Meiotic recombination hotspots activated by PRDM9 are associated with the chromosomal axis and synaptonemal complex via their interaction with other proteins, including CDYL, EHMT2, EWSR1, and CXXC1.
Abstract
In mammals, meiotic recombination occurs at 1- to 2-kb genomic regions termed hotspots, whose positions and activities are determined by PRDM9, a DNA-binding histone methyltransferase. We show that the KRAB domain of PRDM9 forms complexes with additional proteins to allow hotspots to proceed into the next phase of recombination. By a combination of yeast-two hybrid assay, in vitro binding, and coimmunoprecipitation from mouse spermatocytes, we identified four proteins that directly interact with PRDM9’s KRAB domain, namely CXXC1, EWSR1, EHMT2, and CDYL. These proteins are coexpressed in spermatocytes at the early stages of meiotic prophase I, the limited period when PRDM9 is expressed. We also detected association of PRDM9-bound complexes with the meiotic cohesin REC8 and the synaptonemal complex proteins SYCP3 and SYCP1. Our results suggest a model in which PRDM9-bound hotspot DNA is brought to the chromosomal axis by the action of these proteins, ensuring the proper chromatin and spatial environment for subsequent recombination events.
INTRODUCTION
Genetic recombination assures the proper segregation of homologous chromosomes at the first meiotic division, preventing aneuploidy. It also plays an important evolutionary role by facilitating the creation of new, favorable combinations of alleles and the removal of deleterious mutations by unlinking them from surrounding sequences. In mammals, as in yeast, higher plants, and birds, recombination occurs at specialized sites along chromosomes known as hot spots (Paigen and Petkov, 2010; Baudat et al., 2013), typically 1 kb or so in length, separated by tens to hundreds of kilobases that lack recombination.
In mammals, a meiosis-specific protein, PR/SET domain–containing 9 (PRDM9), first identified by our group and others (Baudat et al., 2010; Myers et al., 2010; Parvanov et al., 2010), is the primary determinant of recombination hotspot locations (Baudat et al., 2010; Myers et al., 2010; Parvanov et al., 2010; Hinch et al., 2011; Smagulova et al., 2011). PRDM9 combines domains from two large families of proteins—KRAB zinc finger (ZnF; Lupo et al., 2013) and PR-domain proteins (Fumasoni et al., 2007)—and is the only protein known to contain all three characteristic domains of these families—a KRAB domain implicated in protein–protein interactions, a PR/SET domain with a histone methytransferase activity, and a ZnF domain for DNA recognition and binding. Recombination begins when the C-terminal ZnF domain of PRDM9 recognizes and binds to hotspot-specific DNA sequences (Baudat et al., 2010; Myers et al., 2010; Parvanov et al., 2010; Billings et al., 2013). The PR/SET domain then locally trimethylates histone H3 on lysine 4 (H3K4me3); this results in rearrangement of the local nucleosome pattern, creating a central nucleosome-depleted region (Baker et al., 2014) where the double-strand breaks (DSBs) required for the exchange of DNA sequences between homologous chromatids occur (Brick et al., 2012). The extent of trimethylation of local nucleosomes delimits the span over which the final genetic crossovers can take place (Baker et al., 2014). In the absence of PRDM9, DSBs are formed at other available H3K4me3 sites—mainly promoters—but they cannot be repaired properly, and germ cells undergo apoptosis (Smagulova et al., 2011).
Her, we show that the PRDM9 KRAB domain plays a crucial role in binding and recruiting additional proteins into multiprotein complexes that bring hotspots into the next phase of recombination; these proteins include CXXC domain–containing 1 (CXXC1), a DNA-binding protein with a CXXC domain found in CpG-binding proteins (Illingworth et al., 2010); Ewing sarcoma 1 (EWSR1), which binds single-stranded RNA and DNA (Li et al., 2007; Oakland et al., 2013; Fisher, 2014); euchromatic histone methyltransferase 2 (EHMT2), a histone methyltransferase catalyzing formation of H3K9me1/2 and H3K56me1 (Tachibana et al., 2005; Tachibana et al., 2007); chromodomain-containing Y chromosome–like (CDYL), a methyl reader of H3K9me2/3 (Escamilla-Del-Arenal et al., 2013) and H3K27me3 (Zhang et al., 2011); and a putative histone acetylase (Lahn et al., 2002).
A combination of yeast two-hybrid screens and in vitro pull-downs of purified proteins showed that the four proteins directly bind to the N-terminal region of PRDM9 where the KRAB domain is located. We further confirmed the in vivo interaction between PRDM9 and EWSR1, EHMT2, and CDYL in early prophase spermatocytes by coimmunoprecipitation (coIP), and cytological colocalization. In spermatocytes, we also detected association of PRDM9-bound complexes with the meiotic cohesin REC8 and the chromosomal axis/synaptonemal complex proteins SYCP3 and SYCP1.
These results suggest that PRDM9-bound hotspot DNA is brought to the chromosomal axis by interaction with other proteins serving as a link between PRDM9 and cohesins/SC proteins, thereby assuring a proper spatial environment for DSB initiation and repair.
RESULTS
Yeast two-hybrid assay identifies direct PRDM9 interactors
To search for proteins directly interacting with PRDM9, we performed a yeast two-hybrid (Y2H) screen using cloned full-length Prdm9 as bait and a 6-mo-old mouse testis cDNA library as prey. Screening ∼6 × 106 colonies, we isolated 329 positive clones, which, after sequencing, coalesced to 118 individual open reading frames. Four clones, representing C-terminal portions of Ehmt2 (amino acids 376–1263), Cxxc1 (217-660), Ewsr1 (479–655), and Cdyl (74–593) genes, were confirmed as interacting strongly with full-length PRDM9 by pairwise Y2H under the most discriminating conditions (Figure 1). To identify the positions of their binding sites on the PRDM9 molecule, shorter PRDM9 fragments were cloned as bait constructs and tested by pairwise Y2H with each interacting clone (Figure 1, A and B). All four clones were found to interact with PRDM9 fragments representing the isolated KRAB domain, and the Ehmt2 and Ewsr1 clones interacted with the intervening region between the KRAB and PR/SET domains as well (Figure 1B).
FIGURE 1:
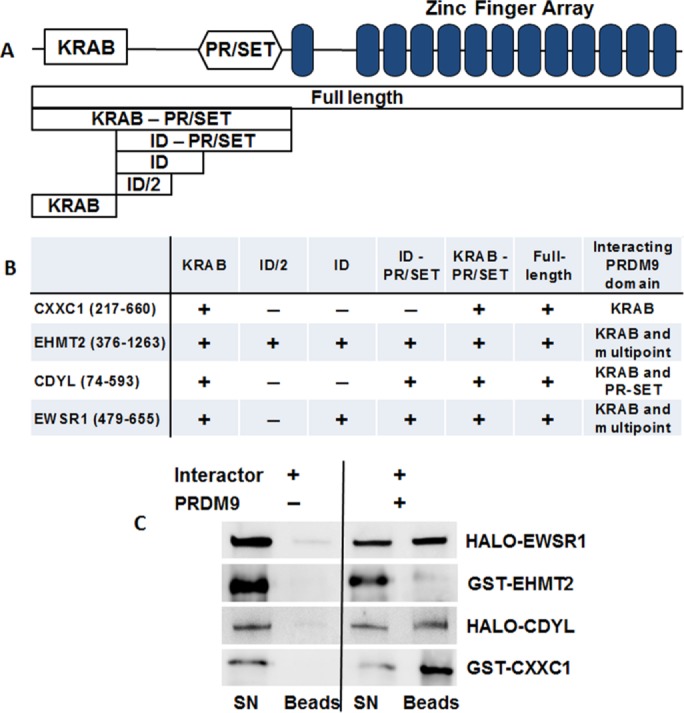
CXXC1, EWSR1, EHMT2, and CDYL directly interact with PRDM9. (A) Scheme of the domain structure of PRDM9 (top), showing the positions of the KRAB, PR/SET, and the zinc finger domains. Below are schematic representations of the full-length and deletion constructs used in the Y2H assay to map the contact points of PRDM9 interacting with the other proteins. (B) Results of the Y2H assay showing that KRAB is the major protein contact domain of PRDM9. The extents of the cloned and expressed fragments of the interactor proteins are in brackets. (C) Purified PRDM9 and its interactors bind to each other in vitro. Purified HaloTag EWSR1 and CDYL or GST-tagged EHMT2 and CXXC1 were immobilized on amylose beads alone (left) or mixed with purified full-length MBP-tagged PRDM9 (right). Specific interactions are detected by the immobilization of interactor protein on amylose beads only in the presence of MBP-tagged PRDM9. All four proteins specifically bind to PRDM9. SN, supernatant; B, beads.
The amino acid sequences of the clones showing positive interactions with PRDM9 provide clues regarding the binding sites of these proteins. The only known functional domain in the cloned fragment of EWSR1 is a C-terminal RanBP2-type zinc finger, which has been implicated in protein binding (Steggerda and Paschal, 2002). The cloned portion of CXXC1 includes acidic, basic, coiled-coil domains, a Set1-interacting domain, and a PHD2 domain (Tate et al., 2009). The cloned portion of EHMT2 contains an NRSF-binding, cysteine-rich domain, an ankyrin domain, and a SET domain (Dillon et al., 2005). The CDYL clone lacks 19 amino acids of its N-terminal chromodomain but contains the remaining part and the C-terminal ClP domain (Wu et al., 2009).
EWSR1, EHMT2, CDYL, and CXXC1 bind PRDM9 in vitro
Each of the interacting proteins detected by Y2H also bound to PRDM9 in vitro. For these tests, we cloned, expressed, and purified N-terminally tagged versions of these proteins from Escherichia coli. Full-length PRDM9 was cloned as a fused construct with an N-terminal maltose-binding protein (MBP) tag. The other four were cloned separately as fused constructs with HaloTag or glutathione S-transferase tag. The tagged proteins were expressed in E. coli and purified. Individual proteins were mixed with PRDM9 and corresponding complexes bound to amylose beads. All four proteins bound PRDM9 in vitro (Figure 1C). Of interest, one of the two histone modifiers, EHMT2, showed relatively weaker binding to PRDM9 (Figure 1C, second row) compared with EWSR1, CDYL, and CXXC1.
Binding of PRDM9 and its interacting proteins in spermatocytes
We next tested whether these proteins also interact with PRDM9 and with each other in mouse spermatocyte lysates by coIP in germ cells of 14-day postpartum (dpp) juvenile mice, which are enriched for the leptotene through early pachytene stages of meiotic prophase I (Figure 2A, left). The coIP with antibodies against PRDM9 also contained EWSR1, EHMT2, and CDYL as interactors, confirming the Y2H data. In vivo binding of PRDM9 to EWSR1 was strong; however, its strength diminished after DNase treatment, suggesting possible bridging of the interaction via DNA. In contrast, PRDM9 interactions with EHMT2 and CDYL were only slightly affected by DNase treatment (Figure 2A, left). We could not test CXXC1 for the lack of specific antibodies. We also carried out reverse coIP experiments in which antibodies against EWSR1 pulled down PRDM9 but not EHMT2, and CDYL (Figure 2A, middle). Antibodies against CDYL showed coIP with PRDM9 and EHMT2, which was retained after DNase treatment but failed to show any signal with EWSR1 (Figure 2A, right). The amount of coimmunoprecipitated PRDM9 actually increased after DNase treatment, probably due to better solubility of the CDYL-PRDM9 complex. The available antibodies against EHMT2 were not suitable for coIP.
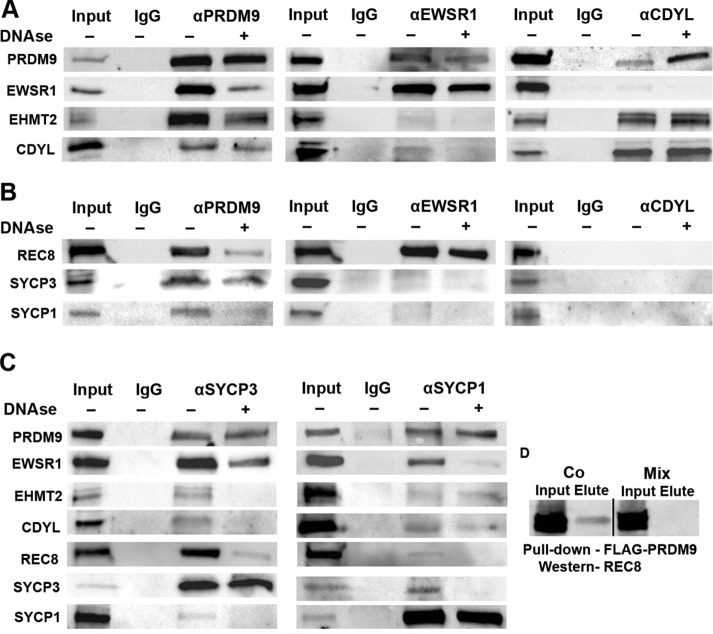
FIGURE 2:
PRDM9, EWSR1, and CDYL coimmunoprecipitate each other and chromosomal axis/synaptonemal complex proteins from wild-type 14-dpp spermatocytes. (A) PRDM9 and its interactors coimmunoprecipitate each other. Left, coIP with anti-PRDM9; middle, coIP with anti-EWSR1; right, coIP with CDYL. Lane 1, input, 10 µg (2.5 µg in the CDYL and EHMT2 blots); lane 2, IgG, 10 µg, coIP with nonimmune IgG; lanes 2 and 3, αPRDM9, αEWSR1, and αCDYL, 10 µg, coIP with the respective antibody, either nontreated (lane 3) or DNase treated (lane 4). The antibodies used to detect specific proteins on Western blots are shown on the left. (B) PRDM9 and its interactors also interact with chromosomal axis/SC proteins. coIP with the same antibodies used in A but probed with antibodies against chromosome axis/SC proteins as marked on the left. (C) Reciprocal coIP shows that SC proteins coimmunoprecipitate PRDM9 and its interactors. coIP with SYCP3 (left) or SYCP1 (right). The antibodies used for detection on Western blots are on the left. (D) Interaction between REC8 and PRDM9 in cultured cells. REC8 was cloned under a V5 tag, and PRDM9 was cloned under a FLAG tag. The two proteins were either coexpressed or separately expressed in HEK 293 cells. Specific interaction between the two proteins was detected in extracts of cells in which the two proteins were coexpressed (Co, left) but not in mixed extracts of cells in which the two proteins were expressed separately (Mix, right). Note that the interacting band is the middle of three bands, indicating that it is a phosphorylated form.
We conclude that PRDM9 forms a separate complex with EWSR1 in vivo, independent of its binding with EHMT2 and CDYL. We also found evidence of strong interaction between EHMT2 and CDYL in vivo. Taken together, these data suggest the possible existence of both a PRDM9-EWSR1 complex and a separate PRDM9-EHMT2-CDYL complex.
Interactions with the synaptonemal complex
Because PRDM9-bound hotspots are translocated to the chromosomal axis where DSBs are subsequently formed and repaired, we asked whether PRDM9 and its interacting proteins bind components of the chromosomal axis and found this to be the case. Specifically, we tested for coIP with the meiotic-specific cohesin REC8, chromosomal axis/SC lateral element protein SYCP3, and the SC central element protein SYCP1. CoIP with anti-PRDM9 pulled down REC8, SYCP3, and SYCP1, and these interactions were partially retained after DNase treatment (Figure 2B, left).
Antibodies against EWSR1 showed a strong coIP signal with REC8, which was retained after DNase treatment, but only a faint signal with SYCP3 and SYCP1 (Figure 2B, middle). The interaction with REC8 appears to be with the phosphorylated form of the protein, as only antibodies able to detect the phosphorylated form of REC8 with apparent electrophoretic mobility corresponding to ∼85 kDa (Fukuda et al., 2012) showed positive signal with PRDM9 and EWSR1. CoIP with antibodies against CDYL did not detect interaction with any of these proteins (Figure 2B, right).
In reverse, antibodies against the lateral element protein SYCP3 strongly pulled down PRDM9 and EWSR1 but showed weak signal with EHMT2 and CDYL, which was not retained after DNase treatment (Figure 2C, left). They also pulled down REC8 and SYCP1, but in a DNase-sensitive manner, confirming that DNA is indeed essential for the integrity of the chromosomal axis. Antibodies against the central element protein SYCP1 pulled down PRDM9, an interaction retained after DNase treatment. They also strongly pulled down EWSR1, but the signal was sensitive to DNase treatment (Figure 2C, right). Weak signals were also detected with EHMT2 and CDYL. Unfortunately, available antibodies against REC8 were not suitable for coIP.
Although these data provide strong evidence that PRDM9 and EWSR1 associate with the meiotic-specific cohesins in vivo, we were unable to detect direct interaction between PRDM9 and REC8 by Y2H or in vitro after expression of the proteins in E. coli (unpublished data ). Consequently, to provide additional confirmation of interactions with the chromosomal axis/SC in vivo, we used a HEK293 mammalian cell expression system and showed coIP of the phosphorylated form of REC8 with PRDM9 when the two were coexpressed in the same cells (the middle band in Figure 2D, second lane) but not when they were expressed separately and the cells mixed before extraction. The lack of interaction between the purified proteins in vitro could therefore be explained by the lack of REC8 phosphorylation when expressed in E. coli. In addition, this interaction may require another protein intermediate, such as EWSR1, which is naturally present in HEK293 cells.
PRDM9 binding to other proteins is dependent on its own binding to DNA
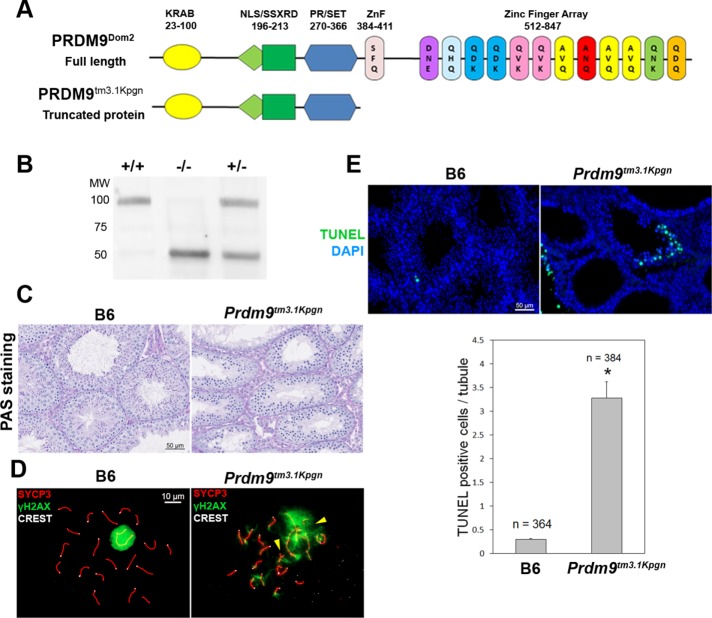
To investigate whether PRDM9 binding to other proteins is dependent on its zinc finger domain and the ability to bind to DNA, we created a new PRDM9 functional knockout (KO) mouse model, B6(Cg)-Prdm9tm3.1Kpgn/Kpgn (designated here as Prdm9tm3.1Kpgn). In this mouse, we placed a mutation creating an alternative splice acceptor site 44 base pairs inside the 5′-end of exon 12 that codes for the entire zinc finger domain, causing a frameshift with a stop codon. As a result, the mutant protein contains amino acids 1–381 of PRDM9 but lacks all of its zinc fingers (Figure 3A) and the ability to bind DNA at specific sequences (Walker et al., 2015). It is stable and appears as a 48-kDa band on Western blots in testes from both heterozygous and homozygous mutant animals (Figure 3B). Both male and female mice homozygous for this mutation are sterile. Spermatogonia and spermatocytes were found in mutant testes, but no postmeiotic spermatids were observed (Figure 3C). Cytological staining of spreads of mutant spermatocytes shows cells arrested at an aberrant pachytene-like stage, with inappropriate γH2AX staining on autosomes, and asynapsis of homologous chromosomes (Figure 3D). Lack of the PRDM9 zinc finger domain also led to increased apoptosis during meiosis compared with wild type (Figure 3E, p < 0.05). This phenotype is very similar to the one described in Prdm9TM1Ymat KO mice, which have no PRDM9 protein (Hayashi et al., 2005; Sun et al., 2015).
FIGURE 3:
Characterization of the Prdm9tm3.1Kpgn mutant. (A) Schematic representation of wild-type PRDM9 and mutant PRDM9Tm3.1Kpgn protein. (B) Western blotting showing that the truncated protein is expressed in both homozygous and heterozygous mice. The bands correspond to the predicted molecular mass of 98 kDa for the wild type and 48 kDa for the truncated protein. The amount of the two forms in the heterozygous testes is approximately equal. Testis extract used: +/+, wild type B6; –/–, homozygous mutant; +/–, heterozygous. (C) PAS staining of testis tubule sections from wild type (left) and mutant mice (right). Note the lack of postmeiotic cells in the mutant, indicating that the germ cells undergo meiotic arrest. (D) SYCP3/γH2AX/CREST staining on germ cell spreads, showing a pachynema cell in the wild type (left), with a γH2AX signal (green) restricted only to the sex body, and the latest stage found in the mutant, representing late zygonema–pachynema transition. Arrowheads, asynapsed regions. (E) Increased levels of apoptosis in the mutant compared with the wild type. Top, TUNEL staining in wild-type (left) and mutant testis (right). Bottom, quantitation of the apoptotic cells in wild-type and mutant testis. *p < 0.05.
The truncated 48-kDa version of PRDM9 was immunoprecipitated from testis lysates using anti-PRDM9 antibodies directed against the remaining portion of the molecule (Figure 4A, left). These coIP experiments showed weak and inconsistent signals with most of the proteins that associate with full-length PRDM9 in three independent experiments. Consistent weak interactions were detected only with EWSR1 and SYCP3 (Figure 4A, left). These results suggest that, although EWSR1, CDYL, and EHMT2 can directly interact with the isolated N-terminal region as well as the full-length protein in vitro, their in vivo interactions require PRDM9 binding to DNA.
FIGURE 4:
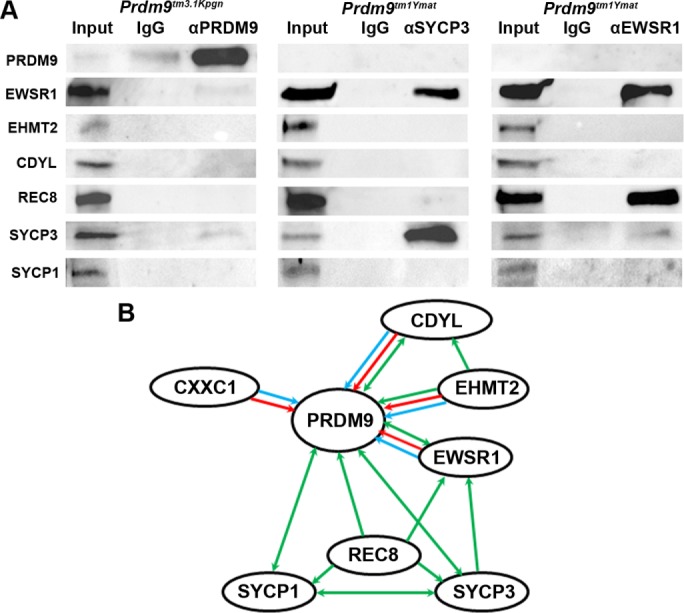
Protein–protein interactions in 14-dpp spermatocytes. (A) Interactions in spermatocytes of PRDM9 mutant mice. Left, coIP with anti-PRDM9 in testes of Prdm9tm3.1Kpgn mice lacking its DNA-binding ZnF domain. The asterisk indicates overlapping signal from the heavy chain of IgG. All protein amounts are as in Figure 2. Middle, coIP with anti-SYCP3 in testes of Prdm9tm1Ymat mice lacking PRDM9 protein. Right, coIP with anti-EWSR1 in testes of Prdm9tm1Ymat mice. Lane 1, input; lane 2, IgG; lane 3, coIP. (B) Summary of protein–protein interactions detected by all methods. Blue line, direct interactions found by Y2H assay; red line, direct interactions detected by mixing purified proteins and isolating their complexes in vitro; green line, interactions detected by coIP. The arrows show the direction of interactions detected. Double-headed arrows show interactions confirmed by reciprocal coIP.
EWSR1 binds to the chromosomal axis in the absence of PRDM9
To determine whether binding of EWSR1, EHMT2, and CDYL to the chromosomal axis depends on their association with PRDM9 or whether these proteins can bind independently, we performed coIP with anti-SYCP3 antibodies in testes of Prdm9tm1Ymat KO mice (Hayashi et al., 2005), which do not express any PRDM9 protein (Sun et al., 2015). EWSR1 showed a strong positive signal (Figure 4A, middle), indicating that it can bind to the synaptonemal complex independently of PRDM9. In contrast, CDYL and EHMT2 were not pulled down by SYCP3 in the absence of PRDM9. In addition, the lack of REC8 and SYCP1 signal indicates chromosomal axis impairment. Together with the coIP data from wild-type testis, this indicates that the CDYL-EHMT2 complex associates with the chromosomal axis only in the presence of PRDM9 bound to hotspot DNA.
CoIP with EWSR1 in Prdm9tm1Ymat KO mice showed a strong signal with REC8 and a weak signal with SYCP3. EHMT2 and CDYL were not detected as EWSR1 interactors (Figure 4A, right). These results suggest that EWSR1 can provide a link between PRDM9 and the chromosomal axis through the meiotic-specific cohesin complexes containing REC8.
A summary of all protein–protein interactions detected is presented in Figure 4B.
PRDM9 is coexpressed with its interactors in mid to late zygonema nuclei
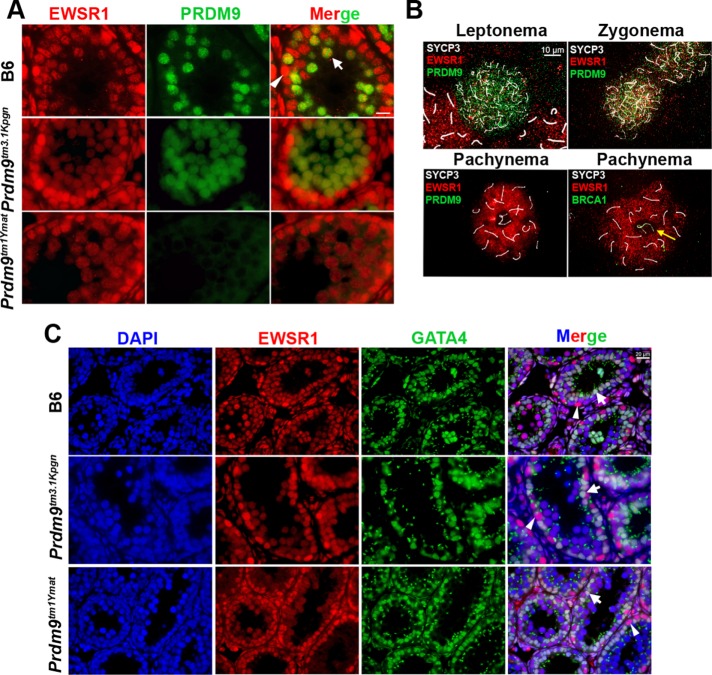
Our previous work established that PRDM9 is present only during meiosis in preleptonema, leptonema, and zygonema, a period of ∼48 h (Sun et al., 2015). To confirm that PRDM9 is coexpressed in the same cells and colocalizes within nuclei with its interactors, we performed double staining with combinations of antibodies against PRDM9 and its interactors in seminiferous tubules and spermatocyte spreads. We first tested for spatial and temporal colocalization of PRDM9 and EWSR1 in seminiferous tubules of 14-dpp juvenile mice (Figure 5A, top). EWSR1 showed high expression in spermatogonia (Figure 5, A and C, top, arrowhead) and Sertoli cells (marked by GATA4; Figure 5C, arrow) located at the base of seminiferous tubule and remained present in the nuclei of spermatocytes located in the tubule lumen (Figure 5A, top, arrow). At 14-dpp, when most spermatocytes are in leptonema and zygonema (Sun et al., 2015), PRDM9 and EWSR1 are clearly coexpressed in those cells (Figure 5A, top, arrow). Because the EWSR1 signal is weaker in PRDM9-positive cells, we sought to confirm that these proteins colocalize in spermatocyte spreads of 14-dpp mice by performing triple staining with EWSR1, PRDM9, and SYCP3, using the chromosomal axis protein SYCP3 as a marker of meiotic progression (Figure 5B). PRDM9 and EWSR1 were clearly coexpressed in preleptonema-to-midzygonema nuclei. The EWSR1 signal increased dramatically in pachynema, a time when PRDM9 has disappeared from the germ cell nuclei. Of interest, the EWSR1 signal was excluded from the sex body in pachynema, as demonstrated by its lack of colocalization with BRCA1 (Figure 5B, yellow arrow).
FIGURE 5:
EWSR1 is coexpressed with PRDM9 in early meiotic prophase. (A) Immunofluorescence analysis of EWSR1 (red)–PRDM9 (green) coexpression in tissue sections from testis tubules of 14-dpp mice. Top, wild-type B6 mice; middle, Prdm9tm3.1Kpgn. Arrowhead, spermatogonia with high EWSR1 expression. Arrow, spermatocytes with EWSR1 and PRDM9 positive signals. Bottom, Prdm9tm1Ymat. (B) Colocalization analysis of EWSR1 and PRDM9 in spermatocyte spreads. Triple staining for EWSR1 (red), SYCP3 (white), and PRDM9 (green) in leptonema, zygonema (top), and pachynema (bottom left) or EWSR1 (red), SYCP3 (white), and BRCA1 (green) in pachynema (bottom right) of B6 mice. EWSR1 is excluded from the sex body, marked by BRCA1 in pachynema (yellow arrow). (C) Double staining with EWSR1 and the Sertoli cell marker GATA4. EWSR1 expression is stronger in spermatogonia (arrowhead) than in Sertoli cells (arrow). Top, wild-type B6 mice; middle, Prdm9tm3.1Kpgn; bottom, Prdm9tm1Ymat. EWSR1 shows a similar expression pattern in mutants and wild type.
In Prdm9tm3.1Kpgn mice lacking the PRDM9 zinc finger domain, only a diffuse EWSR1-PRDM9 colocalization pattern is seen in seminiferous tubules (Figure 5A, second row). This mutant undergoes meiotic arrest around the zygonema-pachynema transition, and the seminiferous tubule lumens contains very few of the EWSR1-positive, PRDM9-negative pachynema germ cells. Very similar EWSR1 staining is found in Prdm9tm1Ymat KO mice, which do not express any PRDM9 protein (Figure 5A, third row).
We conclude that EWSR1 and PRDM9 are coexpressed in early meiotic prophase for the entire time of PRDM9 expression.
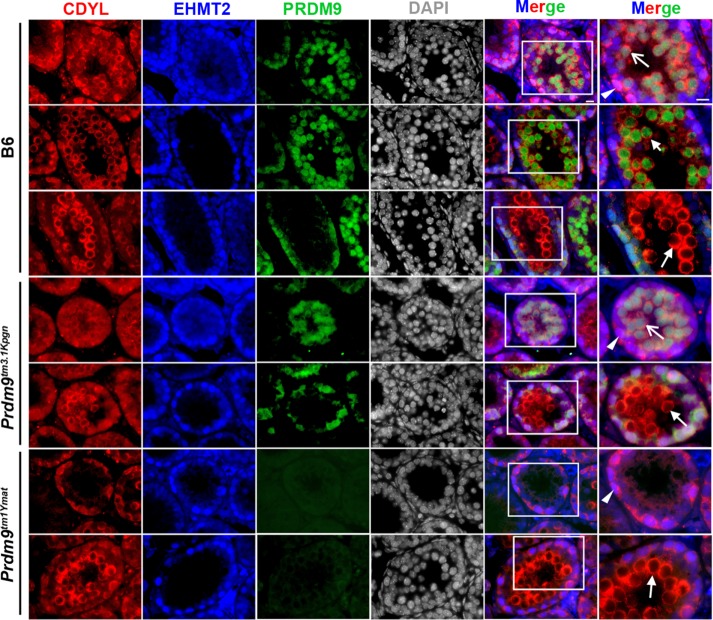
EHMT2 and CDYL signals were not detectable on spermatocyte spreads. For this reason, we tested for coexpression of PRDM9, CDYL, and EHMT2 in seminiferous tubules of 14-dpp mice.
In spermatocytes, CDYL staining in the nucleus persists only until leptonema, after which, it is translocated to the cytoplasm. Strong nuclear and cytoplasmic CDYL signal was detected in some cells close to the basal membrane that were PRDM9 negative (Figure 6, top, arrowhead). Weak nuclear CDYL signals were detected in germ cells that also showed weak PRDM9 signal (Figure 6, top, open arrow). We consider these cells to be spermatocytes in preleptonema or leptonema based on their tubule staging and PRDM9 positivity (Sun et al., 2015). The increase of PRDM9 signal was accompanied by translocation of the CDYL signal to the cytoplasm (Figure 6, top, and short arrows in second row). These cells are most probably in leptonema to zygonema. Pachynema cells abundant in the lumen showed strong cytoplasmic CDYL but not PRDM9 signals (Figure 6, third row, long arrow).
FIGURE 6:
CDYL and EHMT2 are coexpressed with PRDM9 in early meiotic prophase. Coexpression of CDYL (red), EHMT2 (blue), and PRDM9 (green) in tissue sections from testis tubules of 14-dpp mice. Top three rows, coexpression analysis in B6 mice. Fourth and fifth rows, expression in Prdm9tm3.1Kpgn mutant. Sixth and seventh rows, expression in Prdm9tm1Ymat mutant. Arrowhead, Sertoli cells showing strong nuclear and cytoplasmic CDYL and strong EHMT2 signal, PRDM9 negative. Open arrow, preleptonema-to-leptonema cells with weak nuclear CDYL, EHMT2, and PRDM9 signals. Short arrow, late leptonema–to–early zygonema cells with strong PRDM9, cytoplasmic CDYL, and lack of EHMT2 signals. Long arrow, pachynema cells with strong cytoplasmic CDYL but lack of EHMT2 and PRDM9 signals.
Neither the presence of truncated PRDM9 nor the complete loss of PRDM9 affected the localization patterns of CDYL from preleptonema through zygonema/early pachynema–like stages (Figure 6). Triple staining in testes of Prdm9tm3.1Kpgn and Prdm9tm1Ymat mutant mice confirmed the meiotic arrest phenotype found by EWSR1-PRDM9 staining. Prdm9tm3.1Kpgn testes showed CDYL-PRDM9–positive cells extending to the lumen and appearance of a few CDYL cytoplasmic–positive, PRDM9-EHMT2–negative cells (Figure 6, fourth and fifth rows). Similarly, Prdm9tm1Ymat testes, which lack PRDM9 protein, contained some CDYL cytoplasmic-positive cells (Figure 6, sixth and seventh rows).
EHMT2 had a nuclear pattern of expression very similar to CDYL but, unlike CDYL, which was translocated to the cytoplasm, disappeared entirely from the cells in late leptonema– early zygonema. Strong EHMT2-positive but PRDM9- and GATA4-negative spermatogonia (Figures 6, top, arrowhead, and 7, arrowhead) were found close to the basal membrane. Sertoli cells, marked by the presence of GATA4, showed weaker EHMT2 signal (Figure 7, arrow). Weak EHMT2 and weak PRDM9 signals were detected in germ cells attached to the basal membrane (Figure 6, top, open arrow), representing spermatocytes in preleptonema or leptonema. Cells in zygonema were characterized by an increase of the PRDM9 signal and disappearance of the EHMT2 signal (Figure 6, top, and short arrows in second row). Pachynema cells abundant in the lumen had neither EHMT2 nor PRDM9 signals (Figure 6, third row, long arrow). The localization of EHMT2 in Prdm9tm3.1Kpgn and Prdm9tm1Ymat mutant testes followed the same pattern (Figure 6, fourth to seventh rows).
FIGURE 7:
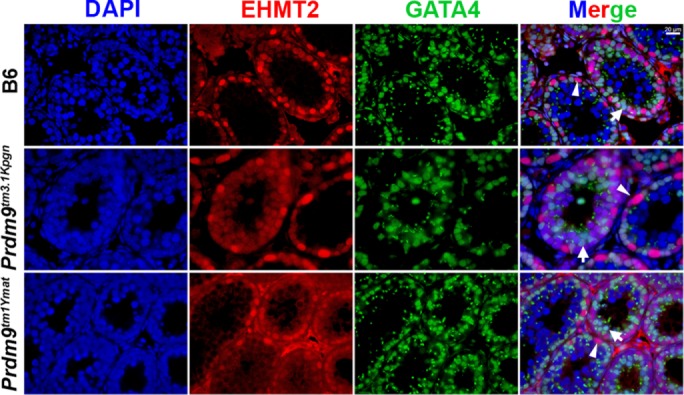
EHMT2 and GATA4 are coexpressed in Sertoli cells. EHMT2 expression is stronger in spermatogonia (arrowhead) compared with Sertoli cells (arrow). Top, wild-type B6 mice; middle, Prdm9tm3.1Kpgn; bottom, Prdm9tm1Ymat. EHMT2 shows similar expression pattern in mutants and wild type.
It appears that EWSR1, CDYL, and EHMT2 coincide with each other and with PRDM9 only in preleptonema to early zygonema, when all four of them are present at low concentrations. After that, CDYL is translocated to the cytoplasm, EHMT2 disappears altogether, and both EWSR1 and PRDM9 increase in expression until the zygonema–pachynema transition. At pachynema, PRDM9 disappears, whereas EWSR1 expression dramatically increases. These findings suggest a temporal order in the formation of the PRDM9-bound complexes. The PRDM9-CDYL-EHMT2 complex is restricted to preleptonema and leptonema, after which it is dissolved, with the disappearance of CDYL and EHMT2 from the nucleus. The PRDM9-EWSR1 complex is probably formed later in leptonema and persists until late zygonema until PRDM9 disappears from the nucleus.
Meiotic progression into zygonema requires PRDM9-bound hotspot translocation to the chromosomal axis
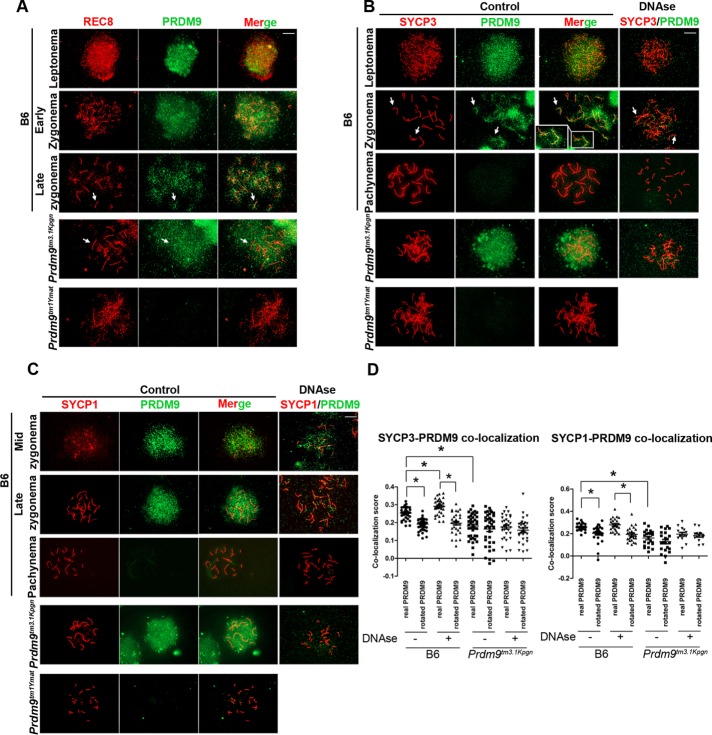
Because the coIP evidence suggested that PRDM9 binds to meiosis-specific cohesins such as REC8, we sought to determine whether colocalization between and PRDM9 and REC8 could be detected in early meiotic prophase on spermatocyte spreads, which can be staged by the appearance of the REC8 signal (Ishiguro et al., 2014). In preleptonema, REC8 staining shows both diffuse and punctate patterns. This is similar to the pattern of PRDM9 at the same stage. In leptonema, REC8 starts forming rod-like structures (Figure 8A, top), which become predominant in zygonema as elements of the chromosomal axes (Figure 8A, second row). The colocalization of these axis structures with some of the PRDM9 foci at late zygonema was apparent (Figure 8A, third row, arrows). Prdm9tm3.1Kpgn testes had a REC8-PRDM9 pattern similar to wild-type zygonema but showed less evidence for colocalization (Figure 8A, fourth row, arrows). The PRDM9 signal in this mutant has a more diffuse and less punctate pattern than in wild-type spermatocytes (Figure 8A, fourth row; see also Figure 8, B and C, fourth rows) suggesting that the punctate pattern reflects PRDM9 binding to hotspot DNA. Prdm9tm1Ymat testes lacking PRDM9 contained mostly cells with REC8 patterns reminiscent of preleptonema to early zygonema (Figure 8A, fifth row), indicating that progression of spermatocytes into zygonema requires an intact PRDM9-bound hotspot translocation to the chromosomal axis.
FIGURE 8:
REC8, SYCP3, and SYCP1 colocalize with PRDM9 in early meiotic prophase. (A) Colocalization of REC8 (red) and PRDM9 (green) in spermatocyte spreads. First three rows, B6 spermatocytes in leptonema (top row), early zygonema (second row), and late zygonema (third row). Arrows show colocalization pattern of the two proteins becoming more prominent at later stages. Fourth row, colocalization pattern in late zygonema–like cells in the Prdm9tm3.1Kpgn mutant. Note the more diffuse pattern of the truncated PRDM9 lacking its DNA-binding domain compared with wild type in the third row. Fifth row, leptonema or early zygonema–like cell in the Prdm9tm1Ymat mutant. (B) Colocalization of SYCP3 (red) and PRDM9 (green) in spermatocyte spreads. Disposition of images is the same as in A. Arrows show colocalization pattern of the two proteins. (C) Colocalization of SYCP1 (red) and PRDM9 (green) in spermatocyte spreads. Disposition of the images is the same as in A. (D) Quantitation of SYCP3/PRDM9 (left) and SYCP1/PRDM9 (right) colocalization in B6 and Prdm9tm3.1Kpgn mutant compared with the colocalization pattern of images in which the two signals were inverted to 180o relative to each other. *p < 0.05.
PRDM9 binding to DNA promotes translocation to the chromosomal axis
The coIP experiments also suggested that PRDM9 binds to SC proteins. For this reason, we tested whether PRDM9 colocalizes with SC proteins in early meiotic prophase by double staining with a combination of PRDM9 and either SYCP3 or SYCP1 antibody on spermatocyte spreads. SYCP3 and SYCP1 show punctate or rod-like staining in early meiosis, similar to that of REC8 (Figure 8, B and C). Both SC protein signals overlapped with PRDM9, with a clearer pattern in late zygonema, when a significant part of the diffuse PRDM9 signal disappears (Figure 8, B and C, second rows, arrows). Because this pattern makes it difficult to determine whether the proteins truly colocalize, we performed a DNase treatment, which removed the loop DNA and resulted in loss of most of the PRM9 signal not associated with the chromosomal axis (Figure 8, B and C, fourth columns). Under these conditions, the association of PRDM9 with both SC proteins was apparent (Figure 8, B and C, second rows, fourth columns, arrows). To determine whether this association was statistically significant, we compared the colocalization of actual signals to images in which one of the signals is inverted 180O relative to the other (Kumar et al., 2015). In wild-type mice, PRDM9 showed colocalization with SYCP3 in zygotene (Li's intensity correlation quotient [ICQ] value, p = 6 × 10−5; Figure 8D, left). PRDM9 also showed colocalization with SYCP1 when the latter first appeared in midzygotene (p = 2.2 × 10−5; Figure 5D, right). The statistical significance of the difference between actual and inverted images was even greater after DNase treatment (Figure 8D, Li's ICQ value, p = 1.33 × 10−12 for SYCP3 and p = 6 × 10−14 for SYCP1).
Of importance, we compared these results with the colocalization pattern of the Prdm9tm3.1Kpgn mutant. At best, we found very weak evidence of significant colocalization between PRDM9 and either SYCP3 or SYCP1 in this mutant (Figure 8D, left, p = 0.13; right, p = 0.16). The lack of colocalization was confirmed after DNase treatment (Figure 8, B and C, fourth rows, fourth columns, and D). The difference in the colocalization patters between wild-type and mutant spermatocytes was statistically significant for both combinations (p < 0.05).
The combined coIP and cytological evidence indicates that the binding of PRDM9 to DNA promotes the translocation of DNA-PRDM9-protein complexes to the chromosomal axis.
DISCUSSION
PRDM9 directly interacts with EWSR1, CXXC1, EHMT2, and CDYL
Yeast two-hybrid screens and in vitro binding assays show that EWSR1, CXXC1, EHMT2, and CDYL can directly bind to PRDM9 via its KRAB domain, as well as by additional contact points extending further to the PR/SET domain (Figure 1). With the exception of CXXC1, for which specific antibodies are not available, we confirmed that these interactions also occur in mouse spermatocytes (Figures 2 and 4). However, the spatial and temporal pattern of expression of each of these proteins in vivo suggests that these interactions probably occur at different stages of meiotic progression and involve two distinct PRDM9 complexes—one with EHMT2 and CDYL and another with EWSR1 and possibly CXXC1. Several lines of evidence show that PRDM9 forms dimers at hotspots (Baker et al., 2015b; Patel et al., 2016), suggesting that PRDM9 can use its KRAB domain to bind two proteins at the same time, with PRDM9-EHMT2-CDYL as one complex and PRDM9-EWSR1-CXXC1 as another.
EHMT2 and CDYL are expressed strongly in both Sertoli cells and spermatogonia, before PRDM9 appears at the onset of meiosis, and they remain present in the subsequent preleptotene and leptotene stages when PRDM9 is first detected in the nuclei of germ cells (Sun et al., 2015). In early zygonema, EHMT2 disappears from the nucleus, and CDYL is translocated out of the nucleus and remains in the cytoplasm into pachynema (Figure 6). CDYL interacts strongly with EHMT2 by coIP (Figure 2), suggesting that their interaction takes place in any cell type in which they are present together—Sertoli cells, spermatogonia, and early spermatocytes. However, the two proteins can each bind to PRDM9 in vitro. Thus, their similar coIP patterns with PRDM9 in spermatocytes could reflect both their binding to each other and independently to PRDM9. CDYL also complexes EHMT2 in mouse embryonic stem cells, but there the two proteins are found bound together in heterochromatin regions, including the inactivated X chromosome in females (Escamilla-Del-Arenal et al., 2013). This is in marked contrast to spermatocytes, in which PRDM9 binding to DNA results in chromatin activation by catalyzing H3K4 and H3K36 trimethylation of nearby nucleosomes (Baker et al., 2014; Powers et al., 2016), suggesting that a triple PRDM9-CDYL-EHMT2 complex may well have a different molecular function than the double CDYL-EHMT2 complex lacking PRDM9.
EWSR1 expression coincides with PRDM9 in leptonema and zygonema. These data, together with the results of Y2H, in vitro binding, and the strong mutual coIP of the two proteins, make a strong case that the two proteins physically interact with each other from leptonema to late zygonema. However, unlike PRDM9, EHMT2, and CDYL, EWSR1 dramatically increases in nuclei at pachynema (Figure 5B). The continued, increased expression of EWSR1 at later stages suggests that EWSR1 likely provides two distinct functions: an earlier one when complexed with PRDM9 and a later one when PRDM9 has disappeared. In humans EWSR1 interacts with BARD1 (Spahn et al., 2002), which, apart from other recombination factors (BRCA1, RAD51, etc.), interacts with SETDB1 (Goehler et al., 2004)—a H3K9 histone methyltransferase—which in turn interacts with CDYL-EHMT2 (Mulligan et al., 2008). PRDM9, which also has a SET domain, may play role equivalent to that of SETDB1 by taking central place as a shared binding partner of EWSR1 and CDYL-EHMT2. This possibility is further supported by the recent findings that PRDM9, like SETDB1, is capable of trimethylating H3K9 in vitro (Wu et al., 2013; Powers et al., 2016), although the presence of H3K9ac at hotspots (Buard et al., 2009) likely prevents this activity in vivo.
CXXC1 binds directly to PRDM9 in Y2H and in vitro. Unfortunately, the lack of appropriate antibodies prevented us from detecting its interactions in vivo. What is known is that in mammals, CXXC1 is part of the Set1 complex responsible for most of H3K4 trimethylation in somatic cells (Lee and Skalnik, 2005; Shilatifard, 2012). In embryonic stem cells, it is required for both H3K4me3 deposition after DNA damage and the subsequent acetylation of H3K9 at the same nucleosomes (Clouaire et al., 2014). With respect to its possible functions in meiosis, studies of Spp1, the yeast homologue of CXXC1, provide clues to its possible role in mammalian meiosis. In Saccharomyces cerevisiae, DSBs initiate at H3K4me3 sites found near promoters (Borde et al., 2009). DSB formation at these sites is promoted when they become tethered to the chromosomal axis by Spp1/CXXC1 (Acquaviva et al., 2013; Sommermeyer et al., 2013). We now show that CXXC1 binds to PRDM9 directly and that REC8 interaction with PRDM9 is indirect, possibly requiring mediation by EWSR1. This suggests that CXXC1 may act cooperatively with EWSR1, providing an additional link between PRDM9-bound H3K4-trimethylated sites and the chromosomal axis to promote proper DSB formation and repair.
PRDM9 interacts and colocalizes with meiotic-specific cohesin REC8 and synaptonemal complex proteins SYCP1 and SYCP3
We found no evidence for direct binding between PRDM9 and the meiotic cohesin REC8 in either our Y2H screen or when the two proteins were tested in vitro after expression in E. coli. However, the two proteins showed ample evidence of interaction in spermatocytes both by pull-down experiments (Figure 2A) and cytological colocalization (Figure 8A) and when they were expressed together in HEK 293 cell cultures. In both cases, the shifted mobility of REC8 and the specificity of the antibody we used suggest that only phosphorylated REC8 interacts with PRDM9. In addition, the substantial reduction in coIP after DNase treatment suggests that these proteins require an additional molecule to provide a link, which needs not to be meiosis specific. Our coIP results suggest EWSR1 as a likely candidate; it is a generally “sticky” protein (Schwartz et al., 2015) and strongly pulls down both PRDM9 and REC8 in spermatocytes independently of the presence of DNA; the other likely candidate is CXXC1, given its known role in bring hotspots to the chromosome axis in yeast, and these are not mutually exclusive possibilities.
These findings open the possibility that complexes including PRDM9, EWSR1, and other proteins, such as CXXC1, play a role in homologue recognition by bringing the homologous hotspot DNA sequences bound to PRDM9 in contact with cohesins and subsequently to the chromosomal axis.
Our findings that the synaptonemal complex proteins REC8 and SYCP3, and to some extent SYCP1, interact not only with each other but also with PRDM9 and EWSR1 support the concept that these proteins play important roles in bringing activated hotspots from out in the DNA loops down to the chromosome axis, stabilizing hotspot location there, and then participating in DSB formation and repair. Of interest, both SYCP3 and SYCP1 showed stronger binding with PRDM9 than with each other (Figure 2C), given that SYCP1 and SYCP3 do not bind directly to each other but through SYCP2. SYCP1 participates in the formation of the SC central element, interacting with intermediates such as SYCP2, SYCE1, SYCE2, and SYCE3, and TEX12 (for review, see Bolcun-Filas and Schimenti, 2012). This raises the possibility that PRDM9 contact with SYCP1 has a function beyond bringing hotspot DNA to the chromosomal axis.
Judging by the properties of its yeast homologue, Spp1, CXXC1 may well function in combination with the histone modifications placed by PRDM9 at hotspots in the transport of hotspots to the chromosome axis. Once DSBs have been formed, EWSR1 is known to promote single-strand DNA invasion into double-strand DNA, forming meiotic Holliday junctions (Guipaud et al., 2006; Schwartz et al., 2015), and has been shown to contribute to DSB repair (Li et al., 2007). Further, EHMT2 and CDYL are involved in the establishment of closed chromatin states (Leung et al., 2011; Xu and Price, 2011; Escamilla-Del-Arenal et al., 2013) such as those evidenced by the presence of γH2AX around as-yet-unrepaired meiotic DNA lesions (Chicheportiche et al., 2007). However, we never detected any presence of closed chromatin marks such as H3K9me2/me3 near active hotspots. Instead, nucleosomes in the immediate vicinity of DSB sites are modified to create an open chromatin state required for repair (Xu and Price, 2011; Baker et al., 2014). In this regard, the temporary presence of EHMT2-CDYL at PRDM9-bound complexes could help restrict the extent of H3K4me3-marked open chromatin to ensure proper space for the subsequent repair before DSBs are initiated by SPO11.
Protein interactions are dependent of PRDM9 binding to DNA
Our characterization of protein–protein interactions in the Prdm9tm3.1Kpgn mutant, which lacks the zinc finger domain, showed that PRDM9 binding to various partners is dependent on the presence of this domain. This seems to be in contrast with the Y2H data showing that PRDM9 binding to EWSR1, EHMT2, CDYL, and CXXC1 occurs through its N-terminal part, including the KRAB and PR/SET domains. A likely explanation of this apparent contradiction is that EWSR1, CDYL, and EHMT2 may be predominantly bound to DNA and/or chromatin in the nuclei, whereas PRDM9 can be present in both the nuclear matrix and bound to hotspot DNA, in which case, the interactions can only occur in the context of DNA packed in chromatin. In addition, each PRDM9 fragment in the Y2H assay is bound to DNA through the GAL4 DNA-binding domain of the vector, which may provide the conditions for proper binding.
EWSR1, but not EHMT2 or CDYL, binds to the SC in the absence of PRDM9
PRDM9 presence is not necessary for binding of EWSR1 to the chromosomal axis but is required for binding of EHMT2-CDYL complexes to the chromosomal axis, as evidenced by our SYCP3 coIP results in testes of Prdm9tm1Ymat KO mutant (Figure 4, middle). EWSR1 binding to SYCP3 and REC8 in this mutant is as strong as in wild type (Figures 2B and 4), suggesting that it is associated with the chromosomal axis from the earlier stages of its formation.
Model of events occurring before recombination initiation
Our results show extensive protein–DNA and protein–protein interactions as part of recombination-related events in early meiotic prophase. Our data suggest the following working model for the mechanisms and dynamics of these events (Figure 9). In early leptonema, a fraction of the PRDM9 molecules present in the nucleus bind to hotspots as dimers in which only one subunit binds to DNA (Baker et al., 2015b). Both subunits participate in the trimethylation of histone 3 at lysine 4 and lysine 36, ensuring the deposition of H3K4me3 and H3K36me3 on both sides of the PRDM9 binding site. The KRAB domains of the PRDM9 subunits bind CDYL and EHMT2, restricting the extent of trimethylation to two to four nucleosomes on each side (Baker et al., 2014; Powers et al., 2016; Figure 9A). By the end of leptonema, CDYL and EHMT2 are removed from the complex and the nucleus, and the hotspot-bound PRDM9 and the adjacent nucleosomes bind EWSR1 and CXXC1 (Figure 9B). These complexes then bind to REC8 and translocate the hotspot DNA from out in chromosome loops down to the chromosomal axis, where REC8 integrates into the axis, with the participation of SYCP3 (Figure 9C). PRDM9 remains bound to the hotspots until DSBs are initiated by SPO11, meanwhile coming into contact with SYCP1 (Figure 9D). PRDM9 then disappears from the nucleus in late zygonema—first the unbound molecules, and later in the ones associated with the SC, whereas EWSR1 further participates in the formation and resolution of Holliday junctions at pachynema and carries out additional functions.
FIGURE 9:
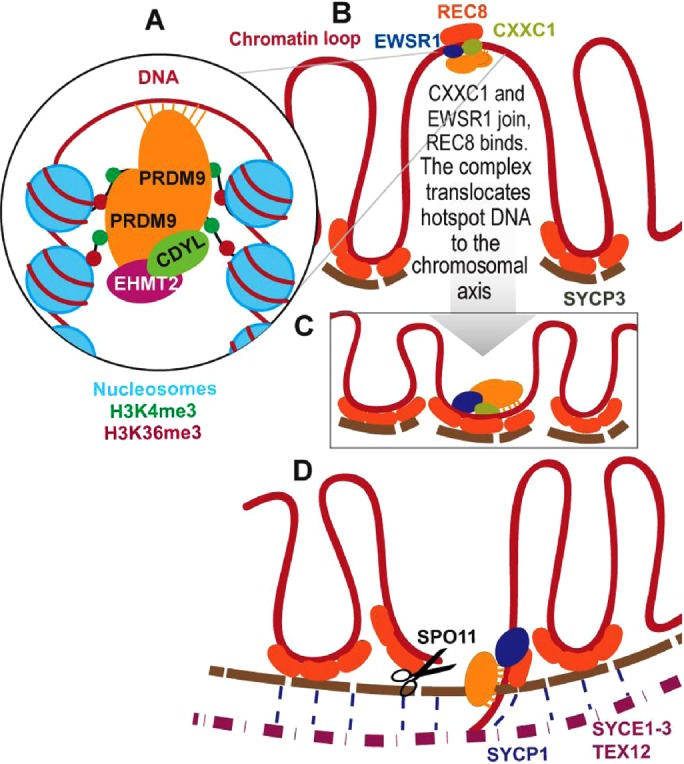
Model of events occurring before recombination initiation (see text for details).
As interesting as this model may be in suggesting new experimental directions, it does lack at least one essential element: what brings the homologous hotspot down to the chromosome axis for precise pairing and DNA exchange? We know from previous studies (Baker et al., 2015a) that precise recombination can occur between homologous hotspots even when one has lost the ability to bind PRDM9, removing PRDM9 binding at the homologue or its consequences as the requisite signal. The model proposed here brings one homologue down to the chromosome axis, but we still lack a model for what brings its homologous partner into apposition.
MATERIALS AND METHODS
Ethics statement
The animal care rules used by the Jackson Laboratory are compatible with the regulations and standards of the U.S. Department of Agriculture and the National Institutes of Health. The protocols used in this study were approved by the Animal Care and Use Committee of the Jackson Laboratory (Summary #04008). Killing of animals for this study was done by cervical dislocation.
Constructs
pBAD-Prdm9 was described in Billings et al. (2013). pGBKT7-Prdm9: whole-length Prdm9 open reading frame from pBAD-Prdm9 was amplified with primers pR638 and pR916 and inserted into pGBKT7 using BamHI-PstI restriction sites. pGBKT7-Prdm9 KRAB (PR-SET): pR638 plus pR608. pGBKT7-Prdm9 KRAB: pR790 plus pR1382. pGBKT7-Prdm9 ID (PR-SET): pR1658 plus pR1661. pGBKT7-Prdm9 ID/2: pR1658 plus pR1383. pGBKT7-Prdm9 ID: pR1658 plus pR1659. pGAD-T7-Prdm9 construct was created by restriction excision by EcoRI-SalI from pBAD-Prdm9 and insertion by the same sites in pGAD-T7 empty vector.
Antibodies
See Table 1. Secondary antibodies (all Life Technologies): goat anti-rabbit immunoglobulin G (IgG; H+L), Alexa Fluor 594 conjugate (A-11037), 1:1000; goat anti-rabbit IgG (H+L), Alexa Fluor 488 conjugate (ab150077), 1:1000; goat anti-mouse IgG (H+L), Alexa Fluor 488 conjugate (ab150113), 1:1000; goat anti-mouse IgG (H+L), Alexa Fluor 647 conjugate (A-21236), 1:1000; goat anti-guinea pig IgG (H+L), Alexa Fluor 594 conjugate (A-11076), 1:1000; goat anti-guinea pig IgG (H+L), Alexa Fluor 488 conjugate (A-11073), 1:1000.
TABLE 1.
Antibodies used.
| Anti-PRDM9 | Guinea pig | Custom made | 1:200 |
| Anti-EHMT2 | PP-A8620A-00 | Perseus Proteomics | 1:500 |
| Anti-EWSR1 | ab54708 | Abcam | 1:200 |
| Anti-SYCP3 | NB300-231 | Novus Biologicals | 1:500 |
| Anti-SYCP1 | NB 300-229 | Novus Biologicals | 1:500 |
| Anti-CDYL | ab5188-50 | Abcam | 1:50 |
| Anti-REC8 | ab38372, lot 3 | Abcam | 1:200 |
| Anti-MBP | E8038S | New England Biolabs | 1:1000 |
Yeast two-hybrid screen
The Y2H screen was performed by cotransformation of pGBKT7-Prdm9 construct and mouse testes cDNA library cloned in pGADT7 (3 × 106 to 107 clones) (638848; Clontech) in PJ69-4α strain. Positive interactions were selected by plating the transformants on three different selective media lacking combinations of –Trp, –Leu, –His, and –Ala and supplemented with 3 mM 3-amino-1,2,4-triazole for detection of more stringent interactions. Each positive clone was tested for autoactivation by crossing to a yeast strain containing empty pGBKT7 plasmid. Plasmid DNA was isolated and sequenced to identify the individual cDNA library clone.
Yeast two-hybrid screen validation and Prdm9-domain mapping
The detected open reading frames interacting with Prdm9 were isolated and transformed in PJ69-4A strain. The shorter pGBKT7-Prdm9 constructs were transformed in PJ69-4α strain. The PJ69-4α strain was crossed with PJ69-4A strain carrying the clone of interest and plated on selective media plates for confirmation of positive interactions.
Protein–protein in vitro pull-downs
Expression of PRDM9 was performed in Arctic DE3 cells. Preculture was grown overnight at 30°C. The cells were reinoculated in the next day; the culture was grown for 3–4 h and shifted for 16–24 h at 14°C at 200 rpm. The cells were collected by centrifugation at 5000 × g for 10 min and the pellet was ground by SPEX SamplePrep 6870 Freezer/Mill and dissolved in 1× CBB buffer (50 mM Tris-HCl, 4 mM EDTA, 200 mM sucrose, pH 7.5) plus 150 mM KCl, 10 ml per 1-g pellet, supplied with protease inhibitor cocktail (aprotinin, chymostatin, leupeptin, pepstatin; Applichem), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM mercaptoethanol, and 0.01% NP40.
The purification of each protein was done in three steps: Sp-Sepharose, amylose beads, and fast-performance liquid chromatography on a MonoS column. The pull-down of the purified PRDM9 protein and the protein of interest were done by affinity beads corresponding to the tag of the protein. The pull-down was performed by mixing 1 µg of each protein for 30 min at 34°C, followed by addition of 30-µl beads washed by K buffer (40 mM K2PO4, 10% glycerol, 0.5 mM EDTA, pH 7.5) plus 150 mM KCl and incubated for another 30 min. The bound proteins were eluted with SDS loading buffer.
Coimmunoprecipitation assays
Testis material from twenty 14-dpp-old C57BL/6J male mice was extracted in cold phosphate-buffered saline (PBS), homogenized by Dounce homogenizer, passed through a 40-µm cell strainer (352340; Falcon BD), and centrifuged for 5 min at 3000 × g. The pellet was resuspended in 1 ml of Pierce IP buffer (87787; ThermoFisher Scientific) with 1 mM PMSF and 1× EDTA-free protease inhibitor cocktail (Roche). The sample was incubated for 30 min with slow rotation and centrifuged at 13,200 × g. For the DNase-treated coIP samples, 100 µl of DNase buffer and 20 U DNase (Ambion) were added, and the samples were incubated for 1 h at room temperature. The coIP was done by protein A or G beads (Dynabeads; Lifesciences), depending on the antibody source. As a negative control, IgG from the same animal species was used. The extract was incubated overnight at 4°C with rotation. After washing of the beads three times with 1 ml of Pierce IP buffer, the complexes were eluted with 200 µl of GST buffer (0.2 M glycine, 0.1% SDS, 1% Tween 20, pH 2.2) for 20 min at room temperature. The sample was neutralized with 40 µl of 1 M Tris-HCl, pH 8. For SDS-PAGE, 40 µl of SDS-loading buffer was added. The samples were heated to 95°C for 5 min and subjected to electrophoresis and Western blotting. Each lane contained 10 µg of protein except for the input in the EHMT2 and CDYL Western blots, which contained 2.5 µg to keep from overwhelming the coIP signal.
Chromosome spreads
For preparation of nuclear spreads from germ cells, the drying-down technique (Peters et al., 1997) was used, followed by double or consecutive immunolabeling with PRDM9/SYCP3, PRDM9/SYCP1, PRDM9/EWSR1/SYCP3, EWSR1/BRCA1/SYCP3, SYCP3/γH2AX/CREST, and PRDM9/REC8. For DNase-treated spread samples, slides were treated with 100 µl of DNase buffer containing 10 U of DNase at 37°C for 2 h, followed by immunolabeling.
Periodic acid–Schiff–diastase staining
For histological evaluation, tissues were dissected out, fixed with Bouin's solution, and embedded in paraffin wax, and 5-μm sections were prepared. Sections were stained with Periodic acid–Schiff–diastase (PAS) using standard techniques.
Immunofluorescence staining
For protein immunolocalization, tissues were dissected out, fixed with 4% paraformaldehyde solution, embedded in paraffin wax, and sectioned at 5 μm. Sections were heated in a microwave in 10 mM sodium citrate buffer, pH 6.0, for 10 min and then treated with PBS containing 0.1% Triton X-100. After blocking of nonspecific binding sites with 10% normal donkey serum (017-000-121; Jackson ImmunoResearch Labs), sections were incubated with primary antibodies overnight at 4°C and secondary antibodies for 2 h at room temperature. The slides were rinsed in PBS, stained for 3 min with 1 µg/ml 4′,6-diamidino-2-phenylindole (28718-90-3 Sigma-Aldrich), rinsed three times in PBS for 5 min each, and mounted in Antifade reagent (S-2828; Life Technologies). Images were photographed with a Microscope Axio Imager.Z2 (Zeiss, Germany).
Terminal deoxynucleotidyl transferase–mediated digoxigenin-dUTP nick-end labeling assay
For detection of apoptosis in tissues, testis sections were subjected to fluorescence labeling of DNA strand breaks by terminal deoxynucleotidyl transferase–mediated digoxigenin-dUTP nick-end labeling (TUNEL) assay, using the In Situ Cell Death Detection Kit (11684795910; Roche) according to the manufacturer's protocol. As a negative control, the TdT enzyme was omitted in parallel reactions.
ACKNOWLEDGMENTS
We thank Anita Hawkins for technical help, Mary Ann Handel for critical reading and helpful suggestions, Neil Hunter for advice on immunofluorescence, and Attila Toth for information about HORMAD proteins. This work was supported by National Institutes of Health Grants R01 GM078452 to P.M.P., P50 GM076468 to Gary Churchill/Project B to P.M.P., and R01 GM078643 to K.P., Cancer Core Grant CA34196 to the Jackson Laboratory, Czech Science Foundation Grants GACR13-26629S and GACR207/12/2323 to L.K., and South Moravian Program Grant 2SGA2773 to E.D.P.
Abbreviations:
- CDYL
chromodomain-containing Y chromosome–like
- CXXC1
CXXC domain–containing 1
- EHMT2
euchromatic histone methyltransferase 2
- EWSR1
Ewing sarcoma 1
- PRDM9
PR/SET domain–containing 9
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-09-0686) on December 8, 2016.
REFERENCES
- Acquaviva L, Szekvolgyi L, Dichtl B, Dichtl BS, de La Roche Saint Andre C, Nicolas A, Geli V. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science. 2013;339:215–218. doi: 10.1126/science.1225739. [DOI] [PubMed] [Google Scholar]
- Baker CL, Kajita S, Walker M, Saxl RL, Raghupathy N, Choi K, Petkov PM, Paigen K. PRDM9 drives evolutionary erosion of hotspots in Mus musculus through haplotype-specific initiation of meiotic recombination. PLoS Genet. 2015a;11:e1004916. doi: 10.1371/journal.pgen.1004916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CL, Petkova P, Walker M, Flachs P, Mihola O, Trachtulec Z, Petkov PM, Paigen K. Multimer formation explains allelic suppression of PRDM9 recombination hotspots. PLoS Genet. 2015b;11:e1005512. doi: 10.1371/journal.pgen.1005512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CL, Walker M, Kajita S, Petkov PM, Paigen K. PRDM9 binding organizes hotspot nucleosomes and limits Holliday junction migration. Genome Res. 2014;24:724–732. doi: 10.1101/gr.170167.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Imai Y, de Massy B. Meiotic recombination in mammals: localization and regulation. Nat Rev Genet. 2013;14:794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- Billings T, Parvanov ED, Baker CL, Walker M, Paigen K, Petkov PM. DNA binding specificities of the long zinc-finger recombination protein PRDM9. Genome Biol. 2013;14 doi: 10.1186/gb-2013-14-4-r35. R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolcun-Filas E, Schimenti JC. Genetics of meiosis and recombination in mice. Int Rev Cell Mol Biol. 2012;298:179–227. doi: 10.1016/B978-0-12-394309-5.00005-5. [DOI] [PubMed] [Google Scholar]
- Borde V, Robine N, Lin W, Bonfils S, Geli V, Nicolas A. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009;28:99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buard J, Barthes P, Grey C, de Massy B. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J. 2009;28:2616–2624. doi: 10.1038/emboj.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicheportiche A, Bernardino-Sgherri J, de Massy B, Dutrillaux B. Characterization of Spo11-dependent and independent phospho-H2AX foci during meiotic prophase I in the male mouse. J Cell Sci. 2007;120:1733–1742. doi: 10.1242/jcs.004945. [DOI] [PubMed] [Google Scholar]
- Clouaire T, Webb S, Bird A. Cfp1 is required for gene expression-dependent H3K4 trimethylation and H3K9 acetylation in embryonic stem cells. Genome Biol. 2014;15:451. doi: 10.1186/s13059-014-0451-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escamilla-Del-Arenal M, da Rocha ST, Spruijt CG, Masui O, Renaud O, Smits AH, Margueron R, Vermeulen M, Heard E. Cdyl, a new partner of the inactive X chromosome and potential reader of H3K27me3 and H3K9me2. Mol Cell Biol. 2013;33:5005–5020. doi: 10.1128/MCB.00866-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher C. The diversity of soft tissue tumours with EWSR1 gene rearrangements: a review. Histopathology. 2014;64:134–150. doi: 10.1111/his.12269. [DOI] [PubMed] [Google Scholar]
- Fukuda T, Pratto F, Schimenti JC, Turner JM, Camerini-Otero RD, Hoog C. Phosphorylation of chromosome core components may serve as axis marks for the status of chromosomal events during mammalian meiosis. PLoS Genet. 2012;8:e1002485. doi: 10.1371/journal.pgen.1002485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumasoni I, Meani N, Rambaldi D, Scafetta G, Alcalay M, Ciccarelli FD. Family expansion and gene rearrangements contributed to the functional specialization of PRDM genes in vertebrates. BMC Evol Biol. 2007;7:187. doi: 10.1186/1471-2148-7-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehler H, Lalowski M, Stelzl U, Waelter S, Stroedicke M, Worm U, Droege A, Lindenberg KS, Knoblich M, Haenig C, et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol Cell. 2004;15:853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Guipaud O, Guillonneau F, Labas V, Praseuth D, Rossier J, Lopez B, Bertrand P. An in vitro enzymatic assay coupled to proteomics analysis reveals a new DNA processing activity for Ewing sarcoma and TAF(II)68 proteins. Proteomics. 2006;6:5962–5972. doi: 10.1002/pmic.200600259. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Yoshida K, Matsui Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature. 2005;438:374–378. doi: 10.1038/nature04112. [DOI] [PubMed] [Google Scholar]
- Hinch AG, Tandon A, Patterson N, Song Y, Rohland N, Palmer CD, Chen GK, Wang K, Buxbaum SG, Akylbekova EL, et al. The landscape of recombination in African Americans. Nature. 2011;476:170–175. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth RS, Gruenewald-Schneider U, Webb S, Kerr AR, James KD, Turner DJ, Smith C, Harrison DJ, Andrews R, Bird AP. Orphan CpG islands identify numerous conserved promoters in the mammalian genome. PLoS Genet. 2010:6, e1001134. doi: 10.1371/journal.pgen.1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro K, Kim J, Shibuya H, Hernandez-Hernandez A, Suzuki A, Fukagawa T, Shioi G, Kiyonari H, Li XC, Schimenti J, et al. Meiosis-specific cohesin mediates homolog recognition in mouse spermatocytes. Genes Dev. 2014;28:594–607. doi: 10.1101/gad.237313.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Ghyselinck N, Ishiguro K, Watanabe Y, Kouznetsova A, Hoog C, Strong E, Schimenti J, Daniel K, Toth A, de Massy B. MEI4 - a central player in the regulation of meiotic DNA double-strand break formation in the mouse. J Cell Sci. 2015;128:1800–1811. doi: 10.1242/jcs.165464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Tang ZL, Zhou JX, Barndt RJ, Parvinen M, Allis CD, Page DC. Previously uncharacterized histone acetyltransferases implicated in mammalian spermatogenesis. Proc Natl Acad Sci U S A. 2002;99:8707–8712. doi: 10.1073/pnas.082248899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem. 2005;280:41725–41731. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- Leung DC, Dong KB, Maksakova IA, Goyal P, Appanah R, Lee S, Tachibana M, Shinkai Y, Lehnertz B, Mager DL, et al. Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proc Natl Acad Sci U S A. 2011;108:5718–5723. doi: 10.1073/pnas.1014660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Watford W, Li C, Parmelee A, Bryant MA, Deng C, O'Shea J, Lee SB. Ewing sarcoma gene EWS is essential for meiosis and B lymphocyte development. J Clin Invest. 2007;117:1314–1323. doi: 10.1172/JCI31222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo A, Cesaro E, Montano G, Zurlo D, Izzo P, Costanzo P. KRAB-zinc finger proteins: a repressor family displaying multiple biological functions. Curr Genomics. 2013;14:268–278. doi: 10.2174/13892029113149990002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan P, Westbrook TF, Ottinger M, Pavlova N, Chang B, Macia E, Shi YJ, Barretina J, Liu J, Howley PM, et al. CDYL bridges REST and histone methyltransferases for gene repression and suppression of cellular transformation. Mol Cell. 2008;32:718–726. doi: 10.1016/j.molcel.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakland TE, Haselton KJ, Randall G. EWSR1 binds the hepatitis C virus cis-acting replication element and is required for efficient viral replication. J Virol. 2013;87:6625–6634. doi: 10.1128/JVI.01006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paigen K, Petkov P. Mammalian recombination hot spots: properties, control and evolution. Nat Rev Genet. 2010;11:221–233. doi: 10.1038/nrg2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov ED, Petkov PM, Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Horton JR, Wilson GG, Zhang X, Cheng X. Structural basis for human PRDM9 action at recombination hot spots. Genes Dev. 2016;30:257–265. doi: 10.1101/gad.274928.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Plug AW, van Vugt MJ, de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res. 1997;5:66–68. doi: 10.1023/a:1018445520117. [DOI] [PubMed] [Google Scholar]
- Powers NR, Parvanov ED, Baker CL, Walker M, Petkov PM, Paigen K. The meiotic recombination activator PRDM9 trimethylates both H3K36 and H3K4 at recombination hotspots in vivo. PLoS Genet. 2016;12:e1006146. doi: 10.1371/journal.pgen.1006146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Cech TR, Parker RR. Biochemical properties and biological functions of FET proteins. Annu Rev Biochem. 2015;84:355–379. doi: 10.1146/annurev-biochem-060614-034325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, Petukhova GV. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011;472:375–378. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommermeyer V, Beneut C, Chaplais E, Serrentino ME, Borde V. Spp1, a member of the Set1 complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol Cell. 2013;49:43–54. doi: 10.1016/j.molcel.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Spahn L, Petermann R, Siligan C, Schmid JA, Aryee DN, Kovar H. Interaction of the EWS NH2 terminus with BARD1 links the Ewing's sarcoma gene to a common tumor suppressor pathway. Cancer Res. 2002;62:4583–4587. [PubMed] [Google Scholar]
- Steggerda SM, Paschal BM. Regulation of nuclear import and export by the GTPase Ran. Int Rev Cytol. 2002;217:41–91. doi: 10.1016/s0074-7696(02)17012-4. [DOI] [PubMed] [Google Scholar]
- Sun F, Fujiwara Y, Reinholdt LG, Hu J, Saxl RL, Baker CL, Petkov PM, Paigen K, Handel MA. Nuclear localization of PRDM9 and its role in meiotic chromatin modifications and homologous synapsis. Chromosoma. 2015;124:397–415. doi: 10.1007/s00412-015-0511-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Nozaki M, Takeda N, Shinkai Y. Functional dynamics of H3K9 methylation during meiotic prophase progression. EMBO J. 2007;26:3346–3359. doi: 10.1038/sj.emboj.7601767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, Ueda J, Fukuda M, Takeda N, Ohta T, Iwanari H, Sakihama T, Kodama T, Hamakubo T, Shinkai Y. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate CM, Lee JH, Skalnik DG. CXXC finger protein 1 contains redundant functional domains that support embryonic stem cell cytosine methylation, histone methylation, and differentiation. Mol Cell Biol. 2009;29:3817–3831. doi: 10.1128/MCB.00243-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M, Billings T, Baker CL, Powers N, Tian H, Saxl RL, Choi K, Hibbs MA, Carter GW, Handel MA, et al. Affinity-seq detects genome-wide PRDM9 binding sites and reveals the impact of prior chromatin modifications on mammalian recombination hotspot usage. Epigenetics Chromatin. 2015;8:31. doi: 10.1186/s13072-015-0024-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Mathioudakis N, Diagouraga B, Dong A, Dombrovski L, Baudat F, Cusack S, de Massy B, Kadlec J. Molecular basis for the regulation of the H3K4 methyltransferase activity of PRDM9. Cell Rep. 2013;5:13–20. doi: 10.1016/j.celrep.2013.08.035. [DOI] [PubMed] [Google Scholar]
- Wu H, Min J, Antoshenko T, Plotnikov AN. Crystal structures of human CDY proteins reveal a crotonase-like fold. Proteins. 2009;76:1054–1061. doi: 10.1002/prot.22472. [DOI] [PubMed] [Google Scholar]
- Xu Y, Price BD. Chromatin dynamics and the repair of DNA double strand breaks. Cell Cycle. 2011;10:261–267. doi: 10.4161/cc.10.2.14543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yang XH, Gui B, Xie GJ, Zhang D, Shang YF, Liang J. Corepressor protein CDYL functions as a molecular bridge between polycomb repressor complex 2 and repressive chromatin mark trimethylated histone lysine 27. J Biol Chem. 2011;286:42414–42425. doi: 10.1074/jbc.M111.271064. [DOI] [PMC free article] [PubMed] [Google Scholar]