Summary
Pseudomonas aeruginosa is a Gram-negative bacterial pathogen associated with acute and chronic infections. The universal c-di-GMP second messenger is instrumental in the switch from a motile lifestyle to resilient biofilm as in the cystic fibrosis lung. The SadC diguanylate cyclase is associated with this patho-adaptive transition. Here we identified an unrecognized SadC partner, WarA, which we show is a methyltransferase in complex with a putative kinase WarB. We established that WarA binds to c-di-GMP, which potentiates its methyltransferase activity. Together, WarA and WarB have structural similarities with the bi-functional Escherichia coli LPS O antigen regulator WbdD. Strikingly, WarA influences P. aeruginosa O antigen modal distribution and interacts with the LPS biogenesis machinery. LPS is known to modulate the immune response in the host, and by using a zebrafish infection model, we implicate WarA in the ability of P. aeruginosa to evade detection by the host.
Pseudomonas aeruginosa is a Gram-negative bacterial pathogen commonly associated with the cystic fibrosis (CF) lung. Upon infection, a switch occurs between two contrasting modes of bacterial growth. Acute infection is characterised by a planktonic lifestyle whereby P. aeruginosa is motile and induces the type III secretion system (T3SS). The transition to chronic infection coincides with a P. aeruginosa behavioural switch to a sessile lifestyle, evident by the loss of motility, a shift in LPS profile and biofilm formation1–3. It has recently emerged that a key signal facilitating this decision making process is the universal eubacterial second messenger cyclic-di-GMP (c-di-GMP). The intracellular levels of c-di-GMP are modulated by two classes of enzymes, diguanylate cyclases (DGC), which synthesize c-di-GMP from two molecules of GTP and phosphodiesterases (PDE), which catabolize c-di-GMP to GMP4–6. Low levels of c-di-GMP are associated with the early stages of acute infection whereas high levels of c-di-GMP are indicative of chronic infection7. Another regulatory pathway that has been shown to play a key role in the acute to chronic switch is the Gac/Rsm cascade, where activation of this pathway leads to a sequestration of the master translational regulator RsmA by the small RNAs RsmZ and RsmY. This leads to an increase in the intracellular levels of c-di-GMP and the activation of chronic infection traits such as biofilm formation1,8. While these two signalling cascades overlap in terms of phenotypic consequence, recent genetic evidence has shown they intersect at a specific membrane bound DGC, SadC. It has been established that SadC is solely responsible for the increased levels of c-di-GMP observed in a P. aeruginosa strain that has the Gac/Rsm cascade constitutively activated9. It has also been shown that RsmA can impact the intracellular levels of SadC most likely at the posttranscriptional level9.
A gap currently exists in the understanding of c-di-GMP as a signalling molecule since for the vast array of phenotypes that are impacted by c-di-GMP levels, comparatively few proteins have been identified that can interact with this dinucleotide. Given that SadC plays a pivotal role in the acute to chronic behavioural change, a bacterial two hybrid screen was performed to identify interacting partners of SadC which play a role in mediating this transition10. The screen identified a number of candidates, some associated with phenotypes such as motility, however one hit was identified repeatedly. The protein, PA4379, is a predicted methyltransferase with no known phenotypic association. In this study we demonstrate that PA4379 directly interacts with SadC and together they modify the LPS O antigen modal distribution of P. aeruginosa. Because of striking homologies with WbdD from Escherichia coli, we called the enzyme WarA for WbdD-like O Antigen Regulator. We also elucidate the molecular mechanism underpinning this LPS phenotype by demonstrating that WarA can bind to c-di-GMP and that this binding influences its enzymatic activity. Finally, we explore the biological significance of the regulation of the LPS O antigen by SadC/WarA on immune evasion using a zebrafish (Danio rerio) model of P. aeruginosa infection.
SadC interacts with the methyltransferase WarA
The Gac/Rsm regulatory cascade and c-di-GMP signalling have been established as the key pathways governing the switch from acute to chronic infection phenotypes in P. aeruginosa1. The molecular basis underpinning the link between the Gac/Rsm network and c-di-GMP has been shown to be the membrane associated DGC, SadC9. Despite the identification of numerous phenotypes influenced by the levels of c-di-GMP within the cell, the mechanisms through which c-di-GMP exerts its phenotypic influence have yet to be fully elucidated. This is largely due to the disparity between the number of enzymes capable of influencing the levels of c-di-GMP within the cell and relatively few proteins that have been shown to interact with c-di-GMP7. To identify the protein-protein interaction network surrounding the central switch component SadC, a P. aeruginosa bacterial two-hybrid (BTH) library was tested10 against sadC cloned in the bait vector pKT25 (Table 1). One of the SadC partners identified multiple times throughout this screen was the predicted methyltransferase PA4379 we called WarA. The warA coding sequence was subsequently amplified from genomic DNA, cloned into pUT18c and re-tested against the sadC-encoding BTH vector, which confirmed that the interaction is significant (Fig. 1a). Far western dot blotting11 using purified proteins was utilized to demonstrate that the cytoplasmic domain of SadC which contains the catalytic GGDEF domain is capable of mediating the interaction with WarA (Fig. 1b).
Table 1. Selected hits from the bacterial two hybrid screen using SadC as a bait.
WarA was identified in 4 separate prey vectors. Strong hits were categorised as those that displayed a blue colour after 2 days incubation on LB X-gal Km/Amp plates. Weak hits were those that did not develop a blue colour until 4 days of incubation.
| Hit | Comments | Gene product |
|---|---|---|
| Strong interactions (after 2 days of incubation) | ||
| FimV (PA3115) | Hits the N-terminus | Motility membrane protein (twitching) |
| PA3926 | Hits the C-terminus | Hypothetical sugar transporter |
| WarA | Hit 4x | Methyltransferase |
| PA4459 | Lipopolysaccharide assembly | |
| Weaker interactions (after 4 days of incubation) | ||
| PA1378 | Hypothetical | |
| PA1891 | Membrane protein | |
| PA3403 | Phosphate starvation inducible protein | |
| cysE (PA3816) | Hits the C-terminus | O-acetylserine synthase |
| WarB | Same operon as warA | Lipopolysaccharide kinase |
| PilS (PA4546) | Hit 2x | Two-component membrane sensor |
| PA4929 | Hits the C-terminus | Diguanylate cyclase |
| AtpF (PA5558) | ATP synthase B chain | |
Figure 1. WarA interacts with SadC.
a, Bacterial two hybrid assay to validate interaction between WarA and the cytoplasmic domain (CD) of SadC. A graphical representation of β-galactosidase activity from E. coli DHM1 cells producing the indicated proteins fused to the adenylate cyclase T25 or T18 subunits. Corresponding spots on LB X-Gal plates are shown. Activity was measured in Miller units. b, Purified SadC (CD)His (16 μg) and WspRHis (16 μg) spotted on a nitrocellulose membrane probed with purified WarAHisGST overnight. A GST antibody was used to detect binding. c, Purified WarAHis (10 μg) and WspRHis (10 μg) spotted on a nitrocellulose membrane probed with purified WarBHisGST overnight. Standard deviation error bars shown and representative images/data of at least 3 replicates shown. Significance testing performed by Student’s t test. *, P<0.05; **, P<0.01.
Interestingly the corresponding genomic neighbour of WarA, PA4378, encoding a predicted kinase we called WarB, was also identified in the BTH library screen, although the interaction was significantly weaker than that of WarA (Sup. Fig. 1). This prompted investigation into whether WarA and WarB could interact. This was demonstrated initially using a BTH approach (Fig. 1a) and biochemically using far western dot blot analysis (Fig. 1c).
WarAB homolog to the LPS biosynthesis enzyme, WbdD
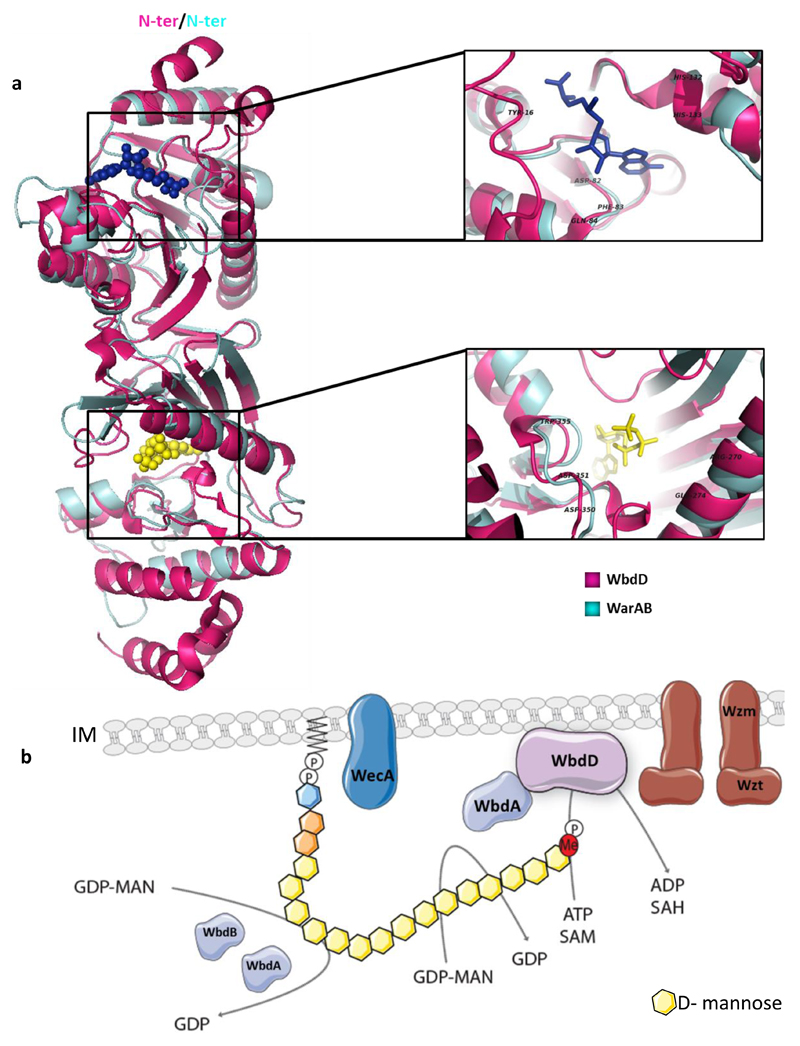
In silico analysis of genes encoding WarA and WarB showed that they were largely conserved in this genomic arrangement throughout the Pseudomonas genus. This high level of synteny, along with the previously described biochemical data, suggests they may function together. To provide additional insight, homology searches were performed using Phyre 2.012.This analysis not only demonstrated the homology of WarA and WarB to known methyltransferases and kinases, respectively, but when both amino acid sequences were combined a predicted three-dimensional model was generated with a very high confidence on the E. coli O9a WbdD protein (19% sequence identity, 69% sequence coverage, 100% confidence) (Fig. 2a, Sup. Fig. 2). WbdD has been shown to play a key role in LPS biogenesis in E. coli O9a, adding a phosphate and a methyl residue to the terminal-reducing end of the polysaccharide chain, which terminates O antigen synthesis and primes for export across the inner membrane13 (Fig. 2b). A WbdD homolog has not been identified in P. aeruginosa although compositional analysis has identified methyl residues on different P. aeruginosa LPS species14,15. Orphan methyltransferases have also been identified that influence the modal distribution but are not associated with a specific kinase16. E. coli O9a is the only currently identified serotype with dual methyltransferase and kinase activity, as in E. coli O8 WbdD lacks the kinase domain and functions only as a methyltransferase17,18. This in silico observation prompted further analysis into the potential role of SadC/WarA in LPS synthesis and assembly in P. aeruginosa.
Figure 2. Structural homology of WbdD and WarAB.
a, Superposition of WbdD (Magenta) secondary structure (PDB:4AZW) with that of the Phyre 2.0 predicted secondary structure of WarAB (Cyan). S-Adenosyl methionine (SAM) in blue (WarA) and ATP in yellow (WarB). Selected residues highlighted. b, Model of LPS O antigen biosynthesis in E. coli O9a. The glycosyltransferases WbdA, WbdB and WbdC are responsible for the synthesis of the O antigen. WbdD adds a phosphoryl and a methyl group to the terminal-reducing end of the O antigen, this terminates synthesis and primes the chain for export through the inner membrane via the ABC transporter Wzm/Wzt. Components of model individually labelled with corresponding names.
WarA has a role in LPS O antigen biogenesis
P. aeruginosa is capable of producing two different glycoforms of LPS via two different systems, a heteropolymeric O antigen composed of up to five different polysaccharides termed O-specific antigen (OSA) assembled by a polymerase/flippase-dependent (Wzy/Wzx-dependent) pathway (Fig. 3a) and a homopolysaccharide O antigen composed of repeating rhamnose residues called the common polysaccharide antigen (CPA) produced by an ABC transporter-dependent pathway19,20 (Fig. 3b). To assess if WarA plays a role in LPS biogenesis, a deletion mutant of warA was generated in a P. aeruginosa PAK background. Preliminary analysis of the LPS profile of a warA mutant using silver-stained SDS page gel did not reveal discernable differences (Sup. Fig. 3). To achieve greater resolution between the two different LPS glycoforms, antibodies specific to P. aeruginosa PAK CPA and OSA were used in western blot analysis. Deletion of warA had no significant impact on the modal distribution of OSA (Fig. 3c) but disrupted the CPA modal distribution when compared to wild-type P. aeruginosa PAK, as did overexpression of warA (Fig. 3d). Remarkably, deletion of warA leads to an increase in average chain length of the LPS indicating a loss of regulation in the termination process, while overexpression results in dysregulation and the production of shorter O antigen chains suggesting premature termination of synthesis. This evidence suggests WarA might play a similar functional role as WbdD in regulating chain length and termination of LPS O antigen synthesis.
Figure 3. WarA impact LPS modal distribution.
a, Model of the heteropolymeric OSA biosynthesis via the Wzy/Wzx-dependent pathway. Individual repeating units are assembled in the cytoplasm and transported via a flippase across the inner membrane to the periplasmic space. Here they are assembled into the O antigen polymer by the Wzy polymerase. Wzz1 and Wzz2 regulate chain length. b, Model of CPA biosynthesis via the ABC transporter pathway. Like LPS biosynthesis in E.coli O9a, synthesis and assembly occur in the cytoplasm via the glycosyltransferases WbpX, WbpY and WbpZ. The assembled O antigen component is then transported via the Wzm/Wzt ABC transporter across the inner membrane. c, Western blot analysis using OSA specific antibody MF83-1 on PAK wild type with an empty vector (+ Ev), PAK wild type overexpressing warA (+ warAOe), a sadC and a warA mutant. d, Western blot analysis using CPA specific antibody (N1F10) on PAK wild type with an empty vector (+ Ev), PAK wild type overexpressing warA (+ warAOe), a sadC and a warA mutant. Representative images of at least two biological replicates shown.
A key feature of WbdD in E. coli O9a is an ability to recruit a specific glycosyltransferase, WbdA to the O antigen assembly interface21–23. Three glycosyltransferases encoded within the central P. aeruginosa CPA biosynthesis operon, WbpX, WbpY and WbpZ, are homologous to the E. coli glycosyltransferases WbdA, WbdB and WbdC, respectively24. All three glycosyltransferases have been shown to be necessary for CPA biosynthesis24–26. Given that deletion of warA is capable of impacting CPA modal distribution, it was assessed whether WarA was capable of interacting with any of the primary glycosyltransferases associated with CPA synthesis. Targeted BTH analysis demonstrated that, like WbdD, WarA interacts specifically with the WbdA homolog, WbpX (Fig. 4a). This provides a further link for WarA to CPA biosynthesis and demonstrates consistency in the interactome homology with WbdD.
Figure 4. WarA is a functional methyltransferase and binds c-di-GMP.
a, Bacterial two hybrid screen of the interaction between WarA and the glycosyltransferases of the primary CPA biosynthesis cluster WbpX, WbpY and WbpZ. A graphical representation of β-galactosidase activity from E. coli DHM1 cells producing the indicated proteins fused to the adenylate cyclase T25 or T18 subunits. Corresponding spots on LB X-Gal plates are shown. Activity was measured in Miller units. b, DRaCALA analysis of WarA using increasing concentrations of WarA. Competition experiment (Middle Panel) using unlabelled c-di-GMP (100 μM), various other nucleotides (100μM) were used to test specificity of WarA (50 μM) binding (bottom panel). c, A fluorescent-based methyltransferase activity assay was used to determine WarA activity in increasing concentrations of LPS. d, Addition of c-di-GMP (15 μM) to the reaction mix at a range of different WarA concentrations. Representative images/data of at least 3 replicates shown. Standard deviation error bars shown. Significance testing performed by Student’s t test. *, P<0.05; **, P<0.01; ***; P<0.001.
WarA is a c-di-GMP-binding methyltransferase
Given that WarA was identified as a SadC interacting partner, we hypothesised that WarA and SadC may function in the same regulatory pathway. In agreement with this statement, deletion of sadC produced an identical impact on the CPA modal distribution as the deletion of warA (Fig. 3d), suggesting that both proteins function together and fine-tune LPS biosynthesis. However, the relevance and molecular consequence of the interaction between SadC and WarA in the process remained to be resolved. This led us to postulate that WarA can bind c-di-GMP. To test this, purified WarAHis was used with 32P-c-di-GMP to perform a differential radial capillary action of ligand assay (DRaCALA)27. WarA was capable of binding to c-di-GMP (Fig. 4b) in the low micromolar range, which is consistent with known biologically relevant c-di-GMP binding concentrations (Sup. Fig. 4a)27,28. While no c-di-GMP binding motif was obvious from the primary amino acid sequence of WarA, an inverted I-site motif was identified (DDRR)29. Mutation of the residues in this motif led to a significant drop in c-di-GMP binding affinity suggesting that these residues may play a role in c-di-GMP binding(Sup. Fig. 4b,c), however binding was not completely abolished suggesting other residues may also be involved. Futher analysis is necessary to conclusively demonstrate the role of this inverted I-site motif in c-di-GMP binding. To test the specificity of binding, competitive DRaCALA assays with unlabelled c-di-GMP and other nucleotides were performed. In all experiments WarA/c-di-GMP binding detection was only shifted by the addition of unlabelled c-di-GMP and by none of the other nucleotides used, including GTP or c-di-AMP (Fig. 4b).
Given the emergent trend of c-di-GMP binding influencing the enzymatic activity of an enzyme28, we hypothesized that binding of c-di-GMP to WarA was capable of influencing its enzymatic activity. Using a fluorescent-based methyltransferase activity assay, LPS was shown to be a substrate for WarA and that increasing the concentration of LPS leads to consequential increase in enzyme activity (Fig. 4c). To test if WarA enzymatic activity was modulated by c-di-GMP binding, c-di-GMP was added across a range of different enzyme concentrations. At every WarA concentration tested, the addition of c-di-GMP led to an increase in the enzymatic activity of WarA (Fig. 4d). This suggests that c-di-GMP is capable of functioning as an agonist, potentiating the enzymatic activity of WarA upon binding. The above-described specific interaction of WarA with SadC suggests that under these circumstances, the spatio-temporal concentrations of c-di-GMP could be high enough to significantly impact WarA activity.
Role of WarA in infection and immune evasion
Numerous studies have demonstrated the physiological relevance of LPS O antigen in P. aeruginosa infections. A shift in the LPS profile is commonly observed in chronic P. aeruginosa isolates and is recognised as playing a key role in the ability to evade host detection and establish chronic infection30–33. To test if the impact of WarA on LPS biogenesis was enough to alter infection progression, two different animal models of infection were utilized, Galleria mellonella and the zebrafish (Danio rerio)34–36. In a Galleria mellonella model of infection, a warA mutant displays attenuated pathogenicity when compared to the wild type strain. A similar attenuation in pathogenicity was observed with a sadC deletion mutant thus supporting the link between these components (Sup. Fig. 5a). LPS is one of the primary pathogen associated molecular patterns (PAMPS) that is capable of triggering the host innate immune response and is known to directly stimulate the production of pro-inflammatory cytokines. To visualize P. aeruginosa interactions with innate immune cells, Galleria were inoculated with GFP-tagged P. aeruginosa strains and the hemolymph, the invertebrate equivalent to blood, was exsanguinated 16 hours post infection (hpi). Analysis by fluorescent microscopy suggested that more hemocytes, a primitive arthropod immune cell, were in direct contact with both sadC and warA mutant bacterial cells as compared to the wild type levels of hemocyte contact. This observation implied that potentially both mutants were more recognisable to the arthropod innate immune system (Sup. Fig. 5b).
To investigate the vertebrate innate immune response to P. aeruginosa, a Pseudomonas- zebrafish hindbrain infection model was developed34,36. Following injection into the hindbrain ventricle, zebrafish larvae showed P. aeruginosa PAK dose-dependent mortality (Sup. Fig. 6). In agreement with the Galleria survival assays, significantly higher mortality rates were observed in zebrafish larvae infected with the wild type strains as compared to either mutant strain (Fig. 5a). Deletion of warA or sadC did not reduce bacterial load (Sup. Fig. 7) suggesting that an altered immune response (and not bacterial replication) is responsible for host susceptibility to P. aeruginosa infection. To directly visualize the innate immune response, we infected Tg(lyz:dsRed)nz50 zebrafish larvae, a transgenic line in which dsRed is expressed specifically in neutrophils37, with GFP-tagged P. aeruginosa strains and recorded neutrophil behavior using fluorescent microscopy (Fig. 5c). Strikingly, a more robust neutrophil response is observed during sadC mutant infections with more neutrophils recruited to the site of infection as compared to larvae infected with wild type or complemented bacteria (Fig. 5b,c) (Sup. Vid.1-3, Sup. Fig. 8a). This phenotype of significantly increased neutrophil recruitment was also observed for a warA deletion mutant, further supporting the link between these two components. In agreement with our results from the Galleria model, our Pseudomonas-zebrafish infection model confirmed that both sadC and a warA mutant are more recognisable to host immune cells than wild type bacteria.
Figure 5. Role of SadC and WarA in infection and immune evasion.
a, Survival analysis of zebrafish 3 days post fertilization (dpf) infected in the hindbrain ventricle with P. aeruginosa at a dose of 5 x 104 CFU. Zebrafish challenged with either mutant displayed a reduced mortality at 12 hpi (12 hpi, ANOVA, *, P<0.05). Data is representative of at least 3 independent biological replicates with a minimum of at least 24 fish per replicate. b, Representative images of lyz:dsRed larvae infected in the hindbrain with PAK::gfp, PAKΔsadC::gfp and PAKΔwarA::gfp from panel C. For each treatment, the same larva was imaged at 0, 6 and 12 hpi using a fluorescent stereomicroscope, where neutrophils were quantified at the site of infection (as defined by the white box, enlarged as inset image) (See also Video S1, S2 and S3).c, Neutrophils were counted around the site of infection at 0 and 6 hours hpi for zebrafish larvae infected in the hindbrain ventricle with 5 x 104 CFU of PAK::gfp, PAKΔsadC::gfp and PAKΔsadC::gfp +psadC. Data is from 4 independent biological replicates with 3-6 larvae per strain per replicate. Standard deviation error bars are shown. Significance testing performed by Student’s t test: *, P<0.05; **, P<0.01. d, TNF-α mRNA levels in larvae infected with 5 x 104 CFU for 6h. TNF-α mRNA are relative to the house keeping gene ef1α. Mean ± SEM (horizontal bars) from 3 independent experiments with 5 pooled larvae per strain per experiment. Significance testing performed by Student’s t test: *, P<0.05; **, P<0.01.
The increased neutrophil recruitment response in both mutants suggests that the shift in the LPS modal distribution can impact the ability of the pathogen to establish infection, and implicated the SadC/WarA complex in regulating how P. aeruginosa is perceived by the host immune system. To explore this altered immune response, and test if it is due to the altered LPS profile displayed by both mutants, RNA was isolated from zebrafish larvae 6 hpi. This analysis revealed that the sadC mutant significantly induced the expression of the LPS associated pro-inflammatory cytokine TNF-α to levels higher than those observed in a wild-type or complemented infected host (Fig. 5d). Infection with a warA mutant also induced TNF-α mRNA levels significantly higher than wild type infected larvae (Sup. Fig. 8b). These data strongly suggest that the regulation of LPS by SadC and WarA plays a critical role in immune cell evasion during infection.
Discussion
The discovery of c-di-GMP as a second messenger molecule was based on key data that eluciated its ability to bind to a specific cellulose synthase and modulate its activity38,39. This interaction with c-di-GMP was shown to occur at a binding interface termed the PilZ domain and until recently it was believed to be one of the only c-di-GMP binding domains outside of those directly associated with a diguanylate cyclase domain40. Recently however, an increasingly diverse array of proteins have been identified that are capable of interacting with nucleotide based second messengers to such an extent that in silico prediction of nucleotide binding sites is no longer feasible28,41–43. The advancement in our understanding of which proteins can bind c-di-GMP and the molecular consequence of this binding is shedding further light on the complexity of c-di-GMP signalling and confirming the long held hypothesis that spatio-temperal fluctuations in the concentrations of this second messenger can have a localized molecular and kinetic impact7,44.
In this study we identified WarA, as a previously unidentified interacting partner of the DGC SadC, an enzyme previously shown to play a central role in the switch from acute to chronic infection phenotypes1,9. WarA was shown to specifically interact with the cytoplasmic part of SadC and form a complex with WarB encoded by a convergently transcribed gene. These two proteins in complex had a striking degree of structural homology to the E. coli O9a LPS O antigen regulator WbdD13. Upon further investigation it was demonstrated that they also maintain a degree of functional homology with WbdD, in that loss or overexpression of WarA had impact on CPA O antigen modal distribution. Intriguingly, the deletion of sadC had an identical impact on CPA O antigen modal distribution as loss of warA suggesting that the interaction of SadC and WarA may be linked in the regulation of LPS biogenesis. Many of the components necessary for CPA O antigen biosynthesis in P. aeruginosa are conserved in E. coli but no WbdD homolog has been identified leaving the question open as to how CPA O antigen biosynthesis is terminated? Based on structural homology, functional analysis and interactome homology it is likely that WarA is performing a similar function to E. coli O9a WbdD (Fig. 6). E.coli O9a WbdD has been shown to be the key enzyme signalling the end of O antigen synthesis and priming the O antigen for export across the inner membrane. WarA appears to have lost some of this functionality as the LPS O antigen is still synthesized in the absence of WarA, albeit at a different average chain length. This is not surprising however, as even within E. coli species the precise nature and level of regulation of LPS length and export priming exhibited by WbdD can vary and little is known about the functionality of WbdD homologs outside E. coli18.
Figure 6. Role of WarA in CPA biosynthesis.
SadC recruits WarA to the inner membrane where it interacts with the CPA O antigen machinery by recruiting WbpX to the assembly interface and also by potentiating the methyltransferase activity of WarA. This fine tuning regulation plays a key role in allowing the cell to escape detection by the host innate immune system.
C-di-GMP is known to promote a wide variety of chronic infection features such as biofilm formation1,9. One specific chronic infection phenotype that has not been previously linked to c-di-GMP levels however, is LPS O antigen biosynthesis. Chronic P. aeruginosa isolates from the CF lung are known to display an altered LPS profile. This altered LPS profile is believed to play a role in allowing these isolates to subvert detection by the host immune system and establish chronic infection, however the molecular mechanisms underpinning this LPS profile shift have not been elucidated and have previously been thought to be exclusively the result of genetic mutation2,45. Given the proximal localization of WarA/SadC with the LPS machinery and the impact of c-di-GMP levels on the activity of WarA (Fig 6), it is plausible that WarA and its enzyme kinetics are playing a key role in modifying the LPS profile of P. aeruginosa to better subvert the host innate immune response. We provide evidence using both Galleria and zebrafish models of infection which suggests this is indeed the case.
Our study has established another layer in the intricate network of physiological impacts mediated by c-di-GMP at the host-pathogen interface, whereby the levels of c-di-GMP can modulate the activity of an enzyme involved in the synthesis of the key PAMP, LPS. The extent of this impact is mediated at the protein-protein level through the interaction of SadC with WarA, likely recruiting WarA to the membrane, and at the kinetic level by the impact of c-di-GMP binding on the enzymatic activity of WarA. The biological consequence of loss of either of these components is highly impacting the success of the infection notably as observed by a perturbation in the ability to avoid neutrophil recruitment and to escape the innate immune response.
Materials & Methods
Bacterial Strains
Primers used in this study are listed in Supplementary Data Table 1, while bacterial strains and plasmids used in this study are listed in Supplementary Data Table 2. Cells were grown in Lysogeny Broth (LB) 46,47 in 100 ml Erlenmeyer flasks filled with 20 ml of medium, with shaking at 180 rpm and at 37°C. LB agar (NA) was used as a solid medium. When required, antibiotics were added to these media at the following concentrations: 100 μg/ml ampicillin, and 10 μg/ml gentamicin for E. coli; and 300 μg/ml carbenicillin, 50 μg/ml gentamicin and 125 μg/ml tetracycline, 2000 μg/ml streptomycin for P. aeruginosa.
Cloning procedures
DNA cloning and plasmid preparation were performed according to standard methods 48. PCR primers were designed with restriction sites at their ends for subsequent digestion and ligation into the specific vector. Restriction and DNA-modifying enzymes were used following the instructions of the manufacturers. Transformation of E. coli DH5α, E. coli TOP10 (for cloning) and P. aeruginosa was carried out by electroporation49. All plasmids were verified by sequencing. All mutations were confirmed by DNA sequencing. The background strains were tagged with GFP by means of four-parental mating with E. coli harbouring pBK-miniTn7-ΩGm as donor strains and with E. coli strains harbouring pUX-BF13 and pRK2013 as helper strains.
Gene Replacement Mutants
For the deletion of the PA4379 (warA) gene in the P. aeruginosa PAK chromosome previously described protocol was followed. Briefly, a fragment containing the upstream region and the first 2 codons of PA4379 and a fragment downstream containing the PA4379 stop codon were amplified by PCR using the primer pairs pPA4379.1/ pPA4379.2 and pPA4379.3/ pPA4379.4, respectively. Mutator fragments were constructed by PCR amplification of upstream and downstream fragments using the primer couple p PA4379.1/p PA4379.4 and cloned into pCR2.1-BLUNT. The mutator fragment was digested from pCR2.1-Blunt and ligated into pKNG101. This construct was transformed into E. coli CC18, was then introduced into P. aeruginosa PAK by triparental mating, using the helper strain E.coli HB101 (pRK2013). Transconjugants were isolated on Pseudomonas Isolation Agar (Difco) supplemented with appropriate antibiotics. Deletion mutants were selected in 5% sucrose after 2 days of incubation at room temperature. Deletions were confirmed by sequencing using external primers pPA4379.5/ pPA4379.6. The resulting strain PAKΔwarA, carried an in-frame ΔPA4379 mutation. Overexpression constructs were generated using the primer combinations p pPA4379OeF/pPA4379OeF and cloned into the isopropyl β-d-thiogalactoside (IPTG) inducible vector pMMB67HE. The mutated WarA construct was generated using the primer combination pWarAMutF/pWarAMutR.
β-Galactosidase assays
β-Galactosidase experiments were performed as described previously 47, with E. coli cells harvested from agar plates and resuspended in PBS . Data are mean values of three independent samples ± standard deviations.
Expression and Protein Purification
The warA (PA4379),warB (PA4378) and sadC genes and were amplified from P. aeruginosa PAK genomic DNA using the following primer combinations pWarAF.1/pWarAF.2, pWarBF.1/pWarBF.2,pSadCCDH.1/pSadCCDH.2 and cloned into pET28a with SadC CD and WarA having an N terminal His Tag and WarB having a C terminal His tag. For GST tagging of WarA and WarB the following primer combinations were used pWarAGST.1/pWarAGST.2 and pWarBGST.1/pWarBGST.2 and cloned into pET-41a-3CD, a vector derived from pET-41a by Novagen to produce fusion proteins with N-terminal GST-His-S tag. In all cases, transformed E. coli B834(DE3) cells were grown at 37 °C to an OD600 of about 0.6 in Tryptic Soy broth. Expression of proteins was induced with 0.5 mM IPTG, and cells were grown overnight at 18 °C before centrifugation (4,000 × g, 20 min at 4 °C). Cell pellets were resuspended in buffer A (50 mM Tris-HCl, 500 mM NaCl, 20 mM imidazole (pH 8.0)) and lysed by sonication after the addition of an anti-protease mixture (Sigma). Cell debris was eliminated by centrifugation (18,000 × g, 50 min).
Proteins were purified by IMAC chromatography using nickel-Sepharose resin (GE Healthcare) equilibrated in buffer A. Proteins were eluted with buffer A containing 500 mM instead of 20 mM imidazole and if necessary were further purified by size exclusion chromatography using a HiLoad Superdex 75 column equilibrated in 50 mM Tris-HCl and 250 mM NaCl (pH 8). All chromatographic steps were performed on an ÄKTAprime Plus system (GE Healthcare) at 4 °C. Protein purity was checked by SDS-PAGE. Proteins were dialyzed overnight in a low sodium buffer (250 mM) with no added imidazole and concentrated using centrifugal concentrators (Millipore) and stored at −80 °C.
Differential Radial Capillary Action of Ligand Assay (DRaCALA)
This assay is based on the ability of nitrocellulose to separate free ligand from bound protein-ligand complexes, allowing the determination of a protein ligand interaction. DRaCALA assays were performed as described by Roelofs et al. 27. Briefly, 32P-labelled c-di-GMP is generated from [α-32P]-GTP using purified WspR. Purified WarAHis in binding buffer was mixed with 4nM 32P-labeled c-di-GMP. 100 μM of unlabelled nucleotides was added to the reaction mixture containing 50 μM WarA in the cold competition experiments. These mixtures were pipetted (2.5 μl) onto dry untreated nitrocellulose (GE Healthcare) in triplicate and allowed to dry for ten minutes. An FLA7100 Fujifilm Life Science PhosphorImager was used to detect luminescence following a 10-min exposure of blotted nitrocellulose to phosphorimager film. Data were quantified using ImageQuant TL Toolbox v8.1. Nonlinear regression analysis determined binding Kd using the saturation binding equation Y = Bmax*X/(Kd + X) as described by Roelofs et al., 2011. Data shown are one representative experiment of three independent experiments.
Bacterial two-hybrid assay
Bacterial two hybrid experiments and cloning strategies were performed as previously described10,50,51. DNA fragments encoding proteins of interest were cloned into pKT25 and PUT18c. DNA regions encoding WarA, WarB, SadC CT, WbpX, WbpY and WbpZ were amplified by using the following primer combinations pSadCCD.1/pSadCCD.2 pPA4378.1/pPA4378.2, pSadCFL.1/pSadCFL.2, pPA4379B2H.1/ pPA4379B2H.2, pWbpY.1/pWbpY.2, pWbpX.1/pWbpX.2, pWbpZ.1/pWbpZ.2. Recombinant pKT25 and pUT18C plasmids were transformed simultaneously into the E. coli DHM1 strain and transformants were spotted onto LB agar plates supplemented with 1mM IPTG in the presence of 100 μg/ml ampicillin, 50 μg/ml kanamycin, and 100 μg/ml 5-bromo-4-chloro-indolyl-β-d-galactopyranoside (X-gal). Positive interactions were identified as blue colonies after 24h incubation at 30°C and quantified by β-galactosidase assays. The positive controls used in the study were pUT18C and pKT25 derivatives encoding the leucine zipper from GCN4, which strongly dimerizes (zip).
LPS Analysis
LPS was prepared as described by Hitchcock and Brown (1983) and visualized on 12% SDS-Page gels using the ultrafast silver staining method 52. For LPS Western Immunoblotting, LPS was transferred onto BioTraceNT nitrocellulose membranes (Pall), and probed using monoclonal antibody (MAb) N1F10 (CPA specific), and MAb MF15-4 (OSA specific). The secondary antibody used was alkaline phosphatase-conjugated goat anti-mouse Fab2 (Jackson ImmunoResearch), and the blots were developed using nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolylphosphate (BCIP) as described previously 53,54. For further LPS purification, total membranes were isolated. In brief, bacterial cells were collected by centrifugation and suspended in 1ml 50 mM Tris pH 8.0 supplemented with complete® protease inhibitor cocktail (Roche) and 1mM EDTA Cells were lysed by sonication. Intact cells were removed by centrifugation 4000 x g for 5 mins at 4 °C, taking uppermost supernatant each time. Soluble and membrane fractions were separated by centrifugation at 100,000g for 1h at 4 °C. Pellet was taken as total membrane and further treated as described by Hitchcock and Brown (1983).
Far Western Dot Blots
Far Western Blots were performed as described in 11. In brief, purified His tagged WarA, SadC Cytoplasmic Domain and WspR were spotted onto a nitrocellulose membrane (GE Healthcare) at comparable concentrations. The spots were let dry before the membrane was blocked with 5% milk in PBST (4 mM KH2PO4, 16 mM Na2HPO4, 115 mM NaCl (pH 7.4) and 0.05% Tween-20) for 1h at room temperature and then incubated overnight at 4C in Protein-binding buffer (100 mM NaCl, 20 mM Tris (pH 7.6), 0.5 mM EDTA, 10% glycerol, 0.1% Tween-20, 2% skim milk powder and 1 mM DTT) with 15 μg of purified GST tagged WarA or WarB. The following day, the membrane was washed three times with TBST to wash off unbound WarAGST /WarBGST. Membrane-bound WarAGST/WarBGST was detected by using monoclonal anti-GST antibody (GE Healthcare) at a dilution 1:1000. Secondary antibody conjugated to horseradish peroxidase was used at a dilution of 1:5000. Western blots were developed using Super-Signal West Pico Chemiluminescent Substrate (Pierce) and visualized on a LAS3000 Fuji Imager.
Methyltransferase Activity Assay
Methyltransferase activity assays were performed using a methyltransferase activity kit (Enzo Life Science, Farmingdale, NY) as per manufacturer’s specifications using S-adenosyl-L-methionine as the methyl group donor and measuring activity in relative fluorescent units. Assays were performed using Ni-NTA affinity column purified WarA that had been dialysed in a low sodium buffer to remove imidazole. This low sodium buffer was also used to resuspended lyophilized c-di-GMP (BioLog) and further purified LPS from a ΔwarA mutant. Data shown are one representative experiment of three independent experiments.
Galleria Assay
The wax moth model Galleria mellonella was used for in vivo pathogenicity assessment. Overnight cultures were diluted 1:100 and subcultured (10ml LB/ 50ml conical flask) at 37°C until early exponential phase. Bacterial cells (OD600= 0.7-0.8) are centrifuged (8,000 rpm, 4°C, 5min) and resuspended to PBS three times to a final concentration OD600=1. The samples were serially diluted (up to 10−8).10μl of the 10–6 dilution were injected into the last abdominal proleg of ten larvae per sample; additional ten larvae were injected with PBS as negative control. 20μl of the dilutions were spotted and enumerated the next day. The larvae mortality was monitored for 48h on an hourly basis (Note: dead larvae turn black due to melanisation and do not respond to tapping). Three biological replicates were produced. For fluorescent microscopy Harding et al,55 was followed with Galleria being inoculated with PAK::gfp, PAKΔwarA::gfp and PAKΔsadC::gfp and exsanguinated 16 hours post infection.
Ethics statement
Animal experiments were performed according to the Animals (Scientific Procedures) Act 1986, and were fully approved by the Home Office (Project license: PPL 70/7446). Animal experiments performed at Imperial College were performed according to the Animals (Scientific Procedures) Act 1986, and were fully approved by the Home Office (Project license: PPL 70/7446). All experimentation involving zebrafish have been performed in larvae up to 5 days post fertilization (ie what is legally considered as in vitro experimentation). To minimize the effects of subjective bias some experiments were performed by different lab members.
Zebrafish care and maintenance
Wild type AB purchased from the Zebrafish International Resource Center (Eugene, OR) and the Tg(lyz:dsRed)nz50 transgenic zebrafish line have been previously described 37. Eggs were obtained by placing egg collection baskets inside the breeding tanks, bleached according to protocols described in 56, and then kept in Petri dishes containing E2 water supplemented with 0.3μg/ml of methylene blue and from 24hpf onwards for microscopy purposes with 0.003% 1-phenyl-2-thiourea (Sigma-Aldrich) to prevent melanization.
Zebrafish bacterial infection
Hindbrain infections were performed as described by Rocker et al.,57 Bacterial strains used in this study were PAK::gfp, PAKΔsadC::gfp and PAKΔwarA::gfp. Bacteria were cultured overnight in TSB, diluted to an A600nm 0.01 and cultured until A600nm = 0.7. For injection of zebrafish larvae, bacteria were recovered by centrifugation, washed and reconstituted at the desired concentration in 1x PBS 0.1% phenol red. 3dpf anesthetized zebrafish larvae were microinjected in the hindbrain ventricle with 1-2nl of bacterial suspension.
Quantification of bacterial burden
At the indicated times, larvae were sacrificed in tricaine, lysed in 200μl of 0.4% Triton X-100 and homogenized mechanically58. Larval homogenates were serially diluted and plated on LB: Gm50 agar and plates were incubated at 37°C. Only larvae having survived the infection were included for CFU enumeration.
Live imaging, image processing and analysis
Whole-animal in vivo imaging was performed on anaesthetized zebrafish larvae immobilized in 1% low melting point agarose in 60mm Petri dishes covered with E2 containing tricaine as previously described 59. Transmission and fluorescence microscopy was done using a Leica M205FA fluorescent stereomicroscope. Imaging was performed at 28°C with a 10x (NA 0.5) dry objective. Multiple-field Z-stacks were acquired every 15min. AVI files were processed and annotated using ImageJ software. Neutrophils were quantified by defining a consistent area around the site of infection (as defined by white box enlarged in inset image) and counting neutrophils within this area.
qRT-PCR
Total RNA from snap-frozen larvae was extracted using RNAqueous Kit (Ambion). cDNA was obtained using QuantiTect reverse transcription kit (Qiagen). Primers used to measure TNF-α mRNA levels were from Stockhammer et al. 60. Quantitative PCR was performed on a Rotor-GeneQ (Qiagen) using SYBR green reaction power mix (Applied Biosystems). Quantifications were performed on triplicate wells. To normalize cDNA amounts, we used the housekeeping gene ef1α 58.
Data Availability
The data that support the findings of this study are available from the corresponding author on reasonable request.
Supplementary Material
Acknowledgements
Work in Alain Filloux’s laboratory was supported by the BBSRC grant BB/L007959/1. Work in the Mostowy laboratory is supported by a Wellcome Trust Research Career Development Fellowship (WT097411MA) and the Lister Institute of Preventive Medicine. Work in the JSL lab is supported by an operating grant from the Canadian Institutes of Health Research (MOP-14687). YH is a recipient of a postdoctoral fellowship from Cystic Fibrosis Canada and JSL holds a Canada Research Chair in Cystic Fibrosis and Microbial Glycobiology funded by the Canadian Foundation of Innovation. We thank Meng Jun for construction of the following plasmids pUT18C_wbpX, pUT18C_wbpY, pUT18C_wbpZ and testing them in the BTH system. We thank Vincent Lee for the gift of the plasmid to express and purify WspR, Anastasia Mylona for the gift of the plasmid pET-41a-3CD and Angelika Grundling for the provision of un-labelled nucleotides.
Footnotes
Author contributions. AF and RMC conceived and designed experiments, and wrote the paper. JSL and YH contributed materials including anti-LPS monoclonal antibodies, performed LPS analysis and helped with editing of the manuscript. JAM and CB performed the bacterial two-hybrid screen. MJMM and SM contributed zebrafish experiments. RMC conducted all experiments.
Competing Interests
The authors declare no competing financial interests.
References
- 1.Moscoso JA, Mikkelsen H, Heeb S, Williams P, Filloux A. The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environmental microbiology. 2011;13:3128–3138. doi: 10.1111/j.1462-2920.2011.02595.x. [DOI] [PubMed] [Google Scholar]
- 2.Lam MY, et al. Occurrence of a common lipopolysaccharide antigen in standard and clinical strains of Pseudomonas aeruginosa. Journal of clinical microbiology. 1989;27:962–967. doi: 10.1128/jcm.27.5.962-967.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Islam ST, Lam JS. Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Canadian journal of microbiology. 2014;60:697–716. doi: 10.1139/cjm-2014-0595. [DOI] [PubMed] [Google Scholar]
- 4.Bellini D, et al. Crystal structure of an HD-GYP domain cyclic-di-GMP phosphodiesterase reveals an enzyme with a novel trinuclear catalytic iron centre. Molecular microbiology. 2014;91:26–38. doi: 10.1111/mmi.12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan RP. Cyclic di-GMP signalling and the regulation of bacterial virulence. Microbiology. 2013;159:1286–1297. doi: 10.1099/mic.0.068189-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sundriyal A, et al. Inherent regulation of EAL domain-catalyzed hydrolysis of second messenger cyclic di-GMP. The Journal of biological chemistry. 2014;289:6978–6990. doi: 10.1074/jbc.M113.516195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiology and molecular biology reviews : MMBR. 2013;77:1–52. doi: 10.1128/mmbr.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mulcahy H, et al. Pseudomonas aeruginosa RsmA plays an important role during murine infection by influencing colonization, virulence, persistence, and pulmonary inflammation. Infection and immunity. 2008;76:632–638. doi: 10.1128/IAI.01132-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moscoso JA, et al. The diguanylate cyclase SadC is a central player in Gac/Rsm-mediated biofilm formation in Pseudomonas aeruginosa. Journal of bacteriology. 2014;196:4081–4088. doi: 10.1128/JB.01850-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Houot L, Fanni A, de Bentzmann S, Bordi C. A bacterial two-hybrid genome fragment library for deciphering regulatory networks of the opportunistic pathogen Pseudomonas aeruginosa. Microbiology. 2012;158:1964–1971. doi: 10.1099/mic.0.057059-0. [DOI] [PubMed] [Google Scholar]
- 11.Wu Y, Li Q, Chen XZ. Detecting protein-protein interactions by Far western blotting. Nature protocols. 2007;2:3278–3284. doi: 10.1038/nprot.2007.459. [DOI] [PubMed] [Google Scholar]
- 12.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protocols. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagelueken G, et al. Structure of WbdD: a bifunctional kinase and methyltransferase that regulates the chain length of the O antigen in Escherichia coli O9a. Molecular microbiology. 2012;86:730–742. doi: 10.1111/mmi.12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yokota S, Kaya S, Kawamura T, Araki Y, Ito E. The structure of the O-specific chain of lipopolysaccharide from Pseudomonas aeruginosa IID 1008 (ATCC 27584) Journal of biochemistry. 1986;99:1551–1561. doi: 10.1093/oxfordjournals.jbchem.a135628. [DOI] [PubMed] [Google Scholar]
- 15.Arsenault TL, et al. Structural Studies on the Polysaccharide Portion of a-Band Lipopolysaccharide from a Mutant (Ak1401) of Pseudomonas-Aeruginosa Strain Pao1. Can J Chem. 1991;69:1273–1280. doi: 10.1139/V91-190. [DOI] [Google Scholar]
- 16.Hao Y, King JD, Huszczynski S, Kocincova D, Lam JS. Five new genes are important for common polysaccharide antigen biosynthesis in Pseudomonas aeruginosa. mBio. 2013;4:e00631–00612. doi: 10.1128/mBio.00631-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke BR, Greenfield LK, Bouwman C, Whitfield C. Coordination of polymerization, chain termination, and export in assembly of the Escherichia coli lipopolysaccharide O9a antigen in an ATP-binding cassette transporter-dependent pathway. The Journal of biological chemistry. 2009;284:30662–30672. doi: 10.1074/jbc.M109.052878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clarke BR, Cuthbertson L, Whitfield C. Nonreducing terminal modifications determine the chain length of polymannose O antigens of Escherichia coli and couple chain termination to polymer export via an ATP-binding cassette transporter. The Journal of biological chemistry. 2004;279:35709–35718. doi: 10.1074/jbc.M404738200. [DOI] [PubMed] [Google Scholar]
- 19.Belanger M, Burrows LL, Lam JS. Functional analysis of genes responsible for the synthesis of the B-band O antigen of Pseudomonas aeruginosa serotype O6 lipopolysaccharide. Microbiology. 1999;145(Pt 12):3505–3521. doi: 10.1099/00221287-145-12-3505. [DOI] [PubMed] [Google Scholar]
- 20.Rocchetta HL, Burrows LL, Lam JS. Genetics of O-antigen biosynthesis in Pseudomonas aeruginosa. Microbiology and molecular biology reviews : MMBR. 1999;63:523–553. doi: 10.1128/mmbr.63.3.523-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Homonylo MK, Wilmot SJ, Lam JS, MacDonald LA, Whitfield C. Monoclonal antibodies against the capsular K antigen of Escherichia coli (O9:K30(A):H12): characterisation and use in analysis of K antigen organisation on the cell surface. Canadian journal of microbiology. 1988;34:1159–1165. doi: 10.1139/m88-204. [DOI] [PubMed] [Google Scholar]
- 22.Hagelueken G, et al. A coiled-coil domain acts as a molecular ruler to regulate O-antigen chain length in lipopolysaccharide. Nature structural & molecular biology. 2015;22:50–56. doi: 10.1038/nsmb.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liston SD, et al. Domain interactions control complex formation and polymerase specificity in the biosynthesis of the Escherichia coli O9a antigen. The Journal of biological chemistry. 2015;290:1075–1085. doi: 10.1074/jbc.M114.622480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King JD, Kocincova D, Westman EL, Lam JS. Review: Lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate immunity. 2009;15:261–312. doi: 10.1177/1753425909106436. [DOI] [PubMed] [Google Scholar]
- 25.Hao Y, Murphy K, Lo RY, Khursigara CM, Lam JS. Single-Nucleotide Polymorphisms Found in the migA and wbpX Glycosyltransferase Genes Account for the Intrinsic Lipopolysaccharide Defects Exhibited by Pseudomonas aeruginosa PA14. Journal of bacteriology. 2015;197:2780–2791. doi: 10.1128/JB.00337-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang S, et al. Biosynthesis of the Common Polysaccharide Antigen of Pseudomonas aeruginosa PAO1: Characterization and Role of GDP-D-Rhamnose:GlcNAc/GalNAc-Diphosphate-Lipid alpha1,3-D-Rhamnosyltransferase WbpZ. Journal of bacteriology. 2015;197:2012–2019. doi: 10.1128/JB.02590-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roelofs KG, Wang J, Sintim HO, Lee VT. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15528–15533. doi: 10.1073/pnas.1018949108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trampari E, et al. Bacterial Rotary Export ATPases Are Allosterically Regulated by the Nucleotide Second Messenger Cyclic-di-GMP. The Journal of biological chemistry. 2015;290:24470–24483. doi: 10.1074/jbc.M115.661439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christen B, et al. Allosteric control of cyclic di-GMP signaling. The Journal of biological chemistry. 2006;281:32015–32024. doi: 10.1074/jbc.M603589200. [DOI] [PubMed] [Google Scholar]
- 30.Pier GB. Pseudomonas aeruginosa lipopolysaccharide: a major virulence factor, initiator of inflammation and target for effective immunity. International journal of medical microbiology : IJMM. 2007;297:277–295. doi: 10.1016/j.ijmm.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cryz SJ, Jr, Pitt TL, Furer E, Germanier R. Role of lipopolysaccharide in virulence of Pseudomonas aeruginosa. Infection and immunity. 1984;44:508–513. doi: 10.1128/iai.44.2.508-513.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldberg JB, Coyne MJ, Jr, Neely AN, Holder IA. Avirulence of a Pseudomonas aeruginosa algC mutant in a burned-mouse model of infection. Infection and immunity. 1995;63:4166–4169. doi: 10.1128/iai.63.10.4166-4169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Priebe GP, et al. The galU Gene of Pseudomonas aeruginosa is required for corneal infection and efficient systemic spread following pneumonia but not for infection confined to the lung. Infection and immunity. 2004;72:4224–4232. doi: 10.1128/IAI.72.7.4224-4232.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterman EM, et al. Neutralization of mitochondrial superoxide by superoxide dismutase 2 promotes bacterial clearance and regulates phagocyte numbers in zebrafish. Infection and immunity. 2015;83:430–440. doi: 10.1128/IAI.02245-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joyce SA, Gahan CG. Molecular pathogenesis of Listeria monocytogenes in the alternative model host Galleria mellonella. Microbiology. 2010;156:3456–3468. doi: 10.1099/mic.0.040782-0. [DOI] [PubMed] [Google Scholar]
- 36.Clatworthy AE, et al. Pseudomonas aeruginosa infection of zebrafish involves both host and pathogen determinants. Infection and immunity. 2009;77:1293–1303. doi: 10.1128/IAI.01181-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hall C, Flores MV, Storm T, Crosier K, Crosier P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC developmental biology. 2007;7:42. doi: 10.1186/1471-213X-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Amikam D, Benziman M. Cyclic diguanylic acid and cellulose synthesis in Agrobacterium tumefaciens. Journal of bacteriology. 1989;171:6649–6655. doi: 10.1128/jb.171.12.6649-6655.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayer R, et al. Polypeptide composition of bacterial cyclic diguanylic acid-dependent cellulose synthase and the occurrence of immunologically crossreacting proteins in higher plants. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:5472–5476. doi: 10.1073/pnas.88.12.5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amikam D, Galperin MY. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics. 2006;22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 41.Roelofs KG, et al. Systematic Identification of Cyclic-di-GMP Binding Proteins in Vibrio cholerae Reveals a Novel Class of Cyclic-di-GMP-Binding ATPases Associated with Type II Secretion Systems. PLoS pathogens. 2015;11:e1005232. doi: 10.1371/journal.ppat.1005232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Grundling A. c-di-AMP is a new second messenger in Staphylococcus aureus with a role in controlling cell size and envelope stress. PLoS pathogens. 2011;7:e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corrigan RM, et al. Systematic identification of conserved bacterial c-di-AMP receptor proteins. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9084–9089. doi: 10.1073/pnas.1300595110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lori C, et al. Cyclic di-GMP acts as a cell cycle oscillator to drive chromosome replication. Nature. 2015;523:236–239. doi: 10.1038/nature14473. [DOI] [PubMed] [Google Scholar]
- 45.Ojeniyi B, Lam JS, Hoiby N, Rosdahl VT. A comparison of the efficiency in serotyping of Pseudomonas aeruginosa from cystic fibrosis patients using monoclonal and polyclonal antibodies. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica. 1989;97:631–636. doi: 10.1111/j.1699-0463.1989.tb00454.x. [DOI] [PubMed] [Google Scholar]
- 46.Sezonov G, Joseleau-Petit D, D'Ari R. Escherichia coli physiology in Luria-Bertani broth. Journal of bacteriology. 2007;189:8746–8749. doi: 10.1128/jb.01368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller JH. Experiments in molecular genetics. 1972 [Google Scholar]
- 48.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. 1989. [Google Scholar]
- 49.Pessi G, Haas D. Transcriptional control of the hydrogen cyanide biosynthetic genes hcnABC by the anaerobic regulator ANR and the quorum-sensing regulators LasR and RhlR in Pseudomonas aeruginosa. Journal of bacteriology. 2000;182:6940–6949. doi: 10.1128/jb.182.24.6940-6949.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bordi C, et al. Regulatory RNAs and the HptB/RetS signalling pathways fine-tune Pseudomonas aeruginosa pathogenesis. Molecular microbiology. 2010;76:1427–1443. doi: 10.1111/j.1365-2958.2010.07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fomsgaard A, Freudenberg MA, Galanos C. Modification of the silver staining technique to detect lipopolysaccharide in polyacrylamide gels. Journal of clinical microbiology. 1990;28:2627–2631. doi: 10.1128/jcm.28.12.2627-2631.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Kievit TR, Dasgupta T, Schweizer H, Lam JS. Molecular cloning and characterization of the rfc gene of Pseudomonas aeruginosa (serotype O5) Molecular microbiology. 1995;16:565–574. doi: 10.1111/j.1365-2958.1995.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 54.Blake MS, Johnston KH, Russell-Jones GJ, Gotschlich EC. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Analytical biochemistry. 1984;136:175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- 55.Harding CR, Schroeder GN, Collins JW, Frankel G. Use of Galleria mellonella as a model organism to study Legionella pneumophila infection. Journal of visualized experiments: JoVE. 2013:e50964. doi: 10.3791/50964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Brachydanio rerio) University of Oregon Press; Eugene: 1993. [Google Scholar]
- 57.Rocker AJ, Weiss AR, Lam JS, Van Raay TJ, Khursigara CM. Visualizing and quantifying Pseudomonas aeruginosa infection in the hindbrain ventricle of zebrafish using confocal laser scanning microscopy. Journal of microbiological methods. 2015;117:85–94. doi: 10.1016/j.mimet.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 58.Mostowy S, et al. The zebrafish as a new model for the in vivo study of Shigella flexneri interaction with phagocytes and bacterial autophagy. PLoS pathogens. 2013;9:e1003588. doi: 10.1371/journal.ppat.1003588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mazon Moya MJ, Colluci-Guyon E, Mostowy S. Use of Shigella flexneri to study autophagy-cytoskeleton interactions. Journal of visualized experiments : JoVE. 2014;91:e51601. doi: 10.3791/51601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stockhammer OW, Zakrzewska A, Hegedus Z, Spaink HP, Meijer AH. Transcriptome profiling and functional analyses of the zebrafish embryonic innate immune response to Salmonella infection. Journal of immunology (Baltimore, Md. : 1950) 2009;182:5641–5653. doi: 10.4049/jimmunol.0900082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author on reasonable request.