ATP8A2, an ATP-dependent phosphatidylserine flippase, generates and maintains membrane lipid asymmetry and has been linked to severe neurodegenerative disorders. The C-terminus of ATP8A2 promotes proper protein folding and regulates phosphatidylserine flippase activity.
Abstract
ATP8A2 is a P4-ATPase that flips phosphatidylserine and phosphatidylethanolamine across cell membranes. This generates membrane phospholipid asymmetry, a property important in many cellular processes, including vesicle trafficking. ATP8A2 deficiency causes severe neurodegenerative diseases. We investigated the role of the C-terminus of ATP8A2 in its expression, subcellular localization, interaction with its subunit CDC50A, and function as a phosphatidylserine flippase. C-terminal deletion mutants exhibited a reduced tendency to solubilize in mild detergent and exit the endoplasmic reticulum. The solubilized protein, however, assembled with CDC50A and displayed phosphatidylserine flippase activity. Deletion of the C-terminal 33 residues resulted in reduced phosphatidylserine-dependent ATPase activity, phosphatidylserine flippase activity, and neurite extension in PC12 cells. These reduced activities were reversed with 60- and 80-residue C-terminal deletions. Unlike the yeast P4-ATPase Drs2, ATP8A2 is not regulated by phosphoinositides but undergoes phosphorylation on the serine residue within a CaMKII target motif. We propose a model in which the C-terminus of ATP8A2 consists of an autoinhibitor domain upstream of the C-terminal 33 residues and an anti-autoinhibitor domain at the extreme C-terminus. The latter blocks the inhibitory activity of the autoinhibitor domain. We conclude that the C-terminus plays an important role in the efficient folding and regulation of ATP8A2.
INTRODUCTION
P4-ATPases comprise a subfamily of P-type ATPases that use the energy from ATP hydrolysis to transport or flip phospholipids from the exoplasmic to the cytoplasmic leaflet of cell membranes (Lopez-Marques et al., 2014; Andersen et al., 2016). This generates and maintains lipid asymmetry, a property that is crucial for many biological processes, such as vesicle budding and transport, cell division, fertilization, phagocytosis, apoptosis, blood coagulation, axonal elongation, and sensory physiology (Tanaka et al., 2011; Sebastian et al., 2012). The importance of P4-ATPases is evident in the finding that a number of these P4-ATPases are associated with severe disorders, including neurodegeneration, intrahepatic cholestasis, sensory loss, diabetes, colorectal cancer, and blood disorders (Folmer et al., 2009; Coleman et al., 2013; Lopez-Marques et al., 2014).
ATP8A2 is a P4-ATPase that is highly expressed in retina, testis, spinal cord, and CNS, where it transports phosphatidylserine (PS) and to a lesser extent phosphatidylethanolamine across membranes (Coleman et al., 2009; Zhu et al., 2012). Loss of ATP8A2 flippase activity causes severe neuronal and axonal degeneration and visual and auditory deficiency in mice and cerebellar ataxia, mental retardation, and disequilibrium syndrome (CAMRQ) in humans (Zhu et al., 2012; Onat et al., 2013; Coleman et al., 2014). Like other P-type ATPases, ATP8A2 consists of a nucleotide-binding domain (N-domain), a phosphorylation domain (P-domain), and an actuator domain (A-domain), which are involved in the ATP reaction cycle, and a membrane domain (M-domain) consisting of 10 predicted transmembrane segments, which comprise the pathway for the translocation of phospholipids across cell membranes (Coleman et al., 2013; Vestergaard et al., 2014; Andersen et al., 2016). ATP8A2 also contains a 100–amino acid C-terminal cytoplasmic segment, the function of which is not known. ATP8A2 associates with its accessory or β-subunit CDC50A (also known as TMEM30A) to form a functional heterodimer that primarily localizes to the Golgi-recycling endosomal network, outer segment disk membranes of photoreceptor cells, and tips of the neurites in nerve growth factor (NGF)–treated PC-12 cells (van der Velden et al., 2010; Coleman and Molday, 2011; Coleman et al., 2014; Lee et al., 2015).
Although the phospholipid-dependent ATPase activity and ATP-dependent phospholipid flippase activity of ATP8A2 have been extensively studied (Coleman et al., 2009, 2012; Coleman and Molday, 2011; Vestergaard et al., 2014), little is known about how ATP8A2 and other mammalian P4-ATPases are regulated. Drs2, a yeast orthologue of ATP8A2, has a C-terminal domain containing autoinhibitory elements (Zhou et al., 2013). Similar autoinhibitory segments have been identified in C-terminal domains of other P-type ATPases, including the plasma membrane Ca2+ ATPase (Brandt et al., 1988; Enyedi et al., 1989), plant plasma membrane H+ ATPase AHA2 (Axelsen et al., 1999; Fuglsang et al., 1999), and yeast plasma membrane H+ ATPase PMA1 (Mason et al., 2006, 2014). To determine whether the C-terminal domain affects the structure or activity of ATP8A2, we generated and characterized the transport function and cellular localization of a series of C-terminal truncation mutants. Our studies indicate that the C-terminus of ATP8A2 plays an important role in promoting efficient protein folding and regulating phospholipid flippase activity.
RESULTS
Expression of ATP8A2 C-terminal deletion mutants
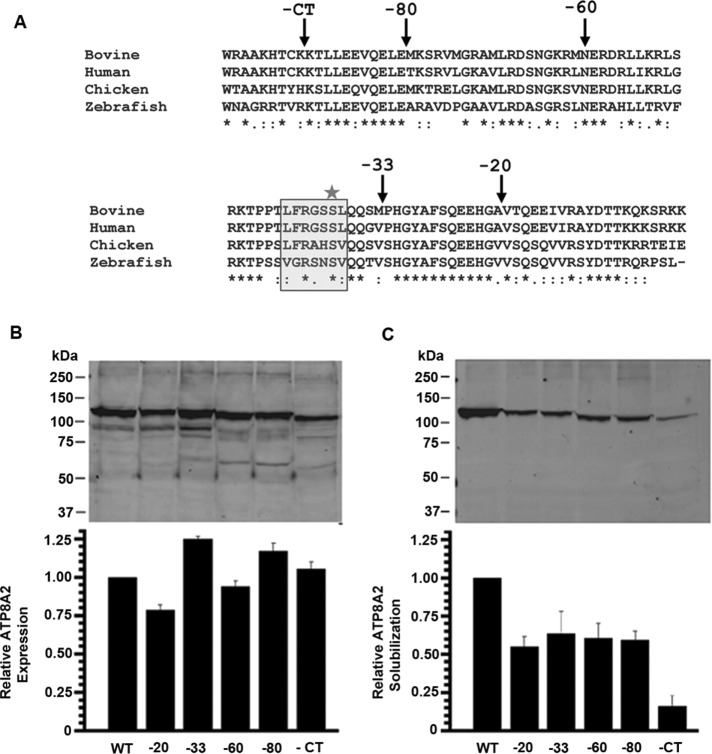
Figure 1A shows the sequence of the ATP8A2 C-terminal domain from different species and the location of sites used to generate the C-terminal truncation mutants used in this study (ATP8A2(–20, –33, –60, –80 and –91 [CT])). The C-terminal deletion mutants containing a nine–amino acid 1D4 tag to facilitate detection and purification were coexpressed with CDC50A in culture cells to examine their effect on the expression, localization, and functional activity of ATP8A2.
FIGURE 1:
The effect of C-terminal truncation mutations on ATP8A2 expression and solubility (A) Multiple sequence alignment of the C-terminal domain from four different species, showing the high degree of conservation. The C-terminal domain of the bovine ATP8A2 (GenBank ACT46164.3) used in this study is 100 amino acids long, from W1077 to K1176. The positions used to generate the C-terminal truncation mutants are marked by arrows. The sequence within the shaded box is the conserved CaMKII recognition motif, with the serine residue marked by a star as the potential target for phosphorylation. The consensus CaMKII target sequence is Φ-x-R-n-x-S/T-Φ (White et al., 1998), where Φ is any hydrophobic amino acid, n is a nonbasic amino acid, and x is any amino acid. (B) The expression profiles of the different ATP8A2 C-terminal truncation mutants in HEK293T cells. Lysates from HEK293T-transfected cells were solubilized in SDS for analysis of ATP8A2 expression on Western blots labeled with the Rho 1D4 antibody (top); ATP8A2 expression is quantified for three independent experiments (bottom). (C) CHAPS solubilization profiles for ATP8A2 C-terminal truncation mutants. Lysates from HEK293T-transfected cells were solubilized in CHAPS. Aggregated protein was removed by high-speed centrifugation; the supernatant was analyzed by Western blotting (top), and ATP8A2 was quantified (bottom); n = 3. The intensities of protein bands from Western blots were normalized to the total protein concentration in the lysate. Error bars represent SEM.
To determine the effect of these mutations on total ATP8A2 expression, cell lysates from transfected HEK293T cells were directly solubilized in SDS and analyzed on Western blots labeled for ATP8A2 with the Rho1D4 antibody. Figure 1B shows that the overall expression level of all of the mutants was comparable to that of the wild-type (WT) protein. To estimate the amount of protein that could be solubilized in a mild detergent, cell lysates were treated with 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) detergent, and the solubilized fraction obtained after removal of aggregated protein by centrifugation was analyzed by Western blotting. The intensity of protein bands from Western blots was normalized to total protein concentration in the lysate and quantified relative to WT ATP8A2. As shown in Figure 1C, deletion of the terminal 20 amino acids (ATP8A2(–20)) reduced the amount of CHAPS-soluble ATP8A2 to ∼50% of the WT level. Longer deletions of –33, –60, and –80 residues did not have any further effect on their solubilization by CHAPS. However, removal of most of the C-terminus (ATP8A2(–CT)) further reduced CHAPS solubilization to only 20% of the WT protein. The low amount of ATP8A2(–CT) prevented us from extensively characterizing this mutant at a biochemical level. Quantitative representations of total protein expression and CHAPS solubility of the various ATP8A2 C-terminal mutants are shown in the bottom half of Figure 1, B and C, respectively. These results indicate that truncation of the C-terminal domain of ATP8A2 results in a significant amount of protein that is refractory to solubilization by a mild detergent relative to the WT protein.
Localization of ATP8A2 C-terminal truncation mutants in PC-12 cells
The 50% decrease in CHAPS solubility of ATP8A2 after truncation of C-terminal domain could be due to an equivalent fraction of misfolded mutant protein. In this case, the misfolded protein would likely be retained in the endoplasmic reticulum (ER) by the quality control machinery. To resolve this possibility, we analyzed the localization of WT and mutant ATP8A2 to the ER by immunofluorescence imaging. Results described in the preceding section show that 20– and 90–amino acid deletions are the inflection points with respect to change in protein conformation. It is only after deletion of these segments from the C-terminal domain that the detergent solubility of ATP8A2 decreases by 50 and 80%, respectively. We therefore analyzed the change in ER and Golgi colocalization of only these mutant proteins relative to WT ATP8A2.
ATP8A2 is expressed in PC12 cells, where it plays a role in promoting neurite growth (Xu et al., 2012). In this experiment, we used PC12 cells to evaluate the effect of the C-terminus of ATP8A2 on its subcellular localization. PC12 cells cotransfected with CDC50A and either the WT or C-terminal ATP8A2 mutants were induced to differentiate in the presence of NGF. The cells were then fixed and double labeled with antibodies against ATP8A2 and either the ER marker calnexin or the Golgi marker GM130 for immunofluorescence imaging.
As previously reported, WT ATP8A2 localized to the neurite terminals and in a punctate pattern to the Golgi/recycling endosomes system in the cell body (Figure 2A; Coleman et al., 2014). A reduced level of ATP8A2 staining was also observed along the neurite outgrowths. From the Manders colocalization coefficient, 22.4 ± 1.95% (n = 13) of WT ATP8A2 colocalized with the ER marker (Figure 2B). The ATP8A2(–20) mutant was also evenly distributed between the neurites and the cell body. However, this mutant showed greater colocalization with the ER marker calnexin (51.5 ± 4.96%; n = 9). ATP8A2(–CT) was mostly localized in the ER (73.02 ± 3.95%; n = 5), with only very faint staining along the neurites (Figure 2). On the other hand, localization of ATP8A2 to Golgi complex decreased after C-terminal truncation. Whereas 11.6 ± 3.3% (n = 4) of WT ATP8A2 localized to the Golgi network, only 6.96 ± 0.57% (n = 9) and 6.45 ± 1.6% (n = 2) of ATP8A2(–20) and ATP8A2CT), respectively, localized to the Golgi.
FIGURE 2:
Immunofluorescence localization of ATP8A2 C-terminal truncation mutants. PC12 cells cotransfected with either WT or mutant ATP8A2 and CDC50A were differentiated with NGF to facilitate formation of neurites. The cells were then labeled for ATP8A2 with Rho1D4 antibody (green) and either (A) the ER marker calnexin (red) or (B) the Golgi marker GM130 (red). Colocalization analysis showed that the WT protein localizes prominently to the terminal regions of neurites and the Golgi membrane, with little presence in the ER membrane (22.4 ± 1.95%, n = 13). (C) Truncation of the C-terminal amino acids increased ER colocalization; ATP8A2(–20) = 51.5 ± 4.9% (n = 9) and ATP8A2(–CT) = 73.02 ± 3.9% (n = 5). Truncation of C-terminal tail also showed decreased localization to the Golgi complex. (D) Whereas 11.6 ± 3.3% of WT protein (n = 4) was found in Golgi, this decreased to 6.96 ± 0.57 and 6.45±1.65% in ATP8A2(–20) (n = 9) and ATP8A2(–CT) (n = 2), respectively. Samples were counterstained with DAPI (blue) nuclear stain. In A and B, arrowheads point to ATP8A2 staining at neurite ends, and arrows point to punctate staining observed in the cell body and neurite extensions. Two-sample t test was used to calculate p values. ****p < 0.0001; N.S., p > 0.05. Bars, 20 µm.
Interaction of ATP8A2 truncation mutants with CDC50A
ATP8A2 requires CDC50A for optimal expression and exit from the ER (van der Velden et al., 2010; Coleman and Molday, 2011). Because the C-terminal deletion mutants showed higher ER retention, we investigated whether these mutants were capable of forming a complex with CDC50A. Toward this goal, we performed two different analyses. In the first set of experiments, WT ATP8A2 or the ATP8A2(–20) mutant was transfected into HEK293T cells alone or together with CDC50A. The overexpressed complex was solubilized with CHAPS and immunoaffinity purified on an anti-Rho1D4 matrix to bind ATP8A2. ATP8A2 expression with and without coexpression with CDC50A was compared by Western blot analysis of the immunoprecipitated protein. The results in Figure 3A show that the expression of both WT ATP8A2 and ATP8A2(–20) increased significantly when coexpressed with CDC50A. The low level of ATP8A2 in the absence of CDC50A heterologous expression is most likely due to small amounts of endogenous CDC50A in HEK293 cells.
FIGURE 3:

Interaction of ATP8A2 C-terminal mutants with CDC50A. (A) Western blots of cell lysates from HEK293T cells transfected with WT and ATP8A2(–20) mutant alone or together with CDC50A. Top, labeled for ATP8A2 with the Rho1D4 antibody; bottom, labeled with the Cdc50-7F4 antibody for CDC50A. Coexpression with CDC50A significantly increased the expression of WT and mutant ATP8A2. (B) Immunoprecipitation followed by Western blot analysis of CHAPS-solubilized lysate from HEK293 cells cotransfected with ATP8A2 and CDC50A. Total ATP8A2 was determined by purification on an anti-Rho1D4 column, which binds ATP8A2. ATP8A2 associated with CDC5A was determined by the amount of ATP8A2 that copurified with CDC50A on a Cdc50-7F4 immunoaffinity column. Top, example of a Western blot labeled with the Rho1D4 antibody to detect ATP8A2 in the bound (B) and unbound (UB) fractions to the immunoaffinity columns. Bottom, extent of WT ATP8A2 and ATP8A2(–20) that interact with CDC50A. The prominent band between 250 and 150 kDa in the Western blots corresponds to the ATP8A2/CDC50A complex and can be visualized using antibodies against both ATP8A2-1D4 and CDC50A. Error bars represent SEM for WT ATP8A2 (n = 3) and ATP8A2(–20) mutant (n = 2).
To examine further the interaction of WT and mutant ATP8A2 with CDC50A, HEK293T cells cotransfected with CDC50A and WT or mutant ATP8A2 were solubilized with CHAPS. Half of the solubilized complex was purified on a Rho-1D4 immunoaffinity column, which binds ATP8A2, and the other half on an anti-Cdc50-7F4 column, which binds CDC50A. Total ATP8A2 and the fraction that bound to the CDC50A immunoaffinity matrix were quantified from Western blots labeled with the Rho 1D4 antibody (Figure 3B, top). These data were used to calculate the fraction of ATP8A2 interacting with CDC50A. Data in Figure 3B (bottom) show that the fraction of ATP8A2(–20) interacting with CDC50A is comparable to that for WT ATP8A2. These results indicate that removal of the C-terminal 20 amino acids does not affect the interaction of ATP8A2 and CDC50A and reinforces the notion that CHAPS-solubilized lysates from cells cotransfected with CDC50A and either WT or mutant ATP8A2 contain the ATP8A2-CDC50A flippase complex.
Phosphatidylserine-activated ATPase activity of C-terminal ATP8A2 mutants
To determine the effect of C-terminal truncation on the functional properties of ATP8A2, we first purified the expressed WT ATP8A2-CDC50A complex and its C-terminal deletion mutants by affinity chromatography on a Rho1D4-Sepharose matrix. As previously shown and confirmed in Figure 4A, the ATP8A2 and deletion mutants run as the predominant Coomassie blue–stained band having an apparent molecular weight of 130 kDa (Coleman and Molday, 2011).
FIGURE 4:
Effect of C-terminal truncation on the ATPase activity of ATP8A2. (A) Coomassie blue–stained SDS gels of ATP8A2-CDC50A complex and C-terminal deletion mutants purified by affinity chromatography on a Rho1D4-Sepharose matrix. ATP8A2 is observed as the major stained band; CDC50A does not stain well with Coomassie blue. (B) The ATPase activity of purified WT and truncated mutants coexpressed with CDC50A was measured as a function of PS concentration. The data were fitted to the Michaelis–Menten equation to calculate the maximum reaction rate (Vmax) of WT or mutant ATP8A2. (C) Vmax for WT and C-terminal mutants immunopurified from an anti-Rho1D4 column; error bars represent SEM for n = 4–18. (D) ATPase activity (Vmax) profile of WT and mutant ATP8A2 purified from anti-Cdc50-7F4 column. Error bars represent SEM. Refer to Table 1 for details of kinetic data. Two-sample t test was used to calculate p values. **p < 0.05; ***p < 0.005; N.S., p > 0.05.
The PS-stimulated ATPase activity of the mutants was compared with that of the WT protein. The resultant data are shown in Figure 4B, and the kinetic parameters derived from a Michaelis–Menten fit are presented in Table 1. Only small differences in the half-maximum activation (KA) of the ATP8A2 mutants by PS were observed. The reaction rates (Vmax) for the mutants, however, were more informative (Figure 4C). Deletion of 20 and 33 amino acids resulted in a significant decrease in PS-stimulated Vmax. However, further deletion of 60 and 80 amino acids resulted in an increased rate of ATP hydrolysis, with ATP8A2(–80) having a Vmax comparable to WT ATP8A2.
TABLE 1:
Parameters of ATP8A2 C-terminal domain mutants determined in this study.
| ATPase activity anti-1D4 column | ATPase activity anti-7F4 column | Neurite length in PC12 cells | [Ca2+] inhibition | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ATP8A2 | n | Vmax (±SEM) μmol Pi/min per mg | KA (±SEM) μM | n | Vmax (±SEM) μmol Pi/min per mg | KA (±SEM) μM | n | Increase in neurite length (±SEM) µm | n | IC50 (±SEM) mM |
| WT | 18 | 136.8 (10.24) | 25.4 (3.11) | 5 | 126.7 (15.08) | 38.5 (5.01) | 121 | 51.21 (3.9) | 6 | 2.72 (0.67) |
| −20 | 6 | 95.03 (16.08) | 22.54 (2.8) | 3 | 55.5 (4.8) | 37.2 (4.6) | 58 | 32.9 (4.2) | 3 | 1.01 (0.02) |
| −33 | 6 | 81.66 (17.32) | 19.51 (2.65) | 3 | 50.36 (2.3) | 29.8 (6.6) | 63 | 4.9 (2.06) | 2 | 0.93 (0.4) |
| −60 | 6 | 108.14 (11.8) | 18.14 (3.15) | 3 | 59.5 (15.8) | 21.6 (0.93) | 105 | 18.4 (4.0) | 2 | 1.37 (0.32) |
| −80 | 4 | 155.93 (12.82) | 20.68 (1.57) | 3 | 85.06 (13.3) | 32.4 (5.9) | 112 | 37.01 (3.53) | 2 | 1.99 (0.4) |
ATPase activity of the different ATP8A2 mutants was titrated against increasing concentration of PS in the presence of 0.5 mM ATP to determine the maximal reaction rate (Vmax) and half-maximum activation constant (KA). Increase in the length of neurites formed by differentiated PC12 overexpressing the different ATP8A2 mutants compared with untransfected cells was measured. IC50 for inhibition of ATPase activity of different ATP8A2 mutants was estimated by titrating activity in the presence of increasing CaCl2 and fixed PS and ATP concentrations. n is the number of independent measurements.
To rule out the possibility that the observed changes in the ATPase activity of ATP8A2 mutants might be an artifact of the presence of a substantial fraction of misfolded protein in the mutant protein purified from anti-Rho1D4 column, we assayed the ATPase activity of protein samples purified from a Cdc7F4 column, which binds CDC50A. Because misfolded ATP8A2 does not complex with CDC50A, only the properly folded ATP8A2 is eluted from this column. As shown in Figure 4D and Table 1, the profile of the ATPase activities for the 7F4 peptide eluted ATP8A2 WT and mutant samples is comparable to that of protein purified from the anti-Rho1D4 column.
Effect of ATP8A2 C-terminal deletions on phosphatidylserine flippase activity
To determine whether the changes observed in ATPase activity of the mutants correlate with their effect on lipid transport activity, we reconstituted the ATP8A2 C-terminal mutants in 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) liposomes containing 3% 1-oleoyl-2-{6-[7-nitro-2-1,3-benzoxadiazol-4-yl)]hexanoyl}-sn-glycero-3-phosphoserine (NBD-PS). Flippase activity of reconstituted proteins was measured as the fraction of total NBD-PS transported to the outer leaflet of liposome in the presence of ATP as previously described (Coleman et al., 2009). Flippase activity data shown in Figure 5 for the different ATP8A2 C-terminal mutants show the same trend as for the PS-activated ATPase activity of the C-terminal mutants. A decrease in flippase activity was observed for ATP8A2(–20) and ATP8A2(–33). This decrease was relieved for ATP8A2(–60) and ATP8A2(–80).
FIGURE 5:

Effect of C-terminal truncation on PS flippase activity of ATP8A2. PS flippase activity of ATP8A2 and the deletion mutants was determined by reconstituting immunopurified protein in liposomes containing NBD-PS. Flippase activity of reconstituted protein was calculated from an increase in the percentage of NBD-PS exposed on the outer leaflet of liposome in the presence of hydrolyzable ATP. Error bars represent SEM. ATP8A2(−20), n = 2; ATP8A2(−33), n = 3; ATP8A2(−60), n = 2; and ATP8A2(–80), n = 2. **p < 0.05.
Effect of ATP8A2 C-terminal deletions on neurite extension of PC12 cells
Earlier studies showed that overexpression of ATP8A2 alone or with CDC50A causes a significant increase in neurite growth of PC12 cells (Xu et al., 2012). We used this system to determine whether the effect of C-terminal truncation on the flippase activity of the purified ATP8A2 complex extends to a cellular system. PC12 cells cotransfected with CDC50A and either WT or mutant ATP8A2 were differentiated in the presence of NGF for 2 d. The length of neurites from cells expressing WT ATP8A2 and mutant proteins was compared with that of untransfected differentiated cells (Figure 6A and Table 1). Neurites from differentiated PC12 cells expressing WT ATP8A2 showed an almost threefold increase over untransfected differentiated cells (90 µm for ATP8A2-expressing cells vs. 38 µm for untransfected cells). A smaller increase in neurite length was observed in cells expressing the C-terminal truncation mutants. Relative to WT ATP8A2 (normalized to 100% increase over untransfected cells), the ATP8A2(–20) and ATP8A2(–33) mutants displayed 64 and 10% increases, respectively (Figure 6B and Table 1). Of interest, this downward trend in neurite length with truncation was reversed with further C-terminal truncation. ATP8A2(–60) showed a 36% increase in neurite length, and ATP8A2(–80) showed an ∼72% increase (Figure 6B and Table 1). This profile was qualitatively similar to that observed for PS-activated ATPase activity and ATP-dependent flippase activity of the purified ATP8A2 C-terminal deletion mutants.
FIGURE 6:

Effect of C-terminal truncation on neurite extension in PC12 cells. Untransfected PC12 cells or PC12 cells cotransfected with CDC50A and either WT or a C-terminal ATP8A2 mutant were differentiated with NGF. After 48 h, the cells were labeled with anti–Cdc50-7F4 primary antibody against CDC50A. (A) Fluorescence images of these cells transfected with the various ATP8A2 mutants. Bars, 20 µm. (B) Graph showing the neurite length of untransfected and ATP8A2 (WT and mutant) transfected cells. Error bars represent SEM. Untransfected cells (Unt), n = 54; cells transfected with WT ATP8A2, n = 121; ATP8A2(–20), n = 58; ATP8A2(–33), n = 63; ATP8A2(–60), n = 105; and ATP8A2(–80), n = 112. Two-sample t test was used to calculate p values. ***p < 0.001, ****p < 0.0001.
Modulation of ATP8A2 and ATP8A1 activity by [Ca2+]
PS translocation activity of erythrocytes is inhibited by submicromolar intracellular calcium concentration, leading to the suggestion that P4-ATPases may be directly regulated by calcium (Bitbol et al., 1987; Daleke and Lyles, 2000). To determine whether Ca2+ alters the activity of the purified ATP8A2-CDC50A complex, possibly through its C-terminal domain, we measured the ATPase activity of WT and mutant ATP8A2-CDC50A complex at increasing CaCl2 concentrations. As shown in Figure 7A, the ATPase activity of WT ATP8A2 was inhibited by Ca2+ but only at high concentrations, with a concentration of Ca2+ resulting in half-maximum inhibition (IC50) = 2.75 ± 0.7 mM (n = 6). The IC50 values for different C-terminal mutants were also in the millimolar range and showed the same trend as observed for the other flippase activity measurements in this study (Figure 7B).
FIGURE 7:
Inhibition of ATPase activity of ATP8A2 and ATP8A1 by Ca2+. (A) ATPase activity of the WT and C-terminal–truncated mutants as a function of increasing concentration of CaCl2. The activity data (micromoles Pi/minute per milligram of protein) were fitted to a four-parameter logistic function to calculate IC50 for Ca2+ inhibition of ATPase activity (see Materials and Methods for the equation). Only the data for ATP8A2 (WT, –33, –80) are shown here for clarity. (B) Bar graph comparing IC50 determined for each C-terminal truncation mutant. The data show that deletion of the terminal-most 33 amino acids from the C-terminal domain renders the protein more susceptible to Ca2+ inhibition. Deletions farther upstream eliminate this Ca2+ sensitivity. (C) Comparison of Ca2+ inhibition of ATPase activity of ATP8A1 and ATP8A2. The IC50 data show that the Ca2+ inhibition profile of WT ATP8A1 is comparable to that of WT ATP8A2. The error bars represent SEM.
Because erythrocytes have been reported to contain the P4-ATPase ATP8A1 (Ding et al., 2000; Soupene and Kuypers, 2006), we also determined the effect of increasing Ca2+ concentrations on the PS-stimulated ATPase activity of this P4-ATPase. The IC50 for Ca2+ inhibition of ATP8A1 activity was 1.56 ± 0.06 mM, comparable to that of ATP8A2 (Figure 7C) but significantly higher than the Ca-mediated inhibitory effect on PS translocation in erythrocytes. This suggests that the inhibitory effect of Ca2+ in erythrocytes may not be due to a direct interaction with the protein. Instead, it may result either through a Ca-dependent intermediary effector or by direct Ca inhibition of PS transport through another calcium-sensitive P4-ATPase in erythrocytes.
Effect of phosphoinositides on the ATPase activity of ATP8A2
In a previous study, we showed that phosphatidylinositol (PI) by itself did not activate the ATPase activity of ATP8A2, suggesting that this phospholipid is not transported by ATP8A2 (Coleman et al., 2009). Previous studies, however, showed that phosphoinositides and in particular phosphatidylinositol 4-phosphate (PI4P) significantly stimulate the PS-activated ATPase and PS-dependent flippase activity of Drs2 in Golgi membranes and when reconstituted into liposomes (Natarajan et al., 2009; Zhou et al., 2013). This regulation occurs through the binding of PI4P to a regulatory domain in the C-terminal segment of Drs2 that has homology to a split PH domain (Figure 8A). Although ATP8A2 and Drs2 show a homologous region within the C-terminal sequence, the C-terminus of ATP8A2 lacks the PH domain required for PI4P binding (Figure 8A). Nonetheless, we investigated whether PI or PI4P altered the PS-stimulated ATPase activity of ATP8A2. As shown in Figure 8B, neither PI nor PI4P had any significant effect on the PS-stimulated ATPase activity of purified ATP8A2.
FIGURE 8:

Effect of phosphoinositides on the PS-activated ATPase activity of ATP8A2. (A) Alignment of the C-terminal region of yeast Drs2 P4-ATPase with ATP8A2, showing the presence of a PH-like domain in Drs2 that binds phosphoinositides. This domain is absent in ATP8A2. (B) PS-stimulated ATPase activity in the absence (control) and presence of 2.5% PI4P or PI and 10% PS.
Phosphorylation of a serine residue within the calcium/calmodulin-dependent protein kinase II binding motif on ATP8A2
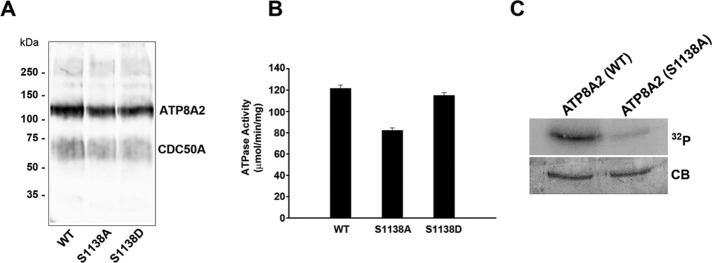
Sequence analysis of the C-terminal segment of ATP8A2 reveals the existence of the conserved calcium/calmodulin-dependent protein kinase II (CaMKII) consensus sequence Φ-x-R-n-x-S/T-Φ, where Φ is a hydrophobic residue, n is a nonbasic residue and x is any residue (Figure 1A). The serine residue in this motif was replaced with either alanine (S1138A) or aspartic acid (S1138D) to determine the effect of substitution in this position on the PS-dependent ATPase activity of ATP8A2. Both mutants expressed at levels similar to WT ATP8A2 and interacted with the CDC50A subunit, as shown by coimmunoprecipitation and Western blot analysis (Figure 9A). The activity of the ATP8A2 S1138A mutant was reduced by 33%, whereas the activity of the S1138D mutant was similar to that of WT ATP8A2 (Figure 9B).
FIGURE 9:
Phosphorylation of serine 1138 (S1138) residue within the CaMKII target sequence defined in Figure 1A. (A) WT ATP8A2 containing S1138 and ATP8A2 mutants S1138A and S1138D were coexpressed with CDC50A and purified on a Rho1D4 affinity column. Western blots double labeled for ATP8A2 with the Rho 1D4 antibody and CDC50A with the Cdc7F4 antibody confirmed the expression and coimmunoprecipitation of ATP8A2 mutants and CDC50A. (B) PS-stimulated ATPase activity of purified WT and S1138A and S1138D mutants. (C) ATP-dependent phosphorylation of WT and S1138A mutants by CaMKII. WT and S1138A mutant were purified on a Rho 1D4 column, reconstituted into liposomes, and phosphorylated with CaMKII in the presence of [γ-32P]ATP. Phosphorylation was measured in a phosphorimager, and ATP8A2 was quantified by Coomassie blue (CB). The amount of 32P in ATP8A2 S1138A was 5–8% that of WT in three independent experiments.
Next we determined whether the CaMKII target sequence is indeed phosphorylated by CaMKII. WT ATP8A2 and the S1138A mutant were coexpressed with CDC50A, purified, and reconstituted into liposomes. CaMKII was added to both samples, and the extent of phosphorylation by [γ-32P]ATP was measured by phosphorimaging after SDS gel electrophoresis, As shown in Figure 9C, the level of phosphorylation of WT ATP8A2 was >10 times higher than that for the S1138A mutant. From these studies, we conclude that S1138 of ATP8A2 serves as a phosphorylation target for CaMKII.
DISCUSSION
Molecular modeling studies indicate that ATP8A2 and Drs2 show high structural homology to sarcoplasmic reticulum Ca-ATPase and Na/K ATPase, for which high-resolution structures are known (Morth et al., 2007; Shinoda et al., 2009; Moller et al., 2010; Baldridge and Graham, 2012; Vestergaard et al., 2014). However, ATP8A2, like most P4-ATPases, has an extended cytoplasmic C-terminal domain of 100 highly conserved amino acids not present in sarcoplasmic Ca-ATPase and Na/K ATPase (Figure 1A). In this study, we used a series of truncation mutants to evaluate the importance of the C-terminal domain in orchestrating the structure and function of ATP8A2. Our results show for the first time that the C-terminus of ATP8A2 contributes to efficient protein folding and regulation of phospholipid flippase activity at a biochemical and cellular level.
To determine the role of the C-terminal domain of ATP8A2 in protein folding, we carried out a series of biochemical and cell localization studies. We first examined the effect of C-terminal deletion mutants on the solubilization of ATP8A2 by CHAPS, a mild detergent that preserves the functional activity of ATP8A2 and other lipid flippases (Coleman et al., 2009; Quazi and Molday, 2013; Lee et al., 2015). Poorly folded membrane proteins typically aggregate in mild detergents and can be subsequently removed by centrifugation. We found that although the ATP8A2 mutants were expressed in HEK293T cells at a similar level to the WT protein, the extent of solubilization of the mutants by CHAPS was reduced by half when up to 80 residues were removed from the C-terminus and 80% when 91 residues were deleted (Figure 1C). Of importance, the mutants solubilized in CHAPS retained both their capacity to interact with their β-subunit CDC50A, as shown by coimmunoprecipitation studies, and their functional properties, as observed by their PS-stimulated ATPase- and ATP-dependent flippase activities. Cellular localization studies supported the role of the C-terminus in protein folding. In previous studies, WT ATP8A2 coexpressed with CDC50A was found to exit the ER and localize to the Golgi/recycling endosome system in COS-1 cells (van der Velden et al., 2010; Coleman and Molday, 2011; Lee et al., 2015). In differentiated PC12 cells, the ATP8A2-CDC50A complex was present in the Golgi/recycling endosomes but also trafficked along the neurites to the tips (Coleman et al., 2014). In the present study, we analyzed the subcellular localization of the two mutants that showed an inflection with respect to detergent solubility, ATP8A2(–20) and ATP8A2(–CT), and found that there was a decrease in Golgi/endosomal and neurite localization for the C-terminal deletion mutants and an increase in ER localization. The latter indicates that a significant fraction of the ATP8A2 C-terminal mutants was misfolded and retained in the ER by the quality control system of the cell.
The C-terminus also has a significant effect on the functional properties of ATP8A2. Deletion of up to the 33 C-terminal amino acids resulted in a significant decrease in both the PS-stimulated ATPase activity and ATP-dependent PS flippase activity. Further deletions of 60 and 80 amino acids showed a reversal, with the 80-residue truncation restoring the activity to a level comparable to that of the full-length protein. Of importance, this trend was also observed in the PC12 cells, where the expression of ATP8A2 with short C-terminal deletions (ATP8A2(–20) and ATP8A2(–33)) resulted in a marked decrease in neurite length relative to those expressing WT ATP8A2, whereas longer deletions of 60 and 80 amino acids reversed this effect (Figure 6). At a cellular level, we speculated that the ATP8A2 flippase activity plays a crucial role in vesicle trafficking, a process essential for neurite extension in PC12 and hippocampal cells and outer segment elongation in photoreceptor cells (Xu et al., 2012; Coleman et al., 2014). Taken together, these biochemical and cell-based studies support the contention that the C-terminal domain of ATP8A2 is involved in the regulation of its phospholipid transport activity.
This is in line with studies showing that the C-terminus of Drs2, the yeast orthologue of ATP8A2, regulates its flippase activity. The binding of the Arf-GEF protein Gea2 and the binding of PI4P to a sequence of basic amino acids (RMKKQR) with homology to a split PH domain activate the flippase activity of Drs2 (Natarajan et al., 2009). Removal of the C-terminus also increases the flippase activity of Drs2 and eliminates activation by PI4P and Gea2 (Zhou et al., 2013). These studies have led to a mechanism in which the C-terminus of Drs2 acts as an autoinhibitory domain (Zhou et al., 2013). The binding of PI4P or Gea2 disengages the interaction of the C-terminus from the catalytic domain, resulting in flippase activity. However, unlike Drs2, purified ATP8A2 has very high PS-stimulated ATPase activity. As shown here, addition of PI or PI4P has no effect on the activity of ATP8A2, consistent with the absence of a targeting motif for phosphoinositide binding. Instead, ATP8A2 harbors a serine residue (S1138) within a CaMKII target motif not found in Drs2. This serine undergoes CaMKII-catalyzed phosphorylation and thus may serve as a mechanism to regulate the flippase activity of ATP8A2, possibly involving protein–protein interactions. These studies indicate that ATP8A2 is regulated by a different mechanism than Drs2.
On the basis of our studies, we propose a model in which the C-terminal segment of ATP8A2 can be divided into two domains—an autoinhibitory domain upstream of 33 residues from the C-terminus and an anti-autoinhibitory domain at the extreme C-terminus (Figure 10). The anti-autoinhibitory domain interacts with the autoinhibitory domain to prevent inhibition of WT ATP8A2 flippase activity. In our model, removal of the anti-autoinhibitory domain as in the ATP8A2(–33) mutant exposes the autoinhibitory domain, enabling it to inhibit ATP8A2 flippase activity, possibly through interaction of this domain with one of the domains responsible for the ATP reaction cycle or the phospholipid translocation pathway (Baldridge and Graham, 2012; Vestergaard et al., 2014). Removal of the autoinhibitory domain together with the anti-autoinhibitory domain, as evident for the ATP8A2(–80) mutant, restores the PS-flippase activity of ATP8A2 to near-WT levels. To the best of our knowledge, the presence of both an autoinhibitor and an anti-autoinhibitor domain is unique and has yet to be described in any other P-type ATPase, although autoinhibitor domains have been reported for a number of P-type ATPases.
FIGURE 10:

Schematic of ATP8A2-CDC50A complex showing the various domains. Topological model of ATP8A2, with a membrane domain containing 10 transmembrane segments, an actuator domain (A), a phosphorylation domain (P), and a nucleotide domain (N) and interacting with its glycosylated β-subunit CDC50A. On the basis of this study, we propose that the C-terminal region is divided into two domains: an autoinhibitor domain and an anti-autoinhibitor domain. WT ATP8A2 expressed in HEK293T cells has the anti-autoinhibitor domain interacting with the autoinhibitor domain to prevent inhibition of ATP8A2 flippase activity. Removal of the C-terminal 33 residues (ATP8A2(–33)) removes the anti-autoinhibitor domain, enabling the autoinhibitor domain to interact with another site on ATP8A2 and inhibit its flippase activity. Removal of the terminal 80 amino acids (ATP8A2(–80)) removes both the anti-autoinhibitor and autoinhibitor domains, leading to WT-like ATP8A2 flippase activity. CDC50A is shown in association with ATP8A2 with two transmembrane segments and a highly glycosylated (hexagons) exoplasmic domain.
How the C-terminal autoinhibitory and anti-autoinhibitory domains regulate ATP8A2 activity at a cell physiology level remains to be investigated. Because elevated Ca2+ concentration activates lipid scramblases (Malvezzi et al., 2013) and at the same time is hypothesized to inhibit P4-ATPases (Daleke and Lyles, 2000; Segawa et al., 2014) of blood cells, we examined the effect of Ca2+ on PS-stimulated ATPase activity of purified ATP8A2 and ATP8A1. The half-maximum of inhibition of Ca2+ in all cases was in the millimolar range, a concentration not generally attainable within cells. Although ATP8A2 and ATP8A1 do not appear to be directly modulated by Ca2+ binding under physiological conditions, they may be modulated by an indirect mechanism such as through a Ca2+ sensor protein. Phosphorylation of S1138 within the CaMKII target site may contribute to regulation of ATP8A2 in a calcium-dependent manner. The CaMKII target site is located just upstream of the –33 deletion site and within the proposed autoinhibitor region (Figure 1A). Site-directed mutagenesis, together with CaMKII-catalyzed phosphorylation studies, confirms that S1138 within this motif is specifically phosphorylated by CaMKII. We suggest that this phosphorylation reaction may occur when intracellular Ca2+ concentration is elevated under physiological conditions. This may directly alter the interaction between the anti-autoinhibitor with the autoinhibitor domain. Alternatively, phosphorylation of S1138 may result in the binding of intracellular proteins to ATP8A2 such that the anti-autoinhibitory domain can no longer bind to the autoinhibitor domain, resulting in suppression of ATP8A2 phospholipid transport. Although the role of CaMKII-catalyzed phosphorylation of ATP8A2 in regulation of its flippase activity requires additional studies, two separate but related observations support this possibility. First, previous work showed that neurite elongation in PC12 cells can be impaired by silencing the expression of ATP8A2 (Xu et al., 2012). Second, two independent studies showed that overexpression of CaMKII also inhibits neurite formation in PC12 cells (Tashima et al., 1996; Masse and Kelly, 1997). These two observations, when taken together, provide circumstantial evidence to support an indirect role of calcium in modulation of ATP8A2 activity. We are investigating the possible role of CaMKII-mediated phosphorylation in regulating the phospholipid flippase function of ATP8A2.
MATERIALS AND METHODS
Reagents
l-α-Phosphatidylcholine (egg, chicken; ePC), DOPC, 1,2-dioleoyl-sn-glycero-3-phosphoserine (DOPS), PI4P, PI, and NBD-PS were purchased from Avanti Polar lipids (Alabaster, AL). ATP, adenylyl-imidodiphosphate (AMP-PNP), and octyl-β-d-glucopyranose (OGP) were purchased from Sigma-Aldrich, sodium hydrosulfite (sodium dithionite) from Fischer Scientific, CHAPS from Anatrace (Maumee, OH), Protease Arrest from G-Biosciences (St. Louis, MO), and 1D4 peptide from Celtek Peptides (Franklin, TN). HEK293T and rat PC12 cells were purchased from ATCC. Rho 1D4 antibody used for preparation of immunoaffinity columns was generated in-house (Hodges et al., 1988; Quazi and Molday, 2013) and purchased from UBC through Flintbox (www.rho1d4.com/); primary antibodies against calnexin, actin, and tubulin were from Abcam; fluorescent-tagged secondary antibodies for immunofluorescence imaging were from Molecular Probes; and anti-Cdc50-7F4 (CDC50A) primary antibodies used in Western blots and immunofluorescence analysis were raised in-house (Coleman and Molday, 2011). Restriction enzymes, T4-DNA ligase, Antarctic Phosphatase, and CaMKII kit were procured from New England Biolabs, DNA polymerase was from Thermo Scientific, and the QuikChange Site-Directed Mutagenesis Kit was from Agilent Technologies.
Preparation of C-terminal mutants
We generated mutants of bovine ATP8A2 (ATP8A2) in which the C-terminal tail was progressively truncated starting with the distalmost 20 amino acids. The truncation points were selected such that five approximately equal-length truncations can be made in the C-terminal domain and keeping in mind that primers can bind efficiently at these points. We previously developed a pcDNA3 plasmid containing ATP8A2 construct with a C-terminal 1D4 tag (Coleman et al., 2009). The 1D4 tag was removed from this construct and used as template to generate five different truncation mutants by PCR. In each mutant, a stretch of 11–27 amino acids was sequentially deleted, starting with the 20 terminal-most amino acids. Ninety-one of the C-terminal 100 amino acids were deleted in the most-truncated mutant. Each mutant had a 9– amino acid 1D4 tag (TETSQVAPA) at the C-terminus. The mutant ATP8A2 constructs were cloned into the pcDNA3 vector using the BamHI and NotI restriction sites.
Cell culture
HEK293T cells were maintained on DMEM (Sigma-Aldrich) supplemented with 8% bovine growth serum (Thermo Scientific), 100 IU/ml penicillin, 100 μg/ml streptomycin, 0.25 μg/ml amphotericin B (Corning), and 2 mM l-glutamine. To induce differentiation and neurite formation in PC12 cells, 2 d after transfection with ATP8A2/CDC50A, cells were changed to a medium of DMEM supplemented with 100 ng/ml NGF, 5% horse serum, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B. After incubation in this medium for 2 d, cells were used for immunofluorescence imaging. PC12 cells were cultured in medium containing DMEM supplemented with 10% horse serum, 5% fetal calf serum, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 0.25 μg/ml amphotericin B (Corning).
Transfection and overexpression of protein
HEK293T cells were grown to ∼30% confluence before transfection. The cells were cotransfected with two pcDNA3 plasmid constructs each carrying either WT or C-terminal truncated ATP8A2 and CDC50A, respectively. A 10-μg amount of each plasmid was used to transfect a 100-mm dish of cells by the calcium phosphate method. Forty-eight hours after transfection, overexpressed protein was harvested from cells. PC12 cells were grown to ∼90% confluence before transfection using Lipofectamine reagent (Invitrogen) following the manufacturer's instructions. Forty-eight hours after transfection, cells were transferred to a low-serum medium containing 1% horse serum and 50 ng/ml NGF (Sigma-Aldrich) to induce differentiation and neurite growth. Cells were used 2 d after induction of neurite growth for immunofluorescence imaging.
Immunoaffinity purification
Cells overexpressing ATP8A2-1D4 were harvested from culture plates and resuspended in 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)/NaOH, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol (DTT), 20 mM CHAPS, 0.5 mg/ml ePC, and 1× Protease Arrest (lysis buffer). Resuspended cells were lysed by stirring at 4°C for 30 min. After lysis, the CHAPS-insoluble fraction was removed by centrifugation at 66,750 × gav for 10 min. The CHAPS-soluble fraction was then loaded on an anti-Rho1D4 or anti-Cdc50-7F4 immunoaffinity column preequilibrated with 10 column volumes of lysis buffer. After binding for 30 min at 4°C, the column was washed six times with 500 μl of buffer containing 50 mM HEPES/NaOH, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 10 mM CHAPS, and 0.5 mg/ml ePC (CHAPS column wash buffer). Bound protein was then eluted from column with 1D4 elution buffer (50 mM HEPES/NaOH, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 10 mM CHAPS, 0.5 mg/ml ePC, and 0.4 mg/ml 1D4 peptide) in two iterations of 30 min each with agitation at 18°C. For immunoaffinity purification of CDC50A on an anti-Cdc50-7F4 column for Western blot analysis of the expression level of different mutants, a buffer containing 50 mM HEPES/NaOH, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 2% (wt/vol) SDS, and 0.5 mg/ml ePC (SDS elution buffer) was used. The concentration of purified protein was measured on a Coomassie blue–stained SDS–PAGE gel with bovine serum albumin of known concentration as standards. Both immunoaffinity columns (anti-1D4 and anti-7F4) were prepared by coupling the respective antibodies to CNBr-activated Sepharose as described previously (Molday et al., 1990).
ATPase activity
The ATPase activity of immunopurified WT and mutant ATP8A2 was measured by a previously described colorimetric method (Chifflet et al., 1988; Coleman et al., 2009). Briefly, ∼1 ng of purified protein was mixed with 0.5 mM ATP and 2.5 mg/ml lipid (a mixture of ePC and DOPS combined at different predetermined ratios) to obtain a final volume of 25 μl. Both ATP and lipids were prepared in ATPase assay buffer containing 50 mM HEPES/NaOH, pH 7.5, 150 mM NaCl, 12.5 mM MgCl2, 1 mM DTT, and 10 mM CHAPS. The reaction was carried out at 37°C for 20 min and terminated by addition of 25 μl of 12% SDS. The phosphate released from ATP hydrolysis was measured by treating the reaction mix with 75 μl of solution I (6% ascorbic acid, 1% ammonium molybdate in 1 N HCl) for 5 min, followed by 120 μl of solution II (2% sodium citrate, 2% sodium meta-arsenite, 2% acetic acid). The intensity of the resultant color was measured from the absorbance at 850 nm in a microplate reader. This measured intensity was compared with those of known phosphate concentrations on a standard curve to calculate the reaction velocity (mircomoles of Pi released/minute/milligram) for each PS concentration used in the assay. The resultant data were fitted to a Michaelis–Menten equation to calculate the maximum reaction velocity (Vmax) and PS-activation constant (KA) for the WT and mutant ATP8A2 alleles. Each measurement was recorded in triplicate, and every experiment was repeated at least four times independently.
To estimate IC50 for [Ca2+] inhibition of ATP8A2 activity, CaCl2 was included in the assay at various concentrations, and PS was used at a fixed concentration. The data were fitted to the equation
 |
The effect of phosphoinositides on the PS-activated ATPase activity of ATP8A2 was determined by adding 2.5% phosphoinositol or PI4P to purified ATP8A2 in the presence of 10% DOPS and 87.5% DOPC. The ATPase reactions were carried out as described.
Flippase activity
To measure the flippase activity, ATP8A2 was reconstituted in liposomes following a previously described protocol (Coleman et al., 2009). During immunopurification of protein intended for reconstitution, the column wash and elution buffers contained 0.75% of OGP instead of CHAPS detergent. After elution, the protein concentration was determined on SDS–PAGE gel. Eluted protein was diluted with liposome reconstitution buffer without lipids to contain ∼250 ng of eluted protein in each reconstitution. The liposome reconstitution buffer contained 50 mM HEPES/NaOH, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 1% OGP, 10% sucrose, and 2.5 mg/ml lipid mixture of 97% DOPC and 3% NBD-PS. A 100-μl amount of eluted protein (∼250 ng) was mixed with 100 μl of reconstitution buffer and stirred gently for 15 min at 4°C. This reconstitution mix was then dialyzed for ∼20 h against 2 l of dialysis buffer (10 mM HEPES/NaOH, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, and 10% sucrose) with at least two buffer changes. After reconstitution, 30 μl of liposomes was mixed with 6 μl of 10 mM ATP or AMP-PNP. The volume was adjusted to 60 μl with dialysis buffer and incubated at room temperature for 1 min. A 540-μl amount of dialysis buffer containing 10 mM EDTA was added to terminate flippase activity. The reaction mix was distributed equally into three wells of a black, clear-bottom, 96-well plate, and total NBD-PS fluorescence (Ft) was measured (470-nm excitation and 538-nm emission wavelengths). A membrane-impermeant reagent, sodium dithionite, which reduces the fluorescent NBD to a nonfluorescent derivative, was used to estimate the fraction of NBD-PS present on the outer liposome leaflet. Liposomes were exposed to 2 mM dithionite (final concentration) for 7.5 min, and fluorescence was measured once again (Ff). The fraction of reconstituted NBD-PS exposed on the outer leaflet of liposomes in the presence of ATP or its nonhydrolyzable analogue AMP-PNP was measured as
 |
and flippase activity was measured as
Immunofluorescence imaging
Cells grown on polylysine-coated glass coverslips were fixed with 4% paraformaldehyde for 15 min. After permeabilization for 1 h with 10% normal goat serum (NGS) and 0.2% Triton X-100, cells were treated for 2 h with primary antibody solution containing 10% NGS, 0.1% Triton X-100, mouse Rho1D4 monoclonal antibody (mAb) or Cdc50-7F4 mAb and rabbit polyclonal anti-calnexin antibody, or anti-GM130 antibody. The coverslips were washed and labeled with 4′,6-diamidino-2-phenylindole (DAPI) and goat Alexa 488 anti-mouse and goat Alexa 594 anti-rabbit secondary antibodies for 1 h. Coverslips were washed and mounted on glass slides for imaging with Mowiol mounting medium. The reagents for fixing, permeabilizing, and labeling were prepared in 100 mM phosphate buffer (pH 7.4), and all antibodies were used at 1:1000 dilution. Fluorescence images were acquired at room temperature using a 20× objective with a numerical aperture of 0.8 on a Zeiss LSM 700 confocal microscope equipped with Zen Image analysis software. Composite figures were prepared using Adobe Photoshop.
To analyze the colocalization of ATP8A2 (WT and mutants) to the ER and Golgi complex, the Manders colocalization coefficient was calculated using ImageJ software. To analyze the increase in neurite length of NGF-differentiated PC12 cells, the images were analyzed using Zen imaging software. We measured the length from base to tip of the longest neurite in each cell labeled by the anti–Cdc50-7F4 antibody. Cells that were not labeled by the antibody were not considered for analysis.
CaMKII-dependent phosphorylation
Ca/calmodulin-dependent protein kinase II and other associated reagents were purchased from New England Biolabs. CaMKII was preactivated by incubating with 1.2 μM calmodulin, 200 μM CaCl2, and 200 μM ATP in protein kinase buffer for 10 min at 30°C as per NEB protocol. Activated CaMKII (10 µl) was then added to 6 µl of 10× protein kinase buffer, 4 µl of [γ-32P]ATP (2 µCi), and 40 µl of ePC liposomes reconstituted with ATP8A2 (WT or S1138A mutant) and incubated for 1 h at 30°C. The amount of 32P-phosphorylated ATP8A2 was determined by phosphorimaging after SDS–PAGE.
Acknowledgments
We thank Theresa Hii and Laurie Molday for technical assistance. This work was supported in whole or part by grants from the National Institutes of Health (EY002422) and the Canadian Institutes of Health Research (MOP-106667). R.S.M. holds a Canada Research Chair in Vision and Macular Degeneration.
Abbreviations used:
- AMP-PNP
adenylyl-imidodiphosphate
- CaMKII
calcium/calmodulin-dependent protein kinase II
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- DOPC
1,2-dioleoyl-sn-glycero-3-phosphocholine
- DOPS
1,2-dioleoyl-sn-glycero-3-phosphoserine
- ePC
phosphatidylcholine (egg, chicken)
- ER
endoplasmic reticulum
- NBD-PS
1-oleoyl-2-{6-[7-nitro-2-1,3-benzoxadiazol-4-yl)]hexanoyl}-sn-glycero-3-phosphoserine
- NGF
nerve growth factor
- OGP
octyl-β-d-glucopyranose
- PI
phosphatidylinositol
- PI4P
phosphatidylinositol 4-phosphate
- PS
phosphatidylserine.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-06-0453) on December 8, 2016.
REFERENCES
- Andersen JP, Vestergaard AL, Mikkelsen SA, Mogensen LS, Chalat M, Molday RS. P4-ATPases as phospholipid flippases-structure, function, and enigmas. Front Physiol. 2016;7:275. doi: 10.3389/fphys.2016.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsen KB, Venema K, Jahn T, Baunsgaard L, Palmgren MG. Molecular dissection of the C-terminal regulatory domain of the plant plasma membrane H+-ATPase AHA2: mapping of residues that when altered give rise to an activated enzyme. Biochemistry. 1999;38:7227–7234. doi: 10.1021/bi982482l. [DOI] [PubMed] [Google Scholar]
- Baldridge RD, Graham TR. Identification of residues defining phospholipid flippase substrate specificity of type IV P-type ATPases. Proc Natl Acad Sci USA. 2012;109:E290–E298. doi: 10.1073/pnas.1115725109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitbol M, Fellmann P, Zachowski A, Devaux PF. Ion regulation of phosphatidylserine and phosphatidylethanolamine outside-inside translocation in human erythrocytes. Biochim Biophys Acta. 1987;904:268–282. doi: 10.1016/0005-2736(87)90376-2. [DOI] [PubMed] [Google Scholar]
- Brandt P, Zurini M, Neve RL, Rhoads RE, Vanaman TC. A C-terminal, calmodulin-like regulatory domain from the plasma membrane Ca2+-pumping ATPase. Proc Natl Acad Sci USA. 1988;85:2914–2918. doi: 10.1073/pnas.85.9.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chifflet S, Torriglia A, Chiesa R, Tolosa S. A method for the determination of inorganic phosphate in the presence of labile organic phosphate and high concentrations of protein: application to lens ATPases. Anal Biochem. 1988;168:1–4. doi: 10.1016/0003-2697(88)90002-4. [DOI] [PubMed] [Google Scholar]
- Coleman JA, Kwok MC, Molday RS. Localization, purification, and functional reconstitution of the P4-ATPase Atp8a2, a phosphatidylserine flippase in photoreceptor disk membranes. J Biol Chem. 2009;284:32670–32679. doi: 10.1074/jbc.M109.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JA, Molday RS. Critical role of the beta-subunit CDC50A in the stable expression, assembly, subcellular localization, and lipid transport activity of the P4-ATPase ATP8A2. J Biol Chem. 2011;286:17205–17216. doi: 10.1074/jbc.M111.229419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JA, Quazi F, Molday RS. Mammalian P4-ATPases and ABC transporters and their role in phospholipid transport. Biochim Biophys Acta. 2013;1831:555–574. doi: 10.1016/j.bbalip.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JA, Vestergaard AL, Molday RS, Vilsen B, Andersen JP. Critical role of a transmembrane lysine in aminophospholipid transport by mammalian photoreceptor P4-ATPase ATP8A2. Proc Natl Acad Sci USA. 2012;109:1449–1454. doi: 10.1073/pnas.1108862109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JA, Zhu X, Djajadi HR, Molday LL, Smith RS, Libby RT, John SW, Molday RS. Phospholipid flippase ATP8A2 is required for normal visual and auditory function and photoreceptor and spiral ganglion cell survival. J Cell Sci. 2014;127:1138–1149. doi: 10.1242/jcs.145052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daleke DL, Lyles JV. Identification and purification of aminophospholipid flippases. Biochim Biophys Acta. 2000;1486:108–127. doi: 10.1016/s1388-1981(00)00052-4. [DOI] [PubMed] [Google Scholar]
- Ding J, Wu Z, Crider BP, Ma Y, Li X, Slaughter C, Gong L, Xie XS. Identification and functional expression of four isoforms of ATPase II, the putative aminophospholipid translocase. Effect of isoform variation on the ATPase activity and phospholipid specificity. J Biol Chem. 2000;275:23378–23386. doi: 10.1074/jbc.M910319199. [DOI] [PubMed] [Google Scholar]
- Enyedi A, Vorherr T, James P, McCormick DJ, Filoteo AG, Carafoli E, Penniston JT. The calmodulin binding domain of the plasma membrane Ca2+ pump interacts both with calmodulin and with another part of the pump. J Biol Chem. 1989;264:12313–12321. [PubMed] [Google Scholar]
- Folmer DE, Elferink RP, Paulusma CC. P4 ATPases—lipid flippases and their role in disease. Biochim Biophys Acta. 2009;1791:628–635. doi: 10.1016/j.bbalip.2009.02.008. [DOI] [PubMed] [Google Scholar]
- Fuglsang AT, Visconti S, Drumm K, Jahn T, Stensballe A, Mattei B, Jensen ON, Aducci P, Palmgren MG. Binding of 14-3-3 protein to the plasma membrane H(+)-ATPase AHA2 involves the three C-terminal residues Tyr(946)-Thr-Val and requires phosphorylation of Thr(947) J Biol Chem. 1999;274:36774–36780. doi: 10.1074/jbc.274.51.36774. [DOI] [PubMed] [Google Scholar]
- Hodges RS, Heaton RJ, Parker JM, Molday L, Molday RS. Antigen-antibody interaction. Synthetic peptides define linear antigenic determinants recognized by monoclonal antibodies directed to the cytoplasmic carboxyl terminus of rhodopsin. J Biol Chem. 1988;263:11768–11775. [PubMed] [Google Scholar]
- Lee S, Uchida Y, Wang J, Matsudaira T, Nakagawa T, Kishimoto T, Mukai K, Inaba T, Kobayashi T, Molday RS, et al. Transport through recycling endosomes requires EHD1 recruitment by a phosphatidylserine translocase. EMBO J. 2015;34:669–688. doi: 10.15252/embj.201489703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Marques RL, Theorin L, Palmgren MG, Pomorski TG. P4-ATPases: lipid flippases in cell membranes. Pflugers Arch. 2014;466:1227–1240. doi: 10.1007/s00424-013-1363-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvezzi M, Chalat M, Janjusevic R, Picollo A, Terashima H, Menon AK, Accardi A. Ca2+-dependent phospholipid scrambling by a reconstituted TMEM16 ion channel. Nat Commun. 2013;4:2367. doi: 10.1038/ncomms3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason AB, Allen KE, Slayman CW. Effects of C-terminal truncations on trafficking of the yeast plasma membrane H+-ATPase. J Biol Chem. 2006;281:23887–23898. doi: 10.1074/jbc.M601818200. [DOI] [PubMed] [Google Scholar]
- Mason AB, Allen KE, Slayman CW. C-terminal truncations of the Saccharomyces cerevisiae PMA1 H+-ATPase have major impacts on protein conformation, trafficking, quality control, and function. Eukaryot Cell. 2014;13:43–52. doi: 10.1128/EC.00201-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse T, Kelly PT. Overexpression of Ca2+/calmodulin-dependent protein kinase II in PC12 cells alters cell growth, morphology, and nerve growth factor-induced differentiation. J Neurosci. 1997;17:924–931. doi: 10.1523/JNEUROSCI.17-03-00924.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molday LL, Cook NJ, Kaupp UB, Molday RS. The cGMP-gated cation channel of bovine rod photoreceptor cells is associated with a 240-kDa protein exhibiting immunochemical cross-reactivity with spectrin. J Biol Chem. 1990;265:18690–18695. [PubMed] [Google Scholar]
- Moller JV, Olesen C, Winther AM, Nissen P. The sarcoplasmic Ca2+-ATPase: design of a perfect chemi-osmotic pump. Q Rev Biophys. 2010;43:501–566. doi: 10.1017/S003358351000017X. [DOI] [PubMed] [Google Scholar]
- Morth JP, Pedersen BP, Toustrup-Jensen MS, Sorensen TL, Petersen J, Andersen JP, Vilsen B, Nissen P. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- Natarajan P, Liu K, Patil DV, Sciorra VA, Jackson CL, Graham TR. Regulation of a Golgi flippase by phosphoinositides and an ArfGEF. Nat Cell Biol. 2009;11:1421–1426. doi: 10.1038/ncb1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onat OE, Gulsuner S, Bilguvar K, Nazli Basak A, Topaloglu H, Tan M, Tan U, Gunel M, Ozcelik T. Missense mutation in the ATPase, aminophospholipid transporter protein ATP8A2 is associated with cerebellar atrophy and quadrupedal locomotion. Eur J Hum Genet. 2013;21:281–285. doi: 10.1038/ejhg.2012.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quazi F, Molday RS. Differential phospholipid substrates and directional transport by ATP-binding cassette proteins ABCA1, ABCA7, and ABCA4 and disease-causing mutants. J Biol Chem. 2013;288:34414–34426. doi: 10.1074/jbc.M113.508812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian TT, Baldridge RD, Xu P, Graham TR. Phospholipid flippases: Building asymmetric membranes and transport vesicles. Biochim Biophys Acta. 2012;1821:1068–1077. doi: 10.1016/j.bbalip.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segawa K, Kurata S, Yanagihashi Y, Brummelkamp TR, Matsuda F, Nagata S. Caspase-mediated cleavage of phospholipid flippase for apoptotic phosphatidylserine exposure. Science. 2014;344:1164–1168. doi: 10.1126/science.1252809. [DOI] [PubMed] [Google Scholar]
- Shinoda T, Ogawa H, Cornelius F, Toyoshima C. Crystal structure of the sodium-potassium pump at 2.4 A resolution. Nature. 2009;459:446–450. doi: 10.1038/nature07939. [DOI] [PubMed] [Google Scholar]
- Soupene E, Kuypers FA. Identification of an erythroid ATP-dependent aminophospholipid transporter. Br J Haematol. 2006;133:436–438. doi: 10.1111/j.1365-2141.2006.06051.x. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Fujimura-Kamada K, Yamamoto T. Functions of phospholipid flippases. J Biochem. 2011;149:131–143. doi: 10.1093/jb/mvq140. [DOI] [PubMed] [Google Scholar]
- Tashima K, Yamamoto H, Setoyama C, Ono T, Miyamoto E. Overexpression of Ca2+/calmodulin-dependent protein kinase II inhibits neurite outgrowth of PC12 cells. J Neurochem. 1996;66:57–64. doi: 10.1046/j.1471-4159.1996.66010057.x. [DOI] [PubMed] [Google Scholar]
- van der Velden LM, Wichers CG, van Breevoort AE, Coleman JA, Molday RS, Berger R, Klomp LW, van de Graaf SF. Heteromeric interactions required for abundance and subcellular localization of human CDC50 proteins and class 1 P4-ATPases. J Biol Chem. 2010;285:40088–40096. doi: 10.1074/jbc.M110.139006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vestergaard AL, Coleman JA, Lemmin T, Mikkelsen SA, Molday LL, Vilsen B, Molday RS, Dal Peraro M, Andersen JP. Critical roles of isoleucine-364 and adjacent residues in a hydrophobic gate control of phospholipid transport by the mammalian P4-ATPase ATP8A2. Proc Natl Acad Sci USA. 2014;111:E1334–E1343. doi: 10.1073/pnas.1321165111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RR, Kwon YG, Taing M, Lawrence DS, Edelman AM. Definition of optimal substrate recognition motifs of Ca2+-calmodulin-dependent protein kinases IV and II reveals shared and distinctive features. J Biol Chem. 1998;273:3166–3172. doi: 10.1074/jbc.273.6.3166. [DOI] [PubMed] [Google Scholar]
- Xu Q, Yang GY, Liu N, Xu P, Chen YL, Zhou Z, Luo ZG, Ding X. P4-ATPase ATP8A2 acts in synergy with CDC50A to enhance neurite outgrowth. FEBS Lett. 2012;586:1803–1812. doi: 10.1016/j.febslet.2012.05.018. [DOI] [PubMed] [Google Scholar]
- Zhou X, Sebastian TT, Graham TR. Auto-inhibition of Drs2p, a yeast phospholipid flippase, by its carboxyl-terminal tail. J Biol Chem. 2013;288:31807–31815. doi: 10.1074/jbc.M113.481986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Libby RT, de Vries WN, Smith RS, Wright DL, Bronson RT, Seburn KL, John SW. Mutations in a p-type ATPase gene cause axonal degeneration. PLoS Genet. 2012;8:e1002853. doi: 10.1371/journal.pgen.1002853. [DOI] [PMC free article] [PubMed] [Google Scholar]