The IFT-A complex is divided into core and peripheral subcomplexes composed of IFT122/IFT140/IFT144 and IFT43/IFT121/IFT139, respectively. IFT139-KO and IFT144-KO cell analyses show that IFT139 is dispensable for IFT-A assembly but essential for retrograde GPCR trafficking, whereas IFT144 is essential for IFT-A assembly and GPCR ciliary entry.
Abstract
Cilia serve as cellular antennae where proteins involved in sensory and developmental signaling, including G protein–coupled receptors (GPCRs), are specifically localized. Intraflagellar transport (IFT)-A and -B complexes mediate retrograde and anterograde ciliary protein trafficking, respectively. Using a visible immunoprecipitation assay to detect protein–protein interactions, we show that the IFT-A complex is divided into a core subcomplex, composed of IFT122/IFT140/IFT144, which is associated with TULP3, and a peripheral subcomplex, composed of IFT43/IFT121/IFT139, where IFT139 is most distally located. IFT139-knockout (KO) and IFT144-KO cells demonstrated distinct phenotypes: IFT139-KO cells showed the accumulation of IFT-A, IFT-B, and GPCRs, including Smoothened and GPR161, at the bulged ciliary tips; IFT144-KO cells showed failed ciliary entry of IFT-A and GPCRs and IFT-B accumulation at the bulged tips. These observations demonstrate the distinct roles of the core and peripheral IFT-A subunits: IFT139 is dispensable for IFT-A assembly but essential for retrograde trafficking of IFT-A, IFT-B, and GPCRs; in contrast, IFT144 is essential for functional IFT-A assembly and ciliary entry of GPCRs but dispensable for anterograde IFT-B trafficking. Thus the data presented here demonstrate that the IFT-A complex mediates not only retrograde trafficking but also entry into cilia of GPCRs.
INTRODUCTION
Cilia are microtubule-based appendages projecting from the surfaces of various eukaryotic cells. Cilia play essential roles in sensing extracellular stimuli and transducing developmental signals, such as Hedgehog (Hh) signaling (Ishikawa and Marshall, 2011; Sung and Leroux, 2013). Therefore a number of proteins are specifically localized in cilia, such as some G-protein–coupled receptors (GPCRs) and ion channels. Defects in ciliary assembly and functions cause a variety of hereditary disorders, generally called ciliopathies, with a broad spectrum of symptoms, including retinal degeneration, polycystic kidney, morbid obesity, and brain and skeletal malformations (Schwartz et al., 2011; Madhivanan and Aguilar, 2014). These disorders include Joubert syndrome, nephronophthisis, Meckel syndrome, Bardet–Biedl syndrome, short-rib thoracic dysplasia (SRTD), and cranioectodermal dysplasia (CED); note that SRTD has phenotypic overlaps with CED.
The assembly and maintenance of cilia by intraflagellar transport (IFT) were first demonstrated in Chlamydomonas reinhardtii (Rosenbaum and Witman, 2002) and subsequently intensively studied in metazoans (Ishikawa and Marshall, 2011; Sung and Leroux, 2013). IFT is mediated by the bidirectional movement of large protein particles, referred to as the IFT trains, along a microtubule-based structure called the axoneme. The IFT train contains the IFT-B complex, which comprises 16 subunits and mediates anterograde trafficking from the ciliary base to the tip with the aid of kinesin-2 motor proteins, and the IFT-A complex, which comprises at least six subunits and mediates retrograde trafficking powered by dynein-2 (Ishikawa and Marshall, 2011; Taschner et al., 2012; Sung and Leroux, 2013; also see Figure 7K later in this article). We and others recently demonstrated the overall architecture of the IFT-B complex, which can be divided into the core subcomplex (composed of 10 subunits) and the peripheral subcomplex (composed of six subunits), which are connected by composite interactions involving the IFT38, IFT52, IFT57, and IFT88 subunits (Boldt et al., 2016; Katoh et al., 2016; Taschner et al., 2016).
FIGURE 7:
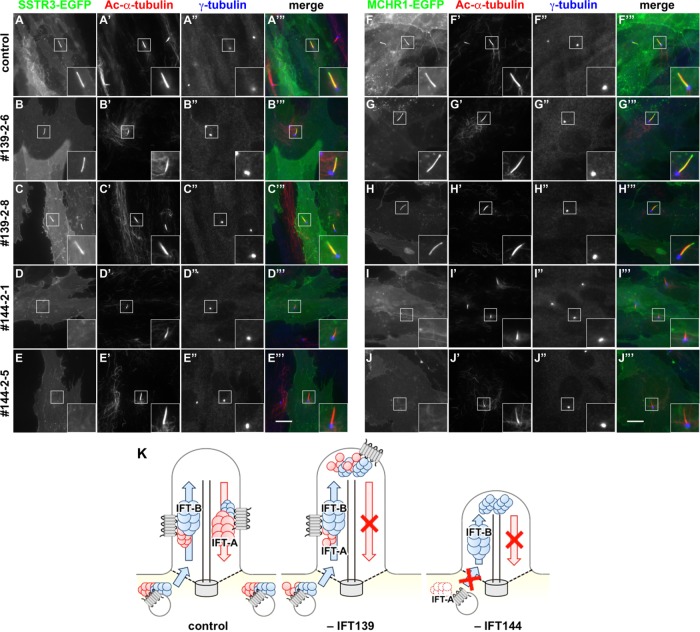
IFT144-KO cells demonstrate defects in the ciliary localization of SSTR3 and MCHR1. Control RPE1 cells (A, F), the IFT139-KO cell lines 139-2-6 (B, G) and 139-2-8 (C, H), and the IFT144-KO cell lines 144-2-1 (D, I) and 144-2-5 (E, J) stably expressing SSTR3-EGFP (A–E) or MCHR1-EGFP (F–J) were serum-starved for 24 h and double immunostained for Ac-α-tubulin (A′–J′) and γ-tubulin (A′′–J′′). (A′′′–D′′′) Merged images. Insets, enlarged images of the boxed regions. Scale bars, 10 µm. (K) Schematic representation of the phenotypes of control, IFT139-KO, and IFT144-KO cells. In control cells, the IFT-A complex is required for ciliary entry of GPCRs and anterogradely trafficked along with the IFT-B complex. Then the IFT-B complex and GPCRs undergo IFT-A–dependent retrograde trafficking. In the absence of IFT139 (−IFT139), other IFT-A subunits can be assembled, mediate ciliary entry of GPCRs, and are anterogradely trafficked along with the IFT-B complex. However, the IFT139-deficient IFT-A complex cannot mediate retrograde trafficking. In the absence of IFT144 (−IFT144), GPCRs cannot enter cilia because functional IFT-A complex cannot be formed. On the other hand, the IFT-B complex can be trafficked anterogradely, although it cannot undergo IFT-A–dependent retrograde trafficking.
In contrast, relatively little is known about the IFT-A complex. A major reason for the delay in biochemical studies of the IFT-A complex compared with those of IFT-B is that most IFT-A subunits (IFT43, IFT121, IFT122, IFT139, IFT140, and IFT144; Figure 1A) are larger than most IFT-B subunits. By combining yeast two-hybrid analysis, recombinant protein expression in Escherichia coli, and genetic analysis, Behal et al. (2012) proposed that Chlamydomonas IFT122, IFT140, and IFT144 create a stable core, with which IFT43 and IFT139 can interact. Others showed that mammalian TULP3, a Tubby family protein, which causes developmental defects through aberrant Hh signaling upon its deficiency (Norman et al., 2009), bridges the mammalian IFT-A complex and ciliary membrane phosphoinositides and also that IFT122, IFT140, and IFT144 constitute the core of IFT-A (Mukhopadhyay et al., 2010).
FIGURE 1:
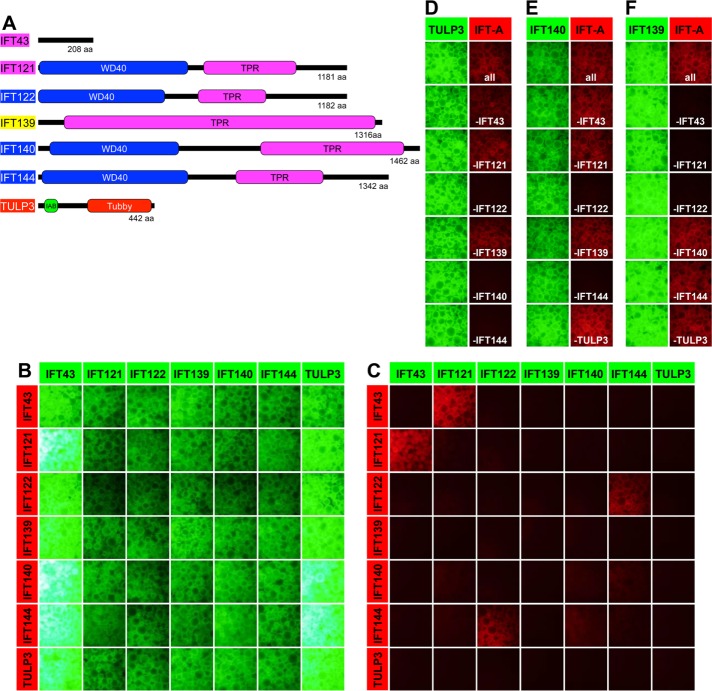
Interactions of IFT-A subunits demonstrated by all-by-all and subtractive VIP assays. (A) Schematic representation of the structure and domain organization of IFT-A proteins. IAB, IFT-A–binding sequence; WD40, WD40 repeat domain; TPR, tetratricopeptide repeat domain; Tubby, Tubby-like domain. (B, C) All-by-all VIP assay. HEK293T cells cultured in six-well plates were transfected with a combination of expression vectors for EGFP-fused and mChe-fused IFT-A proteins as indicated and incubated for 24 h. After confirmation of the expression of the EGFP and mChe fusion proteins in transfected cells under a microscope, lysates were prepared from the cells and precipitated with GST-tagged anti–GFP Nb prebound to glutathione–Sepharose beads. The green (B) and red (C) fluorescence signals on the precipitated beads were observed, and images of the beads were acquired using a BZ-8000 microscope. (D–F) Subtractive VIP assays. HEK293T cells were cotransfected with an expression vector for TULP3 (D), IFT140 (E), or IFT139 (F) fused to EGFP and expression vectors for all but one (as indicated) of the other IFT-A subunits fused to mChe, and lysates prepared from the transfected cells were processed for the VIP assay as described.
Recessive mutations in the genes of IFT-A subunits give rise to SRTD and CED; IFT43 (CED3), IFT121 (SRTD7/CED2), IFT122 (CED1), IFT139 (SRTD4), IFT140 (SRTD9), and IFT144 (SRTD5/CED4; Davis et al., 2011; Huber and Cormier-Daire, 2012; Lin et al., 2013; Miller et al., 2013). Defects in dynein-2 subunits are also known to cause SRTD. In addition, mice defective in their IFT-A genes have morphologically abnormal short cilia generally with bulged tips and exhibited aberrant Hh signaling (Tran et al., 2008; Qin et al., 2011; Liem et al., 2012; Miller et al., 2013).
We recently developed a novel strategy named the visible immunoprecipitation (VIP) assay as a simple, flexible, and versatile method for studying protein–protein interactions (Katoh et al., 2015). The VIP assay can visually detect binary protein interactions by microscopy without the need of electrophoresis and immunoblotting. Furthermore, the VIP assay can detect interactions involving more than two proteins, namely, one-to-many and many-to-many protein interactions. By taking advantage of this strategy, we determined the overall architectures of the exocyst, BBSome, and IFT-B complexes, composed of eight, eight, and 16 subunits, respectively (Katoh et al., 2015, 2016). Furthermore, we determined the way by which ARL13B and KIF17 interact with the IFT-B complex (Funabashi et al., 2017; Nozaki et al., 2017).
In this study, we applied the VIP assay to delineate the overall architecture of the mammalian IFT-A complex, including TULP3. Furthermore, we established human telomerase reverse transcriptase–immortalized retinal pigmented epithelial (hTERT-RPE1) cell lines defective in IFT139 or IFT144 using the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated 9 (Cas9) system with some modifications and found that the ciliary defects observed in the IFT139-knockout (KO) and IFT144-KO cells are different from each other in several aspects that reflect their predicted positioning within the IFT-A complex.
RESULTS
All-by-all VIP assays of IFT-A proteins
To delineate the overall architecture of the IFT-A complex, we first examined binary interactions between the six known constituents of the IFT-A complex, as well as TULP3 (Figure 1A), by applying the VIP assay as described previously (Katoh et al., 2015, 2016). Briefly, we cotransfected any of the 49 possible combinations of expression vectors for the IFT-A proteins fused to enhanced green fluorescent protein (EGFP) and mCherry (mChe) into HEK293T cells, processed the lysates prepared from the transfected cells for immunoprecipitation with glutathione S-transferase (GST)–tagged anti-GFP nanobody (Nb) prebound to glutathione–Sepharose beads, and directly observed green and red fluorescence signals on beads bound to the immunoprecipitates by fluorescence microscopy. Note that images of the beads were acquired under constant conditions in the same sets of experiments. We routinely assess binary interactions as positive when red signals are detected on the precipitated beads in reciprocal combinations of EGFP and mChe fusions under the same conditions because the expression levels and/or stability of certain fluorescent fusion proteins vary and are often affected by coexpressed proteins. The absence of a positive interaction in the VIP assay therefore does not always mean that the two proteins cannot interact with each other, similarly to other protein–protein interaction assays, such as the yeast two-hybrid assay.
Figure 1, B and C, shows signals of EGFP and mChe, respectively, on the beads immunoprecipitated with GST-tagged anti-GFP Nb in all-by-all VIP assays for the IFT-A proteins. Using this assay, we detected a strong interaction between IFT43 and IFT121, which was previously detected by yeast two-hybrid analysis of Chlamydomonas and human IFT-A proteins (Behal et al., 2012). In addition, we found a robust interaction between IFT122 and IFT144; this direct interaction has never been reported, although these two subunits and IFT140 were reported to form a stable subcomplex (Mukhopadhyay et al., 2010; Behal et al., 2012). A weak interaction was also observed between IFT140 and IFT144.
One-to-many subunit interactions demonstrated through subtractive VIP assays
As described, we failed to detect any obvious interactions of IFT139, IFT140, and TULP3 with the other IFT-A subunits. We therefore used a modified version of the flexible VIP assay system, namely, a subtractive VIP assay, with which we previously proved one-to-many and many-to-many subunit interactions in the exocyst and IFT-B complexes (Katoh et al., 2015, 2016; Funabashi et al., 2017; Nozaki et al., 2017). When EGFP-TULP3 was coexpressed with all the other IFT-A subunits fused to mChe in HEK293T cells, immunoprecipitation of lysates prepared from the cells yielded red fluorescence signals on the beads (Figure 1D, top), indicating that some IFT-A subunits interacted with TULP3. We then performed VIP assays by omitting one of the IFT-A subunits fused to mChe and found that the red fluorescence signals were greatly diminished in the absence of mChe-IFT122, mChe-IFT140, or mChe-IFT144. Similar experiments demonstrated that interactions of IFT140 and IFT139 with the other IFT-A subunits were greatly reduced in the absence of coexpressed IFT122 or IFT144 (Figure 1E) and IFT43, IFT121, or IFT122 (Figure 1F), respectively.
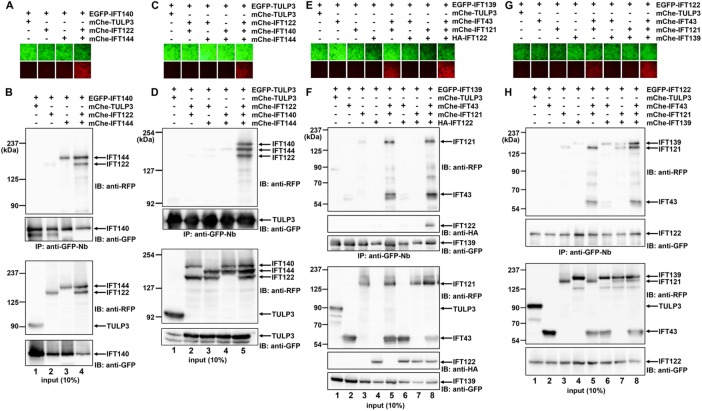
The subtraction assay data shown in Figure 1D suggest that interaction of the IFT-A complex with TULP3 involves IFT122, IFT140, and IFT144. In this context, it is noteworthy that these three IFT-A subunits were suggested to constitute a stable core subcomplex (Mukhopadhyay et al., 2010; Behal et al., 2012). Therefore we first addressed whether this potential subcomplex is actually formed. Because the data shown in Figure 1, B and C, suggest that IFT122 and IFT144 form a stable heterodimer and that IFT140 and IFT144 weakly interact with each other, and the subtraction data shown in Figure 1E suggest that IFT140 interacts with IFT122 and IFT144, we coexpressed EGFP-IFT140 with mChe-IFT122, mChe-IFT144, or their combination in HEK293T cells and subjected lysates from the cells to the VIP assay with GST-tagged anti-GFP Nb followed by conventional immunoblotting analysis. The VIP data indicate that IFT140 interacts robustly with the IFT122–IFT144 dimer to form a stable core subcomplex (Figure 2A). Subsequent immunoblotting analysis using an anti–red fluorescent protein (RFP) antibody that can detect mChe showed that IFT140 interacts strongly with the IFT122–IFT144 dimer (Figure 2B, lane 4) and weakly with IFT144 alone (lane 3).
FIGURE 2:
One-to-many subunit interactions in the IFT-A complex. (A, B) Interaction of IFT140 with IFT122–IFT144. HEK293T cells cultured in 6-cm dishes were transfected with expression vectors for EGFP-IFT140 and mChe-fused IFT-A subunits as indicated. (A) Lysates prepared from the cells were precipitated with GST-tagged anti–GFP Nb prebound to glutathione–Sepharose beads and processed for the VIP assay. (B) Proteins bound to the precipitated beads (top two panels) or input proteins (bottom two panels) were subjected to immunoblotting with an anti-RFP antibody (top and third panels) or anti-GFP antibody (second and bottom panels). (C, D) Interaction of TULP3 with the IFT122–IFT140–IFT144 trimer. Lysates prepared from HEK293T cells cotransfected with expression vectors for EGFP-TULP3 and mChe-fused IFT-A subunits, as indicated, were subjected to the VIP assay (C) and immunoblotting (D) as described. (E, F) Interaction of IFT139 with IFT43–IFT121. Lysates prepared from HEK293T cells coexpressing EGFP-IFT139, HA-IFT122, and mChe-fused IFT-A subunits, as indicated, were subjected to the VIP assay (E) and immunoblotting (F). To distinguish between IFT121 and IFT122, different tags, namely, mChe and HA, were used. (G, H) Interaction of IFT122 with IFT43–IFT121. Lysates prepared from HEK293T cells coexpressing EGFP-IFT122 and mChe-fused IFT-A subunits, as indicated, were subjected to the VIP assay (G) and immunoblotting (H).
We then examined whether TULP3 interacts with the core subcomplex en bloc. As shown in Figure 2, C and D, EGFP-TULP3 did not substantially interact with any two of mChe-IFT122, mChe-IFT140, and mChe-IFT144 (lanes 2–4). In considerable contrast, these three subunits fused to mChe were robustly coimmunoprecipitated with EGFP-TULP3 when they were simultaneously coexpressed (lane 5). Collectively we conclude that TULP3 interacts with the IFT-A complex through the core subcomplex composed of IFT122, IFT140, and IFT144 (see later discussion of Figure 3A).
FIGURE 3:
Validation of the architectural model of IFT-A. (A) Interaction map of IFT-A predicted from the data shown in Figures 1 and 2. (B, C) Interaction of TULP3 with the entire IFT-A complex. HEK293T cells were cotransfected with expression vectors for EGFP-TULP3 and for all but one (IFT43 or IFT140) of the other IFT-A subunits fused to mChe, and (B) lysates prepared from the cells were processed for the VIP assay. (C) Proteins bound to the precipitated beads (lanes 4–6) or input proteins (lanes 1–3) were subjected to immunoblotting with an anti-RFP (top), anti-IFT139 (middle), or anti-GFP antibody (bottom). (D, E) Interaction of IFT139 with the other IFT-A subunits. Lysates prepared from HEK293T cells cotransfected with an expression vector for EGFP-IFT139 and for all but one (IFT43 or IFT144) of the other IFT-A subunits fused to mChe were processed for the VIP assay (D) or immunoblotting (E), as indicated.
Next we addressed the mechanism by which IFT139 interacts with the other IFT-A subunits. Because the subtraction assay data shown in Figure 1F suggest that IFT43, IFT121, and IFT122 interact with IFT139, we coexpressed EGFP-IFT139 with any one, any two, or all three of mChe-IFT43, mChe-IFT121, and HA-IFT122 and subjected the cell lysates to the VIP assay followed by conventional immunoblot analysis; in this set of experiments, we used hemagglutinin (HA)-tagged IFT122 in place of mChe-IFT122 to discriminate between IFT121 and IFT122 because the bands for mChe-IFT121 and mChe-IFT122 overlap with each other on the immunoblot. As shown in Figure 2, E and F, IFT139 demonstrated robust interaction with the other IFT-A subunits when IFT43, IFT121, and IFT122 were simultaneously coexpressed (lane 8) but not when IFT43 plus IFT122 or IFT121 plus IFT122 were coexpressed (lanes 6 and 7). However, somewhat unexpected from the subtraction assay data in Figure 1F, EGFP-IFT139 coprecipitated mChe-IFT43 and mChe-IFT121 in the absence of coexpressed HA-IFT122 (lane 5) to an extent comparable with that in the presence of HA-IFT122 (lane 8). Although we do not know the exact reason for the apparent discrepancy between the results obtained by the subtractive VIP assay and the one-to-many protein VIP assay, IFT122 might be required for the interaction of IFT139 with IFT43–IFT121 within the overall IFT-A complex.
The data in Figure 2, E and F, suggest that IFT139 and IFT122 are indirectly connected by the IFT43–IFT121 dimer (lane 8). To confirm this prediction, we coexpressed EGFP-IFT122 with any one, any two, or all three of mChe-IFT43, mChe-IFT121, and mChe-IFT139 in HEK293T cells and subjected the cell lysates to the VIP assay and immunoblotting. As shown in Figure 2, G and H, IFT122 interacted robustly with the IFT43–IFT121 dimer (lane 5), through which it indirectly interacted with IFT139 (lane 8).
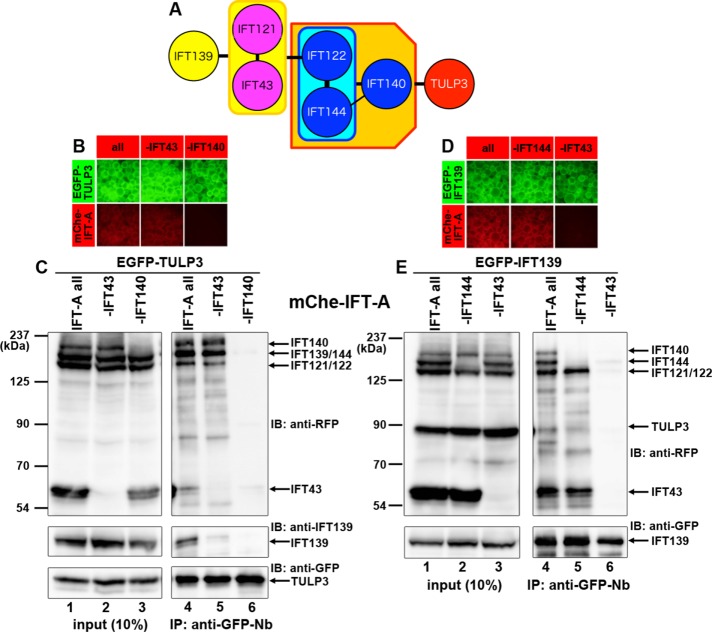
On the basis of all of the data in Figures 1 and 2, we propose a model of the overall architecture of the IFT-A complex (Figure 3A). In this model, TULP3 and IFT139 are located at opposite sides of the IFT-A complex and are connected by the core subcomplex composed of IFT122, IFT140, and IFT144 and the IFT43–IFT121 dimer. To validate this model, we examined whether all the subunits coexpressed can be coimmunoprecipitated en bloc. When EGFP-TULP3 was coexpressed with the other IFT-A subunits fused to mChe in HEK293T cells and the cell lysates processed for the VIP assay using GST–anti-GFP Nb (Figure 3B) followed by immunoblotting analysis with an anti-RFP antibody (Figure 3C), bands corresponding to mChe-IFT140, mChe-IFT139/IFT144, mChe-IFT121/IFT122, and mChe-IFT43 were detected (Figure 3C, top, lane 4). To overcome the overlap of the bands for mChe-IFT139 and mChe-IFT144 on immunoblot, we also detected immunoprecipitated proteins with an anti-IFT139 antibody and confirmed that mChe-IFT139 was indeed coprecipitated with EGFP-TULP3 (Figure 3C, middle, lane 4). When mChe-IFT43 was omitted from the coexpression, the IFT139 band intensity was greatly reduced, whereas the band intensities of the other IFT-A proteins were not substantially changed (lane 5). In striking contrast, bands for the IFT-A proteins were completely abolished when mChe-IFT140 was omitted (lane 6). These results support our model, in which TULP3 and IFT139 are connected through the other IFT-A subunits (Figure 3A).
Reciprocally, when EGFP-IFT139 was coexpressed with the other IFT-A proteins fused to mChe, similar results were obtained (Figure 3, D and E, lanes 4–6). One important point is that the mChe-TULP3 band that was coimmunoprecipitated with EGFP-IFT139 appeared considerably faint (lane 4). These results imply that TULP3 does not always form a complex with the other IFT-A subunits, although we did not further pursue this issue in this study.
IFT139-KO and IFT144-KO cell lines exhibit differential defects in the trafficking of IFT-A and IFT-B proteins
Previous histological and pathophysiological analyses of mutant mice clearly showed the crucial roles of IFT-A proteins in Hh signaling during development (Tran et al., 2008; Stottmann et al., 2009; Mill et al., 2011; Qin et al., 2011; Liem et al., 2012; Miller et al., 2013). In addition, studies on IFT122, IFT139, and IFT144 mutant mice at the cellular level indicated abnormal localization of the IFT-A and IFT-B proteins, as well as components of Hh signaling (Qin et al., 2011; Liem et al., 2012). However, these studies did not systematically address the roles of the IFT-A complex with respect to ciliary protein trafficking. With the aim to deepen our understanding of the roles of IFT-A components in ciliary protein trafficking, we established hTERT-RPE1 cell lines defective in IFT-A components, using the CRISPR/Cas9 system with our original modifications, as described in Materials and Methods; experimental details will be described elsewhere (Katoh, Michisaka, Nozaki, Funabashi, Hirano, Takei, and Nakayama, unpublished data). We chose IFT139 and IFT144 as targets because in our model of the architecture of IFT-A (Figure 3A), the former protein is most peripherally located in the complex, and the latter constitutes the core subcomplex. Among the obtained KO cell lines, we selected two independent cell lines for both IFT139 and IFT144 (Supplemental Figure S1) to use in the subsequent experiments.
When cilia were visualized by staining with antibodies against ARL13B (a marker for the ciliary membrane) and acetylated α-tubulin (Ac-α-tubulin; a marker for the ciliary axoneme), the staining for both markers was observed uniformly along the entire cilia in control RPE1 cells (Supplemental Figure S2, A–A’’’). In the two IFT139-KO cell lines (139-2-6 and 139-2-8), the length of cilia visualized by Ac-α-tubulin staining appeared to be comparable to that of control RPE1 cells (Supplemental Figure S2, B’ and C’). However, in these two cell lines, ARL13B staining often demonstrated bulged structures at the tips (Supplemental Figure S2, B and C, bottom insets), which are suggestive of ciliary membrane expansion, probably caused by excessive protein accumulation. In the two cell lines defective in IFT144 (144-2-1 and 144-2-5), cilia were substantially shorter than those of control cells (Supplemental Figure S2, D’, E’, and F), as observed for cells of IFT144 mutant mice (Liem et al., 2012).
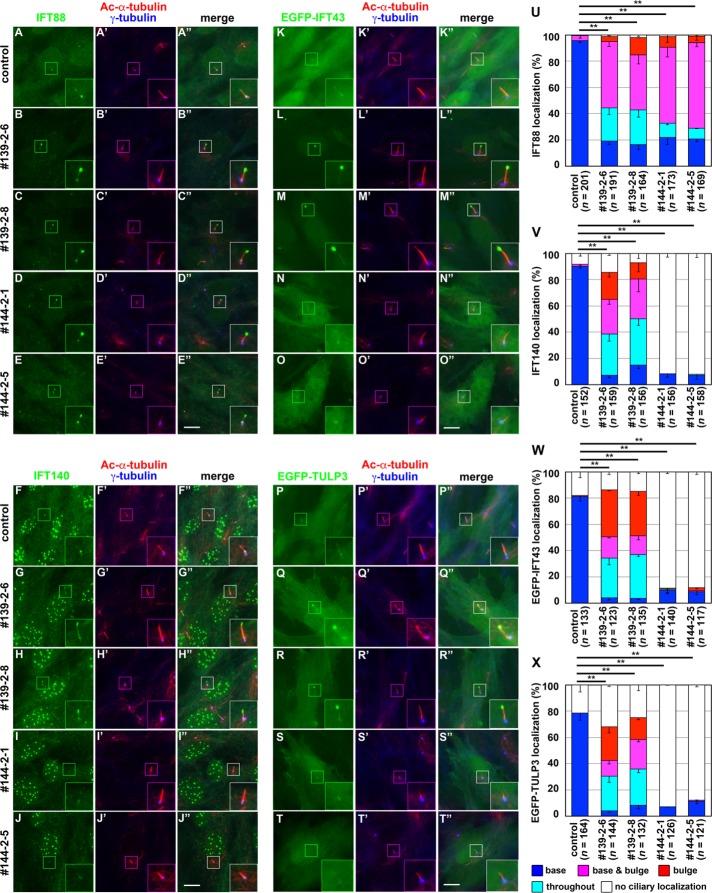
When stained for the IFT-B subunit IFT88, more appreciable differences were observed between control cells and IFT139-KO and IFT144-KO cells. In control RPE1 cells, IFT88 was found mainly around the base of cilia and faintly at ciliary tips (Figure 4A). Cell lines defective in IFT139 (Figure 4, B and C) and IFT144 (Figure 4, D and E) showed prominent IFT88 staining at the bulged tips (also see Figure 4U), which is consistent with previous observations of cells from IFT122, IFT139, and IFT144 mutant mice (Tran et al., 2008; Qin et al., 2011; Liem et al., 2012). The most plausible explanation for these observations is that, in the absence of the IFT-A complex, the IFT-B complex can be trafficked to the ciliary tips but cannot undergo retrograde transport, thereby accumulating at the tips.
FIGURE 4:
IFT139-KO and IFT144-KO cells demonstrate differential defects in the trafficking of IFT-A and IFT-B complexes. (A–J) Control RPE1 cells (A, F), IFT139-KO cell lines 139-2-6 (B, G) and 139-2-8 (C, H), and IFT144-KO cell lines 144-2-1 (D, I) and 144-2-5 (E, J) were serum-starved for 24 h and triple immunostained for IFT88 (A–E) or IFT140 (F–J) and Ac-α-tubulin and γ-tubulin (A′–J′). (K–T) Control RPE1 cells (K, P), the IFT139-KO cell lines 139-2-6 (L, Q) and 139-2-8 (M, R), or the IFT144-KO cell lines 144-2-1 (N, S) and 144-2-5 (O, T), which stably express EGFP-IFT43 (K–J) or EGFP-TULP3 (K–T), were serum-starved for 24 h and double immunostained for Ac-α-tubulin and γ-tubulin (K′–T′). (A′′–T′′) Merged images. Insets, enlarged images of the boxed regions. Scale bars, 10 µm. (U–X) Localization of IFT88 (U), IFT140 (V), EGFP-IFT43 (W), and EGFP-TULP3 (X) in individual control, IFT139-KO, and IFT144-KO cells was classified as mainly ciliary base, mainly ciliary bulge, base and bulge, throughout the cilium, and no ciliary localization and counted. Note that bulges were present at ciliary tips in most of the cells but in the middle of cilia in a small fraction of cells. The percentages of these populations are expressed as stacked bar graphs. Values are means ± SE of three independent experiments. In each set of experiments, 50–75 (U), 45–59 (V), 35–52 (W), and 38–62 (X) ciliated cells were observed, and the total number of ciliated cells observed (n) is shown. **p < 0.0001 (Pearson’s χ2 test).
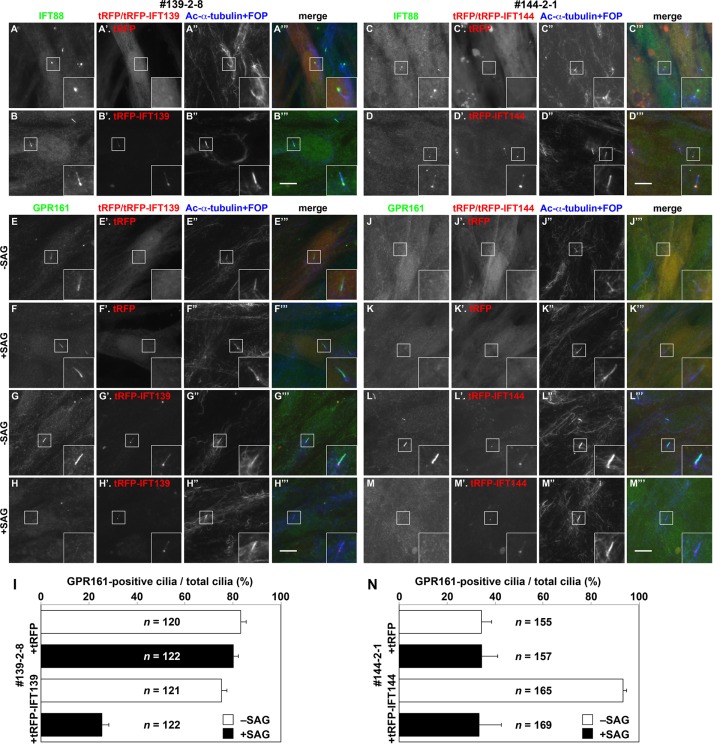
Because one of the most critical concerns with genome editing using the CRISPR/Cas9 system is off-target cleavage, we next performed rescue experiments; IFT139-KO (139-2-8) and IFT144-KO (144-2-1) cells were infected with a lentiviral vector to stably express TagRFP (tRFP), tRFP-IFT139, or tRFP-IFT144. As in noninfected IFT139-KO and IFT144-KO cells (Figure 4, C and D), cells expressing tRFP demonstrated IFT88 staining at the bulged tips (Figure 5, A and C). In contrast, cells expressing tRFP-IFT139 or tRFP-IFT144 demonstrated IFT88 staining mainly at the ciliary base (Figure 5, B and D) as in control RPE1 cells (Figure 4A).
FIGURE 5:
Rescue experiments of IFT139-KO and IFT144-KO cells. (A–D) The IFT139-KO cell line 139-2-8 (A, B) and the IFT144-KO cell line 144-2-1 (C, D) stably expressing tRFP (A′ and C′), tRFP-IFT139 (B′), or tRFP-IFT144 (D′) were serum-starved for 24 h and double immunostained for IFT88 (A–D) and ciliary markers, Ac-α-tubulin (a marker for the ciliary axoneme), and FOP (a marker for the ciliary base; A′′–D′′). (E–N) The IFT139-KO cell line 139-2-8 (E–H) and the IFT144-KO cell line 144-2-1 (J–M) stably expressing tRFP (E′, F′, I′, J′), tRFP-IFT139 (G′, H′), or tRFP-IFT144 (L′, M′) were serum-starved for 24 h, cultured for a further 24 h in the absence (− SAG) or presence (+ SAG) of 200 nM SAG, and immunostained for GPR161 (E–H, J–M) and Ac-α-tubulin and FOP (E′′–H′′, J′′–M′′). (A′′′–H′′′, J′′′–M′′′) Merged images. Insets, enlarged images of the boxed regions. Scale bars, 10 µm. (I, N) Cells with GPR161-positive cilia were counted, and the percentages of ciliated cells with GPR161-positive cilia are represented as bar graphs. Values are means ± SE (error bars) of three independent experiments. In each experiment, 38–41 cells (I) and 49–63 ciliated cells (N) were observed, and the total number of ciliated cells observed (n) is shown in the bar graphs.
We then stained control, IFT139-KO, and IFT144-KO cells with an antibody against IFT140 and observed more complicated differences in IFT140 localization. In control RPE1 cells, IFT140 staining was mainly found at the base, with weak staining along cilia (Figure 4F); note that the available anti-IFT140 antibody also stained undetermined nuclear structures in RPE1 cells, as described on the manufacturer’s website (www.ptglab.com/Products/IFT140-Antibody-17460-1-AP.htm). In IFT139-KO cells, IFT140 staining was mainly observed at the bulged ciliary tips (Figure 4, G and H; also see Figure 4V), similarly to IFT88 staining (Figure 4, B and C). IFT144-KO cells, however, had no apparent IFT140 staining within cilia but had faint staining around the base of cilia (Figure 4, I and J; also see Figure 4V). To address whether the differential effects of the loss of IFT139 and IFT144 on IFT140 localization was specific to this IFT-A core subunit, we established cells stably expressing EGFP-IFT43 and EGFP-TULP3 and examined their localization. Similar to endogenous IFT140, EGFP-IFT43 and EGFP-TULP3 were mainly localized at the ciliary base in control cells (Figure 4, K and P, respectively), predominantly at the bulged tips in IFT139-KO cells (Figure 4, L and M, and Q and R, respectively) and faintly around the ciliary base in IFT144-KO cells (Figure 4, N and O, and S and T, respectively; also see Figure 4, W and X). These observations, together with the data for the IFT-A architecture in Figures 1– 3, suggest that 1) the most peripherally associated subunit, IFT139, is dispensable for the assembly and anterograde transport of the complex containing other IFT-A subunits and the IFT-B complex but essential for the retrograde transport of IFT particles containing the IFT-A and IFT-B complexes; and 2) the core subunit IFT144 is essential for ciliary entry of the IFT-A complex, probably due to its crucial role in assembly of the functional complex.
IFT139-KO and IFT144-KO cell lines demonstrate differential defects in the trafficking of Smoothened and GPR161
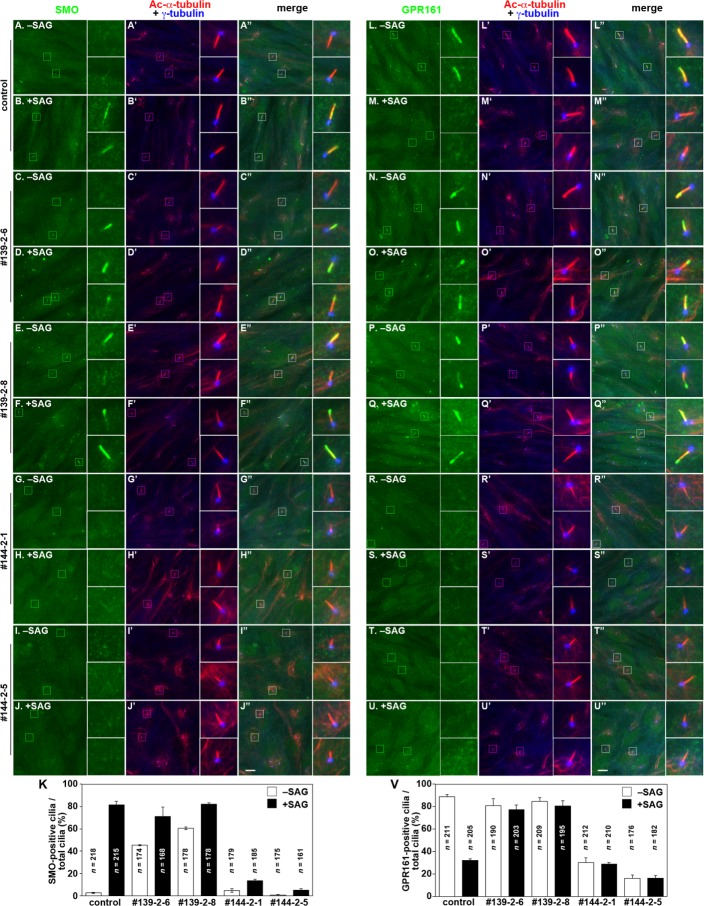
We then examined the effects of the loss of IFT139 and IFT144 on the localization of Smoothened (SMO) and GPR161, both of which are GPCRs involved in Hh signaling. Under basal conditions, SMO is not localized within cilia, whereas GPR161 is localized on the ciliary membrane and negatively regulates the Hh signaling pathway via the activation of protein kinase A (Mukhopadhyay and Rohatgi, 2014). In contrast, on receiving the Hh signal via the Hh receptor Patched 1, SMO is activated by a yet-unknown mechanism and enters cilia, whereas GPR161 is displaced from cilia (Figure 6, A and B, and L and M, respectively).
FIGURE 6:
IFT139-KO and IFT144-KO cells demonstrate differential defects in the trafficking of SMO and GPR161 in response to SAG treatment. Control RPE1 cells (A, B, L, M), the IFT139-KO cell lines 139-2-6 (C, D, N, O) and 139-2-8 (E, F, P, Q), and the IFT144-KO cell lines 144-2-1 (G, H, R, S) and 144-2-5 (I, J, T, U) were serum-starved for 24 h, cultured for a further 24 h in the absence (− SAG) or presence (+ SAG) of 200 nM SAG, and triple immunostained for either SMO (A–J) or GPR161 (L–U), Ac-α-tubulin, and γ-tubulin (A′–J′, L′–U′). (A′′–J′′, L′′–U′′) Merged images. Insets, enlarged images of the boxed regions. Scale bars, 10 µm. (K, V) Cells with SMO-positive cilia (K) and those with GPR161-positive cilia (V) were counted, and the percentages of ciliated cells with SMO- or GPR161-positive cilia are represented as bar graphs. Values are means ± SE (error bars) of three independent experiments. In each experiment, 50–80 cells (K) and 47–78 cells (V) were counted, and the total numbers of ciliated cells observed (n) is shown in the bar graphs.
In the IFT139-KO cell lines, however, SMO was found within ∼50% of cilia even under basal conditions (Figure 6, C and E; also see Figure 6K). When the cells were treated with the Hh pathway activator Smoothened agonist (SAG), a large proportion of SMO was localized within cilia to a level comparable to that in SAG-treated control cells (Figure 6, D, F, and K). These observations suggest that SMO undergoes cycling, albeit at low levels, between the cell body and cilia even under basal conditions and that a block in retrograde transport due to the lack of IFT139 results in the accumulation of SMO within cilia. In clear contrast to the IFT139-KO cell lines, in the IFT144-KO cell lines, SMO was marginally observed in cilia both in the absence and presence of SAG (Figure 6, G–J, and K), indicating that SMO cannot enter cilia in the absence of the functional IFT-A complex due to the lack of the core subunit, IFT144.
In IFT139-KO cells, GPR161 was found in cilia under basal conditions, similarly to in control cells (Figure 6, N and P); however, unlike in control cells, GPR161 did not exit cilia upon SAG treatment (Figure 6, O, Q, and V), indicating a block in the retrograde transport of GPR161. In IFT139-KO (139-2-8) cells exogenously expressing tRFP-IFT139 (Figure 5, G and H) but not in those expressing tRFP (Figure 5, E and F), GPR161 was able to exit cilia in response to SAG treatment (also see Figure 5I), indicating that the abnormal GPR161 localization observed in IFT139-KO cells did not result from off-target effects of the CRISPR/Cas9 system.
In clear contrast to the IFT139-KO cells, GPR161 did not localize within cilia under both basal and SAG-stimulated conditions in IFT144-KO cells (Figure 6, R–U; also see Figure 6V), indicating that the ciliary entry of GPR161, as well as that of SMO, was blocked in the absence of the functional IFT-A complex. The basal ciliary localization and SAG-stimulated exit of GPR161 were restored when tRFP-IFT144 (Figure 5, L and M) but not tRFP (Figure 5, J and K) was exogenously expressed (also see Figure 5N).
GPCRs other than those involved in Hh signaling also fail to enter cilia in the IFT144-KO cell lines
The foregoing data indicate that the IFT-A complex is required not only for the retrograde transport of the GPCRs SMO and GPR161 but also for their ciliary entry. We then set out to address whether the block in ciliary entry of GPCRs in the absence of IFT144 is specific for GPCRs involved in Hh signaling or is a more general event. To this end, we established control, IFT139-KO, and IFT144-KO cells stably expressing SSTR3 and MCHR1 fused to EGFP and examined their localization.
In IFT139-KO cells, both SSTR3-EGFP and MCHR1-EGFP were found throughout the cilia (Figure 7, B and C, and G and H, respectively) similarly to in control cells (Figure 7, A and F). In marked contrast, both SSTR3-EGFP and MCHR1-EGFP failed to localize within cilia in the IFT144-KO cell lines (Figure 7, D and E, and I and J, respectively). Thus our data unequivocally demonstrate that the ciliary entry of GPCRs, including SMO, GPR161, SSTR3, and MCHR1, generally requires the assembly of functional IFT-A complex around the ciliary base.
DISCUSSION
On the basis of the data obtained using a combination of two different approaches (determination of the IFT-A architecture using the VIP assay and analyses of KO cells established using the modified CRISPR/Cas9 system), we here unequivocally showed the differential roles of two IFT-A subunits, which reflect their positioning within the IFT-A complex. Cells defective in IFT139, which is the most peripherally associated subunit, showed accumulation of the IFT-B complex and other IFT-A proteins at the bulged ciliary tips, as well as the ciliary accumulation of GPCRs, including SMO and GPR161. Thus this peripheral subunit is not required for construction of the IFT-A complex by the other subunits or for anterograde ciliary protein trafficking but is essential for retrograde trafficking from the tip to the base (Figure 7K, –IFT139). On the other hand, in cells defective in IFT144, which is a constituent of the core subcomplex, other IFT-A subunits and GPCRs failed to localize within cilia, although the IFT-B complex accumulated at the bulged tips. Therefore these observations indicate that IFT144 is essential for assembly of the functional IFT-A complex and for ciliary entry of GPCRs, including SMO, GPR161, SSTR3, and MCHR1, and that the IFT-B complex can be transported anterogradely in the absence of the IFT-A complex (Figure 7K, –IFT144). Thus the phenotype of IFT139-KO cells appears milder than that of IFT144-KO cells, reflecting the auxiliary role of IFT139 as predicted from the architecture of the IFT-A complex.
Most intriguingly, our data indicate that the IFT-A complex is involved not only in retrograde protein trafficking from the tip to the base of cilia but also in ciliary entry of GPCRs and probably other transmembrane proteins (Figure 7K, control). The role of the IFT-A complex in retrograde ciliary trafficking, as well as the role of the IFT-B complex in anterograde trafficking, is well established (Ishikawa and Marshall, 2011; Sung and Leroux, 2013). However, before our study, fragmentary evidence suggested the roles of the IFT-A complex in anterograde ciliary trafficking or periciliary protein trafficking as follows. 1) On the basis of analyses using cells derived from IFT144 mutant mice, Liem et al. (2012) suggested that this IFT-A subunit is required for anterograde trafficking of some membrane proteins, including SMO and adenylyl cyclase III, although they did not mention the potential involvement of IFT-A in ciliary entry. 2) On the basis of knockdown experiments of the IFT-A subunits, including TULP3, Mukhopadhyay et al. (2010) suggested that the IFT-A complex is involved in some way in ciliary localization of some GPCRs, including SSTR3 and MCHR1, although they observed no appreciable defects in SMO trafficking upon single or double knockdown of any IFT-A subunit. 3) Caparrós-Martín et al. (2015) reported that SMO, EvC, and EvC2, the latter two of which form a ciliary transmembrane protein complex and are mutated in patients of Ellis–van Creveld syndrome, failed to localize within the cilia of cells derived from IFT121-null mice. 4) Finally, on the basis of studies on specialized cilia in Drosophila, Lee et al. (2008) speculated that IFT-A proteins are required for the periciliary trafficking of TRPV channel proteins. Taken our results together with these previous studies, we conclude that the IFT-A complex plays a crucial role in the ciliary entry of transmembrane proteins, including GPCRs.
Although the ciliary membrane is continuous with the plasma membrane, the ciliary transition zone (TZ) partitions the interior and exterior of cilia by serving as a diffusion/permeability barrier (Wei et al., 2015; Verhey and Yang, 2016). Proteins required for the assembly and functions of cilia therefore have to be selectively transported across the TZ from the cell body. Takao et al. (2014) showed that ciliary soluble and membrane proteins enter cilia across the TZ by distinct mechanisms. A study using total internal reflection microscopy to analyze Chlamydomonas flagella suggested that IFT proteins are accumulated at the flagellar base before their entry into flagella (Ludington et al., 2013). Furthermore, a recent study using superresolution microscopy not only clarified the architectural map of the TZ but also indicated that the TZ serves as an assembly or resting site for IFT particles before entering cilia (Yang et al., 2015).
Ciliary transmembrane proteins are believed to be transported from the trans-Golgi network or recycling endosomes by vesicular carriers that target specifically to the periciliary membrane, where transition fibers from the distal appendage of the basal body are attached (Sung and Leroux, 2013; Pedersen et al., 2016). Zhao and Malicki (2011) showed that some TZ proteins interact with IFT proteins and support the ciliary entry of a GPCR (opsin). Furthermore, Wei et al. (2013, 2015) showed that FBF1, a protein located at the transition fiber and the distal appendage of the basal body, interacts directly with IFT54 and is required for the transit of assembled IFT particles across the TZ. On the other hand, activated SMO was shown to enter the ciliary compartment from the periciliary membrane by lateral movement, either via diffusion or active transport (Milenkovic et al., 2009). It is therefore tempting to speculate that binding to preassembled IFT particles is required for the lateral movement of transmembrane proteins across the TZ.
The next issue to be addressed is therefore how the IFT-A complex interacts directly or indirectly with transmembrane proteins and mediates their ciliary entry and retrograde trafficking. It will also be interesting to know how the IFT-A complex interacts with the IFT-B complex to mediate anterograde trafficking and probably the ciliary entry of transmembrane proteins, as well as how the IFT-A complex interacts with dynein-2 to mediate retrograde trafficking. Our VIP assay might pave the way to address these issues by serving as a powerful tool to detect protein–protein interactions.
MATERIALS AND METHODS
Plasmids
The full coding sequences for the IFT-A proteins listed in Supplemental Table S1 were cloned into fluorescent protein vectors as shown in Supplemental Table S2; the IFT122, IFT140, and TULP3 cDNAs were obtained from the RIKEN BRC through the National Bio-Resource Project of the Ministry of Education, Culture, Sports, Science and Technology, Japan (Ota et al., 2004). Expression vectors for mouse SSTR3 and MCHR1 (Nagata et al., 2013) were kindly provided by Yumiko Saito (Hiroshima University, Hiroshima, Japan) and Sen Takeda (University of Yamanashi, Yamanashi, Japan) and modified as described previously (Hamamoto et al., 2016).
Antibodies and reagents
The antibodies used in this study are listed in Supplemental Table S4. GST-tagged anti-GFP Nb prebound to glutathione–Sepharose 4B beads was prepared as described previously (Katoh et al., 2015). SAG and Polyethylenimine Max were purchased from Enzo Life Sciences and Polysciences, respectively.
Establishment of KO cell lines using the CRISPR/Cas9 system
The knockout strategy for genes in hTERT-RPE1 cells (CRL-4000; American Type Culture Collection) by the CRISPR/Cas9 system using homology-independent DNA repair will be described elsewhere in detail (Katoh et al., unpublished data). Single guide RNA (sgRNA) sequences targeting the human IFT139 and IFT144 genes (Supplemental Table S3) were designed using a CRISPR design (Hsu et al., 2013). Double-stranded oligonucleotides for these sequences were separately inserted into a donor knock-in vector, pDonor-tBFP-NLS-Neo (deposited in Addgene, ID 80766), and an all-in-one sgRNA expression vector, pSpCas9(BB)-2A-Puro (Addgene plasmid 48139). hTERT-RPE1 cells cultured to ∼3.0 × 105 cells on a 12-well plate were transfected with 1 µg of the pSpCas9(BB)-2A-Puro and 0.25 µg of the donor vector using X-tremeGENE9 DNA Transfection Reagent (Roche Applied Science). After selection in the presence of G418 (600 µg/ml), the cells with nuclear blue fluorescence were isolated. Genomic DNA was extracted from the isolated cells and subjected to PCR using KOD FX Neo DNA polymerase (Toyobo). Three sets of primers (Supplemental Table S3) were used to distinguish the following three states of integration of the donor knock-in vector: forward integration, reverse integration, and no integration with a small insertion or deletion (Supplemental Figure S1, A and D). Direct sequencing of the PCR products ensured the KO of both alleles of the IFT139 and IFT144 genes, with integration of the donor vector and/or a small deletion/insertion causing a frameshift (Supplemental Figure S1, B, C, E, and F).
Preparation of cells stably expressing EGFP-IFT43, EGFP-TULP3, SSTR3-EGFP, MCHR1-EGFP, tRFP, tRFP-IFT139, or tRFP-IFT144
Lentiviral vectors were prepared as described previously (Takahashi et al., 2012). Briefly, pRRLsinPPT-EGFP-IFT43, pRRLsinPPT-EGFP-TULP3, pRRLsinPPT-SSTR3-EGFP, pRRLsinPPT-MCHR1-EGFP, pRRLsinPPT-tRFP, pRRLsinPPT-tRFP-IFT139, or pRRLsinPPT-tRFP-IFT144 was transfected into HEK293T cells using Polyethylenimine Max together with the packaging plasmids (pRSV-REV, pMD2.g, and pMDL/pRRE; kind gifts from Peter McPherson, McGill University, Montreal, Canada; Thomas et al., 2009). Culture medium was replaced 8 h after transfection and collected at 24, 36, and 48 h after transfection. The medium containing viral particles was filtrated through a 0.45-µm filter and centrifuged at 32,000 × g at 4°C for 4 h. Precipitated lentiviral particles were resuspended in Opti-MEM (Invitrogen) and stored at −80°C until use. Control cells, IFT139-KO cells, and IFT144-KO cells that express EGFP-IFT43, EGFP-TULP3, SSTR3-EGFP, MCHR1-EGFP, tRFP, tRFP-IFT139, and tRFP-IFT144 were prepared by the addition of lentiviral suspension to the culture medium and used for immunofluorescence analysis.
VIP assays
VIP assays were performed as described previously (Katoh et al., 2015, 2016). Briefly, HEK293T cells (∼1.6 × 106 cells in a six-well plate) were transfected with EGFP and mChe fusion constructs (2 µg of each) using Polyethylenimine Max (20 µg) and cultured for 24 h. The cells were then lysed in cell lysis buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid–KOH, pH 7.4, 150 mM NaCl, 10% glycerol, and 0.1% Triton X-100) containing EDTA-free protease inhibitor cocktail (Nacalai Tesque). After 15 min on ice, the cell lysates were centrifuged at 16,100 × g for 15 min at 4°C in a microcentrifuge. The supernatants (200 µl) were incubated with 5 µl of GST-tagged anti–GFP Nb prebound to glutathione–Sepharose beads in 0.2-ml eight-tube strips for 1 h at 4°C. The tube strips were centrifuged at 2000 × g for 30 s at room temperature. The precipitated beads were washed three times with 180 µl of lysis buffer and then transferred to a 96-well plate for observation. Precipitated beads with fluorescent fusion proteins were observed using an all-in-one–type fluorescence microscope (BZ-8000; Keyence) using a 20 ×/0.75 objective lens under constant conditions (sensitivity ISO 400, exposure 1/10 s for green fluorescence; sensitivity ISO 800, exposure 1/10 s for red fluorescence). Image acquisition was performed under constant conditions in the same series of experiments. When indicated, the materials bound to the beads were subjected to immunoblot analysis using an anti-GFP or anti-RFP antibody after image acquisition (see further description).
Immunoblot analysis
Immunoblot analysis was performed as described previously (Katoh et al., 2015, 2016). Proteins in cell lysates prepared as described or on beads after the VIP assay were separated by SDS–PAGE and electroblotted onto an Immobilon-P membrane (EMD Millipore). Membranes were blocked in 5% skim milk and incubated sequentially with a primary antibody (anti-GFP, anti-RFP antibody, anti-HA, or anti-IFT139) and horseradish peroxidase–conjugated secondary antibody. Detection was carried out using the Chemi-Lumi One L Kit (Nacalai Tesque).
Immunofluorescence analysis
hTERT-RPE1 cells were cultured in DMEM/F-12 (Nacalai Tesque) supplemented with 10% fetal bovine serum (FBS) and 0.348% sodium bicarbonate. To induce ciliogenesis, cells were grown to 100% confluence on coverslips and starved for 24 h in Opti-MEM (Invitrogen) containing 0.2% bovine serum albumin. Cells were fixed and permeabilized with 3% paraformaldehyde at 37°C for 5 min and subsequently in methanol at −20°C for 5 min and washed three times with phosphate-buffered saline (PBS). For detection of endogenous IFT140, cells were fixed and permeabilized with methanol at −20°C for 5 min and washed three times with PBS. The fixed/permeabilized cells were blocked with 10% FBS and stained with antibodies diluted with 5% FBS. The stained cells were observed using an Axiovert 200M microscope (Carl Zeiss). Statistical analyses were performed using JMP Pro 12 software (SAS Institute).
Supplementary Material
Acknowledgments
We are grateful to Yumiko Saito and Sen Takeda for providing SSTR3 and MCHR1 cDNAs, Yoshihide Hayashizaki and Sumio Sugano for providing IFT122, IFT140, and TULP3 cDNAs, and Peter McPherson for providing plasmids for recombinant lentivirus production. We also thank Helena Akiko Popiel for a critical reading of the manuscript. This work was supported in part by Grants-in-Aid for Scientific Research on Innovative Areas “Cilia and Centrosome” from the Ministry of Education, Culture, Sports, Science and Technology, Japan (Grants 25113514 and 15H01211 to K.N.), the Japan Society for the Promotion of Science (Grants 15H04370 to K.N. and 25860044 and 15K07929 to Y.K.), and grants from the Uehara Memorial Foundation to K.N. and from the Takeda Science Foundation to Y.K.
Glossary
Abbreviations used:
- Cas9
CRISPR-associated 9
- CED
cranioectodermal dysplasia
- CRISPR
clustered regularly interspaced short palindromic repeats
- EGFP
enhanced green fluorescent protein
- FBS
fetal bovine serum
- GPCR
G protein–coupled receptor
- GST
glutathione S-transferase
- Hh
hedgehog
- IFT
intraflagellar transport
- KO
knockout
- Nb
nanobody
- SAG
smoothened agonist
- SMO
smoothened
- SRTD
short-rib thoracic dysplasia
- TZ
transition zone
- VIP
visible immunoprecipitation
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-11-0813) on December 8, 2016.
REFERENCES
- Behal RH, Miller MS, Qin H, Lucker BF, Jones A, Cole DG. Subunit interactions and organization of the Chlamydomonas reinhardtii intraflagellar transport complex A proteins. J Biol Chem. 2012;287:11689–11703. doi: 10.1074/jbc.M111.287102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldt K, van Reeuwijk J, Lu Q, Koutroumpas K, Nguyen TM, Texier Y, van Beersum SEC, Horn N, Willer JR, Mans D, et al. An organelle-specific protein landscape identifies novel diseases and molecular mechanisms. Nat Commun. 2016;7:11491. doi: 10.1038/ncomms11491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caparrós-Martín JA, De Luca A, Cartault F, Aglan M, Temtamy S, Otaify GA, Mehrez M, Valencia M, Vázquez L, Alessandri JL, et al. Specific variants in WDR35 cause a distinctive form of Ellis-van Creveld syndrome by disrupting the recruitment of the EvC complex and SMO into the cilium. Hum Mol Genet. 2015;24:4126–4137. doi: 10.1093/hmg/ddv152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis EE, Zhang Q, Liu Q, Diplas BH, Davey LM, Hartley J, Stoetzel C, Szymanska K, Ramaswami G, Logan CV, et al. TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat Genet. 2011;43:189–196. doi: 10.1038/ng.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabashi T, Katoh Y, Michisaka S, Terada M., Sugawa M, Nakayama K. Ciliary entry of KIF17 is dependent on its binding to the IFT-B complex via IFT46-IFT56 as well as on its nuclear localization signal. Mol Biol Cell. 2017;28 doi: 10.1091/mbc.E16-09-0648. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamamoto A, Yamato S, Katoh Y, Nakayama K, Yoshimura K, Takeda S, Kobayashi Y, Saito Y. Modulation of ciliary length by melanin-concentrating hormone receptor 1. Cell Signal. 2016;27:572–584. doi: 10.1016/j.cellsig.2016.02.018. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber C, Cormier-Daire V. Ciliary disorder of the skeleton. Am J Med Genet. 2012;160C:165–174. doi: 10.1002/ajmg.c.31336. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF. Ciliogenesis: building the cell’s antenna. Nat Rev Mol Cell Biol. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Nozaki S, Hartanto D, Miyano R, Nakayama K. Architectures of multisubunit complexes revealed by a visible immunoprecipitation assay using fluorescent fusion proteins. J Cell Sci. 2015;128:2351–2362. doi: 10.1242/jcs.168740. [DOI] [PubMed] [Google Scholar]
- Katoh Y, Terada M, Nishijima Y, Takei R, Nozaki S, Hamada H, Nakayama K. Overall architecture of the intraflagellar transport (IFT)-B complex containing Cluap1/IFT38 as an essential component of the IFT-B peripheral subcomplex. J Biol Chem. 2016;291:10962–10975. doi: 10.1074/jbc.M116.713883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Sivan-Loukianova E, Eberl DF, Kernan MJ. An IFT-A protein is required to delimit functionally distinct zones in mechanosensory cilia. Curr Biol. 2008;18:1899–1906. doi: 10.1016/j.cub.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem KF jr, Ashe A, He M, Satir P, Moran J, Beier D, Wicking C, Anderson KV. The IFT-A complex regulates Shh signaling through cilia structure and membrane protein trafficking. J Cell Biol. 2012;197:789–800. doi: 10.1083/jcb.201110049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AE, Traum AZ, Sahai I, Keppler-Noreuil K, Kukolich MK, Adam MP, Westra SJ, Arts HH. Sensenbrenner syndrome (Cranioectodermal dysplasia): clinical and molecular analyses of 39 patients including two new patients. Am J Med Genet. 2013;161A:2762–2776. doi: 10.1002/ajmg.a.36265. [DOI] [PubMed] [Google Scholar]
- Ludington WB, Wemmer KA, Lechtreck K-F, Witman GB, Marshall WF. Avalanche-like behavior in ciliary import. Proc Natl Acad Sci USA. 2013;110:3925–3930. doi: 10.1073/pnas.1217354110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhivanan K, Aguilar RC. Ciliopathies: the trafficking connection. Traffic. 2014;15:1031–1056. doi: 10.1111/tra.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milenkovic L, Scott MP, Rohatgi R. Lateral transport of Smoothened from the plasma membrane to the membrane of the cilium. J Cell Biol. 2009;187:365–374. doi: 10.1083/jcb.200907126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill P, Lockhart PJ, Fitzpatrick E, Mountford HS, Hall EA, Reijns MA, Keighren M, Bahlo M, Bromhead CJ, Budd P, et al. Human and mouse mutations in WDR35 cause short-rib polydactyly syndromes due to abnormal ciliogenesis. Am J Hum Genet. 2011;88:508–515. doi: 10.1016/j.ajhg.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KA, Ah-Cann CJ, Welfare MF, Tan TY, Pope K, Caruana G, Freckmann ML, Savarirayan R, Bertram JF, Dobbie MS, et al. Cauli: a mouse strain with an Ift140 mutation that results in a skeletal ciliopathy modelling Jeune syndrome. PLoS Genet. 2013;9:e1003746. doi: 10.1371/journal.pgen.1003746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Rohatgi R. G-protein-coupled receptors, Hedgehog signaling and primary cilia. Semin Cell Dev Biol. 2014;33:63–72. doi: 10.1016/j.semcdb.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S, Wen X, Chih B, Nelson CD, Lane WS, Scales SJ, Jackson PK. TULP3 bridges the IFT-A complex and membrane phosphoinositides to promote trafficking of G protein-coupled receptors into primary cilia. Genes Dev. 2010;24:2180–2193. doi: 10.1101/gad.1966210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata A, Hamamoto A, Horikawa M, Yoshimura K, Takeda S, Saito Y. Characterization of ciliary targeting sequence of rat melanin-concentrating hormone receptor 1. Gen Comp Endocrinol. 2013;188:159–165. doi: 10.1016/j.ygcen.2013.02.020. [DOI] [PubMed] [Google Scholar]
- Norman RX, Ko HW, Huang V, Eun CM, Abler LL, Zhang Z, Sun X, Eggenschwiler JT. Tubby-like protein 3 (TULP3) regulates patterning in the mouse embryo through inhibition of Hedgehog signaling. Hum Mol Genet. 2009;18:1740–1754. doi: 10.1093/hmg/ddp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki S, Katoh Y, Terada M, Michisaka S, Funabashi T, Takahashi S, Kontani K, Nakayama K. Regulation of ciliary retrograde protein trafficking by the Joubert syndrome proteins ARL13B and INPP5E. J Cell Sci. 2017;130:(in press). doi: 10.1242/jcs.197004. [DOI] [PubMed] [Google Scholar]
- Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- Pedersen LB, Mogensen JB, Christensen ST. Endocytic control of cellular signaling at the primary cilium. Trends Biochem Sci. 2016;41:784–797. doi: 10.1016/j.tibs.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Qin J, Lin Y, Norman RX, Ko HW, Eggenschwiler JT. Intraflagellar transport protein 122 antagonizes Sonic Hedgehog signaling and controls ciliary localization of pathway components. Proc Natl Acad Sci USA. 2011;108:1456–1461. doi: 10.1073/pnas.1011410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- Schwartz R, Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stottmann RW, Tran PV, Turbe-Doan A, Beier DR. Ttc21b is required to restrict sonic hedgehog activity in the developing mouse forebrain. Dev Biol. 2009;335:166–178. doi: 10.1016/j.ydbio.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung C-H, Leroux MR. The roles of evolutionary conserved functional modules in cilia-related trafficking. Nat Cell Biol. 2013;15:1387–1397. doi: 10.1038/ncb2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Kubo K, Waguri S, Yabashi A, Shin H-W, Katoh Y, Nakayama K. Rab11 regulates exocytosis of recycling vesicles at the plasma membrane. J Cell Sci. 2012;125:4049–4057. doi: 10.1242/jcs.102913. [DOI] [PubMed] [Google Scholar]
- Takao D, Dishinger JF, Kee HL, Pinskey JM, Allen BL, Verhey KJ. An assay for clogging the ciliary pore complex distinguishes mechanisms of cytosolic and membrane protein entry. Curr Biol. 2014;24:2288–2294. doi: 10.1016/j.cub.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Bhogaraju S, Lorentzen E. Architecture and function of IFT complex proteins in ciliogenesis. Differentiation. 2012;83:S12–S22. doi: 10.1016/j.diff.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Weber K, Mourão A, Vetter M, Awasthi M, Stiegler M, Bhogaraju S, Lorentzen E. Intraflagellar transport proteins 172, 80, 57, 54, 38, and 20 form a stable tubulin-binding IFT-B2 complex. EMBO J. 2016;35:773–790. doi: 10.15252/embj.201593164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S, Ritter B, Verbich D, Sanson C, Bourbonnière L, McKinney RA, McPherson PS. Intersectin regulates dendritic spine development and somatodendritic endocytosis but not synaptic vesicle recycling in hippocampal neurons. J Biol Chem. 2009;284:12410–12419. doi: 10.1074/jbc.M809746200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PV, Haycraft CJ, Besschetnova TY, Turbe-Doan A, Stottmann RW, Herron BJ, Chesebro AL, Qiu H, Scherz PJ, Shah JV, et al. THM1 negatively modulates mouse sonic hedgehog signal transduction and affects retrograde intraflagellar transport in cilia. Nat Genet. 2008;40:403–410. doi: 10.1038/ng.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey KJ, Yang W. Permeability barriers for generating a unique ciliary protein and lipid composition. Curr Opin Cell Biol. 2016;41:109–116. doi: 10.1016/j.ceb.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Ling K, Hu J. The essential roles of transition fibers in the context of cilia. Curr Opin Cell Biol. 2015;35:98–105. doi: 10.1016/j.ceb.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q, Xu Q, Zhang Y, Li Y, Zhang Q, Hu Z, Harris PC, Torres VE, Ling K, Hu J. Transition fibre protein FBF1 is required for the ciliary entry of assembled intraflagellar transport complexes. Nat Commun. 2013;4:2750. doi: 10.1038/ncomms3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang TT, Su J, Wang W-J, Craige B, Witman GB, Tsou M-FB, Liao J-C. Superresolution pattern recognition reveals the architectural map of the ciliary transition zone. Sci Rep. 2015;5:14096. doi: 10.1038/srep14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Malicki J. Nephrocystins and MKS proteins interact with IFT particle and facilitate transport of selected ciliary cargos. EMBO J. 2011;30:2532–2544. doi: 10.1038/emboj.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.