Nanoscale pulldown (NanoSPD) miniaturizes the concept of affinity pulldown to detect protein–protein interactions in live cells. NanoSPD hijacks the myosin-based intracellular trafficking machinery to assess interactions under physiological buffer conditions and is microscopy-based, allowing for sensitive detection and quantification.
Abstract
Protein–protein interactions (PPIs) regulate assembly of macromolecular complexes, yet remain challenging to study within the native cytoplasm where they normally exert their biological effect. Here we miniaturize the concept of affinity pulldown, a gold-standard in vitro PPI interrogation technique, to perform nanoscale pulldowns (NanoSPDs) within living cells. NanoSPD hijacks the normal process of intracellular trafficking by myosin motors to forcibly pull fluorescently tagged protein complexes along filopodial actin filaments. Using dual-color total internal reflection fluorescence microscopy, we demonstrate complex formation by showing that bait and prey molecules are simultaneously trafficked and actively concentrated into a nanoscopic volume at the tips of filopodia. The resulting molecular traffic jams at filopodial tips amplify fluorescence intensities and allow PPIs to be interrogated using standard epifluorescence microscopy. A rigorous quantification framework and software tool are provided to statistically evaluate NanoSPD data sets. We demonstrate the capabilities of NanoSPD for a range of nuclear and cytoplasmic PPIs implicated in human deafness, in addition to dissecting these interactions using domain mapping and mutagenesis experiments. The NanoSPD methodology is extensible for use with other fluorescent molecules, in addition to proteins, and the platform can be easily scaled for high-throughput applications.
INTRODUCTION
The identification of protein–protein interactions (PPIs) within macromolecular complexes is a powerful approach to understanding cellular biology in normal and disease states. Many methodologies exist to discover PPIs, including yeast two-hybrid (Y2H), mammalian two-hybrid, and affinity purification coupled with mass spectrometry (Fields and Song, 1989; Luo et al., 1997; Shioda et al., 2000; Gingras et al., 2007; Lemmens et al., 2015). Because these methodologies vary in their degree of sensitivity and specificity, no single approach eliminates false positives and false negatives to detect the full landscape of PPIs (Braun et al., 2009; Chen et al., 2010). Replication by independent orthogonal assays is thus critical to build confidence in a putative PPI.
Coimmunoprecipitation (co-IP) and affinity pulldown (AP) assays are commonly used to independently confirm PPIs. While these methodologies are considered gold standards, they are biased toward detecting high-affinity PPIs that remain associated with the affinity matrix throughout high-stringency washing steps (Gingras et al., 2007). Proximity ligase techniques (Roux et al., 2012) or chemical cross-linking can assist detection of low-affinity PPIs by IP/AP. Alternatively, single-molecule pulldown optically interrogates the affinity matrix to visualize transient, low-affinity binding events (Jain et al., 2011). A drawback to these approaches is that PPIs are assessed outside the cell, where buffer conditions can be a significant cause of false-negative interactions (Hakhverdyan et al., 2015).
Microscopy-based techniques, including fluorescence correlation spectroscopy, fluorescence resonance energy transfer, and bimolecular fluorescence complementation can measure PPIs in live cells, with native buffer, temperature, and intramolecular crowding effects (Wallrabe and Periasamy, 2005; Kerppola, 2006; Ries and Schwille, 2012), but require specialized equipment, optimization of fluorescent probe placement, or complex data postprocessing. PPIs can also be detected through spatial correlation (i.e., overlap) of fluorescently labeled bait and prey proteins within a cell (Manders et al., 1992; Costes et al., 2004; Comeau et al., 2006; Dunn et al., 2011). Colocalization techniques are experimentally straightforward but can be confounded when bait or prey localize diffusely throughout the cytosol. Artificially targeting bait–prey complexes to subcellular domains greatly improves specificity of these PPI detection approaches (Bear et al., 2000; Zolghadr et al., 2008; Gallego et al., 2013; Herce et al., 2013). In the case of vesicle-associated proteins, coupling vesicles to microtubule-based molecular motors redirects bait–prey complexes to a distinct cellular location, providing a highly specific readout of the PPI (Jenkins et al., 2012; Bentley and Banker, 2015; Bentley et al., 2015).
Here we describe the nanoscale pulldown (NanoSPD) as a generally applicable technique for studying cytosolic, nuclear, and membrane-associated PPIs within live cells. NanoSPD repurposes myosin motors to capture, traffic (i.e., pull), and actively concentrate fluorescently tagged protein complexes at the tips of filopodia; actin-based membrane protrusions that extend beyond the periphery of cells (Mattila and Lappalainen, 2008). We provide a comprehensive analysis framework and software tool to rigorously quantify and statistically test NanoSPD data sets. Using NanoSPD, we explore a range of macromolecular complexes involved in the detection of sound by auditory hair cells and characterize their binding properties by domain mapping and mutagenesis.
RESULTS
Harnessing myosin-powered intracellular traffic to retarget molecules to filopodia
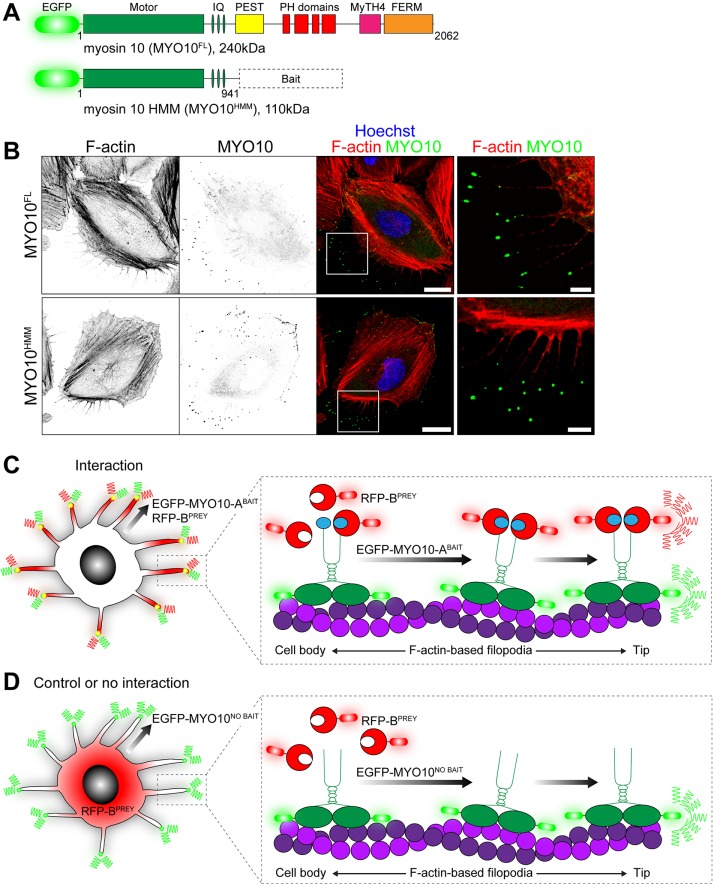
Myosins are a superfamily of ATPase molecular motors that generate force on actin filaments to power a variety of biological processes, including cytokinesis, cell motility, and intracellular trafficking (Sellers, 2000; Sweeney and Houdusse, 2010). We reasoned that myosin-based trafficking could be exploited to artificially displace bait proteins within the cell and hypothesized that interacting prey proteins would be similarly recruited. This idea was explored using the class x myosin molecular motor (MYO10) because of its robust ability to traffic within filopodia and accumulate at their tips (Berg et al., 2000; Berg and Cheney, 2002; Bohil et al., 2006). The MYO10 molecule consists of an ATPase motor domain that binds to actin filaments, a neck domain that associates with regulatory light chains, a coiled-coil motif, and a C-terminal tail domain that binds cargo proteins (Kerber and Cheney, 2011) (Figure 1A). Similar to full-length MYO10, truncated molecules still accumulate at filopodial tips (Berg and Cheney, 2002) and are referred to as heavy meromyosin, MYO10HMM (Figure 1, A and B). The C-terminal tail domain is therefore not required for MYO10 to traffic within filopodia.
FIGURE 1:
Principle of the nanoscale pulldown (NanoSPD). (A) Domain representation of full-length EGFP-tagged MYO10FL with the ATPase motor domain, IQ motifs that bind light chains, a coiled-coil and tail domains that bind cargo. Heavy meromyosin MYO10HMM molecules are identical but truncated after the coiled-coil. Query bait proteins are fused to the C-terminus of MYO10HMM. (B) EGFP-tagged MYO10FL (top panel) and truncated MYO10HMM (bottom panel) traffic to filopodial tips in HeLa cells. Inset white boxes are shown magnified. F-actin is labeled with rhodamine phalloidin (red), and nuclei are stained with Hoechst (blue). Scale bars: 20 µm; 5 µm (inset). F-actin and MYO10 images have been uniformly inverted. (C) Using NanoSPD to test the interaction between ABAIT and BPREY proteins. ABAIT is forcibly trafficked to filopodial tips by MYO10 as part of the EGFP-MYO10-ABAIT fusion protein (green). In the presence of an interaction, RFP-BPREY molecules (red) are actively cotransported along filopodia, resulting in concentration of bait and prey molecules at filopodial tips. (D) In the control, EGFP-MYO10NO BAIT molecules (green) lacking a bait still accumulate at filopodial tips but no longer traffic RFP-BPREY (red). Identical results are expected if RFP-BPREY does not interact with EGFP-MYO10-ABAIT.
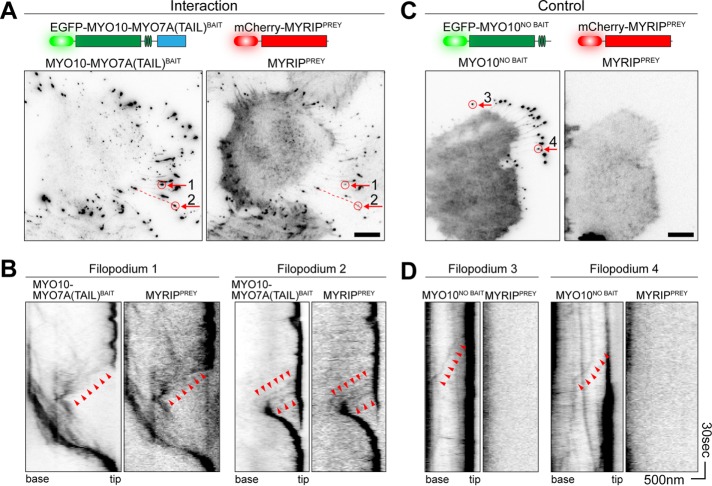
Next we investigated whether replacing the endogenous cargo-binding tail domain of MYO10 with a bait protein could similarly allow chimeric MYO10-bait molecules to traffic along filopodia in complex with interacting prey proteins (Figure 1, C and D). For proof of principle, we tested the known interaction between the tail domain of MYO7A and the MYO7A-Rab interacting protein (MYRIP) (El-Amraoui et al., 2002; Fukuda and Kuroda, 2002). Because trafficking of wild-type MYO10 along filopodia occurs in the single-molecule regime (Kerber et al., 2009), we used time-lapse total internal reflection fluorescence microscopy (TIRFM) to capture these events. Two classes of behavior were observed in HeLa cells expressing a chimeric EGFP-MYO10HMM bait fused to the tail domain of MYO7A, referred to as MYO10-MYO7A(TAIL)BAIT, in addition to the prey mCherry-MYRIP, referred to as MYRIPPREY. First, enhanced green fluorescent protein (EGFP) and mCherry coaccumulated at filopodial tips (Figure 2A). Second, we observed correlated bidirectional motility of EGFP and mCherry puncta along the filopodial shafts (Figure 2B). Bright puncta moved retrogradely toward the cell body, while very dim puncta moved anterogradely toward the filopodial tip (Figure 2B). These trafficking events were qualitatively similar to that reported for wild-type MYO10, where retrograde motility is linked to treadmilling of the underlying actin filaments, and anterograde motion results from active trafficking by the myosin motor (Kerber and Cheney, 2011; Kerber et al., 2009).
FIGURE 2:
MYO10 can be harnessed to traffic bait–prey complexes in live cells. Visualization of bait and prey molecule trafficking in live HeLa cells by time-lapse TIRFM. Imaging was performed at 37°C. Fluorescence images have been uniformly inverted. (A) In HeLa cells expressing MYO10-MYO7A(TAIL)BAIT (left) and MYRIPPREY (right), both molecules accumulate at filopodial tips. A single frame from the time-lapse is shown. Filopodia selected for kymographs are indicated by red arrows. (B) Kymographs from A reveal molecular trafficking along filopodia. Bait and prey molecules accumulate at the filopodial tips, which are visible as a dark vertical line. Dim puncta of MYO10-MYO7A(TAIL)BAIT and MYRIPPREY move synchronously toward the filopodial tip (anterograde, arrowheads). Retrograde flow of bait and prey molecules (toward the cell body) is also observed. (C) MYO10NO BAIT accumulates at filopodial tips but does not traffic MYRIPPREY molecules. This control experiment demonstrates that trafficking of MYRIPPREY is critically dependent on the MYO7A(TAIL) moiety being fused to the myosin motor. (D) Kymographs from C confirm that MYRIPPREY molecules are not trafficked by MYO10NO BAIT. Anterograde (arrowheads) trafficking of MYO10NO BAIT is observed. Filopodia 3 and 4 are indicated by arrows in C. Horizontal scale bars: 10 µm (A and C); 500 nm (B and D); vertical scale bars: 30 s (B and D), with the bottom of each kymograph representing 0 s.
As a control, we performed time-lapse imaging of HeLa cells expressing MYRIPPREY and EGFP-MYO10, lacking the MYO7A-tail moiety (referred to as MYO10NO BAIT) (Figure 2C). EGFP-positive MYO10NO BAIT puncta moved bidirectionally along the shaft and strongly accumulated at filopodial tips. mCherry-positive MYRIPPREY puncta no longer trafficked along the filopodial shaft or accumulated at the tips, showing that they could not bind to MYO10NO BAIT or its associated light chains (Figure 2D). We conclude that the MYO10-MYO7A(TAIL)BAIT chimera specifically cotransported MYRIPPREY and that PPIs can be inferred from correlated motility of bait–prey complexes along filopodia. We call this method nanoscale pulldown (NanoSPD), since bait–prey complexes are forced to traffic, or are pulled, along filopodia by the MYO10 molecular motor.
Quantification of NanoSPD to test binary protein–protein interactions
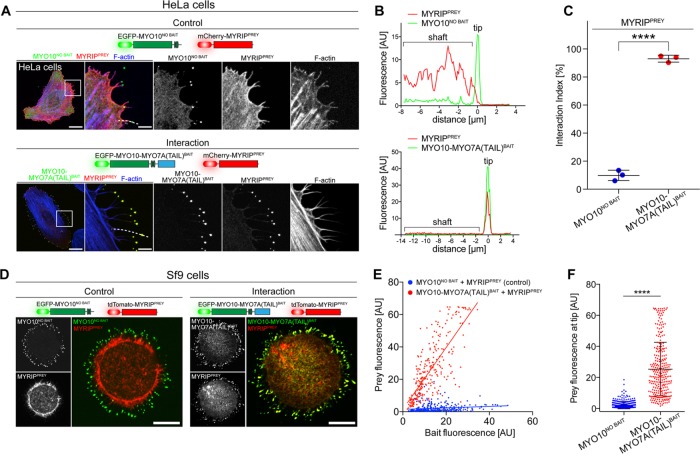
To develop a simple readout for NanoSPD, we quantified the fluorescence intensity of bait–prey complexes at the filopodial tips. The continuous trafficking of bait–prey molecules by MYO10 actively concentrates them at this location, boosting fluorescence intensity and allowing NanoSPD assays to be captured by wide-field or confocal epifluorescence microscopy. NanoSPD was tested in HeLa cells using the MYO7A-MYRIP interaction as a proof of principle. HeLa cells expressing MYRIPPREY and either MYO10-MYO7ABAIT or MYO10NO BAIT were fixed, and images were captured using confocal microscopy (Figure 3A) rather than TIRFM (Figure 2). MYRIPPREY was robustly concentrated at the filopodial tips by MYO10-MYO7ABAIT but not by MYO10NO BAIT (Figure 3A). Similar observations were made in Sf9 insect cells (Figure 3D), demonstrating that NanoSPD is applicable to a range of cell lines. Sf9 insect cells were particularly notable for reproducibly generating large numbers of filopodia when transfected in suspension culture and then seeded onto glass coverslips.
FIGURE 3:
Quantifying PPIs by NanoSPD. NanoSPD concentrates bait–prey complexes into a nanoscopic volume at filopodial tips, providing noise-free fluorescence enhancement and a defined location for imaging away from the cell body. (A) In fixed HeLa cells expressing MYO10-MYO7A(TAIL)BAIT and MYRIPPREY, both bait and prey coaccumulate at filopodial tips. MYO10NO BAIT does not traffic MYRIPPREY in the control, confirming the interaction and that MYRIPPREY cannot traffic autonomously. Scale bars: 20 µm; 5 µm (magnified boxed regions). (B) Using SCA to detect molecular interactions. Pearson’s r coefficient is calculated along the filopodial shaft (dotted line, A) to measure bait and prey fluorescence correlation. The likelihood that the observed correlation can occur by random chance is estimated by bootstrapping (see Materials and Methods). (C) SCA reveals a significantly higher interaction index (t test) when MYRIPPREY is coexpressed with MYO10-MYO7A(TAIL)BAIT (n = 292 filopodia total) than when MYRIPPREY is coexpressed with MYO10NO BAIT (n = 264 filopodia total). Each data point is the average interaction index from a single experiment (three independent determinations). Data are mean ± SD. (D) NanoSPD can be used in nonmammalian Sf9 insect cells that produce large numbers of filopodia. In fixed Sf9 cells, MYRIPPREY accumulates with MYO10-MYO7A(TAIL)BAIT at filopodial tips but not when coexpressed with MYO10NO BAIT. Scale bar: 10 µm. (E) Using ICA to detect PPIs in Sf9 cells. Scatter plot of bait (x-axis) and prey (y-axis) fluorescence at individual filopodia tips (from three independent determinations). MYRIPPREY fluorescence is linearly correlated with MYO10-MYO7A(TAIL)BAIT fluorescence, unlike cells expressing MYO10NO BAIT. (F) Bar graph of MYRIPPREY fluorescence intensities from E. Coexpression with MYO10-MYO7A(TAIL)BAIT significantly increases the average prey fluorescence intensity at filopodial tips, compared with MYO10NO BAIT (Mann-Whitney U-test). Data are mean ± SD. ****, p < 0.0001.
We developed a framework and software tool that uses Pearson’s correlation coefficient (r) to quantify the spatial correlation of bait and prey fluorescence along individual filopodia (Figure 3B). Detailed documentation of this algorithm and user instructions for the MATLAB tool are provided (Materials and Methods). The Pearson’s r-based quantification algorithm was tested on line scans along filopodia of HeLa cells expressing MYRIPPREY and either MYO10-MYO7ABAIT or MYO10NO BAIT (Figure 3B). In cells expressing MYO10-MYO7ABAIT and MYRIPPREY, 93.1 ± 2.4% of filopodia (292 filopodia sampled, three independent trials) had significantly correlated bait–prey fluorescence intensities (Figure 3C and Supplemental Figure S1A). Conversely, in control HeLa cells expressing MYO10NO BAIT and MYRIPPREY, only 9.9 ± 3.7% of filopodia (264 filopodia sampled, three independent trials) were significantly correlated. The ∼10-fold increase in interaction index (percentage of total filopodia with bait–prey correlation) was statistically significant (p < 0.0001, two-tailed t test) and quantitatively confirmed the interaction between these proteins (Figure 3C).
While the Pearson’s r-based algorithm was sensitive to spatial correlations of bait–prey complexes in HeLa cells, it reported a higher level of false-positive correlations in Sf9 cells when used to evaluate control interactions such as MYO10NO BAIT and MYRIPPREY (Figure 3D). The likely cause of this phenomenon was from cytoplasmic volume filling of the filopodial tips in Sf9 cells, which are bulbous. Because Pearson’s r coefficient does not consider the magnitude of fluorescence changes, small increases in prey fluorescence due to volume filling can be detected as an artifactual correlation. To address this, we measured the absolute fluorescence intensities at filopodial tips of Sf9 cells. When examined over a large sample of independent filopodia, bait and prey fluorescence intensities are expected to be correlated in the presence of an interaction, and uncorrelated otherwise. A critical condition of this intensity-based analysis is that imaging conditions are standardized to ensure that data from independent experiments can be combined.
Line scan data from Sf9 cells testing the MYO7A–MYRIP interaction (Figure 3D) were reanalyzed using the intensity-based correlation algorithm. Fluorescence intensities detected at filopodial tips were plotted on a x–y scatter plot, with bait as the independent (x) and prey as the dependent (y) variable (Figure 3E). In both interaction (red, MYO10-MYO7ABAIT + MYRIPPREY) and control (blue, MYO10NOBAIT + MYRIPPREY) data sets, a broad range of bait intensities were observed (Figure 3E). This was consistent with continuous MYO10 trafficking occurring independently in each filopodium, such that at the time of chemical fixation these “snapshot” data contain filopodia in different stages of extension and retraction. Critically, there was a linear correlation between bait and prey fluorescence intensity at the filopodial tips of cells expressing MYO10-MYO7ABAIT and MYRIPPREY (red) (Figure 3E). By comparison, in control cells expressing MYO10NO BAIT and MYRIPPREY, bait and prey fluorescence values were uncorrelated (blue) (Figure 3E). The average fluorescence intensity of MYRIPPREY at filopodial tips was increased by 11.8-fold in the presence of MYO10-MYO7ABAIT compared with MYO10NO BAIT (Figure 3F) (p < 0.0001, Mann-Whitney U-test), providing a separate measure of the interaction by NanoSPD.
We considered the possibility that the difference in MYRIPPREY fluorescence at filopodial tips could be due to differences of trafficking efficiencies between MYO10-MYO7ABAIT and MYO10NO BAIT. While there was a statistically significant difference between the mean fluorescence intensities of MYO10NO BAIT versus MYO10-MYO7ABAIT, the effect size (1.2-fold) was minimal (Supplemental Figure S1B). The large increase in MYRIPPREY fluorescence at filopodial tips was therefore due to the specific interaction with MYO10-MYO7ABAIT. We conclude that bait and prey fluorescence intensities at filopodial tips can be used as a sensitive measure of a PPI in NanoSPD.
NanoSPD can traffic multiple preys to dissect binding hierarchies in macromolecular complexes
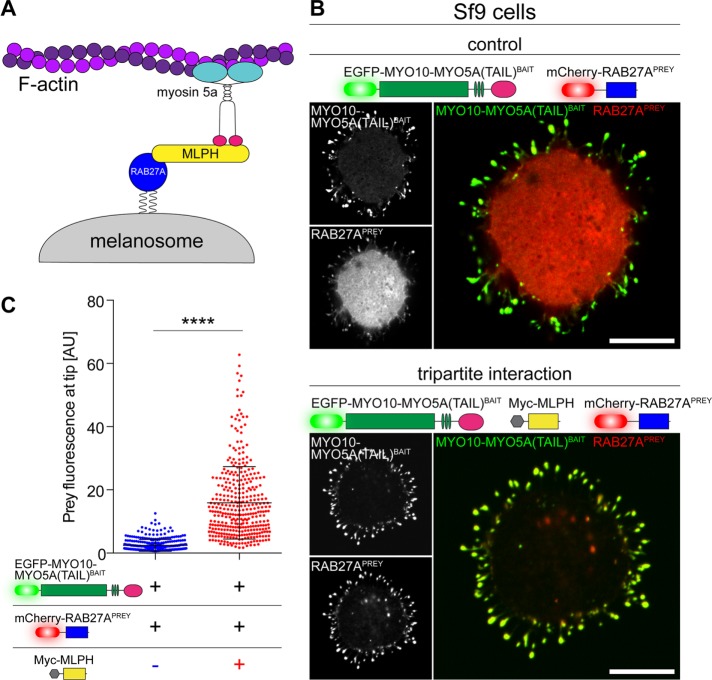
We investigated whether NanoSPD could traffic multiple prey species along a filopodium to probe larger macromolecular assemblies. The tripartite complex (Wu et al., 2002) between the C-terminal tail domain of myosin 5a (MYO5A), the Ras-related GTPase RAB27A, and melanophilin (MLPH) was tested by NanoSPD in Sf9 cells (Figure 4A). RAB27APREY strongly accumulated at filopodial tips when expressed in Sf9 cells along with both MYO10-MYO5A(TAIL)BAIT and a nonfluorescently tagged myc-MLPH (Figure 4B, bottom panel). To test the requirement for MLPH in complex formation, we examined Sf9 cells expressing MYO10-MYO5A(TAIL)BAIT and RAB27APREY alone (Figure 4B, top panel). Qualitatively, we observed a reduction in the intensity of RAB27APREY accumulating at filopodial tips, although small quantities were still detected (Figure 4B, top panel), suggesting the possibility of an endogenous adaptor. These data were analyzed using the intensity-based correlation algorithm. The average fluorescence intensity of MYO10-MYO5A(TAIL)BAIT molecules at filopodial tips was independent of the presence or absence of myc-MLPH, indicating no systematic change in bait-molecule trafficking (Supplemental Figure S1C). Despite the equal trafficking of MYO10-MYO5A(TAIL)BAIT molecules, there was a significant 6.5-fold increase in RAB27APREY fluorescence in the presence of myc-MLPH (Figure 4C). We conclude that the association of RAB27A with MYO5A(TAIL) was critically dependent on the effector MLPH. Our data demonstrate that the MYO10-powered bait can efficiently traffic multiple, interacting prey molecules within filopodia, which can be exploited by NanoSPD to study the composition and hierarchy of larger macromolecules.
FIGURE 4:
Dissecting hierarchies in macromolecular complexes by NanoSPD. (A) Schematic of the tripartite melanosome receptor formed by MYO5A, MPLH, and RAB27A. Active RAB27A (blue) attaches to the melanosome membrane via geranylgeranyl moieties, binds its effector melanophilin (MLPH, yellow), and is recruited to the MYO5A tail domain (pink) to be trafficked along actin filaments by the motor domain (cyan). (B) NanoSPD in Sf9 cells demonstrates the requirement for MLPH in complex formation. RAB27APREY is not robustly trafficked by MYO10-MYO5A(TAIL)BAIT, despite the bait accumulating strongly at filopodial tips (top panel). Inclusion of myc-tagged MLPH (dark) results in coaccumulation of both MYO10-MYO5A(TAIL)BAIT and RAB27APREY at filopodial tips, demonstrating MLPH is required for complex formation (bottom panel). Scale bars: 10 µm. (C) ICA of RAB27APREY fluorescence at filopodial tips from B. RAB27APREY tip fluorescence is significantly increased upon coexpression of Myc-MLPH (Mann-Whitney U-test), confirming the interaction (data points are individual filopodia from three independent determinations). Data are mean ± SD. ****, p < 0.0001.
Using NanoSPD to validate Y2H screens
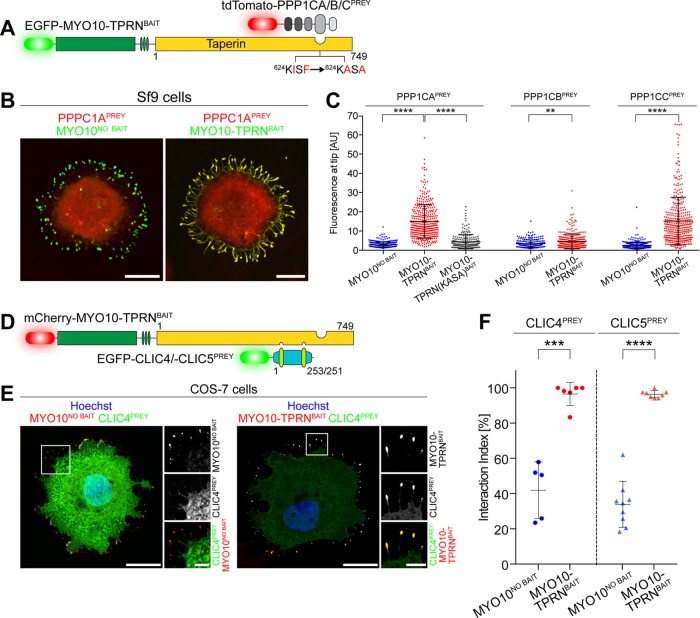
Dissecting mechanisms of disease requires identifying molecular components and understanding how they function within the broader context of cellular biology. As an example, genetic studies of human hereditary hearing loss, in conjunction with proteomic analyses, have identified proteins involved in the detection of sound (Richardson et al., 2011; Barr-Gillespie, 2015). Many of these proteins assemble into macromolecular complexes within hair cell stereocilia, mechanosensory organelles that are the primary transducers of sound (Schwander et al., 2010). We previously reported that taperin (TPRN) concentrates within stereocilia and is mutated in autosomal recessive human deafness, DFNB79 (Li et al., 2010; Rehman et al., 2010); however, its molecular function remains unknown. To discover binding proteins, we screened Y2H libraries using full-length TPRN as bait (Supplemental Table S2). Because Y2H screens can have a high false-positive rate, we validated these interacting proteins by NanoSPD.
Y2H assays identified the alpha (PPP1CA) and gamma (PPP1CC) catalytic subunits of protein phosphatase 1 (PP1) as potential interacting proteins. PP1 is a widely expressed serine/threonine phosphatase and regulates a variety of cellular functions, including glycogen metabolism, transcription, protein synthesis, cell division, and apoptosis (Rebelo et al., 2015). We validated the interaction of the catalytic subunit of PP1 and TPRN by NanoSPD in Sf9 cells expressing MYO10-TPRNBAIT and either PPP1CAPREY (alpha), PPP1CBPREY (beta), or PPP1CCPREY (gamma) subunits (Figure 5, A and B, and Supplemental Figure S2, B and C). Intensity correlation analysis (ICA) revealed that the mean fluorescence intensities of PPP1CAPREY, PPP1CBPREY, and PPP1CCPREY at filopodial tips were significantly increased by 5.2-, 1.3-, and 5.7-fold, respectively, when expressed with MYO10-TPRNBAIT versus MYO10NO BAIT (Figure 5C). Although statistically significant, the fluorescence increase (i.e., the effect size) of PPP1CB was minimal (1.3-fold) and indicated a preference of TPRN to bind PPP1CA (alpha) and PPP1CC (gamma) subunits. These observations confirm the binding of PPP1CA and PPP1CC detected by Y2H (Supplemental Table S2) and AP (Ferrar et al., 2012). We conclude that NanoSPD can successfully detect variations in binding between members of a conserved protein family.
FIGURE 5:
Using NanoSPD to study TPRN binding to PP1 and CLIC proteins. (A) Molecules used to test PPIs between TPRN and catalytic subunits of protein phosphatase 1 (PPP1CA, PPP1CB, PPP1CC) by NanoSPD. Point mutations in TPRN (KISF → KASA) that disrupt the PP1 binding motif are indicated. (B) MYO10-TPRNBAIT traffics PPP1CAPREY to filopodial tips in Sf9 cells (right panel). MYO10NO BAIT is unable to traffic PPP1CAPREY (left panel), confirming the PPI. (C) ICA reveals selective binding of TPRN to PP1 catalytic subunits alpha (PPP1CAPREY) and gamma (PPP1CCPREY) in Sf9 cells expressing MYO10-TPRNBAIT vs. MYO10NO BAIT (PPP1CA, ANOVA; PPP1CB/PPP1CC, Mann-Whitney U-test). Mutating the residues (KISF → KASA) within TPRN significantly reduces the fluorescence intensity of PPP1CAPREY at filopodial tips. Data are mean ± SD, quantified from B and Supplemental Figure S2 (three independent determinations). (D) NanoSPD molecules to test the interaction of TPRN with CLIC family proteins. (E) CLIC4PREY is trafficked to filopodial tips (magnified inset) by MYO10-TPRNBAIT (right panel) in mammalian COS-7 cells but not by MYO10NO BAIT (left panel). (F) Quantification of the TPRN binding to CLIC4/5 by SCA. The interaction index for CLIC4 and CLIC5 was significantly (t test) increased when MYO10-TPRNBAIT was expressed compared with MYO10NO BAIT. Each data point is the average interaction index from one determination (n = 267–516 filopodia total, more than three independent determinations). Scale bars: 10 µm (B); 20 µm (E); 5 µm (E, inset). Data are mean ± SD. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
NanoSPD can study effects of point mutations in conserved binding motifs
TPRN contains a consensus KISF motif (residues 624–627) that binds to a hydrophobic patch within the catalytic domain of PP1 isozymes to inhibit phosphatase activity (Ferrar et al., 2012). Mutation of critical residues in this KISF motif to alanine (KISF to KASA) abolishes TPRN binding to PPP1CA when measured by conventional APs (Ferrar et al., 2012). We examined whether NanoSPD was sensitive enough to detect a change in binding caused by targeted point mutations. Intensity-based analysis in Sf9 cells showed a statistically significant 3.3-fold reduction in PPP1CAPREY accumulation at filopodial tips when coexpressed with MYO10-TPRN(KASA)BAIT versus wild-type MYO10-TPRNBAIT (Figure 5C and Supplemental Figure S2A). Although MYO10-TPRN(KASA)BAIT significantly reduced the accumulation of PPP1CAPREY, we still detected a small 1.6-fold increase in PPP1CAPREY accumulation in Sf9 cells expressing MYO10-TPRN(KASA)BAIT versus MYO10NO BAIT. These data suggest that the KASA mutation did not completely inhibit the interaction, and there was residual binding to TPRN. We conclude that NanoSPD can detect the functional effect of critical amino acid substitutions in a conserved binding motif.
Testing membrane-associated PPIs with NanoSPD
The Y2H screen identified chloride intracellular channels 1, 4, and 5 (CLIC1, CLIC4, CLIC5) as potential TPRN interacting proteins. The CLIC protein family members exist as both soluble and integral membrane proteins and may act as chloride channels (Berryman et al., 2004; Singh and Ashley, 2007), while their function as globular soluble proteins is not well understood (Littler et al., 2010). CLIC4 and CLIC5 are both highly expressed in inner ear hair cells and are detected in the stereocilia bundle proteome (Shen et al., 2015). Furthermore, CLIC5 protein is localized at the base of hair cell stereocilia, in a distribution similar to TPRN (Gagnon et al., 2006; Rehman et al., 2010). Consistent with our Y2H data, CLIC5 was reported to bind TPRN by conventional IP (Salles et al., 2014); however, complex formation was critically dependent on chemical cross-linking, suggesting that the interaction was low affinity and could dissociate from the affinity matrix.
We independently tested the binding of CLIC5 to TPRN using NanoSPD. COS-7 cells were transfected with CLIC5PREY along with either MYO10-TPRNBAIT or MYO10NO BAIT (Figure 5D and Supplemental Figure S3) and analyzed using the Pearson’s r-based correlation algorithm. In cells expressing MYO10-TPRNBAIT, 96.4 ± 2.0% of filopodial tips exhibited significant correlation with CLIC5PREY, but this was significantly reduced to 33.9 ± 13.1% when coexpressed with MYO10NO BAIT (Figure 5F). The interaction between CLIC4PREY and MYO10-TPRNBAIT was confirmed using a similar experimental setup (Figure 5, D–F). Our data demonstrate that NanoSPD can successfully test binding with membrane-associated proteins, in addition to potentially revealing low-affinity interactions in live cells that might otherwise escape detection in a conventional IP.
Using NanoSPD to study interactions with nuclear proteins
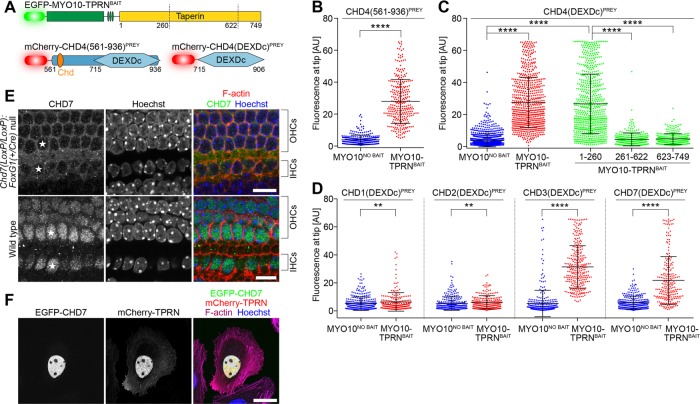
Chromodomain helicase DNA-binding protein 4 (CHD4) was the most abundant interaction with TPRN identified by Y2H. CHD4 is a catalytic subunit of the nucleosome-remodeling and histone deacetylase complex that regulates nuclear DNA damage responses (Larsen et al., 2010; Polo et al., 2010). CHD4 belongs to a superfamily of nine CHD enzymes that contain tandem N-terminal chromodomains (chromatin organization modifier) and a central SNF2-like ATPase domain encompassing a DEAD-box–like helicase superfamily (DEXDc) domain (Durr et al., 2006; Hall and Georgel, 2007). We used NanoSPD in Sf9 cells to test the putative interaction between CHD4 and TPRN (Figure 6A). A CHD4 fragment (aa 561–936) identified in our Y2H screen was used to eliminate all but one of the predicted nuclear localization signals (NLS) to prevent potential accumulation of CHD4 in the nucleus. The fluorescence intensity of CHD4(561-936)PREY at filopodial tips was sevenfold stronger in the presence of MYO10-TPRNBAIT versus MYO10NO BAIT, quantitatively confirming an interaction (Figure 6B). The CHD4(561-936)PREY fragment contains the second chromodomain and the DEXDc domain. We further truncated this CHD4 fragment and found the DEXDc domain alone, CHD4(DEXDc)PREY, was sufficient to bind MYO10-TPRNBAIT and be transported to filopodial tips (Figure 6C). A reciprocal experiment was performed to domain map the region within TPRN that binds the DEXDc domain of CHD4. NanoSPD using CHD4(DEXDc)PREY and a series of truncated MYO10-TPRNBAIT molecules encompassing residues 1–260, 261–622, or 623–749 showed that the CHD4 DEXDc domain specifically interacted with the N-terminal 260 residues of TPRN (Figure 6C).
FIGURE 6:
TPRN interacts with chromodomain (CHD) family proteins in mechanosensory hair cells. (A) NanoSPD molecules to test the interaction between TPRN and CHD4, containing a DEAD-like helicase domain (DEXDc) and chromodomain (Chd). Truncation points for domain-mapping experiments are shown. (B) ICA in Sf9 cells shows that CHD4(561-936)PREY interacts at filopodial tips when expressed with MYO10-TPRNBAIT (Mann-Whitney U-test). Data are mean ± SD. (C) Domain mapping of the CHD4-TPRN interaction by NanoSPD. ICA in Sf9 cells shows the truncated CHD4(DEXDc)PREY containing the DEXDc domain (aa 715–906) by itself bound TPRN. Reciprocal truncations demonstrate that the N-terminal 260 amino acids of TPRN were sufficient to bind and traffic CHD4(DEXDc)PREY. Other truncated regions of TPRN (aa 261–622 + aa 623–749) did not bind CHD4(DEXDc)PREY (ANOVA with Tukey’s post hoc test). Data are mean ± SD from three independent determinations. (D) ICA in Sf9 cells shows binding of TPRN to the DEXDc domains of CHD3 and CHD7 but not to CHD1 or CHD2 (Mann-Whitney U-test). Data are mean ± SD, from three independent determinations. (E) Antibody labeling of CHD7 in P1 cochleae from wild-type and conditionally null Chd7 mutant mice. CHD7 labeling was absent from hair cell nuclei of conditionally null mice (white stars). OHCs, outer hair cell; IHCs, inner hair cells. Scale bars: 10 µm. (F) mCherry-TPRN (red) and EGFP-CHD7 (green) colocalize to the nucleus when overexpressed in HeLa cells. Actin filaments are labeled with Alexa Fluor 633 phalloidin (magenta), and the nucleus is labeled with Hoechst (blue). Scale bar: 25 µm. **, p < 0.01; ****, p < 0.0001.
We investigated whether TPRN might be able to bind other CHD proteins, since the DEXDc domain is broadly conserved across superfamily members. Separate NanoSPD assays were performed in Sf9 cells using CHD1(DEXDc)PREY, CHD2(DEXDc)PREY, CHD3(DEXDc)PREY, or CHD7(DEXDc)PREY in combination with either MYO10-TPRNBAIT or MYO10NO BAIT as a control. Intensity correlation analyses at filopodial tips revealed that TPRN bound to the DEXDc domains of CHD3 and CHD7, but not CHD1 and CHD2 (Figure 6D). We conclude that TPRN can interact with multiple CHD proteins via the DEXDc domain but exhibits selectivity toward specific CHD submembers. More broadly, our results indicate that NanoSPD can be used to study nuclear proteins and to perform domain-mapping experiments to understand structure–function relationships.
CHD7 is expressed in neuroepithelial cells of the developing cochlea at embryonic day 10.5, before hair cells have differentiated (Hurd et al., 2007, 2010). We investigated the localization of CHD7 by immunofluorescence in postnatal day 1 (P1) mouse cochlea. CHD7 was prominently detected in the nucleus of mechanosensory hair cells at P1 (Figure 6E) but absent from nuclei of conditional Chd7-null cochleae (Figure 6E). These data confirm that CHD7 is present in hair cell nuclei and provide a potential new perspective into how mutations in CHD7 cause human CHARGE syndrome (OMIM: #214800, www.omim.org), a pleiotropic developmental disorder that includes deafness among multiple other pathological phenotypes (Martin, 2015). We hypothesized that TPRN and CHD7 may be components of a macromolecular complex critical for hearing. Consistent with this, mCherry-TPRN and EGFP-CHD7 colocalized in the nucleus of HeLa cells, indicating these proteins may act together to regulate gene transcription (Figure 6F). Given that CHD7 can bind to enhancer-like elements and acts as a transcriptional cofactor (Schnetz et al., 2009; Engelen et al., 2011; Van Nostrand et al., 2014), our data suggest that TPRN may similarly influence transcription in sensory hair cells.
Using the GFP-nanotrap to perform cloning-free NanoSPD 2.0 assays
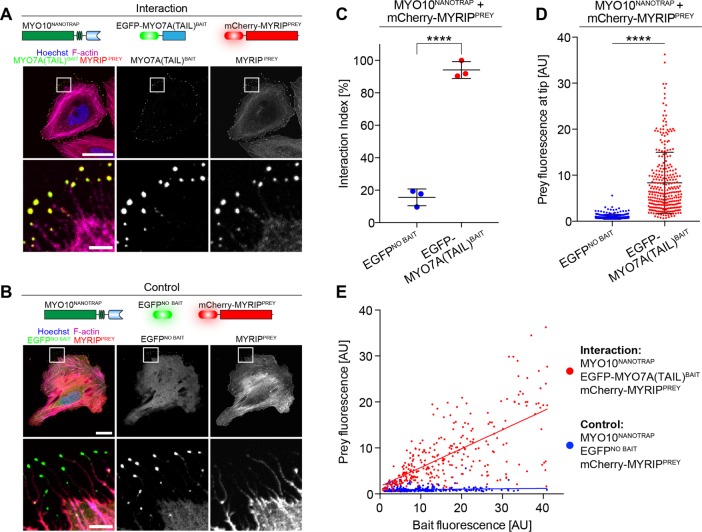
Common to all NanoSPD assays is the forced trafficking of molecular complexes along filopodia by the covalent fusion of MYO10HMM with a bait molecule. Having validated NanoSPD with a range of PPIs, we further streamlined the assay to eliminate initial cloning of the MYO10BAIT fusion construct. We hypothesized that GFP could be used as an affinity tag to noncovalently bind bait proteins to MYO10HMM via the GFP-nanotrap, a 13 kDa single-chain antibody fragment engineered to specifically bind EGFP, but not DsRed, mRFP (monomeric red fluorescent protein), or mCherry variants (Rothbauer et al., 2008). This concept was explored using the MYO7A/MYRIP PPI as a positive control. A fusion between MYO10HMM and the GFP-nanotrap (MYO10NANOTRAP; see Materials and Methods) was expressed in HeLa cells along with EGFP-MYO7A(TAIL)BAIT and mCherry-MYRIPPREY. Robust accumulation of both EGFP and mCherry fluorescence was observed at the filopodial tips of these cells, suggesting that MYO10NANOTRAP trafficked the complex by noncovalently binding to the EGFP affinity tag (Figure 7A). As a control, we repeated the identical experiment but substituted EGFP-MYO7A(TAIL)BAIT with EGFP lacking a bait fusion (EGFPNO BAIT). In these cells, EGFPNO BAIT was robustly trafficked to filopodial tips, while mCherry-MYRIPPREY remained in the cytoplasm (Figure 7B), demonstrating that the GFP-nanotrap does not bind to mCherry-MYRIPPREY directly and that trafficking of mCherry-MYRIPPREY required a specific interaction with EGFP-MYO7A(TAIL)BAIT. Rigorous quantification of these data by spatial correlation analysis (SCA; Figure 7C) and ICA (Figure 7, D and E) confirmed these qualitative observations. Our results demonstrate that NanoSPD 2.0 assays can be performed using the MYO10NANOTRAP coexpressed with candidate GFP-tagged and mCherry/RFP-tagged proteins. Given the widespread availability of fluorescently tagged expression constructs (Kamens, 2014), this strategy eliminates the necessity for cloning and accelerates assay iteration in NanoSPD 2.0.
FIGURE 7:
Simplified NanoSPD 2.0 assays using the GFP-nanotrap. NanoSPD 2.0 uses MYO10HMM fused to the GFP-binding nanotrap (MYO10NANOTRAP) to capture EGFP-tagged bait proteins and transport them along filopodia. (A) Image of fixed HeLa cells expressing nonfluorescent MYO10NANOTRAP, EGFP-MYO7A(TAIL)BAIT, and mCherry-MYRIPPREY. Both bait and prey robustly accumulate at filopodial tips (magnified panels shown below). (B) mCherry-MYRIPPREY does not accumulate at filopodial tips (magnified panels shown below) when coexpressed with mCherry-MYO10NANOTRAP and EGFP (EGFPNO BAIT). This confirms that the GFP-nanotrap does not bind mCherry-MYRIPPREY directly. Scale bars: 25 μm; 5 μm (magnified regions). (C) SCA of data from A and B shows a significantly higher interaction index (t test) in the presence of EGFP-MYO7A(TAIL)BAIT vs. EGFPNO BAIT. Data are mean ± SD with each data point representing the average interaction index from a single experiment. (D) ICA of data from A and B reveals the significant increase (Mann-Whitney U-test) of mCherry-MYRIPPREY fluorescence at filopodial tips in the presence of EGFP-MYO7A(TAIL)BAIT vs. EGFPNO BAIT. Data are mean ± SD with each data point representing a single filopodium. (E) Bait (x-axis) vs. prey (y-axis) scatter plot of fluorescence at filopodial tips from D. Each data point represents a single filopodium. ****, p < 0.0001; all data are from three independent experimental determinations (n = 285–295 filopodia total).
DISCUSSION
NanoSPD uses quantitative fluorescence microscopy to study PPIs within live cells, and complements conventional AP techniques with several distinct advantages. PPIs are assayed within the native cytoplasm, rather than in artificial buffers outside of the cell. NanoSPD does not require protein purification or specific antibodies to capture macromolecular complexes and can assay full-length proteins in addition to protein fragments. In contrast to conventional AP that favors the detection of stable PPIs, macromolecular complexes are rapidly trafficked to filopodial tips in NanoSPD, potentially capturing low-affinity, transient interactions. Finally, each cell projects multiple filopodia, making NanoSPD inherently parallelized for robust quantification and statistical determination of a PPI. Because filopodial production can be strongly stimulated by expressing wild-type MYO10 (Berg and Cheney, 2002), we expect that NanoSPD will be broadly applicable to a range of transfected primary and immortalized cell lines.
NanoSPD uses the continuous delivery of MYO10 to create a “traffic jam” of bait–prey complexes at filopodial tips, providing fluorescence amplification and boosting assay sensitivity. Active concentration of bait–prey complexes separates NanoSPD from other colocalization approaches in which freely diffusing prey are passively captured onto a subcellular bait scaffold (Bear et al., 2000; Zolghadr et al., 2008; Gallego et al., 2013; Herce et al., 2013). Microtubule-based dynein motors have been exploited to detect vesicle-specific PPIs by redirecting traffic of vesicles and associated proteins to the microtubule organizing center (MTOC) (Bentley and Banker, 2015; Bentley et al., 2015). Vesicles are concentrated into a highly variable, poorly defined volume around the MTOC, and a quantification method has not been proposed. In contrast, MYO10 drives accumulation into diffraction-limited volumes that are reproducibly well defined beyond the cell periphery, circumventing fluorescence background from the bulk cytoplasm.
Our analysis pipeline uses these unique features to objectively quantify NanoSPD data sets. Interactions are inferred from bait and prey fluorescence correlation either 1) along the filopodial shaft (SCA), or 2) at filopodial tips (ICA). In both analysis strategies, interactions are inferred by comparing the query data set (using MYO10BAIT) with a matched control (using MYO10NO BAIT). The two analysis techniques have distinct advantages. SCA uses Pearson’s r coefficient to detect correlated changes in bait and prey fluorescence along the filopodial shaft that exceed a bootstrap-derived threshold. An advantage of this approach is that microscope imaging parameters can be freely optimized between experiments, since Pearson’s r measures only relative fluorescence changes from the mean. The disadvantage is that SCA does not quantify the amount of prey transported to filopodial tips. This can result in weak prey accumulations, either from cytoplasmic volume filling or endogenous localization to the filopodial tip, being correlated and detected as false positives. Though uncommon, we observed this phenomenon in the bulbous filopodial tips of Sf9 insect cells and also when assaying CLIC4/5 in COS-7 cells (Figure 5E). As an alternative approach, ICA measures the absolute fluorescence intensity at the filopodial tips and requires that all data be captured with identical imaging parameters. ICA does not use a threshold to detect an interaction, and thus small amounts of prey accumulating in control assays do not cause false positives. A specific advantage of ICA is that it provides a continuous measure of pulldown efficiency, that is, the ratio of bait and prey delivered to the filopodial tips. Whether this ratio might be sensitive to the equilibrium dissociation constant (KD) of a PPI remains to be determined. Irrespective of the algorithm used, both analyses initially require manual tracing of filopodia in ImageJ. Automatic software-based segmentation and skeletonizing of filopodia will significantly streamline this process (Nilufar et al., 2013; Tsygankov et al., 2014).
The sensitivity of NanoSPD to detect low-affinity interactions was not explored in this study. Live-cell TIRF experiments showed that bait–prey complexes were transported along filopodia in ∼30 s (Figure 2), consistent with previous reports of MYO10 intrafilopodial motility (Kerber et al., 2009). For PPIs with slow off rates (koff), this argues that a prey molecule will remain associated with a single MYO10BAIT for the duration of transport. Conversely, a prey with a fast koff rate will likely disassociate during transport. In this latter case, we hypothesize that the prey molecule will still be transported by binding to successive MYO10BAIT molecules as they move anterogradely toward the filopodial tip. For this reason, we suspect that NanoSPD will be able to detect low-affinity interactions, although this has yet to be experimentally tested. Future studies will use NanoSPD to examine a series of PPIs with known affinities. These experiments will also help define the relationship (if any) between ICA pulldown efficiency and KD.
We used NanoSPD to validate a Y2H screen of TPRN-binding proteins to investigate molecular mechanisms of human hearing loss caused by mutations in TPRN. NanoSPD detected PPIs involving molecular motors (Figures 3 and 4), phosphatases (Figure 5), and membrane-associated (Figure 5) and nuclear (Figure 6) proteins, and was used in combination with domain mapping and mutagenesis. We have yet to encounter a PPI incompatible with NanoSPD, although endogenous localization of a prey protein to either the filopodial tip complex or nucleus can initially impede the assay. Truncating or mutating predicted NLS was effective for studying the CHD protein superfamily and allowed these preys to be transported along filopodia (Figure 6). Another strategy for endogenously localized nuclear and filopodial proteins is to present them as bait. The study of proteins that inhibit MYO10-based motility or filopodial production are likely not compatible with NanoSPD. These examples are expected to be rare.
Performing NanoSPD assays within the cytosol has distinct advantages; however, it is not a replacement for conventional AP. The intentional variation of buffer conditions and protein concentrations is a powerful approach to estimating equilibrium constants and binding characteristics by AP (Pollard, 2010; Hakhverdyan et al., 2015). Furthermore, like all intracellular-based assays, NanoSPD cannot infer direct binding between bait and prey, since native proteins can be recruited from the cytosol. In its current form, NanoSPD cannot easily identify these intermediate adaptors. If filopodia could be selectively isolated and enriched, NanoSPD might be a viable platform to couple with mass spectroscopy. Another approach could be to fuse MYO10BAIT with a promiscuous biotin ligase (Roux et al., 2012) and use a brief pulse of biotin before affinity capture of complexes that were tagged at the filopodial tips.
The NanoSPD platform is broadly extensible. We demonstrate that binding hierarchies can be interrogated within a tripartite complex (Figure 4) and speculate that this analysis could be extended further by multiplexing additional fluorescent proteins. Unlabeled proteins can also be introduced to examine their effect upon complex formation (Figure 4). As NanoSPD is based on quantitative fluorescence, it can potentially be used with any fluorescently labeled molecule or in combination with immunocytochemistry to detect endogenously recruited molecules. An exciting possibility is studying the formation of protein–RNA hybrids using fluorescently labeled nucleic acids (Bertrand et al., 1998; Paige et al., 2011; Nelles et al., 2016). Finally, NanoSPD is potentially scalable to perform intracellular pulldowns on a massively parallel scale. An immediate application we envisage is high-throughput screening, wherein cells expressing a validated NanoSPD assay are incubated with a small-molecule library to detect compounds that modulate the interaction. Microscopy-based high-throughput NanoSPD assays are an important area of future development and could help accelerate the discovery of small molecules that bind to clinically important drug targets.
MATERIALS AND METHODS
Expression constructs
PCR amplification and cloning were performed using standard methods. All expression constructs were verified by Sanger sequencing. Site-directed mutagenesis was performed using the QuikChange II SDM Kit (Agilent). Plasmid DNA for transfection was prepared using endotoxin-free purification kits (NucleoBond EF; Clontech). The MYO10 heavy meromyosin (HMM) construct generated for NanoSPD encompasses the ATPase motor, light-chain binding sites, and a coiled-coil region (residues 1–941; NP_062345) and is not a forced dimer (i.e., MYO10HMM was not fused to a leucine zipper or a similar dimerization motif). All expressed molecules are tagged with spectrally orthogonal fluorescent proteins and are not explicitly stated in the main text (see figure captions and Supplemental Table S1). EGFP/mCherry-tagged MYO10HMM and MYO10NANOBODY cloning vectors have been deposited at www.addgene.org.
Animals
All experimental animal procedures were performed in accordance with National Institutes of Health (NIH) guidelines and approved by the respective Institutional Animal Care and Use Committees at the National Institute on Deafness and Other Communication Disorders (NIDCD #1263) and the University of Michigan (PRO00006244). C57BL6/J mice were obtained from the Jackson Laboratories (Bar Harbor, ME). Conditional null Chd7 mice were generated by intercrossing mice carrying the conditional floxed Chd7LoxP allele (Hurd et al., 2010), a gene-trapped null Chd7GT allele (Hurd et al., 2007) and the FoxG1Cre allele (Hebert and McConnell, 2000) to yield Chd7GT/LoxP:FoxG1+/Cre pups. The ROSA26LoxP-STOP-LoxP-ZsGreen1A allele was also included on this mutant background (Madisen et al., 2010).
Y2H screening
Screening of mouse inner ear and kidney prey libraries (Boeda et al., 2002) was performed by Hybrigenics Services SAS (Paris, France) using a N-terminal LexA fusion with full-length mouse TPRN (aa 1–749; NP_780495) as bait.
Cell culture and transfection
COS-7 (American Type Culture Collection [ATCC] CRL-1651) and HeLa (ATCC CCL-2) cells were cultured in DMEM supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS; Atlanta Biologicals) and GlutaMAX (Thermo Fisher Scientific) and incubated at 37°C, 10% CO2. HeLa cell transfections were performed with Lipofectamine 3000 (Thermo Fisher Scientific), and COS-7 transfections were performed using the Neon transfection system (Thermo Fisher Scientific) following the manufacturer’s protocol. Transfected cells were incubated for 12–24 h before being seeded on fibronectin-coated (10 μg/ml; Sigma) glass-bottom culture dishes (#1.5, MatTek). Cells were incubated for another 6–24 h before fixation with 4% paraformaldehyde (PFA) for 15 min. For live-cell imaging, transfected cells were incubated for 1–3 h after seeding and then exchanged into FluoroBrite DMEM (Thermo Fisher Scientific) supplemented with GlutaMAX.
Sf9 (Spodoptera frugiperda) cells (Thermo Fisher Scientific) were cultured in HyClone SFX media (GE Healthcare) supplemented with GlutaMAX and heat-inactivated 2.5% (vol/vol) FBS (Atlanta Biologicals) and maintained at 27°C in a shaking incubator. For transfection, DNA was diluted into phosphate-buffered saline (PBS) at a 1:12 ratio (wt/wt) with polyethylenimine (1 mg/ml; PEI Max, #24765; Polysciences) and incubated for 15 min at room temperature before the complexed DNA was added to cells in suspension culture. At 48 h posttransfection, cells were seeded onto glass-bottom dishes (#1.5; MatTek Corporation) for 1–3 h at 27°C before fixation with 4% PFA for 15 min. Mammalian and insect cells were colabeled with either ATTO 390–conjugated phalloidin (Sigma) or Alexa Fluor–conjugated phalloidin to visualize the filamentous actin cytoskeleton and 4’,6-diamidino-2-phenylindole/Hoechst 33342 (Thermo Fisher Scientific) to stain the nucleus.
Cochlear immunocytochemistry
Mice were killed and temporal bones fixed for 2 h in 4% PFA. Microdissected samples were permeabilized for 30 min in 0.5% Triton X-100 and blocked for 1 h with 2% bovine serum albumin (Roche) and 5% normal goat serum (Sigma) diluted in PBS. Samples were incubated overnight in blocking solution plus a primary antibody recognizing CHD7 (#6505; Cell Signaling Technology). After washing in PBS, primary antibodies were detected using Alexa Fluor–conjugated secondary antibodies (Thermo Fisher Scientific). Samples were colabeled with Alexa Fluor 633 to visualize the actin cytoskeleton, in addition to the nuclear stain Hoechst 33342, before being mounted in Prolong Gold Antifade Reagent (Thermo Fisher Scientific).
Fluorescence microscopy
For live-cell imaging, an inverted microscope (Axiovert 200M; Zeiss) was used to perform objective-based TIRFM with a laser-based illuminator (TIRF3; Zeiss) and a 100×, 1.46 N.A. oil objective (alpha Plan-Apochromat; Zeiss). EGFP and mCherry fluorescence was sequentially excited through a laser clean-up filter (ZET488/561x; Chroma) and a 2 mm dichroic mirror (ZT488/561rpc; Chroma) with 488- and 561-nm laser lines, respectively. Epifluorescence was collected through a laser-blocking filter (ZET488/561m-TRF; Chroma) and a dual-camera adaptor (Zeiss). Split channels were emission filtered for EGFP (ET525/50m; Chroma) and mCherry (ET610/75m), respectively, and captured on individual EM-CCD cameras (Evolve 512; Photometrics) controlled by Zen software (Zeiss). Dual-camera EGFP and mCherry data were reregistered with MATLAB (MathWorks) using fluorescent beads as fiducial markers (Tetraspeck; Thermo Fisher Scientific). Cells were maintained at 37°C in a humidified 10% CO2 environment during all live-cell image acquisitions. Images were captured at 1–2 frames per second.
Fixed cells were imaged using a 63×, 1.4 N.A. oil objective (Plan-Apochromat; Zeiss) and an Axiovert 200M inverted microscope with a confocal scan head (LSM780, Zeiss) controlled by Zen software (Zeiss). Bait and prey signals were captured sequentially to eliminate spectral bleed-through. The LSM780 GaAsP spectral detector was set to capture 500–530 nm for EGFP and 570–620 nm for mCherry/tdTomato.
Analysis of fluorescence colocalization in filopodia
Image analysis was performed on confocal images of fixed cells to quantify bait and prey fluorescence at filopodial tips. Line scans along individual filopodia were prepared in ImageJ (http://rsbweb.nih.gov/ij/) using the bait and/or phalloidin (F-actin) fluorescence signal as a guide. In principle, other markers of filopodia can be used in place of phalloidin, including membrane dyes and phase-contrast techniques. Prey fluorescence was kept hidden during line tracing to avoid selection bias. Filopodia suitable for analysis projected > 2 µm away from the cell body to allow for comparisons between the filopodial tip and shaft. A maximum of 10 filopodia were sampled per cell to avoid any single cell from dominating the overall data set. For each interaction, we typically examined >150 filopodia pooled from three independent experiments. Bait and prey fluorescence line scan values were exported from ImageJ into a tab-delimited text file, and all further data analyses were performed in a custom MATLAB tool with graphical user interface. Compiled binary distributions, source code, and a detailed guide to using this software tool is provided in the Supplemental Material and are available at https://github.com/NanoSPD/NanoSPD.
Within the MATLAB tool, the global maximum of the MYO10BAIT signal was used to determine the position of the filopodial tip. This approximation was valid given the robust accumulation of MYO10 at filopodial tips, even when fused to different bait domains. Line scan data were then screened for basic quality criteria. Filopodia were excluded if the peak bait fluorescence at the tip did not exceed the average bait fluorescence along the shaft by more than 3 SDs. Filopodia were also excluded if the full-width half-maximum of the tip bait fluorescence was too broad, as this indicated the bait was not sufficiently concentrated at the tip. Filopodia line scans passing these quality criteria were analyzed further by one of two algorithms.
Spatial correlation analysis
In the presence of an interaction, we assume the distribution of prey within filopodia is dependent on the MYO10BAIT molecule. Pearson’s coefficient (r) was used to quantify the correlation between bait and prey fluorescence from line scans along individual filopodia, with the expectation these values are correlated in the presence of an interaction (i.e., r = +1), and uncorrelated otherwise (i.e., r = 0). In intermediate cases, where 0 > r < +1, we avoided using an arbitrary threshold to assign significance, since partial correlations may still indicate a significant biological interaction. Instead, we estimated the likelihood that each observed correlation could occur by random chance. This was performed for each filopodium using bootstrapping to randomize the prey signal and calculate the exact distribution of r values for all possible permutations (Lifshitz, 1998; Costes et al., 2004). This approach tested the null hypothesis that a given bait–prey correlation in an individual filopodium occurred randomly. A refinement to this methodology is to perform the randomization in blocks that approximate the point-spread function of the microscope. This preserves inherent correlation that is present in adjacent pixels (Costes et al., 2004). This exact probability distribution was then used to calculate the likelihood (p value) that any observed correlation occurred by chance. A filopodium was scored as a positive bait–prey interaction if 1) the calculated p value < 0.01, and 2) the prey intensity at the filopodial tip exceeded the filopodial shaft signal by 3 SDs. For each experimental determination, an interaction index is calculated as the ratio of positively interacting filopodia to the total number of filopodia assayed. SCA uses relative fluorescence changes so that microscope imaging parameters can vary between independent determinations.
Intensity correlation analysis
In this alternate algorithm, which is strongly recommended for Sf9 cells, we test for the presence of a global correlation between bait and prey fluorescence intensities at filopodial tips. These two quantities are expected to be positively correlated in the presence of an interaction, with the prey fluorescence being dependent on the bait fluorescence. In this approach, statistical significance is determined globally and not on a per filopodium basis (see Spatial correlation analysis). Line scan data from ImageJ were imported into MATLAB and filtered as described earlier. The MATLAB tool exports the maximum bait and prey fluorescence measured at each filopodial tip. Paired bait and prey fluorescence values were plotted on an x–y scatter plot, with each data point representing one filopodium. Because this algorithm depends on absolute fluorescence values, it is critical that imaging conditions (objective, gains/exposures, illumination intensity) are kept constant between independent determinations.
Statistical analyses
All statistical testing was performed in Prism (version 7.0, GraphPad). Data sets were analyzed for normality using the Shapiro-Wilk test. Comparisons between two treatment groups were made using Student’s t test (assuming unequal SD) for normally distributed data and by nonparametric Mann-Whitney U-test otherwise. For multiple comparisons, one-way analysis of variance (ANOVA) was used with Tukey’s post hoc test. All data are mean ± SD and from three independent experimental determinations, unless otherwise stated. Statistical significance is indicated uniformly throughout the article as *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Supplementary Material
Acknowledgments
We thank Giovanni Mann and Yasuharu Takagi for insightful discussions; Dennis Drayna, James Sellers, and Matthias Machner for critical reading; and Joseph Duda for technical assistance. We are grateful to John Hammer, III (National Heart, Lung, and Blood Institute), for the gift of Myo5a and Mlph cDNA and to the animal care staff at both the NIDCD and the University of Michigan. This work is supported (in part) by the Intramural Research Program of the NIH, NIDCD DC000039 (to T.B.F.), NIDCD DC000080 (to T.F.), and extramural funds NIDCD R01 DC009410 (to D.M.M.).
Abbreviations used:
- ANOVA
analysis of variance
- AP
affinity pulldown
- co-IP
coimmunoprecipitation
- DEXDc
DEAD-like helicase domain
- EGFP
enhanced green fluorescent protein
- FBS
fetal bovine serum
- HMM
heavy meromyosin
- ICA
intensity correlation analysis
- IP
immunoprecipitation
- MLPH
melanophilin
- mRFP
monomeric red fluorescent protein
- MTOC
microtubule organizing center
- NanoSPD
nanoscale pulldown
- NIDCD
National Institute on Deafness and Other Communication Disorders
- NIH
National Institutes of Health
- NLS
nuclear localization signals
- PBS
phosphate-buffered saline
- PFA
paraformaldehyde
- PP1
protein phosphatase 1
- PPI
protein–protein interaction
- SCA
spatial correlation analysis
- TIRFM
total internal reflection fluorescence microscopy
- TPRN
taperin
- Y2H
yeast two-hybrid.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-08-0583) on December 8, 2016.
REFERENCES
- Barr-Gillespie PG. Assembly of hair bundles, an amazing problem for cell biology. Mol Biol Cell. 2015;26:2727–2732. doi: 10.1091/mbc.E14-04-0940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Loureiro JJ, Libova I, Fassler R, Wehland J, Gertler FB. Negative regulation of fibroblast motility by Ena/VASP proteins. Cell. 2000;101:717–728. doi: 10.1016/s0092-8674(00)80884-3. [DOI] [PubMed] [Google Scholar]
- Bentley M, Banker G. A novel assay to identify the trafficking proteins that bind to specific vesicle populations. Curr Protoc Cell Biol. 2015;69:13 8 1–12. doi: 10.1002/0471143030.cb1308s69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley M, Decker H, Luisi J, Banker G. A novel assay reveals preferential binding between Rabs, kinesins, and specific endosomal subpopulations. J Cell Biol. 2015;208:273–281. doi: 10.1083/jcb.201408056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JS, Cheney RE. Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat Cell Biol. 2002;4:246–250. doi: 10.1038/ncb762. [DOI] [PubMed] [Google Scholar]
- Berg JS, Derfler BH, Pennisi CM, Corey DP, Cheney RE. Myosin-X, a novel myosin with pleckstrin homology domains, associates with regions of dynamic actin. J Cell Sci. 2000;113:3439–3451. doi: 10.1242/jcs.113.19.3439. [DOI] [PubMed] [Google Scholar]
- Berryman M, Bruno J, Price J, Edwards JC. CLIC-5A functions as a chloride channel in vitro and associates with the cortical actin cytoskeleton in vitro and in vivo. J Biol Chem. 2004;279:34794–34801. doi: 10.1074/jbc.M402835200. [DOI] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- Boeda B, El-Amraoui A, Bahloul A, Goodyear R, Daviet L, Blanchard S, Perfettini I, Fath KR, Shorte S, Reiners J, et al. Myosin VIIa, harmonin and cadherin 23, three Usher I gene products that cooperate to shape the sensory hair cell bundle. EMBO J. 2002;21:6689–6699. doi: 10.1093/emboj/cdf689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohil AB, Robertson BW, Cheney RE. Myosin-X is a molecular motor that functions in filopodia formation. Proc Natl Acad Sci USA. 2006;103:12411–12416. doi: 10.1073/pnas.0602443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun P, Tasan M, Dreze M, Barrios-Rodiles M, Lemmens I, Yu H, Sahalie JM, Murray RR, Roncari L, de Smet AS, et al. An experimentally derived confidence score for binary protein-protein interactions. Nat Methods. 2009;6:91–97. doi: 10.1038/nmeth.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Rajagopala SV, Stellberger T, Uetz P. Exhaustive benchmarking of the yeast two-hybrid system. Nat Methods. 2010;7:667–668. doi: 10.1038/nmeth0910-667. author reply, 668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau JW, Costantino S, Wiseman PW. A guide to accurate fluorescence microscopy colocalization measurements. Biophys J. 2006;91:4611–4622. doi: 10.1529/biophysj.106.089441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn KW, Kamocka MM, McDonald JH. A practical guide to evaluating colocalization in biological microscopy. Am J Physiol Cell Physiol. 2011;300:C723–C742. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durr H, Flaus A, Owen-Hughes T, Hopfner KP. Snf2 family ATPases and DExx box helicases: differences and unifying concepts from high-resolution crystal structures. Nucleic Acids Res. 2006;34:4160–4167. doi: 10.1093/nar/gkl540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Amraoui A, Schonn JS, Kussel-Andermann P, Blanchard S, Desnos C, Henry JP, Wolfrum U, Darchen F, Petit C. MyRIP, a novel Rab effector, enables myosin VIIa recruitment to retinal melanosomes. EMBO Rep. 2002;3:463–470. doi: 10.1093/embo-reports/kvf090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelen E, Akinci U, Bryne JC, Hou J, Gontan C, Moen M, Szumska D, Kockx C, van Ijcken W, Dekkers DH, et al. Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat Genet. 2011;43:607–611. doi: 10.1038/ng.825. [DOI] [PubMed] [Google Scholar]
- Ferrar T, Chamousset D, De Wever V, Nimick M, Andersen J, Trinkle-Mulcahy L, Moorhead GB. Taperin (c9orf75), a mutated gene in nonsyndromic deafness, encodes a vertebrate specific, nuclear localized protein phosphatase one alpha (PP1alpha) docking protein. Biol Open. 2012;1:128–139. doi: 10.1242/bio.2011049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Kuroda TS. Slac2-c (synaptotagmin-like protein homologue lacking C2 domains-c), a novel linker protein that interacts with Rab27, myosin Va/VIIa, and actin. J Biol Chem. 2002;277:43096–43103. doi: 10.1074/jbc.M203862200. [DOI] [PubMed] [Google Scholar]
- Gagnon LH, Longo-Guess CM, Berryman M, Shin JB, Saylor KW, Yu H, Gillespie PG, Johnson KR. The chloride intracellular channel protein CLIC5 is expressed at high levels in hair cell stereocilia and is essential for normal inner ear function. J Neurosci. 2006;26:10188–10198. doi: 10.1523/JNEUROSCI.2166-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego O, Specht T, Brach T, Kumar A, Gavin AC, Kaksonen M. Detection and characterization of protein interactions in vivo by a simple live-cell imaging method. PLoS One. 2013;8:e62195. doi: 10.1371/journal.pone.0062195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Gstaiger M, Raught B, Aebersold R. Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol. 2007;8:645–654. doi: 10.1038/nrm2208. [DOI] [PubMed] [Google Scholar]
- Hakhverdyan Z, Domanski M, Hough LE, Oroskar AA, Oroskar AR, Keegan S, Dilworth DJ, Molloy KR, Sherman V, Aitchison JD, et al. Rapid, optimized interactomic screening. Nat Methods. 2015;12:553–560. doi: 10.1038/nmeth.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JA, Georgel PT. CHD proteins: a diverse family with strong ties. Biochem Cell Biol. 2007;85:463–476. doi: 10.1139/O07-063. [DOI] [PubMed] [Google Scholar]
- Hebert JM, McConnell SK. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev Biol. 2000;222:296–306. doi: 10.1006/dbio.2000.9732. [DOI] [PubMed] [Google Scholar]
- Herce HD, Deng W, Helma J, Leonhardt H, Cardoso MC. Visualization and targeted disruption of protein interactions in living cells. Nat Commun. 2013;4:2660. doi: 10.1038/ncomms3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurd EA, Capers PL, Blauwkamp MN, Adams ME, Raphael Y, Poucher HK, Martin DM. Loss of Chd7 function in gene-trapped reporter mice is embryonic lethal and associated with severe defects in multiple developing tissues. Mamm Genome. 2007;18:94–104. doi: 10.1007/s00335-006-0107-6. [DOI] [PubMed] [Google Scholar]
- Hurd EA, Poucher HK, Cheng K, Raphael Y, Martin DM. The ATP-dependent chromatin remodeling enzyme CHD7 regulates pro-neural gene expression and neurogenesis in the inner ear. Development. 2010;137:3139–3150. doi: 10.1242/dev.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain A, Liu R, Ramani B, Arauz E, Ishitsuka Y, Ragunathan K, Park J, Chen J, Xiang YK, Ha T. Probing cellular protein complexes using single-molecule pull-down. Nature. 2011;473:484–488. doi: 10.1038/nature10016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins B, Decker H, Bentley M, Luisi J, Banker G. A novel split kinesin assay identifies motor proteins that interact with distinct vesicle populations. J Cell Biol. 2012;198:749–761. doi: 10.1083/jcb.201205070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamens J. Addgene: making materials sharing “science as usual.”. PLoS Biol. 2014;12:e1001991. doi: 10.1371/journal.pbio.1001991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerber ML, Cheney RE. Myosin-X: a MyTH-FERM myosin at the tips of filopodia. J Cell Sci. 2011;124:3733–3741. doi: 10.1242/jcs.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerber ML, Jacobs DT, Campagnola L, Dunn BD, Yin T, Sousa AD, Quintero OA, Cheney RE. A novel form of motility in filopodia revealed by imaging myosin-X at the single-molecule level. Curr Biol. 2009;19:967–973. doi: 10.1016/j.cub.2009.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola TK. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat Protoc. 2006;1:1278–1286. doi: 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, Rendtlew Danielsen JM, Menard P, Sand JC, et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol. 2010;190:731–40. doi: 10.1083/jcb.200912135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmens I, Lievens S, Tavernier J. MAPPIT, a mammalian two-hybrid method for in-cell detection of protein-protein interactions. Methods Mol Biol. 2015;1278:447–455. doi: 10.1007/978-1-4939-2425-7_29. [DOI] [PubMed] [Google Scholar]
- Li Y, Pohl E, Boulouiz R, Schraders M, Nurnberg G, Charif M, Admiraal RJ, von Ameln S, Baessmann I, Kandil M, et al. Mutations in TPRN cause a progressive form of autosomal-recessive nonsyndromic hearing loss. Am J Hum Genet. 2010;86:479–484. doi: 10.1016/j.ajhg.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lifshitz LM. Determining data independence on a digitized membrane in three dimensions. IEEE Trans Med Imaging. 1998;17:299–303. doi: 10.1109/42.700742. [DOI] [PubMed] [Google Scholar]
- Littler DR, Harrop SJ, Goodchild SC, Phang JM, Mynott AV, Jiang L, Valenzuela SM, Mazzanti M, Brown LJ, Breit SN, Curmi PM. The enigma of the CLIC proteins: ion channels, redox proteins, enzymes, scaffolding proteins? FEBS Lett. 2010;584:2093–2101. doi: 10.1016/j.febslet.2010.01.027. [DOI] [PubMed] [Google Scholar]
- Luo Y, Batalao A, Zhou H, Zhu L. Mammalian two-hybrid system: a complementary approach to the yeast two-hybrid system. Biotechniques. 1997;22:350–352. doi: 10.2144/97222pf02. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manders EM, Stap J, Brakenhoff GJ, van Driel R, Aten JA. Dynamics of three-dimensional replication patterns during the S-phase, analysed by double labelling of DNA and confocal microscopy. J Cell Sci. 1992;103:857–862. doi: 10.1242/jcs.103.3.857. [DOI] [PubMed] [Google Scholar]
- Martin DM. Epigenetic developmental disorders: CHARGE syndrome, a case study. Curr Genet Med Rep. 2015;3:1–7. doi: 10.1007/s40142-014-0059-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–54. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- Nelles DA, Fang MY, O’Connell MR, Xu JL, Markmiller SJ, Doudna JA, Yeo GW. Programmable RNA tracking in live cells with CRISPR/Cas9. Cell. 2016;165:488–496. doi: 10.1016/j.cell.2016.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilufar S, Morrow AA, Lee JM, Perkins TJ. FiloDetect: automatic detection of filopodia from fluorescence microscopy images. BMC Syst Biol. 2013;7:66. doi: 10.1186/1752-0509-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. A guide to simple and informative binding assays. Mol Biol Cell. 2010;21:4061–4067. doi: 10.1091/mbc.E10-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J. 2010;29:3130–3139. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebelo S, Santos M, Martin F, da Cruz e Silva EF, da Cruz e Silva OA. Protein phosphatase 1 is a key player in nuclear events. Cell Signal. 2015;27:2589–2598. doi: 10.1016/j.cellsig.2015.08.007. [DOI] [PubMed] [Google Scholar]
- Rehman AU, Morell RJ, Belyantseva IA, Khan SY, Boger ET, Shahzad M, Ahmed ZM, Riazuddin S, Khan SN, Riazuddin S, Friedman TB. Targeted capture and next-generation sequencing identifies C9orf75, encoding taperin, as the mutated gene in nonsyndromic deafness DFNB79. Am J Hum Genet. 2010;86:378–388. doi: 10.1016/j.ajhg.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson GP, de Monvel JB, Petit C. How the genetics of deafness illuminates auditory physiology. Annu Rev Physiol. 2011;73:311–334. doi: 10.1146/annurev-physiol-012110-142228. [DOI] [PubMed] [Google Scholar]
- Ries J, Schwille P. Fluorescence correlation spectroscopy. Bioessays. 2012;34:361–368. doi: 10.1002/bies.201100111. [DOI] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salles FT, Andrade LR, Tanda S, Grati M, Plona KL, Gagnon LH, Johnson KR, Kachar B, Berryman MA. CLIC5 stabilizes membrane-actin filament linkages at the base of hair cell stereocilia in a molecular complex with radixin, taperin, and myosin VI. Cytoskeleton (Hoboken) 2014;71:61–78. doi: 10.1002/cm.21159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetz MP, Bartels CF, Shastri K, Balasubramanian D, Zentner GE, Balaji R, Zhang X, Song L, Wang Z, Laframboise T, et al. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009;19:590–601. doi: 10.1101/gr.086983.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwander M, Kachar B, Muller U. Review series: the cell biology of hearing. J Cell Biol. 2010;190:9–20. doi: 10.1083/jcb.201001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers JR. Myosins: a diverse superfamily. Biochim Biophys Acta. 2000;1496:3–22. doi: 10.1016/s0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- Shen J, Scheffer DI, Kwan KY, Corey DP. SHIELD: an integrative gene expression database for inner ear research. Database (Oxford) 2015;2015:bav071. doi: 10.1093/database/bav071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioda T, Andriole S, Yahata T, Isselbacher KJ. A green fluorescent protein-reporter mammalian two-hybrid system with extrachromosomal maintenance of a prey expression plasmid: application to interaction screening. Proc Natl Acad Sci USA. 2000;97:5220–5224. doi: 10.1073/pnas.97.10.5220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H, Ashley RH. CLIC4 (p64H1) and its putative transmembrane domain form poorly selective, redox-regulated ion channels. Mol Membr Biol. 2007;24:41–52. doi: 10.1080/09687860600927907. [DOI] [PubMed] [Google Scholar]
- Sweeney HL, Houdusse A. Structural and functional insights into the myosin motor mechanism. Annu Rev Biophys. 2010;39:539–557. doi: 10.1146/annurev.biophys.050708.133751. [DOI] [PubMed] [Google Scholar]
- Tsygankov D, Bilancia CG, Vitriol EA, Hahn KM, Peifer M, Elston TC. CellGeo: a computational platform for the analysis of shape changes in cells with complex geometries. J Cell Biol. 2014;204:443–460. doi: 10.1083/jcb.201306067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand JL, Brady CA, Jung H, Fuentes DR, Kozak MM, Johnson TM, Lin CY, Lin CJ, Swiderski DL, Vogel H, et al. Inappropriate p53 activation during development induces features of CHARGE syndrome. Nature. 2014;514:228–232. doi: 10.1038/nature13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrabe H, Periasamy A. Imaging protein molecules using FRET and FLIM microscopy. Curr Opin Biotechnol. 2005;16:19–27. doi: 10.1016/j.copbio.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Wu XS, Rao K, Zhang H, Wang F, Sellers JR, Matesic LE, Copeland NG, Jenkins NA, Hammer JA 3rd. Identification of an organelle receptor for myosin-Va. Nat Cell Biol. 2002;4:271–278. doi: 10.1038/ncb760. [DOI] [PubMed] [Google Scholar]
- Zolghadr K, Mortusewicz O, Rothbauer U, Kleinhans R, Goehler H, Wanker EE, Cardoso MC, Leonhardt H. A fluorescent two-hybrid assay for direct visualization of protein interactions in living cells. Mol Cell Proteomics. 2008;7:2279–2287. doi: 10.1074/mcp.M700548-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.