Abstract
Eukaryotic genomes are spatially organized within the nucleus by chromosome folding, interchromosomal contacts, and interaction with nuclear structures. This spatial organization is observed in diverse organisms and both reflects and contributes to gene expression and differentiation. This leads to the notion that the arrangement of the genome within the nucleus has been shaped and conserved through evolutionary processes and likely plays an adaptive function. Both DNA-binding proteins and changes in chromatin structure influence the positioning of genes and larger domains within the nucleus. This suggests that the spatial organization of the genome can be genetically encoded by binding sites for DNA-binding proteins and can also involve changes in chromatin structure, potentially through nongenetic mechanisms. Here I briefly discuss the results that support these ideas and their implications for how genomes encode spatial organization.
SPATIAL ORGANIZATION OF EUKARYOTIC GENOMES
Spatial organization, from organisms to molecules, is a defining feature of life. In multicellular organisms, cells are organized into tissues that are spatially segregated from each other. Cells serve to concentrate and organize biological activities, facilitating metabolism, growth, and division. In eukaryotic cells, membrane-bounded organelles compartmentalize biochemical reactions, permitting greater efficiency and control. Biochemical activities can also be concentrated and regulated through the formation of non–membrane-bounded, phase-separated liquid droplets such as P bodies, germ granules, nucleoli, and others (Courchaine et al., 2016).
Not only is the eukaryotic nucleus segregated from the cytoplasm, but it is also spatially organized within. It has long been appreciated that chromosomes are nonrandomly arranged (Rabl, 1885; Boveri, 1909). During mitosis, chromosome condensation and alignment are critical to facilitate proper segregation (Hirano, 2015). Similarly, during interphase or in postmitotic cells, the spatial organization of decondensed chromosomes is tissue specific, reflecting different functional states of the genome (Parada et al., 2004; Meaburn and Misteli, 2007; Fraser et al., 2015).
Chromosomes fold and occupy characteristic regions of the interphase nucleus. In unicellular eukaryotes such as yeast, as well as in certain metazoan cells during development, chromosomes associate with the nuclear envelope, and their telomeres and centromeres cluster together at opposite sides of the nucleus (Marshall et al., 1996; Heun et al., 2001; Taddei et al., 2010; Zimmer and Fabre, 2011). In differentiated cells, chromosomes tend to fold and occupy distinct, mostly nonoverlapping “territories” (Cremer et al., 2006; Cremer and Cremer, 2010). The position of chromosomes within the nucleus also correlates with cell type; gene-rich chromosomes that are more highly transcribed tend to be positioned toward the center of the nucleus, and gene-poor chromosomes tend to be positioned toward the nuclear periphery (Parada et al., 2004).
The arrangement of chromosomes with respect to nuclear landmarks such as the nuclear periphery reflects their interaction with stable subnuclear structures. Depending on the cell type, a varying but significant fraction of metazoan genomes is associated with the nuclear lamina. These lamina-associated-domains (LADs), up to 10 Mb in length, localize at the nuclear periphery (Figure 1A; Vogel et al., 2007; Guelen et al., 2008). Similarly, in budding yeast and fission yeast, telomeres and centromeres cluster together and associate with the nuclear envelope (Klein et al., 1992; Funabiki et al., 1993; Palladino et al., 1993; Gotta et al., 1996). Such physical interactions provide a significant constraint on chromosome folding and positioning, leading to a nonrandom arrangement of chromosomes within the nucleus.
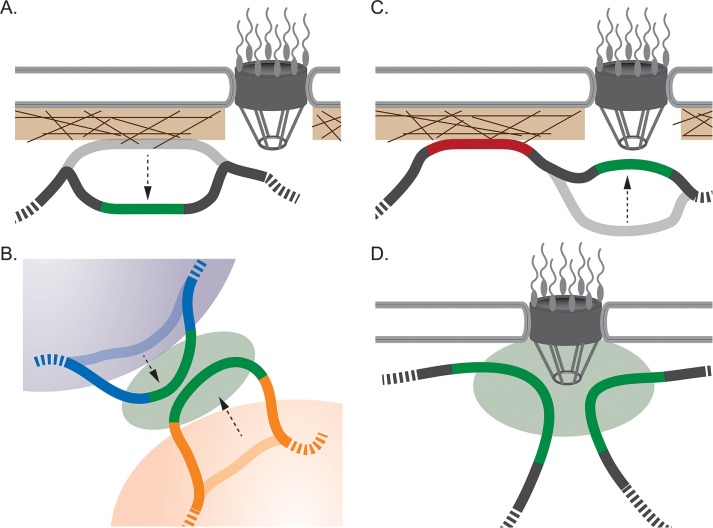
FIGURE 1:
Phenomena that lead to nonrandom spatial arrangement of the genome. (A) During development, genes that are induced often move from a peripheral position in association with the lamina, where they are silenced, to a more internal site, where they are expressed (green locus). (B) Gene activation can lead to looping of genes out of their respective chromosome territories, where they can cluster together. (C) Upon activation (green locus), certain genes physically associate with the NPC, causing them to localize to the nuclear periphery. Thus the nuclear periphery is associated with both silencing (red locus at the lamina) and activation/poising (at the NPC). (D) Genes that are targeted to the NPC in budding yeast frequently undergo interallelic clustering and interchromosomal clustering with other genes that are targeted by the same mechanism.
Individual genes are often nonrandomly positioned with respect to nuclear structures and with respect to each other and their position can change upon transcriptional activation or repression. Silenced loci within LADs localize at the nuclear periphery, but such loci are released from the lamina and move to a more internal site upon activation (Figure 1A; Kosak et al., 2002; Zink et al., 2004; Meister et al., 2010; Peric-Hupkes et al., 2010). Similarly, transcriptional induction can lead to a locus looping out from its chromosome territory (Figure 1B; Noordermeer et al., 2008). Heat shock genes reposition near stress granules upon induction (Khanna et al., 2014). Furthermore, in budding yeast, Drosophila, Caenorhabditis elegans, and mammalian cells, many active genes physically interact with nuclear pore proteins (Casolari et al., 2004, 2005; Brown et al., 2008; Capelson et al., 2010; Kalverda and Fornerod, 2010; Liang et al., 2013; Rohner et al., 2013). Whereas in yeast these interactions apparently occur exclusively at the nuclear periphery (with the nuclear pore complex [NPC]; Figure 1C), in flies and mammals, these interactions can occur either at the nuclear periphery or in the nucleoplasm (with soluble nuclear pore proteins; Capelson et al., 2010; Kalverda et al., 2010; Liang et al., 2013; Light et al., 2013). Interaction with the NPC leads to positioning at the nuclear periphery (Figure 1C). Thus changes in gene expression are frequently coupled to movement of loci with respect to nuclear landmarks.
Coregulated genes sometimes cluster together through interchromosomal interactions (Figure 1, B and D). Clustering has been observed between genes induced during erythropoiesis (Brown et al., 2006; Schoenfelder et al., 2010), genes induced by estrogen (Lin et al., 2009), and genes induced by infection (Apostolou and Thanos, 2008). In yeasts, both tRNA genes and genes that are targeted to the NPC undergo interchromosomal clustering (Figure 1D; Thompson et al., 2003; Noma et al., 2006; Brickner et al., 2012). These observations suggest that the nucleus might be functionally segregated by creating clusters of genes of related function or with related modes of regulation.
Emergent phenomena
Nonrandom spatial organization in some cases represents an emergent phenomenon, a functional outcome of transcription, chromatin structure, or polymer biophysics (Misteli, 2008, 2009). Emergent phenomena resulting from self-organization are common and extremely important in biology, and this is a critical consideration in studying nuclear cell biology. Interpreting genetic perturbation of emergent phenomena is more difficult than interpreting genetic perturbation of active targeting mechanisms. For example, if the biophysical process of transcription, chromatin decompaction, or mRNA processing were more energetically costly in the more densely compacted environment within a chromosome territory, this might result in the movement of active genes to the edge of the territory. This movement, however, would not represent active, specific targeting to the interterritorial space. Indeed, modeling chromosomes as polymers and incorporating simple biophysical differences between euchromatin and heterochromatin can recapitulate formation of chromosome territories and the localization of active genes between chromosome territories (Cook and Marenduzzo, 2009; Heermann, 2011; Wang et al., 2015). Similarly, some subnuclear structures are not stable landmarks but are built around the genes with which they colocalize. For example, nucleoli and histone locus bodies colocalize with the ribosomal DNA and histone genes, respectively. However, such bodies can be initiated to form at an ectopic locus by tethering nucleating factors to that site (Karpen et al., 1988; Kaiser et al., 2008; Mao et al., 2011; Shevtsov and Dundr, 2011). This type of spatial organization, although specific to particular genes and giving rise to a highly biased pattern, reflects an emergent property of certain genes rather than active positioning of genes near nuclear bodies. Distinguishing between nonrandom arrangements that are produced by active targeting mechanisms and those that are produced by emergent phenomena is critical to understanding nuclear architecture.
Here I focus on cases in which the nonrandom positioning of genes and chromosomal domains appears to be conferred by active targeting mechanisms. To define active mechanisms, I draw on the lessons from protein targeting to membrane-bound organelles. Active mechanisms that control subnuclear positioning ought to exhibit specificity and depend on information that is necessary and/or sufficient to promote targeting to stable sites.
DNA-BINDING PROTEINS AND CHROMATIN MODIFICATIONS AFFECT THE SPATIAL ORGANIZATION OF THE GENOME
If spatial organization of the genome is controlled by active mechanisms, then what is the source of the information? Is spatial organization encoded by the DNA sequence, chromatin changes (potentially independent of the DNA sequence), or a combination of both? Work from several experimental systems reveals that both sequence-specific DNA-binding proteins and chromatin modifications play essential roles in controlling the spatial organization of genes and chromosomal domains. Binding sites for sequence-specific DNA-binding proteins such as CTCF and transcription factors serve as the genetic information that affects chromosome folding and spatial positioning. Non–genetic chromatin changes such as histone methylation and variant histones also play essential roles in controlling the positioning of individual genes and chromosomal domains within the nucleus.
Transcription factors
Spatial positioning of individual genes with respect to stable nuclear landmarks can be controlled by cis-acting DNA elements that recruit sequence-specific DNA-binding proteins. In Drosophila, the gypsy insulator element is sufficient to induce targeting of a locus to the nuclear periphery (Gerasimova et al., 2000). For developmentally induced genes that localize at the nuclear periphery in association with the nuclear lamina before induction (Kosak et al., 2002), the positioning at the periphery can be determined by cis-acting DNA sequences. For example, insertion of the IgH and Cyp3a genes at ectopic sites in the genome is sufficient to reposition the ectopic locus to the nuclear periphery (Zullo et al., 2012). The transcription factors cKrox and YY1 are critical for this targeting (Zullo et al., 2012; Harr et al., 2015). Similarly, the β-globin gene possesses cis-acting DNA elements that are necessary for targeting to the nuclear periphery (Bian et al., 2013). This suggests that transcription factor–binding sites play a critical role in targeting to the nuclear lamina (Figure 2A).
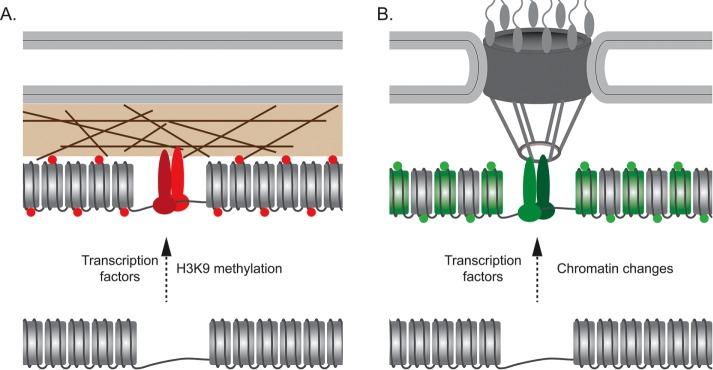
FIGURE 2:
Transcription factors and chromatin function to control interactions with the nuclear periphery. (A) Targeting of genes to the nuclear lamina requires both transcription factors such as c-Krox and YY1 and methylation of histone H3 on lysine 9 (red circles). In C. elegans, this involves a nuclear envelope–localized protein called CEC-4 that binds to this mark. (B) Targeting to the NPC (or interaction with nuclear pore proteins in the nucleoplasm of Drosophila or mammals) requires transcription factors and, in some cases, chromatin changes. Several yeast transcription factors are both necessary and sufficient to cause targeting to the NPC. However, histone acetylation, H3K4 methylation (green circles), and H2A.Z incorporation (green nucleosomes) are also required in certain cases.
Hundreds of genes that interact with nuclear pore proteins have been identified in yeast, flies, and mammalian cells (Casolari et al., 2004, 2005; Brown et al., 2008; Kalverda et al., 2010; Liang et al., 2013). Initially, it was believed that this may represent an emergent product of transcription by which NPC-associated mRNA export factors might create an affinity sink for actively transcribed genes (Casolari et al., 2005; Dieppois et al., 2006; Taddei et al., 2006). However, targeting to the NPC in yeast does not require active transcription (Schmid et al., 2006; Brickner et al., 2007, 2016; Ahmed et al., 2010; Light et al., 2010; Randise-Hinchliff et al., 2016). Furthermore, binding sites for certain transcription factors are enriched among genes that interact with pore proteins (Casolari et al., 2004, 2005; Brown et al., 2008; Liang et al., 2013). In Drosophila, GAGA factor–binding sites are also enriched for nuclear pore protein binding, and the transcription factor MBD-R2 physically interacts with the nuclear pore protein Nup98 and promotes interaction of Nup98 with target genes (Capelson et al., 2010; Kalverda et al., 2010; Pascual-Garcia et al., 2014). Thus transcription factor–binding sites correlate with interactions with nuclear pore proteins. However, in most cases, the functional significance of these transcription factors in controlling interaction with nuclear pore proteins or spatial positioning of target genes has not yet been tested.
In budding yeast, transcription factor–binding sites have been functionally identified as both necessary and sufficient to target genes to the nuclear periphery (Ahmed et al., 2010; Light et al., 2010; Brickner et al., 2012, 2016; D’Urso et al., 2016; Randise-Hinchliff et al., 2016). Not every yeast transcription factor has this ability, suggesting that a subset of transcription factors affects gene positioning. Five transcription factors have been identified (Put3, Cbf1, Gcn4, Ste12, and Sfl1) that are both necessary for targeting endogenous loci to the periphery and whose binding sites function as “DNA zip codes” that are sufficient to target an ectopic site to the periphery (Figure 2B). In several of these cases (Gcn4, Ste12, and Sfl1), the transcription factors cause peripheral targeting when tethered to an ectopic site through a heterologous DNA-binding domain, confirming that they are indeed sufficient to confer positioning at the periphery (D’Urso et al., 2016; Randise-Hinchliff et al., 2016). This suggests that controlling gene positioning is an underappreciated function of some transcription factors (Figure 2B).
Transcription factors and their binding sites also control interchromosomal clustering of coregulated genes. Before differentiation of T helper cells, the interferon γ gene clusters with a locus control region (LCR) on another chromosome, and this is regulated by the DNase-hypersensitive region of the LCR (Spilianakis et al., 2005). Clustering of the Interferon β gene with sites on other chromosomes in response to virus infection requires the NF-κB transcription factor (Apostolou and Thanos, 2008). Clustering of coregulated genes during erythropoiesis requires the Klf1 transcription factor (Schoenfelder et al., 2010), and clustering of bacterial artificial chromosomes carrying the HSP70 gene near nuclear speckles requires the HSP70 promoter (Hu et al., 2010). Similarly, clustering of tRNA genes in both budding yeast and fission yeast requires the sequence-specific transcription factor TFIIIC (Noma et al., 2006; Haeusler et al., 2008). In budding yeast, transcription factors that control positioning to the nuclear periphery are also necessary for specific interchromosomal clustering in association with the nuclear pore (Brickner et al., 2012, 2015; Randise-Hinchliff et al., 2016). Of importance, insertion of their DNA-binding sites at an ectopic site in the genome can promote clustering with endogenous genes that possess the same binding site (Brickner et al., 2012, 2016; Randise-Hinchliff et al., 2016). This suggests that genomes encode both peripheral localization and interchromosomal clustering through cis-acting DNA elements such as transcription factor–binding sites.
Chromatin
Chromatin modifications often correlate with spatial position and, in some cases, are required for the normal spatial arrangement of the genome. Subtelomeric regions are silenced by the yeast histone deacetylase Sir2. Histone deacetylation recruits Sir4, which binds to both hypoacetylated histones and the nuclear envelope membrane protein Esc1, providing a physical link between chromatin state and a subnuclear structure (Taddei and Gasser, 2012). Lamina-associated domains tend to be transcriptionally repressed and methylated on histone H3 lysine 9 (Guelen et al., 2008; Wen et al., 2009; Yokochi et al., 2009; Kind et al., 2013). Loss of the H3K9 methyl transferase G9a leads to loss of peripheral H3K9me2 but does not obviously alter the peripheral localization of all genes (Yokochi et al., 2009). However, association of lamina-targeted BACs or endogenous LADs in human cells or lamina-associated repetitive arrays in C. elegans is dependent on H3K9 methylation (Towbin et al., 2012; Gonzalez-Sandoval et al., 2015). Thus, together with the transcription factors mentioned earlier, H3K9 methylation is required for the interaction with certain loci with the nuclear periphery (Figure 2A).
In C. elegans, H3K9 methylation may function alone in targeting to the lamina. The system in which this requirement was discovered used long arrays of transgenes (Meister et al., 2010). The H3K9 methylation of these arrays is apparently a nonspecific product of their repetitive nature. Targeting to the nuclear lamina and H3K9 methylation are mutually reinforcing events: H3K9 methylation is both promoted by targeting to the nuclear lamina and required for targeting to the lamina. A nuclear envelope–associated protein called CEC-4 binds to methylated H3K9 through a chromodomain to mediate localization of heterochromatic repetitive arrays to the nuclear lamina (Gonzalez-Sandoval et al., 2015). This suggests that H3K9 methylation alone can target such arrays to the nuclear lamina, independent of their sequence and presumably independent of DNA-binding proteins.
The intrachromosomal looping and clustering of Polycomb targets leads to formation of Polycomb bodies. This requires the Polycomb group protein PC (Lanzuolo et al., 2007). However, mutations that disrupt Polycomb response elements, which reduce recruitment of Polycomb group proteins, do not strongly affect either H3K27me3 or Polycomb body formation. This suggests that H3K27 methylation, rather than a DNA-binding protein, may facilitate the formation of Polycomb bodies.
Targeting of certain yeast genes to the NPC requires the SAGA histone acetyltransferase and is inhibited by the Rpd3(L) histone deacetylase (Cabal et al., 2006; Luthra et al., 2007; Randise-Hinchliff et al., 2016). Furthermore, targeting of yeast INO1 to the nuclear periphery after repression (a phenomenon called epigenetic transcriptional memory) requires both local incorporation of H2A.Z and dimethylation of H3K4 (Brickner et al., 2007; Light et al., 2010, 2013). Therefore chromatin modifications also play essential roles in targeting to the nuclear pore complex (Figure 2B).
In addition to active and poised genes, the NPC in budding yeast and Drosophila also interacts with persistent double-strand breaks, eroded telomeres, and collapsed replication forks, and this interaction facilitates DNA repair (Nagai et al., 2008; Kalocsay et al., 2009; Ryu et al., 2015). Relocalization of DSBs to the NPC requires both H2A.Z and SUMOylation of either chromatin or repair factors, leading to a physical interaction with the SUMO-dependent ubiquitin ligase Slx5/Slx8, which interacts with the NPC component Nup84 (Nagai et al., 2008; Kalocsay et al., 2009; Churikov et al., 2016; Horigome et al., 2016). Thus positioning of DNA damage, which is unlikely to be dependent on DNA sequence, is mediated by changes in chromatin and associated proteins.
Coordinating the function of DNA-binding proteins and chromatin
Because transcription factors and chromatin changes influence each other and can have indirect effects, in most cases, it remains to be determined whether either of these mechanisms alone is sufficient to confer spatial positioning. Furthermore, the function of transcription factors themselves might be regulated by posttranslational modifications such as acetylation and SUMOylation (Texari et al., 2013). In cases in which both DNA-binding proteins and chromatin modifications play essential roles in controlling the spatial organization of the genome (i.e., neither is sufficient), they might function in the same linear pathway or by a combinatorial mechanism.
The requirement for histone acetylation or methylation in targeting genes to the NPC (Figure 2B) may represent either a direct role for chromatin in mediating targeting to the NPC or a role for chromatin in regulating transcription factor binding/function. In support of this idea, Rpd3(L) regulates Put3 DNA binding, and SAGA is required for targeting of certain genes to the periphery because histone acetylation promotes binding of transcription factors. In support of this idea, SAGA is required for peripheral targeting of genes whose subnuclear localization is regulated by transcription factor occupancy but is not required for peripheral targeting of genes whose subnuclear localization is regulated downstream of transcription factor occupancy (Randise-Hinchliff et al., 2016). This suggests that histone acetylation (or other functions of SAGA) may regulate transcription factor occupancy by regulating either DNA binding or transcription factor abundance.
The histone modifications associated with transcriptional memory are necessary but not sufficient to confer targeting to the nuclear periphery. Mutants lacking H2A.Z or unable to methylate H3K4 fail to target genes to the NPC during memory (Brickner et al., 2007; Light et al., 2010, 2013). However, neither H2A.Z nor H3K4me2 is sufficient to cause targeting to the NPC (Light et al., 2010; D’Urso et al., 2016). This suggests that targeting to the NPC requires both a transcription factor and the correct chromatin state and that they may function together (Figure 2B). Future work will distinguish a direct role for chromatin in affecting gene positioning in this system from a regulatory role.
CONCLUSION
Nuclear cell biology is an important and challenging area. Future work will seek to develop a more complete understanding of the molecular and evolutionary mechanisms that control the spatial organization of the genome. To make progress, we must first address two important issues. First, we must distinguish spatial organization that is a product of emergent properties of nuclear biology from spatial organization that is the product of active mechanisms. Both types of phenomena contribute to the spatial organization of the genome, and they need to be integrated to understand the whole. Second, we must carefully define the relationship between DNA-binding proteins and chromatin changes in determining spatial organization. Subnuclear positioning of genes and domains is controlled by DNA-binding proteins, chromatin structure, and, in some cases, both. This suggests that both DNA sequence–dependent and DNA sequence–independent mechanisms affect the spatial organization of the genome. Understanding the evolutionary processes that can select for such mechanisms poses an exciting and important problem for future work.
Acknowledgments
Work in my group in this area has been supported by National Institutes of Health Grants R01 GM080484 and R01 GM118712. I am grateful to members of my group for helpful comments on the manuscript and for their exciting contributions to the field.
Abbreviations used:
- BAC
bacterial artificial chromosome
- CTCF
CCCTC-binding factor
- LADs
lamin-associated domains
- LCR
locus control region
- NPC
nuclear pore complex
- H3K4me2
dimethylation of histone H3 on lysine 4
- H3K9me2
dimethylation of histone H3 on lysine 9
- H3K27me3
trimethylation of histone H3 on lysine 27.
Footnotes
REFERENCES
- Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, Volpe T, Brickner JH. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol. 2010;12:111–118. doi: 10.1038/ncb2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolou E, Thanos D. Virus infection induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell. 2008;134:85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- Bian Q, Khanna N, Alvikas J, Belmont AS. beta-Globin cis-elements determine differential nuclear targeting through epigenetic modifications. J Cell Biol. 2013;203:767–783. doi: 10.1083/jcb.201305027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveri T. Die blastermerenkerne von Ascaris megalocephala und die theorie der chromosomenindividualität. Arch Zellforsch. 1909;3:181. [Google Scholar]
- Brickner DG, Ahmed S, Meldi L, Thompson A, Light W, Young M, Hickman TL, Chu F, Fabre E, Brickner JH. Transcription factor binding to a DNA zip code controls interchromosomal clustering at the nuclear periphery. Dev Cell. 2012;22:1234–1246. doi: 10.1016/j.devcel.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Coukos R, Brickner JH. Transcriptional memory leads to DNA zip code-dependent interchromosomal clustering. Microb Cell. 2015;2:481–490. doi: 10.15698/mic2015.12.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brickner DG, Sood V, Tutucci E, Coukos R, Viets K, Singer RH, Brickner JH. Subnuclear positioning and interchromosomal clustering of the GAL1-10 locus are controlled by separable, interdependent mechanisms. Mol Biol Cell. 2016;27:2980–2993. doi: 10.1091/mbc.E16-03-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 2008;22:627–639. doi: 10.1101/gad.1632708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JM, Leach J, Reittie JE, Atzberger A, Lee-Prudhoe J, Wood WG, Higgs DR, Iborra FJ, Buckle VJ. Coregulated human globin genes are frequently in spatial proximity when active. J Cell Biol. 2006;172:177–187. doi: 10.1083/jcb.200507073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, Buc H, Feuerbach-Fournier F, Olivo-Marin JC, Hurt EC, Nehrbass U. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–773. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev. 2005;19:1188–1198. doi: 10.1101/gad.1307205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–439. doi: 10.1016/s0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- Churikov D, Charifi F, Eckert-Boulet N, Silva S, Simon MN, Lisby M, Geli V. SUMO-dependent relocalization of eroded telomeres to nuclear pore complexes controls telomere recombination. Cell Rep. 2016;15:1242–1253. doi: 10.1016/j.celrep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- Cook PR, Marenduzzo D. Entropic organization of interphase chromosomes. J Cell Biol. 2009;186:825–834. doi: 10.1083/jcb.200903083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchaine EM, Lu A, Neugebauer KM. Droplet organelles. EMBO J. 2016;35:1603–1612. doi: 10.15252/embj.201593517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer T, Cremer M, Dietzel S, Muller S, Solovei I, Fakan S. Chromosome territories–a functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Dieppois G, Iglesias N, Stutz F. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol Cell Biol. 2006;26:7858–7870. doi: 10.1128/MCB.00870-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Urso A, Takahashi YH, Xiong B, Marone J, Coukos R, Randise-Hinchliff C, Wang JP, Shilatifard A, Brickner JH. Set1/COMPASS and Mediator are repurposed to promote epigenetic transcriptional memory. Elife. 2016;5:e16691. doi: 10.7554/eLife.16691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser J, Williamson I, Bickmore WA, Dostie J. An overview of genome organization and how we got there: from FISH to Hi-C. Microbiol Mol Biol Rev. 2015;79:347–372. doi: 10.1128/MMBR.00006-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova TI, Byrd K, Corces VG. A chromatin insulator determines the nuclear localization of DNA. Mol Cell. 2000;6:1025–1035. doi: 10.1016/s1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Sandoval A, Towbin BD, Kalck V, Cabianca DS, Gaidatzis D, Hauer MH, Geng L, Wang L, Yang T, Wang X, et al. Perinuclear anchoring of H3K9-methylated chromatin stabilizes induced cell fate in C. elegans embryos. Cell. 2015;163:1333–1347. doi: 10.1016/j.cell.2015.10.066. [DOI] [PubMed] [Google Scholar]
- Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W, van Steensel B. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–951. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 2008;22:2204–2214. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harr JC, Luperchio TR, Wong X, Cohen E, Wheelan SJ, Reddy KL. Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. J Cell Biol. 2015;208:33–52. doi: 10.1083/jcb.201405110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heermann DW. Physical nuclear organization: loops and entropy. Curr Opin Cell Biol. 2011;23:332–337. doi: 10.1016/j.ceb.2011.03.010. [DOI] [PubMed] [Google Scholar]
- Heun P, Laroche T, Raghuraman MK, Gasser SM. The positioning and dynamics of origins of replication in the budding yeast nucleus. J Cell Biol. 2001;152:385–400. doi: 10.1083/jcb.152.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. Chromosome dynamics during mitosis. Cold Spring Harb Perspect Biol. 2015;7:a015792. doi: 10.1101/cshperspect.a015792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horigome C, Bustard DE, Marcomini I, Delgoshaie N, Tsai-Pflugfelder M, Cobb JA, Gasser SM. PolySUMOylation by Siz2 and Mms21 triggers relocation of DNA breaks to nuclear pores through the Slx5/Slx8 STUbL. Genes Dev. 2016;30:931–945. doi: 10.1101/gad.277665.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Plutz M, Belmont AS. Hsp70 gene association with nuclear speckles is Hsp70 promoter specific. J Cell Biol. 2010;191:711–719. doi: 10.1083/jcb.201004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- Kalocsay M, Hiller NJ, Jentsch S. Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell. 2009;33:335–343. doi: 10.1016/j.molcel.2009.01.016. [DOI] [PubMed] [Google Scholar]
- Kalverda B, Fornerod M. Characterization of genome-nucleoporin interactions in Drosophila links chromatin insulators to the nuclear pore complex. Cell Cycle. 2010;9:4812–4817. doi: 10.4161/cc.9.24.14328. [DOI] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Karpen GH, Schaefer JE, Laird CD. A Drosophila rRNA gene located in euchromatin is active in transcription and nucleolus formation. Genes Dev. 1988;2:1745–1763. doi: 10.1101/gad.2.12b.1745. [DOI] [PubMed] [Google Scholar]
- Khanna N, Hu Y, Belmont AS. HSP70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr Biol. 2014;24:1138–1144. doi: 10.1016/j.cub.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kind J, Pagie L, Ortabozkoyun H, Boyle S, de Vries SS, Janssen H, Amendola M, Nolen LD, Bickmore WA, van Steensel B. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153:178–192. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- Klein F, Laroche T, Cardenas ME, Hofmann JF, Schweizer D, Gasser SM. Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J Cell Biol. 1992;117:935–948. doi: 10.1083/jcb.117.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of Ig(Ig) loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol. 2007;9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW. Dynamic association of NUP98 with the human genome. PLoS Genet. 2013;9:e1003308. doi: 10.1371/journal.pgen.1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light W, Freaney J, Sood V, Thompson A, D’Urso A, Horvath C, Brickner JH. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol. 2013;11:e1001524. doi: 10.1371/journal.pbio.1001524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light WH, Brickner DG, Brand VR, Brickner JH. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol Cell. 2010;40:112–125. doi: 10.1016/j.molcel.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, Rosenfeld MG. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthra R, Kerr SC, Harreman MT, Apponi LH, Fasken MB, Ramineni S, Chaurasia S, Valentini SR, Corbett AH. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J Biol Chem. 2007;282:3042–3049. doi: 10.1074/jbc.M608741200. [DOI] [PubMed] [Google Scholar]
- Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the cotranscriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Dernburg AF, Harmon B, Agard DA, Sedat JW. Specific interactions of chromatin with the nuclear envelope: positional determination within the nucleus in Drosophila melanogaster. Mol Biol Cell. 1996;7:825–842. doi: 10.1091/mbc.7.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, Misteli T. Cell biology: chromosome territories. Nature. 2007;445:379–781. doi: 10.1038/445379a. [DOI] [PubMed] [Google Scholar]
- Meister P, Towbin BD, Pike BL, Ponti A, Gasser SM. The spatial dynamics of tissue-specific promoters during C. elegans development. Genes Dev. 2010;24:766–782. doi: 10.1101/gad.559610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Cell biology: nuclear order out of chaos. Nature. 2008;456:333–334. doi: 10.1038/456333a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Self-organization in the genome. Proc Natl Acad Sci USA. 2009;106:6885–6886. doi: 10.1073/pnas.0902010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, Brown GW, Varela E, Hediger F, Gasser SM, Krogan NJ. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 2008;322:597–602. doi: 10.1126/science.1162790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Noordermeer D, Branco MR, Splinter E, Klous P, van Ijcken W, Swagemakers S, Koutsourakis M, van der Spek P, Pombo A, de Laat W. Transcription and chromatin organization of a housekeeping gene cluster containing an integrated beta-globin locus control region. PLoS Genet. 2008;4:e1000016. doi: 10.1371/journal.pgen.1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino F, Laroche T, Gilson E, Pillus L, Gasser SM. The positioning of yeast telomeres depends on SIR3, SIR4, and the integrity of the nuclear membrane. Cold Spring Harb Symp Quant Biol. 1993;58:733–746. doi: 10.1101/sqb.1993.058.01.081. [DOI] [PubMed] [Google Scholar]
- Parada LA, McQueen PG, Misteli T. Tissue-specific spatial organization of genomes. Genome Biol. 2004;5:R44. doi: 10.1186/gb-2004-5-7-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Garcia P, Jeong J, Capelson M. Nucleoporin Nup98 associates with Trx/MLL and NSL histone-modifying complexes and regulates hox gene expression. Cell Rep. 2014;9:433–442. doi: 10.1016/j.celrep.2014.09.002. [DOI] [PubMed] [Google Scholar]
- Peric-Hupkes D, Meuleman W, Pagie L, Bruggeman SW, Solovei I, Brugman W, Graf S, Flicek P, Kerkhoven RM, van Lohuizen M, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38:603–613. doi: 10.1016/j.molcel.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl C. Über Zeillteilung. Morphol Jahrb. 1885;10:214–330. [Google Scholar]
- Randise-Hinchliff C, Coukos R, Sood V, Sumner MC, Zdraljevic S, Meldi Sholl L, Garvey Brickner D, Ahmed S, Watchmaker L, Brickner JH. Strategies to regulate transcription factor-mediated gene positioning and interchromosomal clustering at the nuclear periphery. J Cell Biol. 2016;212:633–646. doi: 10.1083/jcb.201508068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohner S, Kalck V, Wang X, Ikegami K, Lieb JD, Gasser SM, Meister P. Promoter- and RNA polymerase II-dependent hsp-16 gene association with nuclear pores in Caenorhabditis elegans. J Cell Biol. 2013;200:589–604. doi: 10.1083/jcb.201207024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu T, Spatola B, Delabaere L, Bowlin K, Hopp H, Kunitake R, Karpen GH, Chiolo I. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat Cell Biol. 2015;17:1401–1411. doi: 10.1038/ncb3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol Cell. 2006;21:379–391. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. Preferential associations between coregulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13:167–173. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Taddei A, Gasser SM. Structure and function in the budding yeast nucleus. Genetics. 2012;192:107–129. doi: 10.1534/genetics.112.140608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Schober H, Gasser SM. The budding yeast nucleus. Cold Spring Harb Perspect Biol. 2010;2:a000612. doi: 10.1101/cshperspect.a000612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- Texari L, Dieppois G, Vinciguerra P, Contreras MP, Groner A, Letourneau A, Stutz F. The nuclear pore regulates GAL1 gene transcription by controlling the localization of the SUMO protease Ulp1. Mol Cell. 2013;51:807–818. doi: 10.1016/j.molcel.2013.08.047. [DOI] [PubMed] [Google Scholar]
- Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin BD, Gonzalez-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- Vogel MJ, Peric-Hupkes D, van Steensel B. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat Protoc. 2007;2:1467–1478. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- Wang R, Mozziconacci J, Bancaud A, Gadal O. Principles of chromatin organization in yeast: relevance of polymer models to describe nuclear organization and dynamics. Curr Opin Cell Biol. 2015;34:54–60. doi: 10.1016/j.ceb.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–250. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokochi T, Poduch K, Ryba T, Lu J, Hiratani I, Tachibana M, Shinkai Y, Gilbert DM. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proc Natl Acad Sci USA. 2009;106:19363–19368. doi: 10.1073/pnas.0906142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer C, Fabre E. Principles of chromosomal organization: lessons from yeast. J Cell Biol. 2011;192:723–733. doi: 10.1083/jcb.201010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D, Amaral MD, Englmann A, Lang S, Clarke LA, Rudolph C, Alt F, Luther K, Braz C, Sadoni N, et al. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J Cell Biol. 2004;166:815–825. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zullo JM, Demarco IA, Pique-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, Singh H. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell. 2012;149:1474–1487. doi: 10.1016/j.cell.2012.04.035. [DOI] [PubMed] [Google Scholar]