Abstract
The P-glycoprotein (Pgp) transporter plays a central role in drug disposition by effluxing a chemically diverse range of drugs from cells through conformational changes and ATP hydrolysis. A number of drugs are known to activate ATP hydrolysis of Pgp, but coupling between ATP and drug binding is not well understood. The cardiovascular drug verapamil is one of the most widely studied Pgp substrates and therefore, represents an ideal drug to investigate the drug-induced ATPase activation of Pgp. As previously noted, verapamil-induced Pgp-mediated ATP hydrolysis kinetics was biphasic at saturating ATP concentrations. However, at subsaturating ATP concentrations, verapamil-induced ATPase activation kinetics became monophasic. To further understand this switch in kinetic behavior, the Pgp-coupled ATPase activity kinetics was checked with a panel of verapamil and ATP concentrations and fit with the substrate inhibition equation and the kinetic fitting software COPASI. The fits suggested that cooperativity between ATP and verapamil switched between low and high verapamil concentration. Fluorescence spectroscopy of Pgp revealed that cooperativity between verapamil and a non-hydrolyzable ATP analog leads to distinct global conformational changes of Pgp. NMR of Pgp reconstituted in liposomes showed that cooperativity between verapamil and the non-hydrolyzable ATP analog modulate each others interactions. This information was used to produce a conformationally-gated model of drug-induced activation of Pgp-mediated ATP hydrolysis.
Keywords: ABC transporter, nucleotide, drug transport, cardiovascular, nuclear magnetic resonance (NMR), cooperativity
Graphical Abstract
1. INTRODUCTION
P-glycoprotein (Pgp) is an ATP hydrolysis-driven efflux transporter that is part of the ATP-binding cassette (ABC) superfamily of proteins [1, 2]. The transporter effluxes a chemically and structurally diverse range of molecules, including anticancer drugs, neurotherapeutics and cardiovascular drugs, from the cytosol to the extracellular space across cell membranes [1, 2]. The transporter is expressed at relatively high concentrations in the brain, intestines, liver, placenta, and the kidneys [3, 4]. Pgp expression level is also influenced by genetic polymorphisms and disease [5, 6]. The transporter functions to protect tissues from chemical toxicity, but also leads to drug resistance and can significantly affect drug disposition [1, 2]. For example, the transporter protects the brain from chemical insults by effluxing drugs across the blood-brain-barrier (BBB) [7], but also makes cancerous tumors overexpressing the protein resistant to anticancer drugs [8]. As a result, there has been keen interest in unraveling the molecular and structural basis of transport with Pgp. This knowledge is critical for the development of novel transport inhibitors, drugs with desirable transport properties and improving predictions of in vivo drug disposition from in vitro measurements.
Most of our structural understanding of the transporter comes from X-ray crystallography of mouse Pgp (Abcb1a), Caenorhabditis (C.) elegans Pgp and bacterial transporters [9–12]. The X-ray crystal structure of mouse Pgp revealed a 170 kD pseudosymmetric monomer, consisting of two nucleotide-binding domains (NBDs) and 12 transmembrane (TM) helices [9]. The bacterial transporter X-ray crystal structures of MsbA and SAV1866 have only 6 TM helices, but as dimers these proteins resemble the 3-dimensional fold of mammalian Pgps [10, 11]. The bacterial transporter X-ray crystal structures have also been found in different conformations with nucleotide cofactors suggesting that conformational changes play a role in transport [10, 11, 13]. Since both bacterial and mouse Pgp have conserved motifs within the NBDs, including Walker A, Walker B and ABC signature motifs [e.g. 14], they are considered to have similar transport mechanisms. From these bacterial transporter structures, a conformationally gated transport model was proposed [10, 11].
Despite the availability of X-ray crystal structures, our understanding of the coupling between drug binding, ATP hydrolysis and transport remains limited. Cross-linking studies of Pgp in human embryonic kidney (HEK) 293 cell membranes suggest that drug-induced conformational changes can occur with the NBDs or the transmembrane region [15, 16]. A study on human Pgp in nanodiscs with antibodies showed that there are ligand and cofactor-dependent conformational changes [17]. A fluorescence study with mouse Pgp found differences in fluorescence resonance energy transfer (FRET) with drugs, nucleotide cofactors and their analogs suggesting conformational changes [18].
In addition to being substrates for the transporter, a number of drugs are known to activate ATP hydrolysis of Pgp, but little is known of the molecular mechanism or its relationship to transport. One of the most studied drugs is the cardiovascular drug verapamil (Fig. 1A), which is used to treat hypertension, chest pain and arrhythmia [19–22], and can function as both a substrate and an inhibitor of the transporter [23]. The drug is known to activate Pgp-coupled ATP hydrolysis from a number of in vitro studies [e.g. 24, 25]. The kinetics of verapamil-induced Pgp-coupled ATP hydrolysis is biphasic [24–27] suggesting at least two verapamil binding sites. Biphasic drug-induced ATPase activation kinetics has been observed with a chemically diverse range of substrates from amitriptyline to vinblastine [e.g. 24, 28, 29] implying a common mechanism between these types of substrates and Pgp. Despite the large number of studies, the molecular basis for drug-induced ATPase activation of Pgp and the underlying interactions between drugs, ATP and Pgp are not well understood.
FIGURE 1.
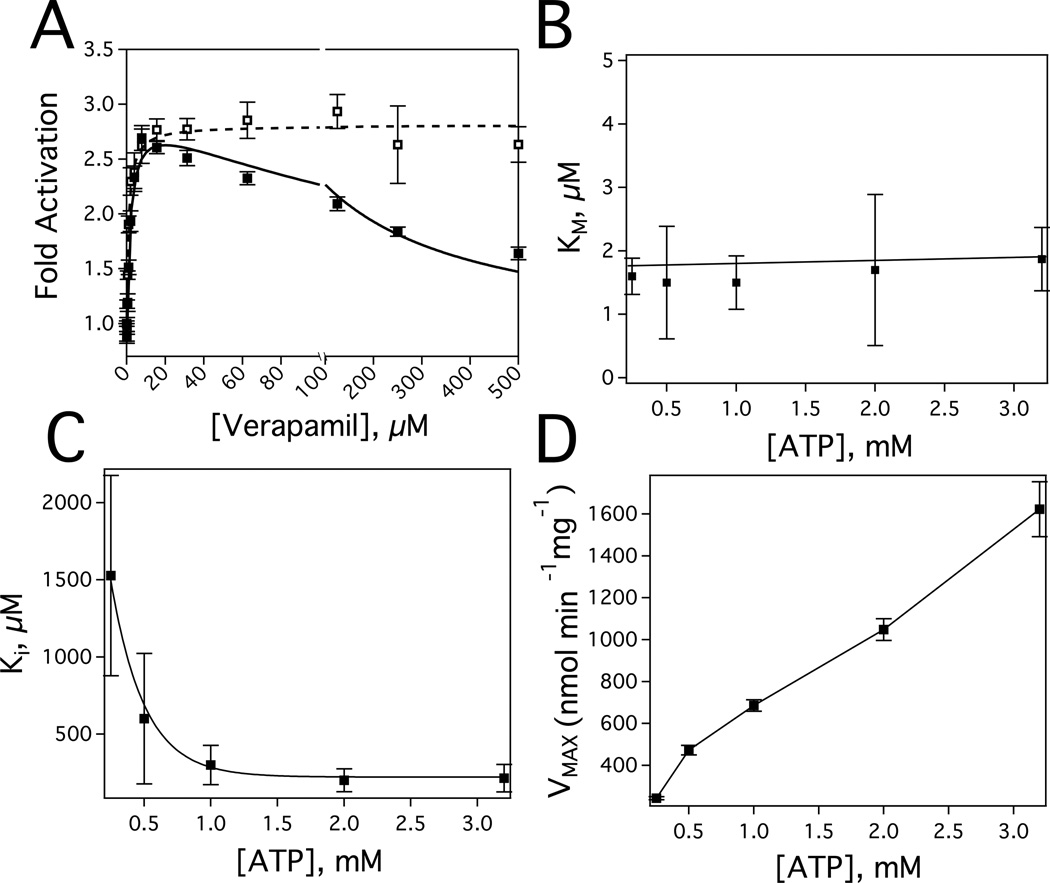
The effect of ATP on verapamil-induced ATPase activation of Pgp. A) The Pgp-coupled ATPase activity as a function of verapamil concentration in the presence of 3.2 mM (closed squares) and 0.25 mM ATP (open squares). The kinetic fits are shown as a dashed and solid line and were fit to equation 1 and 2, respectively. B) The Michaelis-Menten constant (Km), C) the inhibition constant (Ki) and D) the VMAX for verapamil-induced Pgp ATPase activity as a function of different ATP concentrations. Error bars represent the standard deviation and the points represent the average of at least three independent experiments.
In the proposed studies, the interactions of verapamil and ATP were investigated with Pgp reconstituted into liposomes. To investigate the coupling between verapamil and ATP, verapamil-induced activation of ATPase activity was examined with a range of verapamil and ATP concentrations. Acrylamide quenching of intrinsic tryptophan fluorescence spectroscopy was used to investigate drug and nucleotide-induced conformational changes of Pgp. The interactions between verapamil and a non-hydrolyzable ATP analog were investigated by the saturation transfer double difference (STDD) NMR technique. These results were used to build a model of verapamil-induced ATPase activation of verapamil efflux by the transporter. Since similar biphasic drug-induced ATPase activation is observed for several Pgp substrates, this mechanism will likely be generalizable [e.g. 24, 28, 29].
2. MATERIALS AND METHODS
2.1. Materials
Adenosine 5’-(β,γ-imido)triphosphate lithium salt (AMPPNP) was purchased from Sigma Aldrich (Milwaukee, WI) and verapamil hydrochloride was purchased from Fagron (St. Paul, MN). The detergent n-dodecyl-β-D-maltoside (DDM), which is used in protein purification, was purchased from EMD Millipore Corporation (San Diego, CA). Escherichia coli (E. Coli) total lipid extract powder was purchased from Avanti Polar Lipids Inc. (Alabaster, AL) and cholesterol was purchased from Amresco (Solon, OH) for liposome preparations. Disodium ATP (Na2ATP) was purchased from Amresco (Solon, OH) and sodium orthovanadate (Na3VO4) was purchased from Enzo Life Sciences (Farmingdale, NY) for the ATPase activity assays. Acrylamide was purchased from Calbiochem (San Diego, CA) for the acrylamide quenching experiments. Deuterium oxide (D2O) was purchased from Cambridge Isotope Laboratories (Tewksbury, MA) and deuterated (d10) DTT was purchased from CDN Isotopes (Quebec, Canada) for NMR experiments. Dithiothreitol (DTT) was purchased from Gold Biotechnology (St. Louis, MO). HEPES was purchased from Calbiochem (San Diego, CA). Tris-HCl was purchased from Amresco (Solon, OH). MgCl2 and NaCl were purchased from J.T Baker (Center Valley, PA). NH4Cl was purchased from Sigma Aldrich (Milwaukee, WI). MgSO4, NaN3, and potassium phosphate were all purchased from Thermo Fisher Scientific (Waltham, MA).
2.2. Pgp purification and reconstitution
The wild-type his-tagged mouse Pgp transporter was purified from Pichia (P.) pastoris and reconstituted into liposomes as described previously [30–32]. Briefly, detergent solubilized Pgp was reconstituted into liposomes composed of 80% w/v Avanti Escherichia (E.) coli Total Lipid Extract (Avanti Polar Lipids, Alabaster, AL) and 20% w/v cholesterol. The final lipid to protein ratio was 0.16 mg ml−1 liposomes per µM−1 Pgp. Proteoliposomes were stored in aliquots at −80°C in HEPES buffer (20 mM HEPES, 100 mM NaCl, 5 mM MgCl2, 2 mM DTT, pH 7.4). Protein concentration was determined using the extinction coefficient of 1.28 ml mg−1 cm−1 (0.181 µM−1 cm−1) [30] or the DC Protein Assay Kit II (Bio-Rad, Hercules, CA).
2.3. ATPase Activity Measurements
The specific ATPase activity of the Pgp transporter was measured using the Chifflet colorimetric assay as described previously [33]. The rate of ATP hydrolysis was determined by measuring the amount of free inorganic phosphate (Pi) through the formation of Pi and ammonium molybdate. This Pi-molybdenum complex produces a strong absorbance signal at 850 nm and was measured on a 96-well plate in a FlexStation 3 spectrometer (Molecular Devices, Sunnyvale, CA). The ATPase activity with verapamil and ATP was measured with 50 nM liposome reconstituted Pgp in Chifflet buffer (150 mM NH4Cl, 5 mM MgSO4, 0.02% w/v NaN3, 50 mM Tris-HCl, 2 mM DTT, pH 7.4). As a control, 200 µM Na3VO4 was added to the ATP titration to show Pgp-mediated ATPase activity inhibition.
Linear transformations such as the Hans-Woolf, Lineweaver-Burk (double reciprocal) or Eadie-Hofstee plots are classical approaches used to fit enzyme kinetics in the absence of computers [34, 35]. These types of plots suffer from a lack of variable independence across the axes and biasing of the error and the data points [36–38]. These methods have generally been superseded by non-linear regression methods that are significantly more accurate and no longer computationally inaccessible [37]. For monophasic ATP hydrolysis kinetics, the ATP hydrolysis rate (v), the maximum ATP hydrolysis rate (VMAX), the basal ATPase hydrolysis rate (vbasal) and the Michaelis-Menten constant (Km) was estimated with the Michaelis-Menten equation (Equation 1) [35, 39].
| (1) |
In cases where the ATP hydrolysis kinetics was biphasic, the VMAX, Km and the inhibitory constant (Ki) were estimated with the substrate inhibition equation (Equation 2) [35, 39].
| (2) |
For more complicated kinetics, fitting equations have been developed in some cases, but often require specialized numerical methods that result in multiple solutions e.g. [40].
To overcome these challenges, a variety of advanced software modeling packages have been developed to fit arbitrary kinetic models including the free Complex Pathway Simulator (COPASI) and the proprietary Berkeley Madonna (University of California, Berkeley, CA). To estimate the discrete VMAXS and KDS, the ATP hydrolysis kinetics in this study were fit to kinetic models using the evolutionary algorithm in the COPASI software [41].
2.4. Saturation transfer double difference (STDD) NMR
The STDD NMR technique is a method for characterizing ligand interactions with membrane proteins in the presence of lipids e.g. [31, 42, 43]. With this technique, the protein is selectively excited with a radio frequency (RF) outside of the frequency of ligand proton NMR peaks, which disperses the RF throughout the protein by spin diffusion. This RF saturation is transferred from the protein to protons on the bound ligand, ligand is exchanged with the bulk solvent and the 1H STD NMR signal is observed [44]. In the presence of lipids such as Pgp reconstituted in liposomes, there will be significant RF saturation transfer interference from non-specific interactions between the ligand and the liposomes. Utilizing the STDD NMR technique allows the saturation transfer between the lipid and the ligand to be subtracted from the 1H STD NMR signal [42, 45].
The STDD NMR procedure for membrane proteins was performed as described previously [31, 43]. All the STDD NMR samples contained 1 µM Pgp reconstituted into liposomes in 80% deuterated 100 mM KPi buffer, pD 7.4. For the STDD NMR experiments, a saturation transfer difference (STD) pulse sequence was used with a WATERGATE pulse sequence to suppress background water [46], with a 30 ms T1ρ spin lock filter to suppress background NMR signals and a train of 50 ms gaussian shaped selective pulses to excite the protein [47]. A total of 512 off-resonance spectra were subtracted from on-resonance spectra within the pulse program with a 2 sec saturation pulse by phase cycling at 40 and −1.5 ppm, respectively. To produce the STDD NMR spectrum, control samples were performed under identical conditions with the liposomes and the ligands. The 1H STD NMR spectrum with liposomes and ligands was subtracted from the 1H STD NMR spectrum of Pgp reconstituted in liposomes with ligands (ΔI) to isolate specific interactions between the ligand and Pgp. The STDD amplification factor was calculated using the following equation [31, 44]:
| (3) |
where [P] is the protein concentration and I0 is the amplitude of the 1H NMR peaks in the absence of excitation pulses.
All NMR experiments were performed on an 800 MHz Varian INOVA spectrometer at 25oC equipped with a 5 mm z-gradient 1H{13C/15N} cryoprobe. The 1H NMR peaks for verapamil and AMPPNP were assigned using standard 1H 1D and 2D NMR techniques. NMR spectra were processed using the iNMR software package (http://www.inmr.net) and analyzed using Igor Pro 6.2 (Wavemetrics, Tigard, OR). The molecular structure of verapamil and ATP/AMPPNP with the nuclei labeled are shown in Fig. 5A and 5E and 1H NMR peak assignments for verapamil and AMPPNP are shown in Fig. 5B and 5F, respectively, and were essentially identical to previous 1H NMR assignments [48, 49], [50].
FIGURE 5.
Molecular interactions of verapamil and AMPPNP with Pgp probed by saturation transfer double difference (STDD) NMR. Molecular structures of A) verapamil and E) AMPPNP with the nuclei labeled. A representative 1H STDD NMR spectra with B) 1 mM verapamil and F) 3.2 mM AMPPNP with 1 µM Pgp. The 1H STDD NMR amplification factors for C) 1 mM verapamil in the absence of AMPPNP with 1 µM Pgp. D) The STDD amplification factors for 1 mM verapamil in the presence of 0.25 mM (open columns) and 3.2 mM (solid columns) AMPPNP with 1 µM Pgp. E) Molecular structure of AMPPNP with the nuclei labeled F) A representative 1H proton STDD NMR spectrum of 3.2 mM AMMPNP with 1 µM Pgp. G) The STDD amplification factors of 0.25 mM (open columns) and 3.2 mM (solid columns) with 1 µM Pgp. H) The STDD amplification factors for 0.25 mM AMPPNP in the presence of 8 µM (open columns) and 1 mM verapamil (hashed columns) and 3.2 mM AMPPNP in the presence of 8 µM (gray columns) and 1 mM verapamil (solid columns) with 1 µM Pgp. STDD amplitudes in the figure with an asterisk were too small to be accurately measured.
2.5. Intrinsic Tryptophan Fluorescence
Fluorescent quenching of tryptophan residues has been used to measure the binding affinity of drugs and nucleotides to Pgp [31, 51, 52]. The quenching of protein fluorescence with the nucleotide AMPPNP and 1 µM Pgp reconstituted in liposomes was performed as described [31] in Chifflet buffer at pH 7.4. Pgp tryptophan residues were excited at 295 nm and the fluorescence emission maximum was measured at 333 nm on an Olis DM 45 spectrofluorimeter. The fluorescence was measured with an integration time of 0.8 s that produced an average maximum fluorescence signal at 333 nm of ~200,000. AMPPNP-induced fluorescent quenching was corrected (Fcorrected) for background fluorescence, dilution and inner filter effects with the following equation [53]:
| (4) |
where F is the measured protein fluorescence, B is the background and [Q] is the quenching ligand concentration. The extinction coefficients (ε) for excitation and emission are εex and εem, respectively. AMPPNP was essentially transparent above 300 nm and had a ε295 of 0.03 mM−1 cm−1 at 295 nm. The pathlength (b) along the excitation and emission axes are bex and bem, respectively.
Ligand-induced quenching of protein fluorescence can either have a static or a dynamic quenching mechanism. Fluorescence quenching caused by complexation of a ligand to a protein and related to a ligand’s affinity is known as a static quenching [54]. Alternatively, there are instances where random collisions between the ligand and the protein will induce fluorescence quenching by a mechanism known as dynamic quenching [53]. Regardless of the nature of the quenching, monophasic fluorescent quenching curves were fit to [53]:
| (5) |
where Fcorrected,0 is the protein fluorescence in the absence of a quenching ligand and K is the association constant (KA) or the Stern-Volmer quenching constant (KSV) in the case of a static and dynamic quenching processes, respectively. Curves that were biphasic were fit with [55]:
| (6) |
where F0,L and F0,H are the fluorescence amplitudes and KH and KL are the apparent equilibrium constants at low and high concentrations, respectively. The two different quenching mechanisms were differentiated by performing the fluorescence titration experiments at two different temperatures [53]. In the case of a dynamic quenching mechanism, the apparent K value will increase with increasing temperature as a result of increases in collisional frequency. For a static quenching process, the K value will decrease with increasing temperature as a result of decreases in the residence time of the ligand.
2.6. Acrylamide Quenching
Changes in the accessibility of tryptophan residues in proteins can be used as a tool to assess conformational changes upon ligand binding using the collisional quencher acrylamide [51, 56]. Acrylamide quenches intrinsic tryptophan fluorescence in a dynamic fashion and has been used to probe conformational changes of Pgp [51, 56, 57]. For these experiments, fluorescence emission with 1 µM Pgp reconstituted in liposomes was measured at 333 nm following excitation at 295 nm. Fluorescence intensities were corrected for with Equation 4 [53]. The Fcorrected,0/Fcorrected was plotted as a function of acrylamide concentration to produce the Stern-Volmer plots. The extent of dynamic tryptophan quenching was estimated from the slopes of the Stern-Volmer curves, which is related to KSV by Fcorrected,0/Fcorrected=1+KSV[Q] [53].
3. RESULTS
3.1. The effects of saturating and sub-saturating ATP on verapamil-induced Pgp-coupled ATPase activity
Drug-stimulated ATPase activity of Pgp is typically evaluated at saturating ATP concentrations e.g. [24, 58–62]. However, in vivo, intracellular ATP concentrations can vary widely from submillimolar levels up to 10 mM [63–66]. These values vary depending on cell and tissue type [67] and can fluctuate significantly in disease states [68, 69]. Intracellular ATP concentration can also affect cellular function [70–73]. Therefore, the effect of ATP concentration on verapamil-stimulated ATPase activity with Pgp was examined.
Fig. 1 shows the verapamil-induced activation of Pgp ATP hydrolysis in the presence of 3.2 mM (closed squares) and 0.25 mM (open squares) ATP. Kinetics of Pgp-coupled ATP hydrolysis in the presence of saturating ATP (Fig. 1, closed squares) were biphasic with substrate activation and inhibition reaching a maximum activation of 3–4 fold at 8 µM verapamil and decreasing to basal ATPase levels at saturating verapamil. Fitting the kinetics to the substrate inhibition equation (Equation 2) produced values for VMAX, Km, and Ki of 1623 ± 97 nmol min−1 mg−1, 1.9 ± 0.5 µM and 214 ± 52 µM, respectively, which are similar to previous estimates [31]. As suggested previously [31], this characteristic implies a high and a low affinity verapamil binding site on Pgp. Biphasic verapamil-induced ATP hydrolysis kinetics has been observed in hamster [25–27, 29], mouse [28, 31] and human [24] Pgp. Biphasic drug-induced ATPase activation kinetics is also a common feature of Pgp substrates including actinomycin D, paclitaxel, valinomycin, progesterone, quinidine and dipyridamole [12, 25, 74].
Surprisingly, when the verapamil-induced ATPase activity was measured at subsaturating ATP (Fig. 1, open squares) concentrations, the kinetics became almost monophasic and reached a maximum fold-activation close to that observed for Pgp at saturating ATP. Fitting the monophasic ATP hydrolysis kinetics to the Michaelis-Menten equation (Equation 2) gave a VMAX and Km of 233 ± 8 nmol min−1 mg−1 and 0.94 ± 0.2 µM, respectively. The relatively low VMAX is consistent with previous observations at low ATP [75] and the Km at this low ATP concentration is very similar to the Km at saturating ATP concentrations (Fig. 1, closed squares).
To uncover the relationship between ATP and verapamil, the ATP hydrolysis kinetics were measured with a range of verapamil and ATP concentrations and fit to the substrate inhibition equation (Equation 2). The Km and Ki from the substrate inhibition equation were previously attributed to a high and low affinity site on the transporter [31]. Figs. 1B and 1C show the effect of ATP concentration on the Km and Ki for verapamil stimulated Pgp ATPase activity, respectively. In Fig. 1B, the Km for verapamil over a range of ATP concentrations was essentially unaffected, which suggests that verapamil at the Km-associated binding site and ATP are not cooperative. However, the Ki for verapamil dramatically decreases with increasing ATP concentration (Fig. 1C). These results imply that verapamil at the Ki-associated binding site and ATP are cooperative. Fig. 1D shows the apparent VMAX as a function of ATP concentration. As expected [75], the apparent VMAX increased with increasing ATP concentration. Since these are only apparent kinetic parameters, further analysis is required to determine the discrete dissociation constants and the VMAXS for the individual verapamil and ATP bound states
3.2. Modeling Pgp-coupled ATPase activity with verapamil and a panel of ATP concentrations
Fitting the ATP hydrolysis kinetics with the substrate inhibition equation in Fig. 1 implied that there is a high affinity non-cooperative verapamil binding site and a low affinity cooperative verapamil binding site with ATP. To obtain the microscopic dissociation constants and VMAXS, verapamil and ATP were investigated by fitting the verapamil-induced ATP hydrolysis kinetics over a range of verapamil and ATP concentrations using COPASI.
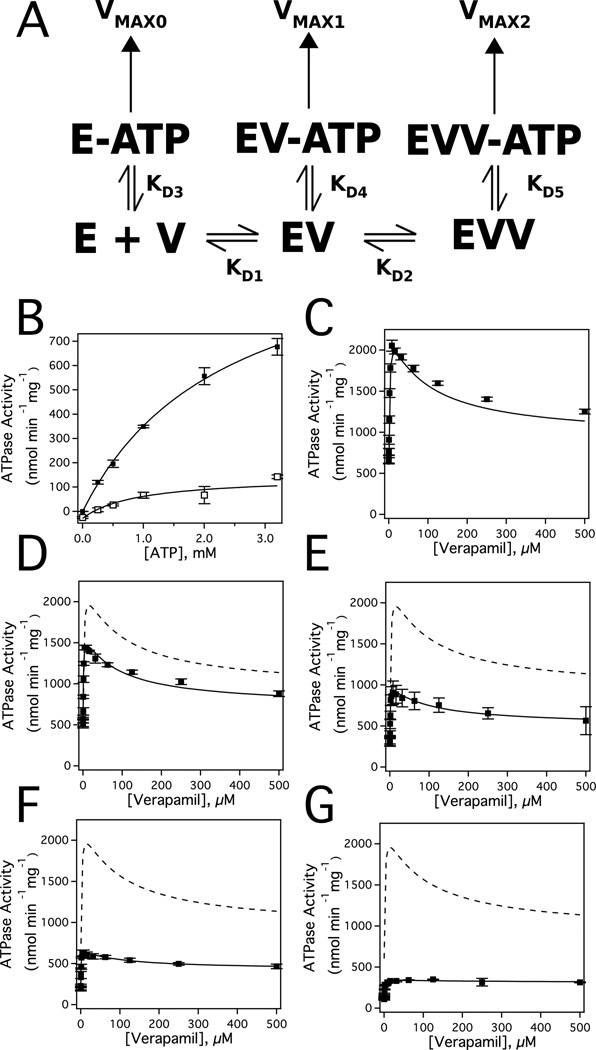
Fig. 2A shows the model used to fit the ATP hydrolysis kinetics and is the simplest that could fit all the data. The verapamil interactions leading to biphasic ATP hydrolysis kinetics in Fig. 1A (closed squares) is represented by EV and EVV for binding of one and two verapamil molecules, respectively. The affinity of verapamil to E and EV is defined by KD1 and KD2, respectively. Nucleotide binding to Pgp is shown as E-ATP, EV-ATP and EVV-ATP with affinities of KD3, KD4 and KD5, respectively. Pgp-mediated ATP hydrolysis from these states is defined by VMAX0, VMAX1 and VMAX2.
FIGURE 2.
Deconvoluting the Pgp-mediated ATP hydrolysis kinetics in the presence of verapamil and ATP. A) The verapamil-nucleotide cooperativity model used to fit the ATP hydrolysis kinetics. Horizontal and vertical left/right arrows denote the equilibria between bound states. The vertical arrows at the top of the panel represent the ATPase activity from the bound states. E and V correspond to Pgp and verapamil, respectively. B) Pgp-mediated ATP hydrolysis activity as a function of ATP concentration. Verapamil-induced activation of Pgp-mediated ATPase activity at C) 3.2, D) 2.0, E) 1.0, F) 0.5, and G) 0.25 mM ATP. For comparison, the kinetics in panel C at 3.2 mM ATP is shown as a dashed line in panels D through G. The error bars and the points represent the standard deviation and average, respectively, of three independent experiments.
Fig. 2B (closed squares) shows the effect of ATP concentration on the basal activity of Pgp in the absence of verapamil and has a hyperbolic shape; therefore, the curve was fit to the Michaelis-Menten equation (Equation 1). The fit gave a VMAX of 619 ± 18 nmol min−1 mg−1 and a Km of 879 ± 69 µM. In the presence of 200 µM of the Pgp inhibitor Na3VO4 and a range of ATP concentrations (Fig. 2B, open squares), the amplitude of the hyperbolic curve was relatively weak. Fitting the ATP hydrolysis kinetics with Equation 1 gives a VMAX of 172 ± 24 nmol min−1 mg−1 and Km of 906 ± 476 µM. The large decreases in Vmax, but no significant change in Km, demonstrate that Na3VO3 is a non-competitive inhibitor of Pgp-mediated ATP hydrolysis as has been previously noted [76]. The difference in VMAXS gives a Na3VO4 sensitive VMAX for Pgp of 447 ± 30 nmol min−1 mg−1. The fitted values are within range of VMAX and KmS observed in the literature of 500 to 2000 nmol min−1 mg−1 and 0.3–1 mM, respectively [27, 51, 59, 61, 77, 78].
Fig. 2C through 2G shows the verapamil-induced Pgp-coupled ATPase activity over a range of ATP concentrations starting at saturating ATP (3.2 mM) and decreasing to sub-saturating ATP (0.25 mM). These kinetics were fit using the COPASI software package. The average basal ATPase activity of Pgp (i.e. VMAX0) determined from fitting with COPASI was 672 ± 93 nmol min−1 mg−1 and the average KD3 for ATP in the absence of ligands was 1163 ± 110 µM. The average fitted KD1 and KD2 values for verapamil to E and EV were 1.11 ± 0.50 µM and 270 ± 45 µM, respectively, which was similar to previous estimates [31].
In the presence of a single bound verapamil molecule (i.e. EV), the average affinity of ATP was not significantly perturbed and had an average KD4 of 1242 ± 217 µM. This result suggests that verapamil and ATP are not cooperative with respect to binding within the EV-ATP complex, which is consistent with the lack of ATP concentration dependence for the verapamil Km in Fig. 1B. However, Pgp mediated ATP hydrolysis was activated about 4-fold in the presence of a single bound verapamil molecule to a VMAX1 of 2561 ± 295 nmol min−1 mg−1. Thus, verapamil and ATP are positively cooperative with respect to ATP hydrolysis within the EV-ATP complex. Non-competitive inhibitors have analogous effects with enzymes, except they inhibit enzyme activity [35] rather than activate it. Therefore, a single verapamil molecule bound to Pgp behaves like a “non-competitive activator” of Pgp.
In the presence of two bound verapamil molecules, the average ATP affinity increased ~4-fold with a decreased KD5 of 257 ± 44 µM. This correlates well with the decreased Ki observed at increasing ATP concentration that is shown in Fig. 1C. The increased affinity of ATP shows that verapamil and ATP are cooperative with respect to binding within the EVV-ATP complex. Fitting also revealed that there was more than a two-fold decrease in the maximum ATP hydrolysis rate with a VMAX2 of 760 ± 147 nmol min−1 mg−1. The decreased VMAX is in line with the reduced ATP hydrolysis activity that was observed at high verapamil concentration (Fig. 1A, closed squares). The decrease in VMAX shows that ATP and verapamil are negatively cooperative with respect to ATP hydrolysis within the EVV-ATP complex. A complete list of kinetic and thermodynamic parameters used to fit the curves is shown in Table 1. In all cases, the R-correlation values for the fits were greater than 0.988.
Table 1.
Kinetic and thermodynamic parameters and the corresponding averages used for fitting the ATPase activity curves with verapamil over a range of ATP concentrations in Figure 2.
| verapamil, µM | 0 – 500 | |||||
|---|---|---|---|---|---|---|
| ATP, mM | 3.2 | 2 | 1 | 0.5 | 0.25 | Average |
| KD1a | 1.93 | 1.00 | 0.89 | 0.61 | 1.12 | 1.11 ± 0.50 |
| KD2a | 299.7 | 299.7 | 249.8 | 200.0 | 300.0 | 269.8 ± 44.6 |
|
KD3a Vmax0b |
1129.6 817.5 |
1000.5 700.0 |
1205.7 608.6 |
1179.0 582.6 |
1299.3 653.1 |
1162.8 ± 109.7 672.4 ± 92.6 |
|
KD4a Vmax1b |
1000.0 3000.0 |
1026.8 2494.5 |
1342.1 2369.4 |
1488.6 2691.7 |
1352.1 2250.8 |
1241.9 ± 216.7 2561.3 ± 294.6 |
|
KD5a Vmax2b |
209.6 1000.0 |
209.6 800.0 |
283.1 644.6 |
284.8 669.5 |
299.9 685.5 |
257.4 ± 44.1 759.9 ± 146.8 |
| R- Correlation | 0.9885 | 0.9925 | 0.9941 | 0.9936 | 0.9949 | |
In units of µM
In units of nmol min−1 mg−1
3.3. The binding affinity of AMPPNP to Pgp by tryptophan fluorescence
The non-hydrolyzable ATP analog AMPPNP has been used as a surrogate for ATP to study nucleotide interactions with Pgp [51, 79–81]. The ATP analog is advantageous for investigating nucleotide interactions with Pgp because several analogous bacterial transporter X-ray crystal structures have been solved with it [10, 82, 83]. In addition, AMPPNP does not undergo ATP hydrolysis like other nucleotide analogs [84].
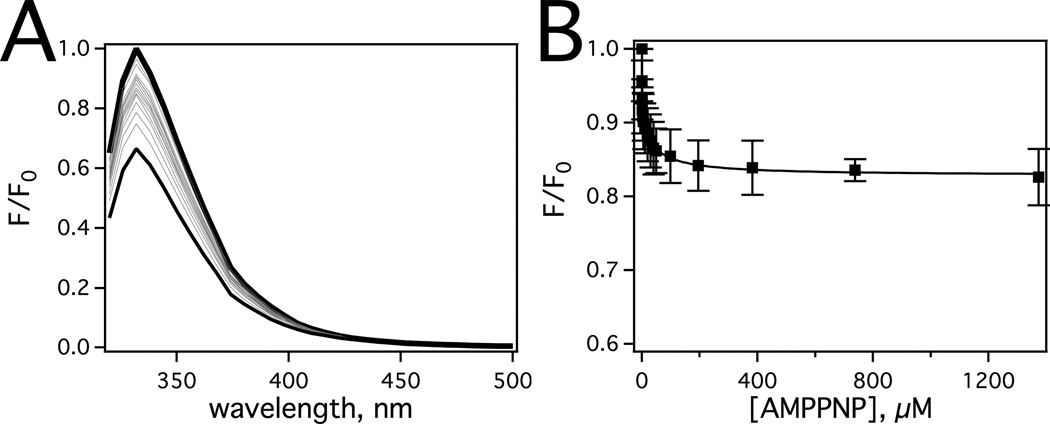
To ensure that saturating AMPPNP concentrations were used in this study, AMPPNP interactions were investigated by nucleotide-induced quenching of Pgp tryptophan fluorescence (Fig. 3). Fig. 3A shows the effect of a range of AMPPNP concentrations on the Pgp tryptophan fluorescence, which decreases about 15% at saturating AMPPNP concentration. The corrected fluorescence amplitude (i.e. Fcorrected) at 333 nm was plotted as a function of AMPPNP concentration in Fig. 3B. The quenching of tryptophan fluorescence was biphasic with a low and a high concentration phase, which was fit to equation 6 (Fig. 3B, solid line). The fraction quenched for the low (FL/F0) and high concentration (FH/F0) phases from the fit were 0.16 and 0.074, respectively. The K1 and K2 values extracted from the fit were 6.66 +/− 3.41 µM −1 and 0.022 +/− 0.006 µM −1, respectively. Ligand-induced quenching can occur by a static mechanism, which correlates to a ligand’s affinity, or by a dynamic mechanism [54]. To determine the quenching mechanism of each phase, the AMPPNP titration with Pgp was repeated at 37 °C as described [31, 53]. At this higher temperature, the K1 associated with the low concentration phase increased to 8 µM−1, while the K2 associated with the high concentration phase decreased to 0.017 µM−1 (data not shown). These results show that the low concentration-quenching phase resulted from dynamic quenching, while the high concentration-quenching phase resulted from static quenching. Therefore, the high concentration phase corresponds to a KD of 45 +/− 13 µM for AMPPNP. This KD value is considerably lower than AMPPNP binding to detergent-solubilized hamster Pgp [80], which likely represents differences between detergent-solubilized Pgp and Pgp reconstituted in liposomes in this study.
FIGURE 3.
Characterization of AMPPNP binding to Pgp by tryptophan fluorescence. A) Pgp fluorescence spectra in the presence of a range of AMPPNP concentrations after exciting at 295 nm. The spectrum at 0 µM and 1.4 mM AMPPNP are shown as thick and thin lines, respectively, while intermediate concentrations of AMPPNP are shown as gray lines. B) Corrected Pgp fluorescence emission at 333 nm as a function of AMPPNP concentration. The average and standard deviations are represented as points and bars, respectively, and reflect at least three independent experiments.
3.4. Verapamil and nucleotide-induced conformational changes of Pgp
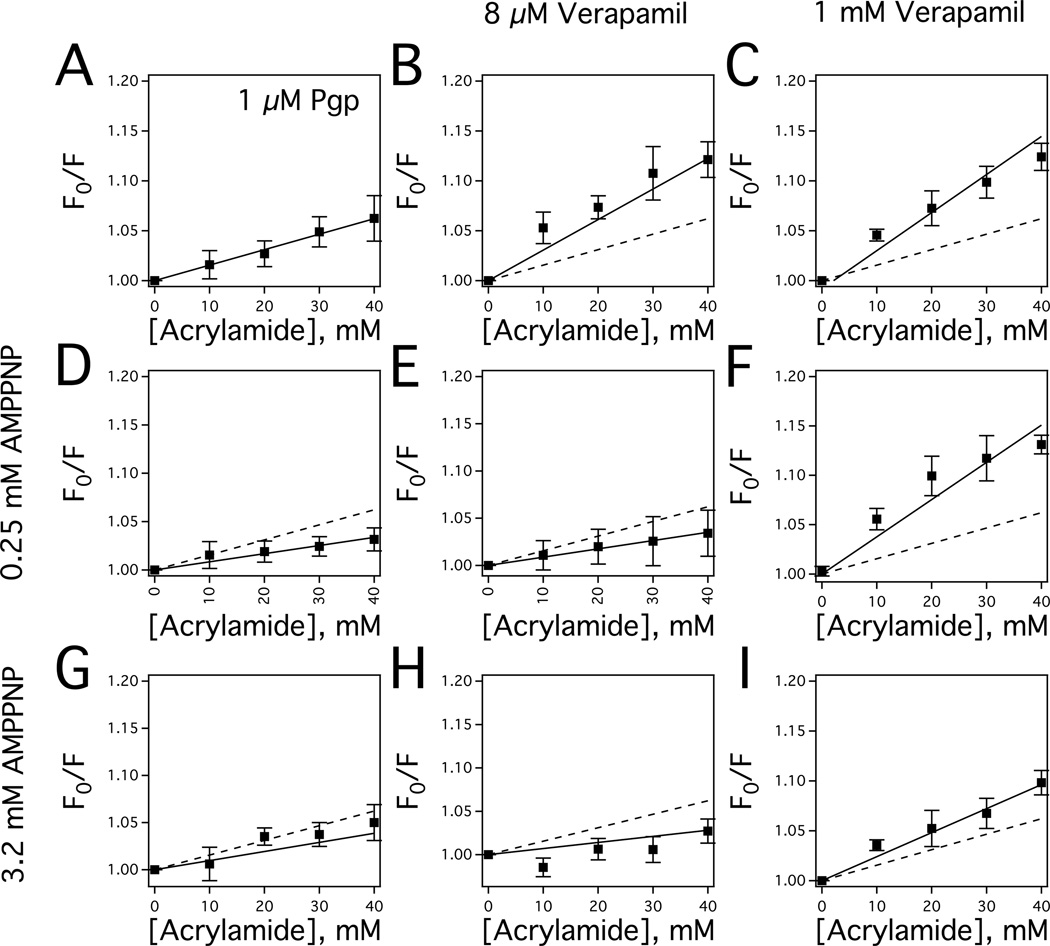
Acrylamide quenching of tryptophan fluorescence has been successfully used to probe changes in tertiary conformation of Pgp with a wide range of ligands and nucleotide cofactors [51, 56, 57, 80, 85]. In this study, this technique was performed with Pgp in the presence of verapamil and the non-hydrolyzable ATP analog AMPPNP to determine the effect of verapamil-nucleotide cooperativity on Pgp conformation.
Fig. 4 shows acrylamide quenching of Pgp with verapamil and AMPPNP represented as Stern-Volmer plots with the slopes reflecting the degree of tryptophan quenching. The Stern-Volmer plot for Pgp without ligands had a KSV value of 1.55 ± 0.04 M−1 (Fig. 4A, closed squares), which is similar to the KSV value for mouse [31] and hamster Pgp [80]. Significant verapamil-induced changes in the KSV values of Pgp in the absence of nucleotides were observed in the Stern-Volmer plots. In Fig. 4B, the addition of 8 µM verapamil increases the KSV value to 3.06 ± 0.21 M−1, while saturating verapamil concentrations (1 mM) increased the KSV value further to 3.81 ± 0.26 M−1 (Fig. 4C). The similarities in the KSV values suggest that their verapamil-bound Pgp conformations are similar, and that the largest conformational shift occurs at low verapamil concentration. This observation is consistent with verapamil-induced Pgp conformational changes deduced from crosslinking of [15, 16, 86], trypsin digestion of [87], and antibody competition with Pgp [88].
FIGURE 4.
Conformational changes of Pgp in the presence of AMPPNP and verapamil characterized by acrylamide quenching of Pgp tryptophan fluorescence. A) The Stern-Volmer plot of Pgp in the absence of drugs. The Stern-Volmer plots of Pgp in the presence of B) 8 µM and C) 1000 µM verapamil. D) Stern-Volmer plot of Pgp in the presence of 0.25 mM AMPPNP. The Stern-Volmer plots of Pgp in the presence of 3.2 mM AMPPNP and E) 8 µM and F) 1000 µM verapamil. G) Stern-Volmer plot of Pgp in the presence of 3.2 mM AMPPNP. The Stern-Volmer plots of Pgp in the presence of 3.2 mM AMPPNP and E) 8 µM and F) 1000 µM verapamil. For comparison, the slope in panel A is presented as a dashed line in panels B through F. The error bars and the points reflect the standard deviation and average, respectively, of at least three independent experiments.
In the presence of 250 µM AMPPNP (Fig. 4D), the KSV value from the slope of the Stern-Volmer plot for Pgp decreased from 1.55 M−1 to 0.84 ± 0.07 M−1, suggesting that the tryptophans became less accessible in the presence of the nucleotide analog and this is similar to the KSV value determined for hamster Pgp with a nucleotide analog [56]. The AMPPNP-induced changes in the KSV value in the presence of 8 µM verapamil were more dramatic in Fig. 4E. The KSV value decreased more than 4-fold from 3.81 M−1 to 0.88 ± 0.03 M−1. In contrast, the KSV value remained relatively constant in the presence of 1 mM verapamil and 250 µM AMPPNP (Fig. 4F) at 3.77 ± 0.33 M−1.
In the presence of saturating (3.2 mM) AMPPNP (Fig. 4G), the slope of the Stern-Volmer plot for Pgp remained relatively constant at 0.97 ± 0.16 M−1, when compared to the Stern-Volmer plot for Pgp with 250 µM AMPPNP in Fig. 4D. The slope of the Stern-Volmer plot also remained relatively constant with the addition of 8 µM verapamil with a KSV value of 0.70 ± 0.11 M−1 (Fig. 4H). In contrast, the KSV value decreased from 3.77 M−1 to 2.28 ± 0.21 M−1 in the presence of 1 mM verapamil and saturating AMPNP (Fig. 4I). The relatively modest AMPPNP-induced shift in KSV values in the presence of 1 mM verapamil, when compared to KSV value shifts with 8 µM verapamil, implying stabilization of the ligand-bound Pgp conformation.
3.5. Verapamil and AMPPNP interactions with Pgp probed by STDD NMR
The STDD NMR technique was used previously to deconvolute the interactions of verapamil and digoxin with Pgp and showed that verapamil competitively displaces digoxin [31]. In Fig. 5, the technique was used to investigate the cooperative molecular interactions of AMPPNP and verapamil with Pgp. To probe the high and low affinity verapamil sites on Pgp, we used 8 µM and 1 mM verapamil, respectively.
Fig. 5A shows the molecular structures of verapamil with the nuclei labeled. Structurally, verapamil has two dimethoxyphenyl groups that are connected together by an alkyl chain. The 1H STDD NMR spectrum of verapamil is shown with the 1H NMR peaks labeled in Fig. 5B. The STDD amplification factors for verapamil were also significantly larger than previously observed [31] as a result of using a higher frequency NMR spectrometer, which allowed us to investigate several weaker verapamil 1H NMR peaks labeled K, I, B, G and H (Fig. 5B). In the NMR spectrum, 1H NMR peaks emanating from the dimethoxyphenyl groups are located downfield above 3.5 ppm, while 1H NMR peaks associated with the connecting alkyl chain are found up field below 3.5 ppm. Protons in the dimethoxyphenyl group closest to the tertiary amine group are labeled M, N, O, and P and those farthest from the tertiary amine are labeled C, D, E, and F. The connecting alkyl chain between the dimethoxyphenyl functional groups consists of 1H NMR proton peaks labeled G, H, I, K, L and Q and a propyl group with protons labeled A’, A, and B.
Fig. 5C shows the Pgp specific 1H STDD amplification factors for 1 mM verapamil in the absence of AMPPNP after subtracting STD NMR contributions from the liposomes. Previously, we showed by STDD NMR that dimethoxyphenyl groups of verapamil were involved in direct interactions with Pgp [31]. The average STDD amplification factor for the dimethoxyphenyl functional groups of verapamil was 163 ± 12. The relative amplitudes of these STDD amplification factors were similar suggesting that all the protons of this functional group have similar interactions with Pgp. STDD NMR of Pgp and 8 µM verapamil was attempted to probe the high affinity verapamil binding site, but no verapamil 1H STDD NMR peaks were observed because of the low concentration and broadening of the verapamil 1H NMR peaks (data not shown). Therefore, the 1H STDD NMR amplitudes and amplification factors of 1 mM verapamil primarily reflect interaction with the low affinity verapamil binding site of Pgp.
Fig. 5D shows the effect of low (open columns) and high (closed columns) concentrations of AMPPNP on the 1H STDD amplification factors of 1 mM verapamil. At low AMPPNP concentration (Fig. 5D), the 1H STDD NMR amplification factors of the dimethoxyphenyl functional group decreased ~43% from the amplification factors in the absence of AMPPPNP (cf. Fig. 5C and Fig. 5D, open columns). There were also significant changes in 1H STDD amplification factors for the protons labeled A and A’. The 1H STDD amplification factor for the proton labeled A increased 13%, and the 1H STDD amplification factor for the proton labeled A’ decreased more than 40%. In the presence of saturating AMPPNP (Fig. 5D, closed columns), the 1H STDD amplification factors for the protons labeled A and A’ were not significantly different from the amplification factors at low AMPPNP concentration. However, the average STDD amplification factor of the dimethoxyphenyl functional group increased more than 2-fold (Fig. 5D, closed columns). There were also large increases in the 1H STDD amplification factors of alkyl protons labeled L and B. These differences in amplification factors between low and high AMPPNP concentration likely reflect differences in AMPPNP occupancy of Pgp. Overall, these AMPPNP-induced changes in the 1H STDD NMR amplification factors of verapamil suggest that nucleotides modulate verapamil interactions when verapamil occupies the low affinity binding site.
Fig. 5E shows the molecular structure of AMPPNP with the nuclei labeled, which is comprised of a ribose and a purine functional group. Fig. 5F shows the 1H STDD NMR spectrum of 3.2 mM with 1H NMR peaks labeled with significant 1H STDD NMR peaks for ribose and purine ring protons. Protons from the aromatic purine functional group are downfield from 6.0 ppm and are labeled g, f and e. The protons emanating from the ribose are upfield from 6.0 ppm and are labeled a, b, c and d.
Fig. 5G shows the 1H STDD NMR amplification factors for 250 µM and 3.2 mM AMPPNP concentration with 1 µM Pgp. At low AMPPNP concentration (open columns), we were not able to reliably estimate 1H STDD NMR amplification factors for the ribose protons (labeled a-d) because the 1H NMR amplitudes were very weak (data not shown). The 1H STDD NMR amplification factors for the aromatic purine protons were considerably stronger with proton f producing the strongest 1H STDD amplification factor. At high AMPPNP concentration (solid columns), the average STDD amplification factor for AMPPNP was 63 ± 51 (Fig. 6D, closed columns). The average STDD amplification factor for the purine base was 79 ± 64 with proton f exhibiting the strongest STDD amplification factor. In contrast, the STDD amplification factors were considerably weaker for the ribose sugar. The average STDD amplification factor for this group was 40 ± 11. These results suggest on average that stronger interactions occur between the purine base and Pgp than the ribose functional group. This is consistent with AMPPNP interactions observed for AMPPNP bound in the X-ray crystal structure of the analogous bacterial transporter SAV1866 [83] (PDB ID: 2ONJ). In the bacterial transporter structure, the purine ring of AMPPNP was sequestered by four residues (i.e. V476, K477, Y359 and Y391), while the ribose functional group interacted with a single I356.
FIGURE 6.
Model of drug-nucleotide cooperativity for verapamil-induced activation of Pgp-mediated ATP hydrolysis. The figure shows cartoons of Pgp in the “open”, “closed” and “intermediate” conformations in the A,C,E) absence and B,D,F) presence of ATP, and in the presence of C,D) one and E,F) two verapamil molecules. Verapamil molecules are shown as gray diamonds and N represents ATP. The proposed high and low affinity verapamil binding sites of Pgp are shown as H and L, respectively. The horizontal arrows reflect equilibria between the verapamil-bound conformational states of Pgp. The middle vertical arrows reflect the degree of ATP binding to Pgp, while the arrows at the top of the figure and their size represent the degree of verapamil transport.
In Fig. 5H, the STDD amplification factors for 250 µM and 3.2 mM AMPPNP concentration are shown in the presence of 8 µM or 1 mM concentrations of verapamil. Because of the weak 1H NMR amplitudes for ribose at low AMPPNP concentration (open and hashed columns), only 1H STDD NMR amplification factors were measured for the purine base. The relative amplitudes of the amplification factors were similar to the amplification factors of 250 µM AMPPNP shown in Fig. 5G (open columns) with the strongest amplification factors emanating from proton f. At high AMPPNP concentration and 8 µM verapamil (gray columns), the average STDD amplification factor decreased 15%. The relative amplitudes of the STDD amplification factors remained relatively unperturbed with respect to the STDD amplification factors in the absence verapamil (cf. Fig. 2G, closed columns and Fig. 2H, gray columns), which is consistent with the lack of verapamil-induced effects on the Km (Fig. 1B) and the KD (Fig. 2). In the presence of saturating 1 mM verapamil (closed columns), the average 1H STDD amplification factors of AMPPNP increased more than 20%. As observed under other conditions in this study, the purine protons had larger STDD amplification factors on average than the ribose protons. A notable difference in the 1H STDD amplification factors of AMPPNP with saturating 1 mM verapamil is that the AMPPNP ribose proton labeled d was more than double the other STDD amplification factors observed under the other conditions. This implies that the Pgp-bound orientation of the ribose functional group of AMPPNP is perturbed in the presence of 1 mM verapamil and might be a contributing factor to the decreased Pgp-mediated ATP hydrolysis at high verapamil concentrations. Interestingly, of the non-exchangeable protons in AMPPNP, this proton is positioned closest to I356 in the AMPPNP-bound X-ray crystal structure of SAV1866 (PDB ID: 2ONJ) [83] that sequesters the ribose.
4. DISCUSSION
A conformationally-gated model of cooperativity between verapamil and nucleotides is presented in Fig. 6. The model is based on our results and the conformational changes that Pgp is known to undergo in the presence of nucleotides and drugs [10, 15, 16, 18, 83]. The model is simply presented by three conformations: “open”, “closed” and “intermediate” as described previously [31]. In reality, the model represents a structural ensemble of Pgp conformations. When the nucleotide binding domains (NBDs) are relatively far apart, Pgp is in the “open” conformation where the cytosolic side is exposed to the bulk solvent. When the NBDs come together, Pgp is in the “closed” conformation and the extracellular side is exposed to the bulk solvent. Instances where the extracellular and cytosolic sides are equally exposed to bulk solvent is the “intermediate” Pgp conformation, which is in-between the “open” and “closed” states.
Ligand-induced changes in tryptophan accessibility deduced from the acrylamide quenching experiments of Pgp with verapamil and the non-hydrolyzable ATP analog AMPPNP in Fig. 4 suggest that their interactions significantly affect the tertiary conformation of Pgp. Ligand-induced decreases in KSV values of Pgp reflect decreases in tryptophan accessibility and imply shifts toward a “closed” Pgp conformation. The similarity of KSV values of Pgp in the presence of low and high verapamil concentration suggest that the verapamil-bound conformations are similar. This information should be used cautiously to assign a specific drug- or nucleotide-bound Pgp conformations, since the fluorescence quenching studies were done with a non-hydrolyzable ATP analog rather than ATP. Therefore, the conformational assignment was additionally weighted on the rates of Pgp-mediated ATP hydrolysis. Our rational was based on the fact that cross-linking studies with Pgp, site-directed mutagenesis of Pgp, cryo-electron microscopy studies with Pgp and structural studies with bacterial transporters with nucleotide analogs have shown that the interaction of the Pgp nucleotide domains with each other is essential for ATP hydrolysis [89–94]. In our model, the average distance between the Pgp NBDs was correlated to the ATP hydrolysis rate. For example, Pgp that has relatively high and low ATP hydrolysis rates is assumed to be in “closed” and “open” conformations, respectively. The ligand-induced changes in tryptophan accessibility of Pgp deduced from acrylamide quenching of tryptophan fluorescence were used to gauge the degree of conformational shift.
The locations of the verapamil binding sites on Pgp have not yet been resolved. Verapamil ATP hydrolysis kinetics is biphasic, which suggests a high and low affinity verapamil-binding site on Pgp as previously reported [32] and shown in Figs. 1 and 2. Several studies of residues within the extracellular side of Pgp [58, 95–97] and G185 (G181 for Pgp in this study) within the transmembrane region of Pgp [98–100] had significant effects on verapamil-induced ATPase kinetics and Pgp-mediated transport of verapamil. Several mutations of residues on the extracellular side of Pgp caused marked decreases in transport of a fluorescent verapamil analog [58]. Dramatic decreases in the Km for ATPase activation of Pgp by verapamil were observed when residues between 78 and 97 were deleted [95]. Labeling an I306C mutant, which is located on the extracellular side of Pgp, with a thiol-reactive verapamil analog lead to permanent verapamil-induced activation of Pgp [96]. Verapamil also inhibited crosslinking between residues C332 and C975 within the extracellular side of Pgp [97]. Mutating the G181 residue to valine within the transmembrane region of Pgp had strong effects on the VMAX of verapamil-induced ATPase activation [98–100]. The G181V mutation had little effect on the Km of verapamil-induced ATPase activation [98, 99], but a recent study showed that it had significant effects on the Ki for substrate inhibition of Pgp [99]. By affecting the Ki from substrate inhibition and not the Km implies that G181 is near an alternate verapamil-binding site. Therefore, we placed the low-affinity (L) verapamil-binding site near G181 in the transmembrane region of Pgp and the high-affinity (H) verapamil-binding site near the extracellular side of Pgp.
The conformation of Pgp in the absence of ligands is known from the X-ray crystal structure to be in the “open” conformation [9] as shown in Fig. 6A. Acrylamide quenching experiments in Fig. 4 showed that nucleotides reduced tryptophan accessibility of Pgp implying a shift toward the “closed” conformation. However, the VMAX deduced from fitting the Pgp-coupled ATPase activation kinetics was inefficient at 500–700 nmol min−1 mg−1. Therefore, we propose that ATP shifts Pgp to an “intermediate” conformation in Fig. 6B, which keeps the NBDs apart and presents an energetic barrier for ATP hydrolysis.
At low verapamil concentrations, the drug binds near the extracellular side of Pgp in our model (Figs. 6C and D) at the H-site. Because the rate of Pgp-mediated ATP hydrolysis was very high at ~3000 nmol min−1 mg−1, we propose that verapamil-bound Pgp is in the “closed” conformation in the presence of ATP in Fig. 6D. In the absence of nucleotides, the acrylamide quenching experiments in Fig. 4 suggested that low concentrations of verapamil induce significant changes in the tertiary conformation of Pgp. There were also significant differences in tryptophan accessibility in the absence (Fig. 4B) and presence (Figs. 4E and H) of AMPPNP at low verapamil concentration suggesting that they are in very distinct conformations. Therefore, we propose that verapamil bound at the H-site of Pgp, but in the absence of ATP, is in an “intermediate” conformation. By shifting to an “intermediate” conformation, the verapamil molecule reduces the energetic barrier for the NBDs to come together and the subsequent Pgp-mediated ATP hydrolysis and transport that follows. By reducing the ATP energetic barrier for hydrolysis, this single bound verapamil molecule is positively cooperative with respect to ATP hydrolysis. With respect to binding, verapamil and ATP are essentially not cooperative as deduced by fitting of the verapamil-induced ATPase activation kinetics in Figs. 1 and 2. The lack of binding cooperativity is consistent with the H-site being far from the NBDs.
At saturating verapamil concentrations, the drug will bind near the L-site that we hypothesize lies within the transmembrane region near G185 (G181 for the Pgp in this study). The activation of Pgp-mediated ATP hydrolysis decreased to about ~700 nmol min−1 mg−1, which was a little bit higher than basal ATPase activity. As deduced from the slopes of the Stern-Volmer plots in Figs. 4B and C, tryptophan accessibility of Pgp in the absence of an ATP analog was just slightly higher at saturating verapamil concentration than low verapamil concentration implying that they have relatively similar conformations. Therefore, Pgp is shown in an “intermediate” conformation in the absence of ATP at low and saturating verapamil concentrations (Figs. 6C and E). Fitting of the ATP hydrolysis kinetics in Fig. 2 revealed that the ATP affinity to Pgp increased 4-fold in the presence of saturating verapamil and that ATP and verapamil binding to Pgp is positively cooperative at saturating verapamil concentrations. Also, addition of an ATP analog only induced a relatively small decrease in tryptophan accessibility in the presence of saturating verapamil (cf. Figs. 4C and 4I) implying that Pgp’s tertiary conformation was not significantly shifted by the ATP analog, when compared to the nucleotide-induced conformational change of Pgp in the presence of low concentrations of verapamil (cf. Figs. 4B and 4I). Therefore, we hypothesize that binding cooperativity between verapamil and ATP stabilizes Pgp in an “intermediate” conformation shown in Fig. 7E and 7F. With the NBDs relatively separated in the “intermediate” conformation compared to the “closed” conformation explains why ATP hydrolysis is lower at high verapamil concentration and is similar to the basal ATP hydrolysis rate. In other words, two Pgp-bound verapamil molecules are negatively cooperative with respect to ATP hydrolysis.
In Figs. 6D and F of the model, verapamil transport rates are shown to correlate to verapamil occupancy of Pgp and verapamil-stimulated ATP hydrolysis rates. This relationship is supported by a study with spin-labeled verapamil and Pgp that found similar Km and Ki values for verapamil transport and verapamil-stimulated ATPase activity [101]. This correlation is also supported by several independent studies that have shown that Pgp-mediated transport of verapamil [102–105] and verapamil-simulated ATP hydrolysis decreased at higher verapamil concentrations [24, 28, 29, 31].
In summary, Fig. 6 shows a model for Pgp-mediated verapamil transport involving conformational changes and cooperativity between verapamil and ATP. In the absence of verapamil, Pgp is in an “open” conformation (Fig. 6A), and is shifted to an “intermediate” conformation by ATP (Fig. 6B). In this conformation, the ATPase activity will be low because the separation in NBDs represents an energetic barrier for ATP hydrolysis. At low concentrations, verapamil will occupy the H-site on Pgp and will shift the transporter into an “intermediate” conformation in Fig. 6C. Since the H-site is within the extracellular domain far from the NBDs, verapamil occupancy at the H-site will not significantly affect the binding affinity of ATP. In other words, verapamil and ATP are not cooperative with respect to binding. However, because Pgp is an “intermediate” rather than an “open” conformation, verapamil reduces the energetic barrier for ATP hydrolysis and transport (Fig. 6D). Therefore, verapamil occupancy at the H-site is positively cooperative with respect to ATP hydrolysis and transport.
At high concentrations, verapamil will occupy both the H and L sites on Pgp as shown in Fig. 6E. Occupancy at both sites of Pgp also shifts Pgp into an “intermediate” conformation. In contrast with the H-site, the L-site on Pgp is located within the transmembrane region closer to the NBDs. Verapamil binding to the L-site increases the affinity of ATP to the NBDs. In this case, verapamil and ATP are positively cooperative with respect to binding to Pgp. Occupancy of verapamil at both sites stabilizes the “intermediate” Pgp conformation and prevents ATP from driving Pgp to the “closed” conformation in Fig. 6F. This reduces verapamil transport and verapamil-stimulated ATP hydrolysis by Pgp. In this case, verapamil occupancy at both sites on Pgp is negatively cooperative with respect to ATP hydrolysis and transport.
Acknowledgments
We would like to give special thanks Dr. Ina L. Urbatsch of Texas Tech University Health Sciences Center for her generous gift of Pichia (P.) pastoris with the wild-type mouse Pgp transporter gene. We would also like to thank her postdoc Dr. Douglas J. Swartz for sending us the materials and providing protocols for genetically manipulating and purifying Pgp from P. pastoris for our laboratory. Without their contribution and generosity, this research would not be possible. This work was funded and supported by the American Heart Association Grant (14GRNT20450044) and the National Institute of Health R15 Area Grant (1R15GM107913-01A1).
ABREVIATIONS
- AMPPNP
adenosine 5’-(β,γ-imido)triphosphate
- COPASI
complex pathway simulator
- DDM
n-dodecyl-β-D-maltoside
- F0,H
initial fluorescence intensity at the high concentration phase
- F0,L
initial fluorescence intensity at the low concentration phase
- KA
association constant
- KD
dissociation constant
- KH
equilibrium constant at high concentration
- KL
equilibrium constant at low concentration
- KSV
Stern-Volmer quenching constant
- L
ligand
- NTA
nickel-nitrilotriacetic acid
- PAGE
polyacrylamide gel electrophoresis
- Pgp
P-glycoprotein
- Pi
inorganic phosphate
- RF
radio frequency
- SDS
sodium dodecylsulfate
- Q
quenching ligand
- STD
saturation transfer difference
- STDD
saturation transfer double difference
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Chemical compounds studied in this article
verapamil hydrochloride (PubChem CID: 62969)
adenosine 5’-( β,γ-imido)triphosphate (AMPPNP, PubChem CID: 33113)
CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest with the contents of this article.
AUTHOR CONTRIBUTIONS
AGR and KVL conceived and coordinated the study and wrote the paper. AGR and KVL designed the experiments for all the figures. KVL prepared the Figures. KVL performed and analyzed the experiments for all the figures. RWB collected fluorescence data for Figure 3. MEG collected fluorescence data for Figure 5. MEG also did some of the proteoliposome preparations for these studies. All the authors approved the final version of the manuscript.
REFERENCES
- 1.Boumendjel An, Boutonnat J, Robert J. ABC transporters and multidrug resistance. Hoboken, N.J: John Wiley & Sons; 2009. [Google Scholar]
- 2.Zhou SF. Structure, function and regulation of P-glycoprotein and its clinical relevance in drug disposition. Xenobiotica. 2008;38:802–832. doi: 10.1080/00498250701867889. [DOI] [PubMed] [Google Scholar]
- 3.Lum BL, Gosland MP. MDR expression in normal tissues. Pharmacologic implications for the clinical use of P-glycoprotein inhibitors. Hematol. Oncol. Clin. North Am. 1995;9:319–336. [PubMed] [Google Scholar]
- 4.Ceckova-Novotna M, Pavek P, Staud F. P-glycoprotein in the placenta: expression, localization, regulation and function. Reprod. Toxicol. 2006;22:400–410. doi: 10.1016/j.reprotox.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Meissner K, Sperker B, Karsten C, MeyerZuSchwabedissen H, Seeland U, Bohm M, Bien S, Dazert P, Kunert-Keil C, Vogelgesang S, Warzok R, Siegmund W, Cascorbi I, Wendt M, Kroemer HK. Expression and localization of P-glycoprotein in human heart: effects of cardiomyopathy. J. Histochem. Cytochem. 2002;50:1351–1356. doi: 10.1177/002215540205001008. [DOI] [PubMed] [Google Scholar]
- 6.Cascorbi I, Paul M, Kroemer HK. Pharmacogenomics of heart failure -- focus on drug disposition and action. Cardiovasc. Res. 2004;64:32–39. doi: 10.1016/j.cardiores.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Ramakrishnan P. The role of p-glycoprotein in the blood-brain barrier. Einstein Q. J. Biol. Med. 2003;19:160–165. [Google Scholar]
- 8.Owens J. Drug resistance: Passing on protection. Nat. Rev. Drug Discov. 2005;4:191. [Google Scholar]
- 9.Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc. Natl. Acad. Sci. U. S. A. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawson RJ, Hollenstein K, Locher KP. Uptake or extrusion: crystal structures of full ABC transporters suggest a common mechanism. Mol. Microbiol. 2007;65:250–257. doi: 10.1111/j.1365-2958.2007.05792.x. [DOI] [PubMed] [Google Scholar]
- 12.Jin MS, Oldham ML, Zhang Q, Chen J. Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature. 2012;490:566–569. doi: 10.1038/nature11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hohl M, Hurlimann LM, Bohm S, Schoppe J, Grutter MG, Bordignon E, Seeger MA. Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter. Proc. Natl. Acad. Sci. U. S. A. 2014;111:11025–11030. doi: 10.1073/pnas.1400485111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Becker JP, Depret G, Van Bambeke F, Tulkens PM, Prevost M. Molecular models of human P-glycoprotein in two different catalytic states. BMC Struct. Biol. 2009;9:3. doi: 10.1186/1472-6807-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loo TW, Bartlett MC, Clarke DM. Drug binding in human P-glycoprotein causes conformational changes in both nucleotide-binding domains. J. Biol. Chem. 2003;278:1575–1578. doi: 10.1074/jbc.M211307200. [DOI] [PubMed] [Google Scholar]
- 16.Loo TW, Bartlett MC, Clarke DM. Substrate-induced conformational changes in the transmembrane segments of human P-glycoprotein. Direct evidence for the substrate-induced fit mechanism for drug binding. J. Biol. Chem. 2003;278:13603–13606. doi: 10.1074/jbc.C300073200. [DOI] [PubMed] [Google Scholar]
- 17.Ritchie TK, Kwon H, Atkins WM. Conformational analysis of human ATP-binding cassette transporter ABCB1 in lipid nanodiscs and inhibition by the antibodies MRK16 and UIC2. J. Biol. Chem. 2011;286:39489–39496. doi: 10.1074/jbc.M111.284554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verhalen B, Ernst S, Borsch M, Wilkens S. Dynamic ligand-induced conformational rearrangements in P-glycoprotein as probed by fluorescence resonance energy transfer spectroscopy. J. Biol. Chem. 2012;287:1112–1127. doi: 10.1074/jbc.M111.301192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleckenstein A. Specific pharmacology of calcium in myocardium, cardiac pacemakers, and vascular smooth muscle. Annu. Rev. Pharmacol. Toxicol. 1977;17:149–166. doi: 10.1146/annurev.pa.17.040177.001053. [DOI] [PubMed] [Google Scholar]
- 20.Gould BA, Mann S, Kieso H, Subramanian VB, Raftery EB. The 24-hour ambulatory blood pressure profile with verapamil. Circulation. 1982;65:22–27. doi: 10.1161/01.cir.65.1.22. [DOI] [PubMed] [Google Scholar]
- 21.Lewis GR, Morley KD, Maslowski AH, Bones PJ. Verapamil in the management of hypertensive patients. Aust. N. Z. J. Med. 1979;9:62–64. doi: 10.1111/j.1445-5994.1979.tb04115.x. [DOI] [PubMed] [Google Scholar]
- 22.Neugebauer G. Comparative cardiovascular actions of verapamil and its major metabolites in the anaesthetised dog. Cardiovasc. Res. 1978;12:247–254. doi: 10.1093/cvr/12.4.247. [DOI] [PubMed] [Google Scholar]
- 23.Wessler JD, Grip LT, Mendell J, Giugliano RP. The P-glycoprotein transport system and cardiovascular drugs. J. Am. Coll. Cardiol. 2013;61:2495–2502. doi: 10.1016/j.jacc.2013.02.058. [DOI] [PubMed] [Google Scholar]
- 24.Aanismaa P, Seelig A. P-Glycoprotein kinetics measured in plasma membrane vesicles and living cells. Biochemistry. 2007;46:3394–3404. doi: 10.1021/bi0619526. [DOI] [PubMed] [Google Scholar]
- 25.Litman T, Zeuthen T, Skovsgaard T, Stein WD. Structure-activity relationships of P-glycoprotein interacting drugs: kinetic characterization of their effects on ATPase activity. Biochim. Biophys. Acta. 1997;1361:159–168. doi: 10.1016/s0925-4439(97)00026-4. [DOI] [PubMed] [Google Scholar]
- 26.Orlowski S, Mir LM, Belehradek J, Jr, Garrigos M. Effects of steroids and verapamil on P-glycoprotein ATPase activity: progesterone, desoxycorticosterone, corticosterone and verapamil are mutually non-exclusive modulators. Biochem. J. 1996;317(Pt 2):515–522. doi: 10.1042/bj3170515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharom FJ, Yu X, Chu JW, Doige CA. Characterization of the ATPase activity of P-glycoprotein from multidrug-resistant Chinese hamster ovary cells. Biochem. J. 1995;308(Pt 2):381–390. doi: 10.1042/bj3080381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Litman T, Nielsen D, Skovsgaard T, Zeuthen T, Stein WD. ATPase activity of P-glycoprotein related to emergence of drug resistance in Ehrlich ascites tumor cell lines. Biochim. Biophys. Acta. 1997;1361:147–158. doi: 10.1016/s0925-4439(97)00025-2. [DOI] [PubMed] [Google Scholar]
- 29.Borgnia MJ, Eytan GD, Assaraf YG. Competition of hydrophobic peptides, cytotoxic drugs, and chemosensitizers on a common P-glycoprotein pharmacophore as revealed by its ATPase activity. J. Biol. Chem. 1996;271:3163–3171. doi: 10.1074/jbc.271.6.3163. [DOI] [PubMed] [Google Scholar]
- 30.Bai J, Swartz DJ, Protasevich II, Brouillette CG, Harrell PM, Hildebrandt E, Gasser B, Mattanovich D, Ward A, Chang G, Urbatsch IL. A gene optimization strategy that enhances production of fully functional P-glycoprotein in Pichia pastoris. PLoS One. 2011;6:e22577. doi: 10.1371/journal.pone.0022577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ledwitch KV, Barnes RW, Roberts AG. Unraveling the Complex Drug-drug Interactions of the Cardiovascular Drugs, Verapamil and Digoxin, with P-glycoprotein. Biosci. Rep. 2016;36 doi: 10.1042/BSR20150317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lerner-Marmarosh N, Gimi K, Urbatsch IL, Gros P, Senior AE. Large scale purification of detergent-soluble P-glycoprotein from Pichia pastoris cells and characterization of nucleotide binding properties of wild-type, Walker A, and Walker B mutant proteins. J. Biol. Chem. 1999;274:34711–34718. doi: 10.1074/jbc.274.49.34711. [DOI] [PubMed] [Google Scholar]
- 33.Chifflet S, Torriglia A, Chiesa R, Tolosa S. A method for the determination of inorganic phosphate in the presence of labile organic phosphate and high concentrations of protein: application to lens ATPases. Anal. Biochem. 1988;168:1–4. doi: 10.1016/0003-2697(88)90002-4. [DOI] [PubMed] [Google Scholar]
- 34.Cook PF, Cleland WW. Enzyme kinetics and mechanism, Garland Science. London; New York: 2007. [Google Scholar]
- 35.Segel IH. Enzyme kinetics : behavior and analysis of rapid equilibrium and steady state enzyme systems. New York: Wiley; 1975. [Google Scholar]
- 36.Martin RB. Disadvantages of Double Reciprocal Plots. J. Chem. Educ. 1997;74:1238. [Google Scholar]
- 37.Leatherbarrow RJ. Using linear and non-linear regression to fit biochemical data. Trends Biochem. Sci. 1990;15:455–458. doi: 10.1016/0968-0004(90)90295-m. [DOI] [PubMed] [Google Scholar]
- 38.Ranaldi F, Vanni P, Giachetti E. What students must know about the determination of enzyme kinetic parameters. Biochem. Educ. 1999;27:87–91. [Google Scholar]
- 39.Roberts AG, Yang J, Halpert JR, Nelson SD, Thummel KT, Atkins WM. The structural basis for homotropic and heterotropic cooperativity of midazolam metabolism by human cytochrome P450 3A4. Biochemistry. 2011;50:10804–10818. doi: 10.1021/bi200924t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davydov DR, Botchkareva AE, Davydova NE, Halpert JR. Resolution of Two Substrate-Binding Sites in an Engineered Cytochrome P450eryF Bearing a Fluorescent Probe. Biophys. J. 2005;89:418–432. doi: 10.1529/biophysj.104.058479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoops S, Sahle S, Gauges R, Lee C, Pahle J, Simus N, Singhal M, Xu L, Mendes P, Kummer U. COPASI--a COmplex PAthway SImulator. Bioinformatics. 2006;22:3067–3074. doi: 10.1093/bioinformatics/btl485. [DOI] [PubMed] [Google Scholar]
- 42.Claasen B, Axmann M, Meinecke R, Meyer B. Direct observation of ligand binding to membrane proteins in living cells by a saturation transfer double difference (STDD) NMR spectroscopy method shows a significantly higher affinity of integrin alpha(IIb)beta3 in native platelets than in liposomes. J. Am. Chem. Soc. 2005;127:916–999. doi: 10.1021/ja044434w. [DOI] [PubMed] [Google Scholar]
- 43.Venkitakrishnan RP, Benard O, Max M, Markley JL, Assadi-Porter FM. Use of NMR Saturation Transfer Difference Spectroscopy to Study Ligand Binding to Membrane Proteins. In: Vaidehi N, Klein-Seetharaman J, editors. Membrane Protein Structure and Dynamics: Methods and Protocols. Totowa, NJ: Humana Press; 2012. pp. 47–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayer M, Meyer B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J. Am. Chem. Soc. 2001;123:6108–6117. doi: 10.1021/ja0100120. [DOI] [PubMed] [Google Scholar]
- 45.Haselhorst T, Munster-Kuhnel AK, Oschlies M, Tiralongo J, Gerardy-Schahn R, von Itzstein M. Direct detection of ligand binding to Sepharose-immobilised protein using saturation transfer double difference (STDD) NMR spectroscopy. Biochem. Biophys. Res. Commun. 2007;359:866–870. doi: 10.1016/j.bbrc.2007.05.204. [DOI] [PubMed] [Google Scholar]
- 46.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 47.Horie A, Ishida K, Shibata K, Taguchi M, Ozawa A, Hirono K, Ichida F, Hashimoto Y. Pharmacokinetic variability of flecainide in younger Japanese patients and mechanisms for renal excretion and intestinal absorption. Biopharm. Drug Dispos. 2014;35:145–153. doi: 10.1002/bdd.1877. [DOI] [PubMed] [Google Scholar]
- 48.Maccotta A, Scibona G, Valensin G, Gaggelli E, Botre F, Botre C. Nuclear magnetic resonance investigations of calcium antagonist drugs. II: Conformational and dynamic features of verapamil in [2H6]DMSO. J. Pharm. Sci. 1991;80:586–589. doi: 10.1002/jps.2600800618. [DOI] [PubMed] [Google Scholar]
- 49.Tetreault S, Ananthanarayanan VS. Interaction of calcium channel antagonists with calcium: structural studies on verapamil and its Ca2+ complex. J. Med. Chem. 1993;36:1017–1023. doi: 10.1021/jm00060a009. [DOI] [PubMed] [Google Scholar]
- 50.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, Nakatani E, Schulte CF, Tolmie DE, Kent Wenger R, Yao H, Markley JL. BioMagResBank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu R, Siemiarczuk A, Sharom FJ. Intrinsic fluorescence of the P-glycoprotein multidrug transporter: sensitivity of tryptophan residues to binding of drugs and nucleotides. Biochemistry. 2000;39:14927–14938. doi: 10.1021/bi0018786. [DOI] [PubMed] [Google Scholar]
- 52.Sharom FJ, Russell PL, Qu Q, Lu P. Fluorescence techniques for studying membrane transport proteins: the P-glycoprotein multidrug transporter. Methods Mol. Biol. 2003;227:109–128. doi: 10.1385/1-59259-387-9:109. [DOI] [PubMed] [Google Scholar]
- 53.Lakowicz JR. Principles of fluorescence spectroscopy. 2nd. New York: Kluwer Academic/Plenum; 1999. [Google Scholar]
- 54.Lakowicz JR. Principles of fluorescence spectroscopy. New York: Plenum Press; 1983. [Google Scholar]
- 55.Doppenschmitt S, Spahn-Langguth H, Regardh CG, Langguth P. Radioligand-binding assay employing P-glycoprotein-overexpressing cells: testing drug affinities to the secretory intestinal multidrug transporter. Pharm. Res. 1998;15:1001–1006. doi: 10.1023/a:1011965707998. [DOI] [PubMed] [Google Scholar]
- 56.Sonveaux N, Vigano C, Shapiro AB, Ling V, Ruysschaert JM. Ligand-mediated tertiary structure changes of reconstituted P-glycoprotein. A tryptophan fluorescence quenching analysis. J. Biol. Chem. 1999;274:17649–17654. doi: 10.1074/jbc.274.25.17649. [DOI] [PubMed] [Google Scholar]
- 57.Russell PL, Sharom FJ. Conformational and functional characterization of trapped complexes of the P-glycoprotein multidrug transporter. Biochem. J. 2006;399:315–323. doi: 10.1042/BJ20060015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hafkemeyer P, Dey S, Ambudkar SV, Hrycyna CA, Pastan I, Gottesman MM. Contribution to substrate specificity and transport of nonconserved residues in transmembrane domain 12 of human P-glycoprotein. Biochemistry. 1998;37:16400–16409. doi: 10.1021/bi980871+. [DOI] [PubMed] [Google Scholar]
- 59.Kerr KM, Sauna ZE, Ambudkar SV. Correlation between steady-state ATP hydrolysis and vanadate-induced ADP trapping in Human P-glycoprotein. Evidence for ADP release as the rate-limiting step in the catalytic cycle and its modulation by substrates. J. Biol. Chem. 2001;276:8657–8664. doi: 10.1074/jbc.M010044200. [DOI] [PubMed] [Google Scholar]
- 60.Loo TW, Clarke DM. Drug-stimulated ATPase Activity of Human P-glycoprotein Requires Movement between Transmembrane Segments 6 and 12. J. Biol. Chem. 1997;272:20986–20989. doi: 10.1074/jbc.272.34.20986. [DOI] [PubMed] [Google Scholar]
- 61.Shapiro AB, Ling V. ATPase activity of purified and reconstituted P-glycoprotein from Chinese hamster ovary cells. J. Biol. Chem. 1994;269:3745–3754. [PubMed] [Google Scholar]
- 62.Sharom FJ, Yu X, Lu P, Liu R, Chu JW, Szabo K, Muller M, Hose CD, Monks A, Varadi A, Seprodi J, Sarkadi B. Interaction of the P-glycoprotein multidrug transporter (MDR1) with high affinity peptide chemosensitizers in isolated membranes, reconstituted systems, and intact cells. Biochem. Pharmacol. 1999;58:571–586. doi: 10.1016/s0006-2952(99)00139-2. [DOI] [PubMed] [Google Scholar]
- 63.Gribble FM, Loussouarn G, Tucker SJ, Zhao C, Nichols CG, Ashcroft FM. A novel method for measurement of submembrane ATP concentration. J. Biol. Chem. 2000;275:30046–30049. doi: 10.1074/jbc.M001010200. [DOI] [PubMed] [Google Scholar]
- 64.Schwiebert EM, Zsembery A. Extracellular ATP as a signaling molecule for epithelial cells. Biochim. Biophys. Acta. 2003;1615:7–32. doi: 10.1016/s0005-2736(03)00210-4. [DOI] [PubMed] [Google Scholar]
- 65.Kennedy HJ, Pouli AE, Ainscow EK, Jouaville LS, Rizzuto R, Rutter GA. Glucose generates sub-plasma membrane ATP microdomains in single islet beta-cells. Potential role for strategically located mitochondria. J. Biol. Chem. 1999;274:13281–13291. doi: 10.1074/jbc.274.19.13281. [DOI] [PubMed] [Google Scholar]
- 66.Manfredi G, Yang L, Gajewski CD, Mattiazzi M. Measurements of ATP in mammalian cells. Methods. 2002;26:317–326. doi: 10.1016/S1046-2023(02)00037-3. [DOI] [PubMed] [Google Scholar]
- 67.Ataullakhanov FI, Vitvitsky VM. What determines the intracellular ATP concentration. Biosci. Rep. 2002;22:501–511. doi: 10.1023/a:1022069718709. [DOI] [PubMed] [Google Scholar]
- 68.Wong R, Lopaschuk G, Zhu G, Walker D, Catellier D, Burton D, Teo K, Collins-Nakai R, Montague T. Skeletal muscle metabolism in the chronic fatigue syndrome. In vivo assessment by 31P nuclear magnetic resonance spectroscopy. Chest. 1992;102:1716–1722. doi: 10.1378/chest.102.6.1716. [DOI] [PubMed] [Google Scholar]
- 69.Zhou Y, Tozzi F, Chen J, Fan F, Xia L, Wang J, Gao G, Zhang A, Xia X, Brasher H, Widger W, Ellis LM, Weihua Z. Intracellular ATP levels are a pivotal determinant of chemoresistance in colon cancer cells. Cancer Res. 2012;72:304–314. doi: 10.1158/0008-5472.CAN-11-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gajewski CD, Yang L, Schon EA, Manfredi G. New Insights into the Bioenergetics of Mitochondrial Disorders Using Intracellular ATP Reporters. Mol. Biol. Cell. 2003;14:3628–3635. doi: 10.1091/mbc.E02-12-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang H, Zhang X, Li S, Liu N, Lian W, McDowell E, Zhou P, Zhao C, Guo H, Zhang C, Yang C, Wen G, Dong X, Lu L, Ma N, Dong W, Dou QP, Wang X, Liu J. Physiological levels of ATP negatively regulate proteasome function. Cell Res. 2010;20:1372–1385. doi: 10.1038/cr.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leist M, Single B, Castoldi AF, Kuhnle S, Nicotera P. Intracellular adenosine triphosphate (ATP) concentration: a switch in the decision between apoptosis and necrosis. J. Exp. Med. 1997;185:1481–1486. doi: 10.1084/jem.185.8.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Soltoff SP. ATP and the regulation of renal cell function. Annu. Rev. Physiol. 1986;48:9–31. doi: 10.1146/annurev.ph.48.030186.000301. [DOI] [PubMed] [Google Scholar]
- 74.Gatlik-Landwojtowicz E, Aanismaa P, Seelig A. Quantification and characterization of P-glycoprotein-substrate interactions. Biochemistry. 2006;45:3020–3032. doi: 10.1021/bi051380+. [DOI] [PubMed] [Google Scholar]
- 75.Ambudkar SV, Lelong IH, Zhang J, Cardarelli CO, Gottesman MM, Pastan I. Partial purification and reconstitution of the human multidrug-resistance pump: characterization of the drug-stimulatable ATP hydrolysis. Proc. Natl. Acad. Sci. U. S. A. 1992;89:8472–8476. doi: 10.1073/pnas.89.18.8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Litman T, Zeuthen T, Skovsgaard T, Stein WD. Competitive, non-competitive and cooperative interactions between substrates of P-glycoprotein as measured by its ATPase activity. Biochim. Biophys. Acta. 1997;1361:169–176. doi: 10.1016/s0925-4439(97)00027-6. [DOI] [PubMed] [Google Scholar]
- 77.Urbatsch IL, Senior AE. Effects of lipids on ATPase activity of purified Chinese hamster P-glycoprotein. Arch. Biochem. Biophys. 1995;316:135–140. doi: 10.1006/abbi.1995.1020. [DOI] [PubMed] [Google Scholar]
- 78.Qu Q, Russell PL, Sharom FJ. Stoichiometry and affinity of nucleotide binding to P-glycoprotein during the catalytic cycle. Biochemistry. 2003;42:1170–1177. doi: 10.1021/bi026555j. [DOI] [PubMed] [Google Scholar]
- 79.Rosenberg MF, Kamis AB, Callaghan R, Higgins CF, Ford RC. Three-dimensional structures of the mammalian multidrug resistance P-glycoprotein demonstrate major conformational changes in the transmembrane domains upon nucleotide binding. J. Biol. Chem. 2003;278:8294–8299. doi: 10.1074/jbc.M211758200. [DOI] [PubMed] [Google Scholar]
- 80.Siarheyeva A, Liu R, Sharom FJ. Characterization of an asymmetric occluded state of P-glycoprotein with two bound nucleotides: implications for catalysis. J. Biol. Chem. 2010;285:7575–7586. doi: 10.1074/jbc.M109.047290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Urbatsch IL, al-Shawi MK, Senior AE. Characterization of the ATPase activity of purified Chinese hamster P-glycoprotein. Biochemistry. 1994;33:7069–7076. doi: 10.1021/bi00189a008. [DOI] [PubMed] [Google Scholar]
- 82.Hohl M, Hurlimann LM, Bohm S, Schoppe J, Grutter MG, Bordignon E, Seeger MA. Structural basis for allosteric cross-talk between the asymmetric nucleotide binding sites of a heterodimeric ABC exporter. Proc. Natl. Acad. Sci. U. S. A. 2014;111:11025–11030. doi: 10.1073/pnas.1400485111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dawson RJ, Locher KP. Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett. 2007;581:935–938. doi: 10.1016/j.febslet.2007.01.073. [DOI] [PubMed] [Google Scholar]
- 84.Resetar AM, Chalovich JM. Adenosine 5’-(gamma-thiotriphosphate): an ATP analog that should be used with caution in muscle contraction studies. Biochemistry. 1995;34:16039–16045. doi: 10.1021/bi00049a018. [DOI] [PubMed] [Google Scholar]
- 85.Sharom FJ, Liu R, Romsicki Y, Lu P. Insights into the structure and substrate interactions of the P-glycoprotein multidrug transporter from spectroscopic studies. Biochim. Biophys. Acta. 1999;1461:327–345. doi: 10.1016/s0005-2736(99)00166-2. [DOI] [PubMed] [Google Scholar]
- 86.Loo TW, Bartlett MC, Clarke DM. Simultaneous binding of two different drugs in the binding pocket of the human multidrug resistance P-glycoprotein. J. Biol. Chem. 2003;278:39706–39710. doi: 10.1074/jbc.M308559200. [DOI] [PubMed] [Google Scholar]
- 87.Wang G, Pincheira R, Zhang JT. Dissection of drug-binding-induced conformational changes in P-glycoprotein. Eur. J. Biochem. 1998;255:383–390. doi: 10.1046/j.1432-1327.1998.2550383.x. [DOI] [PubMed] [Google Scholar]
- 88.Nagy H, Goda K, Arceci R, Cianfriglia M, Mechetner E, Szabo G., Jr P-Glycoprotein conformational changes detected by antibody competition. Eur. J. Biochem. 2001;268:2416–2420. doi: 10.1046/j.1432-1327.2001.02122.x. [DOI] [PubMed] [Google Scholar]
- 89.Beaudet L, Gros P. Functional dissection of P-glycoprotein nucleotide-binding domains in chimeric and mutant proteins. Modulation of drug resistance profiles. J. Biol. Chem. 1995;270:17159–17170. doi: 10.1074/jbc.270.29.17159. [DOI] [PubMed] [Google Scholar]
- 90.Frank G, Shukla S, Rao P, Borgnia MJ, Bartesaghi A, Merk A, Mobin A, Esser L, Earl LA, Gottesman MM, Xia D, Ambudkar SV, Subramaniam S. Cryo-EM analysis of the conformational landscape of human P-glycoprotein (ABCB1) during its catalytic cycle. Mol. Pharmacol. 2016 doi: 10.1124/mol.116.104190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hrycyna CA, Ramachandra M, Germann UA, Cheng PW, Pastan I, Gottesman MM. Both ATP sites of human P-glycoprotein are essential but not symmetric. Biochemistry. 1999;38:13887–13899. doi: 10.1021/bi991115m. [DOI] [PubMed] [Google Scholar]
- 92.Lawson J, O’Mara ML, Kerr ID. Structure-based interpretation of the mutagenesis database for the nucleotide binding domains of P-glycoprotein. Biochim. Biophys. Acta. 2008;1778:376–391. doi: 10.1016/j.bbamem.2007.10.021. [DOI] [PubMed] [Google Scholar]
- 93.Loo TW, Bartlett MC, Detty MR, Clarke DM. The ATPase activity of the P-glycoprotein drug pump is highly activated when the N-terminal and central regions of the nucleotide-binding domains are linked closely together. J. Biol. Chem. 2012;287:26806–26816. doi: 10.1074/jbc.M112.376202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Urbatsch IL, Sankaran B, Bhagat S, Senior AE. Both P-glycoprotein nucleotide-binding sites are catalytically active. J. Biol. Chem. 1995;270:26956–26961. doi: 10.1074/jbc.270.45.26956. [DOI] [PubMed] [Google Scholar]
- 95.Welker E, Szabo K, Hollo Z, Muller M, Sarkadi B, Varadi A. Drug-stimulated ATPase activity of a deletion mutant of the human multidrug-resistance protein (MDR1) Biochem. Biophys. Res. Commun. 1995;216:602–609. doi: 10.1006/bbrc.1995.2665. [DOI] [PubMed] [Google Scholar]
- 96.Loo TW, Bartlett MC, Clarke DM. Permanent activation of the human P-glycoprotein by covalent modification of a residue in the drug-binding site. J. Biol. Chem. 2003;278:20449–20452. doi: 10.1074/jbc.C300154200. [DOI] [PubMed] [Google Scholar]
- 97.Loo TW, Clarke DM. Inhibition of oxidative cross-linking between engineered cysteine residues at positions 332 in predicted transmembrane segments (TM) 6 and 975 in predicted TM12 of human P-glycoprotein by drug substrates. J. Biol. Chem. 1996;271:27482–27487. doi: 10.1074/jbc.271.44.27482. [DOI] [PubMed] [Google Scholar]
- 98.Ramachandra M, Ambudkar SV, Gottesman MM, Pastan I, Hrycyna CA. Functional characterization of a glycine 185-to-valine substitution in human P-glycoprotein by using a vaccinia-based transient expression system. Mol. Biol. Cell. 1996;7:1485–1498. doi: 10.1091/mbc.7.10.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Omote H, Figler RA, Polar MK, Al-Shawi MK. Improved energy coupling of human P-glycoprotein by the glycine 185 to valine mutation. Biochemistry. 2004;43:3917–3928. doi: 10.1021/bi035365l. [DOI] [PubMed] [Google Scholar]
- 100.Rao US. Mutation of glycine 185 to valine alters the ATPase function of the human P-glycoprotein expressed in Sf9 cells. J. Biol. Chem. 1995;270:6686–6690. [PubMed] [Google Scholar]
- 101.Omote H, Al-Shawi MK. A novel electron paramagnetic resonance approach to determine the mechanism of drug transport by P-glycoprotein. J. Biol. Chem. 2002;277:45688–45694. doi: 10.1074/jbc.M206479200. [DOI] [PubMed] [Google Scholar]
- 102.Schwab D, Fischer H, Tabatabaei A, Poli S, Huwyler J. Comparison of in vitro P-glycoprotein screening assays: recommendations for their use in drug discovery. J. Med. Chem. 2003;46:1716–1725. doi: 10.1021/jm021012t. [DOI] [PubMed] [Google Scholar]
- 103.Faassen F, Vogel G, Spanings H, Vromans H. Caco-2 permeability, P-glycoprotein transport ratios and brain penetration of heterocyclic drugs. Int. J. Pharm. 2003;263:113–122. doi: 10.1016/s0378-5173(03)00372-7. [DOI] [PubMed] [Google Scholar]
- 104.Pauli-Magnus C, von Richter O, Burk O, Ziegler A, Mettang T, Eichelbaum M, Fromm MF. Characterization of the major metabolites of verapamil as substrates and inhibitors of P-glycoprotein. J. Pharmacol. Exp. Ther. 2000;293:376–382. [PubMed] [Google Scholar]
- 105.Elsby R, Surry DD, Smith VN, Gray AJ. Validation and application of Caco-2 assays for the in vitro evaluation of development candidate drugs as substrates or inhibitors of P-glycoprotein to support regulatory submissions. Xenobiotica. 2008;38:1140–1164. doi: 10.1080/00498250802050880. [DOI] [PubMed] [Google Scholar]