Abstract
In this study, we aimed to determine the presence as well as the diverse distribution of N-methyl-D-aspartate (NMDA) and non-NMDA glutamate receptor subunits in the rat red nucleus. Using adult Sprague-Dawley rats as the experimental animals, immunohistochemistry was performed on 30 µm thick coronal brain sections with antibodies against α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (GluA1-4), kainate (GluK1, GluK2/3, and GluK5), and NMDA (GluN1 and GluN2A) receptor subunits. The results showed that all ionotropic glutamate receptor subunits are expressed in the red nucleus. Specific staining was localized in the neuron bodies and processes. However, the pattern of immunoreactivity and the number of labeled neurons changed depending on the type of ionotropic glutamate receptor subunits and the localization of neurons in the red nucleus. The neurons localized in the magnocellular part of the red nucleus were particularly immunopositive for GluA2, GluA4, GluK2/3, GluK5, GluN1, and GluN2A receptor proteins. In the parvocellular part of the red nucleus, ionotropic glutamate receptor subunit immunoreactivity of variable intensity (lightly to moderately stained) was detected in the neurons. These results suggest that red nucleus neurons in rat heterogeneously express ionotropic glutamate receptor subunits to form functional receptor channels. In addition, the likelihood of the coexpression of different subunits in the same subgroup of neurons suggests the formation of receptor channels with diverse structure by way of different subunit combination, and the possibility of various neuronal functions through these channels in the red nucleus.
KEY WORDS: Red nucleus, parvocellular, magnocellular, rat, glutamate, neuronal function, GluA2, GluA4, GluK2/3, GluK5, GluN1, GluN2A, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid, kainate, N-methyl-D-aspartate, immunohistochemistry
INTRODUCTION
In vertebrates with limbs, red nucleus (Latin nucleus ruber) is an important part of central motor system and participates in maintaining body posture and controlling limb movements [1-4]. In addition, its participation in the regulation of antinociceptive functions has been proposed [5]. However, in humans, red nucleus has different structure and functional properties when compared to other mammalian species [6]. In rats, red nucleus is located on either side of the midbrain tegmentum and comprised of two parts: The parvocellular part (RPC) and the magnocellular part (RMC). RMC mainly contains large neurons, whereas RPC consists of small to medium neurons [7]. RMC and RPC receive afferent inputs from anterior interposed nucleus and dentate cerebellar nucleus, respectively [2,8,9]. Moreover, projections arisen from sensory-motor cortex supply RPC but do not innervate RMC [2]. The cell projections of magnocellular neurons organize in the rubrospinal tract and extend to the cervical and lumbosacral cord. Axons of the parvocellular neurons form the rubro-olivary pathway and project to the spinal cord [2,9]. RMC and RPC serve different functions in the red nucleus [2,4]. The magnocellular part of the red nucleus is involved in movements such as scratching or locomotion [2], in addition to the control of learned motor skills in rat [8,10]. Although less is known about the role of the parvocellular part, it is suggested that RPC is implicated in the automatization of learned movements [10] or in antinociceptive functions [5].
Glutamate is the major excitatory neurotransmitter of the mammalian central nervous system and it acts by binding to both, ionotropic and metabotropic, glutamate receptors [11]. Ionotropic glutamate receptor family includes N-methyl-D-aspartate (NMDA), α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA), and kainate subtypes [11-13]. While AMPA receptors are composed of four subunits designated as GluA1-4 [13,14], five kainate receptor subunits were identified, named GluK1-5 [13-15]. NMDA receptors consist of a mandatory subunit GluN1 in a combination with GluN2A-2D subunits [16,17]. The combination of different subunits in the formation of functional glutamate receptor channels results in different functional properties of the particular neurons [18].
Red nucleus receives innervation through two major excitatory pathways including the corticorubral and the cerebellorubral projections [2,19-21]. Electrophysiological studies using glutamate, glutamate agonist, and antagonist to investigate the role of excitatory amino acid receptors in excitatory neurotransmission in the red nucleus, have emphasized that glutamate acts as a neurotransmitter in both pathways [9,20-25]. In addition, biochemical [26], morphological [21,27], and immunohistochemical [28] studies have indicated that the glutamatergic system regulates the neurons of the red nucleus and that functional glutamate receptor channels are present in the red nucleus [29].
Considering the fact that the subunits of glutamate receptors are responsible for synaptic efficacy of glutamate, this study aimed to determine the presence and distribution pattern of ionotropic glutamate receptor subunit proteins in the rat red nucleus. Since the red nucleus comprises of two, functionally different, parts that differ both in cellular content and innervations, we conducted differential expression analysis of AMPA, kainate, and NMDA receptor subunits in these parts, using receptor immunohistochemistry.
MATERIALS AND METHODS
Animals
The study protocol was carried out in accordance with the Guide for the Care and Use of Laboratory Animal of the National Institutes of Health and approved by The Experimental Ethical Committee of Uludag University. Ten adult female Sprague-Dawley rats (200-250 g) were used in this study, provided by the Uludag University Experimental Animals Breeding and Research Center. The laboratory animals were housed two per cage on a light-dark cycle (12:12 hours) in temperature controlled environment (21°C). The animals were allowed to access food and water ad libitum.
Anesthetized rats were transcardially perfused with 0.9% saline followed by 4% paraformaldehyde (PFA) [pH 7.4]. The brains were removed, fixed in 10% sucrose prepared in 4% PFA overnight at 4°C and placed in 20% and 30% sucrose in phosphate buffered saline for 2 days at 4°C. Five series of 30 µm thick coronal brain sections throughout the brain stem were obtained on a cryostat. The sections were collected in Tris-HCl buffer (0.05 M, pH 7.6) and kept in cryoprotectant in glass vials at −20°C until being used.
Immunohistochemistry
Tris-HCl buffer was used for all washing steps. Primary and secondary antibodies were diluted in blocking buffer including 10% normal horse serum, 0.1% sodium azide, and 0.2% Triton X-100 in Tris-HCl buffer. All incubation steps were carried out on an orbital shaker with appropriate agitation.
For immunohistochemical staining, free-floating sections were washed 3 times in Tris-HCl buffer to remove cryoprotectant and blocked in blocking buffer for 2 hours. The sections were incubated in rabbit anti-GluA1 (1:250, AB1504, Chemicon, USA), mouse anti-GluA2 (1:500, MAB 397, Chemicon, USA), mouse anti-GluA3 (1:250, MAB5416, Chemicon, USA), rabbit anti-GluA4 (1:250, AB1508, Chemicon, USA), goat anti-GluK1 (1:250, sc-7617, Santa Cruz, USA) rabbit anti-GluK5 (1:2000, Jennes and Eyigor, R52-4) [30], and guinea pig anti-GluK2/3 (1:500, Jennes and Eyigor) [31], mouse anti-GluN1 (1:500, 556308, BD Pharmingen, USA) or mouse anti-GluN2A (1:3000, MAB5216, Millipore, USA) for 48 hours at room temperature. Before the incubation step of some primary antibodies, the sections were pre-treated with 1 mM ethylenediaminetetraacetic acid (pH 8, for GluN1 and GluN2A) and 50 mM trisodium citrate solution (pH 6, for GluK1) for antigen retrieval. The washed sections were processed with suitable biotinylated secondary antibodies raised in donkey (Jackson Immunoresearch Labs, West Grove, PA, USA) for 2 hours. Following the secondary antibody incubation step, the sections were first exposed to avidin-biotin complex (ABC Elite Standard Kit, Vector Labs, Burlingame, CA, USA) and then diaminobenzidine solution (50 mg, in 100 ml Tris-HCl buffer with 5 µl H2O2) was used as a chromogen to visualize the immunocomplex. After thorough washes, the sections were mounted on glass slides, dried, and coverslipped with DPX. Control experiments were performed by the omission of the primary antibody during the primary antibody incubation step and revealed a lack of specific staining. Representative photomicrographs of the control experiments for each primary antibody are shown in Figures 1S-V, 2P-R, and 3M-N.
FIGURE 1.
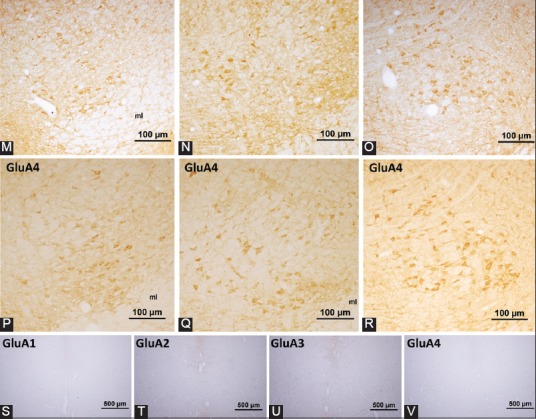
α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor subunit expression in the rat red nucleus. (A-C) represent the coronal levels of the selected sections from the brain atlas [32]. Representative images of these levels with cresyl violet staining are given in (D-F). Note that these images are taken with lower magnification to emphasize the localization and the structure of the red nucleus at a particular level. In (G-I) panels, GluA1-positive neurons can be observed. A small to moderate number of neurons was labeled with GluA1 antibody. GluA2 antibody labeled more neurons and demonstrated higher intensity in both parts of the red nucleus (J-L). Although the labeling intensity was higher, immunoreactivity for GluA3 was detected in a small number of neurons in the rostral part, whereas more neurons were GluA3-positive in the caudal part of the nucleus (M-O). (P-R) panels show the distribution of neurons that expressed GluA4 subunits. More GluA4-positive neurons are preferentially localized in the magnocellular part of the red nucleus. Specific staining was abolished in the control experiments (S-V). ml: Medial lemniscus; RMC: Magnocellular part of the red nucleus; RPC: Parvocellular part of the red nucleus; PaR: Pararubral nucleus; 3N: Oculomotor nucleus.
FIGURE 2.
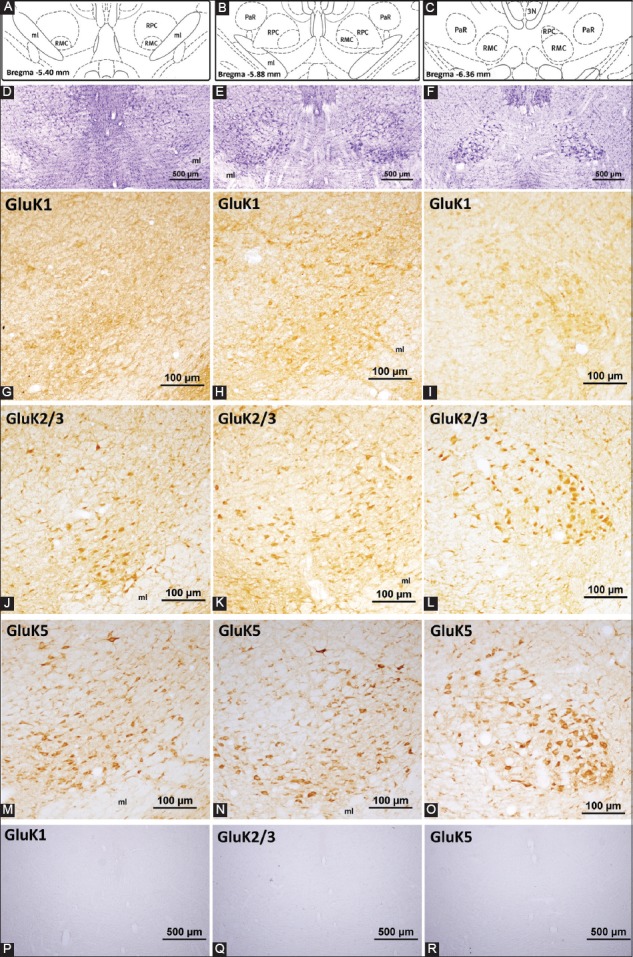
Kainate receptor subunit expression in the rat red nucleus. (A-C) represent the coronal levels of the selected sections from the brain atlas [32]. Representative images of these levels with cresyl violet staining are given in (D-F). Note that these images are taken with lower magnification to emphasize the localization and the structure of the red nucleus at a particular level. (G-I) panels show the GluK1-positive neurons in the red nucleus. Among all the kainate receptor subunits, GluK1 expression was detected in a very small number of neurons and the labeling intensity was the lowest. GluK2/3 antibody recognizes a common sequence in the respective proteins and the staining with this antibody revealed high immunoreactivity in a moderate to high number of neurons (J-L). Very high intensity in the immunoreactivity against GluK5 antibody was detected in both parts of the red nucleus. Moderate to high number of neurons was labeled (M-O). Specific staining was abolished in the control experiments (P-R). ml: Medial lemniscus; RMC: Magnocellular part of the red nucleus; RPC: Parvocellular part of the red nucleus; PaR: Pararubral nucleus; 3N: Oculomotor nucleus.
FIGURE 3.
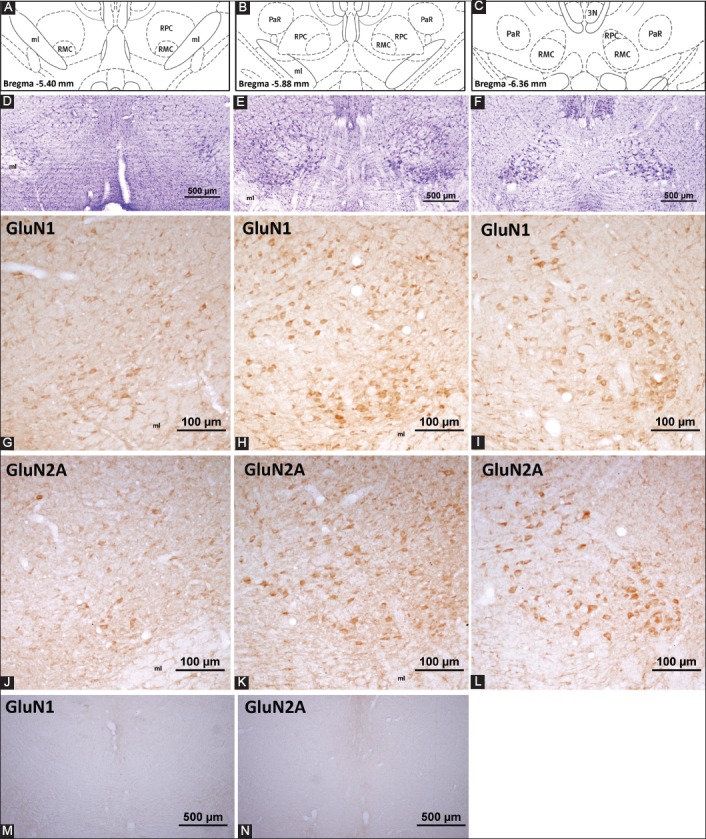
N-methyl-D-aspartate (NMDA) receptor subunit expression in the rat red nucleus. (A-C) represent the coronal levels of the selected sections from the brain atlas [32]. Representative images of these levels with cresyl violet staining are given in (D-F). Note that these images are taken with lower magnification to emphasize the localization and the structure of the red nucleus at a particular level. NMDA receptor subunit antibodies reacted with more neurons in the red nucleus and demonstrated the highest intensity when compared to other ionotropic glutamate receptor subunits. In (G-I) panels, GluN1-positive neurons are observed, note that the most of the neuronal perikarya in the caudal part are GluN1-positive. Neuropil staining was also observed. A moderate to high number of neurons expressed GluN1 in the parvocellular part of the red nucleus. The distribution pattern of the GluN2A-labeled neurons in the red nucleus is represented in panels (J-L). Although not as high as the GluN1 immunoreactivity, GluN2A labeling was dense in the red nucleus. A moderate to high number of GluN2A-positive neurons was detected in both the parvocellular and magnocellular parts of the red nucleus. Specific staining was abolished in the control experiments (M and N). ml: Medial lemniscus; RMC: Magnocellular part of the red nucleus; RPC: Parvocellular part of the red nucleus; PaR: Pararubral nucleus; 3N: Oculomotor nucleus.
Consecutive series of sections were stained with cresyl violet (0.1% cresyl violet acetate in distilled water, C5042, Sigma, St. Louis, MO, USA), to assess the quantity of the neurons in a particular red nucleus area and to make semi-quantitative comparison with the immunohistochemically labeled neurons which are localized in the same part of the nucleus.
Analysis
Red nucleus sections from the rostral-caudal extent (Bregma −5.16 mm and −6.60 mm, respectively [32]) were analyzed for the presence of immunopositive neurons and processed with Olympus BX-50 photomicroscope-attached CCD camera (Olympus DP71 CCD Color Camera, 1.5 million pixels, Olympus Corporation, Japan). The immunohistochemically labeled neurons were semi-quantitatively analyzed by comparing the number of immunopositive neurons with the number of neuronal bodies stained with cresyl violet (Figures 1D-F, 2D-F, and 3D-F) in the sections which represent the same levels of the red nucleus (Figures 1A-C, 2A-C, and 3A-C). The relative quantity of immunolabeled neurons was graded by the following scale: “+” was used for a small number of neurons, “++” for a moderate number of neurons, and “+++” for a high number of neurons. If the quantity of the labeled neurons matched or was close to the quantity of neuronal bodies stained with cresyl violet in the same area, “+++” grade was used. If the number of the labeled neurons was smaller than the number of cresyl violet-stained neurons then “+” grade was assigned. If approximately the half of neuronal population in an area was immunopositive for a particular antibody, then “++” grading was chosen.
RESULTS
The results showed that all ionotropic glutamate receptor subunits were expressed in the red nucleus (Figures 1-3). Specific staining was localized in neuronal perikarya as well as in the neuropil. The pattern of immunoreactivity and the number of stained neurons changed depending on the type of subunits. The neurons localized in the magnocellular part of the red nucleus were particularly immunopositive for GluA2, GluA4, GluK2/3, GluK5, GluN1, and GluN2A antibodies. In the parvocellular part of the red nucleus, the number of neurons labeled with the ionotropic glutamate receptor subunit antibodies varied between few to moderate (Table 1). The number of immunopositive neurons at different levels of the red nucleus (Figures 1A-C, 2A-C, and 3A-C) was compared to the number of neurons stained with cresyl violet (Figures 1D-F, 2D-F, and 3D-F).
TABLE 1.
Localization and numerical density of neurons expressing ionotropic glutamate receptor subunits in the rat red nucleus

AMPA receptor subunits
Dense immunoreactivity was detected for GluA2 (Figure 1J-L) and GluA4 (Figure 1P-R) in the large neurons of the magnocellular part of the red nucleus (Table 1). GluA1-positive neurons were specifically located in the dorsolateral and ventrolateral areas of the magnocellular part of the red nucleus (Figure 1G-I). A moderate number of GluA3-stained neuronal perikarya was detected in the caudal part of the nucleus (Figure 1M-O). In the parvocellular red nucleus, a limited number of neurons was stained with AMPA receptor subunit antibodies and neuropil staining was also detected. When the expression of AMPA receptor subunits in the parvocellular part of the red nucleus was compared, the highest was the number of neurons that expressed GluA1 and GluA2 subunits (Figure 1).
Kainate receptor subunits
The red nucleus was intensely labeled with antibodies against GluK2/3 (Figure 2J-L) and GluK5 (Figure 2M-O) receptor proteins; however, light immunopositivity was detected for GluK1 antibody (Figure 2G-I and Table 1). The labeling with the kainate receptor subunit GluK5 antibody revealed that the immunoreactive neurons were mostly localized in the middle and caudal magnocellular parts of the red nucleus (Figure 2N and O, respectively). GluK5 immunolabeling was apparent in the cell bodies and the proximal dendrites and GluK5-positive neurons were detected in the dorsomedial and ventrolateral neuron subgroups of the magnocellular part of the red nucleus. Furthermore, smaller neurons located in the rostral part of the red nucleus showed moderate GluK5 immunoreactivity.
While the distribution pattern of GluK2/3-positive neurons was similar to the pattern of GluK5-positive neurons, it was observed that GluK2/3 immunoreactivity was more prominent in the ventrolateral subgroup of neurons compared to the dorsomedial subgroup of neurons (Figure 2L). When compared to GluK1, more GluK2/3-positive neurons in the parvocellular part were detected (Figure 2).
NMDA receptor subunits
Many neuron bodies and proximal dendrites were densely labeled with NMDA receptor subunit antibodies (Figure 3). Furthermore, neuropil staining was observed in the red nucleus. In the magnocellular part, a high number of large- and medium-sized neurons were densely stained with GluN1 antibody (Figure 3G-I). The number of GluN2A-positive neurons varied from moderate to high in the red nucleus (Figure 3J-L). When compared to GluN1, GluN2A antibody caused more intense neuropil staining. In the parvocellular part of the red nucleus, a higher number of neurons was labeled with GluN1 and GluN2A antibodies when compared to the number of neurons labeled with other glutamate receptor antibodies used in this study (Figure 3 and Table 1).
DISCUSSION
This study investigated the detailed distribution pattern of neurons expressing ionotropic glutamate receptor subunit proteins in two parts of the rat red nucleus. Our results indicated that these neurons express all of the known ionotropic glutamate receptors subunits in varying quantities and intensities. To the best of our knowledge, this is the first descriptive study on the differential expression pattern of ionotropic glutamate receptors in the rat red nucleus. The previous studies demonstrated the presence of the glutamatergic innervation on red nucleus neurons [19,21,22]. Excitatory effects of glutamate regulate the function of the red nucleus in maintaining the body posture and controlling limb movements. Glutamate binds to and activates two distinct families of receptor channels including metabotropic and ionotropic receptors. Glutamatergic neurotransmission is shown to be via the ionotropic glutamate receptors rather than the metabotropic receptors in the red nucleus [24]. This study was focused on the ionotropic glutamate receptors and the expression pattern of the subunits of these receptors in the red nucleus. In situ hybridization as well as immunohistochemical studies showed that AMPA and NMDA receptor subunits of the ionotropic glutamate receptors are widely expressed in the midbrain and brain stem [28,29,33-36]; however, a report showing a detailed distribution of these receptors in the red nucleus is not available in the literature. Moreover, there is no evidence in the literature about the presence of kainate receptor subunits in the red nucleus. It is important to determine the expression of different glutamate receptor subunits in a particular central nervous system area, since glutamate receptors form heteromeric, in addition to homomeric, receptor channels combining different subunits. Results of this study showed that all ionotropic receptor subunits are expressed in the red nucleus to form functional heteromeric channels. This is important because different combinations of subunits result in different functions of the glutamate receptors.
Other studies showed that glutamate agonists activate AMPA receptors in the red nucleus [23], whereas antagonists of non-NMDA glutamate receptors block the neuronal activation [20,23,24]. This suggests the presence of functional AMPA receptors in the red nucleus which is confirmed by the results of our study, showing the expression of all four subtypes of AMPA receptors in the red nucleus. We found that the expression pattern of GluA2 and GluA3 subunits in the red nucleus is in agreement with previous reports [34]; however, a comparison of the distribution pattern of GluA1 and GluA4 subunits reported in this study with the pattern of immunolabeling demonstrated in the previous studies showed some differences. According to Petralia and Wenthold [34], the immunoreactivity for GluA1 antibody was intense. Moreover, another study reported light immunoreactivity for GluA4 subunit [28]. These discrepancies could be due to the different experimental animals used, i.e. the former study used male rats and the latter study utilized turtles compared to female rats used in our study. Contrary to the results of our study, Sato et al. [29] reported that GluA1 mRNA was not expressed in parvocellular neurons, while GluA2 mRNA was weakly detected in the neurons of both parts. This inconsistency could be because of the different molecular characterization techniques used in the two studies, i.e., Sato et al. [29] detected the mRNA expression, while in our study, the protein expression was analyzed.
Previous studies reported similar results with regard to the number of GluN1- and GluN2A-positive neurons and neuropil staining in the red nucleus [28,35,36]. Electrophysiological studies showed that NMDA receptors play an important role in conveying glutamatergic signals from cortical or cerebellar areas to the red nucleus, particularly to the magnocellular part [20,21]. Similar studies also showed that functional NMDA receptor channels are present in the neurons of the red nucleus [22-24]. The results of our study suggest that NMDA receptors, comprised of GluN1 and GluN2A subunits, may be the target of the glutamatergic innervation in the red nucleus.
For the first time in the literature, our study demonstrated the presence of kainate-preferring glutamate receptor subunit expression in the red nucleus. Kainate receptors are distinct receptor channels with different functional properties when compared to other glutamate receptors. We showed that all kainate receptor subunits are present in the red nucleus, to form heteromeric receptor channels. Further functional studies are necessary to determine the differential functions of these receptors in the red nucleus.
Since dual immunofluorescence labeling was not used, the results of this study lack information on the coexpression of subunits in a particular neuron in the red nucleus. Nevertheless, it is reasonable to suggest that the neurons in the red nucleus can coexpress different subunits, since our results showed that all of the subunit proteins are synthesized in the red nucleus neurons.
CONCLUSION
The results of this study showed that all the ionotropic glutamate receptor subunit proteins are synthesized by the neurons of the red nucleus. In addition to the NMDA and AMPA receptors, kainate-preferring glutamate receptor subunits are also present in the red nucleus. Together, this data suggests that homomeric or heteromeric functional receptor channels can be formed by these neurons to mediate the glutamatergic signals arising from the higher central nervous system areas. Further functional and pharmacological studies should determine diverse properties of these receptor channels with different combinations of their subunits.
DECLARATION OF INTERESTS
The authors declare no conflict of interests.
ACKNOWLEDGMENTS
We are grateful for the excellent technical assistance by Ayse Akbas, BSBio. GluK2/3 and GluK5 antibodies were kindly provided by Prof. Lothar Jennes, University of Kentucky (retired).
REFERENCES
- [1].Houk JC. Red nucleus: Role in motor control. Curr Opin Neurobiol. 1991;1(4):610–5. doi: 10.1016/s0959-4388(05)80037-6. [DOI] [PubMed] [Google Scholar]
- [2].Ruigrok TJH. Precerebellar nuclei and red nucleus. In: Paxinos G, editor. The Rat Nervous System. 3rd ed. Amsterdam: Elsevier Academic Press; 2004. pp. 167–204. http://dx.doi.org/10.1016/B978-012547638-6/50009-2. [Google Scholar]
- [3].Herter TM, Takei T, Munoz DP, Scott SH. Neurons in red nucleus and primary motor cortex exhibit similar responses to mechanical perturbations applied to the upper-limb during posture. Front Integr Neurosci. 2015;9:29. doi: 10.3389/fnint.2015.00029. http://dx.doi.org/10.3389/fnint.2015.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Morris R, Vallester KK, Newton SS, Kearsley AP, Whishaw IQ. The differential contributions of the parvocellular and the magnocellular subdivisions of the red nucleus to skilled reaching in the rat. Neuroscience. 2015;295:48–57. doi: 10.1016/j.neuroscience.2015.03.027. http://dx.doi.org/10.1016/j.neuroscience.2015.03.027. [DOI] [PubMed] [Google Scholar]
- [5].Peschanski M, Mantyh PW. Efferent connections of the subfascicular area of the mesodiencephalic junction and its possible involvement in stimulation-produced analgesia. Brain Res. 1983;263(2):181–90. doi: 10.1016/0006-8993(83)90311-6. http://dx.doi.org/10.1016/0006-8993(83)90311-6. [DOI] [PubMed] [Google Scholar]
- [6].Hicks TP, Onodera S. The mammalian red nucleus and its role in motor systems, including the emergence of bipedalism and language. Prog Neurobiol. 2012;96(2):165–75. doi: 10.1016/j.pneurobio.2011.12.002. http://dx.doi.org/10.1016/j.pneurobio.2011.12.002. [DOI] [PubMed] [Google Scholar]
- [7].Reid JM, Gwym DG, Flumerfelt BA. A cytoarchitectonic and Golgi study of the red nucleus in the rat. J Comp Neurol. 1975;162(3):337–61. doi: 10.1002/cne.901620305. http://dx.doi.org/10.1002/cne.901620306. http://dx.doi.org/10.1002/cne.901620305. [DOI] [PubMed] [Google Scholar]
- [8].Gruber P, Gould DJ. The red nucleus: Past, present, and future. Neuroanatomy. 2010;9:1–3. [Google Scholar]
- [9].Satoh Y, Tsuji K, Tsujimura T, Ishizuka K, Inoue M. Suppression of the swallowing reflex by stimulation of the red nucleus. Brain Res Bull. 2015;116:25–33. doi: 10.1016/j.brainresbull.2015.05.007. http://dx.doi.org/10.1016/j.brainresbull.2015.05.007. [DOI] [PubMed] [Google Scholar]
- [10].Kennedy PR. Corticospinal, rubrospinal and rubro-olivary projections: A unifying hypothesis. Trends Neurosci. 1990;13(12):474–9. doi: 10.1016/0166-2236(90)90079-p. http://dx.doi.org/10.1016/0166-2236(90)90079-P. [DOI] [PubMed] [Google Scholar]
- [11].Hollmann M, Heinemann S. Cloned glutamate receptors. Annu Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. http://dx.doi.org/10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- [12].Wisden W, Seeburg PH. Mammalian ionotropic glutamate receptors. Curr Opin Neurobiol. 1993;3(3):291–8. doi: 10.1016/0959-4388(93)90120-n. http://dx.doi.org/10.1016/0959-4388(93)90120-N. [DOI] [PubMed] [Google Scholar]
- [13].Collingridge GL, Olsen RW, Peters J, Spedding M. A nomenclature for ligand-gated ion channels. Neuropharmacology. 2009;56(1):2–5. doi: 10.1016/j.neuropharm.2008.06.063. http://dx.doi.org/10.1016/j.neuropharm.2008.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bettler B, Mulle C. Review: Neurotransmitter receptors. II. AMPA and kainate receptors. Neuropharmacology. 1995;34(2):123–39. doi: 10.1016/0028-3908(94)00141-e. http://dx.doi.org/10.1016/0028-3908(94)00141-E. [DOI] [PubMed] [Google Scholar]
- [15].Lerma J, Paternain AV, Rodríguez-Moreno A, López-García JC. Molecular physiology of kainate receptors. Physiol Rev. 2001;81(3):971–98. doi: 10.1152/physrev.2001.81.3.971. [DOI] [PubMed] [Google Scholar]
- [16].Sucher NJ, Awobuluyi M, Choi YB, Lipton SA. NMDA receptors: From genes to channels. Trends Pharmacol Sci. 1996;17(10):348–55. http://dx.doi.org/10.1016/S0165-6147(96)80008-3. [PubMed] [Google Scholar]
- [17].Paoletti P, Neyton J. NMDA receptor subunits: Function and pharmacology. Curr Opin Pharmacol. 2007;7(1):39–47. doi: 10.1016/j.coph.2006.08.011. http://dx.doi.org/10.1016/j.coph.2006.08.011. [DOI] [PubMed] [Google Scholar]
- [18].Seeburg PH. The TINS/TiPS lecture. The molecular biology of mammalian glutamate receptor channels. Trends Neurosci. 1993;16(9):359–65. doi: 10.1016/0166-2236(93)90093-2. http://dx.doi.org/10.1016/0166-2236(93)90093-2. [DOI] [PubMed] [Google Scholar]
- [19].Nieoullon A, Vuillon-Cacciuttolo G, Dusticier N, Kerkérian L, André D, Bosler O. Putative neurotransmitters in the red nucleus and their involvement in postlesion adaptive mechanisms. Behav Brain Res. 1988;28(1-2):163–74. doi: 10.1016/0166-4328(88)90093-9. http://dx.doi.org/10.1016/0166-4328(88)90093-9. [DOI] [PubMed] [Google Scholar]
- [20].Billard JM, Daniel H, Pumain R. Sensitivity of rubrospinal neurons to excitatory amino acids in the rat red nucleus in vivo. Neurosci Lett. 1991;134(1):49–52. doi: 10.1016/0304-3940(91)90506-o. http://dx.doi.org/10.1016/0304-3940(91)90506-O. [DOI] [PubMed] [Google Scholar]
- [21].Jiang MC, Alheid GF, Nunzi MG, Houk JC. Cerebellar input to magnocellular neurons in the red nucleus of the mouse: Synaptic analysis in horizontal brain slices incorporating cerebello-rubral pathways. Neuroscience. 2002;110(1):105–21. doi: 10.1016/s0306-4522(01)00544-9. http://dx.doi.org/10.1016/S0306-4522(01)00544-9. [DOI] [PubMed] [Google Scholar]
- [22].Davies J, Miller AJ, Sheardown MJ. Amino acid receptor mediated excitatory synaptic transmission in the cat red nucleus. J Physiol. 1986;376:13–29. doi: 10.1113/jphysiol.1986.sp016139. http://dx.doi.org/10.1113/jphysiol.1986.sp016139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Davies J, Qume M, Harris NC. Pharmacological characterisation of excitatory synaptic transmission in the cat red nucleus in vivo. Brain Res. 1994;649(1-2):43–52. doi: 10.1016/0006-8993(94)91047-2. http://dx.doi.org/10.1016/0006-8993(94)91047-2. [DOI] [PubMed] [Google Scholar]
- [24].Yang JC, Fan XL, Song XA, Li Q. The role of different glutamate receptors in the mediation of glutamate-evoked excitation of red nucleus neurons after simulated microgravity in rat. Neurosci Lett. 2008;448(3):255–9. doi: 10.1016/j.neulet.2008.10.044. http://dx.doi.org/10.1016/j.neulet.2008.10.044. [DOI] [PubMed] [Google Scholar]
- [25].Satoh Y, Yajima E, Ishizuka K, Nagamine Y, Iwasaki S. Modulation of two types of jaw-opening reflex by stimulation of the red nucleus. Brain Res Bull. 2013;97:24–31. doi: 10.1016/j.brainresbull.2013.05.007. http://dx.doi.org/10.1016/j.brainresbull.2013.05.007. [DOI] [PubMed] [Google Scholar]
- [26].Nieoullon A, Kerkerian L, Dusticier N. High affinity glutamate uptake in the red nucleus and ventrolateral thalamus after lesion of the cerebellum in the adult cat: Biochemical evidence for functional changes in the deafferented structures. Exp Brain Res. 1984;55(3):409–19. doi: 10.1007/BF00235271. http://dx.doi.org/10.1007/BF00235271. [DOI] [PubMed] [Google Scholar]
- [27].Bromberg MB, Pamel G, Stephenson BS, Young AB, Penney JB. Evidence for reactive synaptogenesis in the ventrolateral thalamus and red nucleus of the rat: Changes in high affinity glutamate uptake and numbers of corticofugal fiber terminals. Exp Brain Res. 1987;69(1):53–9. doi: 10.1007/BF00247028. http://dx.doi.org/10.1007/BF00247028. [DOI] [PubMed] [Google Scholar]
- [28].Keifer J, Carr MT. Immunocytochemical localization of glutamate receptor subunits in the brain stem and cerebellum of the turtle Chrysemys picta. J Comp Neurol. 2000;427(3):455–68. doi: 10.1002/1096-9861(20001120)427:3<455::aid-cne11>3.0.co;2-x. http://dx.doi.org/10.1002/1096-9861(20001120)427:3<455::AID-CNE11>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- [29].Sato K, Kiyama H, Tohyama M. The differential expression patterns of messenger RNAs encoding non-N-methyl-D-aspartate glutamate receptor subunits (GluR1-4) in the rat brain. Neuroscience. 1993;52(3):515–39. doi: 10.1016/0306-4522(93)90403-3. http://dx.doi.org/10.1016/0306-4522(93)90403-3. [DOI] [PubMed] [Google Scholar]
- [30].Eyigor O, Jennes L. Identification of glutamate receptor subtype mRNAs in gonadotropin-releasing hormone neurons in rat brain. Endocrine. 1996;4(2):133–9. doi: 10.1007/BF02782758. http://dx.doi.org/10.1007/BF02782758. [DOI] [PubMed] [Google Scholar]
- [31].Eyigor O, Jennes L. Identification of kainate-preferring glutamate receptor subunit GluR7 mRNA and protein in the rat median eminence. Brain Res. 1998;814(1-2):231–5. doi: 10.1016/s0006-8993(98)01056-7. http://dx.doi.org/10.1016/S0006-8993(98)01056-7. [DOI] [PubMed] [Google Scholar]
- [32].Paxinos G, Watson C. The rat brain in stereotaxic coordinates. 6th ed. Amsterdam: Elsevier Academic Press; 2006. [Google Scholar]
- [33].Martin LJ, Blackstone CD, Levey AI, Huganir RL, Price DL. AMPA glutamate receptor subunits are differentially distributed in rat brain. Neuroscience. 1993;53(2):327–58. doi: 10.1016/0306-4522(93)90199-p. http://dx.doi.org/10.1016/0306-4522(93)90199-P. [DOI] [PubMed] [Google Scholar]
- [34].Petralia RS, Wenthold RJ. Light and electron immunocytochemical localization of AMPA-selective glutamate receptors in the rat brain. J Comp Neurol. 1992;318(3):329–54. doi: 10.1002/cne.903180309. DOI: 10.1002/cne.903180309. [DOI] [PubMed] [Google Scholar]
- [35].Petralia RS, Wang YX, Wenthold RJ. The NMDA receptor subunits NR2A and NR2B show histological and ultrastructural localization patterns similar to those of NR1. J Neurosci. 1994;14(10):6102–20. doi: 10.1523/JNEUROSCI.14-10-06102.1994. http://dx.doi.org/10.1002/cne.903180309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Petralia RS, Yokotani N, Wenthold RJ. Light and electron microscope distribution of the NMDA receptor subunit NMDAR1 in the rat nervous system using a selective anti-peptide antibody. J Neurosci. 1994;14(2):667–96. doi: 10.1523/JNEUROSCI.14-02-00667.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]


