Abstract
Soluble αβ-tubulin heterodimers are maintained at high concentration inside eukaryotic cells, forming pools that fundamentally drive microtubule dynamics. Five conserved tubulin cofactors and ADP ribosylation factor–like 2 regulate the biogenesis and degradation of αβ-tubulins to maintain concentrated soluble pools. Here I describe a revised model for the function of three tubulin cofactors and Arl2 as a multisubunit GTP-hydrolyzing catalytic chaperone that cycles to promote αβ-tubulin biogenesis and degradation. This model helps explain old and new data indicating these activities enhance microtubule dynamics in vivo via repair or removal of αβ-tubulins from the soluble pools
INTRODUCTION
Dynamic microtubules (MTs) are essential force generators inside eukaryotes that modulate cell shape and organization and promote cell division (Akhmanova and Steinmetz, 2015). MTs assemble from heterodimers of α- and β-tubulins (termed αβ-tubulin). Assembly of α-and β-tubulins into one form of αβ-tubulin is critical for their ability to form head-to-tail polymers, which is a feature that is fundamental to MT polarity and dynamic instability (Akhmanova and Steinmetz, 2015). Inside most eukaryotic cells so far studied, soluble αβ-tubulins are concentrated into pools that drive the polymerization at MT plus ends. Despite extensive knowledge gained from decades of studying MT regulators, organizers, and motors, our current understanding of how soluble αβ-tubulin pools are formed and maintained remains poor and neglected despite its importance for dynamic MT function.
An abundance of α- and β-tubulin polypeptides is generated as result of tubulin mRNAs being stabilized by cotranslational regulation mediated by nascent tubulin peptides (Theodorakis and Cleveland, 1992). Tubulin polypeptides fold into globular α and β-tubulin monomers through cycles of ATP hydrolysis inside type 2 chaperonins (CCT/TRIC; Melki and Cowan, 1994; Lewis et al., 1996). Five highly conserved tubulin cofactors, also termed tubulin chaperones (TBCA, TBCB, TBCC, TBCD, TBCE), assemble folded α- and β-tubulin into a single topology, a process termed biogenesis (Figure 1A; Lewis et al., 1997). The disassembly of αβ-tubulin into monomers is termed degradation and is presumed to be the reverse process. The tubulin cofactors ensure a high concentration of active αβ-tubulin through biogenesis and degradation, a collective process termed homeostasis. An extensive body of in vitro studies suggests that the soluble αβ-tubulin concentration fundamentally regulates MT polymerization rates and frequency of dynamic instability transitions (Al-Bassam and Chang, 2011; Akhmanova and Steinmetz, 2015). Specifically, αβ-tubulin concentration regulates the association and dissociation rates of individual soluble αβ-tubulins with those polymerized at MT ends.
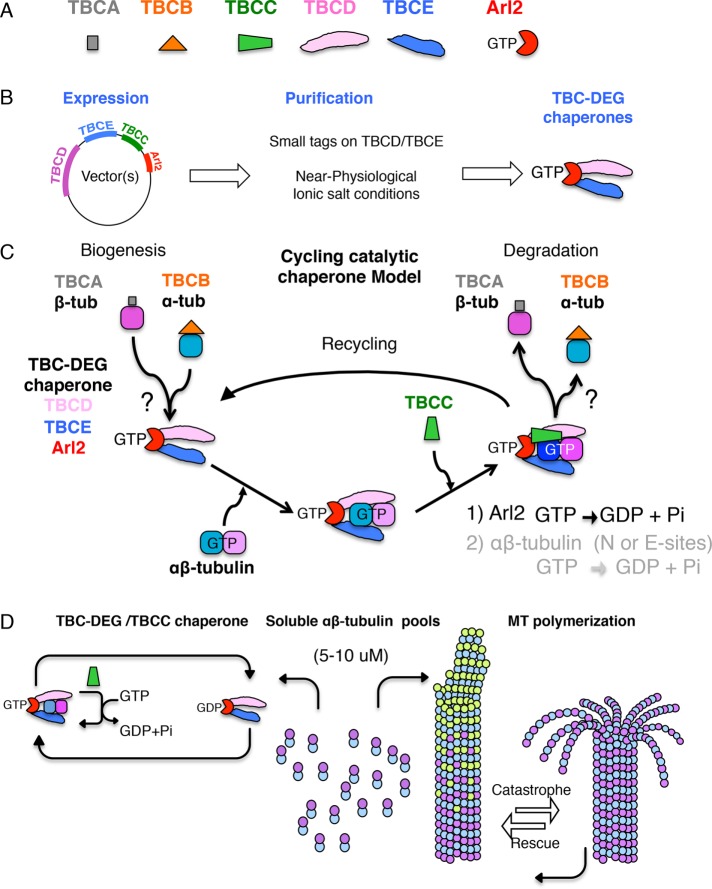
FIGURE 1:
The “cycling catalytic chaperone” model for tubulin factors and Arl2 function, based on recombinant reconstitution and in vivo cell biology studies. (A) The tubulin cofactors (TBCA, TBCB, TBCC, TBCD, and TBCE) and the Arl2 GTPase shown in schematic format. (B) The biochemical reconstitution approach that led to identifying TBC-DEG chaperones. Expression was accomplished using vectors with multiple genes. Purification was accomplished with small tags at the TBCD and TBCE N-termini in near-physiological ionic strength conditions. These approaches led to isolation of TBC-DEG chaperones, which consist of TBCD, TBCE, and Arl2. (C) The revised “cycling chaperone” model for the activity of tubulin cofactors and Arl2 in tubulin biogenesis and degradation (adapted from Nithianantham et al., 2015). (D) The role of the cycling TBC-DEG/TBCC chaperone in regulating the homeostasis of soluble αβ-tubulin pools leading to improved MT polymerization in vivo (adapted from Nithianantham et al., 2015).
Here, I present the current knowledge for the functions of tubulin cofactors in the homeostasis of soluble αβ-tubulin pools. I present a revised model for the activities of tubulin cofactors and Arl2 GTPase in the regulation of soluble αβ-tubulin pools and the consequences of this regulation on MT dynamic polymerization (Nithianantham et al., 2015; Chen et al., 2016).
DISCOVERY OF THE TUBULIN COFACTORS THROUGH GENETICS AND BIOCHEMISTRY
The cin1, cin2, cin4, and pac2 genes are important for mitosis in Saccharomyces cerevisiae (Hoyt et al., 1990, 1997; Stearns et al., 1990). Defects or deletions in these genes are not essential in yeast but lead to chromosomal instability (CIN) phenotypes in which mutant cells fail to segregate all sets of duplicated chromosomes, leading to unstable chromosome numbers. Mutant cells lose dynamic MTs very rapidly upon depletion or overexpression of these genes and become hypersensitive to MT drugs. On the other hand, overexpression of α- or β-tubulin individually leads to defects that can be rescued by the overexpression of Alf1 or Rbl2 genes, respectively; their loss results in defects involving poor MT dynamics (Archer et al., 1995; Alvarez et al., 1998; Feierbach et al., 1999). The orthologues of the budding yeast Alf1, Rbl2, Cin2, Cin1, and Pac2 were later identified to be highly conserved throughout eukaryotes and are essential in Schizosaccharomyces pombe, Caenorhabditis elegans, Drosophila melanogaster, and Arabidopsis thaliana (Hirata et al., 1998; Radcliffe et al., 1999; Szymanski, 2002). In conjunction with biochemical work (described later), these proteins were renamed tubulin cofactors TBCA, TBCB, TBCC, TBCD, and TBCE, respectively (Figure 1A; Lewis et al., 1997). Tubulin cofactors are essential in human development, and their mutations are linked to inherited human disorders: TBCB mutations are linked with giant axonal neuropathy (Wang et al., 2005), TBCE mutations lead to hyperparathyroidism and facial dysmorphism (Parvari et al., 2002), and TBCD missense mutations lead to inherited early-onset encephalopathy (Flex et al., 2016; Miyake et al., 2016). These familial disorders result from severe neurological and developmental defects due to poorly polymerizing tubulin pools.
Cowan and colleagues carried out pioneering biochemical studies to identify α- and β-tubulin translation and folding factors. They purified TBCA, TBCB, TBCC, TBCD, and TBCE from bovine testis extracts, revealing a collective function in αβ-tubulin biogenesis in extracts (Tian et al., 1996; Cowan and Lewis, 2001). In a series of studies, they described how folding of newly translated α- and β-tubulins requires ATP-dependent folding in CCT/TRIC chaperonins (Lewis et al., 1996). Furthermore, five tubulin cofactors promote assembly of folded α- and β-tubulins into αβ-tubulin heterodimers or its disassembly (Lewis et al., 1997): TBCA and TBCB bind monomeric β and α tubulins, respectively. The biogenesis of α- and β-tubulin into αβ-tubulin heterodimer requires TBCC, TBCD, TBCE, and a cycle of GTP hydrolysis. Cowan and colleagues suggested that GTP hydrolysis occurs in the β-tubulin exchangeable site (E-site). Nonhydrolyzable GTP analogues such as GTPγS trap α-and β-tubulins in a 300-kDa assembly bound to TBCC, TBCD, and TBCE.
A “LINEAR” MODEL FOR αβ-TUBULIN BIOGENESIS AND DEGRADATION
Cowan and colleagues assembled data from genetics, cell biology, and biochemistry into a “linear” tubulin cofactor model (Tian et al., 1996). This linear model is characterized by a biosynthetic-like equilibrium state in which α and β-tubulins are handed between different proteins: 1) TBCA and TBCB sequester β and α-tubulin and are then replaced by TBCD and TBCE, respectively. 2) TBCE–α-tubulin and TBCD–β-tubulin assemble with TBCC to form a transient supercomplex (TBCC, TBCD, TBCE) onto which a αβ-tubulin heterodimer is assembled or possibly degraded. The tubulin cofactor “linear” model was posed about two decades ago, and yet it remains poorly understood how the tubulin cofactors mediate αβ-tubulin biogenesis and degradation, which are activities that occur in different directions. It is unclear how the linear model modulates soluble αβ-tubulin homeostasis. Furthermore, we lack validation of the tubulin cofactor linear model using modern biochemical reconstitution with recombinant components.
Arl2 REGULATES THE TUBULIN COFACTORS AND MODULATES αβ-tubulin BIOGENESIS AND DEGRADATION
Genetics studies in various organisms identified ADP-ribosylation factor- 2 like–related GTPase (Arl2) genes as regulators of tubulin cofactors and αβ-tubulin biogenesis degradation (Figure 1A; Antoshechkin and Han, 2002; Radcliffe et al., 2000; Price et al., 2010). Among a diverse group of Arl GTPases that regulate organelle structure and membrane traffic, Arl2 is uniquely associated with MT function. Initially, inactivation of the Arl2 S. cerevisiae orthologue, cin4, lead to an identical phenotype to the loss of dynamic MTs observed for TBCD, TBCE, and TBCC inactivation (Hoyt et al., 1997; Fleming et al., 2000). Based on these data, Arl2 was postulated to be a soluble regulator of tubulin cofactors through its interaction with TBCC (Mori and Toda, 2013). TBCC may activate Arl2 GTP hydrolysis because TBCC bears strong homology to retinitis pigmentosa 2, which is a GTPase-activating protein (GAP) for Arl3, an Arl2 paralogue (Mori and Toda, 2013). The molecular role of Arl2 and its GTPase activity in regulating the tubulin cofactors and the role of Arl2 in MT function are unknown.
TUBULIN COFACTORS AND Arl2 FORM GTP-POWERED CYCLING CATALYTIC αβ-TUBULIN CHAPERONES
We reconstituted the tubulin cofactors using recombinant methods (Nithianantham et al., 2015). TBCA , TBCB, and TBCC can be purified in isolation; however, recombinant TBCD and TBCE could not be purified as soluble entities from any expression system, including reconstitution by coexpression with TBCA, TBCB, and TBCC (Nithianantham et al., 2015). We then considered the possibility that an additional subunit, likely Arl2, was missing (Tian et al., 2010). When we coexpressed TBCA–E and Arl2 using a multisubunit expression system (Figure 1B, left), we were able to isolate stable assemblies that contain TBCD-TBCE-Arl2 at a 1:1:1 molar ratio; we termed these TBC-DEG chaperones (where G stands for Arl2 GTPase). We observe that only small purification tags at specific sites and the use of moderate ionic conditions allowed purification of intact TBC-DEG chaperones (Figure 1B, middle). Other groups can only isolate very small amounts of recombinant TBCD and TBCE if expressed in isolation or TBCD-Arl2 complexes. In the prior studies, the use of large purification tags and/or high–ionic strength purification conditions likely dissociates these complexes, leading to isolated TBCD and TBCE proteins, which show poor solubility (Kortazar et al., 2006; Tian and Cowan, 2013; Nithianantham et al., 2015). Using this approach, TBC-DEG chaperones can now be purified from additional species using the same strategy, suggesting a consistent chaperone organization (Al-Bassam, Bodrug, and Nithianantham, unpublished results).
Reconstitution of TBC-DEG chaperones with TBCC and αβ-tubulin revealed a unique cycling GTP hydrolysis activity. TBCC activates rapid Arl2 GTP hydrolysis in TBC-DEG chaperones after soluble αβ-tubulin binding onto these assemblies (Nithianantham et al., 2015). TBCC affinity for TBC-DEG Arl2 is increased when αβ-tubulin is bound (Nithianantham et al., 2015). Nonhydrolyzable GTP analogues such as GTPγS and GTP-locked Arl2 (Glu73Leu) mutation interfere with the GTP hydrolysis cycle and inhibit TBCC and αβ-tubulin dissociation from TBC-DEG chaperones (Nithianantham et al., 2015). Our data reveal TBC-DEG chaperone GTP hydrolysis as the first example of a protein-substrate–specific GAP. Our data show that Arl2 is indeed the missing GTPase catalytic subunit in TBC-DEG chaperone and αβ-tubulin biogenesis. Arl2 was not considered as a tubulin cofactor, despite its clear genetic involvement in tubulin biogenesis. Our data indicate that TBC-DEG chaperones do not disassemble in each cycle but instead remain assembled as they rebind αβ-tubulin and TBCC. Previous studies, in contrast, suggest that tubulin cofactors dissociate after each cycle (Tian et al., 1999). The GTP hydrolysis dependence in those previous studies suggests that Arl2 may have been partially present but not identified. Our data indicate that Arl2 is the primary catalytic GTPase in the TBC-DEG chaperone as activated by TBCC in response to αβ-tubulin binding. It is unclear whether β-tubulin E- or N-site GTPases may still become activated, as previously suggested (Tian et al., 1997, 1999).
A “CYCLING CATALYTIC CHAPERONE” MODEL FOR αβ-tubulin BIOGENESIS AND DEGRADATION
Our data suggest a new model for the function of the tubulin cofactors and Arl2. We term this the “cycling catalytic chaperone” model (Figure 1C; Nithianantham et al., 2015). Three subunits TBCD, TBCE, and Arl2 form stable TBC-DEG chaperones. These TBC-DEG chaperones catalytically drive the biogenesis and degradation of αβ-tubulins through TBCC binding on loaded chaperones to activate Arl2 GTP hydrolysis (Figure 1C). The chaperone activity is cyclic and would allow TBCA and TBCB access to load and unload α- and β-tubulins into the complex, but it is unclear how this occurs. The collective activities of the cycling TBC-DEG chaperones assemble concentrated soluble αβ-tubulin pools. In this model, TBC-DEG chaperones likely recognize and bind α- and β-tubulin in unique manner, arranging them into a single αβ-tubulin heterodimeric topology (Figure 1C). It is unclear how TBC-DEG chaperones set this heterodimer organization. TBCC-activated GTP hydrolysis likely accelerates and/or controls the direction of this process, which likely evolved to decrease intracellular toxicity of isolated α- and β-tubulin monomers on MT polymerization. A chaperone-driven, GTP-dependent catalysis may stabilize or destabilize intradimer interfaces of the αβ-tubulin heterodimer, presumably allowing TBCA and TBCB access. The extremely slow natural low dissociation rate of α and β-tubulins from heterodimers is consistent with this, as it ranges between 10–6 and 10–9 M–1 (Caplow and Fee, 2002).
HOMEOSTASIS OF αβ-tubulin POOLS MODULATES MT DYNAMICS IN VIVO
Our “cycling catalytic chaperone” model suggests that TBC-DEG chaperones regulate the biogenesis and degradation of soluble αβ-tubulins to and from large intracellular pools through repeating catalytic cycles (Figure 1D). However, how does this regulation influence MT dynamics in vivo? Studies by Chen et al. (2016) and our group (Nithianantham et al. (2015) recently addressed this question, using two different organisms and completely unique approaches, arriving at fairly consistent models. We identified a GTP-locked Arl2 (cin4 Glu73Leu) mutant that traps TBC-DEG chaperones in a single step of the catalytic cycle while in a high-affinity state for TBCC and αβ-tubulin. The GTP-locked cin4 severely interferes with dynamic MT polymerization, which resulted in dominant-negative defects on MT function in both the presence and absence of endogenous cin4. In vivo MT dynamics in yeast cells expressing GTP-locked cin4 at very low levels for short periods suggest that this mutant dramatically decreases MT rescues, which are reversals of depolymerization, and an increase in MT pausing (Nithianantham et al., 2015). In parallel, Chen et al. (2016) used a genetic RNAi screen to search for genes that regulate asymmetric mitotic cell division in early D. melanogaster neural stems. They identified Arl2 is an essential regulator for mitotic spindle polarity and asymmetric assembly (Chen et al., 2016). A GDP-locked Arl2 mutant led to a loss of mitotic spindles due to poor MT polymerization, whereas a GTP-locked Arl2 mutant promoted an overabundance of MTs, leading to overgrown and stable mitotic spindles and also to defective mitotic cell division (Chen et al., 2016). These data shed new light on soluble αβ-tubulin regulation and explain much of the previous studies on how Arl2 GTP hydrolysis cycles might affect MT polymerization (Figure 1D).
WHY DOES HOMEOSTASIS OF αβ-tubulin POOLS MODULATE MT DYNAMICS?
Soluble αβ-tubulin homeostasis by TBC-DEG chaperones has a substantial effect on MT dynamics in vivo (Figure 1D). Unlike in vitro studies using polymerization-cycled pools of αβ-tubulin, soluble αβ-tubulin pools within the cytoplasm are subject to aging or damage, leading to defective or inconsistent MT dynamics. Damage of αβ-tubulins may accumulate in MT polymerization due mechanical deformation or defects, as recently demonstrated by Aumeier et al. (2016). MTs with damaged αβ-tubulins show decreased depolymerization, and damage sites promote MT rescues (Aumeier et al., 2016). Alternatively, soluble αβ-tubulins may age in the cytoplasm, leading to defects in its E-site GTPase and resulting in polymerization defects. Defective αβ-tubulin can poison the polymerization activities of soluble αβ-tubulin pools (Figure 1D). Collectively the polymerization health of αβ-tubulin pools can be strongly influenced by a few defective αβ-tubulins amplifying defects in MT dynamics. Thus TBC-DEG chaperones may either degrade or recycle damaged or aging αβ-tubulins from the soluble pool via catalytic activity cycles and thus improve MT dynamics in vivo.
FUTURE QUESTIONS: MOLECULAR BASIS FOR αβ-tubulin BIOGENESIS AND DEGRADATION
Many pressing questions remain regarding αβ-tubulin biogenesis and degradation via TBC-DEG chaperones and how their activities improve the homeostasis of soluble αβ-tubulin pools. The whereabouts and lifetimes of folded monomeric α-tubulin and β-tubulin in the cytoplasm are enigmatic. The roles of TBCA and TBCB in binding these intermediates and loading them onto TBC-DEG chaperones remain poorly studied and not completely known. How do TBC-DEG chaperones catalyze αβ-tubulin biogenesis and degradation? Higher-resolution structural studies of TBC-DEG chaperones in different GTP hydrolysis and αβ-tubulin– and TBCC-bound states will reveal the nature of biogenesis and degradation. Deeper questions remain regarding how the homeostasis of soluble αβ-tubulin pools may modulate MT dynamics in vivo. Does the soluble αβ-tubulin concentration increase or decrease during the lifespans of eukaryotic cells? How are the soluble αβ-tubulin pools modulated during MT polymerization–intensive cellular phases such as cell division or cellular expansions in development? How does the in vivo soluble αβ-tubulin concentration modulate MT polymerization dynamics? What are the origins of αβ-tubulin damage and aging, and how do TBC-DEG chaperones repair or recognize such damage? Furthermore, even bigger questions remain about how soluble αβ-tubulin concentration modulates the expression and translation of αβ-tubulin mRNAs inside eukaryotic cells. This an exciting time for understanding soluble αβ-tubulin homeostasis, which has suffered from neglect despite extensive interest in the MTs and their regulators, motors, and organizers.
Acknowledgments
I thank Richard McKenney and Stanley Nithianantham (Molecular Cellular Biology, University of California, Davis) for critical reading of the manuscript and a reviewer for the very helpful suggestions. Research on the tubulin cofactors and Arl2 is supported by National Institutes of Health Grant 1R01-GM110283.
Abbreviations used:
- Arl2
ADP-ribosylation factor like–related 2
- CIN
chromosomal instability
- GTP
gaunosine 3,5 triphosphate
- MT
microtubule
- TBC
tubulin cofactor or chaperone
- TBC-DEG
TBC-D-E-Arl2
Footnotes
REFERENCES
- Akhmanova A, Steinmetz MO. Control of microtubule organization and dynamics: two ends in the limelight. Nat Rev Mol Cell Biol. 2015;16:711–726. doi: 10.1038/nrm4084. [DOI] [PubMed] [Google Scholar]
- Al-Bassam J, Chang F. Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 2011;21:604–614. doi: 10.1016/j.tcb.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez P, Smith A, Fleming J, Solomon F. Modulation of tubulin polypeptide ratios by the yeast protein Pac10p. Genetics. 1998;149:857–864. doi: 10.1093/genetics/149.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoshechkin I, Han M. The C. elegans evl-20 gene is a homolog of the small GTPase ARL2 and regulates cytoskeleton dynamics during cytokinesis and morphogenesis. Dev Cell. 2002;2:579–591. doi: 10.1016/s1534-5807(02)00146-6. [DOI] [PubMed] [Google Scholar]
- Archer JE, Vega LR, Solomon F. Rbl2p, a yeast protein that binds to beta-tubulin and participates in microtubule function in vivo. Cell. 1995;82:425–434. doi: 10.1016/0092-8674(95)90431-x. [DOI] [PubMed] [Google Scholar]
- Aumeier C, Schaedel L, Gaillard J, John K, Blanchoin L, Thery M. Self-repair promotes microtubule rescue. Nat Cell Biol. 2016;18:1054–1064. doi: 10.1038/ncb3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplow M, Fee L. Dissociation of the tubulin dimer is extremely slow, thermodynamically very unfavorable, and reversible in the absence of an energy source. Mol Biol Cell. 2002;13:2120–2131. doi: 10.1091/mbc.E01-10-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Koe CT, Xing ZB, Tian X, Rossi F, Wang C, Tang Q, Zong W, Hong WJ, Taneja R, et al. Arl2- and Msps-dependent microtubule growth governs asymmetric division. J Cell Biol. 2016;212:661–676. doi: 10.1083/jcb.201503047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan NJ, Lewis SA. Type II chaperonins, prefoldin, and the tubulin-specific chaperones. Adv Protein Chem. 2001;59:73–104. doi: 10.1016/s0065-3233(01)59003-8. [DOI] [PubMed] [Google Scholar]
- Feierbach B, Nogales E, Downing KH, Stearns T. Alf1p, a CLIP-170 domain-containing protein, is functionally and physically associated with alpha-tubulin. J Cell Biol. 1999;144:113–124. doi: 10.1083/jcb.144.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming JA, Vega LR, Solomon F. Function of tubulin binding proteins in vivo. Genetics. 2000;156:69–80. doi: 10.1093/genetics/156.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flex E, Niceta M, Cecchetti S, Thiffault I, Au MG, Capuano A, Piermarini E, Ivanova AA, Francis JW, Chillemi G, et al. Biallelic mutations in TBCD, encoding the tubulin folding cofactor D, perturb microtubule dynamics and cause early-onset encephalopathy. Am J Hum Genet. 2016;99:962–973. doi: 10.1016/j.ajhg.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata D, Masuda H, Eddison M, Toda T. Essential role of tubulin-folding cofactor D in microtubule assembly and its association with microtubules in fission yeast. EMBO J. 1998;17:658–666. doi: 10.1093/emboj/17.3.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Macke JP, Roberts BT, Geiser JR. Saccharomyces cerevisiae PAC2 functions with CIN1, 2 and 4 in a pathway leading to normal microtubule stability. Genetics. 1997;146:849–857. doi: 10.1093/genetics/146.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt MA, Stearns T, Botstein D. Chromosome instability mutants of Saccharomyces cerevisiae that are defective in microtubule-mediated processes. Mol Cell Biol. 1990;10:223–234. doi: 10.1128/mcb.10.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortazar D, Carranza G, Bellido J, Villegas JC, Fanarraga ML, Zabala JC. Native tubulin-folding cofactor E purified from baculovirus-infected Sf9 cells dissociates tubulin dimers. Protein Expr Purif. 2006;49:196–202. doi: 10.1016/j.pep.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Lewis SA, Tian G, Cowan NJ. The alpha- and beta-tubulin folding pathways. Trends Cell Biol. 1997;7:479–484. doi: 10.1016/S0962-8924(97)01168-9. [DOI] [PubMed] [Google Scholar]
- Lewis SA, Tian G, Vainberg IE, Cowan NJ. Chaperonin-mediated folding of actin and tubulin. J Cell Biol. 1996;132:1–4. doi: 10.1083/jcb.132.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melki R, Cowan NJ. Facilitated folding of actins and tubulins occurs via a nucleotide-dependent interaction between cytoplasmic chaperonin and distinctive folding intermediates. Mol Cell Biol. 1994;14:2895–2904. doi: 10.1128/mcb.14.5.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake N, Fukai R, Ohba C, Chihara T, Miura M, Shimizu H, Kakita A, Imagawa E, Shiina M, Ogata K, et al. Biallelic TBCD mutations cause early-onset neurodegenerative encephalopathy. Am J Hum Genet. 2016;99:950–961. doi: 10.1016/j.ajhg.2016.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori R, Toda T. The dual role of fission yeast Tbc1/cofactor C orchestrates microtubule homeostasis in tubulin folding and acts as a GAP for GTPase Alp41/Arl2. Mol Biol Cell. 2013;24:1713–1724. doi: 10.1091/mbc.E12-11-0792. S1–S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nithianantham S, Le S, Seto E, Jia W, Leary J, Corbett KD, Moore JK, Al-Bassam J. Tubulin cofactors and Arl2 are cage-like chaperones that regulate the soluble alphabeta-tubulin pool for microtubule dynamics. Elife. 2015:4. doi: 10.7554/eLife.08811. doi: 10.7554/eLife.08811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvari R, Hershkovitz E, Grossman N, Gorodischer R, Loeys B, Zecic A, Mortier G, Gregory S, Sharony R, Kambouris M, et al. Mutation of TBCE causes hypoparathyroidism-retardation-dysmorphism and autosomal recessive Kenny-Caffey syndrome. Nat Genet. 2002;32:448–452. doi: 10.1038/ng1012. [DOI] [PubMed] [Google Scholar]
- Price HP, Peltan A, Stark M, Smith DF. The small GTPase ARL2 is required for cytokinesis in Trypanosoma brucei. Mol Biochem Parasitol. 2010;173:123–131. doi: 10.1016/j.molbiopara.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe PA, Hirata D, Vardy L, Toda T. Functional dissection and hierarchy of tubulin-folding cofactor homologues in fission yeast. Mol Biol Cell. 1999;10:2987–3001. doi: 10.1091/mbc.10.9.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radcliffe PA, Vardy L, Toda T. A conserved small GTP-binding protein Alp41 is essential for the cofactor-dependent biogenesis of microtubules in fission yeast. FEBS Lett. 2000;468:84–88. doi: 10.1016/s0014-5793(00)01202-3. [DOI] [PubMed] [Google Scholar]
- Stearns T, Hoyt MA, Botstein D. Yeast mutants sensitive to antimicrotubule drugs define three genes that affect microtubule function. Genetics. 1990;124:251–262. doi: 10.1093/genetics/124.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski D. Tubulin folding cofactors: half a dozen for a dimer. Curr Biol. 2002;12:R767–R769. doi: 10.1016/s0960-9822(02)01288-5. [DOI] [PubMed] [Google Scholar]
- Theodorakis NG, Cleveland DW. Physical evidence for cotranslational regulation of beta-tubulin mRNA degradation. Mol Cell Biol. 1992;12:791–799. doi: 10.1128/mcb.12.2.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Bhamidipati A, Cowan NJ, Lewis SA. Tubulin folding cofactors as GTPase-activating proteins. GTP hydrolysis and the assembly of the alpha/beta-tubulin heterodimer. J Biol Chem. 1999;274:24054–24058. doi: 10.1074/jbc.274.34.24054. [DOI] [PubMed] [Google Scholar]
- Tian G, Cowan NJ. Tubulin-specific chaperones: components of a molecular machine that assembles the alpha/beta heterodimer. Methods Cell Biol. 2013;115:155–171. doi: 10.1016/B978-0-12-407757-7.00011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Huang Y, Rommelaere H, Vandekerckhove J, Ampe C, Cowan NJ. Pathway leading to correctly folded beta-tubulin. Cell. 1996;86:287–296. doi: 10.1016/s0092-8674(00)80100-2. [DOI] [PubMed] [Google Scholar]
- Tian G, Lewis SA, Feierbach B, Stearns T, Rommelaere H, Ampe C, Cowan NJ. Tubulin subunits exist in an activated conformational state generated and maintained by protein cofactors. J Cell Biol. 1997;138:821–832. doi: 10.1083/jcb.138.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G, Thomas S, Cowan NJ. Effect of TBCD and its regulatory interactor Arl2 on tubulin and microtubule integrity. Cytoskeleton. 2010;67:706–714. doi: 10.1002/cm.20480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Ding J, Allen E, Zhu P, Zhang L, Vogel H, Yang Y. Gigaxonin interacts with tubulin folding cofactor B and controls its degradation through the ubiquitin-proteasome pathway. Curr Biol. 2005;15:2050–2055. doi: 10.1016/j.cub.2005.10.052. [DOI] [PubMed] [Google Scholar]