. doi: 10.18632/oncotarget.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stampfer MR, Yaswen P. Culture models of human mammary epithelial cell transformation. J Mammary Gland Biol Neoplasia. 2000;5:365–378. doi: 10.1023/a:1009525827514. [DOI] [PubMed] [Google Scholar]
- 12.Bazarov AV, et al. Telomerase activation by c-Myc in human mammary epithelial cells requires additional genomic changes. Cell Cycle. 2009;8:3373–3378. doi: 10.4161/cc.8.20.9856. [DOI] [PubMed] [Google Scholar]
- 13.Bazarov AV, et al. p16(INK4a) -mediated suppression of telomerase in normal and malignant human breast cells. Aging Cell. 2010;9:736–746. doi: 10.1111/j.1474-9726.2010.00599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nawijn MC, Alendar A, Berns A. For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat Rev Cancer. 2010;11:23–34. doi: 10.1038/nrc2986. [DOI] [PubMed] [Google Scholar]
- 15.Mikkers H, et al. Mice deficient for all PIM kinases display reduced body size and impaired responses to hematopoietic growth factors. Mol Cell Biol. 2004;24:6104–6115. doi: 10.1128/MCB.24.13.6104-6115.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Lohuizen M, et al. Predisposition to lymphomagenesis in pim-1 transgenic mice: cooperation with c-myc and N-myc in murine leukemia virus-induced tumors. Cell. 1989;56:673–682. doi: 10.1016/0092-8674(89)90589-8. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, et al. Pim1 kinase synergizes with c-MYC to induce advanced prostate carcinoma. Oncogene. 2010;29:2477–2487. doi: 10.1038/onc.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim J, Roh M, Abdulkadir SA. Pim1 promotes human prostate cancer cell tumorigenicity and c-MYC transcriptional activity. BMC Cancer. 2010;10:248. doi: 10.1186/1471-2407-10-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Lohuizen M, et al. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, et al. Pim1 kinase is required to maintain tumorigenicity in MYC-expressing prostate cancer cells. Oncogene. 2012;31:1794–1803. doi: 10.1038/onc.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirschner AN, et al. PIM Kinase Inhibitor AZD1208 for Treatment of MYC-Driven Prostate Cancer. J Natl Cancer Inst. 2015;107:dju407–dju407. doi: 10.1093/jnci/dju407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie Y, et al. The 44 kDa Pim-1 kinase directly interacts with tyrosine kinase Etk/BMX and protects human prostate cancer cells from apoptosis induced by chemotherapeutic drugs. Oncogene. 2005 doi: 10.1038/sj.onc.1209058. [DOI] [PubMed] [Google Scholar]
- 23.Malinen M, et al. Proto-oncogene PIM-1 is a novel estrogen receptor target associating with high grade breast tumors. Mol Cell Endocrinol. 2013;365:270–276. doi: 10.1016/j.mce.2012.10.028. [DOI] [PubMed] [Google Scholar]
- 24.Chen LS, Redkar S, Bearss D, Wierda WG, Gandhi V. Pim kinase inhibitor, SGI-1776, induces apoptosis in chronic lymphocytic leukemia cells. Blood. 2009 doi: 10.1182/blood-2009-03-212852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mumenthaler SM, et al. Pharmacologic inhibition of Pim kinases alters prostate cancer cell growth and resensitizes chemoresistant cells to taxanes. Molecular Cancer Therapeutics. 2009;8:2882–2893. doi: 10.1158/1535-7163.MCT-09-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeRose YS, et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat Med. 2011;17:1514–1520. doi: 10.1038/nm.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia PD, et al. Pan-PIM kinase inhibition provides a novel therapy for treating hematologic cancers. Clinical Cancer Research. 2014;20:1834–1845. doi: 10.1158/1078-0432.CCR-13-2062. [DOI] [PubMed] [Google Scholar]
- 28.Lu J, et al. Pim2 is required for maintaining multiple myeloma cell growth through modulating TSC2 phosphorylation. Blood. 2013;122:1610–1620. doi: 10.1182/blood-2013-01-481457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burger MT, et al. Identification of N-(4-((1R,3S,5S)-3-Amino-5-methylcyclohexyl)pyridin-3-yl)-6-(2,6-difluorophenyl)-5-fluoropicolinamide (PIM447), a Potent and Selective Proviral Insertion Site of Moloney Murine Leukemia (PIM) 1, 2, and 3 Kinase Inhibitor in Clinical Trials for Hematological Malignancies. J Med Chem. 2015;58 doi: 10.1021/acs.jmedchem.5b01275. acs.jmedchem.5b01275–8386. [DOI] [PubMed] [Google Scholar]
- 30.D’Cruz CM, et al. c-MYC induces mammary tumorigenesis by means of a preferred pathway involving spontaneous Kras2 mutations. Nat Med. 2001;7:235–239. doi: 10.1038/84691. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Wang Z, Li X, Magnuson NS. Pim kinase-dependent inhibition of c-Myc degradation. Oncogene. 2008;27:4809–4819. doi: 10.1038/onc.2008.123. [DOI] [PubMed] [Google Scholar]
- 32.Sears RC. The life cycle of C-myc: from synthesis to degradation. Cell Cycle. 2004;3:1133–1137. [PubMed] [Google Scholar]
- 33.Pulverer BJ, et al. Site-specific modulation of c-Myc cotransformation by residues phosphorylated in vivo. Oncogene. 1994;9:59–70. [PubMed] [Google Scholar]
- 34.Zhang Y, Wang Z, Magnuson NS. Pim-1 kinase-dependent phosphorylation of p21Cip1/WAF1 regulates its stability and cellular localization in H1299 cells. Mol Cancer Res. 2007;5:909–922. doi: 10.1158/1541-7786.MCR-06-0388. [DOI] [PubMed] [Google Scholar]
- 35.Morishita D, Katayama R, Sekimizu K, Tsuruo T, Fujita N. Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer Res. 2008;68:5076–5085. doi: 10.1158/0008-5472.CAN-08-0634. [DOI] [PubMed] [Google Scholar]
- 36.Bachmann M, et al. The oncogenic serine/threonine kinase Pim-1 directly phosphorylates and activates the G2/M specific phosphatase Cdc25C. Int J Biochem Cell Biol. 2006;38:430–443. doi: 10.1016/j.biocel.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 37.Mochizuki T, et al. Physical and functional interactions between Pim-1 kinase and Cdc25A phosphatase. Implications for the Pim-1-mediated activation of the c-Myc signaling pathway. J Biol Chem. 1999;274:18659–18666. doi: 10.1074/jbc.274.26.18659. [DOI] [PubMed] [Google Scholar]
- 38.Yan B, et al. The PIM-2 kinase phosphorylates BAD on serine 112 and reverses BAD-induced cell death. J Biol Chem. 2003;278:45358–45367. doi: 10.1074/jbc.M307933200. [DOI] [PubMed] [Google Scholar]
- 39.Aho TLT, et al. Pim-1 kinase promotes inactivation of the pro-apoptotic Bad protein by phosphorylating it on the Ser112 gatekeeper site. FEBS Lett. 2004;571:43–49. doi: 10.1016/j.febslet.2004.06.050. [DOI] [PubMed] [Google Scholar]
- 40.Fox CJ, et al. The serine/threonine kinase Pim-2 is a transcriptionally regulated apoptotic inhibitor. Genes Dev. 2003;17:1841–1854. doi: 10.1101/gad.1105003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang J, et al. eIF4B phosphorylation by pim kinases plays a critical role in cellular transformation by Abl oncogenes. Cancer Res. 2013;73:4898–4908. doi: 10.1158/0008-5472.CAN-12-4277. [DOI] [PubMed] [Google Scholar]
- 42.Wang X, et al. Phosphorylation regulates c-Myc’s oncogenic activity in the mammary gland. Cancer Res. 2011;71:925–936. doi: 10.1158/0008-5472.CAN-10-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 44.Mohanty AR, et al. Successive phosphorylation of p27(KIP1) protein at serine-10 and C terminus crucially controls its potency to inactivate Cdk2. J Biol Chem. 2012;287:21757–21764. doi: 10.1074/jbc.M112.346254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujita N, Sato S, Katayama K, Tsuruo T. Akt-dependent phosphorylation of p27Kip1 promotes binding to 14-3-3 and cytoplasmic localization. J Biol Chem. 2002;277:28706–28713. doi: 10.1074/jbc.M203668200. [DOI] [PubMed] [Google Scholar]
- 46.Vervoorts J, Lüscher B. Post-translational regulation of the tumor suppressor p27(KIP1) Cell Mol Life Sci. 2008;65:3255–3264. doi: 10.1007/s00018-008-8296-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Czabotar PE, Lessene G, Strasser A, Adams JM. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol. 2014;15:49–63. doi: 10.1038/nrm3722. [DOI] [PubMed] [Google Scholar]
- 48.Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat Rev Cancer. 2008;8:253–267. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- 49.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1–pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Narita M, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 54.Yau C, et al. A multigene predictor of metastatic outcome in early stage hormone receptor-negative and triple-negative breast cancer. Breast Cancer Res. 2010;12:R85. doi: 10.1186/bcr2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Esserman LJ, et al. Chemotherapy response and recurrence-free survival in neoadjuvant breast cancer depends on biomarker profiles: results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657) Breast Cancer Res Treat. 2012;132:1049–1062. doi: 10.1007/s10549-011-1895-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hatzis C, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305:1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 58.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Langfelder P, Horvath S. Fast R Functions for Robust Correlations and Hierarchical Clustering. J Stat Softw. 2012;46 [PMC free article] [PubMed] [Google Scholar]
- 60.Wickham H. ggplot2. Springer Science & Business Media; 2009. [DOI] [Google Scholar]
- 61.Therneau TM. Modeling Survival Data: Extending the Cox Model. Springer Science & Business Media; 2000. [Google Scholar]
- 62.Chandriani S, et al. A Core MYC Gene Expression Signature Is Prominent in Basal-Like Breast Cancer but Only Partially Overlaps the Core Serum Response. PLoS ONE. 2009;4:e6693. doi: 10.1371/journal.pone.0006693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neve RM, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen LS, Redkar S, Taverna P, Cortes JE, Gandhi V. Mechanisms of cytotoxicity to Pim kinase inhibitor, SGI-1776, in acute myeloid leukemia. Blood. 2011;118:693–702. doi: 10.1182/blood-2010-12-323022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. List of the genes identified that induce MYC dependent synthetic lethality. Only genes targeted by at least two independent shRNA clones that selectively induced cell death in the MYC activated (i.e., +TAM) HMECs were identified as MYC synthetic lethal genes. Positive/total refers to the number of shRNA clones that induced MYC-dependent cell death/total number of shRNA clones available in the human kinome shRNA library that target a given gene. Decimals rounded to ones.
Supplementary Table 2. Clinical information on a panel of previously reported patient derived orthotopic breast tumor xenografts used in this study.
Supplementary Figure 1. PIM2 expression in human primary tumor samples and its prognostic significance.
(a) PIM2 mRNA expression in primary breast tumor samples from TCGA, pooled node-negative chemotherapy naïve, pooled neoadjuvant chemotherapy (taxane-anthracycline) treated, and I-SPY1 cohorts, respectively, stratified by receptor status. Values are log2-transformed and median-centered. Bars representing patient groups are given a number that indicates sample size. Error bars represent means +/− S.E.M. ***** indicates P < 0.00001 as determined by pairwise two-tailed t-tests between the respective groups. N.S. = not significant.
(b) Kaplan-Meier graphs of the patients with hormone receptor-negative tumors, dichotomized by PIM2 mRNA expression at an optimal threshold, from node-negative chemotherapy-naïve (top), pooled neoadjuvant chemotherapy treated (middle), and I-SPY (bottom) cohorts. Samples with elevated PIM2 expression are represented with red lines.
(c) Univariate analysis indicating the effect of PIM2 mRNA expression on survival in pooled node-negative chemotherapy-naïve, pooled neoadjuvant chemotherapy (taxane-anthracycline) treated, and I-SPY1 cohorts, specifically in hormone receptor-positive and receptor-negative subsets. PR status was not available for the pooled node-negative chemotherapy-naïve dataset, and as such ER−/ER+ subsets were considered instead. Hazard ratio is associated with a 1 standard deviation in PIM2 expression. P values are based on the likelihood ratio test.
Supplementary Figure 2. PIM3 expression in human primary tumor samples and its prognostic significance.
(a) PIM3 mRNA expression in primary breast tumor samples from TCGA and I-SPY1 cohorts, respectively, stratified by receptor status. Values are log2-transformed and median-centered. Bars representing patient groups are given a number that indicates sample size. Error bars represent means +/− S.E.M. * indicates P < 0.05 as determined by pairwise two-tailed t-tests between the respective groups. N.S. = not significant.
(b) Kaplan-Meier graphs of the patients with hormone receptor-negative tumors, dichotomized by PIM3 mRNA expression at an optimal threshold, from I-SPY cohort. Samples with elevated PIM3 expression are represented with red lines.
(c) Univariate analysis indicating the effect of PIM3 mRNA expression on survival in I-SPY1 cohort, specifically in hormone receptor-positive and receptor-negative subsets. PR status was not available for the pooled node-negative chemotherapy-naïve dataset, and as such ER−/ER+ subsets were considered instead. Hazard ratio is associated with a 1 standard deviation in PIM3 expression. P values are based on the likelihood ratio test.
Supplementary Figure 3. Correlation of MYC mRNA expression and sensitivity to PIM inhibition (t ratio) in triple-negative and receptor-positive cancer cell lines used in Figure 3a except HBL100 for which expression data was not publicly available. MYC mRNA expression data was extracted from the Cancer Cell Line Encyclopedia50 using cBioPortal (cbioportal.org)51,52. Pearson correlation and two-tailed t-test were used to generate the correlation coefficients and associated P values.
Supplementary Figure 4. siRNA mediated knock-down of PIM1 is accompanied by acute up-regulation of PIM2 in MDAMB 231 cells.
(a) The effects of knocking-down PIM1 and PIM2, respectively, on protein expression of one another, on (b) cell proliferation as assessed by cell count, and (c) induction of apoptosis as assessed by Annexin V/7-AAD staining in MDAMB 231 cells. The experiment in (b and c) was independently repeated three times in triplicate. Error bars represent means +/− S.E.M. P values were calculated by two-tailed t-test comparing the control siRNA treated group to each of the experimental groups. **P < 0.01, and N.S. = not significant.
Supplementary Figure 5. The effects of small molecule PIM kinase inhibitors on the induction of cell death in PDX tumors in vivo. HCI-002 tumors were treated with either SGI-1776 at 75mg/kg or NVP-LGB321 at 100mg/kg, or with respective control diluent, for 2 weeks and TUNEL staining was performed on the tumor samples harvested 24 hours after the final drug treatment. Representative images of control or NVP-LGB321 treated HCI-002 PDX tumors that were TUNEL stained are shown (top). Scale bar represents 100μm. The number of TUNEL-positive cells was counted in 10 high-power fields (40x) per tumor/animal (bottom). N=3 in each experimental group. Error bars represent means +/− S.E.M. P values were calculated by two-tailed t-test.
Supplementary Figure 6. Relative tumor volume of orthotopic HCI-004 PDX tumors in NOD/SCID mice that were treated with vehicle (N=5) or with NVP-LGB321 at 100 mg/kg (N=3), daily for 14 d. Vehicle-treated (N=4) and NVP-LGB321-treated (N=2) xenograft-bearing mice were followed for an additional 36 d. Error bars represent means +/− S.E.M. P values were calculated by two-tailed t-test comparing the two treatment groups.
Supplementary Figure 7. Protein expression levels and phosphorylation-status of the indicated PIM1 substrates as well as select CDKs in PDX tumors harvested before (Day 0) and after 2 weeks of treatment with either SGI-1776 (75mg/kg) or NVP-LGB321 (100mg/kg), or with respective control diluent, daily, via oral gavage. The tumors were harvested 24 hours after the final drug treatment. Three tumor samples from three independent mice are represented for each time point and treatment condition. The Day 0 samples shown are identical across the two columns. Actin serves as a loading control.
Supplementary Figure 8. Relative expression of miR-18a, miR-19b, and miR-20a in PDX tumors treated with SGI-1776, NVP-LGB321, or respective control diluents. Three tumor samples from three independent mice are represented in each treatment group. Error bars represent means +/− S.E.M. P values were calculated by two-tailed t-test.
Supplementary Figure 9. Sensitivity of non-tumorigenic and tumorigenic mammary cells to PIM kinase inhibitors.
(a) Sensitivity of HMEC-MycER cells to pan PIM kinase inhibitors, SGI-1776 and NVP-LGB321, in the presence or absence of exogenous MYC activation via the addition of 4-Hydroxytamoxifen. The cells were treated with the indicated drugs for 48 hours. Error bars represent means +/− S.E.M.
(b–c) Sensitivity of a panel of human breast cancer cell lines to PIM kinase inhibitors, SGI-1776 (a) and NVP-LGB321 (b). The cells were treated with the indicated drugs for 72 hours. Error bars represent means +/− S.E.M.
Supplementary Figure 10. P27 mRNA expression in primary breast tumor samples from TCGA, pooled node-negative chemotherapy naïve, pooled neoadjuvant chemotherapy (taxane-anthracycline) treated, and I-SPY1 cohorts, respectively, stratified by receptor status. Values are log2-transformed and median-centered. Bars representing patient groups are given a number that indicates sample size. Error bars represent means +/− S.E.M. *P < 0.05, **P < 0.01, ***P < 0.001, and N.S. = not significant as determined by pairwise two-tailed t-tests between the respective groups.
Supplementary Figure 11. A panel of breast cancer cell lines with elevated MYC expression that are sensitive to PIM1 inhibition are similarly sensitive to MYC inhibition.
(a) Protein expression levels of PIM and MYC in a panel of breast cancer cell lines treated with indicated siRNA for 60hrs. Actin serves as a loading control.
(b–c) The effects of knocking down MYC on the (b) proliferation as assessed by cell count, and the (c) induction of apoptosis as assessed by Annexin V/7-AAD staining, in a panel of breast cancer cell lines. The experiment was independently repeated three times in triplicate. Error bars represent means +/− S.E.M. P values were calculated by two-tailed t-test comparing the control siRNA treated group to each of the experimental groups. **P < 0.01, ***P < 0.001, and N.S. = not significant.
Supplementary Figure 12. Time dependent effects of PIM kinase inhibitor NVP-LGB321 on MYC mRNA expression in MDAMB231 and T47D cell lines.
A triple-negative cell line MDAMB231 and a receptor-positive cell line T47D were treated with NVP-LGB321 at 10μM for the indicated amount of time. The effect of PIM inhibition on MYC mRNA expression was determined using Real-Time PCR. The samples are normalized to time point 0 (hrs). The experiment was independently repeated at least three times. Error bars represent means +/− S.E.M. Statistical significance was evaluated by two-tailed t-test comparing the inhibitor treated samples collected at different time points as indicated. N.S. = not significant.
Supplementary Figure 13. The effects of siRNA mediated PIM1 knock down on p27 protein expression in a panel of TNBC cell lines. The indicated cell lines were treated with either control or PIM1 specific scrambled siRNA for 60 hours and were tested for protein expression of PIM1 and p27. Actin serves as a loading control.
Supplementary Figure 14. Correlation between protein expression levels of p27 in a panel of breast cancer cell lines, shown in Fig. 3a and Fig. 4e, and their sensitivity to PIM1 inhibition as determined by Pearson correlation.